Abstract
Clostridium perfringens type B and D isolates produce epsilon-toxin, the third most potent clostridial toxin. The epsilon-toxin gene (etx) is plasmid borne in type D isolates, but etx genetics have been poorly studied in type B isolates. This study reports the first sequencing of any etx plasmid, i.e., pCP8533etx, from type B strain NCTC8533. This etx plasmid is 64.7 kb, carries tcp conjugative transfer genes, and encodes additional potential virulence factors including beta2-toxin, sortase, and collagen adhesin but not beta-toxin. Interestingly, nearly 80% of pCP8533etx open reading frames (ORFs) are also present on pCPF5603, an enterotoxin-encoding plasmid from type A isolate F5603. Pulsed-field gel electrophoresis and overlapping PCR indicated that a pCP8533etx-like etx plasmid is also present in most, if not all, other type B isolates and some beta2-toxin-positive, cpe-negative type D isolates, while other type D isolates carry different etx plasmids. Sequences upstream of the etx gene vary between type B isolates and some type D isolates that do not carry a pCP8533etx-like etx plasmid. However, nearly all type B and D isolates have an etx locus with an upstream IS1151, and those etx loci typically reside near a dcm ORF. These results suggest that pCPF5603 and pCP8533etx evolved from insertion of mobile genetic elements carrying enterotoxin or etx genes, respectively, onto a common progenitor plasmid.
Clostridium perfringens is an important pathogen of humans and livestock (17). Isolates of this gram-positive, anaerobic bacterium are classified into one of five types (A to E), based upon their production of four typing toxins (α-, β-, ɛ-, or ι-toxin). By definition, type B and D isolates must produce ɛ-toxin, which is the third most potent of all clostridial toxins and thus listed as a U.S. Department of Agriculture/CDC overlap class B select toxin. In addition to ɛ-toxin, type D isolates always produce alpha-toxin, while type B isolates consistently express both β-toxin and alpha-toxin. Besides their typing toxins, some type B or D isolates produce additional lethal toxins that are not used in the typing classification scheme. For example, some type B isolates produce perfringolysin O and/or beta2-toxin, while some type D isolates express perfringolysin O, beta2-toxin, and/or C. perfringens enterotoxin (8, 21, 22).
Beyond their biodefense significance, epsilon-toxin-producing C. perfringens type B and D isolates are also important natural pathogens of domestic animals (17). Type B isolates cause enterotoxemias in sheep, and possibly other livestock, particularly in the United Kingdom (25). Type B enterotoxemias initiate with proliferation of these bacteria in the gut. Toxins are then produced that initially act on the intestines but are later absorbed through the intestinal mucosa to act on internal organs. Similar enterotoxemias are caused by type D isolates in sheep and goats, where circulating toxins lead to elevated blood pressure, fluid accumulation in body cavities, and edema in brain, heart, lungs, liver, and kidney. Studies using a mouse intravenous injection model have strongly suggested that ɛ-toxin is a major contributor to the lethality associated with both type B and D enterotoxemias (8, 21, 22).
Recently, there has been increased research focus on the role of plasmids in C. perfringens virulence. It is now established that the genes encoding the enterotoxin (cpe), beta2-toxin (cpb2), ɛ-toxin (etx), iota-toxin (iap/ibp), and β-toxin (cpb) can each be carried by plasmids (5, 15, 16, 22). To date, only a few of those toxin plasmids have been studied in any detail; for example, among etx plasmids, only those in type D isolates have received even partial characterization (22). Southern blot analyses of pulsed-field gels showed that type D etx plasmids exhibit considerable heterogeneity, ranging in apparent size from ∼50 kb to ∼110 kb (22). Those Southern blot analyses also indicated that the etx plasmids of type D isolates vary considerably in their carriage of genes encoding additional toxins or other potential accessory virulence factors. In contrast, nearly all type D etx plasmids were shown (22) to carry the tcp locus that mediates conjugative transfer of C. perfringens tetracycline resistance plasmid pCW3 (20). Consistent with their carriage of a tcp locus, recent studies formally demonstrated conjugative transfer of the etx plasmid from two type D isolates (13).
Although still incompletely understood, organization of the etx locus itself also varies among type D isolates. Type D isolates carrying neither the cpe nor the cpb2 genes have been shown to carry an IS1151 sequence immediately upstream of their etx gene (GenBank accession number X60694) (7, 22). However, an upstream IS1151 sequence was not detected near the etx gene of those type D isolates carrying either the cpb2 or cpe genes (22). Sequences with homology to a mutator-type transposase were consistently identified immediately downstream of the etx gene in type D isolates (GenBank accession number X60694) (22), but it has remained unclear whether the entire mutator-type transposase open reading frame (ORF) is present in type D isolates, since the available sequence stops in the middle of that mutator-type transposase ORF. No information has been available for the region further downstream of the type D etx gene.
For type B isolates, etx genetics have been even less well examined. An older study (15) did show that the etx gene is plasmid borne in one type B isolate (CN3922); that study also suggested that the CN3922 etx plasmid is larger than the etx plasmid in type D isolate NCTC2062. Assuming that the etx gene is also plasmid borne in other type B isolates, the extent of etx plasmid diversity among type B isolates remains unknown, as is the relationship (if any) between the etx plasmids in type B isolates and the etx plasmids of type D isolates. In addition, organization of the etx locus of type B isolates is unclear, although unpublished Southern blotting results (15) have suggested that IS1151 sequences are present near the etx gene of a few surveyed type B isolates.
To help address the incomplete understanding of etx genetics, the current study examined the diversity among type B and D isolates with respect to their etx plasmids and their etx locus organization. Included in this work was the first complete sequencing of an etx plasmid. Findings from these studies facilitate understanding of the virulence, toxin plasmid diversity, and virulence evolution of C. perfringens.
MATERIALS AND METHODS
Bacterial strains, media, and reagents.
C. perfringens type B or D isolates described in Table 1 were grown overnight at 37°C in either FTG (fluid thioglycolate; Difco Laboratories, Michigan) or TGY (3% tryptic soy broth [Becton Dickinson and Company, Maryland], 2% glucose, 1% yeast extract [Difco], and 0.1% sodium thioglycolate [Sigma Chemical, Missouri]). All isolates were maintained as stock cultures in cooked meat medium (Oxoid, Basingstoke, England) and stored at −20°C.
TABLE 1.
List of C. perfringens isolates used in the study
| Strain | Type | Source and dated |
|---|---|---|
| Type B | ||
| NCTC8533 | etx (65), cpb2 (65), cpba | Lamb dysentery, UK, 1950s |
| Bar2 | etx (65), cpb2 (65), cpba | Sheep, Australia, 1980 |
| NCTC3110 | etx (65), cpb2 (65), cpba | NCTC, UK, unknown |
| CN677 | etx (65), cpb2 (65), cpba | Acute lamb dysentery, UK, 1942 |
| CN3447 | etx (65), cpb2 (65), cpba | Lamb dysentery, unknown, 1951 |
| CN1793 | etx (65), cpb2 (65), cpba | Toxigenic, UK, 1947 |
| CN2414 | etx (65), cpb2 (65), cpba | 3-day-old lamb, UK, 1948 |
| CN3425 | etx (65), cpb2 (65), cpba | Jejunum, 5-day-old lamb, UK, 1948 |
| JGS1984 | etx (65), cpb2 (65), cpba | Lamb, dysentery, unknown, 1996 |
| Type D | ||
| CN1020 | etx (48)b | Connaught Labs, Canada, 1944 |
| CN1675 | etx (NDc) | Ewe's udder, UK, 1946 |
| CN2062 | etx (48)b | Goat, fatal diarrhea, unknown, 1948 |
| CN3977 | etx (48)b | Lamb, carcass, UK, 1956 |
| CN3978 | etx (48)b | Lamb, carcass, UK, 1956 |
| CN3841 | etx (48)b | Ewe intestinal contents, unknown, 1955 |
| CN3793 | etx (48)b | NCTC8503, vaccine production strain, Kenya, 1955 |
| CN4031 | etx (65)b | Sheep carcass, UK, 1956 |
| CN1634 | etx (65)b | Lamb with suspected dysentery, UK, 1945 |
| CN2068 | etx (60), cpb2 (60)b,e | Lamb stomach, UK, 1948 |
| JGS1240 | etx (65), cpb2 (65)b,e | Sheep, bronchopneumonia and enterotoxemia, USA, 1992 |
| JGS1182 | etx (ND), cpb2 (ND)e | Sheep, sudden death and enterotoxemia, Canada, 1992 |
| CN1183 | etx (65), cpb2 (65), cpe (65)b | Lamb, UK, 1942 |
| CN3842 | etx (85), cpb2 (85), cpe (85)b | Ewe, small intestine, Spain, 1955 |
| CN4003 | etx (65), cpb2 (45), cpe (110)b | 3-day-old lamb, unknown, 1956 |
| CN3948 | etx (65), cpb2 (65), cpe (110)b | Sheep abomasums, Iran, 1956 |
| JGS1902 | etx (110), cpb2 (65), cpe (110)b | Sheep, enterotoxemia, USA, 1999 |
| JGS4138 | etx (110), cpb2 (65), cpe (110)b | Goat, sudden death, USA, 2002 |
| JGS4139 | etx (65), cpb2 (65), cpe (110)b | Goat, sudden death, USA, 2002 |
| JGS4152 | etx (110), cpb2 (65), cpe (110)b | Lamb, pulpy kidney, USA, 2002 |
The cpb plasmid size in type B or C isolates has not yet been evaluated.
Numbers in parentheses indicate the size of the plasmid (in kb) carrying the indicated toxin gene, as determined in a previous study (22), except the previous designations of some plasmids as 75 kb was changed to 65 kb to reflect their comigration with pCP8533etx, which was determined in this study to be 65 kb.
ND, not determined.
Abbreviations: UK, United Kingdom; USA, United States.
Carries type B etx plasmid (this study).
PFGE.
C. perfringens DNA plugs were prepared as described previously (16, 22). Briefly, overnight TGY cultures were washed, pelleted, and resuspended in 2% pulsed-field gel electrophoresis (PFGE)-certified agarose (Bio-Rad Laboratories, California), for a final agarose concentration of 1%. Those plugs were then electrophoresed in a CHEF-DR II PFGE system (Bio-Rad Laboratories), as previously described (16, 22).
In some experiments, DNA plugs from each of nine type B isolates were digested with the rare-cutting restriction enzyme ApaI, AvaI, or KpnI, according to manufacturer's instructions (New England Biolabs, Massachusetts). The digested DNA samples were then subjected to PFGE and stained with ethidium bromide, as described previously (2).
Southern blot analysis of pulsed-field gels.
Digoxigenin (DIG)-labeled probes were constructed using the PCR DIG probe synthesis kit (Roche, New Jersey). Primers used to PCR amplify internal etx, cpb2, rep, tcpH, and lam sequences for constructing these DIG-labeled probes were described previously (16, 22). Primers used for constructing DIG-labeled probes specific for cna, tcpF, dam, dcm, and the ABC transporter ORF are listed in Table S1 in the supplemental material. After hybridization of a probe, as described previously (22), the pulsed-field Southern blots were developed using reagents from the DIG DNA labeling and detection kit (Roche).
Southern blot analyses of conventional agarose gels.
C. perfringens DNA was isolated from type D strain CN1183 or from type B strains NCTC8533, NCTC3110, and Bar2, using the MasterPure gram-positive DNA purification kit (Epicentre, Wisconsin). Each isolated DNA sample was then digested overnight with ClaI, HindIII, or NcoI, according to the manufacturer's (New England Biolabs) instructions. The digested DNA samples were electrophoresed on a conventional 1% agarose gel, and the separated DNA digestion products were then transferred onto nylon membranes (Roche) for hybridization with an etx probe, as described above.
Sequencing of the NCTC8533 etx plasmid.
Ethidium bromide staining of pulsed-field gels demonstrated the presence of ∼90-kb, ∼80-kb, and ∼70-kb large plasmids in type B strain NCTC8533 (data not shown). Previous studies (16, 18, 22) have reported that different large plasmids in a single C. perfringens isolate can share some common regions, such as the tcp locus, which might complicate sequencing of the NCTC8533 etx plasmid. Therefore, two non-etx plasmids were cured from NCTC8533 prior to initiation of etx plasmid sequencing. This was accomplished by electroporating pJIR750ai (4, 23) into NCTC8533 and then selecting transformants on brain heart infusion agar plates containing 15 μg/ml of chloramphenicol, as described previously (4, 23). PCR demonstrated (data not shown) that 11 of 12 transformants obtained from this electroporation had lost their lambda-toxin (lam) gene, which was later localized by Southern blotting of pulsed-field gels to an ∼80-kb, non-etx large plasmid in NCTC8533 (data not shown), possibly indicating incompatibility between that NCTC8533 plasmid and pJIR750ai. From one of those transformants, pJIR750ai was cured by repeated subculture (4, 23), creating NCTC8533B4. This clone was then further subcultured until an ∼90-kb non-etx plasmid was spontaneously lost (data not shown) to create clone NCTC8533B4D. As confirmed by PFGE and etx PCR (data not shown), NCTC8533B4D still carried the ∼70-kb etx plasmid along with a non-lam large plasmid of ∼80 kb.
To sequence the etx plasmid in NCTC8533B4D, short-range PCRs were first performed using previously described (18) primers for internal sequences for each of the following ORFs found in pCPF5603: dcm, dam, cna, rep, cpb2, tcpF, and a putative ABC transporter ORF. After positive amplification results were obtained for each of those PCRs (data not shown), NCTC8533B4D DNA was subjected to PFGE and then sequentially hybridized with a DIG-labeled probe specific for each one of the above-listed pCPF5603 ORFs or with a DIG-labeled probe specific for etx. Those Southern blots localized (data not shown) each of the seven ORFs also found in pCP5603 to the 70-kb etx plasmid of NCTC8533B4D, while none of those seven ORFs hybridized to the ∼80-kb non-etx plasmid also present in this derivative. Therefore, selected primers for pCPF5603 sequences were used in long-range PCRs. Construction of these PCR primers was based upon previous pCPF5603 sequencing analyses (18), and the conditions used for the long- and short-range PCRs were as previously described (18). Each long-range PCR product amplified from NCTC8533 was then sequenced using either (i) primers described previously for pCPF5603 plasmid sequencing or (ii) new primers developed specifically for NCTC8533, as listed in Table S2 in the supplemental material.
Once a putative complete etx plasmid sequence for NCTC8533B4D became available, it was confirmed by long-range PCR approaches using primers corresponding to this obtained sequence, as listed in Table S1 in the supplemental material.
Completion of etx locus sequencing for cpe-negative, cpb2-negative type D isolate.
DNA was isolated from type D strain CN1675 by using the Master-Pure gram-positive DNA purification kit. Long-range PCRs were performed using the long-range PCR kit from Roche. Primers dcmF1 (5′-GGTAATAGTATCGTAATCCAGG-3′) and etx2R (5′-CATTTCATTAGATACTGTATTAG-3′) were used for these PCR amplifications. The PCRs were conducted in a Techne (Burkhardstdorf, Germany) thermocycler using the following conditions: 94°C for 2 min; 40 cycles of 92°C for 15 s, 55°C for 30 s, and 68°C for 8 min; and a single extension at 68°C for 15 min. The resultant ∼6-kb long-range PCR product was then cloned into Topo 2.1 (Invitrogen, California), and this insert was sequenced at the University of Pittsburgh Core Sequencing facility.
Overlapping PCR analyses to determine whether type B or D isolates carry a pCP8533etx-like etx plasmid.
For these short-range PCRs, template DNA was obtained, as described previously (26), from C. perfringens colony lysates. Each PCR mixture contained 2 μl of template DNA, 10 μl of TAQ Complete 2× mix (New England Biolabs), and 1 μl of each primer pair (1 μM final concentration). Overlapping primers for adjacent pCP8533etx ORFs, which were used in a PCR battery to investigate whether type B or D isolates carry a pCP8533etx-like etx plasmid, are described in Tables S3 and S4 in the supplemental material. The reaction mixtures, with a total volume of 20 μl, were placed in a thermocycler (Techne) and subjected to the following amplification conditions: one cycle of 95°C for 2 min; 35 cycles of 95°C for 30 s, 55°C for 40 s, and 68°C for 100 s; and a single extension at 68°C for 10 min. PCR products were then electrophoresed on a 1% agarose gel, which was stained with ethidium bromide for product visualization.
To compare the etx locus in type B isolates with that in type D isolates, PCRs were performed that used overlapping primers for adjacent ORFs present in either the etx locus of NCTC8533 or the etx locus of type D isolate CN1675 (see Table S5 in the supplemental material); these primers spanned from the second mutator-type transposase ORF located furthest downstream of the etx gene to the dcm ORF in each etx locus (see Results). The design of these primers was based upon sequencing results obtained for the etx locus of NCTC8533 or CN1675 (see above). Template DNA for these short-range overlapping PCRs was obtained from colony lysates (26). Each PCR mixture contained the same amount of reagents, and used the same PCR conditions, as described above for the pCP8533etx overlapping PCRs.
Nucleotide sequence accession numbers.
Results from these sequencing analyses, including the entire pCP8533etx sequence and the CN1675 etx locus sequence, are located in GenBank under accession numbers AB444205 and EU852100.
RESULTS
Pulsed-field and conventional agarose gel electrophoresis comparisons of etx plasmids in type B isolates with those in type D isolates.
By using electrophoresis conditions that allow plasmid DNA (but not chromosomal DNA) to enter a pulsed-field gel, previous etx Southern blotting studies had demonstrated considerable heterogeneity among etx plasmids from different type D isolates (22). Specifically, the etx plasmids of type D isolates were found to range in apparent size from ∼50 kb to ∼110 kb. Therefore, the current study first confirmed those previous reports of etx plasmid migration differences among type D isolates by using similar etx Southern blotting analyses of pulsed-field gels (Fig. 1A).
FIG. 1.
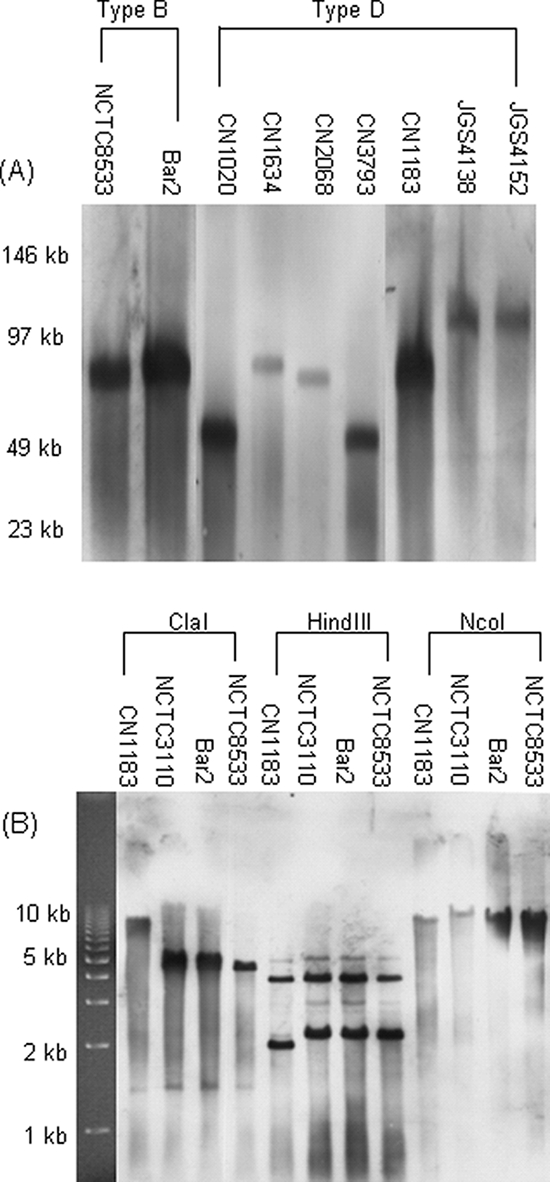
Southern blot analyses of type B and D isolates. (A) DNA from type B or D isolates was subjected to PFGE prior to Southern blotting and hybridization with a DIG-labeled, etx-specific probe. (B) DNA from type B or D isolates was digested with ClaI, HindIII, or NcoI prior to conventional agarose gel electrophoresis and Southern blotting with an etx-specific probe. Numbers at left of blots in panels A and B indicate migration of size markers in kb.
Since the diversity of etx plasmids among type B isolates had not yet been evaluated, similar etx Southern blotting of pulsed-field gels was performed using DNA from nine type B isolates (Fig. 1A shows representative results). For each of those type B isolates, the etx probe apparently hybridized to a single plasmid. However, in contrast to the etx plasmid migration differences noted among type D isolates in this (Fig. 1A) and previous (22) studies, the etx plasmids present in each of the nine examined type B isolates exhibited similar migration, with an apparent size of ∼70 kb (Fig. 1A and data not shown).
The similar migration of etx plasmids among all nine type B isolates surveyed suggested that these type B isolates might share a common etx plasmid. To further test this hypothesis, ClaI-, HindIII-, or NcoI-digested DNA from three randomly selected type B isolates was subjected to conventional agarose electrophoresis, followed by etx Southern blot analyses (Fig. 1B). For all three tested restriction enzymes, the digested DNA from the three type B isolates (NCTC3110, Bar2, and NCTC8533) showed the same pattern of etx probe hybridization, which is consistent with these three isolates carrying a similar etx plasmid. In contrast, the digested DNA from type D isolate CN1183 showed an etx probe hybridization pattern that was distinctly different from that of the three type B isolates surveyed.
Since the Fig. 1 Southern blotting results were consistent with carriage of a similar etx plasmid among the surveyed type B isolates, the clonality of these nine type B isolates was examined (Fig. 2). Ethidium bromide staining of pulsed-field gels run with KpnI-, AvaI-, or ApaI-digested DNA from these type B isolates showed, for all three restriction digests, band pattern differences that strongly suggested that the examined type B isolates are not all clonally related.
FIG. 2.
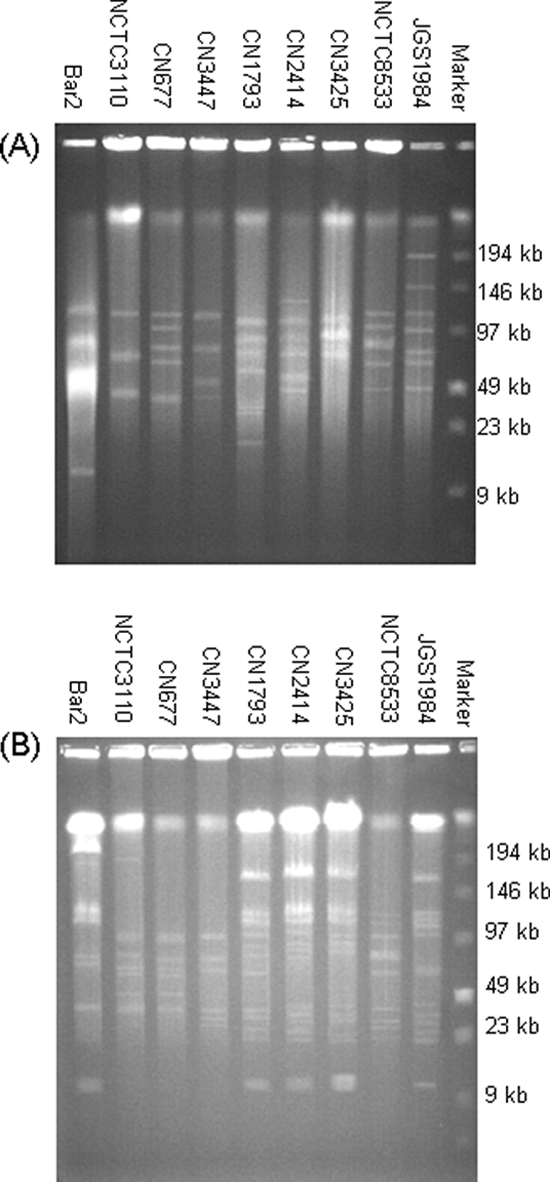
PFGE of type B isolates. Agarose plugs containing DNA from each specified isolate were digested with KpnI (A) or ApaI (B) and then subjected to PFGE and staining with ethidium bromide. Numbers at right of each blot indicate migration of size markers in kb.
Sequencing of the etx plasmid in type B isolate NCTC8533.
The availability of etx plasmid sequence information would facilitate a more definitive evaluation of etx plasmid diversity among type B and type D isolates. Since no etx plasmid sequence has yet been published, we sequenced the etx plasmid of the well-characterized type B isolate NCTC8533; this isolate was chosen because it produces ɛ-toxin (unpublished data) and served as the source strain for the original cloning and sequencing of the type B etx gene (14).
Ethidium bromide staining of a pulsed-field gel run with NCTC8533 DNA revealed (data not shown) that this isolate naturally carries large plasmids of ∼90 kb, ∼80 kb, and ∼70 kb. The presence of several other large plasmids might become problematic for closing the etx plasmid sequence, so NCTC8533 was first cured of its ∼90-kb plasmid and an ∼80-kb lam-carrying plasmid (see Materials and Methods). The resultant derivative, which carries the ∼70-kb etx plasmid and a second ∼80-kb non-etx plasmid, was named NCTC8533B4D and used for etx plasmid sequencing.
To date, sequences have been published for only two C. perfringens toxin-encoding plasmids (18), i.e., pCPF5603 (∼75.3 kb) and pCPF4969 (∼70.5 kb), which are both plasmids from type A isolates that carry the cpe gene and share an ∼35-kb conserved region carrying (among other ORFs) a tcp conjugative transfer locus. Therefore, individual PCRs were performed to test whether the etx plasmid retained by NCTC8533B4D might exhibit some similarity to the two sequenced cpe plasmids. This initial survey successfully amplified products for several pCPF5603 ORFs from the cured type B strain. Southern blotting of pulsed-field gels then demonstrated (data not shown) that probes specific for cpb2, rep, tcpF, tcpH, dcm, dam, and an ABC transporter ORF of pCPF5603 hybridized with the 70-kb etx plasmid, but not with the ∼80-kb plasmid, of the cured type B strain. Therefore, long-range PCR approaches using pCPF5603 primers were employed to completely sequence the etx plasmid of NCTC8533B4D (see Materials and Methods).
To confirm that the obtained sequence was correct, primers specific for the putative etx plasmid sequence were used to perform additional long-range PCRs with NCTC8533B4D DNA. Consistent with the putative NCTC8533B4D etx plasmid sequence, those long-range PCRs amplified (data not shown) an ∼5.5-kb product using a cnaR-repF primer pair, an ∼5.8-kb product using a repR-cpb2F primer pair, an ∼15.6-kb product using a cpb2R-ABC transporter ORFF primer pair, an ∼6.2-kb product using an ABC transporter ORFR-etxF primer pair, an ∼11.5-kb product using an etxR-dcmF primer pair, an ∼9.4-kb product using a dcmR-tcpFF primer pair, an ∼8.3-kb product using a tcpFR-damF primer pair and an ∼5.1-kb product using a damR-cnaF primer pair. Each of those products matched the expected product size from the etx plasmid sequence obtained for the cured NCTC8533 derivative.
Bioinformatic analysis of the sequence obtained for the NTCT8533B4D etx plasmid, which is being named pCP8533etx, showed that it consists of 64,753 bp, which is in good agreement with the ∼70-kb size estimated by the Fig. 1A PFGE-Southern blotting results. Using the NCBI ORF Finder (http://www.ncbi.nim.nih.gov/projects/gorf/), 63 ORFs were identified on pCP8533etx, for a coding density of 72.3%. Of those 63 ORFs, 49.2% were classified as encoding hypothetical proteins with unknown functions. The GC content of pCP8533etx was found to be 25.9%, which is similar to those of other sequenced C. perfringens plasmids (1, 18, 24) but slightly lower than the ∼28.4% GC content of the C. perfringens genome (19, 24).
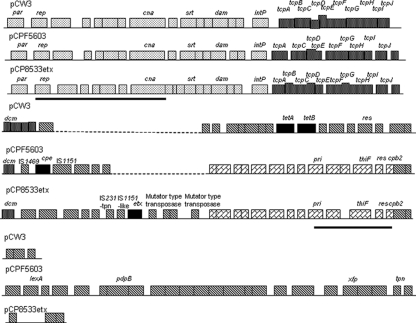
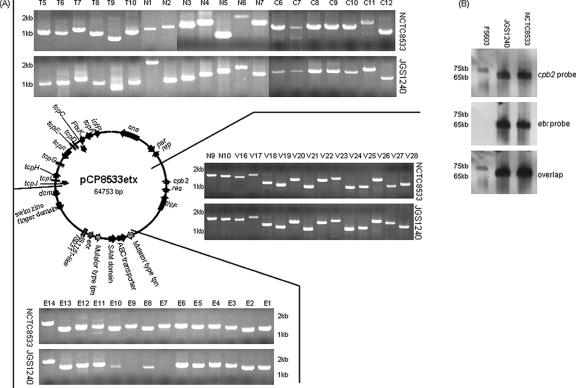
Sequence comparisons confirmed the PCR results suggesting considerable similarity between pCP8533etx and pCPF5603. Specifically, 79.4% of the pCP8533etx ORFs were also found on pCPF5603. Those shared ORFs common to both pCP8533etx and pCPF5603 account in total for ∼43.4 kb of sequence and include nearly the entire conserved region found in both pCPF5603 and pCPF4969, plus some additional variable-region sequence present only in pCPF5603. As shown in Fig. 3, notable ORFs common to pCP8533etx and pCPF5603 include the tcp conjugative transfer locus as well as ORFs encoding known or potential virulence factors such as beta2-toxin, sortase, and a potential collagen adhesin.
FIG. 3.
Comparative alignment of pCP8533etx versus two other sequenced C. perfringens plasmids, i.e., pCW3 and pCPF5603. Each box represents an ORF; ORF boxes filled with white dots represent the conserved tcp locus and dcm ORF shared by all three plasmids; ORF boxes filled with black dots represent other conserved ORFs shared by these three plasmids; ORF boxes filled with diagonal brickwork are shared by both pCP8533etx and pCPF5603; black ORF boxes are unique ORFs on each plasmid encoding either toxins or tetracycline resistance; ORF boxes filled with diagonal stripes are in regions unique to each plasmid. The two black bars depict regions found on pCP13. Sequences of pCPF5603, pCW3, and pCP13 have been reported previously (1, 18, 24).
These bioinformatic comparisons also revealed several major differences between pCPF5603 and pCP8533etx. First, the 5.6-kb cpe locus of pCPF5603 is missing from pCP8533etx, as expected since multiplex toxin PCR assays (11) had demonstrated (data not shown) that NCTC8533 does not carry the cpe gene. Second, the ∼16.8-kb pCP8533etx region that includes the etx gene is absent from pCPF5603 (further analysis below), which is consistent with pCPF5603 originating from a type A isolate that lacks the etx gene (18). Third, pCP8533etx contains an additional small ∼5-kb region that is not found on pCPF5603. Finally, pCP8533etx lacks a large (∼25.8-kb) portion of the pCPF5603 variable region that contains several putative metabolic ORFs usually associated with the C. perfringens chromosome. The presence of this ∼25.8-kb region, which is also missing from pCPF4969, is mainly responsible for the larger size of pCPF5603 (∼75.3 kb) than of pCP8533etx (∼64.7 kb).
The etx locus of pCP8533etx.
The Fig. 3 pCP8533etx sequencing results provided the first complete information regarding the organization of an etx locus in a type B or D isolate. Several sequences with homology to insertion sequence elements were identified both upstream and downstream of the etx gene in this type B isolate. The first of these is an IS1151-like sequence, disrupted by a premature stop codon, that is located immediately upstream of the etx gene on pCP8533etx. This pCP8533etx IS1151-like sequence lies in the opposite orientation from that reported for the IS1151 sequence near the etx gene of type D isolate NCTC2026 (GenBank accession number X60694) (7, 22). A second insertion sequence-related sequence upstream of the NCTC8533 etx gene exhibits strong similarity to an IS231-related transposase. Downstream of the etx gene in pCP8533etx are two identical copies, in opposite orientations, of an ORF with homology to a mutator-type transposase. These transposase ORFs, which might represent an integrated transposon, flank two ORFs potentially encoding an ABC transporter and a member of the radical SAM protein superfamily. The etx locus on pCP8533etx is proximal to a dcm gene, which is notable since previous studies (16, 18, 22) have identified the dcm region of plasmids as a possible hot spot for insertion of mobile genetic elements carrying C. perfringens toxin genes (see Discussion).
Overlapping PCR survey to evaluate the presence of a pCP8533etx-like plasmid among other type B isolates.
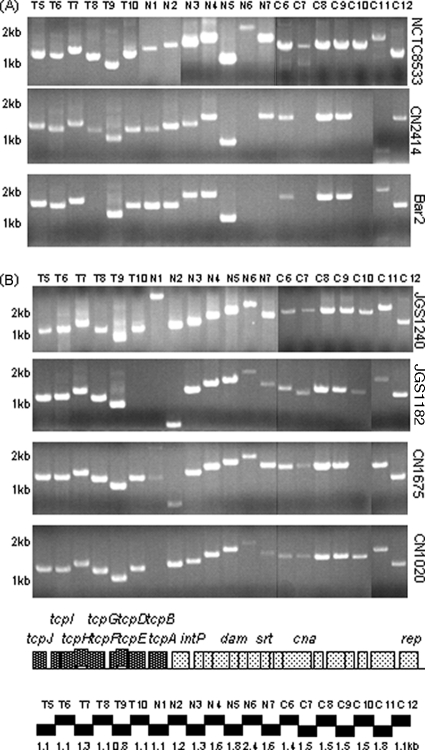
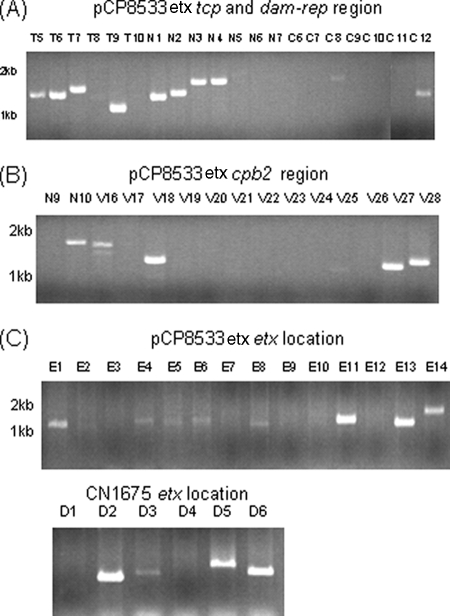
The Fig. 1 results suggested that a similar etx plasmid might be present among other type B isolates. In previous studies (10, 16, 18), overlapping PCR batteries using primers that overlap ORFs on sequenced cpe plasmids have proven to be invaluable tools for evaluating plasmid diversity among type A and E isolates. Therefore, the pCP8533etx sequencing results were used to develop a PCR battery with primers that overlap pCP8533etx ORFs in order to more definitively evaluate the diversity of etx plasmids among type B isolates. This pCP8533etx PCR battery amplified nearly all of the expected products from NCTC8533 and five other tested type B isolates (Fig. 4A and 5A and data not shown), strongly suggesting that these type B isolates all carry an etx plasmid similar to that of NCTC8533.
FIG. 4.
Overlapping PCR amplification of the pCP8533etx tcp transfer region (reactions T5 to T10 and N1 to N3) and dam-rep region (reactions N4 to N7 and C6 to C12) using the primer battery shown in Table S3 in the supplemental material. (A) Products of these reactions using DNA from three type B isolates: NCTC8533, CN2414, and Bar2. (B) Products of these reactions using DNA from four type D isolates: JGS1240, JGS1182, CN1675, and CN1020. Shown under panels A and B are maps depicting the relationship between ORFs and reactions in this overlapping PCR battery. Numbers at left of each gel indicate migration of size markers in kb.
FIG. 5.
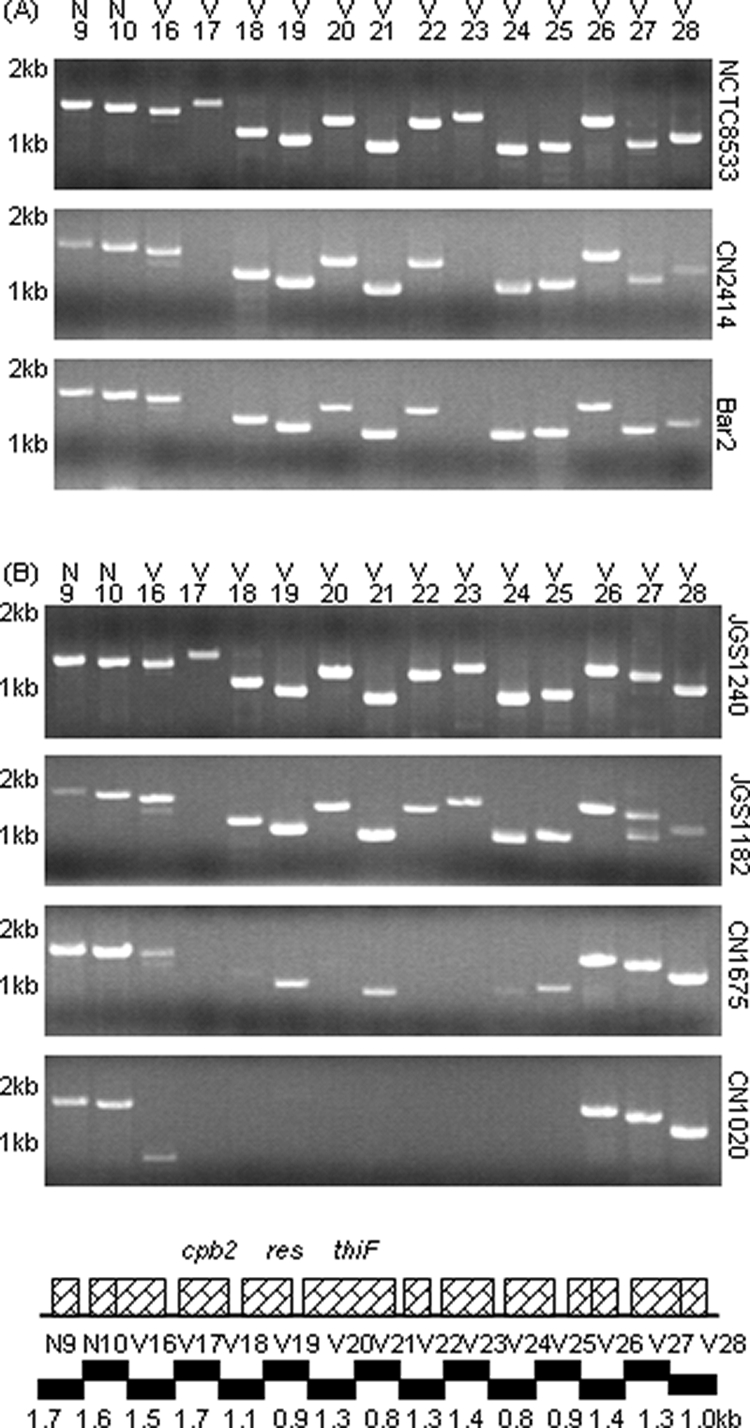
Overlapping PCR amplification of the pCP8533etx cpb2 region of pCP8533etx using the primer battery shown in Table S4 in the supplemental material. (A) Products of these reactions using DNA from three type B isolates: NCTC8533, CN2414, and Bar2. (B) Products of these reactions using DNA from four type D isolates: JGS1240, JGS1182, CN1675, and CN1020. Numbers at left of each gel indicate migration of size markers in kb. Shown under panels A and B are maps depicting the relationship between ORFs and reactions in this overlapping PCR battery.
Sequencing analysis of the etx locus in type D isolate CN1675.
We next wanted to compare the organization of the etx locus in type B isolates with that in type D isolates, but only limited sequence data were available for a type D etx locus prior to our study (12, 22). That limited previous sequencing had revealed the presence, downstream of the etx gene in type D isolate NCTC2026, of sequences homologous to an ORF encoding a mutator-type transposase (although the available sequence stops in the middle of this ORF). PCR approaches later showed that those same mutator-type transposase sequences are also present downstream of the etx gene in many other type D isolates. Previous overlapping PCR analyses based upon the partial NCTC2026 etx locus sequencing had identified heterogeneity in the region upstream of the etx gene in type D isolates, but the nature and extent of this diversity have remained unknown (22). Specifically, type D isolates carrying neither the cpe nor the cpb2 gene were found to possess, upstream of their etx gene, an IS1151 sequence (in the same orientation as that of the etx ORF) and an ORF homologous to a Tn3-family transposase. However, overlapping PCR based upon the NCTC2026 etx locus organization did not amplify products using DNA from type D isolates carrying either the cpb2 or the cpe gene. Finally, this earlier work had also not resolved whether the etx gene in some or all type D isolates might reside near a dcm gene, as is true for many other plasmid-borne C. perfringens toxins (16, 18, 22).
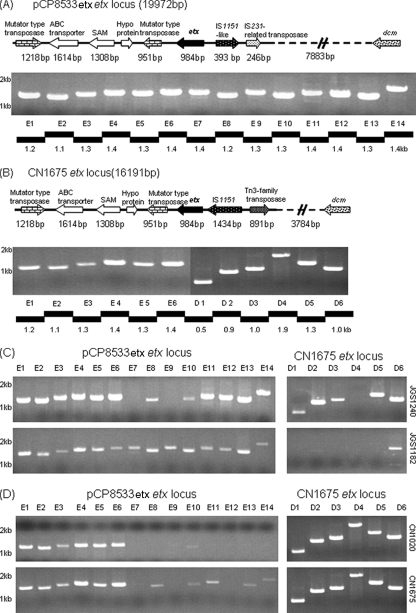
Therefore, the current study first completed sequencing of the etx locus in a type D isolate. Since our isolate collection did not include NCTC2026, another cpb2-negative, cpe-negative type D isolate (CN1675) was chosen that, according to overlapping PCR results (data not shown), possesses an etx locus very similar to that reported for NCTC2026. Sequencing of long-range PCR products (Fig. 6B), obtained using primers for sequences in the etx locus of pCP8533etx, revealed that CN1675 and type B isolate NCTC8533 share identical sequences immediately downstream of their etx genes, including the presence of the two oppositely oriented, mutator-type transposase ORFs that possibly represent an integrated transposon. Sequencing of this long-range PCR product also identified a dcm ORF ∼6 kb upstream of the CN1675 etx gene.
FIG. 6.
Characterization of the etx locus in type B and D isolates. Overlapping PCR batteries were developed based upon sequencing results for the etx locus in pCP8533etx or cpb2-negative type D isolate CN1675 and applied to nine type B isolates and 18 type D isolates. (A and B) Results for NCTC8533 DNA using the pCP8533etx battery (A) (results for other type B isolates were identical; data not shown) and CN1675 DNA using the CN1675 battery (B). (C) Results using the pCP8533etx etx locus battery (left) or CN1675 etx locus battery (right) for two type D isolates carrying an etx plasmid found in the experiments shown in Fig. 4 and 5 to strongly resemble pCP8533etx (Fig. 8); these results indicate that JGS1240 has a hybrid etx locus with aspects of both the pCP8533 and CN1675 etx locus and that JGS1182 has an etx locus like pCP8533etx. (D) Results using the pCP8533etx etx locus battery (left) or CN1675 etx locus battery (right) for two cpb2-negative, cpe-negative type D isolates (CN1020 and CN1675) that carry an etx plasmid lacking the cpb2 region of pCP8533etx; these results indicate that these two type D isolates carry an etx locus indistinguishable from that of CN1675. Numbers at left of each gel indicate migration of size markers in kb.
Comparison of etx locus organization in type B isolates with that in type D isolates using overlapping PCR analyses.
With the availability of complete sequence for the CN1675 type D etx locus (GenBank accession number EU852100), a second overlapping PCR battery, specific for this variant etx locus organization, was developed. That new CN1675 etx locus overlapping PCR battery and the portion of the overlapping PCR battery covering the etx locus of pCP8533etx were then applied to a collection of type D isolates.
DNA from 7 of 18 surveyed type D isolates amplified a product pattern indicative of a CN1675-like etx locus. Those seven type D isolates with a CN1675-like etx locus were each negative for carriage of either the cpb2 or the cpe gene. Interestingly, DNA from 10 of the 18 surveyed type D isolates that failed to amplify products with the CN1675 etx locus PCR battery was able to support amplification of nearly all the expected products using the pCP8533etx etx locus battery (Fig. 6 and data not shown). Those type D isolates with a type B-like etx locus were each found to carry the cpb2 gene, cpe gene, or both the cpb2 and cpe genes, although on a variety of different-sized toxin plasmids (22; also data not shown). Interestingly, DNA from one cpe-positive, cpb2-positive type D isolate (JGS4139) did not consistently support amplification of either upstream or downstream products using the overlapping PCR battery for the etx locus of either NCTC8533 or CN1675 (Fig. 7), suggesting the existence of a third, relatively uncommon, variant etx locus organization.
FIG. 7.
Overlapping PCR amplification of products using DNA from type D isolate JGS4139. Shown are results using pCP8533etx batteries for the tcp and dam-rep region (A), the cpb2 region (B), and the etx locus (C, top). Also shown in panel C (bottom) are results using the overlapping PCR battery for the CN1675 etx locus. Numbers at left of each gel indicate migration of size markers in kb.
Detection of a pCP8533etx-like plasmid among some type D isolates.
DNA from the same 18 type D isolates was also subjected to portions of the pCP8533etx overlapping PCR battery covering sequences beyond the etx locus. DNA from 17 of these 18 isolates supported amplification of products for reactions covering the tcp locus and the dam-to-rep ORF region also found in several other large C. perfringens plasmids (24). JGS4139 DNA failed to support amplification of most products from either the tcp locus or the dam-rep ORF region (Fig. 7).
However, the ability of DNA from the surveyed type D isolates to support amplification of products using primers to the cpb2 region of pCP8533etx was more variable. DNA from JGS4139 and 14 other type D isolates failed to support amplification of products in this region. In contrast, DNA from three type D isolates (JGS1240, CN2068, and JGS1182) supported amplification of pCP8533etx PCR products from the cpb2 region, as well as all other regions of pCP8533etx (Fig. 4B and 8A and data not shown), strongly suggesting that these three isolates carry a pCP8533etx-like etx plasmid.
FIG. 8.
Some type D isolates carry a plasmid nearly identical to pCP8533etx. (A) Comparative overlapping PCR assay results for DNA from type D JGS1240 versus DNA from type B isolate NCTC8533. (B) Southern blotting results of pulsed-field gels electrophoresed with DNA from NCTC8533 (type B), JGS1240 (type D), and F5603 (type A) and then hybridized with etx probe. That same blot was then stripped and hybridized with a cpb2 probe. An overlay of the cpb2 and etx blots is also shown (note comigration of cpb2 and etx for JGS1240 and NCTC8533 and the slightly larger cpb2 plasmid pCPF5603 in F5603). Numbers at left or right of each blot indicate migration of size markers in kb.
To further confirm similarity between pCP8533etx and the etx plasmid in one of these type D isolates, i.e., JGS1240, PFGE and etx Southern blotting were performed. These analyses demonstrated identical migration patterns for the etx plasmids of NCTC8533 and JGS1240. When this blot was stripped and rehybridized with a cpb2 probe, the cpb2 signal comigrated with the etx signal for both NCTC8533 and JGS1240. These analyses also demonstrated that the cpb2- and etx-carrying plasmids of NCTC8533 and JGS1240 are smaller than the 75-kb cpb2- and cpe-carrying plasmid of type A isolate F5603, consistent with pCP8533etx and pCPF5603 sequencing results (Fig. 8B).
Type D isolate CN2068 also amplified nearly all the products in the pCP8533etx overlap PCR assays. However, the etx plasmid in CN2068 appears slightly smaller on pulsed-field gels than pCP8533etx (Fig. 1), possibly due to loss of a few ORFs in the dam-rep region, as suggested by overlapping PCR results (data not shown). A cpb2 probe cohybridized with this etx plasmid when CN2068 DNA was analyzed by Southern blotting of pulsed-field gels (22; also data not shown).
Similar etx and cpb2 Southern blot analyses of pulsed-field gels were attempted with the third type D isolate, i.e., cpb2-positive isolate JGS1182, apparently carrying a pCP8533etx-like plasmid. However, those efforts for JGS1182 were unsuccessful due to its high endonuclease levels causing smearing on the pulsed-field gel, a common problem among C. perfringens isolates (5, 6).
DISCUSSION
Despite an increasing appreciation for their importance in virulence, the toxin plasmids of C. perfringens have only recently come under intensive study. In particular, significant knowledge gaps currently exist for the toxin plasmids of type B to E isolates, where plasmid-encoded toxins are critically important for pathogenesis (9, 16, 21, 22). For example, the only C. perfringens toxin plasmid sequences published (18) prior to the current study were those for pCPF5603 and pCPF4969, prototypes for the two enterotoxin-encoding plasmid families that predominate among the C. perfringens type A isolates causing human non-food-borne gastrointestinal diseases (such as antibiotic-associated diarrhea). The lack of type B to E toxin plasmid sequence information has limited insights into important issues such as the carriage of potential accessory virulence ORFs among these toxin plasmids, as well as the diversity and evolution of C. perfringens toxin plasmids.
Complete sequencing of C. perfringens toxin plasmids has been difficult because these toxin plasmids are present at only a few copies/genome in C. perfringens cells often containing several other large plasmids (16, 18, 22). By curing a type B isolate of two other large plasmids, the current work overcame this problem to completely sequence the first toxin plasmid of a type B to E isolate, i.e., the etx plasmid (pCP8533etx) from a type B isolate NCTC8533 derivative. The availability of pCP8533etx sequence has provided several new insights into C. perfringens virulence genetics. First, the pCP8533etx sequence revealed that NCTC8533 carries its cpb gene (encoding β-toxin, another important lethal toxin of type B isolates) on a different plasmid than its etx plasmid; studies are now under way to investigate the still poorly studied cpb plasmids of type B and C isolates. Sequencing also demonstrated that, in addition to the etx gene, pCP8533etx carries several additional ORFs potentially encoding factors (e.g., beta2-toxin, sortase, and an adhesin) that could contribute to virulence. Another potentially important finding for understanding C. perfringens virulence is the presence of a tcp locus on pCP8533etx. Since a nearly identical tcp region mediates transfer of C. perfringens plasmid pCW3 (20), the presence of a tcp locus strongly predicts that pCP8533etx, like the etx plasmids in two type D isolates (13), should be conjugative (further discussion below).
The pCP8533etx sequence was used to develop a powerful overlapping PCR battery for surveying etx plasmid diversity among a number of type B isolates predominantly originating from diseased sheep or lambs in the United Kingdom, which is currently considered (25) the primary epidemiologic setting for type B disease (future studies might examine type B isolates from additional sources). Interestingly, results of this PCR survey strongly suggested that an etx plasmid nearly identical to pCP8533etx is present in many, if not all, other type B isolates. These PCR results were further supported by (i) etx Southern blot analyses of pulsed-field gels showing that a similar-size etx plasmid is present in nine different type B isolates and (ii) conventional etx Southern blotting results supporting the sequence similarity of etx plasmids among three type B isolates. A possible explanation for the presence of a nearly identical etx plasmid in many nonclonal type B isolates could involve conjugative transfer of this plasmid via the tcp locus.
The overlapping pCP8533etx PCR battery also allowed assessment of etx plasmid diversity among type D isolates, which revealed that many type D isolates also carry both the tcp locus and the dam-rep region of pCP8533etx. However, only 3 of 18 surveyed type D isolates were found to carry an etx plasmid nearly identical to pCP8533etx. The presence of cpb2 sequences on the etx plasmid in those three cpb2-positive type D isolates is consistent with current and previous etx Southern blotting results (22) for two of those three isolates, which showed comigration of the etx and cpb2 genes on pulsed-field gels (the third type D isolate could not be analyzed by pulsed-field gels due to high nuclease levels). Previous pulsed-field analyses (22) had also suggested that one of these three type D isolates (JGS1240) might be cpe positive, but multiplex analyses conducted during the current study determined that (like most or all type B isolates) the three type D isolates carrying a pCP8533etx-like plasmid are each cpe negative (data not shown). Additional support for the presence of a pCP8533etx-like plasmid in one of these three type D isolates (JGS1240) was provided by etx Southern blot analyses of pulsed-field gels, which demonstrated comigration between the JGS1240 etx plasmid and pCP8533etx. However, those Southern blots also indicated that the etx plasmid in CN2068, another type D isolate with an pCPF8533etx-like plasmid, is slightly smaller (Fig. 1A); overlapping PCR results (data not shown) suggest that this limited size difference may reflect loss of some ORFs from the dam-rep region of the CN2068 etx plasmid. A few other small differences were also observed between the JGS1240 and CN2068 etx plasmids and pCP8533etx; most notably, the IS1151 sequences located in the etx locus of JGS1240 and CN2068 are oriented oppositely from those in the etx locus of pCP8533etx (Fig. 6C and 8A).
While all of the surveyed cpb2-positive, cpe-negative type D isolates were found to carry an pCP8533etx-like plasmid, none of the cpe-positive type D isolates or cpb2-negative, cpe-negative type D isolates examined in this study possessed an etx plasmid closely resembling pCP8533etx. The absence of pCP8533etx-like plasmids from type D isolates carrying a cpe plasmid raises the possibility that certain toxin plasmid combinations may be incompatible within a single C. perfringens type D cell. However, our results also indicate that many, or all, type B isolates maintain a second toxin plasmid carrying a cpb gene, implying that pCP8533etx must be compatible with at least some cpb plasmids. The sharing of a similar CN1675-like etx locus by all surveyed cpb2-negative, cpe-negative type D isolates might suggest that their etx plasmid (usually ∼50 kb in size) is incompatible with other toxin plasmids. Plasmid compatibility has not yet been examined in C. perfringens, but the current findings suggest that it may be a subject deserving of future study.
As mentioned above, the pCP8533etx-like etx plasmids differ from the etx plasmids of cpb2-negative, cpe-negative type D isolates with respect to their etx locus organization. For example, the IS1151 sequences in the etx locus of pCP8533etx-like plasmids are oriented oppositely from those in the etx locus of cpb2-negative, cpe-negative type D isolates. This opposite orientation of IS1151 sequences in isolates with a pCP8533etx-like etx locus explains why our previous overlapping PCR survey using primers for the etx locus of NCTC2026 had failed to detect IS1151 sequences in the etx locus of some type D isolates (22). More importantly, the current results establish that IS1151 sequences are present immediately upstream of most, if not all, etx genes in both type B and type D isolates. This finding adds further support to the emerging view that IS1151 sequences may have mobilized many C. perfringens toxin genes, including the cpe gene (18), the iota-toxin genes (2, 16), and the etx genes (this study and references 7 and 22). Possible IS1151-mediated mobilization of C. perfringens toxin genes is further supported by studies detecting possible circular transposition intermediates containing IS1151 and various toxin genes, including the etx gene of cpb2-negative, cpe-negative type D isolates (3, 16, 22). Notably, the current results also identified a dcm ORF near most or all etx genes, which is significant since the dcm region of C. perfringens plasmids may represent a hot spot for integration of mobile genetic elements carrying toxin genes (18), as well as some antibiotic resistance genes (e.g., a dcm ORF is located near the pCW3 tet resistance genes in Fig. 3).
The current results identify significant similarity between pCP8533etx and the cpe plasmids of type A isolates, particularly the pCPF5603 family of cpe plasmids. Previous studies (16) had found homology between pCPF5603 and some iota-toxin plasmids of type E isolates, but it was suggested (1, 2, 16) that those pCPF5603-like type E iota-toxin plasmids arose merely from integration, near the pCPF5603 cpe gene of a type A isolate, of a mobile genetic element carrying the iota-toxin genes. Therefore, the current finding of similarity between pCPF5603 and the pCP8533etx-like etx plasmids, which lack cpe genes, significantly extends the virulence importance of this toxin plasmid family. Since pCPF5603 was sequenced before pCP8533etx was, we propose naming this toxin plasmid family the pCPF5603-like toxin plasmids. It is also notable that some C. perfringens antibiotic resistance plasmids, i.e., pCW3 (Fig. 3), also share relatively extensive similarity with the pCPF5603-like toxin plasmids (1). However, the only other sequenced large C. perfringens plasmid, i.e., pCP13, displays only limited similarity to the pCPF5603-like plasmids (24), indicating that sequence diversity exists among large C. perfringens plasmids.
The new and previous findings suggest a possible evolutionary scenario for the pCPF5603-like toxin plasmid family. Initially, a Tn916 transposon carrying the tcp locus may have integrated onto a plasmid bearing the dam-rep region present in many large C. perfringens plasmids (18). The resultant plasmid, now conjugative, could then have acquired a variety of additional sequences. Sometimes these acquired sequences may have encoded antibiotic resistance, giving rise to plasmids like pCW3 (1). Other plasmids, like pCPF4969 (18), could have acquired sequences encoding bacteriocins and an IS1470-like mobile genetic element carrying a cpe gene. A pCPF5603 family progenitor plasmid may have arisen when the acquired sequences included a cpb2 locus (18), some chromosomal genes, and an IS1151 element carrying the cpe gene. Later, an IS1151 element carrying the iota-toxin genes could have inserted near the cpe gene of a pCPF5603 plasmid, creating the pCPF5603-like iota-toxin plasmids currently found in some type E isolates (2, 16). A final evolutionary event may have involved the insertion (near dcm) of an IS1151-based mobile element carrying the etx gene onto the pCPF5603 family progenitor plasmid, creating the pCP8533etx plasmid family that is now present in most/all type B isolates and some type D isolates. Isolates carrying pCP8533etx may then have given rise to type B isolates, possibly by transfer of this etx plasmid via tcp-mediated conjugation to a type C isolate carrying a cpb plasmid.
The pCPF5603-like toxin plasmid family is now emerging as a major contributor to the virulence of some C. perfringens type A, B, D, and E isolates. However, it is also clear that pCP8533etx-like plasmids are not the only etx plasmids carried by C. perfringens type D isolates. Therefore, additional studies should sequence and evaluate the diversity of the type D toxin etx plasmids not closely resembling pCP8533etx. In addition, future studies should elucidate whether the cpb toxin gene is also carried by pCPF5603-related plasmids in type B and C isolates.
Supplementary Material
Acknowledgments
We thank Glenn Songer for providing some isolates used in these studies.
This research was generously supported by grants R01 AI054177-05 and R37 AI19844-25 from the National Institute of Allergy and Infectious Diseases.
Footnotes
Published ahead of print on 5 September 2008.
Supplemental material for this article may be found at http://jb.asm.org/.
REFERENCES
- 1.Bannam, T. L., W. L. Teng, D. Bulach, D. Lyras, and J. I. Rood. 2006. Functional identification of conjugation and replication regions of the tetracycline resistance plasmid pCW3 from Clostridium perfringens. J. Bacteriol. 1884942-4951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Billington, S. J., E. U. Wieckowski, M. R. Sarker, D. Bueschel, J. G. Songer, and B. A. McClane. 1998. Clostridium perfringens type E animal enteritis isolates with highly conserved, silent enterotoxin sequences. Infect. Immun. 664531-4536. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 3.Brynestad, S., B. Synstad, and P. E. Granum. 1997. The Clostridium perfringens enterotoxin gene is on a transposable element in type A human food poisoning strains. Microbiology 1432109-2115. [DOI] [PubMed] [Google Scholar]
- 4.Chen, Y., B. A. McClane, D. J. Fisher, J. I. Rood, and P. Gupta. 2005. Construction of an alpha-toxin gene knockout mutant of Clostridium perfringens type A by use of a mobile group II intron. Appl. Environ. Microbiol. 717542-7547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Collie, R. E., and B. A. McClane. 1998. Evidence that the enterotoxin gene can be episomal in Clostridium perfringens isolates associated with nonfoodborne human gastrointestinal diseases. J. Clin. Microbiol. 3630-36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Cornillot, E., B. Saint-Joanis, G. Daube, S. Katayama, P. E. Granum, B. Carnard, and S. T. Cole. 1995. The enterotoxin gene (cpe) of Clostridium perfringens can be chromosomal or plasmid-borne. Mol. Microbiol. 15639-647. [DOI] [PubMed] [Google Scholar]
- 7.Daube, G., P. Simon, and A. Kaeckenbeeck. 1993. IS1151, an IS-like element of Clostridium perfringens. Nucleic Acids Res. 21352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Fernandez-Miyakawa, M. E., D. J. Fisher, R. Poon, S. Sayeed, V. Adams, J. I. Rood, B. A. McClane, and F. A. Uzal. 2007. Both epsilon-toxin and beta-toxin are important for the lethal properties of Clostridium perfringens type B isolates in the mouse intravenous injection model. Infect. Immun. 751443-1452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Fisher, D. J., M. E. Fernandez-Miyakawa, S. Sayeed, R. Poon, V. Adams, J. I. Rood, F. A. Uzal, and B. A. McClane. 2006. Dissecting the contributions of Clostridium perfringens type C toxins to lethality in the mouse intravenous injection model. Infect. Immun. 745200-5210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Fisher, D. J., K. Miyamoto, B. Harrison, S. Akimoto, M. R. Sarker, and B. A. McClane. 2005. Association of beta2 toxin production with Clostridium perfringens type A human gastrointestinal disease isolates carrying a plasmid enterotoxin gene. Mol. Microbiol. 56747-762. [DOI] [PubMed] [Google Scholar]
- 11.Garmory, H. S., N. Chanter, N. P. French, D. Busechel, J. G. Songer, and R. W. Titball. 2000. Occurrence of Clostridium perfringens β2-toxin amongst animals, determined using genotyping and subtyping PCR assays. Epidemiol. Infect. 12461-67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Havard, H. L., S. E. Hunter, and R. W. Titball. 1992. Comparison of the nucleotide sequence and development of a PCR test for the epsilon toxin gene of Clostridium perfringens type B and type D. FEMS Microbiol. Lett. 7677-81. [DOI] [PubMed] [Google Scholar]
- 13.Hughes, M. L., R. Poon, V. Adams, S. Sayeed, J. Saputo, F. A. Uzal, B. A. McClane, and J. I. Rood. 2007. Epsilon-toxin plasmids of Clostridium perfringens type D are conjugative. J. Bacteriol. 1897531-7538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hunter, S. E. C., I. N. Clarke, D. C. Kelley, and R. W. Titball. 1992. Cloning and nucleotide sequencing of the Clostridium perfringens epsilon-toxin gene and its expression in Escherichia coli. Infect. Immun. 60102-110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Katayama, S. I., B. Dupuy, G. Daube, B. China, and S. T. Cole. 1996. Genome mapping of Clostridium perfringens strains with I-Ceu I shows many virulence genes to be plasmid-borne. Mol. Gen. Genet. 251720-726. [DOI] [PubMed] [Google Scholar]
- 16.Li, J., K. Miyamoto, and B. A. McClane. 2007. Comparison of virulence plasmids among Clostridium perfringens type E isolates. Infect. Immun. 751811-1819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.McClane, B. A., F. A. Uzal, M. F. Miyakawa, D. Lyerly, and T. Wilkins. 2006. The enterotoxic clostridia, p. 698-752. In M. Dworkin, S. Falkow, E. Rosenburg, K. H. Schleifer, and E. Stackebrandt (ed.) The prokaryotes, 3rd ed. Springer-Verlag, New York, NY.
- 18.Miyamoto, K., D. J. Fisher, J. Li, S. Sayeed, S. Akimoto, and B. A. McClane. 2006. Complete sequencing and diversity analysis of the enterotoxin-encoding plasmids in Clostridium perfringens type A non-food-borne human gastrointestinal disease isolates. J. Bacteriol. 1881585-1598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Myers, G. S., D. A. Rasko, J. K. Cheung, J. Ravel, R. Seshadri, R. T. DeBoy, Q. Ren, J. Varga, M. M. Awad, L. M. Brinkac, S. C. Daugherty, D. H. Haft, R. J. Dodson, R. Madupu, W. C. Nelson, M. J. Rosovitz, S. A. Sullivan, H. Khouri, G. I. Dimitrov, K. L. Watkins, S. Mulligan, J. Benton, D. Radune, D. J. Fisher, H. S. Atkins, T. Hiscox, B. H. Jost, S. J. Billington, J. G. Songer, B. A. McClane, R. W. Titball, J. I. Rood, S. B. Melville, and I. T. Paulsen. 2006. Skewed genomic variability in strains of the toxigenic bacterial pathogen, Clostridium perfringens. Genome Res. 161031-1040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Parsons, J. A., T. L. Bannam, R. J. Devenish, and J. I. Rood. 2007. TcpA, an FtsK/SpoIIIE homolog, is essential for transfer of the conjugative plasmid pCW3 in Clostridium perfringens. J. Bacteriol. 1897782-7790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Sayeed, S., M. E. Fernandez-Miyakawa, D. J. Fisher, V. Adams, R. Poon, and J. I. Rood. 2005. Epsilon-toxin is required for most Clostridium perfringens type D vegetative culture supernatants to cause lethality in the mouse intravenous injection model. Infect. Immun. 737413-7421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Sayeed, S., J. Li, and B. A. McClane. 2007. Virulence plasmid diversity in Clostridium perfringens type D isolates. Infect. Immun. 752391-2398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Sayeed, S., F. A. Uzal, D. J. Fisher, J. Saputo, J. E. Vidal, Y. Chen, P. Gupta, J. I. Rood, and B. A. McClane. 2008. Beta toxin is essential for the intestinal virulence of Clostridium perfringens type C disease isolate CN3685 in a rabbit ileal loop model. Mol. Microbiol. 6715-30. [DOI] [PubMed] [Google Scholar]
- 24.Shimizu, T., K. Ohtani, H. Hirakawa, K. Ohshima, A. Yamashita, T. Shiba, N. Ogasawara, M. Hattori, S. Kuhara, and H. Hayashi. 2002. Complete genome sequence of Clostridium perfringens, an anaerobic flesh-eater. Proc. Natl. Acad. Sci. USA 99996-1001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Songer, J. G. 1996. Clostridial enteric diseases of domestic animals. Clin. Microbiol. Rev. 9216-234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Wen, Q., and B. A. McClane. 2004. Detection of enterotoxigenic Clostridium perfringens type A isolates in American retail foods. Appl. Environ. Microbiol. 702685-2691. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.