Abstract
Cytoplasmic protein reduction via generalized thiol/disulfide exchange reactions and maintenance of cellular redox homeostasis is mediated by the thioredoxin superfamily of proteins. Here, we describe the characterization of the thioredoxin system from Mycobacterium tuberculosis, whose genome bears the potential to encode three putative thioredoxins from the open reading frames designated trxAMtb, trxBMtb, and trxCMtb. We show that all three thioredoxins, overproduced in Escherichia coli, are able to reduce insulin, a model substrate, in the presence of dithiothreitol. However, we observe that thioredoxin reductase is not capable of reducing TrxAMtb in an NADPH-dependent manner, indicating that only TrxBMtb and TrxCMtb are the biologically active disulfide reductases. The absence of detectable mRNA transcripts of trxAMtb observed when M. tuberculosis strain H37Rv was cultivated under different growth conditions suggests that trxAMtb expression may be cryptic. The measured redox potentials of TrxBMtb and TrxCMtb (−262 ± 2 mV and −269 ± 2 mV, respectively) render these proteins somewhat more oxidizing than E. coli thioredoxin 1 (TrxA). In E. coli strains lacking components of cytoplasmic protein reduction pathways, heterologous expression of the mycobacterial thioredoxins was able to effectively substitute for their function.
The cytoplasm in living cells is maintained in the reduced state by the activities of specialized enzyme systems comprising members of the oxidoreductase family that perform generalized protein reduction (2). In the well-studied example of Escherichia coli, the thioredoxin (Trx) system comprises two thioredoxins and thioredoxin reductase, while the glutaredoxin (Grx) system is composed of glutathione (GSH), three glutaredoxins, and GSH oxidoreductase. Trx, with an approximate mass of 12,000 Da, is found ubiquitously in nature. All thioredoxins share similar three-dimensional structures and possess a conserved WCXXC catalytic motif, buried within a protein fold known as the thioredoxin fold (27). Thioredoxins are able to cycle between the oxidized disulfide and the reduced dithiol forms. Oxidized Trx is in turn reduced by a flavoenzyme, thioredoxin reductase, via a redox-active cysteine pair and flavin adenine dinucleotide as a cofactor consuming cellular NADPH. Reduced Trx generated in this manner is thus poised for reducing disulfides of the target proteins in the cellular milieu (20).
The NADPH-dependent disulfide reduction mechanism is required for a variety of physiological functions. For example, the two E. coli thioredoxins TrxA and TrxC and the glutaredoxin GrxA are known to be required for cellular DNA synthesis by acting as electron donors for the essential enzyme ribonucleotide reductase (14, 30). The other physiological functions that are accomplished by reduced thioredoxins include protein disulfide reduction, sulfur assimilation, detoxification of reactive oxygen species, protein repair, and redox regulation of enzymes and transcription factors (2, 6, 31). Apart from these functions, E. coli TrxA functions as a processivity factor for the bacteriophage T7-encoded DNA polymerase (26). This property of Trx, though, is independent of its redox activity and depends upon a structural interaction between T7 DNA polymerase and the Trx domain (21).
The oxidation/reduction function of the thiol/disulfide oxidoreductase family of proteins depends upon two important determinants, the pKa values of the cysteine residues in the CXXC motif and the standard-state redox potential. The members of the oxidoreductase family of proteins have a wide range of redox potentials. Among the most-reducing members are those from the Trx superfamily, which have the lowest redox potential and function as general protein reductants in the cytoplasm. The redox potential of E. coli TrxA is −270 mV, while those of glutaredoxins range from −233 to −198 mV (4). In contrast, the oxidizing members, like DsbA and DsbC, found in the periplasm of prokaryotes, which perform formation and isomerization of disulfide bonds in newly translocated proteins (5), have redox potentials ranging from −120 mV to −200 mV (10, 41, 42).
Mycobacterium tuberculosis, the causative agent of tuberculosis, is an intracellular pathogen that resides in mononuclear phagocytes. The mechanisms by which M. tuberculosis and other mycobacteria resist oxidative killing by mononuclear phagocytes are somewhat unclear. Several mechanisms for resistance to intracellular killing have been proposed, one of which is the scavenging of free radicals produced by mononuclear phagocytes upon infection (38). The M. tuberculosis genome lacks the potential to code for components of the GSH system, though M. tuberculosis has the capacity to synthesize low-molecular-weight thiols, like mycothiol (8, 32). The absence of the GSH system suggests that M. tuberculosis Trx and TrxR are likely to resist oxidative killing in a manner similar to their involvement against oxidative stress in other prokaryotes. The genome of M. tuberculosis encodes three thioredoxins and bears a single copy of trxR, the gene encoding thioredoxin reductase. M. tuberculosis thioredoxins and TrxR have been shown to be involved in reduction of peroxides and dinitrobenzenes (43) and also to detoxify hydroperoxides in vitro (22). Thus, thioredoxins and TrxR are considered to be attractive targets for antitubercular drug design studies. Toward achieving this, the crystal structure of TrxCMtb provides an opportunity for designing drugs against this target (16).
In this report, we describe the biochemical properties of the thioredoxins from M. tuberculosis, wherein we show that the three M. tuberculosis thioredoxins function as bona fide disulfide reductases. Furthermore, we obtain estimates of their intrinsic redox potentials and show that two out of the three M. tuberculosis thioredoxins display redox potential values closer to that of E. coli TrxA. We also show that expression of trxAMtb, whose recombinant product appears to be a low-activity thioredoxin, is cryptic and that it alone is not an effective substrate of M. tuberculosis TrxR. Heterologous expression of M. tuberculosis thioredoxins is shown to complement specific phenotypes in E. coli strains devoid of TrxA. Expression of trxBMtb and trxCMtb under a variety of oxidative conditions is suggestive of the principal role played by these thioredoxins in the M. tuberculosis cytoplasm and may offer an explanation for the natural resistance of M. tuberculosis toward oxidative killing.
MATERIALS AND METHODS
Bacterial strains/plasmids.
All the bacterial strains and plasmids used in this study are listed in Table 1. The sequences of the oligonucleotide primers used for PCR amplification of trx open reading frames (ORFs) are listed in Table S1 in the supplemental material.
TABLE 1.
Bacterial strains and plasmids used in this study
| Strain or plasmid | Description | Source or referencea |
|---|---|---|
| Strains | ||
| DH5α | λ− φ80dlacZΔM15 Δ(lacZYA-argF)U169 recA1 endA hsdR17 (rk− mk−) supE44 thi-1 gyrA relA1 | 17 |
| BL21(DE3) | F−ompT hsdSB (rB− mB−) gal dcm (DE3) | 40 |
| DHB4 | Δ(ara-leu)7697 araD139 ΔlacX74 galE galK rpsL phoR Δ(phoA) pvuII ΔmalF3 thi | 7 |
| GJ8025 | DHB4 ΔtrxA grxA::kan zbi::Tn10 | |
| Plasmids | ||
| pET23a | PT7-based expression vector | Novagen |
| pBAD33 | l-Arabinose-based expression vector with p15A origin of replication | 15 |
| pSCM1101 | pET23a bearing trxAMtb (Rv1470) within NdeI/HindIII restriction sites | |
| pSCM1102 | pET23a bearing trxBMtb (Rv1471) within NdeI/HindIII restriction sites | |
| pSCM1103 | pET22b bearing trxCMtb (Rv3914) within NdeI/HindIII restriction sites | |
| pSCM1104 | pET23a bearing trxAEc within NdeI/HindIII restriction sites | |
| pSCM1105 | pET23a bearing trxRMtb (Rv3913) within NdeI/HindIII restriction sites | |
| pHYD3058 | pBAD33 bearing trxAMtb within XbaI/HindIII restriction sites | |
| pHYD3059 | pBAD33 bearing trxBMtb within XbaI/HindIII restriction sites | |
| pHYD3060 | pBAD33 bearing trxCMtb within XbaI/HindIII restriction sites | |
| pHYD3064 | pBAD33 bearing trxAEc within XbaI/HindIII restriction sites |
Unless otherwise indicated, the source of the listed plasmids is the present study.
Construction of Trx expression plasmids.
The two ORFs carrying trx genes (trxAMtb [Rv1470] and trxBMtb [Rv1471]) were PCR amplified from a cosmid library of M. tuberculosis strain H37Rv by using the oligonucleotide primers listed in Table S1 in the supplemental material. In each case, the reverse primer was designed so that the 3′ end of any given trx ORF bears a DNA sequence encoding hexahistidine. The amplified trx genes (trxAMtb and trxBMtb) were cloned in the pET23a expression vector (Novagen) via restriction sites NdeI and HindIII at the 5′ and 3′ ends, respectively. The resultant plasmids were designated pSCM1101 and pSCM1102, respectively. trxCMtb (Rv3914), cloned in pET22b, was a gift from Yossef Av-Gay and has been designated pSCM1103. The E. coli trxA (trxAEc, encoding thioredoxin 1) ORF was PCR amplified using genomic DNA of E. coli K-12 strain MG1655 as a template, and the resultant product was cloned into the NdeI and HindIII sites of pET23a to obtain plasmid pSCM1104. Plasmid pSCM1105 bears M. tuberculosis trxR under the expression control of the T7 promoter and has been described previously (1). The integrity of all the plasmid clones was confirmed by DNA sequencing.
Overproduction and purification of thioredoxins and TrxR.
Plasmids pSCM1102 and pSCM1103 were transformed into BL21(DE3) cells, and transformants were grown in 500 ml Terrific broth with 100 μg/ml ampicillin. Isopropyl β-d-1-thiogalactopyranoside (IPTG) was added to mid-log-phase culture (at 0.5 mM), which was further incubated for 6 h at 37°C. pSCM1101 was transformed into BL21 DE3/pLysS cells, expression of trxAMtb was induced by addition of IPTG (at 0.5 mM) to the culture, and the culture was incubated further for 12 h at 18°C. Cells were harvested by centrifugation, resuspended, and sonicated in 20 ml buffer containing 50 mM Tris, pH 8, 300 mM NaCl, 5% glycerol, 5 mM imidazole, and 0.1 mM phenylmethylsulfonyl fluoride. A second round of centrifugation at 12,000 rpm for 15 min was carried out to remove cell debris. The resulting supernatant was applied to a Ni-nitrilotriacetic acid column preequilibrated with lysis buffer. The supernatant containing the mixture of soluble proteins was allowed to bind on the Ni-nitrilotriacetic acid column. The column was washed with 5 bed volumes of wash buffer (50 mM Tris, pH 8, 300 mM NaCl, 5% glycerol, and 20 mM imidazole), followed by elution with lysis buffer containing 200 mM imidazole (elution buffer). Eluted recombinant proteins were concentrated using an Amicon concentrator with 3,000-Da-cutoff membranes and further purified by loading them on a Superdex 75 gel filtration column (Pharmacia) equilibrated with 10 mM Tris (pH 8) and 20 mM NaCl. The purified protein fractions were analyzed on 15% sodium dodecyl sulfate-polyacrylamide gel electrophoresis gel.
The protein concentrations were determined by absorption at 280 nm, using extinction coefficients of 11,460, 13,980, 11,000, 13,980, and 14,440 M−1 cm−1 for TrxAMtb, TrxBMtb, TrxCMtb, TrxAEc, and TrxR, respectively. The extinction coefficients for thioredoxins were calculated using the online program ProtParam (http://ca.expasy.org/tools/protparam.html) (13). Overproduction and purification of TrxR were done as reported previously (1).
Size exclusion chromatography.
The purity levels and molecular weights of the recombinant proteins were determined by size exclusion chromatography. The chromatography experiments were performed with Superdex 75 FPLC columns from the Bio-Rad system, using 10 mM Tris, pH 8, and 100 mM NaCl as the running buffer. The void volume of the column was determined using Blue Dextran 200. The elution times of all the recombinant proteins were recorded, and the molecular weights were calculated by estimating the elution volumes of standards of known molecular weights.
Trx activity assay.
The activity of thioredoxins was determined by reduction of DTNB [5,5′-dithiobis(2-nitrobenzoic acid)] and insulin. Insulin was used to determine the protein disulfide reductase activities of thioredoxins as described previously (18). The DTNB assay was performed as described elsewhere (19). Briefly, the reaction mixture contained 200 mM phosphate buffer, pH 7.0, 2 mM EDTA, 2 μM bovine serum albumin, 1 mM DTNB, and 0.5 mM NADPH. The reaction mixture also contained 0.5 to 14 μM each of TrxAMtb, TrxBMtb, TrxCMtb, and TrxAEc. The reaction was started by addition of 10 nM TrxR. The progress of the reactions was monitored at 412 nm against a blank control for 15 min at 25°C with a final volume of 100 μl. The rate of DTNB reduction was calculated from the increase in absorbance at 412 nm by using a molar extinction coefficient of 27,000 M−1 cm−1. Reduction of DTNB by 1 mol of reduced Trx yields 2 mol of TNB (1,3,5-trinitrobenzene) with a molar extinction coefficient of 13,600 M−1 cm−1.
RT-PCR.
M. tuberculosis strain H37Rv was cultured in Middlebrook 7H9 supplemented with 0.5% glycerol, 0.05% Tween 80, and 1× albumin dextrose catalase to mid-log phase. Fifty milliliters of mid-log-phase-grown bacterial cultures was harvested and washed once in phosphate-buffered saline. Bacterial cells were further resuspended in 50 ml of fresh Middlebrook 7H9 supplemented with albumin dextrose catalase and Tween 80 and were individually treated with H2O2, diamide, cumene hydroperoxide, nitroprusside, and tert-butylhydroperoxide, each at a final concentration of 2 mM, and cultures were incubated for 2 h at 37°C under aerobic conditions. Total RNA extraction from the treated culture of M. tuberculosis H37Rv was carried out by the Trizol method. For this, 50 ml of mycobacterial cultures was harvested by centrifugation, washed with phosphate-buffered saline, and resuspended in 1 ml Trizol solution (Sigma). Bacterial cells were lysed by bead beating and treated with chloroform. Total RNA was precipitated by adding isopropanol, washed with 70% ethanol, and finally resuspended in diethyl pyrocarbonate-treated DNase-RNase-free water. RNA concentration was determined spectrophotometrically at 260 nm. Total RNA was treated with DNase (USB) to remove the contaminating DNA. Further, 2 μg total RNA was reverse transcribed using Moloney murine leukemia virus RT Superscript III (Invitrogen). Reverse transcriptase PCR (RT-PCR) was performed, and the expression level of sigA was used as an internal control. The primers used for this study are listed in Table S1 in the supplemental material. In all instances, no amplification was observed for mRNA samples in the absence of RT treatment.
Determination of equilibrium constant and redox potential with GSH.
To determine the redox equilibrium constant between thioredoxins (TrxAMtb, TrxBMtb, TrxCMtb, or TrxAEc) and GSH, 1.6 μM of purified thioredoxins was incubated in a nitrogen-purged solution of 100 mM Na-phosphate buffer, pH 7, at 30°C for 12 h in the presence of 0.1 mM oxidized GSH (GSSG) and different concentrations (0 mM to 120 mM) of GSH. The change in fluorescence intensity caused by exciting the protein at 295 nm was recorded at 355 nm, where the difference in the fluorescence intensity between the oxidized and the reduced forms for a given thioredoxin was maximal. Equilibrium constant and redox measurements were done as described previously (29). The equilibrium constants were determined by fitting the original fluorescence data according to the following equations:
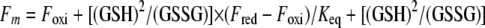 |
(1) |
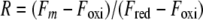 |
(2) |
where Fm is the measured fluorescence intensity and Foxi and Fred are the fluorescence intensities of the oxidized and reduced proteins, respectively. R is the fraction of reduced protein at equilibrium. A value of −240 mV was used for the standard redox potential (Eo) of GSH (34) to calculate the redox potentials of the thioredoxins by applying the equilibrium constant (Keq) in equation 3.
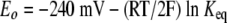 |
(3) |
Construction of M. tuberculosis trx ORFs under the expression control of the PBAD promoter.
The ORFs carrying trxAMtb, trxBMtb, trxCMtb, and trxAEc were cloned under the expression control of the l-arabinose-inducible PBAD promoter in the plasmid pBAD33 (15). In each construct, any given trx ORF was preceded by an optimally placed ribosome-binding site and bore at its 3′ terminus a DNA sequence encoding hexahistidine. The resultant plasmids were designated pHYD3058 (trxAMtb), pHYD3059 (trxBMtb), pHYD3060 (trxCMtb), and pHYD3064 (trxAEc) and are listed in Table 1.
Heterologous complementation studies with M. tuberculosis trx ORFs.
A trxA grxA (encoding glutaredoxin 1) double mutant strain of E. coli GJ8025 was constructed via bacteriophage P1-mediated transduction and transformed with pBAD33 and its derivatives containing trx ORFs from M. tuberculosis and E. coli. Restoration of the cysteine prototrophy of the trxA grxA double mutant was assayed by examining the extent of growth conferred by a given trx ORF to strain GJ8025 in minimal medium containing 0.2% (wt/vol) glucose supplemented with leucine and isoleucine (each at 40 μg/ml) in the presence of 0.2% arabinose. Leucine and isoleucine addition was necessitated by the presence of the (ara-leu)7697 deletion in strain GJ8025, which leads to an auxotrophy for leucine and isoleucine.
RESULTS
Thioredoxins from M. tuberculosis function as disulfide reductases.
The ORFs encoding the three M. tuberculosis thioredoxins, namely, Rv1470 (trxAMtb), Rv1471 (trxBMtb), and Rv3914 (trxCMtb), were PCR amplified and cloned in suitable expression vectors, and the corresponding polypeptides were overproduced in BL21(DE3) cells. In size exclusion chromatography experiments performed in the absence of a reductant, TrxAMtb, TrxBMtb, and TrxCMtb displayed protomer masses of 14,500 Da, 14,000 Da, and 17,800 Da, which are marginally higher than the predicted masses of 13,500 Da, 13,300 Da, and 12,500 Da, respectively. Some extent of anomalous migration on denaturing acrylamide gels, despite consideration of the contribution of the hexahistidine tag appended to the carboxy termini of the M. tuberculosis thioredoxins, was also apparent, especially for TrxCMtb (data not shown). When a similar approach was used, E. coli TrxA (TrxAEc) was also overproduced and purified, and its protomer mass was estimated to be 12,000 Da (data not shown).
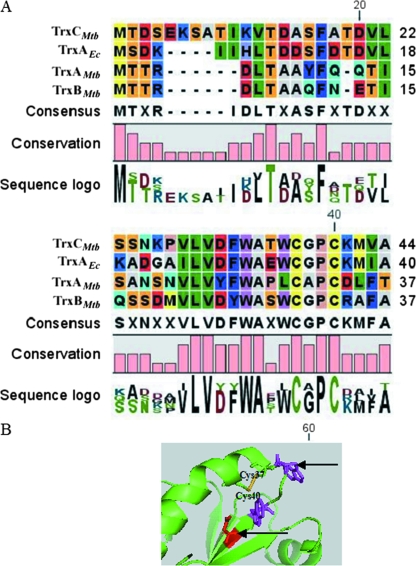
Sequence alignment was generated between all three M. tuberculosis thioredoxins and TrxAEc, with special attention to the residues in the vicinity of the active Cys site, which are known to maintain appropriate pKa and redox values (29). The sequence alignment so generated showed two major changes in the highly conserved region of TrxAMtb near the redox-active site (Fig. 1A and B). The Trp32 and Asp27 residues in TrxAEc (which are conserved in all other bacterial thioredoxins) as well as TrxBMtb and TrxCMtb are replaced with Leu (29th residue in TrxAMtb) and Tyr (24th residue in TrxAMtb), respectively. Moreover, TrxAMtb also has an Asp residue on the C-terminal side of the resolving Cys residue, while TrxAEc, TrxBMtb, and TrxCMtb have basic residues, either Arg or Lys. Thus, the sequence comparisons suggested that TrxAMtb possesses unusual sequence features near its putative redox-active site (Fig. 1B).
FIG. 1.
Multiple sequence alignment of M. tuberculosis thioredoxins and E. coli TrxA. (A) Trp 32 and Asp 27 are highly conserved in the Trx superfamily but are replaced by Leu and Tyr in TrxAMtb at positions 29 and 24, respectively. A histogram depicting amino acid conservation among the indicated thioredoxins was generated with CLC Protein Workbench 2.0.2. (B) Structure of the redox-active region of TrxCMtb (Protein Data Bank accession no. 2I1U) (20). Tryptophan residues are depicted in magenta, aspartic acid residues are depicted in red, and the disulfide between two cysteine residues is depicted in yellow. The amino acid variation in TrxAMtb is marked with arrows.
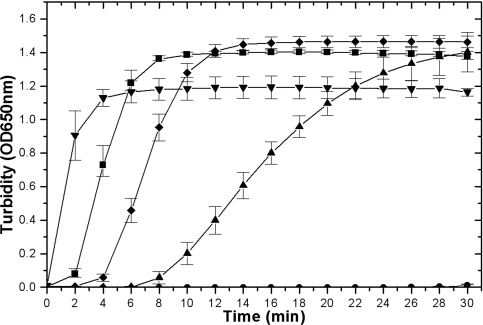
Members of the oxidoreductase family of proteins display the ability to reduce disulfide bonds in insulin, a model substrate, leading to precipitation of its β chain (18). The three M. tuberculosis thioredoxins were proficient to various degrees in reducing insulin in the presence of the nonphysiological reductant dithiothreitol (DTT). TrxAMtb, TrxBMtb, and TrxCMtb displayed low, high, and moderate insulin β-chain precipitation abilities, respectively (Fig. 2). TrxAEc displayed an activity comparable to those of TrxBMtb and TrxCMtb. It is thus likely that the subtle changes observed in the sequence analysis might affect the active-site electrostatics and thereby the pKa values of the Cys residues, offering a possible explanation for the lower disulfide reductase activity of TrxAMtb in the presence of DTT.
FIG. 2.
Thioredoxin-catalyzed reduction of insulin by DTT. Insulin turbidity due to thioredoxin-promoted precipitation of the insulin β chain by DTT was measured at 650 nm and is plotted as a function of time. The assay mixture contained 130 μM insulin, 1 mM DTT in 100 mM potassium phosphate, 2 mM EDTA, (pH 7.5), and 8 μM of thioredoxins. OD, optical density; •, control lacking thioredoxins; ▴, TrxAMtb; ▾, TrxBMtb; ♦, TrxCMtb; and ▪, TrxAEc.
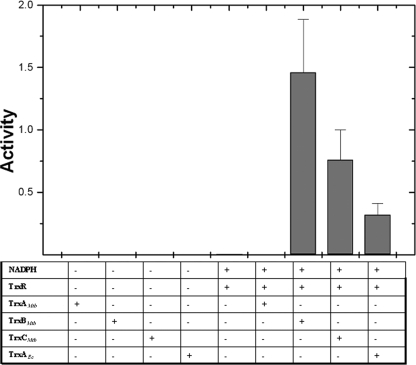
In order to assess which thioredoxins serve as substrates for TrxR, a NADPH-dependent insulin reduction assay was performed. TrxR was able to mediate reduction of TrxBMtb and TrxCMtb but not TrxAMtb, suggesting that TrxBMtb and TrxCMtb might be functionally active disulfide reductases in vivo (Fig. 3). However, TrxR was more efficient in reducing TrxBMtb than TrxCMtb. The inability of TrxR to reduce TrxAMtb suggests that TrxAMtb may not be a natural substrate of TrxR.
FIG. 3.
TrxR-dependent M. tuberculosis thioredoxin activity. The activities of thioredoxins as measured by insulin β-chain precipitation in the presence of TrxR and NADPH are depicted as a histogram. The assay mixture contained 130 μM insulin, 200 μM NADPH and 2 μM of M. tuberculosis TrxA/B/C or TrxAEc in 100 mM potassium phosphate, 1 mM EDTA (pH 6.5), and 2 μM bovine serum albumin.
Further, DTNB, a generic disulfide substrate, was used to estimate the kinetic parameters for thioredoxins with M. tuberculosis TrxR. In this assay, a low concentration (10 nM) of M. tuberculosis TrxR was used to obtain saturation Michaelis-Menten kinetics and calculate Km for thioredoxins at pH 7.0 and 25°C. The values of Km and Vmax obtained were used to calculate kcat as well as kcat/Km for the reaction between TrxR and reduced thioredoxins. The Km and kcat values for TrxBMtb and TrxCMtb are almost similar and are comparable to the values for TrxAEc. No activity was detected for TrxAMtb. The comparative kinetic parameters are shown in Table 2. Thus, these results suggest that the redox system in M. tuberculosis could be operative with either of the two functional thioredoxins, namely, TrxBMtb or TrxCMtb.
TABLE 2.
Kinetic parameters of M. tuberculosis thioredoxinsa
| Protein | Km (μM) | kcat (s−1) | kcat (s−1)/Km (M) |
|---|---|---|---|
| TrxAMtb | ND | ND | ND |
| TrxBMtb | 2.65 ± 0.07 | 1.79 ± 0.13 | (6.7 ± 0.03) × 105 |
| TrxCMtb | 2.05 ± 0.5 | 1.17 ± 0.24 | (5.75 ± 0.02) × 105 |
| TrxAEc | 2.8 ± 0.14 | 1.34 ± 0.12 | (4.7 ± 0.06) × 105 |
The assay was carried out as described in Materials and Methods. The apparent Km and apparent turnover (kcat) values were calculated by nonlinear regression using the Michaelis-Menten equation. Data are represented as means ± standard deviations. ND, not detected.
Equilibrium constant with GSH and redox potential of M. tuberculosis thioredoxins.
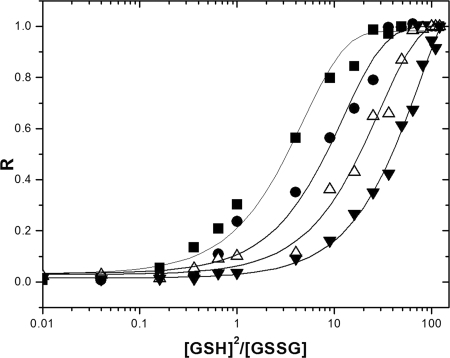
Thiol/disulfide catalysis mediated by thioredoxins is dependent upon the intrinsic redox potential and pKa values of the cysteines of the CXXC motif. Redox potential is known to be a determinant of the extent of function of Trx (4). To address the functional properties of multiple thioredoxins from M. tuberculosis, redox potential was determined using GSH as the redox buffer. The redox equilibrium constants of TrxAMtb, TrxBMtb, TrxCMtb, and TrxAEc with GSH were measured according to the redox state-dependent fluorescence changes of proteins, with the assumption that an insignificant concentration of mixed disulfide between Trx and GSH is not a contributor to redox-dependent changes in intrinsic fluorescence (41). The equilibrium measurements and deduced redox potentials of TrxAMtb, TrxBMtb, TrxCMtb, and TrxAEc are shown in Fig. 4 and Table 3. The estimated redox potential of TrxAEc (−280 mV) is close to the reported value of −278 ± 3 mV (4), and thus, TrxBMtb (−262 ± 2 mV) and TrxCMtb (−269 ± 2 mV) show modest decreases in reducing ability in comparison to TrxAEc. The redox potential of TrxAMtb was estimated to be −248 ± 3 mV.
FIG. 4.
Redox equilibrium curves of M. tuberculosis thioredoxins and TrxAEc with GSH. The plots show the fractions of reduced protein (R) at equilibrium under various concentrations of GSH-GSSG buffer, obtained by recording the changes in the redox state-dependent fluorescence emission spectra at the appropriate emission maxima, for the oxidized versions of indicated thioredoxins. ▪, TrxAMtb; •, TrxBMtb; ▵, TrxCMtb; and ▾, TrxAEc.
TABLE 3.
Redox potentials of M. tuberculosis thioredoxins and other oxidoreductasesa
| Protein | Eo (mV) |
|---|---|
| M. tuberculosis thioredoxin A (TrxAMtb) | −248 ± 3 |
| M. tuberculosis thioredoxin B (TrxBMtb) | −262 ± 2 |
| M. tuberculosis thioredoxin C (TrxCMtb) | −269 ± 2 |
| E. coli thioredoxin A (TrxAEc) | −278 ± 3 |
| E. coli glutaredoxin (4) | −233 |
| T4 bacteriophage thioredoxin (22) | −230 |
The redox potentials of M. tuberculosis thioredoxins and other prototype members of the thioredoxin family are shown. The equilibrium constant and redox potential (Eo) values for TrxAMtb, TrxBMtb, TrxCMtb, and TrxAEc were obtained using equations 1, 2, and 3 as described in the text. Known redox potentials for some members are denoted as reported previously (4, 23). In the current experiments, the redox potential of TrxA was recorded at −280 mV, close to that reported by Åslund et al. (4).
M. tuberculosis trx gene expression under various growth conditions.
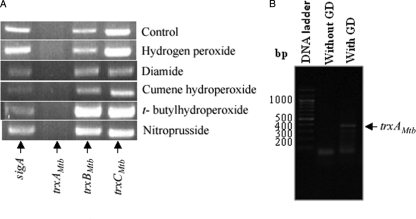
We attempted to observe trxAMtb mRNA in M. tuberculosis strain H37Rv under conditions of steady-state growth and in media containing a variety of agents that induce oxidative stress. Simultaneously, we also undertook studies to estimate the mRNA expression levels of other trx genes present in M. tuberculosis under these growth conditions to test for their expression. sigA, an essential housekeeping gene, was used as an internal control. The results shown in Fig. 5A indicate that mRNA expression for trxBMtb and trxCMtb is observed under all the employed conditions. However, no mRNA expression for trxAMtb was detected under these conditions, suggesting that trxAMtb expression may be cryptic. We excluded the possibility that the trxAMtb primer pair could have been noncompetent for PCR by showing that an expected-size DNA fragment of trxAMtb could be obtained when H37Rv genomic DNA was used as a template (Fig. 5B).
FIG. 5.
RT-PCR analysis of M. tuberculosis trx transcripts under various growth regimens. (A) Cultures of M. tuberculosis H37Rv were grown aerobically to mid-exponential phase (optical density at 600 nm, ∼0.9) and treated individually with the indicated oxidants, each at a concentration of 2 mM, for 2 h. Total RNA was isolated from grown cultures, and RT-PCR was performed with primers specific for the indicated trx transcripts. Total RNA extracted from a nontreated culture was used as a control. A primer pair specific to the sigA transcript was used as an internal control. (B) Control experiment showing that the trxAMtb gene is amplified using M. tuberculosis genomic DNA (GD).
M. tuberculosis thioredoxins can compensate for lack of TrxA in E. coli.
Mutations in the genes involved in pathways of cytosolic protein reduction in E. coli, such as trxA, trxB, and grxA (encoding glutaredoxin 1), lead to specific phenotypes (3, 39). For example, E. coli strains deficient for TrxAEc and GrxA exhibit cysteine auxotrophy because an enzyme of the cysteine biosynthetic pathway PAPS (3′-phosphoadenosine-5′-phosphosulfate) reductase requires either TrxAEc or GrxA for catalytic activity (35). The abilities of M. tuberculosis thioredoxins to complement this defect of an E. coli trxA grxA double mutant were gauged. A suitable strain, GJ8025, was constructed and transformed with plasmids expressing from the PBAD promoter trxMtb ORFs and trxAEc. Expression of trxBMtb and trxCMtb, but not trxAMtb, by supplementation of minimal medium with 0.2% l-arabinose allowed growth of strain GJ8025. GJ8025 derivatives bearing a vector and a plasmid expressing trxAEc were rendered auxotrophic and prototrophic for cysteine, respectively. It may be noted that in a standard strain, like DH5α, robust production of TrxAMtb was detected in the presence of l-arabinose and that expression (even at basal levels) of trxAEc was sufficient to promote fairly robust growth of GJ8025 (Fig. 6A and B). These studies show that M. tuberculosis-derived thioredoxins can functionally substitute for TrxAEc in PAPS reduction.
FIG. 6.
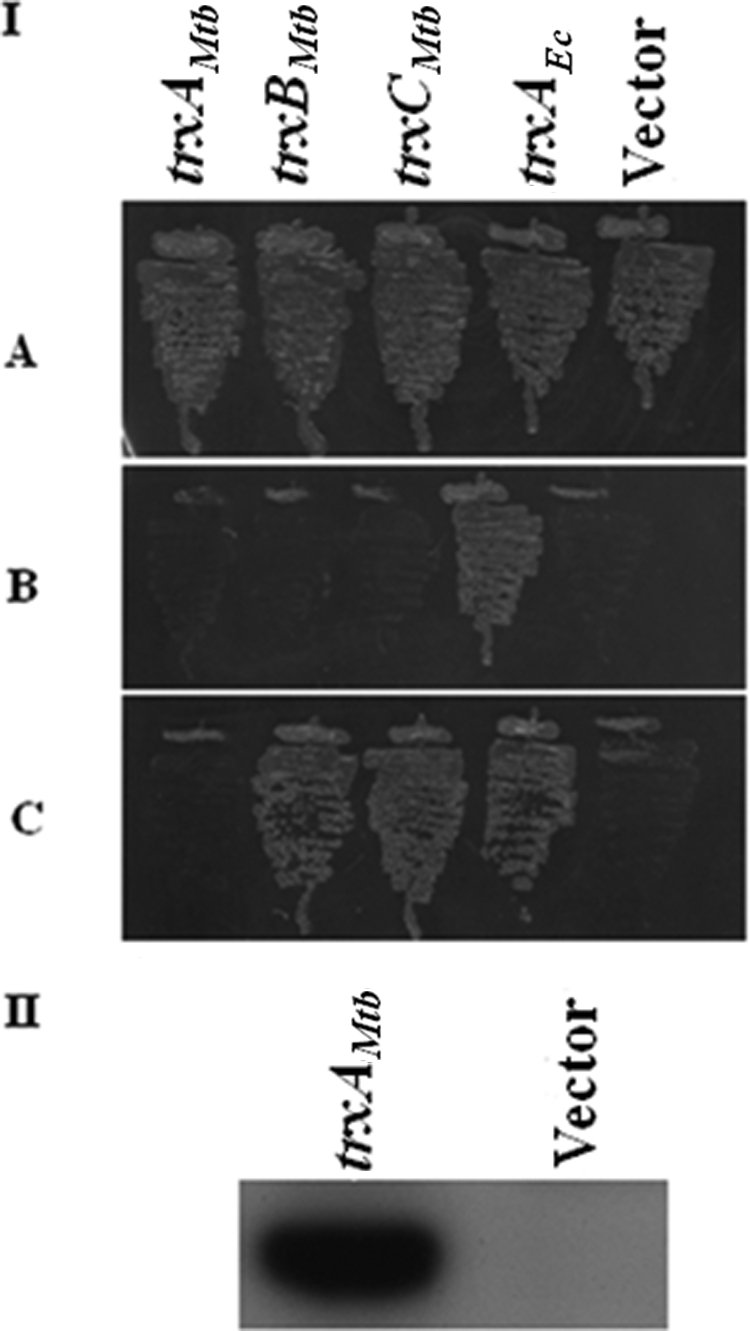
Rescue of the cysteine auxotrophy of an E. coli trxA grxA double mutant by M. tuberculosis trx expression. (I) Transformants of strain GJ8025 (ΔtrxA grxA::kan) bearing indicated trx genes under the expression control of the PBAD promoter in plasmids pHYD3058 (trxAMtb), pHYD3059 (trxBMtb), pHYD3060 (trxCMtb), pHYD3064 (trxAEc), and pBAD33 (vector) were streaked on minimal agar plates containing 0.2% d-glucose and 40 μg/ml l-cysteine (A), 0.2% d-glucose (B), and 0.2% d-glucose and 0.2% arabinose (C). An appropriate amount of l-leucine-l-isoleucine supplement was added to the indicated plates. (II) Immunodetection of TrxAMtb in lysates of DH5α bearing the indicated plasmids pHYD3058 (trxAMtb) and pBAD33 (vector). Whole-cell extracts of transformants of DH5α bearing plasmids pHYD3058 (trxAMtb) and pBAD33 (vector) after cultivation of the cells in LB broth with 0.2% d-glucose and 0.2% l-arabinose, prepared after normalization for cell number, were probed with a monoclonal antibody directed against hexahistidine, an epitope appended to the carboxy terminus of TrxAMtb.
DISCUSSION
A characteristic feature of the bacterial cytoplasm (and the cytoplasm from eukaryotes) is its reduced state. Member proteins of the thioredoxin family are involved in maintaining cysteine residues of cytoplasmic proteins in the reduced state and do so via a pair of conserved cysteine residues (20). In this study, we have examined biochemical, physicochemical, and genetic features of three M. tuberculosis thioredoxins to show that they are active disulfide reductases. Of the three, TrxAMtb exhibited a weak capacity, whereas TrxBMtb and TrxCMtb displayed high and intermediate capacities, respectively, to reduce the disulfides of insulin.
In TrxAEc, mutation of Asp27, a conserved, buried, and charged residue located near the redox-active center, to Ala is known to impair disulfide reduction (11). It is thought that Asp27 is critical for modulating the ionization of the active-site thiol group. Similarly, Trp32, preceding the CXXC motif, is also a conserved residue whose replacement by Ala in TrxAEc renders TrxAEc inefficient in disulfide reduction (24). Variation of these critical residues in the analogous positions of TrxAMtb may be contributing to the weak disulfide oxidoreductase activity of this thioredoxin (Fig. 1 and 2). Moreover, only TrxBMtb and TrxCMtb were found to be substrates of TrxR, suggesting that they may be the major cellular thioredoxins. The differences in activity between M. tuberculosis thioredoxins may be thus attributed to their intrinsic redox potentials (Fig. 4 and Table 3).
The observation that, of the three trx ORFs, expression of trxAMtb was found to be cryptic, despite the variety of growth conditions tested, leads to a questioning of the existence of cellular TrxAMtb activity. We are hesitant to bracket trxAMtb as a pseudogene, however, because in the M. tuberculosis genome, trxA is present in an apparent operonic arrangement with trxB, located downstream, and the gene upstream of trxA, ctpD, codes for a probable cation transporter, the p-type ATPase. This gene arrangement is conserved in mycobacterial species like Mycobacterium ulcerans, M. bovis, and M. marinum, which bear multiple trx ORFs, thus leading to synteny for this region. However, the Mycobacterium leprae genome possesses only two trx genes, and the conserved ctpD gene is arranged downstream of one of the trx genes. The synteny in the gene cluster consisting of ctpD, trxA, and trxB is suggestive of a need for retention of trxAMtb despite its apparent crypticity. It is possible that there may be other environmental conditions under which trxAMtb is expressed. At the same time, the observation that trxBMtb gene expression is observed under many different conditions can be explained by the presence of at least one promoter of the gene within the trxAMtb gene (25).
In these studies, we have employed the surrogate system of E. coli strains defective in components of cytoplasmic protein reduction to perform an in vivo test of Trx function, namely, that of PAPS reductase reduction (Fig. 6). The abilities of M. tuberculosis thioredoxins, namely, TrxBMtb and TrxCMtb, to functionally compensate for a deficiency of E. coli TrxA in this test were apparent, suggesting that these thioredoxins may perform analogous functions in vivo. In the in vivo test of Trx function, TrxAMtb displayed a negligible propensity to compensate for lack of TrxAEc. It is possible to rationalize this behavior of TrxAMtb based on the premise that TrxAMtb is defective in its own oxidation but proficient in its reduction. Thus, the inability of TrxAMtb to donate electrons to PAPS reductase may account for its failure to complement the cysteine auxotrophy of a trxAEc grxAEc double mutant. This premise may also account for its weakened disulfide reductase activity and high redox potential, since formally the redox potential of a given thioredoxin can be influenced by rate constants governing its own reduction or oxidation (28).
M. tuberculosis is known to be devoid of GSH, instead possessing a biosynthetic capacity for production of mycothiol, a small molecule thiol (12, 32). Recent studies have shown that two genes involved in mycothiol biosynthesis, mshA and mshC, are essential in M. tuberculosis, whereas deletion of mshC in Mycobacterium smegmatis leads to sensitivity to hydrogen peroxide (33, 36). It therefore appears that M. tuberculosis may rely more upon mycothiol as a protective measure against oxidative stress.
Multiple thioredoxins are found in many organisms. For example, E. coli possesses two functional thioredoxins and three glutaredoxins. However, Arabidopsis thaliana possesses at least 35 Trx isoforms (9). It appears that multiple thioredoxins may endow an organism with a backup measure for coping with redox stresses. As an alternative, some cellular process(es), such as reduction of ribonucleotide reductase, may have evolved to utilize one thioredoxin preferentially. For A. thaliana, there is evidence in vitro that some Trx isoforms exhibit substrate specificity (9). Evidence for Trx substrate specificity has not been obtained, at least in the current studies, and the two M. tuberculosis thioredoxins TrxBMtb and TrxCMtb seem to function with near-equal potencies as generalized disulfide reductases. One feature of the biology of M. tuberculosis pathogenesis is its ability to survive intracellular redox stress. It appears that M. tuberculosis mutants missing any of the three trx ORFs are not significantly impaired for survival in vivo (37). This observation, coupled with the finding, obtained in this study (Fig. 5), that M. tuberculosis trx genes are fairly well expressed (transcribed) under a variety of oxidative stress conditions, suggests that, in M. tuberculosis, multiple thioredoxins may provide a cover for coping with metabolic demands and conditions of oxidative stress.
Supplementary Material
Acknowledgments
We thank J. Beckwith, S. Cole, and J. Gowrishankar for providing strains and plasmids and N. Ahmad for helpful discussions.
This work was supported by grants from the Department of Biotechnology, India. M.A. and G.K. were supported by fellowships from the Council for Scientific and Industrial Research, India, and A.A.S. was supported by a fellowship from the Department of Science and Technology, India. S.C.M. is a Wellcome Trust International Senior Research Fellow. We declare no financial conflict of interest.
Footnotes
Published ahead of print on 22 August 2008.
Supplemental material for this article may be found at http://jb.asm.org/.
REFERENCES
- 1.Akif, M., K. Suhre, C. Verma, and S. C. Mande. 2005. Conformational flexibility of Mycobacterium tuberculosis thioredoxin reductase: crystal structure and normal-mode analysis. Acta Crystallogr. D 611603-1611. [DOI] [PubMed] [Google Scholar]
- 2.Arner, E. S. J., and A. Holmgren. 2000. Physiological functions of thioredoxin and thioredoxin reductase. Eur. J. Biochem. 2676102-6109. [DOI] [PubMed] [Google Scholar]
- 3.Åslund, F., and J. Beckwith. 1999. The thioredoxin superfamily: redundancy, specificity, and gray-area genomics. J. Bacteriol. 1811375-1379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Åslund, F., K. D. Berndt, and A. Holmgren. 1997. Redox potentials of glutaredoxins and other thiol-disulfide oxidoreductases of the thioredoxin superfamily determined by direct protein-protein redox equilibria. J. Biol. Chem. 27230780-30786. [DOI] [PubMed] [Google Scholar]
- 5.Bardwell, J. C. A., K. McGovern, and J. Beckwith. 1991. Identification of a protein required for disulfide bond formation in-vivo. Cell 67581-589. [DOI] [PubMed] [Google Scholar]
- 6.Becker, K., S. Gromer, R. H. Schirmer, and S. Muller. 2000. Thioredoxin reductase as a pathophysiological factor and drug target. Eur. J. Biochem. 2676118-6125. [DOI] [PubMed] [Google Scholar]
- 7.Boyd, D., C. Manoil, and J. Beckwith. 1987. Determinants of membrane protein topology. Proc. Natl. Acad. Sci. USA 848525-8529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Cole, S. T., R. Brosch, J. Parkhill, T. Garnier, C. Churcher, D. Harris, S. V. Gordon, K. Eiglmeier, S. Gas, C. E. Barry III, F. Tekaia, K. Badcock, D. Basham, D. Brown, T. Chillingworth, R. Connor, R. Davies, K. Devlin, T. Feltwell, S. Gentles, N. Hamlin, S. Holroyd, T. Hornsby, K. Jagels, A. Krogh, J. McLean, S. Moule, L. Murphy, K. Oliver, J. Osborne, M. A. Quail, M. A. Rajandream, J. Rogers, S. Rutter, K. Seeger, J. Skelton, R. Squares, S. Squares, J. E. Sulston, K. Taylor, S. Whitehead, and B. G. Barrell. 1998. Deciphering the biology of Mycobacterium tuberculosis from the complete genome sequence. Nature 393537-544. [DOI] [PubMed] [Google Scholar]
- 9.Collin, V., E. Issakidis-Bourguet, C. Marchand, M. Hirasawa, J. M. Lancelin, D. B. Knaff, and M. Miginiac-Maslow. 2003. The Arabidopsis plastidial thioredoxins: new functions and new insights into specificity. J. Biol. Chem. 27823747-23752. [DOI] [PubMed] [Google Scholar]
- 10.Darby, N. J., and T. E. Creighton. 1995. Characterization of the active site cysteine residues of the thioredoxin-like domains of protein disulfide isomerase. Biochemistry 3416770-16780. [DOI] [PubMed] [Google Scholar]
- 11.Dyson, H. J., M. Jeng, L. L. Tennant, I. Slaby, M. Lindell, D. Cui, S. Kuprin, and A. Holmgren. 1997. Effect of buried charged groups on cysteine thiol ionization and reactivity in Escherichia coli thioredoxin: Structural and functional characterization of mutant of Asp 26 and Lys 57. Biochemistry 362622-2636. [DOI] [PubMed] [Google Scholar]
- 12.Fahey, R. C. 2001. Novel thiols of prokaryotes. Annu. Rev. Microbiol. 55333-356. [DOI] [PubMed] [Google Scholar]
- 13.Gasteiger, E., A. Gattiker, C. Hoogland, I. Ivanyi, R. D. Appel, and A. Bairoch. 2003. ExPASy: the proteomics server for in-depth protein knowledge and analysis. Nucleic Acids Res. 313784-3788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Gleason, F. K., and A. Holmgren. 1988. Thioredoxin and related proteins in procaryotes. FEMS Microbiol. Rev. 4271-297. [DOI] [PubMed] [Google Scholar]
- 15.Guzman, L. M., D. Belin, M. J. Carsonand, and J. Beckwith. 1995. Tight regulation, modulation, and high-level expression by vectors containing the arabinose PBAD promoter. J. Bacteriol. 1774121-4130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hall, G., M. Shah, P. A. McEwan, C. Laughton, M. Stevens, A. Westwell, and J. Emsley. 2006. Structure of Mycobacterium tuberculosis thioredoxin C. Acta Crystallogr. D 621453-1457. [DOI] [PubMed] [Google Scholar]
- 17.Hanahan, D. 1983. Studies on transformation of Escherichia coli with plasmids. J. Mol. Biol. 166557-580. [DOI] [PubMed] [Google Scholar]
- 18.Holmgren, A. 1979. Thioredoxin catalyzes the reduction of insulin disulfides by ithiothreitol and dihydrolipoamide. J. Biol. Chem. 2549627-9632. [PubMed] [Google Scholar]
- 19.Holmgren, A., and M. Björnstedt. 1995. Thioredoxin and thioredoxin reductase. Methods Enzymol. 252199-208. [DOI] [PubMed] [Google Scholar]
- 20.Holmgren, A. 2000. Antioxidant function of thioredoxin and glutaredoxin systems. Antioxid. Redox Signal. 2811-820. [DOI] [PubMed] [Google Scholar]
- 21.Huber, H. E., M. Russel, P. Model, and C. C. Richardson. 1986. Interaction of mutant thioredoxins of Escherichia coli with the gene 5 protein of phage T7. The redox capacity of thioredoxin is not required for stimulation of DNA polymerase activity. J. Biol. Chem. 26115006-15012. [PubMed] [Google Scholar]
- 22.Jaeger, T., H. Budde, L. Flohe, U. Menge, M. Singh, M. Trujillo, and R. Radi. 2004. Multiple thioredoxin-mediated routes to detoxify hydroperoxides in Mycobacterium tuberculosis. Arch. Biochem. Biophys. 423182-191. [DOI] [PubMed] [Google Scholar]
- 23.Joelson, T., B. M. Sjöberg, and H. Eklund. 1990. Modifications of the active center of T4 thioredoxin by site-directed mutagenesis. J. Biol. Chem. 2653183-3188. [PubMed] [Google Scholar]
- 24.Krause, G., and A. Holmgren. 1991. Substitution of the conserved tryptophan 31 in Escherichia coli thioredoxin by site-directed mutagenesis and structure-function analysis. J. Biol. Chem. 2664056-4066. [PubMed] [Google Scholar]
- 25.Manganelli, R., M. I. Voskuil, G. K. Schoolnik, E. Dubanu, M. Gomez, and I. Smith. 2002. Role of extracytoplasmic function σ factor σH in Mycobacterium tuberculosis global gene expression. Mol. Microbiol. 45365-374. [DOI] [PubMed] [Google Scholar]
- 26.Mark, D. F., and C. C. Richardson. 1976. Escherichia coli thioredoxin: a subunit of bacteriophage T7 DNA polymerase. Proc. Natl. Acad. Sci. USA 73780-784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Martin, J. L. 1995. Thioredoxin—a fold for all reasons. Structure 3245-250. [DOI] [PubMed] [Google Scholar]
- 28.Mossner, E., M. Huber-Wunderlich, A. Rietsch, J. Beckwith, R. Glockshuber, and F. Åslund. 1999. Importance of redox potential for the in vivo function of the cytoplasmic disulfide reductant thioredoxin from Escherichia coli. J. Biol. Chem. 27425254-25259. [DOI] [PubMed] [Google Scholar]
- 29.Mossner, E., M. Huber-Wunderlich, and R. Glockshuber. 1998. Characterization of Escherichia coli thioredoxin variants mimicking the active sites of other thiol/disulfide oxidoreductases. Protein Sci. 71233-1244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ortenberg, R., S. Gon, A. Porat, and J. Beckwith. 2004. Interactions of glutaredoxins, ribonucleotide reductase, and components of the DNA replication system of Escherichia coli. Proc. Natl. Acad. Sci. USA 1017439-7444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Prinz, W. A., F. Aslund, A. Holmgren, and J. Beckwith. 1997. The role of the thioredoxin and glutaredoxin pathways in reducing protein disulfide bonds in the Escherichia coli cytoplasm. J. Biol. Chem. 27215661-15667. [DOI] [PubMed] [Google Scholar]
- 32.Rawat, M., and Y. Av-Gay. 2007. Mycothiol-dependent proteins in actinomycetes. FEMS Microbiol. Rev. 31278-292. [DOI] [PubMed] [Google Scholar]
- 33.Rawat, M., G. L. Newton, M. Ko, G. J. Martinez, R. C. Fahey, and Y. Av-Gay. 2002. Mycothiol-deficient Mycobacterium smegmatis mutants are hypersensitive to alkylating agents, free radicals, and antibiotics. Antimicrob. Agents Chemother. 463348-3355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Rost, J., and S. Rapoport. 1964. Reduction-potential of glutathione. Nature 201185. [DOI] [PubMed] [Google Scholar]
- 35.Russel, M., P. Model, and A. Holmgren. 1990. Thioredoxin or glutaredoxin in Escherichia coli is essential for sulfate reduction but not for deoxyribonucleotide synthesis. J. Bacteriol. 1721923-1929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Sareen, D., G. L. Newton, R. C. Fahey, and N. A. Buchmeier. 2003. Mycothiol is essential for growth of Mycobacterium tuberculosis Erdman. J. Bacteriol. 1856736-6740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Sassetti, C. M., and E. J. Rubin. 2003. Genetic requirements for mycobacterial survival during infection. Proc. Natl. Acad. Sci. USA 10012989-12994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Shinnick, T. M., H. King, and F. D. Quinn. 1995. Molecular biology, virulence, and pathogenicity of mycobacteria. Am. J. Med. Sci. 30992-98. [DOI] [PubMed] [Google Scholar]
- 39.Stewart, E. J., F. Åslund, and J. Beckwith. 1998. Disulfide bond formation in the Escherichia coli cytoplasm: an in vivo role reversal for the thioredoxins. EMBO J. 175543-5550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Studier, F. W., and B. A. Moffatt. 1986. Use of bacteriophage T7 RNA polymerase to direct selective high-level expression of cloned genes. J. Mol. Biol. 189113-130. [DOI] [PubMed] [Google Scholar]
- 41.Wunderlich, M., and R. Glockshuber. 1993. Redox properties of protein disulfide isomerase (DsbA) from Escherichia coli. Protein Sci. 2717-726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Zapun, A., D. Missiakas, S. Raina, and T. E. Creighton. 1995. Structural and functional characterization of DsbC, a protein involved in disulfide bond formation in Escherichia coli. Biochemistry 345075-5089. [DOI] [PubMed] [Google Scholar]
- 43.Zhang, Z., P. J. Hillas, and P. R. Ortiz de Montellano. 1999. Reduction of peroxides and dinitrobenzenes by Mycobacterium tuberculosis thioredoxin and thioredoxin reductase. Arch. Biochem. Biophys. 36319-26. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.