Abstract
The SecA nanomotor promotes protein translocation in eubacteria by binding both protein cargo and the protein-conducting channel and by undergoing ATP-driven conformation cycles that drive this process. There are conflicting reports about whether SecA functions as a monomer or dimer during this dynamic process. Here we reexamined the roles of the amino and carboxyl termini of SecA in promoting its dimerization and functional state by examining three secA mutants and the corresponding proteins: SecAΔ8 lacking residues 2 to 8, SecAΔ11 lacking residues 2 to 11, and SecAΔ11/N95 lacking both residues 2 to 11 and the carboxyl-terminal 70 residues. We demonstrated that whether SecAΔ11 or SecAΔ11/N95 was functional for promoting cell growth depended solely on the vivo level of the protein, which appeared to govern residual dimerization. All three SecA mutant proteins were defective for promoting cell growth unless they were highly overproduced. Cell fractionation revealed that SecAΔ11 and SecAΔ11/N95 were proficient in membrane association, although the formation of integral membrane SecA was reduced. The presence of a modestly higher level of SecAΔ11/N95 in the membrane and the ability of this protein to form dimers, as detected by chemical cross-linking, were consistent with the higher level of secA expression and better growth of the SecAΔ11/N95 mutant than of the SecAΔ11 mutant. Biochemical studies showed that SecAΔ11 and SecAΔ11/N95 had identical dimerization defects, while SecAΔ8 was intermediate between these proteins and wild-type SecA in terms of dimer formation. Furthermore, both SecAΔ11 and SecAΔ11/N95 were equally defective in translocation ATPase specific activity. Our studies showed that the nonessential carboxyl-terminal 70 residues of SecA play no role in its dimerization, while increasing the truncation of the amino-terminal region of SecA from 8 to 11 residues results in increased defects in SecA dimerization and poor in vivo function unless the protein is highly overexpressed. They also clarified a number of conflicting previous reports and support the essential nature of the SecA dimer.
Prokaryotes and eukaryotes contain a phylogenetically conserved protein-conducting channel in their major translocation-competent membrane systems that interacts with different cytosolic components in order to promote protein translocation (26). In bacteria, the SecYEG channel utilizes primarily the signal recognition particle-dependent pathway for cotranslational insertion of integral membrane proteins, while it utilizes the SecB chaperone and SecA ATPase nanomotor for posttranslational translocation of secretory preproteins (10, 27). SecA has a dual function in the latter pathway; first it binds preprotein and SecYEG in the correct topological configuration, and then it undergoes ATP-driven conformational cycles that drive protein translocation in a stepwise fashion.
The oligomeric state of SecA during protein translocation has been a matter of considerable controversy recently (29). Originally, SecA was found to exist in a temperature- and salt-dependent monomer-dimer equilibrium at the micromolar concentration range (33). It has been shown that many of SecA's translocation ligands affect its oligomeric state; phospholipid and SecYEG binding promoted monomerization of SecA, whereas there have been conflicting reports concerning the effect of signal peptides and adenylate nucleotides on SecA's oligomeric state in soluble or membranous environments (1, 3, 24, 30).
Several approaches have been taken to determine whether SecA functions as a monomer or dimer or alternates between these two states in order to promote protein translocation. Both in vivo and in vitro protein cross-linking techniques have been utilized to assess the fraction of the SecA dimer in soluble and membrane-bound states. Unfortunately, the studies have produced equivocal results (17, 23, 24, 32) that are of limited value; on the one hand, cross-linking traps proteins in the oligomeric state and thus distorts a natural equilibrium, while on the other hand, suboptimal cross-linking conditions (e.g., nonideal cross-linker chemistry, insufficient cross-linker concentration, or nonphysiological buffer or salt conditions) can lead to underrepresentation of the true oligomeric population. Genetic techniques have been utilized to engineer SecA dimers that were linked in a head-to-tail fashion, and such modified proteins were found to be functional both in vivo and in vitro (22, 32). The findings of the latter studies are consistent with the findings of parallel biochemical studies in which disulfide cross-linking was utilized to create covalently bonded SecA dimers that were found to be functional in vitro (however, see reference 25 for a description of a contradictory study) (7, 16). One limitation of the latter approaches, however, is that they cannot rule out the possibility that covalently bonded SecA dimers actually function as tethered monomers. However, the findings are consistent with additional findings supporting the hypothesis that the dimer is the active state of SecA. For example, secA mutants display intragenic complementation, often a hallmark of proteins that function as oligomers (5, 32). Furthermore, it has been shown that in other mutant situations, membrane-bound SecA heterodimers comprised of functional and nonfunctional subunits could be produced and were inactive (9, 17), a result that is difficult to rationalize if membrane-bound SecA functions solely as a monomer.
Studies of SecA monomer-biased mutants have been used to specifically assess the functionality of the SecA monomer. The amino-terminal 12 residues of SecA are an important interprotomer contact site for stabilizing the antiparallel SecA dimer structure described by Hunt and colleagues (15). However, conflicting results have been obtained previously for the functional state of a SecA mutant protein lacking residues 2 to 11 (SecAΔ11) or a similar protein that also lacks the dispensable carboxyl-terminal 70-amino-acid residues of SecA (SecAΔ11/N95). Or et al. reported that secAΔ11/N95 complemented a secA conditional null mutant and that the mutant protein exhibited significant in vitro protein translocation activity, particularly when it was assayed with membranes or proteoliposomes derived from a prlA4 mutant (23). In contrast, we reported that secAΔ11 was unable to complement a secA conditional null mutant and that the SecAΔ11 mutant protein did not have in vitro protein translocation activity and had poor SecA-dependent translocation ATPase activity (17), consistent with the result of Randall and coworkers (28).
Characterization of secAΔ11, secAΔ11/N95, and secAΔ8 mutants.
In order to clarify the important issue described above, we reexamined it. For this purpose an Escherichia coli strain carrying a chromosomal secA amber allele and a temperature-sensitive amber suppressor, BL21.19 [secA13(Am) supF(Ts) trp(Am) zch::Tn10 recA::CAT clpA::KAN (λDE3)], and an isogenic derivative of this stain, BL21.20 [secA13(Am) supF(Ts) trp(Am) zch::Tn10 recA::KAN (λDE3)], were used as hosts for secA-containing plasmids. pT7secA-his and pT7secAΔ11-his, which contain wild-type secA encoding a His-tagged protein and a derivative missing secA codons 2 to 11, respectively, have been described previously (17). pT7secAΔ11/N95 is similar to pT7secAΔ11 but lacks the last 70 codons of secA and was a kind gift from P. C. Tai (32). Sequence analysis of the latter plasmid revealed that it contained a nucleotide substitution at codon 700 of secA that resulted in a methionine-to-valine substitution. However, when this codon was changed back to the wild-type codon (QuikChange; Stratagene) and the growth properties of the relevant strains were compared, no significant differences were found. Finally, we constructed pT7secAΔ8his missing secA codons 2 to 8 by using the QuikChange procedure, appropriate mutagenic primers (Integrated DNA Technologies), and pT7secAΔ11his as the template. All mutations were confirmed by DNA sequence analysis (University of Pennsylvania DNA Sequencing Facility).
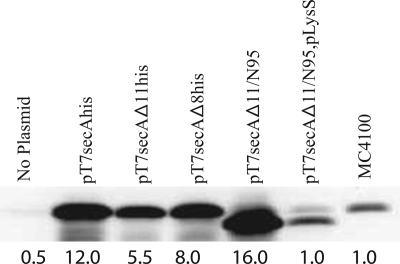
We first tested the growth properties of the relevant strains by comparing colony counts for serial dilutions of overnight cultures grown at 30°C in LB (Sigma) supplemented with 100 μg/ml ampicillin (Amp) and 25 μg/ml chloramphenicol, where appropriate, and plated on LB plates containing Amp or LB plates containing Amp and chloramphenicol, which were subsequently incubated overnight at either 42°C (at which only the plasmid-borne secA gene was expressed) or 30°C (at which both chromosomal and plasmid-borne secA genes were expressed). The ratio of the two values obtained was the plating efficiency for a given strain. In our tests, the colony counts for all strains at 30°C were comparable. Consistent with previous reports, we found that secAΔ11/N95 was functional, while secAΔ11his was not functional, since both BL21.19(pT7secA-his) and BL21.19(pT7secAΔ11/N95) had good plating efficiencies (0.91 and 0.93, respectively), while BL21.19(pT7secAΔ11his) was not viable under these conditions (plating efficiency, 5 × 10−5) (17, 23, 32). The viability of BL21.19(pT7secAΔ8his) was also low under our conditions (plating efficiency, 3 × 10−5), despite the fact that SecAΔ8 has been reported previously to be fully functional both in vivo and in vitro (18). In order to assess SecA levels under these conditions, Western blotting was performed (Fig. 1). We found that the level of SecA protein was nearly three times higher in BL21.19(pT7secAΔ11/N95) than in BL21.19(pT7secAΔ11his). The overproduction was likely to be due to the stabilizing effect of truncation of the carboxyl terminus of SecA, which is weakly structured and therefore is a target for in vivo proteolysis (6, 15). Alternatively, truncation of the 3′ end of secA may stabilize secA mRNA in the strain used. By contrast, the level of the SecA protein was only ∼50% higher in BL21.19(pT7secAΔ8his) than in BL21.19(pT7secAΔ11his). When we constructed and analyzed BL21.19(pT7secAΔ11/N95, pLysS), which contained the pLysS plasmid that reduces the endogenous level of T7 RNA polymerase in this expression system (31), we found that the level of the SecAΔ11/N95 protein was equivalent to the level of chromosomally produced SecA, and this strain was not viable at 42°C (the plating efficiency was 2 × 10−6). Collectively, our results indicate that the specific activities of the SecAΔ8his, SecAΔ11his, and SecAΔ11/N95 proteins in vivo are low, underscoring the importance of the amino-terminal region of SecA for promotion of normal cell growth. In addition, they suggest that the likely mechanism of suppression of the SecAΔ11 defect by SecAΔ11/N95 is SecA overproduction (see below for the results of a specific test of this hypothesis). Such overproduction would obviously help to populate the SecA dimer pool within the cell.
FIG. 1.
SecAΔ11/N95 is functional in vivo due to overproduction of SecA protein. BL21.19 containing the indicated plasmids was grown in LB supplemented with appropriate antibiotics at 30°C to an A600 of 0.2, and then it was shifted to 42°C for an additional 2 h. The A600 of all cultures were adjusted to 1.0 by dilution of LB, and then cells were chilled, harvested by sedimentation at 4°C, and resuspended in sample buffer (2% sodium dodecyl sulfate, 125 mM Tris-HCl [pH 6.8], 5% 2-mercaptoethanol, 15% glycerol, 0.005% bromophenol blue) for analysis by sodium dodecyl sulfate-polyacrylamide gel electrophoresis followed by Western blotting with SecA antisera and visualization by enhanced chemiluminescence utilizing SuperSignal West Pico (Pierce) and a Syngene Gelbox system as described previously (17). Prior to sodium dodecyl sulfate-polyacrylamide gel electrophoresis analysis, the protein concentration in each sample was measured utilizing the Bradford assay, and equivalent amounts of total protein were loaded onto the gel. SecA levels were determined using a Syngene Gelbox system; the amount of SecA present in wild-type strain MC4100 lacking any plasmid was arbitrarily defined as 1.0, and the amounts determined are indicated under the lanes.
Characterization of secAΔ11 suppressor mutants.
Since the carboxyl-terminal 70 residues of SecA have been shown to contain SecB- and phospholipid-binding sites (4, 12), we wanted to specifically examine whether the dramatically different growth properties of BL21.19(pT7secAΔ11/N95) and BL21.19(pT7secAΔ11his) were related to the presence of the carboxyl terminus of SecA. We reasoned that if the essential properties of the two SecA proteins of these strains were biochemically equivalent, then only modest overproduction of the SecAΔ11 protein would be needed to produce good cell viability similar to that of BL21.19(pT7secAΔ11/N95). In particular, mutations within the lac operator region that partially derepress the T7 promoter driving the secA gene may be common (pT7secAΔ11his is a pET29b derivative [Novagen] that contains a lac operator immediately downstream of the consensus T7 promoter, as well as a lacI gene in order to provide adequate repressor levels [31]).
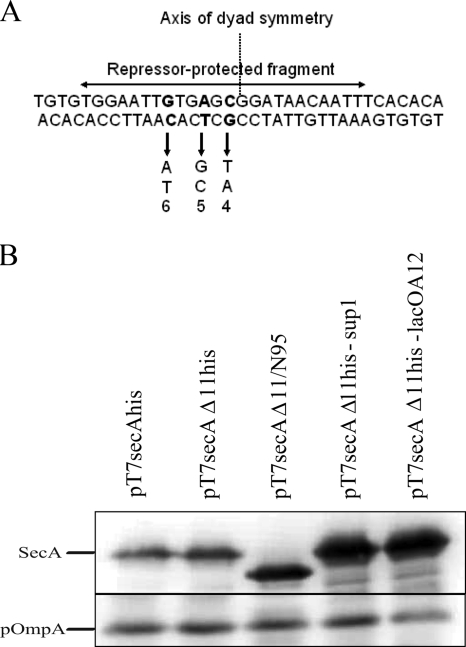
In order to isolate suppressor mutants, single colonies of BL21.19(pT7secAΔ11his) were inoculated into 5 ml of LB containing Amp and grown overnight at 30°C. One hundred microliters of a 1-to-40 dilution of each overnight culture was plated on an LB plate containing Amp, which was incubated overnight at 42°C. Colonies that grew under these conditions were picked and purified twice at 42°C, and plasmid DNA was extracted (Qiagen) and used to retransform BL21.19 at 30°C. The transformants were purified and tested for growth at 30°C and 42°C in order to check the plasmid linkage of the suppressor mutation. We found that approximately one-half of all potential suppressor mutations were plasmid linked. The plasmid-linked mutants were analyzed by Western blotting in order to identify suppressors that overproduced SecA protein. Approximately one-half of the mutants screened clearly overproduced SecA protein compared to BL21.19(pT7secAΔ11his). Sequence analysis of plasmid DNA isolated from nine such strains showed that four of them contained previously described lac operator constitutive [lacO(Con)] mutations in three of the most important nucleotides determining repressor binding activity and retained the secAΔ11 deletion (Fig. 2A) (2). All three substitutions occurred in the left half of the operator, which has been shown to be critical for tight repressor binding. In order to verify that the lacO(Con) mutation was solely responsible for growth suppression as well as SecA overproduction, site-directed mutagenesis (QuikChange) was utilized to reconstruct one of the lacO(Con) mutations (the A at position 12 [Fig. 2A]) in pT7secAΔ11his, and the properties of the relevant strains were compared (Fig. 2B). The original suppressor mutant, BL21.19(pT7secAΔ11his-sup1), and the reconstructed mutant, BL21.19(pT7secAΔ11his-lacOA12), showed similar plating efficiencies (1.1 and 0.64, respectively), and they both overproduced SecA protein at a level that was more comparable to the level produced by BL21.19(pT7secAΔ11/N95) than the level produced by BL21.19(pT7secAΔ11his). Our results show that overproduction of the SecAΔ11his protein at a level comparable to the SecAΔ11/N95 level is sufficient to suppress the observed growth defect. Furthermore, they imply that the lack of the carboxyl-terminal 70 amino acid residues of SecAΔ11/N95 has no special role in promoting the functional state of the protein; rather, the protein is functional due to its overproduction.
FIG. 2.
lacO(Con) mutants restore SecAΔ11his function by overproduction. (A) Sequence of the lac operator region with the lacO(Con) suppressor mutations indicated below the sequence. The lacO(Con) mutation at position 12 was present in two independent suppressor mutants, whereas the mutation at position 15 and the mutation at position 17 were each present in a single suppressor mutant. The number below each lacO(Con) mutation was obtained from a previous study (2), and these numbers indicate the corresponding repressor affinities for the mutant operator compared to the wild-type operator, which was defined as 100%. The lac repressor-protected fragment is indicated above the sequence, and the axis of dyad symmetry of the sequence is also indicated. (B) Western blot of BL21.19 containing the indicated plasmids. The analysis was performed like the analysis described in the legend to Fig. 1 except that both SecA and OmpA antisera were used to probe the blot. OmpA was utilized as a control protein to verify that equivalent levels of total protein were loaded on the gel.
Subcellular fractionation of secAΔ11 and secAΔ11/N95 mutants.
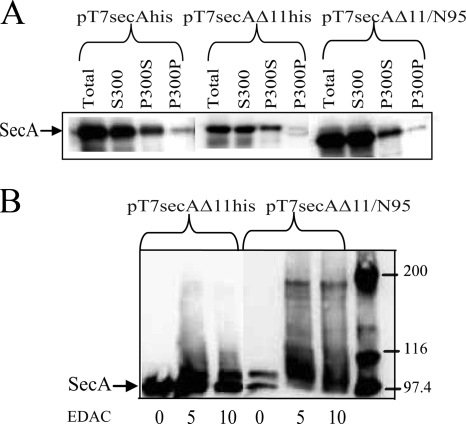
In order to continue to elucidate the different growth properties of BL21.19(pT7secAΔ11his) and BL21.19(pT7secAΔ11/N95), we performed subcellular fractionation experiments to assess SecA membrane association, which is critical for the SecA function (11, 14). We found that both SecAΔ11his and SecAΔ11/N95 associated with the cellular membrane fraction (fraction P300S) (Fig. 3A) and that the level of SecAΔ11/N95 was 12% higher than the level of SecAΔ11his, consistent with the higher level of expression of SecAΔ11/N95. In addition, the levels of both mutant proteins were reduced in the integral membrane fraction (fraction P300P). These results are consistent with the results of our previous study, where we demonstrated that SecAΔ11his displayed normal SecYEG binding activity but that the reduced amount of this protein in the integral membrane fraction suggested that dimer formation was needed to promote the SecA membrane insertion reaction (17). In order to detect the presence of membrane-bound SecA dimer, chemical cross-linking experiments were performed using the total membrane fraction (fraction P300) from the two mutants. In this case we detected the presence of a modest level of the SecAΔ11/N95 dimer but not the SecAΔ11his dimer in the membrane fraction (Fig. 3B), consistent with the growth properties of the two strains, as well as the importance of the SecA dimer for the functional state of this protein.
FIG. 3.
Subcellular fractionation and cross-linking studies support the in vivo results. BL21.19 strains containing the indicated plasmids were grown and harvested as described in the legend to Fig. 1. Cell pellets were resuspended in 0.02 volume of ice-cold (A) TKMDP (10 mM Tris-Cl [pH 7.5], 50 mM KCl, 10 mM magnesium acetate, 1 mM dithiothreitol, 0.5 mM phenylmethylsulfonyl fluoride, 1× protease inhibitor cocktail [Sigma]) or (B) HKM (50 mM HEPES-KOH [pH 7.2], 50 mM KCl, 1 mM magnesium acetate, 1× protease inhibitor cocktail) and broken by two passages at 8,000 lb/in2 in a French pressure cell. Unbroken cells were removed by two successive centrifugations at 13,000 × g for 10 min at 4°C, which resulted in the total cleared lysate (Total). (A) For subcellular fractionation, soluble (S300) and membrane (P300) fractions were obtained by centrifugation at 150,000 × g for 25 min at 4°C in a Sorvall Discovery M120 microultracentrifuge. S300 was carefully removed, and P300 was resuspended in the original volume of 0.2 M sodium carbonate (pH 11.5), incubated on ice for 30 min, and resedimented. The carbonate-treated supernatant (P300S) was carefully removed, and the integral membrane pellet (P300P) was resuspended in the original volume of TKMD. (B) For protein cross-linking, the P300 pellet was briefly washed and resuspended in the original volume of HKM. Ten microliters of P300 was subjected to chemical cross-linking with the indicated concentration of EDAC [1-ethyl-3-(3-dimethyl-aminopropyl)carbodiimide] in HKM supplemented with 50 mM potassium acetate at room temperature for 15 min in a 40-μl reaction mixture, and then the reaction mixtures were quenched on ice by addition of 3 μl of 1 M Tris-HCl (pH 7.5). All samples were heated to 100°C for 2 min after addition of sample buffer and analyzed by (A) 11.3% or (B) 5.5% sodium dodecyl sulfate-polyacrylamide gel electrophoresis followed by Western blotting and visualization of SecA using appropriate antisera by enhanced chemiluminescence utilizing SuperSignal West Pico (Pierce) and a Syngene Gelbox system.
Biochemical analysis of the SecAΔ11, SecAΔ11/N95, and SecAΔ8 proteins.
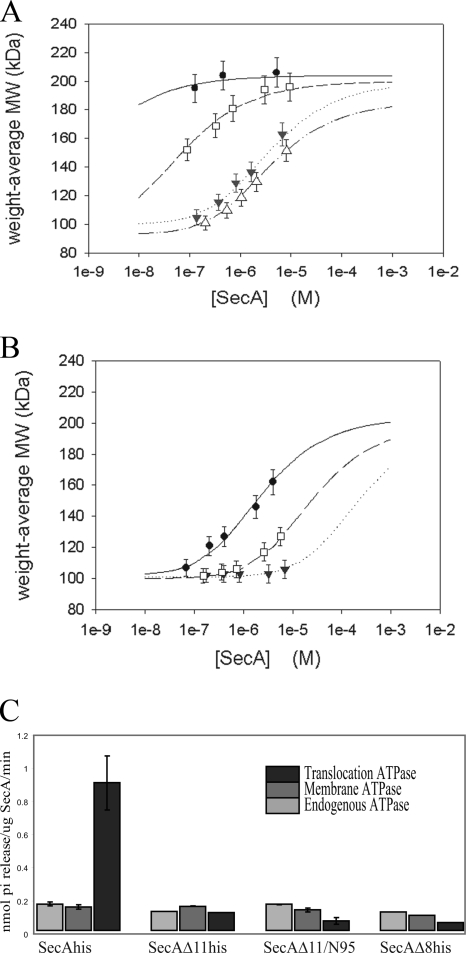
Our genetic and physiologic results suggested that the biochemical properties of the SecAΔ11his and SecAΔ11/N95 proteins are similar and that expression differences in the relevant strains were responsible for the remarkable differences in growth properties described above. To explicitly examine this, we purified these two proteins, as well as SecAΔ8, and determined the monomer-dimer equilibrium constants by using size exclusion chromatography and static light scattering (Fig. 4). Since it has been reported previously that the SecA monomer-dimer equilibrium is highly salt dependent (33), two different salt concentrations were employed to more accurately measure differences in SecA interprotomer affinity. We found that the SecAΔ11his and SecAΔ11/N95 proteins had essentially identical monomer-dimer equilibrium constants (3.5 × 10−6 ± 0.2 × 10−6 M and 3.87 × 10−6 ± 0.09 × 10−6 M, respectively) (Fig. 4A) with 100 mM KCl, while the value for SecAΔ8his (7 × 10−8 ± 1 × 10−8 M) was intermediate between these values and the value for the wild-type SecA-his protein (<1 × 10−9 M). A recent study utilizing equilibrium sedimentation also showed that the SecAΔ11/N95 protein was a dimer at a concentration of 1 to 13 μM with 50 mM KCl (13). In order to more accurately compare the interprotomer affinity of SecAΔ8his with that of wild-type SecA-his, we increased the salt concentration to 300 mM KCl. Under these conditions the monomer-dimer equilibrium constants for the wild-type SecA-his, SecAΔ8his, and SecAΔ11his proteins increased with decreasing length of the stabilizing amino terminus of SecA protein (2.2 × 10−6 ± 0.2 × 10−6 M, 2.41 × 10−5 ± 0.05 × 10−5 M, and >2.3 × 10−4 M, respectively) (Fig. 4B). Our results demonstrate that the SecAΔ11his and SecAΔ11/N95 proteins are essentially identical in terms of their monomer-dimer equilibrium defects, and they also show that SecAΔ8his has an intermediate defect that is commensurate with the length of its amino-terminal region. The results are also consistent with the observed poor plating efficiency of BL21.19(pT7secAΔ8his). Although the latter results differ from those reported previously by Karamanou et al. (18), these investigators utilized only a single concentration of SecAΔ8his protein in their gel filtration study and the salt concentration utilized was not reported. In addition, although these investigators observed good viability for their secAΔ8his mutant strain, we believe that modest differences in the level of expression of the SecAΔ8his protein in the different strains (similar to our observations described above) could readily account for the apparent discrepancy in this case.
FIG. 4.
The monomer-dimer equilibrium of SecAΔ11, SecAΔ11/N95, and SecAΔ8 proteins is altered, and these proteins have defects in their SecA-dependent translocation ATPase activities. SecA proteins were overproduced and purified using either His-bind resin (Novagen) or SP Sepharose (Sigma) as previously described (17). (A and B) Light scattering data were collected using a Superose 6 10/30 HR size exclusion chromatography column (GE Healthcare) connected to a high-performance liquid chromatography system (Alliance 2965; Waters Corp.) equipped with an autosampler. The eluate from the size exclusion chromatography column was monitored by using a photodiode array UV/Vis detector (996 PDA; Waters Corp.), a differential refractometer (OPTI-Lab or OPTI-rEx; Wyatt Corp.), and a static, multiangle laser light scattering detector (DAWN-EOS; Wyatt Corp.). The system was equilibrated with a solution containing 10 mM Tris-HCl (pH 7.5), 100 mM or 300 mM KCl, 10 mM magnesium acetate, 1 mM dithiothreitol, and 1× protease inhibitor cocktail at a flow rate of 0.3 ml/min. The weight average molecular mass (MW) for SecA protein was determined for the concentration ranges shown with either (A) 100 mM KCl or (B) 300 mM KCl. Two software packages were used for data collection and analysis; the Millennium software (Waters Corp.) controlled the high-performance liquid chromatograph operation and data collection from the multiwavelength UV/Vis detector, while the ASTRA software (Wyatt Corp.) collected data from the refractive index detector and the light scattering detector and recorded the UV trace at 280 nm sent from the 996 PDA detector. The lines indicate the nonlinear least-square fits of weight average molecular mass determined for the apex of each eluting peak at various concentrations for a monomer-dimer association model; the error bars indicate 5% uncertainties for molecular mass determinations expected from measurement of light scattering. Symbols: filled circles, SecA-his; open squares, SecAΔ8his; filled triangles, SecAΔ11his; open triangles, SecAΔ11/N95. (C) Endogenous, membrane, and translocation ATPase activities of the indicated SecA mutant proteins, determined as described previously (17).
In order to assess the biochemical activities of the different mutant SecA proteins, we performed ATPase assays. SecA has a basal ATPase activity in solution (endogenous ATPase) that is mildly stimulated upon binding to SecYEG protein present in inverted membrane vesicles (membrane ATPase), and its ATPase activity is substantially stimulated in the presence of the SecYEG protein and an export-competent preprotein (translocation ATPase) (20). While the SecAΔ11his, SecAΔ11/N95, and SecAΔ8his proteins had endogenous and membrane ATPase activities that were roughly similar to those of wild-type SecA-His, all three mutant proteins had very low translocation ATPase activities at equivalent concentrations of protein (Fig. 4C). These results indicate that the essential activities of SecAΔ11his and SecAΔ11/N95 are biochemically equivalent when equivalent concentrations of the two proteins are compared. In addition, these results are consistent with the low plating efficiencies observed for BL21.19(pT7secAΔ11his), BL21.19(pT7secAΔ8his), and BL21.19(pT7secAΔ11/N95, pLysS), as well as the need for high levels of secA expression in BL21.19(pT7secAΔ11/N95) and BL21.19(pT7secAΔ11his-sup1) in order to obtain good growth.
The simplest interpretation of our results is that the monomer-biased secAΔ11his and secAΔ11/N95 mutants require SecA overproduction in order to populate the dimer pool and achieve in vivo function. Indeed, while the SecAΔ11his and SecAΔ11/N95 proteins have been described as defective for dimerization (17, 23), our study clearly shows that the dimeric state can be populated at higher protein concentrations (Fig. 4A and B). Or and Rapoport estimated that the SecA concentration in a normal E. coli cell is 5 μM (25), while we calculated that the concentration is approximately 10-fold higher (based on 2,500 to 5,000 SecA molecules per cell [21] and the volume of a cylinder that is 1.8 μm long and 0.3 μm in diameter [8]). Since the relevant strains discussed here overproduce SecA by 5- to 15-fold and effective intracellular protein concentrations should be at least 10-fold higher than this concentration owing to excluded volume effects produced by the crowding of macromolecules (19), such calculations suggest that the ∼200 μM monomer-dimer equilibrium constant for the SecAΔ11his protein with 300 mM KCl may well be physiologically relevant. However, such calculations cannot take into account the fact that SecA translocation ligands dramatically effect this equilibrium, so true assessment of the oligomeric state of SecA in its translocationally active state is a much more complex matter that requires further study. In this regard de Keyzer et al. have argued that the SecA dimer is the normal SecYEG-binding form based on assessment of the parameters of binding of a chemically cross-linked SecA dimer to inverted membrane vesicles, as well as an increase in SecYEG-bound SecA dimer during SecYEG overproduction (7). By contrast, our present study and a previous study of the properties of the SecAΔ11his protein indicated that this monomer-biased mutant protein is capable of binding to SecYEG with normal affinity but that the SecA insertion reaction is defective and appears to require the dimeric state of SecA protein (Fig. 3A) (17).
Our study underscores the fact that close attention needs to be paid to secA expression levels in vivo, as well as SecA concentrations and conditions utilized for in vitro assays for proper assessment of SecA function. This is particularly true of the monomer-biased SecA mutant proteins studied here, where different expression levels and different biochemical assay conditions created seemingly conflicting results that are more readily reconcilable in light of our studies. We determined that the large changes in the stability of the SecA dimer observed for the amino-terminal deletion mutants, SecAΔ11his and SecAΔ8his, are comparable to destabilization of the SecA dimer caused by a modest change in the salt concentration in the assay buffer. For example, increasing the KCl concentration from 100 mM to 300 mM decreased the stability of the SecA dimer by 4.6 kcal/mol, nearly mimicking the effect caused by deletion of 11 amino-terminal residues (which decreased the stability by 4.9 kcal/mol) and exceeding the effect caused by deletion of 8 amino-terminal residues (which decreased the stability by 2.6 kcal/mol). These remarkable results underscore the difficulty of comparing studies done under different salt conditions that can dramatically affect the SecA monomer-dimer equilibrium.
Acknowledgments
We thank Stanley Lin and Jia Liu for assistance with use of the Syngene Gelbox system.
This work was supported by grant GM42033 from NIGMS to D.O.
Footnotes
Published ahead of print on 22 August 2008.
REFERENCES
- 1.Alami, M., K. Dalal, B. Lelj-Garolla, S. Sligar, and F. Duong. 2007. Nanodiscs unravel the interaction between the SecYEG channel and its cytosolic partner SecA. EMBO J. 261995-2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Barkley, M. D., and S. Bourgeois. 1980. Repressor recognition of operator and effectors, p. 177-220. In J. H. Miller and W. S. Reznikoff (ed.), The operon, 2nd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY.
- 3.Benach, J., Y.-T. Chou, J. J. Fak, A. Itkin, D. D. Nicolae, P. C. Smith, G. Wittrock, D. L. Floyd, C. M. Golsaz, L. M. Gierasch, and J. F. Hunt. 2003. Phospholipid-induced monomerization and signal-peptide-induced oligomerization of SecA. J. Biol. Chem. 2783628-3638. [DOI] [PubMed] [Google Scholar]
- 4.Breukink, E., N. Nouwen, A. van Raalte, S. Mizushima, J. Tommassen, and B. de Kruijff. 1995. The C terminus of SecA is involved in both lipid binding and SecB binding. J. Biol. Chem. 270:7902-7907. [DOI] [PubMed] [Google Scholar]
- 5.Cabelli, R. J. 1991. Biochemical characterization of the role of SecA protein in protein export in Escherichia coli. Ph.D. thesis. State University of New York, Stony Brook, NY.
- 6.Chou, Y.-T., J. Swain, and L. Gierasch. 2002. Functionally significant mobile regions of Escherichia coli SecA ATPase identified by NMR. J. Biol. Chem. 27750985-50990. [DOI] [PubMed] [Google Scholar]
- 7.de Keyzer, J., E. van der Sluis, R. Spelbrink, N. Nijstad, B. de Kruijff, N. Nouwen, C. van der Does, and A. Driessen. 2005. Covalently dimerized SecA is functional in protein translocation. J. Biol. Chem. 28035255-35260. [DOI] [PubMed] [Google Scholar]
- 8.Donachie, W., and A. Robinson. 1987. Cell division: parameter values and process, p. 1578-1593. In F. Neidhardt (ed.), Escherichia coli and Salmonella typhimurium: cellular and molecular biology. ASM Press, Washington, DC.
- 9.Driessen, A. 1993. SecA, the peripheral subunit of the Escherichia coli precursor protein translocase, is functional as a dimer. Biochemistry 3213190-13197. [DOI] [PubMed] [Google Scholar]
- 10.Driessen, A., and N. Nouwen. 2008. Protein translocation across the bacterial plasma membrane. Annu. Rev. Biochem. 77:2.1-2.25. [DOI] [PubMed] [Google Scholar]
- 11.Economou, A., and W. Wickner. 1994. SecA promotes preprotein translocation by undergoing ATP-driven cycles of membrane insertion and deinsertion. Cell 78835-843. [DOI] [PubMed] [Google Scholar]
- 12.Fekkes, P., C. van der Does, and A. Driessen. 1997. The molecular chaperone SecB is released from the carboxy-terminus of SecA during initiation of precursor protein translocation. EMBO J. 166105-6113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Gold, V., A. Robson, A. Clarke, and I. Collinson. 2007. Allosteric regulation of SecA: magnesium-mediated control of conformation and activity. J. Biol. Chem. 28217424-17432. [DOI] [PubMed] [Google Scholar]
- 14.Hartl, F.-U., S. Lecker, E. Schiebel, J. P. Hendrick, and W. Wickner. 1990. The binding cascade of SecB to SecA to SecY/E mediates preprotein targeting to the E. coli plasma membrane. Cell 63269-279. [DOI] [PubMed] [Google Scholar]
- 15.Hunt, J. F., S. Weinkauf, L. Henry, J. J. Fak, P. McNicholas, D. B. Oliver, and J. Deisenhofer. 2002. Nucleotide control of interdomain interactions in the conformational reaction cycle of SecA. Science 2972018-2026. [DOI] [PubMed] [Google Scholar]
- 16.Jilaveanu, L. B., and D. Oliver. 2006. SecA dimer cross-linked at its subunit interface is functional for protein translocation. J. Bacteriol. 188335-338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Jilaveanu, L. B., C. R. Zito, and D. Oliver. 2005. Dimeric SecA is essential for protein translocation. Proc. Natl. Acad. Sci. USA 1027511-7516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Karamanou, S., G. Sianidis, G. Gouridis, C. Pozidis, Y. Papanikolau, E. Papanikou, and A. Economou. 2005. Escherichia coli SecA truncated at its termini is functional and dimeric. FEBS Lett. 2791267-1271. [DOI] [PubMed] [Google Scholar]
- 19.Konopka, M., I. Shkel, S. Cayley, M. T. Record, and J. Weisshaar. 2006. Crowding and confinement effects on protein diffusion in vivo. J. Bacteriol. 1886115-6123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lill, R., K. Cunningham, L. A. Brundage, K. Ito, D. Oliver, and W. Wickner. 1989. SecA protein hydrolyzes ATP and is an essential component of the protein translocation ATPase of Escherichia coli. EMBO J. 8961-966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Matsuyama, S.-I., Y. Fujita, K. Sagara, and S. Mizushima. 1992. Overproduction, purification and characterization of SecD and SecF, integral membrane components of the protein translocation machinery of Escherichia coli. Biochim. Biophys. Acta 112277-84. [DOI] [PubMed] [Google Scholar]
- 22.McFarland, L., O. Francetic, and C. Kumamoto. 1993. A mutation of Escherichia coli SecA protein that partially compensates for the absence of SecB. J. Bacteriol. 1752255-2262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Or, E., D. Boyd, S. Gon, J. Beckwith, and T. A. Rapoport. 2005. The bacterial ATPase SecA functions as a monomer in protein translocation. J. Biol. Chem. 2809097-9105. [DOI] [PubMed] [Google Scholar]
- 24.Or, E., A. Navon, and T. Rapoport. 2002. Dissociation of the dimeric SecA ATPase during protein translocation across the bacterial membrane. EMBO J. 214470-4479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Or, E., and T. A. Rapoport. 2007. Cross-linked SecA dimers are not functional in protein translocation. FEBS Lett. 5812616-2620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Osborne, A. R., T. A. Rapoport, and B. van den Berg. 2005. Protein translocation by the Sec61/SecY channel. Annu. Rev. Cell Dev. Biol. 21529-550. [DOI] [PubMed] [Google Scholar]
- 27.Papanikou, E., S. Karamanou, and A. Economou. 2007. Bacterial protein secretion through the translocase nanomachine. Nat. Rev. Microbiol. 5839-851. [DOI] [PubMed] [Google Scholar]
- 28.Randall, L., J. Crane, A. Lilly, G. Liu, C. Mao, and S. Hardy. 2005. Asymmetric binding between SecA and SecB, two symmetric proteins: implications for function in export. J. Mol. Biol. 348479-489. [DOI] [PubMed] [Google Scholar]
- 29.Rusch, S., and D. Kendall. 2007. Oligomeric states of SecA and SecYEG core components of the bacterial Sec translocon. Biochim. Biophys. Acta 17685-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Shin, J. Y., M. Kim, and T. Ahn. 2006. Effects of signal peptide and adenylate on the oligomerization and membrane binding of soluble SecA. J. Biochem. Mol. Biol. 39319-328. [DOI] [PubMed] [Google Scholar]
- 31.Studier, W. F., A. H. Rosenberg, J. J. Dunn, and J. W. Dubendorff. 1990. Use of T7 RNA polymerase to direct expression of cloned genes. Methods Enzymol. 18560-89. [DOI] [PubMed] [Google Scholar]
- 32.Wang, H., B. Na, H. Yang, and P. C. Tai. 2008. Additional in vivo and in vitro evidence for SecA functioning as dimers in the membrane: dissociation into monomers is not essential for protein translocation in Escherichia coli. J. Bacteriol. 1901413-1418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Woodbury, R. L., S. Hardy, and L. Randall. 2002. Complex behavior in solution of homodimeric SecA. Protein Sci. 11875-882. [DOI] [PMC free article] [PubMed] [Google Scholar]