Abstract
Renibacterium salmoninarum is the causative agent of bacterial kidney disease and a significant threat to healthy and sustainable production of salmonid fish worldwide. This pathogen is difficult to culture in vitro, genetic manipulation is challenging, and current therapies and preventative strategies are only marginally effective in preventing disease. The complete genome of R. salmoninarum ATCC 33209 was sequenced and shown to be a 3,155,250-bp circular chromosome that is predicted to contain 3,507 open-reading frames (ORFs). A total of 80 copies of three different insertion sequence elements are interspersed throughout the genome. Approximately 21% of the predicted ORFs have been inactivated via frameshifts, point mutations, insertion sequences, and putative deletions. The R. salmoninarum genome has extended regions of synteny to the Arthrobacter sp. strain FB24 and Arthrobacter aurescens TC1 genomes, but it is approximately 1.9 Mb smaller than both Arthrobacter genomes and has a lower G+C content, suggesting that significant genome reduction has occurred since divergence from the last common ancestor. A limited set of putative virulence factors appear to have been acquired via horizontal transmission after divergence of the species; these factors include capsular polysaccharides, heme sequestration molecules, and the major secreted cell surface antigen p57 (also known as major soluble antigen). Examination of the genome revealed a number of ORFs homologous to antibiotic resistance genes, including genes encoding β-lactamases, efflux proteins, macrolide glycosyltransferases, and rRNA methyltransferases. The genome sequence provides new insights into R. salmoninarum evolution and may facilitate identification of chemotherapeutic targets and vaccine candidates that can be used for prevention and treatment of infections in cultured salmonids.
Infection of salmon and trout by Renibacterium salmoninarum causes bacterial kidney disease (BKD), a progressive, multifocal, granulomatous disease that involves all internal organs, particularly the kidney (21, 26, 67). This gram-positive bacterium causes morbidity and mortality in both farmed and wild fish in nearly all regions of the world where salmonids are found (66). Despite the fact that R. salmoninarum has been recognized as a major fish pathogen for over 70 years, the mechanisms of R. salmoninarum transmission, pathogenesis, and immune evasion are poorly understood. The disease is chronic in nature, and mortality occurs most often in 6- to 12-month-old juvenile salmonids and prespawning adults (17). The spread of BKD has followed the rapid expansion of salmonid culture, and to date, most recorded outbreaks of BKD have occurred in fish culture facilities; the losses have been as high as 80% in stocks of Pacific salmon and 40% in stocks of Atlantic salmon (Salmo salar) (21). The chronic nature of the disease has hindered accurate estimation of fish losses in feral fish populations. In the Pacific Northwest of the United States, the prevalence of R. salmoninarum in juvenile Chinook salmon can vary from 60 to 100% in different populations, although the pathogen burden is usually low in the majority of the fish (16, 52). BKD is also the greatest cause of infectious disease-related mortality in restoration and conservation programs for several endangered species (25, 35).
R. salmoninarum is one of the few vertebrate bacterial pathogens known to be vertically transmitted by intraovum infection during egg maturation (7, 19, 20). This pathogen can also be transmitted horizontally from infected fish sharing a water supply (1, 40). Prophylaxis or prevention of R. salmoninarum infection is challenging, due in part to the ability of this bacterium to survive phagocytosis and possibly replicate within macrophages (2, 33, 62, 73). For Pacific salmon, there are no efficacious vaccines, and antibiotic therapy never fully eliminates the pathogen (15, 55). For Atlantic salmon, however, which has greater innate resistance, immunization with live Arthrobacter davidanieli (commercially known as Renogen) can provide significant cross-protection (32, 57). The ability of A. davidanieli to protect against BKD is presumably a function of the overall genetic relationship between the genus Arthrobacter and R. salmoninarum (Fig. 1A). Both taxa are high-G+C-content gram-positive members of the Micrococcaceae (26).
FIG. 1.
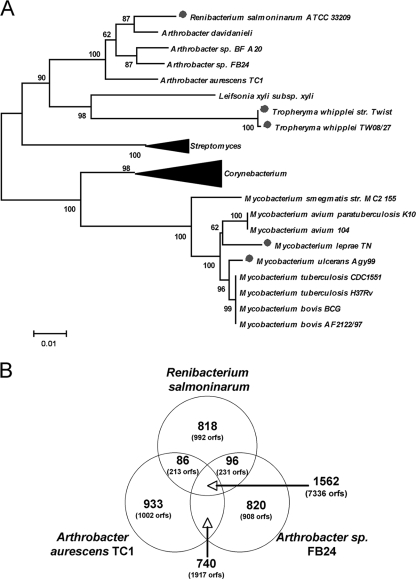
R. salmoninarum is closely related to Arthrobacter spp. based on 16S rRNA sequence analysis (A) and protein cluster analysis (B). (A) 16S rRNA sequences were aligned using ClustalW, and a tree was constructed using the neighbor-joining method with 1,000 bootstrap replications. The dots indicate pathogenic bacterial species with evidence of genome reduction. Bar = 0.01 nucleotide substitution. The large arrowheads indicate collapsed branches containing the 16S rRNA sequences of the following Streptomyces and Corynebacterium strains: Corynebacterium diphtheriae NCTC 13129 (accession number NC_002935), Corynebacterium jeikeium K411 (accession number NC_007164), Corynebacterium efficiens YS-314 (accession number NC_004369), Corynebacterium glutamicum ATCC 13032 (accession number NC_006958), Streptomyces coelicolor A3(2) (accession number NC_003888), and Streptomyces avermitilis MA-4680 (accession number NC_003155). Ten additional 16S rRNA sequences from completed genomes were included in the alignment but are not shown. (B) Venn diagram showing the numbers of common and unique protein clusters for R. salmoninarum and the sequenced Arthrobacter spp. The numbers of protein clusters and the total numbers of ORFs in the clusters are indicated.
There are many challenges in studying the biology and pathogenesis of R. salmoninarum. This bacterium grows extremely slowly in vitro, and up to 6 weeks is required for primary isolation (26). While genetic tools for studying R. salmoninarum are available, the process used to introduce or inactivate specific genes is arduous and time-consuming (51). Because of these challenges to research with this organism, we completed and analyzed the R. salmoninarum genome sequence. The results of this sequencing study demonstrate that this pathogen evolved, via genomic reduction and horizontal gene acquisition, from members of the nonpathogenic genus Arthrobacter.
MATERIALS AND METHODS
Clone isolation.
R. salmoninarum ATCC 33209 was originally isolated from a yearling Chinook salmon (Oncorhynchus tshawytscha) at a salmon hatchery in western Oregon (58). Although other workers have reported that this strain is modestly attenuated compared to other R. salmoninarum isolates (39, 50, 60), challenge experiments carried out prior to extraction of DNA for sequencing showed that it had not lost virulence due to laboratory passage (see Fig. S1 in the supplemental material). A single colony of the pathogen was isolated and cultured in KDM-II medium (18) supplemented with 10% bovine serum for production of DNA. Genomic DNA was prepared as described previously, and the DNA was used for construction of small- and large-insert libraries for sequencing (70).
Genome sequencing.
A whole-genome shotgun sequencing approach was used to determine the sequence of the R. salmoninarum strain ATCC 33209 genome, as described previously (34, 71). Randomly picked small-insert plasmids (average insert size, 2.6 kbp), cloned into the blunt-ended pUC19 vector, were sequenced at both ends using universal forward and reverse sequencing primers and standard protocols established at the University of Washington Genome Center. Altogether, 52,183 reads were performed, which provided 9.7× sequence coverage with an average Q20 (22) of 590 bases/read. A Sau3A partial fosmid library was also constructed in the pCCFOS vector digested with BamHI using the standard protocol (48). A total of 768 independent fosmid clones were picked and paired-end sequenced, and mated paired-end sequences were obtained for 91.8% of the clones. BigDye terminator chemistry and capillary DNA sequencers (model 3700; Applied Biosystems) were used for generating the sequence data (34, 71). The sequence data were assembled and visualized using Phred/Phrap/Consed software tools (22, 23, 28). The sequence quality and contiguity were improved by carrying out six rounds of experiments designed by using the Autofinish tool in Consed (29). The Autofinish-designed experiments utilized primarily small-insert clones to improve the sequence quality and/or to close some gaps. Following six rounds of Autofinish, both small-insert and fosmid clones were utilized for manual finishing. This involved (i) using specialized sequencing chemistries to sequence difficult regions, (ii) PCR amplification and sequencing of specific targeted regions, (iii) transposon mutagenesis of the small-insert clones, followed by sequencing to fix misassembled or difficult-to-assemble regions, and (iv) shotgun sequencing of the targeted fosmid clones to fix long-range misassemblies in the assembled genome. Altogether, 8,968 finishing reads were attempted during the course of this project. In addition, 52 small-insert clones were mutagenized by transposon mutagenesis, and independent clones were picked to generate a consensus sequence for these clones. Twenty-nine fosmid clones that spanned misassembled or large gap regions were sequenced and assembled independently. The consensus sequences for transposon mutagenized and fosmid clones were used as backbones in the main R. salmoninarum genome assembly to resolve misassembled regions. The final R. salmoninarum ATCC 33209 genome assembly was oriented so that the genome coordinates started at the origin of replication (ori) at position 1, as defined by the GC skew, the characteristic nucleotide composition, and the presence of dnaA immediately following the ori.
Genome assembly validation.
The final genome assembly was validated by two independent methods. The gross-scale long-range validity of the genome assembly was established by complete agreement between the virtual fingerprint pattern of the genome in PmeI, PacI, or SwaI restriction enzyme domains and the experimentally determined restriction fragment sizes determined by pulsed-field gel electrophoresis (data not shown). For 1-kb and larger-scale validation, fingerprint data were generated from the paired-end sequences of 768 fosmid clones by digestion with three independent restriction enzymes, BglII, HindIII, and PstI, generating 2,256 fingerprints or 97.9% of the theoretical 2,304 fingerprints. The fosmid paired-end sequence and experimentally derived fingerprint data were used for validation of the assembly by comparison with the virtual fingerprint patterns for the assembled genome using the SeqTile software tools developed for this purpose at the University of Washington Genome Center (34, 71). Thus, the assembly was validated at 1-kb resolution by comparing fosmid-based fingerprints for BglII, HindIII, and PstI restriction enzyme domains with the virtual patterns for the finished genome assembly. A complete correspondence between the virtual and experimentally derived fingerprint patterns of the genome in the BglII, HindIII, and PstI restriction enzyme domains was observed, which validated the genome assembly.
Phylogenetic trees.
16S rRNA sequences were obtained using an NCBI taxonomy browser (http://www.ncbi.nlm.nih.gov) search for Actinobacteria (high-G+C-content gram-positive bacteria) with known genome sequences. 16S rRNA sequences were compared using a predetermined alignment from the NAST alignment tool (11). A manually inspected, multiple alignment was used as the input for generation of phylogenetic trees, using the program package MEGA v 3.1 (37). Both neighbor-joining and maximum parsimony methods, performed using default settings in the MEGA program, were employed to obtain character-based trees. The two methods yielded similar clusters and arrangements of taxa. The genome sequences of Arthrobacter sp. strain BF A20 and A.davidanieli have not been determined, but the 16S RNA sequences were included for comparative purposes.
ORF identification and annotation.
Open reading frame (ORF) calling was performed as described previously (46). Two rounds of automated annotation were initially performed utilizing the ERGO genome analysis suite (Integrated Genomics, Chicago, IL) during genome assembly and closure (46). The final finished genome sequence was also subjected to automated ORF calling and annotation on 23 August 2006 using The Institute for Genomics Research (TIGR) automated annotation software (65). Subsequently, each ORF was manually examined using a number of bioinformatics tools, including BLAST and PSI-BLAST (http://www.ncbi.nlm.nih.gov:80/blast/Blast.cgi?CMD=Web&PAGE_TYPE=BlastHome), Pfam (http://pfam.janelia.org/), TMPred (http://www.ch.embnet.org/software/TMPRED_form.html), and ProSite (http://myhits.isb-sib.ch/cgi-bin/index). During the annotation process, a large number of partial genes were noted. Most of these ORFs were also flagged by ERGO and TIGR ORF-calling software as possible frameshifts and point mutations. The sequence data for candidate frameshifts were manually inspected to rule out sequencing or assembly errors. These ORFs were also compared with putative orthologues in the Arthrobacter sp. strain FB24 and Arthrobacter aurescens TC1 genomes. Candidate partial ORFs and the protein-encoding regions were designated either “partial,” “partial N terminus” or “partial C terminus.” The partial ORFs were then categorized using the TIGR cellular main role list, as shown in the supplemental material.
Sequencing of selected R. salmoninarum genes from different sources.
DNAs from laboratory strains R. salmoninarum MT239 and R. salmoninarum 684 were prepared as previously described (70). DNA was also prepared from kidney tissue of naturally infected Upper Columbia River spring Chinook salmon (Onchorhyncus tschawytscha) brood stock being reared at the Bonneville Fish Hatchery, Bonneville, OR. These fish had recently died of R. salmoninarum infection, and their kidneys were heavily infected with the pathogen. Each of these DNAs was used as a template for PCR amplification of selected genes, using Pfx DNA polymerase and oligonucleotide primers shown in Table 1. Products of the amplification reactions were cloned into the pCR-Blunt II-TOPO vector according to the manufacturer's directions (Invitrogen, CA). Plasmids were purified, and the nucleotide sequence of each cloned insert was determined in both orientations. Each of the sequences was compared to the corresponding gene sequence in the completed R. salmoninarum ATCC 33209 genome sequence using ClustalW in the MacVector DNA analysis program (MacVector, Inc., Cary, NC). Any gene sequences that differed from those in the ATCC 33209 genome were reamplified, recloned, and resequenced for confirmation.
TABLE 1.
Oligonucleotide primers used for amplification of four fragmented R. salmoninarum ORFs from recent clinical isolates and clinically infected Chinook salmon
| Oligonucleotide | Gene | Oligonucleotide sequence |
|---|---|---|
| dppD/F (F) | RSal33209_1660 | 5′-GGCCGCGATGTGTCGATGGTTTT-3′ |
| dppD/F (R) | RSal33209_1659 | 5′-GCCCAAGTGCGGCACTGCAGC-3′ |
| Citrate synthase (F) | RSal33209_2900 | 5′-GCGCGGAGAGAAGTTCTTGTG-3′ |
| Citrate synthase (R) | RSal33209_2898 | 5′-CGATGCGGTGCGACGTTTT-3′ |
| tetP (F) | RSal33209_1874 | 5′-GCCTAGCGACGCAAAAG-3′ |
| tetP (R) | RSal33209_1873 | 5′-ATAGTGACTAAGCAATCGGTG-3′ |
| fbp (F) | RSal33209_1552 | 5′-CTGACGCCAACGGTAAATACACC-3′ |
| fbp (R) | RSal33209_1551 | 5′-GGCGGATTCTCAACACTCACG-3′ |
Comparative genomics of R. salmoninarum, A. aurescens TC1, and Arthrobacter sp. strain FB24.
ERGO Workbench was used to determine common and unique Role/IDs for R. salmoninarum, A. aurescens TC1, and Arthrobacter sp. strain FB24. Clusters of similar Role/IDs were generated using the Build Clusters function and a protein similarity threshold of 1.0e−10, and subset clusters were extracted by organism sets containing either individual genomes, paired combinations of genomes, or all three genomes.
Nucleotide sequence accession number.
The R. salmoninarum ATCC 33209 genome sequence has been deposited in the GenBank database under accession number NC_010168.
RESULTS
General genome features.
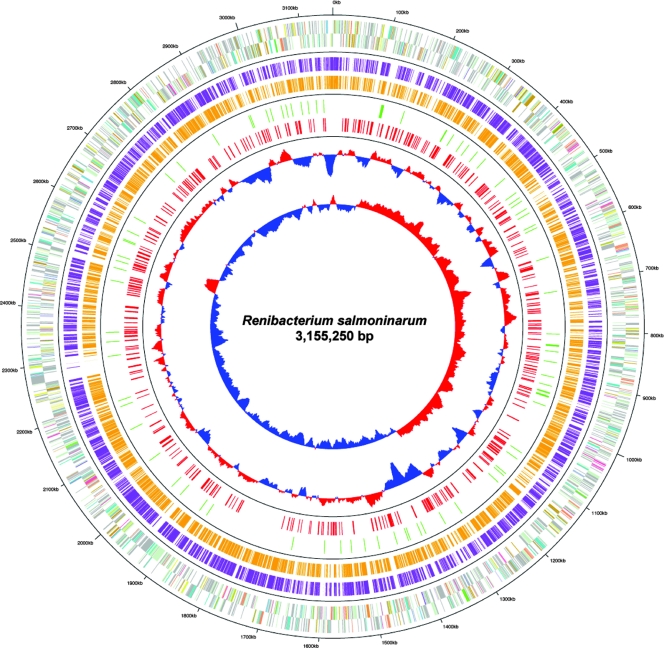
The final R. salmoninarum ATCC 33209 genome was assembled as a single circular chromosome consisting of 3,155,250 bases (Table 2), and 51,799 shotgun reads were included in the final genome assembly. Cumulative GC skew analysis identified a transition in the G+C content close to dnaA (Fig. 2). There are 46 tRNAs and two rRNA operons located at bp 364,813 to 370,362 and bp 674,875 to 680,424 in relation to the putative origin of replication. The G+C content is 56.3%, in agreement with previous measurements (3). This value is 6 to 9% lower than the value for either sequenced species of Arthrobacter (41) (Table 2). The ATCC 33209 strain lacks any associated plasmids or apparent integrated phage. The genome was predicted to contain 2,777 protein-encoding sequences, and manual annotation identified 730 “partial” protein-encoding sequences that represent putative pseudogenes; 360 of the partial gene sequences were disrupted by frameshifts, 208 were disrupted by point mutations, and 162 were disrupted by uncharacterized changes, including insertion sequences and putative deletions (Fig. 2; see Table S1 in the supplemental material). Determination of the precise number of deleted genes is complicated by the large number of hypothetical genes that appear to be disrupted, and therefore, the number should serve as an estimate of the total number of disrupted genes. The categories with the highest percentages of disrupted ORFs were the membrane transporter category, in which 43% of the ORFs were identified as partial ORFs, and the fatty acid/phospholipid metabolism category, in which 31% of the ORFs were identified as partial ORFs (Table 3). In contrast, the lowest numbers of partial ORFs were in the protein synthesis, transcription, and mobile genetic element categories, and the percentage for each of these categories was less than 8%.
TABLE 2.
Genome statistics for R. salmoninarum strain ATCC 33209, A. aurescens TC1, and Arthrobacter sp. strain FB24
| Data category | R. salmoninarum strain ATCC 33209a | A. aurescens TC1 | Arthrobacter sp. strain FB24 |
|---|---|---|---|
| Total no. of DNA bases sequenced | 3,155,250 (100)b | 5,226,648 (100) | 5,070,478 (100) |
| No. of bases in DNA coding sequence | 2,854,595 (90.5) | 4,587,903 (87.8) | 4,538,003 (89.5) |
| No. of G+C bases in DNA | 1,775,501 (56.3) | 3,262,989 (62.4) | 3,315,507 (65.4) |
| Total no. of ORFs | 3,507 (100) | 4,587 (100) | 4,506 (100) |
| No. of ORFs with assigned function | 2,620 (74.7) | 2,891 (63.0) | 2,986 (66.3) |
| No. of ORFs with no assigned function | 887 (25.3) | 1,696 (37.0) | 1,520 (33.7) |
| No. of ORFs with no assigned function or similarity | 240 (6.8) | 262 (5.7) | 147 (3.3) |
| No. of ORFs with no assigned function but with similarity | 647 (18.5) | 1,434 (31.3) | 1,373 (30.5) |
| No. of ORFs in paralog clusters | 592 (16.9) | 1,000 (21.8) | 1,067 (23.7) |
| No. of ORFs in COGs | 2,428 (69.2) | 3,382 (73.7) | 3,426 (76.0) |
| No. of ORFs with Pfam matches | 2,002 (57.1) | 3,273 (71.4) | 2,700 (59.9) |
| No. of COGs | 1,064 | 1,262 | 1,287 |
ORF data were compiled from the ERGO Light website (http://www.ergo-light.com/index.cgi/).
The numbers in parentheses are percentages.
FIG. 2.
Circular diagram of the R. salmoninarum ATCC 33209 genome showing genes and gene similarity to Arthrobacter spp. The circles show (from the inside to the outside) (i) GC skew (blue, low; red, high), (ii) G+C content (red, ≥50%; blue, <50%), (iii) disrupted ORFs (red), (iv) insertion elements (green), (v) ORFs with similarity to Arthrobacter sp. strain FB24 ORFs with a score of <1e−10 (orange), (vi) ORFs with similarity to A. aurescens TC1 ORFs with a score of <1e−10 (purple), and (vii) R. salmoninarum ORFs on different strands (different colors indicate different assigned functions [see Fig. S4 in the supplemental material for an explanation of the colors]).
TABLE 3.
Categories, numbers, and percentages of disrupted R. salmoninarum ORFs
| Category | Total no. of ORFs | No. of partial ORFs | % of partial ORFs |
|---|---|---|---|
| Transport and binding proteins | 346 | 152 | 43.9 |
| Fatty acid and phospholipid metabolism | 100 | 31 | 31.0 |
| Central intermediary metabolism | 170 | 48 | 28.2 |
| Energy metabolism | 304 | 86 | 28.3 |
| Regulatory functions | 240 | 53 | 22.1 |
| DNA metabolism | 108 | 23 | 21.3 |
| Cellular processes | 90 | 18 | 20.0 |
| Biosynthesis of cofactors and prosthetic groups | 109 | 24 | 22.0 |
| Protein fate | 152 | 28 | 18.4 |
| Amino acid biosynthesis | 111 | 18 | 16.2 |
| Cell envelope | 215 | 27 | 12.6 |
| Signal transduction | 6 | 1 | 16.7 |
| Hypothetical, unclassified, andunknown functions | 1,182 | 198 | 16.8 |
| Nucleotides | 61 | 6 | 9.8 |
| Protein synthesis | 128 | 9 | 7.0 |
| Transcription | 34 | 2 | 5.9 |
| Mobile and extrachromosomal elements | 151 | 6 | 4.0 |
| Total | 3,507 | 730 | 20.8 |
Analysis of selected partial genes in clinical R. salmoninarum strains.
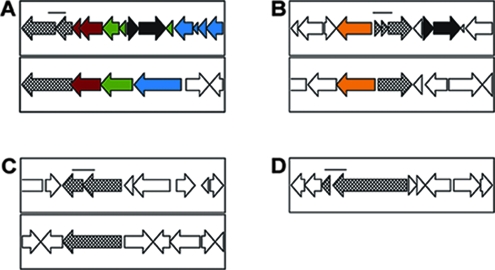
The high percentage of partial ORFs in the sequenced R. salmoninarum genome suggests that the strain used for sequence analysis may have undergone mutagenesis during its extended culture in the laboratory. To examine this possibility, genomic DNA from the ATCC 33209 reference strain, from two different clinical isolates, and from infected kidney tissue from naturally infected fish in Oregon were used as templates to amplify DNA surrounding four selected regions that appear to have frameshifts in the R. salmoninarum ATCC 33209 genome sequence (Table 1 and Fig. 3). These genomic regions represent the types of gene inactivation that occur throughout the chromosome. The targeted chromosomal regions included (i) a dipeptide permease operon (RSal33209_1659 through RSal33209_1671) that appears to be severely degenerate, perhaps as a function of integration of an IS994 element into the center of the operon (RSal33209_0105 and RSal33209_0106); (ii) a citrate synthase gene (RSal33209_2899, RSal33209_2900, and RSal33209_2901) that contains two apparent frameshifts compared to a homolog in Streptomyces spp.; (iii) tetP (RSal33209_1873 and RSal33209_1874), a gene encoding an efflux pump with an apparent frameshift in its coding region; and (iv) a putative fibronectin binding protein gene (RSal33209_1552) that is separated by a mutation from a short ORF containing a gram-positive anchor domain (RSal33209_1551) (Fig. 3). A 500-bp fragment of each target gene with sequences that might be mutated near the center was amplified and sequenced for each of the strains. In all 20 sequences examined, the frameshifts found in the completed genome were also found in the amplified DNA samples, and only two sequences had unique single-base changes compared to the ATCC 33209 genome. These data support the conclusion that the observed frameshifts were highly conserved in clinical (wild-type) strains of the pathogen and did not accumulate during laboratory culture of the ATCC strain.
FIG. 3.
Examples of disrupted ORFs in R. salmoninarum. The panels show ORF maps for different regions of the genome compared to syntenous regions of the Arthrobacter sp. strain FB24 chromosome (A and C) or the chromosome of S. coelicolor (B). The R. salmoninarum sequence is shown in the top box in panels A to C, and homologous ORFs are indicated by the same colors in each panel. Nonhomologous ORFs are indicated by open arrows in each panel. (A) Dipeptide permease operon in which each ORF in R. salmoninarum is inactivated by frameshifts. There is also a complete IS994 sequence (black arrow) that is inserted into the center of the operon. (B) Two divergently oriented citrate synthase genes that are arranged similarly in the related species S. coelicolor. One of the citrate synthase ORFs is truncated by serial frameshift mutations. An IS994 sequence is indicated by a black arrow. (C) tetP sequence encoding a candidate tetracycline resistance protein, which is interrupted in R. salmoninarum by a frameshift mutation. (D) ORF encoding a candidate fibronectin binding protein that may have been a sortase (SrtA) substrate, in which the sortase signal is separated from the majority of the ORF by a frameshift mutation. The bar in each panel indicates a 500-bp region of the chromosome that was sequenced for five different R. salmonarum samples, including two samples amplified directly from diseased fish collected at an Oregon hatchery. In 18/20 samples, the genome sequences of these regions were identical to the assembled R. salmoninarum ATCC 33209 genome sequences.
Genome evolution.
In order to determine the origin of R. salmoninarum and to better understand the evolutionary process shaping the genome, phylogenetic analysis and whole-genome alignment were performed. Analyses of the 16S rRNA sequence with reference to the high-G+C-content bacteria, as well as phylogenetic analysis of common genes, identified Arthrobacter spp. as the closest known relatives of R. salmoninarum (Fig. 1A; see Fig. S2 in the supplemental material). Two Arthrobacter genomes, the TC1 and FB24 genomes, have recently been published or released (41). The R. salmoninarum genome is 1.44 Mb smaller than the chromosome of TC1 and 1.55 Mb smaller than the chromosome of FB24. In addition, these two Arthrobacter strains have several large plasmids that are not present in the ATCC 33209 strain, nor are sequences with high levels of similarity to Arthrobacter plasmids present in the R. salmoninarum chromosome. Whole-genome nucleotide alignment using Mauve (Multiple Genome Alignment Tool; http://gel.ahabs.wisc.edu/mauve/) was used to identify large blocks of synteny between the two Arthrobacter genomes and rearrangements within the R. salmoninarum genome (see Fig. S3 in the supplemental material).
The R. salmoninarum chromosome and the sequences of the two Arthrobacter genomes share 1,562 protein ORF clusters (defined as clusters having a similarity threshold of >1 × 10e−10), including a combined total of 7,336 ORFs consistent with phylogenetic grouping of R. salmoninarum in the Arthrobacter clade (Fig. 1B; see Table S2 in the supplemental material). Approximately equal numbers of ORFs are part of the core set of similar genes (2,273 R. salmoninarum ORFs, 2,507 Arthrobacter sp. strain FB24 ORFs, and 2,556 A. aurescens ORFs). The two Arthrobacter species share 740 protein ORF clusters (1,917 ORFs) not found in R. salmoninarum that may have been lost in the course of genome reduction. Similar numbers of unique ORF clusters were identified in the three microorganisms (range, 818 to 933 clusters), suggesting that the levels of genomic divergence are similar. Compared to all sequenced microorganisms, a total of 240 R. salmoninarum ORFs were without function or similarity and are thus unique to R. salmoninarum (Table 2). Analysis of predicted protein functions for the Arthrobacter and R. salmoninarum genomes demonstrated that the evolutionary processes shaping R. salmoninarum evolution were skewed toward certain functional groups (Table 4). For example, compared to the chromosome of A. aurescens TC1, the R. salmoninarum genome has lower percentages of ORFs involved in energy metabolism (8.86% versus 17.87%), cellular processes (2.65% versus 4.5%), and transcription (1.11% versus 2.15%) but has high percentages of ORFs involved in central intermediary metabolism (5.13% versus 2.83%) and mobile and extrachromosomal element function (4.48% versus 0.85%) (Table 4).
TABLE 4.
Comparison of ORF functional role categories in R. salmoninarum and A. aurescens TC1
| Functional role category |
R. salmoninaruma
|
A. aurescens TC1b
|
||
|---|---|---|---|---|
| No. of ORFs | % | No. of ORFs | % | |
| Transport and binding proteins | 372 | 10.60 | 517 | 12.50 |
| Energy metabolism | 311 | 8.86 | 739 | 17.87 |
| Regulatory functions | 255 | 7.27 | 332 | 8.03 |
| Cell envelope | 220 | 6.27 | 340 | 8.22 |
| Central intermediary metabolism | 180 | 5.13 | 117 | 2.83 |
| Protein fate | 158 | 4.50 | 167 | 4.04 |
| Mobile and extrachromosomal element function | 158 | 4.50 | 35 | 0.85 |
| Protein synthesis | 135 | 3.85 | 123 | 2.97 |
| Synthesis of cofactors, prosthetic groups, and carriers | 112 | 3.19 | 141 | 3.41 |
| Amino acid biosynthesis | 112 | 3.19 | 136 | 3.29 |
| DNA metabolism | 110 | 3.14 | 111 | 2.68 |
| Cellular processes | 93 | 2.65 | 186 | 4.50 |
| Fatty acid and phospholipid metabolism | 107 | 3.05 | 123 | 2.97 |
| Purines, pyrimidines, nucleosides, and nucleotides | 62 | 1.77 | 82 | 1.98 |
| Transcription | 34 | 0.97 | 89 | 2.15 |
| Signal transduction | 7 | 0.20 | 54 | 1.31 |
| Total | 3,507 | 4,136 | ||
ORF categories were determined by using Manatee and manual annotation (see Table S1 in the supplemental material). Partial ORFs were also included in the totals, but it is unclear whether they are functional.
ORF numbers and categories were obtained from reference 41.
The genome of R. salmoninarum contains three recognized insertion sequences, IS994 (69 copies), ISRs2 (10 copies), and ISRs3 (one copy). The genome of the related organism A. aurescens TC1 contains 46 ORFs encoding functions consistent with transposons or insertion sequence elements, 23 of which are on the main chromosome. None of the insertion sequence elements in either sequenced Arthrobacter genome are present in the R. salmoninarum genome. As originally described by Rhodes et al. (53), the IS994 element consists of two ORFs, designated orfA and orfB, which encode candidate transposases consisting of approximately 120 and 360 amino acids, respectively. Analysis of the genome sequence revealed that there are 69 and 67 copies of IS994 orfA and orfB, respectively. This insertion sequence is distributed relatively randomly throughout the genome in both orientations. There is a single stand-alone copy of orfB, and there are two copies of orfA that are separated from orfB. There are three places where orfB is interrupted by a common frameshift, but the ORFs in most copies of IS994 are generally highly conserved and intact. Rhodes et al. (53) previously demonstrated that each of the seven copies of IS994 sequenced contained 24-nucleotide inverted repeats at each end of the element. The same 24-nucleotide inverted repeat is found in all intact copies of IS994 in the genome.
ISRs2 is a 1,343-bp insertion sequence element with a conserved 17-bp inverted repeat at each end and with a single 1,071-bp ORF encoding a protein product consisting of 357 amino acids. The predicted protein is most similar to transposases from Rhodococcus and Gordonia spp. The sequence of ISRs2 is highly conserved, with variations at only six nucleotide positions resulting in only two amino acid substitutions in the nine full-size copies. A tenth copy of ISRs2 is truncated and encodes only the carboxy-terminal 139 amino acids of the predicted protein.
The third R. salmoninarum insertion sequence element, ISRs3, is represented in the chromosome by an ORF encoding a single candidate transposase that is inactivated by a central frameshift. The interrupted sequence encodes a short IS200-like transposase with identity to transposases in Streptomyces spp. and Mycobacterium spp., among others. ISRs3 is located between msa1 and a copy of IS994 (RSal33209_0135 and RSal33209_0136).
Information processing.
Despite the large numbers of ORFs that are truncated in R. salmoninarum, all housekeeping genes, such as genes encoding large and small ribosomal protein subunits, are present and intact. R. salmoninarum also possesses most of the aminoacyl tRNA synthetases; the exceptions are the aminoacyl tRNA synthetases for arginine and glutamine. Likewise, R. salmoninarum contains a full complement of factors for translation initiation (e.g., IF-1, IF-2, and IF-3), translation elongation (e.g., EF-G, EF-TU, EF-P, and EF-Ts), and peptide release factors (e.g., RF-1 and RF-2). The chromosomal DNA replication machinery is also highly homologous to that in the sequenced Arthrobacter genomes, including a DNA polymerase with αβδδ'γ/τɛ subunits.
DNA modification and repair.
The R. salmoninarum genome appears to have very poor representation of genes encoding DNA restriction and modification enzymes. No classical type I, II, or III restriction-modification system genes are in the genome. R. salmoninarum lacks any ORFs encoding a DNA polymerase V function, while the Arthrobacter genomes encode both subunits of this protein. DNA polymerase V (homologous to Escherichia coli UmuDC) is involved in the bacterial SOS response. DNA polymerase V and RecA are involved in translesion synthesis (TLS) to avoid DNA replication-blocking lesions. Since TLS is highly inaccurate, SOS induction leads to mutagenic replication bypass by DNA polymerase V. The absence of DNA polymerase V in the R. salmoninarum genome indicates that this bacterium does not perform TLS in a RecA/PolV-dependent manner.
Central carbohydrate metabolism.
The R. salmoninarum genome encodes most of the enzymes participating in core central metabolic pathways, including glycolysis, pentose phosphate, tricarboxylic acid (or Kreb's) cycle, pyruvate cycle, and anaplerotic reactions. R. salmoninarum is probably able to utilize several sugars and polyols. For instance, this organism is predicted to import and utilize glucose, fructose, arabinose, gluconate, glycerol, and citrate.
Amino acid metabolism.
R. salmoninarum is able to synthesize de novo the following amino acids: histidine, aspartate, lysine, threonine, valine, leucine, isoleucine, d- and l-alanine, glutamate, glutamine, arginine, tyrosine, phenylalanine, and tryptophan. Conversely, R. salmoninarum is unable to synthesize serine, glycine, cysteine, asparagine, and methionine de novo. The inability to synthesize these amino acids is frequently manifested by the presence of truncated ORFs encoding putative enzymatic functions. For instance, in the three-step phosphorylated serine biosynthetic pathway that converts 3-phosphoglycerate to l-serine, the phosphoserine transaminase function is likely absent since the ORFs encoding this function are truncated (e.g., RSal33209_0497 and RSal33209_0496). Since the biosynthesis of cysteine is dependent upon serine availability, this might limit intracellular de novo cysteine synthesis, despite the fact that R. salmoninarum has an intact pathway that converts serine to cysteine. Other routes for cysteine production from either sulfate or homocysteine are similarly hindered. However, the R. salmoninarum genome appears to encode an ABC-type alkylphosphonate transport system (e.g., RSal33209_1533, RSal33209_1531, RSal33209_1530, and RSal33209_1529) that is likely able to transport external O-phosphoserine that is present in fish tissue (36) into the bacterial cell. The O-phosphoserine can then be transformed into serine using the canonical serine biosynthetic pathway using phosphoserine phosphatase (e.g., RSal33209_0497 and RSal33209_0496). In the case of tryptophan, one caveat is that although phosphoribosylanthranilate synthase is absent, it is likely that another function substitutes for this missing enzyme (4). Interestingly, inspection of the R. salmoninarum genome identified a gene encoding tryptophan-2,3-dioxygenase (RSal33209_2215), an enzyme that transforms tryptophan into N-formyl-l-kynurenine. Some intracellular pathogens (e.g., Lawsonia) are able to utilize tryptophan breakdown products via N-formyl kynurenine to produce anthranilate that can be utilized in the classic tryptophan biosynthetic pathway. However, the absence of an arylformamidase encoded by ORFs in the R. salmoninarum genome suggests that this bacterium is unable to degrade this compound to l-kynurenine, potentially leading to accumulation of the compound.
Cofactor biosynthesis.
R. salmoninarum appears to have a complete set of genes encoding enzymes for biosynthesis of riboflavin, flavin adenine dinucleotide, flavin mononucleotide, pyridoxine, pantothenate (including 4′-phosphopantetheine), coenzyme A, lipoate, and menoquinone. In addition, genes encoding enzymes for the biosynthesis and recycling of folate, which is essential for purine and thymidylate metabolism, are present in the genome. Moreover, the machinery to synthesize heme a, d, and o is also encoded in the genome.
Fatty acid and lipid metabolism.
The absence of discernible homologs of the fabA and fabZ genes in the R. salmoninarum genome suggests that this organism cannot synthesize both unsaturated and saturated fatty acids and, therefore, has to scavenge these compounds from the host. Moreover, the absence of the phospholipid biosynthetic genes suggests that R. salmoninarum is dependent upon the host for acquiring lipid building blocks and in this way resembles rickettsial pathogens. Although R. salmoninarum lacks any ORFs encoding a phospholipid-degrading capability, the R. salmoninarum genome does encode a triacylglycerol lipase, which hydrolyzes the carboxyl ester bonds in mono-, di-, and triglycerides to liberate fatty acids and alcohols. This lipase is likely essential for maintaining the lipolytic activity and is responsible for management of fatty acid availability in the bacterium. In addition, R. salmoninarum possesses genes encoding enzymes capable of performing beta-oxidation, suggesting that this organism can metabolize fatty acids and use them as carbon and nitrogen sources.
Bioenergetics.
R. salmoninarum is capable of microaerophilic growth and respiration. This organism appears to be able to utilize a variety of carbon substrates for energy, including pyruvate, lactate, succinate, malate, glycerol-3-phosphate, proline, butanoyl coenzyme A, and fatty acids. The utilization of fatty acids is particularly noteworthy since R. salmoninarum likely scavenges host-derived fatty acids to complement its fatty acid biosynthetic deficiencies. R. salmoninarum possesses genes responsible for biosynthesis of menaquinone (e.g., RSal33209_2700 to RSal33209_2702 and RSal33209_2638), which is the electron carrier for the R. salmoninarum electron transport chain, and the electron acceptor prior to oxygen is a cytochrome c oxidase. The presence of the full complement of enzymes for the tricarboxylic acid cycle and glycolytic components suggests that ATP generation in R. salmoninarum takes place via substrate-level phosphorylation. The R. salmoninarum genome also contains a full complement of ORFs encoding the proton-transporting F1F0 ATP synthase complex.
R. salmoninarum is not capable of anaerobic respiration. This organism does possess related but truncated enzymatic systems for anaerobiosis, suggesting that there was an anaerobic respiratory chain in its evolutionary past.
R. salmoninarum resuscitation-promoting factors.
Resuscitation-promoting factors are family of proteins that are common in high-G+C-content bacteria and are involved in reactivation of bacterial growth from a dormant state. R. salmoninarum contains two genes, RSal33209_2532 and RSal33209_2995, that have a resuscitation-promoting factor domain. These genes encode predicted proteins consisting of 220 and 384 amino acids, respectively. Both proteins have predicted signal peptide sequences, suggesting that they are exported. The predicted protein product encoded by R. salmoninarum ORF RSal33209_2532 has high sequence similarity to the original Rpf protein discovered in Micrococcus luteus (42), while the predicted protein encoded by R. salmoninarum ORF RSal33209_2995 is most similar to Mycobacterium tuberculosis RpfB (43). In addition to the resuscitation-promoting factor domains, R. salmoninarum Rpf contains a peptidoglycan binding motif, LysM, while R. salmoninarum RpfB contains three domains with unknown functions (DUF 348), as well as a single G5 domain identical to M. tuberculosis RpfB.
Secondary metabolite synthesis or sequestration.
The R. salmoninarum genome contains one fragmented cluster of genes that may be responsible for siderophore synthesis (RSal33209_0770 to RSal33209_0772). This set of genes is highly conserved in the Arthobacter spp. and R. salmoninarum, and the genes are in a highly syntenous region of the chromosome. R. salmoninarum also contains multiple operons associated with sequestration of siderophores, including genes encoding ferric siderophore binding proteins and permeases associated with transport of siderophores across the cytosolic membrane (49). There are three clusters of these genes. One cluster (ORFs RSal33209_2862 through RSal33209_2859) is present in both Arthrobacter species and is degenerate in R. salmoninarum. A second cluster containing apparently intact coding sequences (RSal33209_1684 through RSal33209_1681) has a homologous cluster only in A. aurescens TC1, although there are intact reading frames in R. salmoninarum. The final gene cluster is largely degenerate but contains an intact fepG homolog (RSal33209_3347). Therefore, R. salmoninarum encodes enzymes comprising an entire ferric siderophore import system, but no single operon encodes all components and the process may be complicated by inactivation of certain genes.
Two very large ORFs (RSal33209_2539 and RSal33209_2540) appear to contain modules for polyketide synthesis systems, and products of adjacent ORFs may also participate in the process. These ORFs are not found in either Arthrobacter species. While it is not possible to predict an end product of this pathway, this set of genes may be involved in synthesis of unique secondary metabolites that protect the pathogen from competing microorganisms.
Stress response genes.
R. salmoninarum possesses several ORFs encoding enzymes that confer resistance to oxygen radicals. The encoded enzymes comprise superoxide dismutase, catalase, peroxidases, and thioredoxin peroxidase. These enzymes constitute a bacterial defense mechanism against bactericidal radical agents generated by host neutrophils and macrophages. Some of these enzymes may also provide resistance to nitrogen compounds and radicals produced by the host. One aspect of intermediate resistance to nitrogen involves the reduction of reactive nitrogen intermediates to less toxic products. R. salmoninarum encodes one enzyme (ferridoxin nitrite reductase; RSal33209_2621) that may be involved in reduction of nitrite produced by macrophages. Interestingly, this protein is homologous to an enzyme found in many plants, including Arabidopsis thaliana. R. salmoninarum also encodes a signal-transducing histidine kinase, a nitrate/nitrite sensor protein (narX; RSal33209_0978), suggesting that it is capable of sensing and destroying nitric oxide produced by host macrophages. The products of these genes may function in resistance to host defenses, but homologs are present in the Arthrobacter genomes (see Table S1 in the supplemental material). While genes involved in the stress response within a host are conserved, ORFs putatively involved in the response to environmental stressors have not been found. Overall, of the 78 stress-related ORFs identified in A. aurescens TC1, only 36 (46%) are present in R. salmoninarum. Interestingly, R. salmoninarum appears to be particularly deficient in cupin-related genes, and only 2 of 15 ORFs identified in Arthrobacter have been found in R. salmoninarum. Cupin-related genes are involved in Mn accumulation and morphogenesis (41) but appear to be dispensable for R. salmoninarum host survival.
Heme acquisition.
R. salmoninarum acquired genes encoding several proteins involved in heme and hemin acquisition. There are two different gene clusters that share identity with heme acquisition loci of different bacterial species. ORFs RSal33209_2547 through RSal33209_2552 encode proteins involved in these processes in several other systems. One of these proteins, heme oxygenase (RSal33209_26547), may also be important in stress response, as homologs in mammalian cells are upregulated under stress conditions (8). ORFs RSal33209_2063 through RSal33209_2061 also encode putative hemin binding and transport proteins and exhibit the highest levels of similarity to similar genes in Streptomyces coelicolor. Neither of the R. salmoninarum gene clusters shares identity with ORFs present in Arthrobacter, and therefore these clusters appear to have been acquired after species divergence. Arthrobacter species have ORFs encoding proteins with similarity to these gene products, but they are in gene clusters associated with ferric siderophore acquisition instead of heme acquisition.
Capsular synthesis.
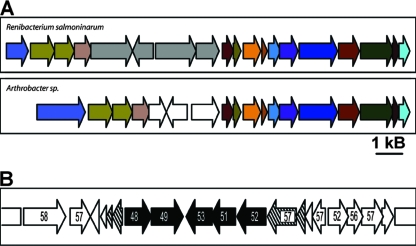
Immunological and microscopic approaches were used to demonstrate that R. salmoninarum produces a capsule and/or soluble extracellular carbohydrate (14, 59). The nature of this product is not well characterized. The R. salmoninarum genome sequence encodes a set of proteins that form the core of a capsular polysaccharide assembly pathway. These proteins include homologs of at least seven proteins encoded in Staphylococcus species capsular synthesis genetic clusters (45). However, in contrast to the Staphylococcus genes, the R. salmoninarum capsular synthesis genes are not present at a single locus. For example, ORFs RSal33209_1611 and RSal33209_1612 are homologous to cap5G and cap5H but are physically separated from other genes involved in a putative capsular synthesis pathway. These genes share no significant identity with sequences from Arthrobacter but are adjacent to genes involved in extracellular carbohydrate synthesis and transport (RSal33209_1613 through RSal33209_1618), which are syntenous with homologs in both Arthrobacter spp. (Fig. 4).
FIG. 4.
Comparison of R. salmoninarum and Arthrobacter sp. strain FB24 capsular biosynthesis genes suggests that there was horizontal gene acquisition. (A) Set of capsular synthesis genes (RSal33209_1436 to RSal33209_1439) (gray arrows) that differ from Arthrobacter sp. strain FB24 ORFs (open arrows) in an otherwise fully conserved region of the genome. Arrows that are the same color indicate ORFs that are homologous in the organisms. (B) Evidence for horizontal acquisition of a set of capsular synthesis genes (RSal33209_1338 to RSal33209_1342) (black arrows) located between complete and truncated IS994 sequences (cross-hatched arrows) inserted into a region of the genome that is otherwise syntenous with the Arthrobacter genome (open arrows). The G+C contents (expressed as percentages) of the ORFs in the region are indicated in the ORFs.
The regions encoding capsular synthesis proteins suggest that there have been multiple mechanisms of evolutionary divergence from the Arthrobacter group of bacteria. First, ORFs RSal33209_1436 to RSal33209_1439 are located in a region of the chromosome that is highly syntenous with regions in both Arthrobacter species (Fig. 4A). Only one of the candidate capsular synthesis ORFs (RSal33209_1436) is shared with one of the Arthrobacter species (strain TC1). The other candidate capsular synthesis genes are not found in the sequenced Arthrobacter spp., even though each genome has genes for extracellular polysaccharide synthesis at the same locus. It is possible that this region has been a locus of divergence as R. salmoninarum and Arthrobacter spp. have evolved to occupy different niches. A different mechanism is found in ORFs RSal33209_1338 to RSal33209_1342. This region may represent a genomic island acquired by R. salmoninarum via a horizontal transmission event. These ORFs encode proteins that can be placed in capsular synthesis pathways, have low G+C contents (48 to 51%) compared to the general R. salmoninarum genome (56.3%), and are flanked by complete and partial IS994 insertion elements (Fig. 4B). Therefore, at some point in the evolution away from a progenitor species, R. salmoninarum may have lost selected pathways associated with environmental exopolysaccharide synthesis, modified existing genes encoding exopolysaccharide synthesis proteins, and gained a pathway associated with synthesis of a protective capsule.
Gene products encoded by RSal33209_1750 and RSal33209_2984 are similar to enzymes involved in UDP-glucose metabolism, and homologs of these gene products participate in capsular synthesis in a diverse collection of bacteria (12, 45). These gene products also share sequence identity with proteins that participate in carbohydrate metabolism in many different organisms, and the genes are represented in the Arthrobacter sp. strain FB24 genome.
Protein secretion machinery and the sortase pathway.
R. salmoninarum contains a complete Sec-dependent secretion pathway with the exception of a YajC homolog (protein translocase). Four homologs of signal peptidase I (RSal33209_1086, RSal33209_1087, RSal33209_1255, and RSal33209_2979) and one homolog of signal peptidase II (lipoprotein signal peptidase; RSal33209_2485) are present. A SignalP and TmPred search demonstrated the presence of at least 444 ORF products having a typical Sec secretion leader peptide (see Table S1 in the supplemental material).
The R. salmoninarum genome also contains an ORF encoding a sortase, a member of a group of cysteine transpeptidases in gram-positive bacteria that promote covalent anchoring of proteins to the peptidoglycan surface of the cell envelope (38). Sortases are interesting in that they represent a new target for novel anti-infective therapeutic drugs (38). The R. salmoninarum sortase (RSal33209_2896) is a member of subfamily 5 or class D of bacterial sortases (10, 13) and shares 65 and 62% identity with the sortases in A. aurescens TC1 and Arthrobacter sp. strain FB24, respectively. Using the tripartite pattern search method of Boekhorst et al. (5), as well as tools such as Pfam, SignalP, TMpred, and ProDom, we identified seven ORFs in the R. salmoninarum ATCC 33209 genome specifying potential sortase substrates containing the LAxTG motif (RSal33209_2105, RSal33209_3407, RSal33209_2525, RSal33209_3273, RSal33209_0619, RSal33209_1326, and RSal33209_0402), some of which were predicted to be cell surface proteinases or adhesins (62). Two of the seven potential SrtD substrates identified through bioinformatics analysis are annotated as products of pseudogenes (RSal33209_0619 and RSal33209_1326), and the current search may have failed to identify additional complete sortase substrates. Of the seven ORFs with SrtD recognition signals, three are syntenous with Arthrobacter species homologs (RSal33209_3407, RSal33209_1326, and RSal33209_0402), and three are unique to R. salmoninarum (RSal33209_2105, RSal33209_2525, and RSal33209_3273). The remaining putative sortase substrate (RSal33209_0619) shares sequence identity with candidate acid phosphatases in gram-negative bacteria.
In addition to the probable functional SrtD, there is a second ORF product containing a sortase catalytic domain. This 123-amino-acid ORF product (RSal33209_1799) appears to be encoded by a remnant of a srtA gene. Two potential SrtA substrates are encoded in the genome; one of them is complete and syntenous with a hypothetical protein in Arthrobacter sp. strain FB24 (RSal33209_1929), and the other is a unique ORF product encoded by a pseudogene (RSal33209_1551). The latter ORF is adjacent to a large ORF encoding a putative fibronectin binding protein that lacks a sortase recognition motif (Fig. 3D).
Antibiotic resistance genes.
A search of the R. salmoninarum genome yielded 68 ORFs homologous to genes encoding factors known to be involved in resistance to different antibiotics. These factors include a variety of multidrug transporters, several β-lactamases (e.g., RSal33209_0207, RSal33209_0436, and RSal33209_0920), and tetracycline (tetA [RSal33209_0457] and tetB [RSal33209_0459]) and macrolide resistance factors. Macrolide resistance is an especially important concern since erythromycin (and to a lesser extent azithromycin) is commonly used to control R. salmoninarum infections in both supplementation and conservation hatcheries. Several classes of macrolide resistance genes are represented in the genome, including genes encoding two 23S rRNA methyltransferases (rlmAII [RSal33209_2019] and spoU [RSal33209_3401]), a macrolide efflux factor (mefA [RSal33209_2224]), a multidrug resistance efflux pump (pvsC [RSal33209_1691]), and a 16S rRNA dimethylase (ksgA [RSal33209_2994]). The genome also contains an ORF with similarity to a gene encoding macrolide glycosyltransferase (mgtA [RSal33209_2959]), but the ORF is truncated and likely nonfunctional. The rlmAII, spoU, mefA, and ksgA ORFs show high synteny with homologs in Arthrobacter spp., while pvsC and mgtA have other ancestral origins.
MSA.
The R. salmoninarum major soluble antigen (MSA) (also referred to as major surface antigen) is a unique protein that makes up approximately 70% of the bacterial cell surface protein in R. salmoninarum (72) and is secreted into fish tissues at concentrations up to mg per ml (64). MSA is found in culture supernatants, in fish tissues, and on the cell surface as an intact 57-kDa protein, and as defined fragments of the full-length molecule (69). This protein has immunosuppressive properties and is considered a major virulence factor (9). Previous work demonstrated that the MSA gene is duplicated in the genomes of many R. salmoninarum strains, including ATCC 33209 (44, 70), and this was confirmed by genome sequencing. The entire msa coding sequence and more than 700 downstream base pairs are exactly duplicated in each of the two copies. Both copies of msa are linked to nearby IS994 sequences, and one msa copy is adjacent to the single degenerate ISRs3 sequence. Interestingly, a third copy is present in many clinical isolates, and these strains exhibit increased virulence compared to strains carrying two copies of msa (50). No homolog of the MSA gene has been found in any other sequenced bacterial genome.
Serine proteinases.
As mentioned above, MSA is targeted by an unknown serine proteinase activity in bacterial culture supernatants. The enzymatic activity is associated with a >100-kDa enzymatically active nonreduced protein species (56). The R. salmoninarum genome encodes several candidate serine proteinases, including some that are conserved in R. salmoninarum and Arthrobacter spp. and several that are not found in Arthrobacter spp. Included in this group are a pair of subtilisinlike enzymes that are encoded by sequential ORFs (RSal33209_0104 to RSal33209_0106). One of this pair is interrupted by a frameshift. The serine proteinases that do not have homologs in Arthrobacter spp. group into four families, including representatives of ORFs RSal33209_0898, RSal33209_1246, and RSal33209_0571 and the subtilisin homologs. Several of the ORFs encode proteins that have secretory signal sequences and therefore might have a role in the extracellular digestion of MSA. However, none of these proteins has a predicted molecular mass greater than 60 kDa, and none of them has been directly implicated as the proteinase responsible for this activity.
Hemolysins.
It was reported previously that the R. salmoninarum genome encodes at least two specific hemolysins (21, 30, 31). The previous investigators expressed random R. salmoninarum DNA fragments in E. coli and looked for hemolysis by plating preparations on blood agar plates. One of the clones obtained was sequenced and identified in the genome as ORF RSal33209_3168. As noted by Grayson et al. (30), the product of this gene contains peptidase and proteinase motifs and may function in digestion of proteins following secretion from R. salmoninarum. Three additional candidate hemolysins are represented in the genome, encoded by ORFs RSal33209_0811, RSal33209_3195, and RSal33209_3047. Each candidate hemolysin has clear homologs in Arthrobacter, indicating that they were not horizontally acquired as an adaptation to the piscine host. It is possible that other reported hemolysins whose sequences are not present in the GenBank database may be encoded by one of these ORFs.
DISCUSSION
The genome sequence of R. salmoninarum ATCC 33209, coupled with the results of a comparative genomic analysis of related microorganisms, supports the hypothesis that R. salmoninarum evolved from an Arthrobacter-like precursor, largely via genome reduction. While R. salmoninarum is a fastidious obligate pathogen of salmonid fish, members of the genus Arthrobacter are soil inhabitants that are noted for their ability to survive many types of environmental stress. However, there is no evidence that R. salmoninarum grows for extended periods outside the salmonid host, and no member of the Arthrobacter family colonizes any host species. Therefore, this is an interesting model for analysis of pathogen evolution.
Mechanisms for evolution of pathogens from commensal or environmental organisms are being elucidated now (47). Stinear et al. (61) noted six genomic features that characterize rapid adaptation of pathogens to the eukaryotic host environment. These features include (i) expansion of insertion elements, (ii) the presence of abundant pseudogenes, (iii) smaller genome size, (iv) chromosomal rearrangements, (v) acquisition of foreign genes, and (vi) a high degree of genetic relatedness or clonality within the species. Each of these features is represented in the R. salmoninarum genome. There are a total of 80 copies of three insertion elements, including 69 highly conserved copies of the bicistronic element IS994. There are also 10 conserved copies of ISRs2 and a single degenerate copy of the IS200-like element ISRs3. There are many apparent pseudogenes in R. salmoninarum (n = 730), and many pathways are apparently reduced via this strategy. The lack of restriction-modification systems encoded in the R. salmoninarum genome may contribute to the extensive disruption of the ORFs. The genome is approximately 1.9 Mbp smaller than the Arthrobacter genomes, and there are concomitant reductions in functional capability and level of adaptability. There are examples of horizontally acquired genomic islands in the R. salmoninarum chromosome, including a group of capsular synthesis genes flanked by partial and complete IS994 sequences (Fig. 4). Finally, strains of R. salmoninarum are highly homogeneous with respect to the overall genomic structure, biochemical properties, and surface antigens (24, 27, 53, 68).
The differences between the Arthrobacter species and R. salmoninarum genomes are reflected in the reduction in the relative number of specific classes of bacterial proteins, including the proteins involved with energy metabolism, transcription, and signal transduction (Table 4). In addition, there is an increased number of frameshifted genes in certain functional classes, including the transport, central intermediary metabolism, fatty acid synthesis, and energy metabolism classes (Table 3). This nonrandom reduction in genetic function may reflect the unique needs of R. salmoninarum during infection of the host compared to the needs of the environmental Arthrobacter spp.
While the major force in R. salmoninarum evolution was likely genome reduction, there are several examples of apparently horizontally acquired genes associated with pathogenesis. First and foremost, the pathogen acquired msa1 and msa2 via horizontal transmission and subsequent duplication. In agreement with previous analyses, the msa genes are identical (44, 50, 70). MSA binds leukocytes, mammalian erythrocytes, and fish sperm cells and is the predominant protein produced in culture and in infected fish tissues (63, 64, 67, 69). This protein is also the only conclusively proven virulence factor in the species, and no orthologs are present in the two Arthrobacter species or in any other microorganism. One copy of the gene is adjacent to the degenerate ISRs3, and both copies are adjacent to or very near a copy of IS994. At present, the original source of msa is unknown.
The genome contains a number of ORFs encoding likely surface proteins, including eight ORFs that contain sortase cleavage motifs, and it encodes one intact sortase. Based on recent classification schemes, the R. salmoninarum sortase (RSal33209_2896) is most closely related to the subfamily 5 or class D bacterial sortases (SrtD) (10, 13). Members of this subfamily recognize an LAxTG motif. The R. salmoninarum SrtD shares 65 and 62% identity with the sortases in A. aurescens TC1 and Arthrobacter sp. strain FB24, respectively, while most gram-positive bacteria possess SrtA or subfamily 3 sortases that recognize proteins with LPxTG and LPxTGG motifs, respectively. Three of the seven sortase cleavage motif-encoding ORFs are also present in Arthrobacter, and two of these ORFs appear to be pseudogenes in R. salmoninarum. Treatment of R. salmoninarum with sortase inhibitors decreases the adherence of R. salmoninarum to fibronectin and fish cells in vitro, suggesting that these proteins have a role in attachment (62). Recent drug discovery efforts have focused on sortases as a possible therapeutic target, and it is possible that such efforts will be important in preventing R. salmoninarum infection (62). While R. salmoninarum SrtD is the only apparent active sortase, the presence of a degenerate srtA gene and at least one degenerate SrtA substrate with homologs in Arthrobacter may represent additional examples of specific genome decay in a pathway not needed for the intracellular lifestyle of R. salmoninarum.
As might be expected for a microorganism related to bacteria that survive in soil, the R. salmoninarum genome contains many genes encoding antibiotic resistance factors, ranging from multidrug transporters to factors that specifically modify either an antibiotic or its substrate. The number of macrolide-resistant factors is particularly interesting in that erythromycin is often the antibiotic of choice in treatment of BKD. Expression of at least two identified macrolide resistance genes (rlmAII and mefA) was increased upon exposure of R. salmoninarum to erythromycin or azithromycin, and this increased expression correlates with true resistance to these two macrolides (P. S. Sudheesh and M. S. Strom, unpublished data). The identification and subsequent characterization of these genes through analysis of the R. salmoninarum genome demonstrated a molecular mechanism for the generalized failure of macrolides to completely clear R. salmoninarum from an infected fish population, despite the observed low MIC (54).
A total of 1,562 ORF clusters were similar in R. salmoninarum and Arthrobacter spp. This high level of genomic similarity may explain the efficacy of the commercial vaccine Renogen, which is composed of a live, lyophilized Arthrobacter sp. that is injected into young fish. However, this efficacy extends only to Atlantic salmon (S. salar), and the vaccine does not provide significant protection to Pacific salmonids (Oncorhynchus spp.); thus, vaccine coverage remains low (6, 55). The genome sequence may be useful in identifying novel candidate vaccine targets, which may lead to more efficacious vaccines that will stimulate long-lasting cellular immunity against R. salmoninarum.
R. salmoninarum contains genes encoding selected horizontally acquired virulence factors, including capsular synthesis genes, heme acquisition operons, genes encoding possible hemolysins, and the poorly characterized msa genes. Most of these genes do not have homologs in Arthrobacter spp., and thus, they may form the basis of the niche differences between these bacteria. While the classic BKD lesion is analogous to the granuloma found in piscine mycobacterial infections, R. salmoninarum does not produce mycolic acids or other mycobacterial surface virulence factors. Therefore, the genome sequence increases our understanding of the pathogenesis of R. salmoninarum infection, but most of the host-microbe interactions in this system remain poorly understood. It is anticipated that the R. salmoninarum genome sequence will be a valuable tool for research scientists working to identify potential R. salmoninarum diagnostic, immunogenic, and therapeutic targets, with the goal of assisting fishery professionals in prevention and control of BKD.
Supplementary Material
Acknowledgments
This project was funded by NSF/USDA Microbial Genome Sequencing Program agreement 2004-35600-14173 awarded to M.S.S., G.D.W., and D.D.R. G.D.W. was supported by Agricultural Research Service CRIS project 1930-32000-002 (Host-Pathogen and Environmental Interactions in Cool and Cold Water Aquaculture). M.S.S. was also supported by the NOAA FCRPS Biological Opinion Implementation Project and the NOAA Fisheries Service.
We thank Eric Haugen, Donald Bovee, Karen Phelps, Regina Lim, Will Gillett, Hillary Hayden, and Don Guenthener of the University of Washington Genome Center for sequencing support. Mark Zabriskie of the Oregon State University College of Pharmacy provided valuable analysis of genes involved in secondary metabolism, and Theresa Walunas of Integrated Genomics was instrumental in generation of genome maps and preparation of the genome sequence for submission to GenBank. We thank Walton W. Dickhoff for a critical review of the manuscript.
Mention of trade names or commercial products is solely for the purpose of providing specific information and does not imply recommendation or endorsement by the U.S. Department of Agriculture and the U.S. Department of Commerce.
Footnotes
Published ahead of print on 22 August 2008.
Supplemental material for this article may be found at http://jb.asm.org/.
REFERENCES
- 1.Balfry, S. K., L. J. Albright, and T. P. T. Evelyn. 1996. Horizontal transfer of Renibacterium salmoninarum among farmed salmonids via the fecal-oral route. Dis. Aquat. Org. 2563-69. [Google Scholar]
- 2.Bandin, I., A. E. Ellis, J. L. Barja, and C. J. Secombes. 1993. Interaction between the rainbow trout macrophages and Renibacterium salmoninarum in vitro. Fish Shellfish Immunol. 325-33. [Google Scholar]
- 3.Banner, C. R., J. S. Rohovec, and J. L. Fryer. 1991. A new value for mol percent guanine + cytosine of DNA for the salmonid fish pathogen Renibacterium salmoninarum. FEMS Microbiol. Lett. 6357-59. [DOI] [PubMed] [Google Scholar]
- 4.Barona-Gomez, F., and D. A. Hodgson. 2003. Occurrence of a putative ancient-like isomerase involved in histidine and tryptophan biosynthesis. EMBO Rep. 4296-300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Boekhorst, J., M. W. de Been, M. Kleerebezem, and R. J. Siezen. 2005. Genome-wide detection and analysis of cell wall-bound proteins with LPxTG-like sorting motifs. J. Bacteriol. 1874928-4934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bravo, S., and P. J. Midtlyng. 2007. The use of fish vaccines in the Chilean salmon industry 1999-2003. Aquaculture 27036-42. [Google Scholar]
- 7.Bruno, D. W., and A. L. S. Munro. 1986. Observations on Renibacterium salmoninarum and the salmonid egg. Dis. Aquat. Org. 183-87. [Google Scholar]
- 8.Choi, A. M., and J. Alam. 1996. Heme oxygenase-1: function, regulation, and implication of a novel stress-inducible protein in oxidant-induced lung injury. Am. J. Respir. Cell Mol. Biol. 159-19. [DOI] [PubMed] [Google Scholar]
- 9.Coady, A. M., A. L. Murray, D. G. Elliott, and L. D. Rhodes. 2006. Both msa genes in Renibacterium salmoninarum are needed for full virulence in bacterial kidney disease. Appl. Environ. Microbiol. 722672-2678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Comfort, D., and R. T. Clubb. 2004. A comparative genome analysis identifies distinct sorting pathways in gram-positive bacteria. Infect. Immun. 722710-2722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.DeSantis, T. Z., Jr., P. Hugenholtz, K. Keller, E. L. Brodie, N. Larsen, Y. M. Piceno, R. Phan, and G. L. Andersen. 2006. NAST: a multiple sequence alignment server for comparative analysis of 16S rRNA genes. Nucleic Acids Res. 34W394-W399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Dillard, J. P., and J. Yother. 1994. Genetic and molecular characterization of capsular polysaccharide biosynthesis in Streptococcus pneumoniae type 3. Mol. Microbiol. 12959-972. [DOI] [PubMed] [Google Scholar]
- 13.Dramsi, S., P. Trieu-Cuot, and H. Bierne. 2005. Sorting sortases: a nomenclature proposal for the various sortases of Gram-positive bacteria. Res. Microbiol. 156289-297. [DOI] [PubMed] [Google Scholar]
- 14.Dubreuil, D., R. Lallier, and M. Jacques. 1990. Immunoelectron microscopic demonstration that Renibacterium salmoninarum is encapsulated. FEMS Microbiol. Lett. 54313-315. [DOI] [PubMed] [Google Scholar]
- 15.Elliott, D. G., R. J. Pascho, and G. L. Bullock. 1989. Developments in the control of bacterial kidney disease of salmonid fishes. Dis. Aquat. Org. 6201-215. [Google Scholar]
- 16.Elliott, D. G., R. J. Pascho, L. M. Jackson, G. M. Matthews, and J. R. Harmon. 1997. Renibacterium salmoninarum in spring-summer chinook salmon smolts at dams on the Columbia and Snake Rivers. J. Aquati. Anim. Health 9114-126. [Google Scholar]
- 17.Evelyn, T. 1993. Bacterial kidney disease—BKD, p. 177-195. In V. Inglis, R. J. Roberts, and N. R. Bromage (ed.), Bacterial diseases of fish. Blackwell Scientific Publications, Oxford, United Kingdom.
- 18.Evelyn, T. P. T. 1977. An improved growth medium for the kidney disease bacterium and some notes on using the medium. Bull. Off. Int. Epizoot. 87511-513. [Google Scholar]
- 19.Evelyn, T. P. T., J. E. Ketcheson, and L. Prosperi-Porta. 1984. Further evidence for the presence of Renibacterium salmoninarum in salmonid eggs and for the failure of povidone-iodine to reduce the intra-ovum infection rate in water-hardened eggs. J. Fish Dis. 7173-182. [Google Scholar]
- 20.Evelyn, T. P. T., L. Prosperi-Porta, and J. E. Ketcheson. 1986. Experimental intra-ovum infection of salmonid eggs with Renibacterium salmoninarum and vertical transmission of the pathogen with such eggs despite their treatment with erythromycin. Dis. Aquat. Org. 1197-202. [Google Scholar]
- 21.Evenden, A. J., T. H. Grayson, M. L. Gilpin, and C. B. Munn. 1993. Renibacterium salmoninarum and bacterial kidney disease—the unfinished jigsaw. Annu. Rev. Fish Dis. 387-104. [Google Scholar]
- 22.Ewing, B., and P. Green. 1998. Base-calling of automated sequencer traces using phred. II. Error probabilities. Genome Res. 8186-194. [PubMed] [Google Scholar]
- 23.Ewing, B., L. Hillier, M. C. Wendl, and P. Green. 1998. Base-calling of automated sequencer traces using phred. I. Accuracy assessment. Genome Res. 8175-185. [DOI] [PubMed] [Google Scholar]
- 24.Fiedler, F., and R. Draxl. 1986. Biochemical and immunochemical properties of the cell surface of Renibacterium salmoninarum. J. Bacteriol. 168799-804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Flagg, T. A., C. V. W. Mahnken, and J. J. Hard. 1995. An assessment of the status of captive broodstock technology of Pacific salmon, 1995 final report. DOE/BP-55064-1. Bonneville Power Administration, Portland, OR.
- 26.Fryer, J. L., and J. E. Sanders. 1981. Bacterial kidney disease of salmonid fish. Annu. Rev. Microbiol. 35273-298. [DOI] [PubMed] [Google Scholar]
- 27.Getchell, R. G., J. S. Rohovec, and J. L. Fryer. 1985. Comparison of Renibacterium salmoninarum isolates by antigenic analysis. Fish Pathol. 20149-159. [Google Scholar]
- 28.Gordon, D., C. Abajian, and P. Green. 1998. Consed: a graphical tool for sequence finishing. Genome Res. 8195-202. [DOI] [PubMed] [Google Scholar]
- 29.Gordon, D., C. Desmarais, and P. Green. 2001. Automated finishing with autofinish. Genome Res. 11614-625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Grayson, T. H., A. J. Evenden, M. L. Gilpin, K. L. Martin, and C. B. Munn. 1995. A gene from Renibacterium salmoninarum encoding a product which shows homology to bacterial zinc-metalloproteases. Microbiology 1411331-1341. [DOI] [PubMed] [Google Scholar]
- 31.Grayson, T. H., M. L. Gilpin, A. J. Evenden, and C. B. Munn. 2001. Evidence for the immune recognition of two haemolysins of Renibacterium salmoninarum by fish displaying clinical symptoms of bacterial kidney disease (BKD). Fish Shellfish Immunol. 11367-370. [DOI] [PubMed] [Google Scholar]
- 32.Griffiths, S. G., K. J. Melville, and K. Salonius. 1998. Reduction of Renibacterium salmoninarum culture activity in Atlantic salmon following vaccination with avirulent strains. Fish Shellfish Immunol. 8607-619. [Google Scholar]
- 33.Gutenberger, S. K., R. J. Duimstra, J. S. Rohovec, and J. L. Fryer. 1997. Intracellular survival of Renibacterium salmoninarum in trout mononuclear phagocytes. Dis. Aquat. Org. 2893-106. [Google Scholar]
- 34.Hendrickson, E. L., R. Kaul, Y. Zhou, D. Bovee, P. Chapman, J. Chung, E. Conway de Macario, J. A. Dodsworth, W. Gillett, D. E. Graham, M. Hackett, A. K. Haydock, A. Kang, M. L. Land, R. Levy, T. J. Lie, T. A. Major, B. C. Moore, I. Porat, A. Palmeiri, G. Rouse, C. Saenphimmachak, D. Soll, S. Van Dien, T. Wang, W. B. Whitman, Q. Xia, Y. Zhang, F. W. Larimer, M. V. Olson, and J. A. Leigh. 2004. Complete genome sequence of the genetically tractable hydrogenotrophic methanogen Methanococcus maripaludis. J. Bacteriol. 1866956-6969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Hoffnagle, T. L., R. W. Carmichael, and W. T. Noll. 2002. Grande Ronde Basin spring Chinook salmon captive broodstock program, 1995-2002 project status report. Oregon Fish and Wildlife Department, La Grande, OR.
- 36.Ingles, C. J., and G. H. Dixon. 1967. Phosphorylation of protamine during spermatogenesis in trout testis. Proc. Natl. Acad. Sci. USA 581011-1018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Kumar, S., K. Tamura, and M. Nei. 2004. MEGA3: integrated software for molecular evolutionary genetics analysis and sequence alignment. Brief Bioinform. 5150-163. [DOI] [PubMed] [Google Scholar]
- 38.Marraffini, L. A., A. C. Dedent, and O. Schneewind. 2006. Sortases and the art of anchoring proteins to the envelopes of gram-positive bacteria. Microbiol. Mol. Biol. Rev. 70192-221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.McIntosh, D., E. Flano, T. H. Grayson, M. L. Gilpin, B. Austin, and A. J. Villena. 1997. Production of putative virulence factors by Renibacterium salmoninarum grown in cell culture. Microbiology 1433349-3356. [DOI] [PubMed] [Google Scholar]
- 40.Mitchum, D. L., and L. E. Sherman. 1981. Transmission of bacterial kidney disease from wild to stocked hatchery trout. Can. J. Fish. Aquat. Sci. 38547-551. [Google Scholar]
- 41.Mongodin, E. F., N. Shapir, S. C. Daugherty, R. T. DeBoy, J. B. Emerson, A. Shvartzbeyn, D. Radune, J. Vamathevan, F. Riggs, V. Grinberg, H. Khouri, L. P. Wackett, K. E. Nelson, and M. J. Sadowsky. 2006. Secrets of soil survival revealed by the genome sequence of Arthrobacter aurescens TC1. PLoS Genet. 2e214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Mukamolova, G. V., O. A. Turapov, K. Kazarian, M. Telkov, A. S. Kaprelyants, D. B. Kell, and M. Young. 2002. The rpf gene of Micrococcus luteus encodes an essential secreted growth factor. Mol. Microbiol. 46611-621. [DOI] [PubMed] [Google Scholar]
- 43.Mukamolova, G. V., O. A. Turapov, D. I. Young, A. S. Kaprelyants, D. B. Kell, and M. Young. 2002. A family of autocrine growth factors in Mycobacterium tuberculosis. Mol. Microbiol. 46623-635. [DOI] [PubMed] [Google Scholar]
- 44.O'Farrell, C. L., and M. S. Strom. 1999. Differential expression of the virulence-associated protein p57 and characterization of its duplicated gene msa in virulent and attenuated strains of Renibacterium salmoninarum. Dis. Aquat. Org 38115-123. [DOI] [PubMed] [Google Scholar]
- 45.O'Riordan, K., and J. C. Lee. 2004. Staphylococcus aureus capsular polysaccharides. Clin. Microbiol. Rev. 17218-234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Overbeek, R., N. Larsen, T. Walunas, M. D'Souza, G. Pusch, E. Selkov, Jr., K. Liolios, V. Joukov, D. Kaznadzey, I. Anderson, A. Bhattacharyya, H. Burd, W. Gardner, P. Hanke, V. Kapatral, N. Mikhailova, O. Vasieva, A. Osterman, V. Vonstein, M. Fonstein, N. Ivanova, and N. Kyrpides. 2003. The ERGO genome analysis and discovery system. Nucleic Acids Res. 31164-171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Pallen, M. J., and B. W. Wren. 2007. Bacterial pathogenomics. Nature 449835-842. [DOI] [PubMed] [Google Scholar]
- 48.Raymond, C. K., S. Subramanian, M. Paddock, R. Qiu, C. Deodato, A. Palmieri, J. Chang, T. Radke, E. Haugen, A. Kas, D. Waring, D. Bovee, R. Stacy, R. Kaul, and M. V. Olson. 2005. Targeted, haplotype-resolved resequencing of long segments of the human genome. Genomics 86759-766. [DOI] [PubMed] [Google Scholar]
- 49.Raymond, K. N., E. A. Dertz, and S. S. Kim. 2003. Enterobactin: an archetype for microbial iron transport. Proc. Natl. Acad. Sci. USA 1003584-3588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Rhodes, L. D., A. M. Coady, and R. K. Deinhard. 2004. Identification of a third msa gene in Renibacterium salmoninarum and the associated virulence phenotype. Appl. Environ. Microbiol. 706488-6494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Rhodes, L. D., A. M. Coady, and M. S. Strom. 2002. Expression of duplicate msa genes in the salmonid pathogen Renibacterium salmoninarum. Appl. Environ. Microbiol. 685480-5487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Rhodes, L. D., C. Durkin, S. L. Nance, and C. A. Rice. 2006. Prevalence and analysis of Renibacterium salmoninarum infection among juvenile Chinook salmon Oncorhynchus tshawytscha in North Puget Sound. Dis. Aquat. Org. 71179-190. [DOI] [PubMed] [Google Scholar]
- 53.Rhodes, L. D., T. H. Grayson, S. M. Alexander, and M. S. Strom. 2000. Description and characterization of IS994, a putative IS3 family insertion sequence from the salmon pathogen, Renibacterium salmoninarum. Gene 24497-107. [DOI] [PubMed] [Google Scholar]
- 54.Rhodes, L. D., O. T. Nguyen, R. K. Deinhard, T. M. White, L. W. Harrell, and M. C. Roberts. 2008. Characterization of Renibacterium salmoninarum with reduced susceptibility to macrolide antibiotics by a standardized antibiotic susceptibility test. Dis. Aquat. Org. 80173-180. [DOI] [PubMed] [Google Scholar]
- 55.Rhodes, L. D., C. K. Rathbone, S. C. Corbett, L. W. Harrell, and M. S. Strom. 2004. Efficacy of cellular vaccines and genetic adjuvants against bacterial kidney disease in chinook salmon (Oncorhynchus tshawytscha). Fish Shellfish Immunol. 16461-474. [DOI] [PubMed] [Google Scholar]
- 56.Rockey, D. D., P. S. Turaga, G. D. Wiens, B. A. Cook, and S. L. Kaattari. 1991. Serine proteinase of Renibacterium salmoninarum digests a major autologous extracellular and cell-surface protein. Can. J. Microbiol. 37758-763. [DOI] [PubMed] [Google Scholar]
- 57.Salonius, K., C. Siderakis, A. M. MacKinnon, and S. G. Griffiths. 2005. Use of Arthrobacter davidanieli as a live vaccine against Renibacterium salmoninarum and Piscirickettsia salmonis in salmonids. Dev. Biol. 121189-197. [PubMed] [Google Scholar]
- 58.Sanders, J. E., and J. L. Fryer. 1980. Renibacterium salmoninarum gen. nov., sp. nov., the causative agent of bacterial kidney disease in salmonid fishes. Int. J. Syst. Bacteriol. 30496-502. [Google Scholar]
- 59.Sorum, U., B. Robertsen, and L. Kenne. 1998. Structural studies of the major polysaccharide in the cell wall of Renibacterium salmoninarum. Carbohydr. Res. 306305-314. [DOI] [PubMed] [Google Scholar]
- 60.Starliper, C. E., D. R. Smith, and T. Shatzer. 1997. Virulence of Renibacterium salmoninarum to salmonids. J. Aquat. Anim. Health 91-7. [Google Scholar]
- 61.Stinear, T. P., T. Seemann, S. Pidot, W. Frigui, G. Reysset, T. Garnier, G. Meurice, D. Simon, C. Bouchier, L. Ma, M. Tichit, J. L. Porter, J. Ryan, P. D. Johnson, J. K. Davies, G. A. Jenkin, P. L. Small, L. M. Jones, F. Tekaia, F. Laval, M. Daffe, J. Parkhill, and S. T. Cole. 2007. Reductive evolution and niche adaptation inferred from the genome of Mycobacterium ulcerans, the causative agent of Buruli ulcer. Genome Res. 17192-200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Sudheesh, P. S., S. Crane, K. D. Cain, and M. S. Strom. 2007. Sortase inhibitor phenyl vinyl sulfone inhibits Renibacterium salmoninarum adherence and invasion of host cells. Dis. Aquat. Org. 78115-127. [DOI] [PubMed] [Google Scholar]
- 63.Turaga, P. S. D., G. D. Wiens, and S. L. Kaattari. 1987. Analysis of Renibacterium salmoninarum antigen production in situ. Fish Pathol. 22209-214. [Google Scholar]
- 64.Turaga, P. S. D., G. D. Wiens, and S. L. Kaattari. 1987. Bacterial kidney diseases: the potential role of soluble protein antigen(s). J. Fish Biol. 31191-194. [Google Scholar]
- 65.White, O. 2004. Bacterial genome annotation at TIGR, p. 33-46. In C. M. Fraser, T. D. Read, and K. E. Nelson (ed.), Microbial genomes. Humana Press, Totowa, NJ.
- 66.Wiens, G. D. 2006. Bacterial kidney disease. CAB International, Wallingford, United Kingdom. http://www.cabicompendium.org/ac.
- 67.Wiens, G. D., and S. L. Kaattari. 1999. Bacterial kidney disease (Renibacterium salmoninarum), p. 269-301. In P. T. K. Woo and D. W. Bruno (ed.), Fish diseases and disorders: viral, bacterial and fungal infections, vol. 3. CAB International, Wallingford, United Kingdom. [Google Scholar]
- 68.Wiens, G. D., and S. L. Kaattari. 1989. Monoclonal antibody analysis of common surface protein(s) of Renibacterium salmoninarum. Fish Pathol. 241-7. [Google Scholar]
- 69.Wiens, G. D., and S. L. Kaattari. 1991. Monoclonal antibody characterization of a leukoagglutinin produced by Renibacterium salmoninarum. Infect. Immun. 59631-637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Wiens, G. D., R. Pascho, and J. R. Winton. 2002. A single Ala139-to-Glu substitution in the Renibacterium salmoninarum virulence-associated protein p57 results in antigenic variation and is associated with enhanced p57 binding to Chinook salmon leukocytes. Appl. Environ. Microbiol. 683969-3977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Wood, D. W., J. C. Setubal, R. Kaul, D. E. Monks, J. P. Kitajima, V. K. Okura, Y. Zhou, L. Chen, G. E. Wood, N. F. Almeida, Jr., L. Woo, Y. Chen, I. T. Paulsen, J. A. Eisen, P. D. Karp, D. Bovee, Sr., P. Chapman, J. Clendenning, G. Deatherage, W. Gillet, C. Grant, T. Kutyavin, R. Levy, M. J. Li, E. McClelland, A. Palmieri, C. Raymond, G. Rouse, C. Saenphimmachak, Z. Wu, P. Romero, D. Gordon, S. Zhang, H. Yoo, Y. Tao, P. Biddle, M. Jung, W. Krespan, M. Perry, B. Gordon-Kamm, L. Liao, S. Kim, C. Hendrick, Z. Y. Zhao, M. Dolan, F. Chumley, S. V. Tingey, J. F. Tomb, M. P. Gordon, M. V. Olson, and E. W. Nester. 2001. The genome of the natural genetic engineer Agrobacterium tumefaciens C58. Science 2942317-2323. [DOI] [PubMed] [Google Scholar]
- 72.Wood, P. A., and S. L. Kaattari. 1996. Enhanced immunogenicity of Renibacterium salmoninarum in chinook salmon after removal of the bacterial cell surface-associated 57 kDa protein. Dis. Aquat. Org. 2571-79. [Google Scholar]
- 73.Young, C. L., and G. B. Chapman. 1978. Ultrastructural aspects of the causative agent and renal histopathology of bacterial kidney disease in brook trout (Salvelinus fontinalis). J. Fish Res. Board Can. 351234-1248. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.