Abstract
While remaining a major problem in hospitals, Staphylococcus aureus is now spreading in communities. Strain MW2 (USA400 lineage) and other community methicillin-resistant S. aureus strains most commonly cause skin infections with abscess formation. Multidrug resistance (MDR) efflux pumps contribute to antimicrobial resistance but may also contribute to bacterial survival by removal of environmental toxins. In S. aureus, NorA, NorB, NorC, and Tet38 are chromosomally encoded efflux pumps whose overexpression can confer MDR to quinolones and other compounds (Nor pumps) or tetracyclines alone (Tet38), but the natural substrates of these pumps are not known. To determine the role of these efflux pumps in a natural environment in the absence of antibiotics, we used strain MW2 in a mouse subcutaneous abscess model and compared pump gene expression as determined by reverse transcription-PCR in the abscesses and in vitro. norB and tet38 were selectively upregulated in vivo more than 171- and 24-fold, respectively, whereas norA and norC were downregulated. These changes were associated with an increase in expression of mgrA, which encodes a transcriptional regulator known to affect pump gene expression. In competition experiments using equal inocula of a norB or tet38 mutant and parent strain MW2, each mutant exhibited growth defects of about two- to threefold in vivo. In complementation experiments, a single-copy insertion of norB (but not a single-copy insertion of tet38) in the attB site within geh restored the growth fitness of the norB mutant in vivo. Our findings indicate that some MDR pumps, like NorB, can facilitate bacterial survival when they are overexpressed in a staphylococcal abscess and may contribute to the relative resistance of abscesses to antimicrobial therapy, thus linking bacterial fitness and resistance in vivo.
Staphylococcus aureus, which readily colonizes the nasal mucosa and skin, is one of the most important causes of human infections, and these infections range from mild skin infections to potentially lethal bacteremia and endocarditis. S. aureus has become a major public health concern as a result of the steadily increasing incidence of antimicrobial resistance. Before the 1950s, staphylococcal infections could often be cured with penicillin. Soon after this time, penicillin was progressively less often active as S. aureus strains acquired a penicillinase. Methicillin, which is resistant to the action of penicillinases, was introduced for treatment of staphylococcal infections in 1960, but methicillin-resistant S. aureus (MRSA) strains were soon isolated, and they emerged in the early 1980s as a major cause of hospital-acquired infections (9). MRSA strains now commonly circulate and cause nosocomial infections in hospitals and long-term care facilities, and the occurrence of MRSA infections has been further amplified by the rapid and clonal spread of community-associated MRSA strains (CA-MRSA) in the United States (11). Strain MW2 was identified early as a CA-MRSA strain that was responsible for severe soft tissue and bloodstream infections. Genome sequence analysis has identified multiple virulence determinants, some of which contribute to the ability of this organism to form subcutaneous abscesses.
Genes encoding efflux pumps have been found to be part of the normal genetic makeup of S. aureus and other human pathogens, and resistance or reduced susceptibility to a number of antimicrobials can result when these genes are overexpressed (20, 21). Several genes in the S. aureus genome that encode efflux pumps have been characterized (8, 17, 27). NorA, NorB, and NorC are multidrug resistance (MDR) pumps that confer resistance to quinolones and other agents, and Tet38 is specific for resistance to tetracyclines. Although a number of antimicrobial agents are natural products, quinolones are not, and in many cases the natural roles of MDR pumps are uncertain. The expression of staphylococcal efflux pumps is controlled in part by the transcriptional regulators MgrA and NorG and by the ArlRS two-component regulatory system (8, 19). MgrA and ArlRS also regulate expression of multiple other genes, including the genes responsible for capsule biosynthesis and autolysin production, thus linking efflux pump expression to the coordination of other central cellular metabolic activities. Mutations in MgrA and NorG and the overexpression of these proteins from plasmids alter pump expression patterns and can variously cause resistance to quinolones, chloramphenicol, some β-lactams, and certain dyes. Substrate-level induction of expression of these staphylococcal pumps has not been demonstrated, and thus the environmental triggers of expression remain unknown.
In order to assess the roles of efflux pump expression in vivo, we used a subcutaneous abscess model with S. aureus strain MW2 to evaluate the expression of four established staphylococcal efflux pumps and the contribution of overexpressed pumps to bacterial survival and replication in the abscess environment. Expression of the NorB and Tet38 pumps was selectively increased in abscesses relative to the expression in laboratory media in association with an increase in MgrA expression, establishing that a different pattern of pump expression was related to the abscess environment. In addition, norB and tet38 mutants were shown to have a selective growth defect in the abscess environment that could, in the case of norB, be complemented by insertion of the intact gene into the chromosome, thus identifying a role for an MDR pump that contributes to bacterial fitness in an abscess.
MATERIALS AND METHODS
Bacterial strains and growth conditions.
Bacterial strains and plasmids used in this study are listed in Table 1. All of the S. aureus strains except RN4220 (r-) were derivatives of CA-MRSA strain MW2 (2, 3). S. aureus strains were cultivated in a rich medium, brain heart infusion (BHI) broth (Difco, Sparks, MD), and Escherichia coli strains were grown in Luria-Bertani (LB) broth (Difco) at 37°C, unless otherwise indicated. Antibiotics were obtained from Fisher (Sewanee, GA) or Sigma (St. Louis, MO) and used at the following concentrations: ampicillin, 100 μg/ml; chloramphenicol, 5 μg/ml and 10 μg/ml for S. aureus strains having one and two copies of the cat gene, respectively, and 15 μg/ml for strains carrying a cat plasmid; kanamycin, 50 μg/ml; and tetracycline, 3 μg/ml. 5-Bromo-4-chloro-3-indolyl-β-d-galactopyranoside (X-Gal) (Sigma, St. Louis, MO) was used at 260 μg/ml in LB agar. Lysostaphin (Sigma, St. Louis, MO) was used to facilitate the lysis of S. aureus. All primers used in this study (Table 2) were synthesized at the MGH DNA Core Facility, Boston, MA.
TABLE 1.
Bacterial strains and plasmids used in this study
| Strain or plasmid | Genotype or characteristicsa | Reference(s) or source |
|---|---|---|
| S. aureus strains | ||
| RN4220 (r-) | Restriction-deficient mutagenized RN450 | 12 |
| MW2 | CA-MRSA (USA400 lineage) | 1, 3 |
| MW2 norB | MW2 norB::cat | This study |
| MW2 tet38 | MW2 tet38::cat | This study |
| MW2 geh::pSK950 | MW2 with pSK950 insert in geh | This study |
| MW2 norB geh::pSK950 | MW2 norB with pSK950 insert in geh | This study |
| MW2 tet38 geh::pSK950 | MW2 tet38 with pSK950 insert in geh | This study |
| MW2 norB geh::pSKnorB | MW2 norB with pSKnorB insert in geh | This study |
| MW2 tet38 geh::pSKtet38 | MW2 tet38 with pSKtet38 insert in geh | This study |
| E. coli DH5α | General host for pUC vector | Gibco-BRL |
| Plasmids | ||
| pSK950 | Spr (E. coli), Tcr and Emr (S. aureus), temperature sensitive, L54a attP site | 18 |
| pYL112Δ19 | Apr (E. coli) Cmr (S. aureus), pYL112 without attP site | 13, 31 |
| pCL52.2 | Tcr, E. coli-S. aureus shuttle vector, temperature sensitive | 23 |
| pYO1 | pCL52.2 with norB::cat fragment | This study |
| pYO2 | pCL52.2 with tet38::cat fragment | This study |
| pSKnorB | pSK950 with norB complementary fragment, temperature sensitive | This study |
| pSKtet38 | pSK950 with tet38 complementary fragment, temperature sensitive | This study |
| pUC19 | Apr, general-purpose cloning vector | 30 |
Ap, ampicillin; Cm, chloramphenicol; Em, erythromycin; Sp, spectinomycin; Tc, tetracycline.
TABLE 2.
Primers used for PCR or RT-PCR
| Primer | Sequencea |
|---|---|
| NorBF | 5′-CCCGAATTCTAATAACAGCACTTGGTATAC |
| NorBR | 5′-CCCGAATTCAAAAACCAATCCCTATGTTC |
| Tet38F | 5′-CCCGGATCCGTATGGGTATTTTAGGTGTA |
| Tet38R | 5′-CCCGGATCCCGTGTTATATAATCTCTTTA |
| TET-7 | 5′-AGGTATGGGGATCCCATCTATTGG |
| TET-8 | 5′-CCTGTTGAAGTTTTAACGAAGTCCG |
| SANB-3 | 5′-GCTATCATCATGATGTCAATGGGAG |
| SANB-4 | 5′-CTTAATAATAGACAAAGTTGCAGGC |
| SANB-5 | 5′-AAGTAGGCGTTGCTGCAGGTATC |
| SANB-6 | 5′-CTGTTTAACCACGCTTGCTGGTTG |
| norA-RTf | 5′-ATCGGTTTAGTAATACCAGTCTTGC |
| norA-RTr | 5′-GCGATATAATCATTTGAGATAACGC |
| norB-RTf | 5′-ATGGAAAAGCCGTCAAGAGA |
| norB-RTr | 5′-AACCAATGATTGTGCAAATAGC |
| norC-RTf | 5′-ATGAATGAAACGTATCGCGG |
| norC-RTr | 5′-GTCTGCACCAAAACTTTGTTGTAAA |
| tet38-RTf | 5′-AAATTGATTTCTGTAGCCATTGCTG |
| tet38-RTr | 5′-ACAGCGCCAATACCAATTACT |
| mgrA-RTf | 5′-GAACAGCTATGCTTTAGTTTGTACA |
| mgrA-RTr | 5′-GACAAGAAATTGTGGGTATGTTAGA |
| gmk-RTf | 5′-TATCAGGACCATCTGGAGTAGG |
| gmk-RTr | 5′-CATCAACTTCACCTTCACGC |
| attRf | 5′-GCTGTCTTTTTCGTGACATTCA |
| attRr | 5′-TTTGAAATCCAGGAAGTCCAC |
| attLf | 5′-CTGCCTATGATTTAACGTTAGATG |
| attLr | 5′-GCAGTTACAGAGTGGAATAGAGCAT |
Appended nucleotides with restriction endonuclease sites are underlined.
Strain and plasmid construction.
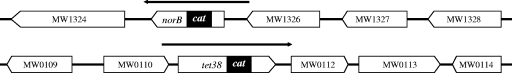
MW2 tet38 was generated after partial deletion of the tet38 coding sequence and insertion of a cat gene, which confers resistance to chloramphenicol. Allelic exchange was performed using the temperature-sensitive shuttle plasmid pYO1, a pCL52.2 derivative, as described previously (23). To partially replace the tet38 coding sequence with a functional cat gene (Fig. 1), tet38 with 520 bp of upstream sequence and 544 bp of downstream sequence was amplified from MW2 chromosomal DNA by performing PCR with primers TET-7 and TET-8 and cloned in pUC19 at the BamHI and HindIII sites. A PCR yielded a 800-bp DNA fragment with the cat gene (28), which was used to replace the fragment between PstI and BglII sites within tet38 in pUC19 by blunt-end ligation. The resultant ∼2.4-kb tet38::cat fragment in pUC19 was then subcloned in pCL52.2 at BamHI and HindIII sites to obtain pYO1. After electroporation into RN4220 (r-), pYO1 was introduced into MW2 by electroporation, and transformants were allowed to grow at 30°C in the presence of chloramphenicol and tetracycline. MW2 harboring pYO1 was then propagated at 42°C for 24 h on BHI agar (Difco) containing chloramphenicol. A colony was picked and grown at 30°C without antibiotics for four successive subcultures. Chloramphenicol-resistant, tetracycline-sensitive colonies, representing possible double-crossover events, were screened for the cat insert in tet38 by performing PCR and sequence analysis of the PCR product.
FIG. 1.
Structure of the tet38 and norB loci on the MW2 chromosome. The filled rectangles show the extent of the in-frame deletion and replacement by the cat gene. The horizontal arrows indicate the chromosomal fragments that were cloned in pSK950 to complement the norB and tet38 deletions.
MW2 norB was constructed by using a procedure similar to the procedure used to construct MW2 tet38, with the following exceptions (Fig. 1). Two DNA fragments flanking norB were amplified from the MW2 chromosome by PCR. Primers SANB-3 and SANB-4 were used to amplify an 843-bp fragment containing upstream sequences and the 5′ region of norB (5′-norB fragment), and primers SANB-5 and SANB-6 were used to amplify a 765-bp fragment containing the 3′ region and a region downstream of norB (3′-norB fragment). The 3′-norB fragment was digested with HindIII and PstI, and the fragment was cloned into pUC19. The resulting plasmid was digested with PstI and made blunt; an 800-bp DNA fragment containing the cat gene was inserted. Then the plasmid was digested with BamHI and SalI, and the 5′-norB fragment digested with BamHI and SalI was inserted. The resultant plasmid contained a ∼2.4-kb norB::cat fragment, which was subsequently inserted into pCL52.2 at BamHI and HindIII sites to generate pYO2. This temperature-sensitive plasmid was employed to generate MW2 norB.
Complementation of the norB and tet38 mutant strains.
An E. coli-S. aureus shuttle vector, pSK950 (24), was used to generate complementation strains with single copies of norB and tet38 (see Fig. 4). This vector contains an attP site from phage L54a, and thus it can integrate at the attB site of the lipase gene (geh) on the MW2 chromosome when cells are grown at the nonpermissive temperature (42°C) for plasmid replication and in the presence of the L54a integrase gene provided by plasmid pYL112Δ19 (13). The entire 1,357-bp tet38 and 1,392-bp norB coding sequences along with 191 bp and 216 bp of upstream sequences were amplified from MW2 chromosomal DNA using primer NorBF with primer NorBR and primer Tet38F with primer Tet38R, respectively (Fig. 1). PCR fragments were cloned into pSK950 at EcoRI or BamHI sites to generate pSKnorB and pSKtet38, respectively. After electroporation into RN4220 (r-), these plasmids were subsequently introduced by electroporation into MW2 norB and MW2 tet38 which already harbored pYL112Δ19, respectively. BHI agar plates containing 3 μg/ml tetracycline (for selection for pSK950 and its derivatives) and 15 μg/ml chloramphenicol (for selection for pYL112Δ19) were used to screen transformants at 30°C. A temperature shift from 30°C to 42°C induced integration of pSK950 derivatives at the attB site in transformants grown in BHI broth without antibiotics. After plasmid pYL112Δ19 was cured from host cells by successive growth at 42°C, complemented strains MW2 norB geh::pSKnorB and MW2 tet38 geh::pSKtet38 were obtained. Strain MW2 and its norB and tet38 mutants were also transformed with pSK950 lacking inserts in order to generate strains MW2 geh::pSK950, MW2 norB geh::pSK950, and MW2 tet38 geh::pSK950. The authenticity of the single-copy integration at the attB site of chromosomal geh was confirmed by DNA sequence analysis using PCR products of attL and attR sites.
FIG. 4.
Diagram showing single-copy integration in the geh gene in the chromosome using pSK950. The filled triangle indicates the attB site in geh at which complementing plasmids pSKnorB and pSKtet38 were integrated.
Mouse abscess model and in vivo growth.
Swiss Webster male mice that were 4 to 6 weeks old were used for the subcutaneous abscess model (4, 7). Exponential-phase cells were prepared by diluting overnight cultures 1:100 into BHI broth and incubating the cultures at 37°C with rotation until the optical density at 600 nm (OD600) was 0.8. MW2 alone or a 1:1 mixture of MW2 and either its norB mutant or its tet38 mutant was washed with and diluted 1:20 in phosphate-buffered saline (PBS). The cell suspension was then mixed with an equal volume of autoclaved Cytodex-1 beads (131 to 220 μm; Sigma, St. Louis, MO) in PBS. A 0.2-ml suspension of S. aureus cells and beads was injected subcutaneously into each shaved flank of a mouse anesthetized with ketamine and xylazine. The inoculum for each abscess typically contained 1 × 106 to 3 × 106 CFU S. aureus. In experiments in which a single strain was inoculated, this protocol was used with one strain instead of two strains. After 48 h, the mice were euthanized, and the subcutaneous abscesses were removed aseptically and homogenized in 4 ml PBS. After 10-fold serial dilutions in PBS were prepared, the total numbers of bacteria recovered from the abscesses were determined by plating preparations on LB agar and LB agar containing 5 μg/ml chloramphenicol. The number of MW2 cells was calculated by subtracting the number of colonies growing on the chloramphenicol-containing agar from the number of colonies growing on the antibiotic-free agar. For in vitro competition assays, 100 μl of a suspension of cells and beads was inoculated into 4 ml of BHI broth and grown for ∼24 h at 37°C with rotation.
Total bacterial RNA isolation.
Total S. aureus RNA was isolated using an RNeasy mini kit (Qiagen, Valencia, CA) by following the manufacturer's protocol. For early-stationary- and stationary-phase bacterial RNA, MW2 grown in BHI broth at 37°C was collected when the OD600 of the culture reached ∼7.0 (after 6 h) and ∼15.0 (after 26 h), respectively, and the cells were lysed with lysostaphin. Total RNA of MW2 collected from abscesses was isolated as follows. Abscess tissue (a mixture of S. aureus cells, host cells, and Cytodex-1 beads) was removed aseptically from mice and was suspended in 0.5 ml RLT buffer (RNeasy mini kit; Qiagen) in a 1.5-ml microcentrifuge tube (6). The tube was immediately applied to an amalgamator and vigorously shaken for 1 min. The supernatant was discarded after a brief centrifugation (1 min at 16,000 × g). After addition to the tube of 0.5 ml RLT buffer and 2 capfuls (0.2-ml PCR tube) of 0.1-mm zirconia-silica beads (BioSpec, Bartlesville, OK), the bacterial cells were disrupted in the amalgamator for 1 min. After centrifugation (3 min at 16,000 × g), the supernatant of the cell lysate was collected and then mixed with an equal volume of 70% ethanol. Bacterial RNA was isolated by using the manufacturer's protocols. Following this procedure the preparation contained predominantly bacterial RNA, as assessed by visualization of bacterial rRNA bands without visible eukaryotic rRNA bands on native agarose gels stained by ethidium bromide (data not shown). After treatment with DNase I (TURBO DNA-free; Ambion, Austin, TX), RNA samples contained no DNA detectable by PCR.
Real-time RT-PCR.
cDNA synthesis was performed using Verso cDNA (ABgene, Epsom, Surrey, United Kingdom) with gene-specific reverse primers according to the manufacturer's instructions. Using the resulting cDNAs as templates, quantitative PCR amplification was conducted in Chromo 4 on PTC-200 (Bio-Rad, Hercules, CA) using SYBR green master mixture from ABgene. The cycling parameters were 94°C for 15 min, followed by 40 cycles of 95°C for 15 s, 55°C for 15 s, and 72°C for 15 s. The primers used for reverse transcription (RT) and quantitative PCR are shown in Table 2. The relative expression of efflux pump genes was calculated by using the ΔΔCT method, in which the amount of target cDNA, normalized to an endogenous reference (housekeeping gene gmk [29]) and relative to an in vitro calibrator, is given by the variable 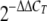 , where CT is the cycle number of the detection threshold.
, where CT is the cycle number of the detection threshold.
Statistical methods.
For the fitness competition assays using norB and tet38 mutants grown together with the parent MW2 strain, the statistical significance of differences was assessed as previously described (14). Briefly, the ratio of the numbers of CFU of the competing strains (mutant and parent strains) in the inoculum and harvested from an abscess was transformed to the natural logarithm. The difference between the ln ratios for the inoculum and abscess was compared to zero, representing no difference, and the significance was estimated by using a two-tailed, paired Student's t test.
RESULTS
Mouse subcutaneous abscess model.
Strain MW2 was chosen for the mouse model because it commonly causes skin and soft tissue infections, including subcutaneous abscesses, in humans. In the mouse model MW2 reliably formed abscesses with an inoculum containing ∼106 CFU together with Cytodex beads, which potentiate infection (16). During the 48-h abscess formation period, the level of the strain consistently increased to 1 × 108 to 8 × 108 CFU, indicating that MW2 was able to replicate approximately 100-fold in tissues. The number of CFU peaked between 24 and 72 h (data not shown). In addition, a visibly localized abscess with a thick layer of surrounding fibrous tissue formed by 48 h after inoculation. At later time points some abscesses (about one-fourth of the abscesses by 72 h) drained spontaneously, and for this reason 48 h was chosen as the time point for harvesting bacteria in abscesses. The Cytodex beads are abscess-promoting agents in the mouse model, as discussed in previous studies (4, 7, 16). Without them, ∼108 CFU of MW2 was needed to provoke formation of an abscess (data not shown) (5). The use of Cytodex beads allowed us to use an inoculum that was 100-fold smaller and to measure bacterial gene expression as bacteria replicated in vivo.
Staphylococcal NorB and Tet38 are upregulated in abscesses.
We hypothesized that the pattern of S. aureus efflux pump expression in an abscess would differ from the patterns in the exponential and stationary phases of staphylococcal growth in vitro. To examine this hypothesis, we measured the transcript levels of the genes for four known efflux pumps, NorA, NorB, NorC, and Tet38, which are encoded on the chromosome of S. aureus and have been shown to confer resistance to quinolones (NorA, NorB, and NorC) or tetracycline (Tet38). Additionally, we evaluated the transcript levels of MgrA, a transcriptional regulator involved in the expression of all four pumps. Compared to the transcript levels for bacteria grown to stationary phase in BHI broth (Fig. 2), the transcript levels of norB and tet38 in the abscesses were 171- and 24-fold greater, respectively (Table 3). In contrast, norA and norC were downregulated in the abscesses (4- and 1.6-fold, respectively). Similar patterns were confirmed by performing RT-PCR using different samples and primers and measuring the intensities of the PCR amplicon bands on gels stained by ethidium bromide compared to the intensities for 16S rRNA expression (data not shown).
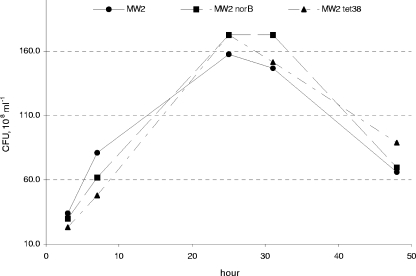
FIG. 2.
Growth curves for MW2 and norB and tet38 mutants in vitro. Cells were grown in BHI broth at 37°C.
TABLE 3.
Expression of MW2 efflux pump genes in murine subcutaneous abscessesa
| Gene | Mean relative expression | SEM |
|---|---|---|
| mgrA | 4.99 | 2.79 |
| norA | 0.24 | 0.10 |
| norB | 171.49 | 61.47 |
| norC | 0.62 | 0.30 |
| tet38 | 24.80 | 15.55 |
The relative expression of efflux pumps in abscesses was normalized by using expression of gmk and was compared to the expression in vitro in the stationary phase. At least four sets of samples were assayed independently, and the means and standard errors of the means were determined.
Expression of mgrA also increased ∼5-fold in the abscesses compared to growth to stationary phase in BHI broth (Table 3), suggesting that MgrA functions as a positive regulator of norB and tet38 but a negative regulator of norA and norC in strain MW2. This regulation pattern is similar to that described for strain Newman, in which MgrA upregulates norB and downregulates norA but has no effect on tet38 (15).
Expression of efflux pumps in different growth phases.
In order to assess further whether the pattern of efflux pump gene expression observed in the abscesses could be attributed to the bacterial growth phase, total bacterial RNA was extracted from MW2 grown in BHI broth in the exponential phase (OD600, ∼0.3; ∼2 h), early stationary phase (OD600, ∼8.0; ∼7 h), and stationary phase (OD600, ∼15.0; ∼26 h). For the most part the transcript levels of norA, norB, norC, and tet38 as measured by real-time RT-PCR (Table 4) increased in the early stationary and stationary phases compared to the exponential phase; the only exceptions were the levels of tet38 (level not altered in early stationary phase) and norB (level decreased almost twofold in stationary phase). NorC showed the most striking increase in expression during early stationary phase (84-fold) and stationary phase (31-fold). The pattern and extent of the expression were distinctly different from those in an abscess environment, suggesting that the upregulation of norB and tet38 in abscesses (Table 3) is specifically triggered by the abscess conditions and is not attributable to reaching the stationary phase of growth in the abscesses.
TABLE 4.
Expression of MW2 efflux pump genes in different growth phasesa
| Gene | Avg relative expression (SEM)
|
|
|---|---|---|
| Early stationary phase | Stationary phase | |
| mgrA | 9.30 (1.82) | 2.92 (1.28) |
| norA | 6.59 (1.53) | 4.97 (1.38) |
| norB | 5.44 (1.75) | 0.57 (0.39) |
| norC | 84.65 (16.89) | 31.28 (26.01) |
| tet38 | 1.06 (0.23) | 9.85 (6.77) |
MW2 was grown in BHI broth at 37°C. gmk was used as an internal reference gene. Averages and standard errors of the means were calculated based on the results for three sets of samples.
S. aureus norB or tet38 mutants show specific growth defects in an abscess.
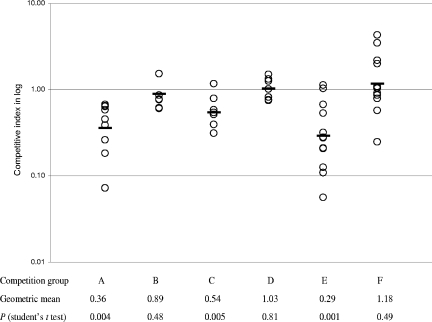
norB and tet38 in MW2 were upregulated in abscesses, suggesting that their increased expression may be important for bacterial growth and survival in abscesses. To test this hypothesis, we constructed norB and tet38 mutants and tested their fitness in the abscess model of S. aureus infection. When inoculated individually, each of the mutants was able to provoke abscess formation in mice by 48 h after bacterial challenge. The abscesses were similar in size and bacterial load to those caused by wild-type parent strain MW2 (data not shown). Thus, we performed competition experiments in which each mutant was inoculated with parent strain MW2. Using a mutant/parent input ratio of ∼1:1, we measured changes in the bacterial strain ratios with growth under in vivo and in vitro conditions and calculated a competitive index (CI). In the in vivo competition experiments, the CIs were 0.36 for MW2 norB versus MW2 and 0.54 for MW2 tet38 versus MW2, and in both cases the differences were highly statistically significant (Fig. 3). In contrast, the CIs in the in vitro competition experiments performed with the mutants and the parent strain, 0.89 and 1.03, respectively, were not statistically different. In addition, the growth of both mutants in BHI broth was similar to that of MW2, and both mutants showed no growth defect by at least 48 h (Fig. 2). These data indicate that neither norB nor tet38 mutations obviate abscess formation, but the mutant strains exhibited a distinctive growth defect in vivo that was not seen in vitro, suggesting that NorB and Tet38 contribute specifically to fitness for S. aureus survival and growth in abscesses.
FIG. 3.
Competitive growth of norB and tet38 mutants with wild-type strain MW2. The CI is defined as the mutant/wild-type output CFU ratio normalized by using the input CFU ratio. A CI of <1.0 indicates a mutant with a growth disadvantage compared to the wild type. Each open circle indicates a CI for either an abscess (in vivo) or a BHI culture (in vitro). The geometric mean of CIs for each group is indicated by a horizontal bar. Competition experiments were repeated separately at least three times for each group. Competition group A, MW2 norB versus MW2 in vivo; competition group B, MW2 norB versus MW2 in vitro; competition group C, MW2 tet38 versus MW2 in vivo; competition group D, MW2 tet38 versus MW2 in vitro; competition group E, MW2 norB geh::pSK950 versus MW2 geh::pSK950 in vivo; competition group F, MW2 norB geh::pSKnorB versus MW2 geh::pSK950 in vivo.
A single copy of norB restores abscess fitness to the MW2 norB mutant.
To determine whether the fitness defects of the norB and tet38 mutants seen in the abscess model could be attributed specifically to the disruption of norB or tet38, we generated mutants complemented with chromosomal insertions of intact copies of the corresponding wild-type genes (Fig. 1 and 4). The single copy of norB, carried by pSK950, was specifically integrated at the MW2 attB site for phage L54a, which is located within the chromosomal lipase gene, geh (Fig. 4). The transcript levels of norB in trans in the norB-complemented strain were about 2.43 and 0.57 times the levels in the wild-type parent strain in the exponential and early stationary phases, respectively, suggesting that norB expression in the norB-complemented strain was similar to that in the wild-type parent strain. To confirm the virulence defect of the norB strain after integration into the geh gene, we repeated the competition experiments in mice using MW2 norB geh::pSK950 and MW2 geh::pSK950 and obtained a CI of 0.29, a statistically significant difference consistent with the results of previous experiments. In vivo competition of MW2 norB geh::pSKnorB and MW2 geh::pSK950, however, generated a CI of 1.18, which did not differ statistically from 1, suggesting that a single copy of intact norB restores abscess fitness to the norB mutant. Thus, intact norB contributes specifically to bacterial survival in mouse abscesses, indicating the importance of NorB efflux pump gene expression in this environment.
Efforts to demonstrate complementation of the weaker in vivo growth defect of the tet38 mutant were not successful (data not shown), and thus the role of tet38 in bacterial fitness in an abscess is uncertain. It is unlikely due to direct polar effects of disruption of geh, since geh and its downstream gene (MW0298) are orientated in opposite directions. One noticeable finding was that the abundance of the tet38 transcript was about 225- and 7.6-fold higher in the tet38 complemented strain than in the wild-type parent strain in the exponential and early stationary phase, respectively, suggesting that the complemented tet38 gene in trans was highly upregulated, which might lead to excess production of Tet38. Particularly high levels of Tet38 may disturb S. aureus physiology and thus be toxic to the bacterial cells.
DISCUSSION
Efflux pumps are widespread in free-living bacterial species, and many bacteria have multiple efflux pumps. It is estimated that efflux pumps constitute more than 10% of the total number of transporters per organism (19). Such pumps may have a variety of functions, such as export of bacterial products and removal of environmental substances that the organism encounters, including natural antibiotics or other natural toxins. MDR pumps have broad substrate profiles, and many have been studied largely to determine their abilities to confer levels of resistance to antimicrobial agents and disinfectants used in human and veterinary medicine (20, 21); however, in most cases their native functions have not been defined. In the case of E. coli, the AcrAB-TolC pump is induced by and provides resistance to bile salts, a property that likely underlies the ability of this organism to be present at high levels in the human intestinal tract (22). For S. aureus, however, no such natural function for its MDR efflux pumps is known.
In S. aureus, the chromosomally encoded Tet38 pump and the MDR pumps NorA, NorB, and NorC are widely present in different strains and are identified based on their ability to confer resistance to tetracyclines and quinolones, respectively. Studies have shown that both types of efflux pumps belong to one group of the major facilitator superfamily (25, 26). In addition to hydrophilic (norfloxacin and ciprofloxacin) and hydrophobic (moxifloxacin and sparfloxacin) quinolones, Nor family pumps can also extrude other chemical compounds, such as ethidium bromide, cetrimide, and tetraphenylphosphonium.
Among the substrates, the tetracyclines are natural products and might have provided evolutionary selection pressure in nature, but the quinolones are synthetic agents and did not create an evolutionary selection pressure prior to their introduction into clinical medicine in the 1960s. In an effort to understand the roles of the pumps in the natural functions of S. aureus, we focused on subcutaneous abscesses, a type of infection commonly caused by S. aureus and an environment in which bacterial cells, in order to be successful as pathogens, must survive a variety of host defense mechanisms that include toxic antimicrobial substances. We found that a specific pattern of expression of the four pumps examined occurs in response to growth in an abscess, which is distinct from the pattern of expression that occurs in bacteria growing to high density in laboratory media. This pattern appears to be mediated in part by changes in expression of the transcriptional regulator MgrA, which has been shown to have direct and indirect effects on pump gene expression in cells grown in vitro. Because the pattern of efflux pump expression in an abscess is distinct from the pattern seen when cells reach stationary phase in vitro, there are likely specific environmental triggers that determine the cellular response. The overexpression of norB and tet38 is also not likely due to the staphylococcal general response to stress stimuli. When challenged by different stress conditions (cold shock, heat shock, stringent conditions, and SOS), the expression of only one of the four efflux pump genes (norA, norB, norC, and tet38), norA, was induced (10-fold) as part of the stringent response of S. aureus (1). Thus, we postulate that specific elements of the abscess environment trigger the observed pattern of efflux pump expression. These elements have not been defined yet, but they are the subject of ongoing work.
In this context, the overexpression of norB and tet38 in the abscesses suggests that these genes and their products may have important functions in bacteria in infection, even in the absence of antibiotic exposure, and thereby contribute to staphylococcal pathogenesis. Competition experiments clearly demonstrated that norB and presumably the NorB protein provide fitness to S. aureus in an abscess. The fitness defect of a norB mutant was consistent, specific for abscesses, and complemented by a single copy of norB provided in trans. We postulate that the fitness advantage provided by increased expression of norB in an abscess is related to the ability of NorB to remove antibacterial toxins produced by host defense mechanisms. The broad substrate profiles of MDR pumps like NorB document the ability of the pumps to remove structurally diverse molecules from the cell. Which host-derived antistaphylococcal substance(s) in an abscess is a substrate for NorB remains to be defined, and we cannot yet exclude the possibility that NorB exports a cellular substance that itself contributes to pathogenesis. The concept that efflux pumps may contribute to bacterial pathogenesis is supported by the results of work with Neisseria gonorrhoeae, in which the MtrCDE efflux pump system enhances bacterial survival in a mouse genital tract infection (10).
Drug resistance is a serous problem for staphylococcal infections in both hospital and community settings, and efflux pump expression in S. aureus can contribute to resistance to disinfectants and several antimicrobials, including macrolides, trimethoprim, and fluoroquinolones. The substrate profile of NorB includes levofloxacin and moxifloxacin, which are marketed fluoroquinolones with antistaphylococcal activities. Thus, physiologic overexpression of NorB or other drug resistance pumps in an abscess environment might contribute in part to the reduced responses to antibacterial agents seen during treatment of abscesses and might contribute to the discordance between determinations of bacterial susceptibility in vitro and responses to antimicrobials in vivo. Thus, in S. aureus and other bacteria some efflux pumps may provide a link between bacterial pathogenesis and antibiotic resistance.
Acknowledgments
This work was supported in part by Public Health Service grant AI093288 from the National Institutes of Health (to D.C.H).
We thank Chia Y. Lee for providing plasmid pYL112Δ19 and technical assistance.
Footnotes
Published ahead of print on 22 August 2008.
REFERENCES
- 1.Anderson, K. L., C. Roberts, T. Disz, V. Vonstein, K. Hwang, R. Overbeek, P. D. Olson, S. J. Projan, and P. M. Dunman. 2006. Characterization of the Staphylococcus aureus heat shock, cold shock, stringent, and SOS responses and their effects on log-phase mRNA turnover. J. Bacteriol. 1886739-6756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Anonymous. 1999. Four pediatric deaths from community-acquired methicillin-resistant Staphylococcus aureus—Minnesota and North Dakota, 1997-1999. JAMA 2821123-1125. [PubMed] [Google Scholar]
- 3.Baba, T., F. Takeuchi, M. Kuroda, H. Yuzawa, K. Aoki, A. Oguchi, Y. Nagai, N. Iwama, K. Asano, T. Naimi, H. Kuroda, L. Cui, K. Yamamoto, and K. Hiramatsu. 2002. Genome and virulence determinants of high virulence community-acquired MRSA. Lancet 3591819-1827. [DOI] [PubMed] [Google Scholar]
- 4.Bunce, C., L. Wheeler, G. Reed, J. Musser, and N. Barg. 1992. Murine model of cutaneous infection with gram-positive cocci. Infect. Immun. 602636-2640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Chan, P. F., S. J. Foster, E. Ingham, and M. O. Clements. 1998. The Staphylococcus aureus alternative sigma factor σB controls the environmental stress response but not starvation survival or pathogenicity in a mouse abscess model. J. Bacteriol. 1806082-6089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Di Cello, F., Y. Xie, M. Paul-Satyaseela, and K. S. Kim. 2005. Approaches to bacterial RNA isolation and purification for microarray analysis of Escherichia coli K1 interaction with human brain microvascular endothelial cells. J. Clin. Microbiol. 434197-4199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ford, C. W., J. C. Hamel, D. Stapert, and R. J. Yancey. 1989. Establishment of an experimental model of a Staphylococcus aureus abscess in mice by use of dextran and gelatin microcarriers. J. Med. Microbiol. 28259-266. [DOI] [PubMed] [Google Scholar]
- 8.Fournier, B., R. Aras, and D. C. Hooper. 2000. Expression of the multidrug resistance transporter NorA from Staphylococcus aureus is modified by a two-component regulatory system. J. Bacteriol. 182664-671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Grundmann, H., M. Ires-de-Sousa, J. Boyce, and E. Tiemersma. 2006. Emergence and resurgence of meticillin-resistant Staphylococcus aureus as a public-health threat. Lancet 368874-885. [DOI] [PubMed] [Google Scholar]
- 10.Jerse, A. E., N. D. Sharma, A. N. Simms, E. T. Crow, L. A. Snyder, and W. M. Shafer. 2003. A gonococcal efflux pump system enhances bacterial survival in a female mouse model of genital tract infection. Infect. Immun. 715576-5582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Klevens, R. M., M. A. Morrison, J. Nadle, S. Petit, K. Gershman, S. Ray, L. H. Harrison, R. Lynfield, G. Dumyati, J. M. Townes, A. S. Craig, E. R. Zell, G. E. Fosheim, L. K. McDougal, R. B. Carey, S. K. Fridkin, and the Active Bacterial Core surveillance (ABCs) MRSA investigators. 2007. Invasive methicillin-resistant Staphylococcus aureus infections in the United States. JAMA 2981763-1771. [DOI] [PubMed] [Google Scholar]
- 12.Kreiswirth, B. 1983. The toxic shock syndrome exotoxin structural gene is not detectably transmitted by a prophage. Nature 305709-712. [DOI] [PubMed] [Google Scholar]
- 13.Lee, C. Y., S. L. Buranen, and Y. Zhi-Hai. 1991. Construction of single-copy integration vectors for Staphylococcus aureus. Gene 103101-105. [DOI] [PubMed] [Google Scholar]
- 14.Lipsitch, M., C. T. Bergstrom, and B. R. Levin. 2000. The epidemiology of antibiotic resistance in hospitals: paradoxes and prescriptions. Proc. Natl. Acad. Sci. USA 971938-1943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Luong, T. T., P. M. Dunman, E. Murphy, S. J. Projan, and C. Y. Lee. 2006. Transcription profiling of the mgrA regulon in Staphylococcus aureus. J. Bacteriol. 1881899-1910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.McLoughlin, R. M., R. M. Solinga, J. Rich, K. J. Zaleski, J. L. Cocchiaro, A. Risley, A. O. Tzianabos, and J. C. Lee. 2006. CD4+ T cells and CXC chemokines modulate the pathogenesis of Staphylococcus aureus wound infections. Proc. Natl. Acad. Sci. USA 10310408-10413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ng, E. Y., M. Trucksis, and D. C. Hooper. 1994. Quinolone resistance mediated by norA: physiologic characterization and relationship to flqB, a quinolone resistance locus on the Staphylococcus aureus chromosome. Antimicrob. Agents Chemother. 381345-1355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Niemeyer, D. M., M. J. Pucci, J. A. Thanassi, V. K. Sharma, and G. L. Archer. 1996. Role of mecA transcriptional regulation in the phenotypic expression of methicillin resistance in Staphylococcus aureus. J. Bacteriol. 1785464-5471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Paulsen, I. T. 2003. Multidrug efflux pumps and resistance: regulation and evolution. Curr. Opin. Microbiol. 6446-451. [DOI] [PubMed] [Google Scholar]
- 20.Poole, K. 2000. Efflux-mediated resistance to fluoroquinolones in gram-negative bacteria. Antimicrob. Agents Chemother. 442233-2241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Poole, K. 2000. Efflux-mediated resistance to fluoroquinolones in gram-positive bacteria and the mycobacteria. Antimicrob. Agents Chemother. 442595-2599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Rosenberg, E. Y., D. Bertenthal, M. L. Nilles, K. P. Bertrand, and H. Nikaido. 2003. Bile salts and fatty acids induce the expression of Escherichia coli AcrAB multidrug efflux pump through their interaction with Rob regulatory protein. Mol. Microbiol. 481609-1619. [DOI] [PubMed] [Google Scholar]
- 23.Sau, S., J. Sun, and C. Y. Lee. 1997. Molecular characterization and transcriptional analysis of type 8 capsule genes in Staphylococcus aureus. J. Bacteriol. 1791614-1621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Sharma, V. K., C. J. Hackbarth, T. M. Dickinson, and G. L. Archer. 1998. Interaction of native and mutant MecI repressors with sequences that regulate mecA, the gene encoding penicillin binding protein 2a in methicillin-resistant staphylococci. J. Bacteriol. 1802160-2166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Truong-Bolduc, Q. C., P. M. Dunman, J. Strahilevitz, S. J. Projan, and D. C. Hooper. 2005. MgrA is a multiple regulator of two new efflux pumps in Staphylococcus aureus. J. Bacteriol. 1872395-2405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Truong-Bolduc, Q. C., and D. C. Hooper. 2007. Transcriptional regulators NorG and MgrA modulate resistance to both quinolones and β-lactams in Staphylococcus aureus. J. Bacteriol. 1892996-3005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Truong-Bolduc, Q. C., J. Strahilevitz, and D. C. Hooper. 2006. NorC, a new efflux pump regulated by MgrA of Staphylococcus aureus. Antimicrob. Agents Chemother. 501104-1107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Truong-Bolduc, Q. C., X. Zhang, and D. C. Hooper. 2003. Characterization of NorR protein, a multifunctional regulator of norA expression in Staphylococcus aureus. J. Bacteriol. 1853127-3138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Vandecasteele, S. J., W. E. Peetermans, R. Merckx, and J. Van Eldere. 2001. Quantification of expression of Staphylococcus epidermidis housekeeping genes with Taqman quantitative PCR during in vitro growth and under different conditions. J. Bacteriol. 1837094-7101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Yanisch-Perron, C., J. Vieira, and J. Messing. 1985. Improved M13 phage cloning vectors and host strains: nucleotide sequences of the M13mp18 and pUC19 vectors. Gene 33103-119. [DOI] [PubMed] [Google Scholar]
- 31.Ye, Z. H., and C. Y. Lee. 1989. Nucleotide sequence and genetic characterization of staphylococcal bacteriophage L54a int and xis genes. J. Bacteriol. 1714146-4153. [DOI] [PMC free article] [PubMed] [Google Scholar]