Abstract
The universal stress proteins (Usps) UspK (PA3309) and UspN (PA4352) of Pseudomonas aeruginosa are essential for surviving specific anaerobic energy stress conditions such as pyruvate fermentation and anaerobic stationary phase. Expression of the respective genes is under the control of the oxygen-sensing regulator Anr. In this study we investigated the regulation of uspN and three additional P. aeruginosa usp genes: uspL (PA1789), uspM (PA4328), and uspO (PA5027). Anr induces expression of these genes in response to anaerobic conditions. Using promoter-lacZ fusions, we showed that PuspL-lacZ, PuspM-lacZ, and PuspO-lacZ were also induced in stationary phase as described for PuspN-lacZ. However, stationary phase gene expression was abolished in the P. aeruginosa triple mutant Δanr ΔrelA ΔspoT. The relA and spoT genes encode the regulatory components of the stringent response. We determined pppGpp and ppGpp levels using a thin-layer chromatography approach and detected the accumulation of ppGpp in the wild type and the ΔrelA mutant in stationary phase, indicating a SpoT-derived control of ppGpp accumulation. Additional investigation of stationary phase in LB medium revealed that alkaline pH values are involved in the regulatory process of ppGpp accumulation.
Bacteria are well prepared to adapt to a variety of growth-inhibiting stress conditions (reviewed in reference 48). One major factor controlling bacterial growth is the availability of oxygen. Pseudomonas aeruginosa prefers aerobic respiration and is capable of using nitrate or nitrite as a terminal electron acceptor under anaerobic growth conditions (10). Additionally, respiration-independent pathways like arginine fermentation (53) and pyruvate fermentation (15) support survival of P. aeruginosa in the absence of any terminal electron acceptors. Anaerobic growth and survival are essential for chronic P. aeruginosa infections in immunocompromised individuals, including those affected by the genetic disorder cystic fibrosis (1, 37, 46, 57, 61).
In previous studies we investigated the regulation of two anaerobically induced P. aeruginosa genes, uspK (PA3309) and uspN (PA4352), and demonstrated their significance for a successful adaptation to anaerobic energy stress (6, 44). Both genes encode proteins containing domains with a universal stress protein (Usp) signature (Pfam accession number PF0582). Members of the Usp family have been shown to confer survival to a variety of stress conditions in Escherichia coli (20, 26, 35) and have been additionally linked to motility, adhesion, and iron metabolism (34). In contrast, the P. aeruginosa UspK and UspN proteins increase survival under specific anaerobic stress conditions. UspK supports long-term survival during pyruvate fermentation (44), and UspN increases survival in anaerobic stationary phase (6). In addition to UspK and UspN, three additional Usp-encoding genes, uspL (PA1789), uspM (PA4328), and uspO (PA5027), are expressed in response to oxygen limitation (1; also N. Boes and M. Schobert, unpublished data). In contrast to the UspK protein, which contains a single Usp domain, UspN and the three additional Usp-type stress proteins, UspL, UspM, and UspO, contain two Usp domains in tandem. They share similar molecular masses and an overall moderate identity from 23 to 31% to UspN (see Fig. S1 in the supplemental material). But no information about gene regulation or function of these three proteins is available.
We demonstrated that anaerobic induction of uspK and uspN gene expression is dependent on the Anr regulator (6, 44). Anr is a homolog of the E. coli oxygen-sensing Fnr regulator and mediates the adaptation process from aerobic to anaerobic conditions (17, 43). In addition to Anr-dependent induction of uspK and uspN, we observed that gene expression of uspK and uspN is induced in stationary phase by a yet unknown regulator. In E. coli, the stringent response was shown to induce usp genes in stationary phase (20).
The stringent response is one of the global regulatory networks in bacteria, providing a rapid adaptation to a variety of growth-inhibiting stress conditions (8). The regulatory components of the stringent response are the guanosine nucleotides pppGpp and ppGpp. The accumulation of pppGpp and ppGpp in the bacterial cell alters the transcriptional profile (13, 16, 51) and promotes growth arrest by repressing transcription of genes involved in protein biosynthesis. In E. coli the stringent response has recently been shown to induce a large-scale restructuring of metabolic gene expression including genes involved in central metabolism (51). Two distinct enzymes, RelA and SpoT, control the accumulation of ppGpp in E. coli. In response to amino acid deprivation, pppGpp synthesis of RelA is activated (8). The second regulatory mechanism of the stringent response controls the balance between the synthetase and hydrolase activity of ppGpp by SpoT (8, 59). In contrast to RelA, SpoT-controlled ppGpp accumulation is triggered by a variety of stimuli, e.g., inhibition of fatty acid metabolism (19, 45), carbon deprivation (59), or membrane-perturbating agents (50).
In P. aeruginosa research on the stringent response focused primarily on RelA. It has been shown that overexpression of relA increases rpoS transcription and additionally activates quorum sensing (52). Moreover, a ΔrelA mutant displayed decreased levels of the quorum-sensing-dependent elastase LasB and showed a reduced virulence in a Drosophila melanogaster infection model (14, 52).
In this study we investigated the transcriptional regulation of three new usp genes, PA1789 (uspL), PA4328 (uspM), and PA5027 (uspO). All three genes encode proteins with tandem Usp domains, and transcriptional regulation of all three is similar to that of uspN. They are induced in response to oxygen limitation in an Anr-dependent manner and in stationary phase. We determined that stationary phase gene expression is caused by the stringent response and can be abolished only in a ΔrelA ΔspoT double mutant but not in a ΔrelA single mutant. Further characterization of the ΔrelA ΔspoT double mutant identified a SpoT-mediated response triggered by alkaline pH. To our knowledge this is the first research in P. aeruginosa indicating an alkaline pH-mediated SpoT-controlled stringent response.
MATERIALS AND METHODS
Bacterial strains and growth conditions.
The bacterial strains and plasmids used in this study are shown in Table 1. For standard molecular biology protocols, E. coli and P. aeruginosa strains were grown in Luria Bertani (LB) medium as described previously (44).
TABLE 1.
Strains and plasmids used in this study
| Bacterial strain or plasmid | Genotype or phenotype | Reference or source |
|---|---|---|
| Strains | ||
| P. aeruginosa | ||
| PAO1 | Wild type | 12 |
| PA14 | Wild type | 27 |
| PAO6261 | PAO1 Δanr | 60 |
| NB021 | PAO6261 attB::(PuspL-lacZ) | This study |
| NB022 | PAO6261 attB::(PuspM-lacZ) | This study |
| NB023 | PAO6261 attB::(PuspN-lacZ) | 6 |
| NB024 | PAO6261 attB::(PuspO-lacZ) | This study |
| NB134 | PAO6261 attB::(PuspL-lacZ) ΔrelA | This study |
| NB135 | PAO6261 attB::(PuspN-lacZ) ΔrelA | This study |
| NB136 | PAO6261 attB::(PuspO-lacZ) ΔrelA | This study |
| NB146 | PAO6261 attB::(PuspM-lacZ) ΔrelA | This study |
| NB147 | PAO6261 attB::(PuspL-lacZ) ΔrelA ΔspoT | This study |
| NB148 | PAO6261 attB::(PuspN-lacZ) ΔrelA ΔspoT | This study |
| NB149 | PAO6261 attB::(PuspO-lacZ) ΔrelA ΔspoT | This study |
| NB152 | PAO6261 attB::(PuspM-lacZ) ΔrelA ΔspoT | This study |
| NB159 | PAO1 ΔrelA | This study |
| NB170 | PAO1 ΔrelA ΔspoT | This study |
| PA0149::MAR2xT7a | ΔPA0149 (mutant ID 26541)b | 27 |
| PA0472::MAR2xT7 | ΔPA0472 (mutant ID 25794) | 27 |
| PA1912::MAR2xT7 | ΔPA1912 (mutant ID 5297) | 27 |
| PA2050::MAR2xT7 | ΔPA2050 (mutant ID 35909) | 27 |
| PA2896::MAR2xT7 | ΔPA2896 (mutant ID 23265) | 27 |
| PA0762::MAR2xT7 | ΔPA0762 (mutant ID 40799) | 27 |
| PA1455::MAR2xT7 | ΔPA1455 (mutant ID 28485) | 27 |
| PA2426::MAR2xT7 | ΔPA2426 (mutant ID 34241) | 27 |
| PA4462::MAR2xT7 | ΔPA4462 (mutant ID 44482) | 27 |
| PA3622::MAR2xT7 | ΔPA3622 (mutant ID 32095) | 27 |
| NB202 | PA0149::MAR2xT7 attB::(PuspNΔAnr-lacZ) | This study |
| NB203 | PA0472::MAR2xT7 attB::(PuspNΔAnr-lacZ) | This study |
| NB205 | PA1912::MAR2xT7 attB::(PuspNΔAnr-lacZ) | This study |
| NB208 | PA2050::MAR2xT7 attB::(PuspNΔAnr-lacZ) | This study |
| NB210 | PA2896::MAR2xT7 attB::(PuspNΔAnr-lacZ) | This study |
| NB216 | PA0762::MAR2xT7 attB::(PuspNΔAnr-lacZ) | This study |
| NB217 | PA1455::MAR2xT7 attB::(PuspnΔAnr-lacZ) | This study |
| NB218 | PA2426::MAR2xT7 attB::(PuspnΔAnr-lacZ) | This study |
| NB219 | PA4462::MAR2xT7 attB::(PuspnΔAnr-lacZ) | This study |
| NB220 | PA3622::MAR2xT7 attB::(PuspnΔAnr-lacZ) | This study |
| NB104 | PA14 attB::(PuspnΔAnr-lacZ) | This study |
| NB071 | PAO1 attB::(PuspnΔANR-lacZ) | 6 |
| NB160 | PAO1 ΔrelA attB::(PuspnΔAnr-lacZ) | This study |
| NB171 | PAO1 ΔrelA ΔspoT, attB::(PuspnΔAnr-lacZ) | This study |
| KS11 | PAO1 attB::(mini-CTX-lacZ) | 44 |
| E. coli | ||
| DH10B | F−mcrA Δ(mrr-hsdRMS-mcrBC) φ80dlacZΔM15 ΔlacX74 deoR recA1 endA1 araD139 Δ(ara leu)7697 galU galK λ−rpsL nupG | GibcoBRL (Invitrogen) |
| S17 λpir | pro thi hsdR+ Tpr Smr chromosome::RP4-2 Tc::Mu-Km::Tn7 λpir | 11 |
| ST18 | S17 λpir ΔhemA | Sabrina Thoma (unpublished) |
| SM10 | thi-1 thr leu tonA lacY supE recA::RP4-2-Tc::Mu (Kmr) | 11 |
| Plasmids | ||
| pEX18Ap | AproriT+ sacB+; gene replacement vector with MCS from pUC18 | 22 |
| mini-CTX-lacZ | Tcr; promoterless lacZ gene | 4 |
| pPS858 | Apr Gmr; source of gentamicin cassette | 22 |
| pFLP2 | Apr; source of FLP recombinase | 22 |
| pNB001 | Tcr; mini-CTX-lacZ containing a 462-bp fragment of the putative promoter region of the PA1789 gene between EcoRI and BamHI | This study |
| pNB002 | Tcr; mini-CTX-lacZ containing a 448-bp fragment of the putative promoter region of the PA4352 gene between EcoRI and BamHI | This study |
| pNB003 | Tcr; mini-CTX-lacZ containing a 465-bp fragment of the putative promoter region of the PA4352 gene between EcoRI and BamHI | 6 |
| pNB004 | Tcr; mini-CTX-lacZ containing a 453-bp fragment of the putative promoter region of the PA4352 gene between EcoRI and BamHI | This study |
| pNB54 | Apr Gmr; pEX18Ap with a 636-bp promoter spot; Gmr-gfp fragment from pPS858 and 622 bp downstream of the coding region of spoT between SacI and HindIII | This study |
| pKS18 | Apr Gmr; pEX18Ap with 796-bp promoter of relA (PA0934); Gmr-gfp fragment from pPS858, and 824 bp of the 3′ coding region of relA between EcoRI and HindIII | 44 |
| pNB19 | Tcr; mini-CTX-lacZ containing a 465-bp fragment of the putative promoter region, with a mutation in the Anr-box (TTGATGTGCATCAA → TTGATGTGCATACG) of the PA4352 gene between EcoRI and BamHI | 6 |
Construction and testing of promoter-lacZ reporter gene fusions.
Chromosomal promoter-lacZ reporter gene fusions were constructed using the mini-CTX-lacZ vector (4). To analyze the uspL (PA1789) promoter, a 462-bp PCR product upstream of the translational start of uspL (PA1789) was generated using primer oKS20 (5′-GGAATTCTCTTCAGGATCGCCAGCAC-3′) and primer oKS21 (5′-CGCGGATCCACGAGGATGCTGCGAATG-3′). Furthermore, a 448-bp PCR product upstream of the translational start of uspM (PA4328) was generated using primer oKS22 (5′-GGAATTCACAGCAGAACCTCGATCTC-3′) and primer oKS23 (5′-CGCGGATCCACCACCAGCAGGTTATG-3′), and a 453-bp PCR product upstream of the translational start of uspO (PA5027) was generated using primer oKS24 (5′-GGAATTCACCACATCGGCAGCATGTA-3′) and primer oKS25 (5′-CGCGGATCCATCATGGCGGACTCCGT-3′). Primers oKS20, oKS22 and oKS24 contained an EcoRI restriction site at the 5′ end (underlined), and oKS21, oKS23, and oKS25 contained a restriction site for BamHI also at the 5′ end (underlined). The EcoRI- and BamHI-digested PCR products were cloned between the EcoRI and BamHI sites of mini-CTX-lacZ to generate pNB001 (PuspL-lacZ), pNB002 (PuspM-lacZ), and pNB004 (PuspO-lacZ).
To monitor the expression of the stringent response-controlled promoter of PuspN, we used PuspNΔAnr, which was constructed in previous studies (6) and which contains a nonfunctional Anr box. Transfer of promoter-lacZ fusions harboring plasmids in P. aeruginosa was carried out by a diparental mating using E. coli S17 λpir as the donor. The CTX integrase of the parental mini-CTX-lacZ vector promoted integration of the plasmid into the attB site of the P. aeruginosa genome. Constructed plasmids were transferred into P. aeruginosa PAO1 wild type and the anr mutant strain PAO6261 to generate the P. aeruginosa strains NB005 (PAO1 with PuspL-lacZ), NB006 (PAO1 with PuspM-lacZ), NB008 (PAO1 with PuspO-lacZ), NB021 (PAO1 Δanr with PuspL-lacZ), NB022 (PAO1 Δanr with PuspM-lacZ), and NB024 (PAO1 Δanr with PuspO-lacZ) (Table 1). pNB19 was introduced in PA14 wild type, PAO1 ΔrelA ΔspoT, and in diverse transposon mutants to generate the strains NB104 (PA14 wild type), NB171 (ΔrelA ΔspoT), NB202 (ΔPA0149), NB203 (ΔPA0472), NB205 (ΔPA1912), NB208 (ΔPA2050), NB210 (ΔPA2896), NB216 (ΔPA0762), NB217 (ΔPA1455), NB218 (ΔPA2426), NB219 (ΔPA4462), and NB220 (ΔPA3622). In these strains parts of the mini-CTX-lacZ vector containing the tetracycline resistance cassette were deleted using a FLP recombinase encoded on the pFLP2 plasmid (22). β-Galactosidase assays were performed as outlined before in detail (15, 42), and activity is reported in Miller units (29).
To analyze the activation of the usp promoters under anaerobic conditions, cells were incubated aerobically in LB medium and were transferred to anaerobic flasks at an optical density at 578 nm (OD578) of 0.7. Activities were determined before and 15, 30, 60, and 180 min after the cultures were shifted to anaerobic conditions.
β-Galactosidase activities of aerobically grown cultures were monitored from exponential phase to stationary phase in LB medium.
Construction of P. aeruginosa ΔrelA and ΔrelA ΔspoT mutant strains.
Unmarked gene deletion mutants were obtained using well-established strategies based on sacB counter-selection and FLP recombinase excision (22). The suicide vector pKS18 used to replace relA has been described previously (44). To achieve a deletion of spoT, the suicide vector pNB54 was constructed. The BamHI-digested gentamicin resistance cassette of pPS858 was cloned between two PCR fragments of spoT in the multiple cloning site of pEX18Ap. The two PCR fragments contained DNA homologous to the upstream and downstream areas of spoT. A 636-bp fragment containing the upstream promoter region of the spoT was amplified using primer oNB33 (5′-CGAGCTCAGGACAGCGACGAGGTGAT), containing a SacI restriction site at the 5′ end (restriction sites are underlined), and oNB34 (5′-CGCGGATCCTTCACCCCCTGCCCGTA-3′), containing a BamHI site. The primers oNB35 (5′-CGCGGATCCACGGCAACAT CGAGAAG-3′), containing a BamHI site, and oNB36 (5′-CCAAGCTTCCGGCTTA CTCGAGGACG-3′), containing a HindIII site, amplified 622 bp of the corresponding downstream region of spoT. In order to create ΔrelA and ΔrelA ΔspoT mutant strains, we used pKS18 to replace relA in NB021, NB022, NB023, NB024, NB071, and PAO1 wild type to create NB134, NB146, NB135, NB136, NB160, and NB159, respectively. We used the suicide vector pKS18 to replace relA with a gentamicin cassette by sacB-based counter-selection. FLP recombinase encoded on the pFLP2 plasmid removed the Flp recognition target (FRT)-flanked gentamicin cassette. To introduce ΔspoT in the created ΔrelA mutant strains, we used the suicide vector pNB54 to replace spoT with a gentamicin cassette by sacB-based counter-selection to create the strains NB147, NB152, NB148, NB149, NB171, and NB170. FLP recombinase encoded on the pFLP2 plasmid removed the FRT-flanked gentamicin cassette once more.
Culture conditions for pppGpp/ppGpp assays.
In order to monitor the accumulation of ppGpp during the transition from exponential to stationary phase, cultures were inoculated to an initial OD578 of 0.05 and were incubated aerobically at 37°C. For each ppGpp assay, aliquots of cultures were labeled with 32Pi (0.1 mCi/ml) for 1 h prior to harvest.
To investigate the accumulation of both ppGpp and pppGpp during stationary phase in LB medium, cultures of P. aeruginosa (wild-type PAO1, ΔrelA, and ΔrelA ΔspoT) were grown for 8 h aerobically to stationary phase (OD578 of 5.0 to 7.0). Aliquots of 60 μl were labeled with 32Pi (0.2 mCi/ml) for 1 h prior to harvest. In order to determine ppGpp and pppGpp levels in stationary phase with defined pH values, cultures were buffered with 0.1 M morpholinepropanesulfonic acid (MOPS) at either pH 7.0 or 8.5 prior to labeling with 32Pi (0.2 mCi/ml) for 30 min.
ppGpp and pppGpp accumulation in response to alkaline pH values or serine hydroxamate (SHX) was carried out in exponential growth phase under anaerobic conditions. Cultures of P. aeruginosa were inoculated from overnight cultures to an initial OD578 of 0.05. Aliquots of 60 μl were incubated at 37°C in LB medium with 50 mM nitrate. To achieve anaerobic conditions, cultures were overlaid with mineral oil. Cells were incubated for 4 h to reach exponential phase with an OD578 of 0.5 and were labeled with 32Pi (2 mCi/ml) for 15 min prior to treatment with either SHX (3 mM) or NaOH (20 mM, pH 9) and were incubated for an additional 15 min before harvest.
In control experiments we performed ppGpp assays with longer equilibration periods. We equilibrated stationary-phase cultures for 4 h and exponential-phase cultures for 2 h prior to NaOH or SHX treatment and obtained the same results as with short incubation periods. However, longer equilibration times resulted in higher background activity caused by the incorporation of radioactive phosphate in cell components such as membrane, RNA, DNA, etc. (data not shown).
Detection of ppGpp and pppGpp using a TLC approach.
To obtain the same cell quantities of each growth phase for each ppGpp assay, appropriate volumes were harvested by centrifugation for 1 min at 12,000 × g (100 μl of a culture with an OD578 of 1.0, 20 μl of a culture with an OD578 of 5.0). Pellets were resuspended in 10 μl of 1 M formic acid and were frozen at −20°C. To extract pppGpp and ppGpp, acidic mixtures were frozen at −20°C and thawed three times; cell debris was removed by centrifugation at 14,000 rpm for 2 min, and 5 μl of supernatant was spotted onto a polyethylenimine cellulose plate (PEI cellulose-F; Merck). Separation of pppGpp and ppGpp by thin-layer chromatography (TLC) was performed with 1.5 M KH2PO4 (pH 3.4) as running solvent. Dried plates were exposed to a K-Screen (Kodak) and were analyzed with a phosphorimager (FX-Scanner; Bio-Rad).
RESULTS
Anaerobic induction of PuspL-lacZ, PuspM-lacZ, and PuspO-lacZ reporter gene expression is dependent on the oxygen-sensing regulator Anr.
Previously we demonstrated Anr-dependent gene expression of uspK (PA3309) and uspN (PA4352) in response to anaerobiosis (6, 44). Investigations of the P. aeruginosa transcriptome under anaerobic conditions identified three additional usp genes, uspL (PA1789), uspM (PA4328), and uspO (PA5027), to be expressed in response to anaerobiosis (1; also Boes and Schobert, unpublished). Promoter analyses of all putative usp genes in P. aeruginosa using the Virtual Footprint tool of the PRODORIC database (32) indicated the presence of conserved Anr binding sites in the promoter regions of uspL, uspM, and uspO (Fig. 1). In order to investigate Anr-dependent gene expression of uspL, uspM, and uspO, we used transcriptional promoter-lacZ fusions.
FIG. 1.
Promoter regions of uspL, uspM, uspN, and uspO. Promoter regions of uspL, uspM, uspN, and uspO upstream of the translational start codon are shown (italic). The putative Anr box determined by the Virtual Footprint tool (32) is underlined, and conserved bases are shown in bold. The published consensus sequence (47) of the Anr homologue Fnr is given on top of each putative Anr box.
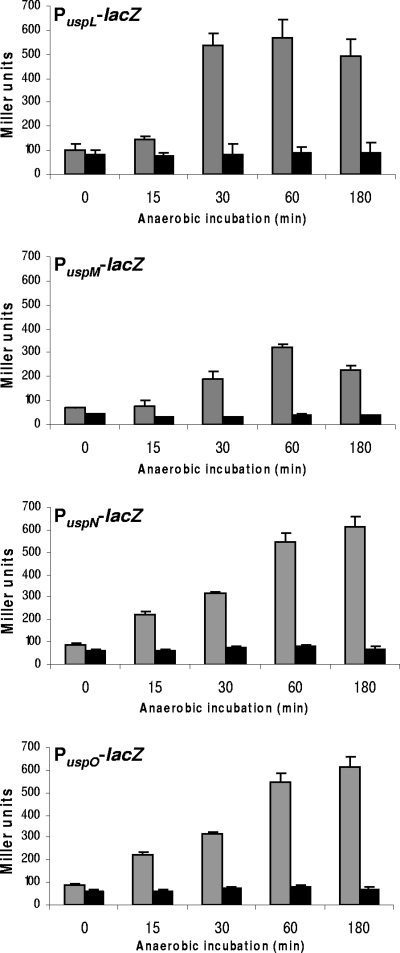
We introduced PuspL-lacZ, PuspM-lacZ, and PuspO-lacZ separately in the P. aeruginosa wild type and a Δanr mutant strain to monitor β-galactosidase activities under aerobic and anaerobic conditions. Since a Δanr mutant strain fails to grow anaerobically, we performed a shift experiment. P. aeruginosa wild-type and Δanr mutant strains were grown in LB medium aerobically to the mid-log phase (OD578 of 0.7). Immediate transfer of the cultures to hermetically sealed serum bottles generated anaerobic conditions within 3 min (15). During aerobic exponential growth, P. aeruginosa had doubling times of about 30 min. Transfer to anaerobic conditions resulted in a growth arrest of both strains, wild type and the Δanr mutant, since no nitrate was added to the growth medium. Aliquots for β-galactosidase activity determination were taken prior to the shift to anaerobic incubation and after 15, 30, 60, and 180 min of anaerobic incubation (Fig. 2). β-Galactosidase activities of all strains harboring promoter-lacZ constructs increased during anaerobic conditions in the wild type, while no increase was detected in Δanr mutant strains, demonstrating Anr-dependent gene expression of PuspL-lacZ, PuspM-lacZ, and PuspO-lacZ in vivo (Fig. 2) and confirming our previous results for PuspN-lacZ (6).
FIG. 2.
β-Galactosidase activities in P. aeruginosa wild type (gray bars) and the Δanr mutant (black bars) containing the respective PuspL-lacZ, PuspM-lacZ, PuspN-lacZ, and PuspO-lacZ fusions during anaerobiosis. Strains were grown aerobically at 37°C in LB medium to an OD578 of 0.7 (time point 0) and transferred to anaerobic conditions. Since the medium contained no alternative electron acceptor, the shift to anaerobic conditions resulted in a growth arrest for 4 h. Aliquots after 15, 30, 60, and 180 min were taken for β-galactosidase assays. All experiments were repeated three times; standard deviations are indicated.
Induction of PuspL-lacZ, PuspM-lacZ, PuspN-lacZ, and PuspO-lacZ reporter gene expression in aerobic stationary phase is mediated by the stringent response.
In our previous studies, we monitored increased promoter activities using primer extension and PuspN-lacZ reporter gene fusions during aerobic and anaerobic stationary phase. However, we excluded the involvement of RpoS, RelA, RhlR, or LasR in stationary-phase induction of PuspN-lacZ (6).
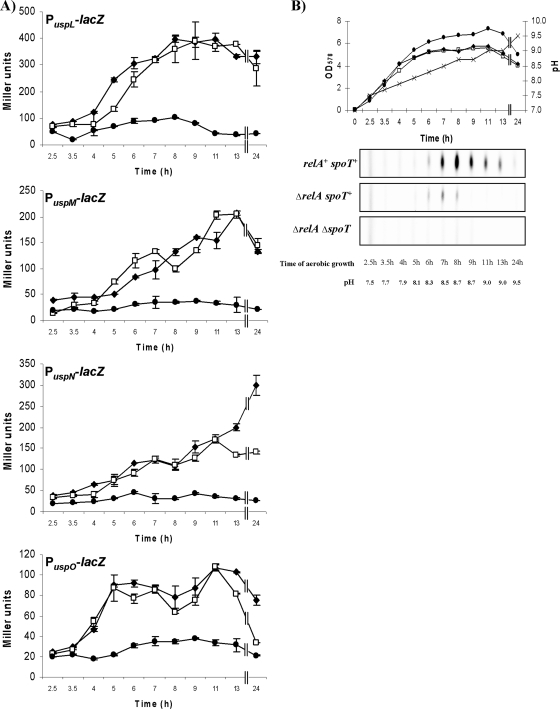
We have already demonstrated that Anr contributes in part to aerobic stationary-phase induction of PuspN (6) since respiration of P. aeruginosa at high cell densities in stationary phase results in an oxygen-limited environment (9). To exclude the impact of Anr, we determined β-galactosidase activities of PuspL-lacZ, PuspM-lacZ, PuspN-lacZ, and PuspO-lacZ in Δanr mutant during aerobic stationary phase. All promoter-lacZ reporter gene fusions were induced in a Δanr mutant strain during stationary phase, confirming Anr-independent induction (Fig. 3A). To investigate the involvement of the stringent response in promoter activation, we deleted both genes encoding the enzymes regulating ppGpp concentration, RelA and SpoT, in a Δanr strain. First, we introduced a relA deletion to create Δanr ΔrelA double mutants harboring the respective four promoter-lacZ fusions. Deletion of relA results in a strong decrease in ppGpp levels in E. coli (33). While PuspL-lacZ and PuspM-lacZ showed almost no decrease in β-galactosidase activities as a consequence of a relA deletion in stationary phase, after 13 h we observed a decrease in the induction patterns of PuspN-lacZ and PuspO-lacZ in the Δanr ΔrelA double mutant compared to the Δanr strain (Fig. 3A). To abolish residual ppGpp accumulation in ΔrelA strains of E. coli, deletion of spoT is required (59). Consequently, we deleted spoT in P. aeruginosa to create Δanr ΔrelA ΔspoT triple mutant strains and determined β-galactosidase activities of PuspL-lacZ, PuspM-lacZ, PuspN-lacZ, and PuspO-lacZ harboring strains in stationary phase. Activation of all tested usp promoters ceased in the absence of Anr, RelA, and SpoT. These results clearly demonstrated a stringent response-controlled induction of PuspL-lacZ, PuspM-lacZ, PuspN-lacZ, and PuspO-lacZ in stationary phase.
FIG. 3.
β-Galactosidase activities of Pusp-lacZ (A) and accumulation of ppGpp (B) in stationary phase in various P. aeruginosa mutant strains. (A) β-Galactosidase activities of PuspL-lacZ, PuspM-lacZ, PuspN-lacZ, and PuspO-lacZ were determined in the following P. aeruginosa mutant strains: Δanr mutant (filled diamonds), Δanr ΔrelA double mutant (open squares), and Δanr ΔrelA ΔspoT triple mutant (filled circles). Cells were grown in LB medium aerobically at 37°C. β-Galactosidase activities were determined every hour from exponential to stationary phase (see Materials and Methods). All experiments were repeated three times; standard deviations are indicated. (B) Growth curves, pH of cultures, and ppGpp accumulation shown as an example of P. aeruginosa strains harboring PuspL-lacZ, described in panel A, during the transition from exponential to stationary phase. PuspL-lacZ harboring strains NB021 (Δanr) (filled diamonds), NB134 (Δanr ΔrelA) (open squares), and NB146 (Δanr ΔrelA ΔspoT) (filled circles) were inoculated to an initial OD578 of 0.05 in LB medium and were grown at 37°C aerobically to the stationary phase. pH (crosses), OD578, and ppGpp accumulation were monitored from exponential phase to stationary phase. Aliquots of cultures for ppGpp determination were taken and labeled for 1 h with 32Pi. OD578 and pH values of cultures were determined prior to harvesting the cells for a ppGpp assay. Nucleotides were separated on a PEI cellulose-F TLC plate using 1.5 M K2HPO4, pH 3.6, as a solvent. Dried plates were exposed to a K-Screen (Kodak) and analyzed by a phosphorimager (FX-Scanner; Bio-Rad).
Accumulation of ppGpp in P. aeruginosa during stationary phase requires relA and spoT.
The activation of all tested stringent response-controlled usp promoters in relA-deficient strains was abolished by the introduction of a spoT deletion, indicating a residual ppGpp accumulation in ΔrelA single mutants in stationary phase. To confirm these results, we determined ppGpp levels simultaneously in the β-galactosidase samples during the transition from exponential to stationary phase, using a TLC approach.
For each ppGpp assay, samples were labeled with 32Pi (0.1 mCi/ml) for 1 h, and identical cell numbers were harvested. Cell lysates were applied to a PEI cellulose-F TLC plate. ppGpp was separated using 1.5 M K2HPO4 as a running solvent. Results of ppGpp accumulation in correlation with growth phase, time of cultivation, and pH of cultures are shown in Fig. 3B. Accumulation of ppGpp was detected after 6 h of incubation dependent on the presence of SpoT, when the pH of cultures reached a value of 8.3. At this time point similar amounts of ppGpp were detected either in the absence or presence of RelA, indicating that ppGpp accumulation is strictly initiated by SpoT. Introducing a spoT deletion in a ΔrelA mutant abolished any ppGpp accumulation during stationary phase, as expected. A longer time course of ppGpp accumulation revealed clearly lower ppGpp levels in the absence of RelA. The accumulation pattern of ppGpp during 24 h of cultivation is characterized by a peak after 8 h of cultivation in the presence of RelA and SpoT and a peak after 7 h of cultivation in the absence of RelA.
P. aeruginosa accumulates ppGpp but not pppGpp during stationary phase.
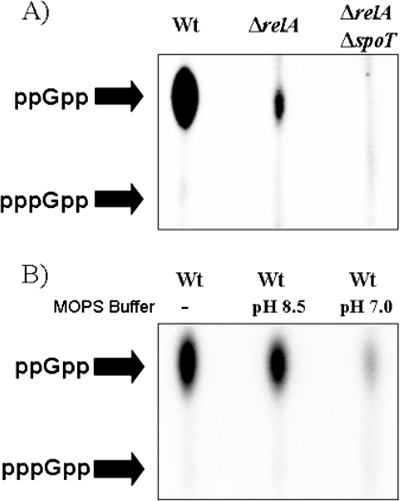
In order to analyze both ppGpp and pppGpp levels during stationary phase, we applied samples in parallel on a TLC plate. Wild-type, ΔrelA, and ΔrelA ΔspoT strains were labeled for 1 h prior to harvest with 32Pi from stationary phase after 8 h of cultivation (compare growth curves in Fig. 3B). As shown in Fig. 4A, we detected accumulation of ppGpp in stationary phase in the wild type and in a ΔrelA single mutant, as already shown in Fig. 3B. Interestingly, no corresponding accumulation of the precursor pppGpp was detected in the wild-type or ΔrelA strain.
FIG. 4.
Accumulation of ppGpp in P. aeruginosa during stationary phase confirmed by TLC. Cells were grown to stationary phase (OD578 of 5.0 to 7.0) and were labeled with 32Pi for 1 h. Nucleotides were separated on a PEI cellulose-F TLC plate using 1.5 M K2HPO4, pH 3.6, as a solvent. Dried plates were exposed to a K-Screen (Kodak) and analyzed by a phosphorimager (FX-Scanner; Bio-Rad). (A) ppGpp accumulation in P. aeruginosa wild type, ΔrelA, and ΔrelA ΔspoT during stationary phase in LB medium. (B) ppGpp accumulation in P. aeruginosa wild type grown in unbuffered LB medium or LB medium buffered at pH 8.5 or pH 7.0. Wt, wild type.
Alkaline pH triggers the stringent response during stationary phase in P. aeruginosa.
To elucidate conditions that might be involved in controlling an accumulation of ppGpp in stationary phase, we assumed one of these conditions to be alkaline pH. Increased pH in stationary phase of cultures grown in LB medium has been described previously (21). To confirm our assumption that alkaline pH triggers ppGpp accumulation during stationary phase, we buffered stationary phase cultures of the wild type with 0.1 M MOPS buffer at either pH 7.0 or 8.5 prior to labeling with 32Pi for 30 min (Fig. 4B). Neutralizing the growth medium to pH 7 resulted in a strongly decreased accumulation of ppGpp compared to alkaline pH values of 8.5, indicating a pH-regulated accumulation of ppGpp. We observed no accumulation of the precursor pppGpp in stationary phase, indicating a typical SpoT-controlled accumulation of ppGpp (see Discussion).
Accumulation of ppGpp/pppGpp in response to alkaline pH or SHX treatment.
To examine the effect of alkaline pH values on the stringent response, we treated cells during exponential growth with 20 mM NaOH to induce a pH shift from 7.5 to 9. During exponential growth, cells without SHX or NaOH treatment showed only low to undetectable levels of ppGpp. A pH shift from 7.5 to 9 was followed by accumulation of ppGpp in the wild type (Fig. 5), supporting our assumption that high pH values do induce the stringent response in P. aeruginosa. Again, no corresponding accumulation of the precursor pppGpp could be detected, indicating a SpoT-controlled accumulation of ppGpp (see discussion). To demonstrate the difference between RelA- and SpoT-controlled accumulation of ppGpp, we induced the stringent response using SHX, commonly used to simulate amino acid limitation and to initiate a RelA-controlled accumulation of ppGpp. In this case we observed accumulation of both the precursor pppGpp and ppGpp (Fig. 5).
FIG. 5.
Accumulation of ppGpp and pppGpp induced by either NaOH or SHX during exponential growth phase. Cells were grown anaerobically to exponential growth phase (OD578 of 0.5) and were labeled with 32Pi for 15 min prior to treatment with NaOH (20 mM; pH 9) or SHX (3 mM) as indicated. Nucleotides were separated as described in Materials and Methods. Wt, wild type.
Involvement of alternative sigma factors in ppGpp-mediated gene expression of the PuspNΔAnr-lacZ reporter gene.
Positive effects of ppGpp on gene expression can be mediated by the housekeeping sigma factor σ70 (38) or by the increased competitiveness of alternative sigma factors during the stringent response (23). In order to identify alternative sigma factors mediating ppGpp-dependent gene expression of PuspN in stationary phase, we tested 10 transposon mutants of putative sigma factors (Table 2) using strains from the P. aeruginosa PA14 Insertion Mutant Library (27). To monitor exclusively the stringent response-activated promoter of uspN in strains from the PA14 Insertion Mutant Library, it was again necessary to exclude gene expression induced by Anr of PuspN-lacZ. Previously, we showed that the mutation of the Anr binding site in PuspN did not influence stationary-phase gene expression (6). Hence, we used a mutated promoter of uspN, designated PuspNΔAnr, with a nonfunctional Anr box (6). We introduced PuspNΔAnr-lacZ in a wild-type strain, in a ΔrelA ΔspoT double mutant, and in various transposon mutant strains listed in Table 2. We observed a loss of activation of PuspNΔAnr-lacZ in stationary phase in the ΔrelA ΔspoT double mutant, confirming a stringent response dependence of PuspNΔAnr-lacZ (Table 2). All tested transposon mutants showed induction patterns of PuspNΔAnr-lacZ in stationary phase similar to the pattern of the wild type, indicating that none of the tested sigma factors was acting with ppGpp in stationary phase to coregulate induction of PuspN.
TABLE 2.
β-Galactosidase activities of PuspNΔAnr-lacZ in diverse mutants deficient in given putative sigma factors or regulatory components
| Genotype of parent straina | Description or function of disrupted geneb | β-Galactosidase activity (Miller units) with PuspNΔAnr-lacZ in stationary phasec |
|---|---|---|
| Wild type | 280.0 (± 20.0) | |
| ΔPA0762 (ΔalgU) | Sigma factor AlgU | 500.6 (± 35.0) |
| ΔPA1455 (ΔfliA) | Sigma factor FliA | 472.9 (± 8.0) |
| ΔPA2426 (ΔpvdS) | Sigma factor PvdS | 437.3 (± 20.0) |
| ΔPA4462 (ΔrpoN) | Sigma 54 factor/sigma factor RpoN | 329.5 (± 27.9) |
| ΔPA3622 (ΔrpoS) | Sigma factor RpoS | 425.7 (± 6.6) |
| ΔPA0472 (ΔfiuI) | Sigma factor FiuI | 376.6 (± 25.8) |
| ΔPA0149 | Probable sigma 70 factor, ECF subfamily | 441.5 (± 5.9) |
| ΔPA1912 | Probable sigma 70 factor, ECF subfamily | 372.3 (± 11.3) |
| ΔPA2050 | Probable sigma 70 factor, ECF subfamily | 434.3 (± 29.9) |
| ΔPA2896 | Probable sigma 70 factor, ECF subfamily | 399.1 (± 10.7) |
| ΔPA0934/ΔPA5338 (ΔrelA ΔspoT) | GTP pyrophosphokinase/guanosine-3′,5′-bis (diphosphate) 3′-pyrophosphohydrolases | 60.3 (± 2.5) |
PuspNΔAnr-lacZ was integrated in transposon mutant strains from the P. aeruginosa PA14 Insertion Mutant Library (27) in the wild-type and in the constructed ΔrelA ΔspoT strain (this study).
ECF, extracytoplasmic function.
Background β-galactosidase activity of the donor vector mini-CTX-lacZ was determined to be 70.2 (±14.5) Miller units in the wild type.
DISCUSSION
Expression of usp genes in P. aeruginosa.
Expression of genes encoding the universal stress proteins UspK, UspL, UspM, UspN, and UspO in response to oxygen limitation has been observed using transcriptome analysis for P. aeruginosa under microaerobic conditions and in biofilm-grown cells (1, 28, 54). Extensive phenotypic screenings of P. aeruginosa uspL, uspM, and uspO mutants in our laboratory revealed no stress-related phenotype similar to usp mutants in E. coli or anaerobic phenotypes as described for uspK or uspN mutants (6, 44) in P. aeruginosa (data not shown). Nevertheless, we found identical transcriptional control of the uspL, uspM, and uspO promoters compared to the uspN promoter. Using promoter-lacZ fusions, we demonstrated Anr-dependent gene expression of uspL, uspM, and uspO under anaerobic conditions, similar to uspN (Fig. 2), confirming bioinformatics predictions of Anr binding sites in the corresponding promoter regions (Fig. 1). Usp-type stress proteins were also reported to be produced under oxygen-limiting conditions in Mycobacterium (36), indicating that Usps might be important for adaptation to anaerobic environments in other pathogenic bacterial species.
We observed increased activities of the tested usp promoter-lacZ fusions PuspL-lacZ, PuspM-lacZ, and PuspO-lacZ in stationary phase that were similar to the results for PuspN-lacZ. We showed that the expression of these genes is dependent on the presence of RelA and SpoT, the two regulatory components of the stringent response. The positive control by pppGpp and ppGpp has also been reported for uspA, uspC, uspD, and uspE in E. coli (20, 25) and uspA3 in Corynebacterium glutamicum (7). In E. coli regulation of usp gene expression by the stringent response seems plausible since the corresponding usp mutant phenotypes indicate that Usps contribute to survival in response to a variety of different stress conditions (5, 20, 34, 35). A recent publication indicates that E. coli uspA is additionally regulated by fructose-6-phosphate, which points to additional levels of regulation via metabolic intermediates (39). Whether metabolic intermediates also regulate usp gene expression in P. aeruginosa is an interesting question. Although a broad range of phenotypes has been described for usp deletion strains of E. coli and P. aeruginosa, the exact biological function of Usps remains unknown. It is also interesting that phenotypic characterization of P. aeruginosa uspN and uspK mutants indicates that these proteins are essential for surviving specific stress conditions. In contrast, E. coli Usp-type proteins contribute to survival in response to more universal stress conditions. This specific role of the P. aeruginosa Usps might also explain why even an extensive screening did not result in identification of a phenotype for mutants defective in uspL, uspM, or uspO genes although they are transcriptionally regulated in a manner similar to uspN.
SpoT-controlled stringent response.
Depending on growth conditions and medium, a variety of regulators and sigma factors contribute to stationary-phase gene expression in P. aeruginosa. We showed that the oxygen-sensing Anr regulator contributes to stationary-phase gene expression in a culture incubated under aerobic conditions (6). This Anr-dependent gene expression is due to oxygen-limiting conditions caused by respiration of P. aeruginosa at high cell densities (9).
Published research on the stringent response in P. aeruginosa has focused on RelA and amino acid limitation-induced stringent response (14, 52). RelA is well studied in E. coli, and was described as a ribosome-associated protein that catalyzes the synthesis of pppGpp. This synthetase activity is greatly enhanced by unloaded tRNAs that enter the ribosome (56), leading to a stringent response evoked by amino acid limitation (Fig. 6A and C). However, accumulation of pppGpp and ppGpp is additionally controlled by SpoT, which is present in a number of gram-negative bacteria (30).
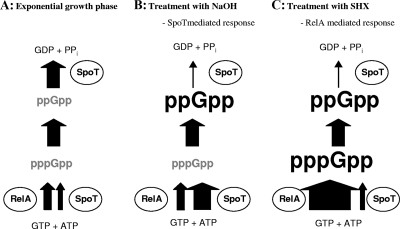
FIG. 6.
Regulation of the stringent response in P. aeruginosa wild type during exponential growth (A) and in response to either NaOH (B) or SHX (C) treatment. Accumulation of nucleotides is indicated by bold letters, undetectable nucleotide levels are represented by gray letters, and synthesis or degradation rates are indicated by the size of arrows. Conversion of pppGpp to ppGpp is mediated by a polyphosphate kinase (Ppx). (A) ppGpp and pppGpp metabolism during exponential phase is characterized by an equal rate of pppGpp synthesis and ppGpp degradation. (B) Treatment with NaOH induces a SpoT-regulated stringent response. The accumulation of ppGpp is achieved by a decrease in the ppGpp degradation process, resulting in lower ppGpp degradation rates than synthesis rates. (C) Treatment with SHX induces mainly a RelA-mediated stringent response, resulting in increased synthesis rates of pppGpp. In this case both, pppGpp and ppGpp accumulate in the cell. However, a reduction in ppGpp degradation by SpoT was also described under these conditions in E. coli (41).
A SpoT-controlled stringent response appears to be more complex since SpoT was shown to respond to a broad range of stress factors and to be associated with enzymes of seemingly unrelated function. On one hand, it was shown to be linked by CgtA to ribosomes (58), and, on the other hand, it was shown to interact with the acyl carrier protein, a protein involved in fatty acid biosynthesis (3).
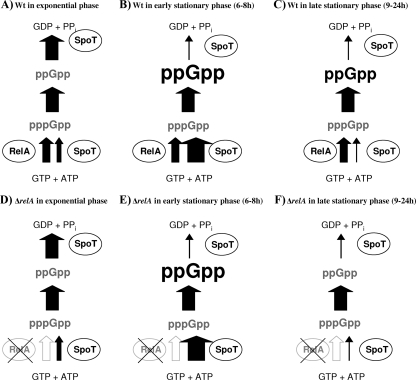
Several lines of evidence indicate that in our experimental setup the stringent response is triggered by SpoT and, to a minor extent, by RelA. Only a Δanr ΔrelA ΔspoT mutant but not a Δanr ΔrelA mutant completely lost the ability to induce lacZ reporter gene fusions of the uspL, uspM, uspN, and uspO promoters in aerobic stationary phase. This result is in accordance with our TLC-based ppGpp determination. We detected ppGpp accumulation in P. aeruginosa during stationary phase in a ΔrelA single mutant (Fig. 3B and 4A). After 6 h of aerobic incubation, we observed similar ppGpp concentrations in the wild type and the ΔrelA single mutant, demonstrating a ppGpp accumulation strictly accomplished by SpoT (Fig. 3B). From these results we assume that the balance between the synthetase and hydrolase activity of SpoT is shifted toward pppGpp synthesis at this time point, initiating the accumulation of ppGpp completely independent of RelA (Fig. 7, compare B and E). Further incubation in stationary phase results in a decrease in ppGpp in the wild type and a reduction to almost undetectable levels in a ΔrelA single mutant (Fig. 7C and F). This observation correlates with the functionally unstable character of SpoT synthetase activity shown in E. coli (33). Murray and Bremer reported that SpoT synthetase activity is dependent on continuous protein biosynthesis since synthetase activity of SpoT in E. coli has an average functional lifetime of about 40 s or less after translation of spoT mRNA, whereas the ppGpp degradation activity and its control are independent of the SpoT protein's lifetime. Taking into account that translation is generally downregulated during stationary phase, it is plausible that ppGpp accumulation decreases over the course of time during stationary phase because of the loss of SpoT-dependent synthetase activity (compare Fig. 7B and C). Prolonged cultivation during stationary phase results in higher ppGpp levels in the wild type than in the ΔrelA mutant. This observation is clearly dependent on the basic production of pppGpp by RelA, which is consequently absent in the ΔrelA mutant.
FIG. 7.
Scheme of ppGpp accumulation in P. aeruginosa wild type (A to C) and the ΔrelA mutant (D to F) during growth from exponential to stationary phase. Accumulation of nucleotides is indicated by bold letters, undetectable nucleotide levels are represented in gray letters, and synthesis or degradation rates are indicated by the size of arrows. During exponential growth a balance between synthesis and degradation of ppGpp is regulated by SpoT (A and D). During early stationary phase (6 to 8 h of cultivation) (compare Fig. 3 B), SpoT increases its synthesis activity and decreases the degradation rates for ppGpp (indicated by arrow size), resulting in the accumulation of ppGpp (bold letters) (B and E). In late stationary phase, pppGpp synthesis by SpoT ceases, resulting in the accumulation of ppGpp, which is dependent on a complex interplay of the pppGpp synthesis rates of RelA and diminished degradation activity of SpoT in the wild type (C and F). Loss of SpoT-dependent pppGpp synthesis results in levels of ppGpp that are lower in late stationary phase than in early stationary phase, indicated by letter size (compare with Fig. 3B).
The lowered ppGpp levels in the ΔrelA mutant still allowed almost wild-type induction of the promoter-lacZ reporter gene fusions in stationary phase between 5 and 11 h of incubation (Fig. 3A). Interestingly, we observed a decrease in induction of PuspO-lacZ and PuspN-lacZ in the ΔrelA mutant after 13 h of incubation.
An additional indication for a SpoT-triggered stringent response during stationary phase is the increase in ppGpp but not pppGpp levels. A RelA-triggered stringent response usually results in ppGpp and pppGpp accumulation (Fig. 6C). This is caused by a massive increase in pppGpp synthesis activity by RelA itself and additionally by a reduction in ppGpp degradation by SpoT (Fig. 6C) (41). In a SpoT-triggered stringent response, SpoT itself is presumed to control ppGpp levels mainly on the level of ppGpp degradation since synthesis activity is unstable in the absence of protein biosynthesis (18, 33). This, in turn, leads to the accumulation of ppGpp and not its precursor pppGpp. We observed the accumulation of ppGpp alone in stationary phase (Fig. 4A), while a RelA-controlled stringent response induced by SHX results clearly in an accumulation of both ppGpp and pppGpp (Fig. 5 and 6C).
Alkaline pH elicits a SpoT-controlled stringent response.
We observed an increase in pH up to 8.3 at the end of the exponential growth phase and usually measured pH values of 8.5 in early stationary phase, as previously described by others (21). The following two experiments indicated that alkaline pH is the trigger for SpoT-controlled ppGpp accumulation in stationary phase. First, ppGpp levels in LB medium buffered at pH 7.0 were drastically reduced compared to levels in LB medium buffered at pH 8.5 or unbuffered LB medium (pH 8.7) (Fig. 4B). Second, we also demonstrated clearly that a shift to alkaline pH induces an accumulation of ppGpp during exponential growth (Fig. 5). Induction of ppGpp accumulation during exponential growth in response to alkaline pH is not limited to LB medium but could also be initiated in a defined minimal medium containing succinate as the sole carbon source (see Fig. S2 in the supplemental material). The accumulation of ppGpp but not the precursor pppGpp in response to alkaline pH again indicated a SpoT-controlled accumulation of ppGpp (Fig. 5 and 6B).
Induction of the stringent response as a consequence of a pH shift was also reported for Helicobacter pylori (31, 55). Despite the fact Wells et al. (55) used acidic pH stress to induce the stringent response in H. pylori, they observed the same accumulation pattern for only ppGpp and not the precursor pppGpp, as we showed with alkaline stress in P. aeruginosa. The mechanism triggering the accumulation of ppGpp in response to pH stress might be linked to the pH-dependent perturbation of the proton gradient since treatment with the protonophor carbonyl cyanide m-chlorophenylhydrazone has also been shown to induce a SpoT-regulated stringent response by a reduction of the rate of ppGpp degradation in E. coli (50). Nevertheless, the capability of SpoT to detect and respond to a variety of apparently unrelated stress factors remains a highly complex and greatly unsolved network, which now can be additionally linked to environmental alkaline pH.
Stringent response-mediated induction of PuspNΔAnr in stationary phase is independent of 10 tested alternative sigma factors.
Gene expression by ppGpp occurs either directly by an interplay of DksA, a transcriptional cofactor of stringent response, and the housekeeping sigma factor σ70 (38) or indirectly by an increase in competitiveness of other alternative sigma factors for the core of RNA polymerase (2). Additionally, expression of genes transcribed by alternative sigma factors like RpoS and RpoN has been shown to be dependent on the presence of ppGpp (24, 49). In order to identify sigma factors involved in ppGpp-mediated induction of PuspNΔAnr in stationary phase, we screened for reduced β-galactosidase activities in diverse transposon mutants deficient in known alternative sigma factors (algU, fliA, pvdS, fiuI, rpoN, and rpoS) or putative sigma factors (PA0149, PA1912, PA2050, and PA2896) (for a review, see reference 40). All tested mutant strains showed similar or even higher expression levels of PuspNΔAnr-lacZ than the wild type, which excluded the tested sigma factors as essential cofactors in ppGpp-mediated induction of PuspNΔAnr-lacZ. That the expression values of PuspNΔAnr in the tested mutant strains (329 to 500 Miller units) (Table 2) were higher than the wild type (280 Miller units) supports the assumption that a different sigma factor is controlling PuspN in concert with ppGpp. From these results we assume that ppGpp induction of PuspN might be directly mediated by σ70 and DksA. However, we cannot exclude the possibility that other unknown regulatory components or sigma factors coregulate ppGpp-mediated induction of PuspNΔAnr during stationary phase. This question is currently under investigation in our laboratory.
Supplementary Material
Acknowledgments
We thank Sabrina Thoma from our laboratory for providing E. coli ST18.
The study was sponsored by grants of the Deutsche Forschungsgemeinschaft. K.S. was supported by the DFG-European Graduate College, “Pseudomonas—Pathogenicity and Biotechnology” project no. 653.
Footnotes
Published ahead of print on 5 September 2008.
Supplemental material for this article may be found at http://jb.asm.org/.
REFERENCES
- 1.Alvarez-Ortega, C., and C. S. Harwood. 2007. Responses of Pseudomonas aeruginosa to low oxygen indicate that growth in the cystic fibrosis lung is by aerobic respiration. Mol. Microbiol. 65153-165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Barker, M. M., T. Gaal, and R. L. Gourse. 2001. Mechanism of regulation of transcription initiation by ppGpp. II. Models for positive control based on properties of RNAP mutants and competition for RNAP. J. Mol. Biol. 305689-702. [DOI] [PubMed] [Google Scholar]
- 3.Battesti, A., and E. Bouveret. 2006. Acyl carrier protein/SpoT interaction, the switch linking SpoT-dependent stress response to fatty acid metabolism. Mol. Microbiol. 621048-1063. [DOI] [PubMed] [Google Scholar]
- 4.Becher, A., and H. P. Schweizer. 2000. Integration-proficient Pseudomonas aeruginosa vectors for isolation of single-copy chromosomal lacZ and lux gene fusions. BioTechniques 29948-950, 952. [DOI] [PubMed] [Google Scholar]
- 5.Bochkareva, E. S., A. S. Girshovich, and E. Bibi. 2002. Identification and characterization of the Escherichia coli stress protein UP12, a putative in vivo substrate of GroEL. Eur. J. Biochem. 2693032-3040. [DOI] [PubMed] [Google Scholar]
- 6.Boes, N., K. Schreiber, E. Härtig, L. Jaensch, and M. Schobert. 2006. The Pseudomonas aeruginosa universal stress protein PA4352 is essential for surviving anaerobic energy stress. J. Bacteriol. 1886529-6538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Brockmann-Gretza, O., and J. Kalinowski. 2006. Global gene expression during stringent response in Corynebacterium glutamicum in presence and absence of the rel gene encoding (p)ppGpp synthase. BMC Genomics 7230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Cashel, M., D. R. Gentry, V. J. Hernandez, and J. Vinella. 1996. The stringent response, p. 1458-1496. In F. C. Neidhardt, R. Curtiss III, J. L. Ingraham, E. C. C. Lin, K. B. Low, B. Magasanik, W. S. Reznikoff, M. Riley, M. Schaechter, and H. E. Umbarger (ed.), Escherichia coli and Salmonella typhimurium: cellular and molecular biology. ASM Press, Washington, DC.
- 9.Cooper, M., G. R. Tavankar, and H. D. Williams. 2003. Regulation of expression of the cyanide-insensitive terminal oxidase in Pseudomonas aeruginosa. Microbiology 1491275-1284. [DOI] [PubMed] [Google Scholar]
- 10.Davies, K. J., D. Lloyd, and L. Boddy. 1989. The effect of oxygen on denitrification in Paracoccus denitrificans and Pseudomonas aeruginosa. J. Gen. Microbiol. 1352445-2451. [DOI] [PubMed] [Google Scholar]
- 11.de Lorenzo, V., and K. N. Timmis. 1994. Analysis and construction of stable phenotypes in gram-negative bacteria with Tn5- and Tn10-derived minitransposons. Methods Enzymol. 235386-405. [DOI] [PubMed] [Google Scholar]
- 12.Dunn, N. W., and B. W. Holloway. 1971. Pleiotrophy of p-fluorophenylalanine-resistant and antibiotic hypersensitive mutants of Pseudomonas aeruginosa. Genet. Res. 18185-197. [DOI] [PubMed] [Google Scholar]
- 13.Durfee, T., A. M. Hansen, H. Zhi, F. R. Blattner, and D. J. Jin. 2008. Transcription profiling of the stringent response in Escherichia coli. J. Bacteriol. 1901084-1096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Erickson, D. L., J. L. Lines, E. C. Pesci, V. Venturi, and D. G. Storey. 2004. Pseudomonas aeruginosa relA contributes to virulence in Drosophila melanogaster. Infect. Immun. 725638-5645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Eschbach, M., K. Schreiber, K. Trunk, J. Buer, D. Jahn, and M. Schobert. 2004. Long-term anaerobic survival of the opportunistic pathogen Pseudomonas aeruginosa via pyruvate fermentation. J. Bacteriol. 1864596-4604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Eymann, C., G. Homuth, C. Scharf, and M. Hecker. 2002. Bacillus subtilis functional genomics: global characterization of the stringent response by proteome and transcriptome analysis. J. Bacteriol. 1842500-2520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Galimand, M., M. Gamper, A. Zimmermann, and D. Haas. 1991. Positive FNR-like control of anaerobic arginine degradation and nitrate respiration in Pseudomonas aeruginosa. J. Bacteriol. 1731598-1606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Gentry, D. R., and M. Cashel. 1996. Mutational analysis of the Escherichia coli spoT gene identifies distinct but overlapping regions involved in ppGpp synthesis and degradation. Mol. Microbiol. 191373-1384. [DOI] [PubMed] [Google Scholar]
- 19.Gong, L., K. Takayama, and S. Kjelleberg. 2002. Role of spoT-dependent ppGpp accumulation in the survival of light-exposed starved bacteria. Microbiology 148559-570. [DOI] [PubMed] [Google Scholar]
- 20.Gustavsson, N., A. Diez, and T. Nystrom. 2002. The universal stress protein paralogues of Escherichia coli are co-ordinately regulated and co-operate in the defence against DNA damage. Mol. Microbiol. 43107-117. [DOI] [PubMed] [Google Scholar]
- 21.Heurlier, K., V. Denervaud, M. Haenni, L. Guy, V. Krishnapillai, and D. Haas. 2005. Quorum-sensing-negative (lasR) mutants of Pseudomonas aeruginosa avoid cell lysis and death. J. Bacteriol. 1874875-4883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hoang, T. T., R. R. Karkhoff-Schweizer, A. J. Kutchma, and H. P. Schweizer. 1998. A broad-host-range Flp-FRT recombination system for site-specific excision of chromosomally-located DNA sequences: application for isolation of unmarked Pseudomonas aeruginosa mutants. Gene 21277-86. [DOI] [PubMed] [Google Scholar]
- 23.Jishage, M., K. Kvint, V. Shingler, and T. Nystrom. 2002. Regulation of sigma factor competition by the alarmone ppGpp. Genes Dev. 161260-1270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kvint, K., A. Farewell, and T. Nystrom. 2000. RpoS-dependent promoters require guanosine tetraphosphate for induction even in the presence of high levels of sigma(s). J. Biol. Chem. 27514795-14798. [DOI] [PubMed] [Google Scholar]
- 25.Kvint, K., C. Hosbond, A. Farewell, O. Nybroe, and T. Nystrom. 2000. Emergency derepression: stringency allows RNA polymerase to override negative control by an active repressor. Mol. Microbiol. 35435-443. [DOI] [PubMed] [Google Scholar]
- 26.Kvint, K., L. Nachin, A. Diez, and T. Nystrom. 2003. The bacterial universal stress protein: function and regulation. Curr. Opin. Microbiol. 6140-145. [DOI] [PubMed] [Google Scholar]
- 27.Liberati, N. T., J. M. Urbach, S. Miyata, D. G. Lee, E. Drenkard, G. Wu, J. Villanueva, T. Wei, and F. M. Ausubel. 2006. An ordered, nonredundant library of Pseudomonas aeruginosa strain PA14 transposon insertion mutants. Proc. Natl. Acad. Sci. USA 1032833-2838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Mikkelsen, H., Z. Duck, K. S. Lilley, and M. Welch. 2007. Interrelationships between colonies, biofilms, and planktonic cells of Pseudomonas aeruginosa. J. Bacteriol. 1892411-2416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Miller, J. M. 1992. A short course in bacterial genetics: a laboratory manual and handbook for Escherichia coli and related bacteria. Cold Spring Harbor Laboratory Laboratory Press, Cold Spring Harbor, NY.
- 30.Mittenhuber, G. 2001. Comparative genomics and evolution of genes encoding bacterial (p)ppGpp synthetases/hydrolases (the Rel, RelA and SpoT proteins). J. Mol. Microbiol. Biotechnol. 3585-600. [PubMed] [Google Scholar]
- 31.Mouery, K., B. A. Rader, E. C. Gaynor, and K. Guillemin. 2006. The stringent response is required for Helicobacter pylori survival of stationary phase, exposure to acid, and aerobic shock. J. Bacteriol. 1885494-5500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Münch, R., K. Hiller, A. Grote, M. Scheer, J. Klein, M. Schobert, and D. Jahn. 2005. Virtual Footprint and PRODORIC: an integrative framework for regulon prediction in prokaryotes. Bioinformatics 214187-4189. [DOI] [PubMed] [Google Scholar]
- 33.Murray, K. D., and H. Bremer. 1996. Control of spoT-dependent ppGpp synthesis and degradation in Escherichia coli. J. Mol. Biol. 25941-57. [DOI] [PubMed] [Google Scholar]
- 34.Nachin, L., U. Nannmark, and T. Nystrom. 2005. Differential roles of the universal stress proteins of Escherichia coli in oxidative stress resistance, adhesion, and motility. J. Bacteriol. 1876265-6272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Nystrom, T., and F. C. Neidhardt. 1994. Expression and role of the universal stress protein, UspA, of Escherichia coli during growth arrest. Mol. Microbiol. 11537-544. [DOI] [PubMed] [Google Scholar]
- 36.O'Toole, R., M. J. Smeulders, M. C. Blokpoel, E. J. Kay, K. Lougheed, and H. D. Williams. 2003. A two-component regulator of universal stress protein expression and adaptation to oxygen starvation in Mycobacterium smegmatis. J. Bacteriol. 1851543-1554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Palmer, K. L., S. A. Brown, and M. Whiteley. 2007. Membrane-bound nitrate reductase is required for anaerobic growth in cystic fibrosis sputum. J. Bacteriol. 1894449-4455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Paul, B. J., M. B. Berkmen, and R. L. Gourse. 2005. DksA potentiates direct activation of amino acid promoters by ppGpp. Proc. Natl. Acad. Sci. USA 1027823-7828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Persson, O., A. Valadi, T. Nystrom, and A. Farewell. 2007. Metabolic control of the Escherichia coli universal stress protein response through fructose-6-phosphate. Mol. Microbiol. 65968-978. [DOI] [PubMed] [Google Scholar]
- 40.Potvin, E., F. Sanschagrin, and R. C. Levesque. 2008. Sigma factors in Pseudomonas aeruginosa. FEMS Microbiol. Rev. 3238-55. [DOI] [PubMed] [Google Scholar]
- 41.Richter, D. 1980. Uncharged tRNA inhibits guanosine 3′,5′-bis (diphosphate) 3′-pyrophosphohydrolase [ppGppase], the spoT gene product, from Escherichia coli. Mol. Gen. Genet. 178325-327. [DOI] [PubMed] [Google Scholar]
- 42.Rompf, A., C. Hungerer, T. Hoffmann, M. Lindenmeyer, U. Romling, U. Gross, M. O. Doss, H. Arai, Y. Igarashi, and D. Jahn. 1998. Regulation of Pseudomonas aeruginosa hemF and hemN by the dual action of the redox response regulators Anr and Dnr. Mol. Microbiol. 29985-997. [DOI] [PubMed] [Google Scholar]
- 43.Sawers, R. G. 1991. Identification and molecular characterization of a transcriptional regulator from Pseudomonas aeruginosa PAO1 exhibiting structural and functional similarity to the FNR protein of Escherichia coli. Mol. Microbiol. 51469-1481. [DOI] [PubMed] [Google Scholar]
- 44.Schreiber, K., N. Boes, M. Eschbach, L. Jaensch, J. Wehland, T. Bjarnsholt, M. Givskov, M. Hentzer, and M. Schobert. 2006. Anaerobic survival of Pseudomonas aeruginosa by pyruvate fermentation requires an Usp-type stress protein. J. Bacteriol. 188659-668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Seyfzadeh, M., J. Keener, and M. Nomura. 1993. spoT-dependent accumulation of guanosine tetraphosphate in response to fatty acid starvation in Escherichia coli. Proc. Natl. Acad. Sci. USA 9011004-11008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Son, M. S., W. J. Matthews, Jr., Y. Kang, D. T. Nguyen, and T. T. Hoang. 2007. In vivo evidence of Pseudomonas aeruginosa nutrient acquisition and pathogenesis in the lungs of cystic fibrosis patients. Infect. Immun. 755313-5324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Spiro, S., and J. R. Guest. 1990. FNR and its role in oxygen-regulated gene expression in Escherichia coli. FEMS Microbiol. Rev. 6399-428. [DOI] [PubMed] [Google Scholar]
- 48.Storz, G., and R. Hengge-Aronis. 2000. Bacterial stress response. ASM Press, Washington DC.
- 49.Szalewska-Palasz, A., L. U. Johansson, L. M. Bernardo, E. Skarfstad, E. Stec, K. Brannstrom, and V. Shingler. 2007. Properties of RNA polymerase bypass mutants: implications for the role of ppGpp and its co-factor DksA in controlling transcription dependent on σ54. J. Biol. Chem. 28218046-18056. [DOI] [PubMed] [Google Scholar]
- 50.Tetu, C., E. Dassa, and P. L. Boquet. 1980. The energy-dependent degradation of guanosine 5′-diphosphate 3′-diphosphate in Escherichia coli. Lack of correlation with ATP levels in vivo and role of the transmembrane proton gradient. Eur. J. Biochem. 103117-124. [DOI] [PubMed] [Google Scholar]
- 51.Traxler, M. F., S. M. Summers, H. T. Nguyen, V. M. Zacharia, G. A. Hightower, J. T. Smith, and T. Conway. 2008. The global, ppGpp-mediated stringent response to amino acid starvation in Escherichia coli. Mol. Microbiol. 681128-1148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.van Delden, C., R. Comte, and A. M. Bally. 2001. Stringent response activates quorum sensing and modulates cell density-dependent gene expression in Pseudomonas aeruginosa. J. Bacteriol. 1835376-5384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Vander Wauven, C., A. Pierard, M. Kley-Raymann, and D. Haas. 1984. Pseudomonas aeruginosa mutants affected in anaerobic growth on arginine: evidence for a four-gene cluster encoding the arginine deiminase pathway. J. Bacteriol. 160928-934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Waite, R. D., A. Paccanaro, A. Papakonstantinopoulou, J. M. Hurst, M. Saqi, E. Littler, and M. A. Curtis. 2006. Clustering of Pseudomonas aeruginosa transcriptomes from planktonic cultures, developing and mature biofilms reveals distinct expression profiles. BMC Genomics 7162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Wells, D. H., and E. C. Gaynor. 2006. Helicobacter pylori initiates the stringent response upon nutrient and pH downshift. J. Bacteriol. 1883726-3729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Wendrich, T. M., G. Blaha, D. N. Wilson, M. A. Marahiel, and K. H. Nierhaus. 2002. Dissection of the mechanism for the stringent factor RelA. Mol. Cell 10779-788. [DOI] [PubMed] [Google Scholar]
- 57.Worlitzsch, D., R. Tarran, M. Ulrich, U. Schwab, A. Cekici, K. C. Meyer, P. Birrer, G. Bellon, J. Berger, T. Weiss, K. Botzenhart, J. R. Yankaskas, S. Randell, R. C. Boucher, and G. Doring. 2002. Effects of reduced mucus oxygen concentration in airway Pseudomonas infections of cystic fibrosis patients. J. Clin. Investig. 109317-325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Wout, P., K. Pu, S. M. Sullivan, V. Reese, S. Zhou, B. Lin, and J. R. Maddock. 2004. The Escherichia coli GTPase CgtAE cofractionates with the 50S ribosomal subunit and interacts with SpoT, a ppGpp synthetase/hydrolase. J. Bacteriol. 1865249-5257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Xiao, H., M. Kalman, K. Ikehara, S. Zemel, G. Glaser, and M. Cashel. 1991. Residual guanosine 3′,5′-bispyrophosphate synthetic activity of relA null mutants can be eliminated by spoT null mutations. J. Biol. Chem. 2665980-5990. [PubMed] [Google Scholar]
- 60.Ye, R. W., D. Haas, J. O. Ka, V. Krishnapillai, A. Zimmermann, C. Baird, and J. M. Tiedje. 1995. Anaerobic activation of the entire denitrification pathway in Pseudomonas aeruginosa requires Anr, an analog of Fnr. J. Bacteriol. 1773606-3609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Yoon, S. S., R. F. Hennigan, G. M. Hilliard, U. A. Ochsner, K. Parvatiyar, M. C. Kamani, H. L. Allen, T. R. DeKievit, P. R. Gardner, U. Schwab, J. J. Rowe, B. H. Iglewski, T. R. McDermott, R. P. Mason, D. J. Wozniak, R. E. Hancock, M. R. Parsek, T. L. Noah, R. C. Boucher, and D. J. Hassett. 2002. Pseudomonas aeruginosa anaerobic respiration in biofilms: relationships to cystic fibrosis pathogenesis. Dev. Cell 3593-603. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.