Abstract
Proteins of the WXG100 family represent the prototypical substrates of bacterial type VII secretion systems that typically encompass 100 residues, lack canonical signal peptides, and form helix-turn-helix hairpin structures with WXG positioned in the turn element. Bacillus anthracis encodes six WXG100 proteins, herein referred to as EsxB, EsxL, EsxP, EsxQ, EsxV, and EsxW. With the exception of EsxB, B. anthracis proteins harbor C-terminal extensions that are appended to canonical WXG domains. When cultured in liquid broth, B. anthracis secretes two substrates, EsxB and EsxW, into the extracellular environment. EsxB is required for the stability and secretion of EsxW; however, EsxW is dispensable for EsxB secretion. In agreement with the hypothesis that EsxB binding to substrates promotes recognition and secretion by the type VII pathway, EsxB is reported to interact with EsxB and EsxW. Unlike deletions in mycobacterial EsxB, deletion of five N- or C-terminal residues does not affect the ability of mutant B. anthracis EsxB to travel the type VII pathway and initiate secretion of EsxW. Translational fusion of ubiquitin to the N or C terminus of EsxB also had no effect, while ubiquitin insertion into the center turn abrogated secretion. Anthrax-infected guinea pigs mounted humoral immune responses to EsxB, EsxP, and EsxW, which suggests that B. anthracis activates the type VII secretion pathway during infection.
Bacterial pathogens transport polypeptides across their envelope as a mechanism of survival during infection (14). The bulk of these proteins are secreted by the Sec pathway and provide housekeeping functions such as hydrolysis of complex macromolecules and nutrient uptake (17, 21). A subset of these proteins, typically designated effectors, targets dedicated host pathways to circumvent innate or acquired immune responses during infection. Often, such effectors are secreted via a dedicated secretion system (14, 20). In contrast to the general secretory (Sec) pathway, alternate secretion systems are dispensable for bacterial growth in culture but contribute important virulence functions once bacteria enter their host. The recently recognized type VII secretion system (T7SS) appears to fulfill the aforementioned criteria (1). Genes for type VII secretion are found in actinobacteria and firmicutes but are conspicuously lacking from the genomes of gram-negative bacteria (19). The T7SS includes one or more ATPase-containing FtsK-SpoIIIE-like domains (FSDs). These membrane proteins may be involved in protein secretion, and their designated substrates belong to the WXG100 family of proteins (Fig. 1A). Prompted by the close proximity and clustering of WXG100 and FSD genes in bacterial chromosomes, Pallen was the first to propose a new secretion system, now designated type VII (19). This prediction has been validated for Mycobacterium tuberculosis (12, 23, 29) and Staphylococcus aureus (7).
FIG. 1.
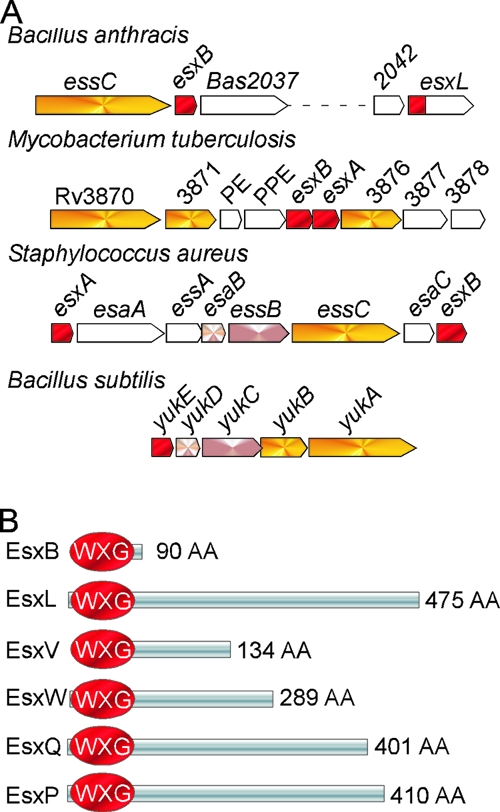
Schematic representations of ESAT-6 genetic loci and B. anthracis WXG100 proteins. (A) Clusters encoding known and putative T7SSs in the sequenced genomes of B. anthracis, M. tuberculosis, S. aureus, and Bacillus subtilis. WXG100 factors (red) encoded in the vicinity of FtsK-SpoIIIE protein homologues (yellow) are shown. (B) B. anthracis proteins with predicted WXG100 domains are depicted in red. The proteins have been named EsxB, EsxL, EsxP, EsxQ, EsxV, and EsxW and are encoded by genes BAS2036, BAS2043, pXO1-98, BAS2159, BAS1183, and BAS1184, respectively. AA, amino acids.
Mycobacterial ESAT-6 and CFP-10 represent the prototypic substrates of the type VII pathway (3, 19, 28). Both proteins encompass approximately 100 amino acids, lack canonical signal peptides, and dimerize to form a four-helix bundle complex (24, 25). The sequence motif WXG is positioned in the middle of this domain, providing the characteristic turn in the α-helical structure and the eponym of the WXG100 protein family. The attributes of ESAT-6 and CFP-10 are clearly shared by EsxA and EsxB, two ESAT-6-like proteins of S. aureus. Although the structure of these proteins has not yet been solved, each polypeptide is predicted to adopt a helical hairpin structure. In S. aureus, the type VII pathway has been termed ESS for ESAT-6 secretion system; it is important for host-pathogen interaction and establishment of persistent infections (6, 7).
M. tuberculosis encodes 23 ESAT-6 homologues, 11 of which are encoded within five gene clusters that also specify large soluble and membrane-bound ATPases with two or more FSDs (11). Two of these clusters, ESX-1, which includes ESAT-6 and CFP-10, as well as ESX-5, are known to be important for mycobacterial virulence (2, 12, 22, 29). The type VII pathway also provides for the secretion of non-ESAT-6-like proteins (9, 16). In silico predictions for type VII substrates in bacterial genomes has thus far not been reported, presumably because non-WXG100 substrates appear to lack sequence similarity. Here, we examined the genome of Bacillus anthracis for the presence of WXG100 proteins and identified six putative substrates for the type VII pathway. Remarkably, five WXG100 proteins harbor large C-terminal domains appended to the WXG domain. Bacilli secrete some of these polypeptides during growth in liquid broth or during anthrax infection.
MATERIALS AND METHODS
Growth medium.
Bacilli cultures were grown overnight in Luria broth with 0.5% glucose and 0.85% sodium bicarbonate (when indicated) at 37°C and diluted in fresh medium at 37°C. Antibiotics were added to cultures for plasmid selection as follows: 100 μg/ml ampicillin and 50 μg/ml kanamycin for E. coli strains and 20 μg/ml kanamycin and 10 μg/ml chloramphenicol for B. anthracis.
Bacterial strains and plasmids.
B. anthracis Sterne 34F2 (30) was used as a parent strain. Plasmid pTS1 with a thermosensitive replicon was used for allelic replacement (15). Plasmid pOS1 was used for complementation studies, as well as for expression of EsxB truncated variants and ubiquitin fusions (26). Plasmids used in this study are listed in Table 1.
TABLE 1.
Plasmids used in this study
| Plasmid | Relevant description or genotypea | Reference or source |
|---|---|---|
| pOS1 | Multicopy shuttle vector | 26 |
| pTS1 | Temperature-sensitive shuttle vector | 15 |
| pKKUb-βGal | Ub14-Sacc | 4 |
| pKK172 | pTS1 carrying 1-kbp 3′ esxB-Km cassette and 1-kbp 5′ esxB | This study |
| pGG12 | pTS1 carrying 1-kbp 3′ esxW and 1-kbp 5′ esxW | This study |
| pGG42 | pTS1 carrying 1-kbp 3′ essC and 1-kbp 5′ essC | This study |
| pGG119 | pOS1 carrying pesxB and Ub-EsxB1-90 | This study |
| pGG120 | pOS1 carrying pesxB and EsxB1-90-Ub | This study |
| pGG141 | pOS1 carrying pesxB and Ub-EsxB1-85 | This study |
| pGG142 | pOS1 carrying pesxB and Ub-EsxB11-90 | This study |
| pGG165 | pOS1 carrying pesxB and Ub-EsxB1-81 | This study |
| pGG167 | pOS1 carrying pesxB and Ub-EsxB6-90 | This study |
| pGG196 | pOS1 carrying pesxB and EsxB1-44-Ub-EsxB45-90 | This study |
| pGG256 | pOS1 carrying pesxB and EsxB6-90 | This study |
| pGG258 | pOS1 carrying pesxB and EsxB11-90 | This study |
| pGG261 | pOS1 carrying pesxB and EsxB1-81 | This study |
| pGG262 | pOS1 carrying pesxB and EsxB1-85 | This study |
| pGADT7 | GAL4AD768-881 | Clontech |
| pGBKT7 | GAL4 DNA-BD1-147 | Clontech |
| pGG153 | pGADT7-GAL4 DNA-BD::esxW | This study |
| pGG155 | pGADT7-GAL4 DNA-BD::esxB | This study |
| pGG156 | pGADT7-GAL4AD::esxB | This study |
| pGG157 | pGADT7-GAL4AD::esxW | This study |
| pGG158 | pGADT7-GAL4AD::esxV | This study |
| pGG159 | pGBKT7-GAL4BD::esxV | This study |
| pGG182 | pGADT7-GAL4AD::esxL | This study |
| pGG183 | pGADT7-GAL4AD::esxP | This study |
| pGG184 | pGBKT7-GAL4BD::esxL | This study |
| pGG185 | pGBKT7-GAL4BD::esxP | This study |
| pGG157 | pGADT7-GAL4AD::esxW | This study |
| pEB199 | pOS1 carrying pesxB and EsxB | This study |
| pEB255 | pOS1 carrying pesxB and EsxB-His | This study |
| pUTE618 | Kan cassette | T. Koehler |
Numbers in subscripts indicate first and last amino acid encoded by DNA fragment cloned.
Cloning procedures for allelic replacement.
For allelic replacement, bacillus template DNA was isolated by lysing cells with 10 mg/ml lysozyme and extracted using a Wizard Genomic DNA purification kit (Promega). Using primer pairs listed in Table S1 in the supplemental material, 5′ and 3′ 1-kbp flanking sequences of esxB, esxL, esxW, and essC were PCR amplified from B. anthracis Sterne template DNA. PCRs were performed with Pfu DNA polymerase (Stratagene). Ligation products were transformed into E. coli K1077 (dam dcm mutant), and purified (nonmethylated) plasmid DNA was transformed into B. anthracis following a previously developed protocol (27). Transformants were selected on LB agar with chloramphenicol (pTS1) antibiotics at 30°C (permissive temperature). Allelic exchange was induced with a temperature shift to 43°C, as described previously (10). B. anthracis esxL, esxW, and essC variants lacked the entire coding sequence of each of the respective genes. In case of esxB, the coding region of the gene was replaced with the kanamycin resistance cassette of pUT618 containing the aphA3 gene (gift of T. Koehler). Nucleic acid sequences of wild-type and mutant alleles were verified by DNA sequencing.
Cloning procedures for complementation studies with esxB and esxB protein hybrids.
Chromosomal DNA isolated from the Sterne strain was used as a template to amplify all esxB fragments along with flanking sequences. Primers used for the study are listed in Table S1 in the supplemental material. Plasmid pKKUb-βGal (where Gal is galactosidase) served as a template for amplification of the ubiquitin-encoding gene (4). All protein hybrids and truncated variants were cloned into the pOS1 plasmid carrying the promoter region of esxB (pesxB). For complementation studies using esxB with and without a histidine tag, clones were generated by amplifying DNA along with pesxB. Primers hybridizing at the 3′ end of the esxB gene were designed to include or not include six codons for histidine before the stop codon of the predicted open reading frame.
Yeast two-hybrid analysis.
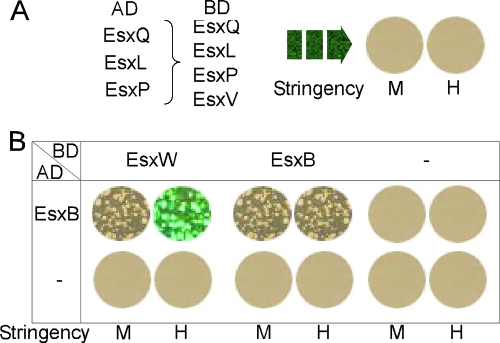
A Matchmaker Two-Hybrid System 3 (Clontech) was used for all yeast two-hybrid analysis. Bait and prey genes were expressed as a fusion to the GAL4 DNA-binding domain (DNA-BD) and GAL4 activation domain (AD), using plasmids pGBKT7 and pGADT7, respectively. All six putative WXG100 encoding genes (BAS1183, BAS1184, BAS2036, BAS2043, BAS2159, and pXO1-98) were amplified by PCR using genomic DNA of B. anthracis strain Sterne as a template and inserted into the multiple cloning sites of plasmids pGBKT7 and pGADT7. Interactions between bait and prey proteins bring the DNA-BD and AD into proximity and may lead to the transcriptional activation of three reporter genes in Saccharomyces cerevisiae strain AH109—ADE2, HIS3, and MEL1 (or lacZ)—under the control of distinct GAL4 upstream activating sequences (UAS) and TATA boxes. Yeast transformation was carried out as described previously (13). Yeast transformants were selected and cultivated on SD synthetic medium (2% glucose and 0.67% yeast nitrogen base without amino acids) supplemented with essential amino acids and nucleotides. The yeast strain AH109 (MATa ura3-52 his3-200 ade2-101 lys2-801 trp1-901 leu2-3,112 gal4Δ gal80Δ cyhr2 LYS2::GAL1UAS-HIS3TATA-HIS3 URA3::GAL1UAS-GAL1TATA-lacZ [Clontech]) was used for two-hybrid analyses. To screen for protein interactions, transformants were selected on SD plates lacking Leu and Trp (low stringency); SD plates lacking His, Leu, and Trp (medium stringency); and SD plates lacking Ade, His, Leu, and Trp and supplemented with X-α-Gal (where X-α-Gal is 5-bromo-4-chloro-3-indolyl-α-D-galactopyranoside; high stringency) at 30°C for 2 days. Positive interactions can be identified with a simple blue/white colony screen by adding X-α-Gal, the substrate of α-galactosidase, directly to the selection plate.
Culture fractionation and Western blotting.
B. anthracis strains were grown overnight in LB medium with or without 0.5% glucose and 0.8% sodium bicarbonate as indicated. Overnight cultures were diluted 1:100 in fresh medium and grown to an optical density of 3 at 600 nm (A600). Total proteins in the cell culture were obtained by precipitating 1 ml of the culture with 7.5% trichloroacetic acid (TCA), thus lysing the bacilli. To assay for protein secretion in the medium, 3 ml of the culture was spun for 5 min at 6000 × g. Proteins in 1 ml of supernatant were precipitated with 7.5% TCA. All TCA precipitates were washed with ice-cold acetone, solubilized in 50 μl of 0.5 M Tris-HCl (pH 8.0)-4% sodium dodecyl sulfate (SDS), and heated at 90°C for 10 min. Proteins were separated by SDS-polyacrylamide gel electrophoresis (PAGE) and transferred to polyvinylidene difluoride membrane for immunoblot analysis with appropriate rabbit polyclonal antibodies. Immunoreactive signals were revealed by using a secondary antibody coupled to horseradish peroxidase and by chemiluminescence.
Protein purification.
B. anthracis Sterne producing EsxB-His was grown in LB medium with glucose to an optical density of 3 at 600 nm. The culture (250 ml) was spun, and proteins in the supernatant were precipitated with ammonium sulfate, recovered by centrifugation, and dialyzed against buffer A (50 mM Tris-HCl buffer, pH 7.5, 10 mM imidazole). Bacilli from the original 250-ml culture were washed twice in fresh medium, suspended in buffer A, and lysed with a bead beater. Insoluble material in the supernatant and cellular fractions was removed by ultracentrifugation (for 2 h at 100,000 × g). Soluble proteins were purified over Ni-nitrilotriacetic acid beads and eluted with an imidazole step gradient (10 to 500 mM). Proteins in the elution fractions were separated by SDS-PAGE and visualized by staining the gel with Coomassie blue prior to mass measurement by mass spectrometry.
B. anthracis infection of guinea pigs.
To examine whether bacillus WXG100 proteins are expressed during anthrax disease, B. anthracis strain Ames infection of Guinea pigs was performed. Briefly, B. anthracis spores were prepared by growing the Ames strain in sporulation medium for 6 days to induce sporulation. Spores were treated at 65°C for 60 min to kill vegetative cells, and the spore suspension was examined for colony formation. A dilution of 10 to 100 spores in 0.1 ml of phosphate-buffered saline was injected subcutaneously into the right hind leg of male, 8-week-old guinea pigs, and progression to acute disease was monitored over a 14-day time period. At day 5 and day 10, ciprofloxacin was administered to ensure animal survival. At day 14, animals were bled, and serum was isolated. Serum was examined by enzyme-linked immunosorbent assay (ELISA) for immunoglobulin G (IgG) titers with specific antigen-binding activity. Animal experiments were performed in accordance with institutional guidelines following experimental protocol review and approval by the Institutional Biosafety Committee and the Institutional Animal Care and Use Committee.
RESULTS
WXG100 proteins encoded by the B. anthracis genome.
The genome sequence of B. anthracis strain Sterne was examined for the presence of gene products with WXG100 motifs, using mycobacterial EsxA (ESAT-6), EsxB (CFP-10), and staphylococcal EsxA and EsxB as queries in BLAST searches. BAS2036 was identified as a close homolog of mycobacterial EsxB (29% identity) and staphylococcal EsxA (26% identity); BAS2036 is here designated B. anthracis EsxB, a 90-residue polypeptide with a WXG100 motif (also referred to as domain of unknown function DUF909; Pfam accession number PF06013). BAS2035, a gene positioned immediately upstream of EsxB, encodes an FtsK-SpoIIIE-type ATPase. These enzymes are known to be involved in type VII secretion of Esx proteins in either mycobacteria or staphylococci. BAS2035 was therefore designated B. anthracis EssC. Four additional products of genes on the B. anthracis Sterne chromosome contained WXG100 (DUF909) domains. BAS2043 (esxL) is located in close proximity to esxB and essC, whereas BAS2159 (esxQ) and two adjacent genes, BAS1183 (esxV) and BAS1184 (esxW), are positioned elsewhere on the chromosome of B. anthracis. Identification of these gene products as members of the WXG100 family was facilitated by using EsxB (BAS2036) as the query in BLAST searches. The reason for this is that Bacillus family members are more closely related than WXG100 members of other microbes (Table 2). EsxV shares little similarity with other WXG100 proteins, and only searches with the EsxW query identified the esxV product. Bioinformatics analysis of B. anthracis virulence plasmids, pXO1 and pXO2, revealed a fifth WXG100 protein, EsxP (pXO1-98). All five WXG100 proteins of B. anthracis (EsxL, EsxP, EsxQ, EsxV, and EsxW) harbor large C-terminal domains, a feature not observed in either mycobacteria or staphylococci (Fig. 1B). All five proteins appear to be a signature of the Bacillus cereus group that encompasses close relatives of B. anthracis. Of note, the C-terminal domain of EsxL shares homology with a non-WXG100 protein of staphylococci encoded downstream of the type VII gene cluster. Additional database searches identified some degree of homology between distinct subdomains of EsxL and EsxV and the C-terminal domains of several gram-negative outer membrane proteins that possess a Pfam Haemagg_act domain (http://pfam.sanger.ac.uk/family?acc = PF05860) (see Table S2 in the supplemental material). BLAST searches with the C-terminal domain of pX01-98 revealed homology to the extreme C terminus of several large and secreted gram-negative proteins (possibly outer membrane proteins) containing several YD repeats (see Table S2 in the supplemental material). In Neisseriaceae, a small gene appears to encode a protein that consists exclusively of this C terminus domain and for which BLink search in the reverse direction confirms the homology with pXO1-98. Clearly, more work needs to be done to establish the function of WXG100 proteins of B. anthracis.
TABLE 2.
BLAST searches for WXG100 proteins in B. anthracis
| Locus tag in databanka | Protein nameb | Predicted ORF (aa)c | % Identity (no. of shared aa/total no. of aa) | E value |
|---|---|---|---|---|
| BAS2036 | EsxBBa | 90 | 100 (90/90) | 6 e-47 |
| BAS2043 | EsxLBa | 475 | 39 (35/88) | 1 e-13 |
| BAS2159 | Ba-EsxQ | 401 | 39 (35/88) | 6 e-11 |
| Rv3874 | CFP-10/EsxBMt | 100 | 29 (26/87) | 7 e-09 |
| BAS1184 | EsxWBa | 289 | 33 (28/84) | 2 e-06 |
| pXO1-98 | EsxPBa | 410 | 32 (29/88) | 2 e-06 |
| SAV0282 | EsxASa | 97 | 26 (24/91) | 2 e-05 |
| BAS1183 | EsxVBa | 134 | 33 (11/33) | 9.8 |
BAS, pXO1, SAV and Rv designate gene numbers in the genomes of B. anthracis Sterne, pXO1, S. aureus Mu50, and M. tuberculosis H37Rv, respectively.
Subscripts identifying proteins are as follows: Ba, B. anthracis; Mt, M. tuberculosis; and Sa, S. aureus.
ORF, open reading frame; aa, amino acid.
B. anthracis secretes EsxB and EsxW.
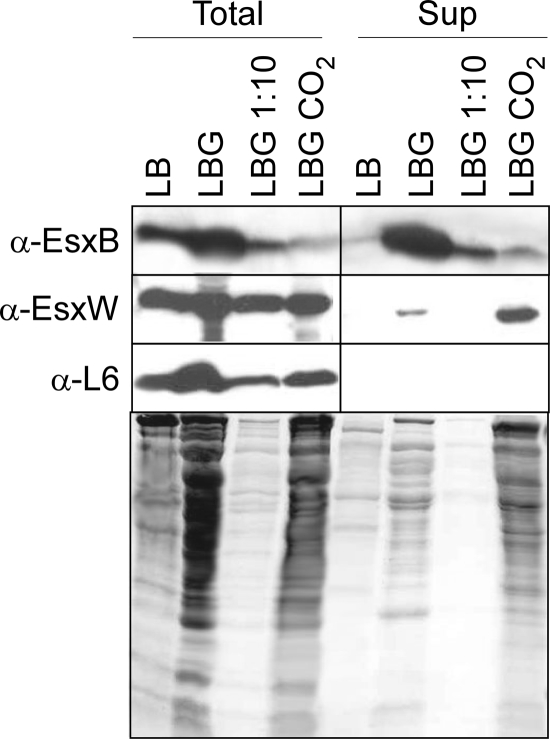
EsxB, EsxL, EsxP, EsxV, and EsxW were expressed in Escherichia Coli, and recombinant proteins with fused N-terminal affinity tags (six-histidine) were purified by affinity chromatography. Purified proteins were used to immunize rabbits and thereby raise specific serum antibodies. B. anthracis Sterne was grown in LB broth supplemented with glucose (0.5%) or sodium bicarbonate (0.85%) to late log phase. Proteins in entire cultures were precipitated with TCA, sedimented by centrifugation, separated by SDS-PAGE, and analyzed by immunoblotting with specific antibodies (Fig. 2). In laboratory broth, bacilli expressed EsxB and EsxW in both the presence and absence of glucose or sodium bicarbonate, known inducers of capsule and toxin genes, suggesting that expression of the Bacillus Esx pathway is not coordinately controlled by the atxA master regulator. Immunoblotting failed to detect EsxL, EsxP, or EsxV in Bacillus extracts of early-, mid-, or late-exponential-phase cultures, in agreement with the conjecture that these WXG100 genes are not expressed under laboratory conditions (data not shown). Addition of glucose to laboratory cultures increased Bacillus replication about 10-fold, generating greater abundance of Coomassie blue-stained proteins on SDS-PAGE (Fig. 2, lower panel) and of immunoreactive signals for ribosomal protein L6. Addition of sodium bicarbonate to LB glucose cultures reduced EsxB expression 10-fold (Fig. 2); however, the abundance of EsxW was unaffected. As expected, immunoblot analysis of cultures derived from esxB and esxW mutant strains failed to detect the corresponding WXG100 proteins.
FIG. 2.
Detection and localization of EsxB and EsxW by Western blotting. Cultures of B. anthracis Sterne were grown to A600 nm of 3 in either Luria broth alone (LB) or complemented with glucose 0.5% (LBG) or glucose 0.5% and sodium bicarbonate 0.85% (LBG NaCO2). A volume of 1 ml was precipitated with TCA to examine total proteins in the culture (Total) fraction, while 1 ml of supernatant was recovered after centrifugation of a 3-ml volume culture. Proteins in the supernatant (Sup) fraction were submitted to TCA precipitation. TCA pellets were washed with acetone and solubilized in SDS-PAGE loading buffer before separation on a 15% SDS-PAGE gel. The gels were either stained with Coomassie blue or used for Western blot analysis. The presence of EsxB and EsxW was revealed by immunoblotting using polyclonal antibodies. The ribosomal protein L6 was used as a nonsecreted cytosolic marker. The same sample volumes were loaded in the gels. For quantitative comparison, samples grown in LBG were loaded undiluted and diluted 1:10. α, anti.
To measure secretion, cultures were centrifuged, separating extracellular medium (supernatant) from the bacterial sediment (pellet). Proteins were precipitated with TCA, separated by SDS-PAGE, and analyzed by immunoblotting (Fig. 2). B. anthracis Sterne secreted EsxB into the extracellular medium when cultures were supplemented with 0.5% glucose, whereas the addition of sodium bicarbonate, an inducer of toxin and capsule genes, had little effect (Fig. 2). Glucose also enabled bacilli to secrete EsxW; however, EsxW secretion was increased even further when bacilli were grown in LB medium supplemented with both glucose and sodium bicarbonate (Fig. 2). Under laboratory conditions, EsxW secretion ranges from 40% to 10% of the total amount of EsxW, considerably less than the level of secretion measured for EsxB (about 60%). More efficient secretion of EsxW may require environmental cues in vivo, i.e., during anthrax infection. If so, the cues and their molecular nature have not yet been recapitulated in vitro.
Host immune responses to WXG100 proteins during anthrax infection.
Immunoblot analysis failed to detect EsxL, EsxP, or EsxV in extracts of bacilli that had been grown under laboratory conditions. We wondered whether these proteins are produced during infection and, if so, whether infected hosts mount immune responses against B. anthracis WXG100 proteins. Guinea pigs were infected with a lethal dose of spores of B. anthracis strain Ames and then treated with antibiotic to clear the infection (without antibiotic treatment, animals die within 4 days, a period of time that is too short to observe humoral immune responses). Blood was collected from infected and control (mock infected) animals on days 0 and 14 following inoculation with B. anthracis. The presence of specific IgG in serum samples on day 14 was tested by ELISA, using purified immobilized EsxB, EsxL, EsxP, EsxV, and EsxW and protective antigen (PA) as a control. Data in Fig. 3 show that animals infected with B. anthracis strain Ames developed IgG-type antibodies against EsxB, EsxP, and EsxW (Fig. 3). As expected, anthrax-infected guinea pigs mounted humoral immune responses against PA, the secreted translocation factor of lethal and edema toxins. Humoral immune responses against EsxL and EsxV were not detected, suggesting that the immune system of anthrax-infected guinea pigs had recognized some, but not all, members of the WXG100 family.
FIG. 3.
Guinea pigs infected with B. anthracis strain Ames generate EsxB, EsxP, and EsxW IgG-specific antibodies. Spores, suspended in 0.1 ml of sterile water, were injected subcutaneously into the loose subdermal tissue of the hind leg of animals. At 48-h postinfection animals were treated with antibiotics to prevent the development of an acute disease. Serum was collected 14 days postinfection and analyzed for the presence of EsxB-, EsxP-, EsxW-, and PA-reactive antibodies; IgG titers were determined in triplicate by ELISA and are reported as absorbance values at 405 nm.
EsxB is not processed upon secretion.
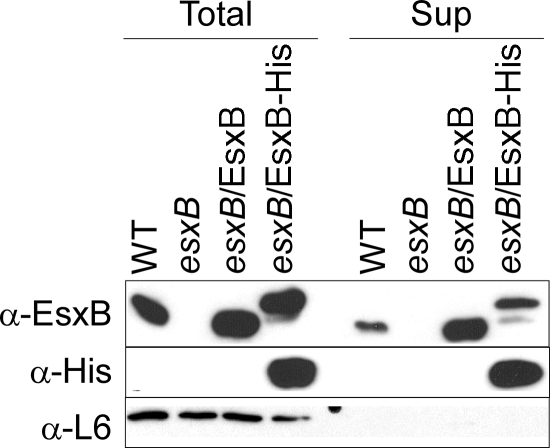
N-terminal sequencing of ESAT-6 and CFP-10 from culture filtrates of M. tuberculosis demonstrated that these proteins lack their N-terminal methionine residues but are otherwise not cleaved (5, 28). This finding led to the earlier, erroneous conclusions that the presence of ESAT-6 and CFP-10 in the extracellular milieu could be the result of regulated bacterial lysis. We wondered whether B. anthracis secretion of EsxB may be accompanied by a posttranslational modification of the polypeptide chain. To test this, wild-type esxB or esxB with an appended 3′ coding sequence for a six-His affinity tag were cloned on plasmids and expressed from the native esxB promoter (EsxB+ and EsxB-His, respectively) encompassed in 5′ upstream sequences. Transformation of plasmids encoding EsxB+ or EsxB-His into esxB mutant bacilli restored EsxB expression and secretion. Immunoblotting with EsxB or His-specific reagents revealed that EsxB-His migrated more slowly than wild-type EsxB and that secreted EsxB-His had retained its C-terminal His tag (Fig. 4). EsxB-His was purified by affinity chromatography on Ni-nitrilotriacetic acid Sepharose using lysate of bacilli (cytoplasmic EsxB-His) as well as conditioned culture medium (secreted EsxB-His). EsxB-His purified from both compartments was analyzed by matrix-assisted laser desorption ionization-time of flight mass spectrometry. Predominant ions with m/z 10,774.3 and 10,775.1 were identified for cytoplasmic and secreted EsxB-His, respectively. Assuming that the N-terminal methionine of B. anthracis EsxB is also removed prior to secretion, the calculated mass of full-length EsxB-His is 10,780.15. As the difference between observed and calculated masses of EsxB-His is within the error rate for matrix-assisted laser desorption ionization-time of flight analysis, these data suggest that EsxB-His is secreted as a full-length polypeptide and without posttranslational modifications.
FIG. 4.
EsxB produced in trans is secreted with or without a C-terminal histidine tag. Wild-type or esxB mutant bacilli with no vector, vector carrying esxB (esxB/EsxB+) or a modified esxB with six histidine residues (esxB/EsxB-His) were grown to the same optical density (A600 nm of 3) in LB medium with 0.5% glucose (LBG). Cells cultures (Total) or an equivalent volume of culture supernatant (Sup) were precipitated with TCA, separated by SDS-PAGE, and detected by immunoblotting with specific antibodies (anti-EsxB, anti-His, and anti-L6 as a loading control). Extracts from cells carrying plasmids expressing esxB were diluted 10 times compared to extracts from wild-type and esxB mutant strains. α, anti; WT, wild type.
Genetic requirements for EsxB and EsxW secretion.
M. tuberculosis Rv3870 and Rv3871 (Snm1/Snm2) as well as S. aureus EssC encode FtsK-SpoIIIE-type ATPases (Fig. 1) that are required for stable expression and secretion of EsxA and EsxB. BAS2035, located immediately adjacent to esxB, encodes a 1,342-amino-acid residue membrane protein and close B. anthracis homolog of EssC with three FSDs. Allelic replacement was used to delete the coding region of BAS2035, and the resulting essC mutant strain was assayed for EsxB and EsxW secretion. Surprisingly, essC was dispensable for B. anthracis in vitro secretion of EsxB and EsxW (Fig. 5). The B. anthracis genome encodes three additional FstK-SpoIIIE ATPases with sequence similarity to mycobacterial Snm1/Snm2 and staphylococcal EssC. A fifth B. anthracis ATPase, FstK-SpoIIIE, is required for chromosome partitioning during sporulation. We think it is plausible that more than one FtsK-SpoIIIE-type ATPase can fuel WXG100 protein secretion; if so, genetic redundancy could not be addressed with mutations in a single gene but likely requires future characterization of FtsK-SpoIIIE-type ATPases by multiple allelic replacements.
FIG. 5.
Genetic requirements for EsxB and EsxW secretion. Wild-type and isogenic mutants esxB, esxW, esxL, and essC were grown to an A600 nm of 3 in LB medium with glucose (LBG) or LB medium with glucose-sodium bicarbonate (LBG CO2). One-milliliter cell cultures (Total) or equivalent volumes of culture supernatants (Sup) were precipitated with TCA. Proteins in extracts were solubilized in SDS-PAGE sample buffer prior to separation on 15% acrylamide gels and transfer to a polyvinylidene difluoride membrane for immunoblotting with specific antibodies (anti-EsxB, anti-EsxW, and anti-L6 as a loading control). Immunoreactive signals were revealed using a secondary antibody coupled to horseradish peroxidase and chemiluminescence and captured on film. The intensity of bands corresponding to EsxB and EsxW immune species was measured by scanning the films. All samples were compared to the wild-type total extract set as 100%. α, anti; WT, wild type.
We sought to test whether EsxB is required for the secretion of other WXG100 family members and vice versa. For example, M. tuberculosis EsxA (ESAT-6) and EsxB (CFP-10) form a complex prior to secretion. As only EsxB (CFP-10) is thought to carry the sequence element that targets the complex for secretion, mutations that abrogate esxB (CFP-10) expression abolish M. tuberculosis EsxA (ESAT-6) secretion (8). To examine EsxB and EsxW secretion, B. anthracis mutants carrying deletions in esxB, esxL, esxW, or essC were grown in LB cultures supplemented with glucose or with glucose-sodium bicarbonate, and protein secretion was measured by immunoblotting. Data shown in Fig. 5 indicate that deletions of esxL, esxW, or essC do not affect synthesis and secretion of EsxB in LB medium-glucose. EsxW was expressed and secreted when bacilli were grown in LB medium supplemented with glucose-sodium bicarbonate. Expression and secretion of EsxW were not affected by deletion of esxL or essC. However, deletion of esxB greatly diminished the abundance of EsxW, which could no longer be detected in the supernatant of bacillus cultures (Fig. 5). As a control, expression of ribosomal protein L6 was not affected by mutations in the B. anthracis esx genes. L6 was also not detected in culture supernatants, indicating that the presence of EsxB and EsxW in the extracellular medium is a feature of the type VII secretion pathway and is not caused by lysis of bacilli. In mid-log cultures, EsxW degradation products could be revealed in esxB mutant bacilli (data not shown). This observation is in agreement with the conjecture that EsxW must be unstable and rapidly degraded in the cytoplasm of bacilli unless the polypeptide is bound to EsxB.
Association of substrates for the type VII secretion pathway.
If B. anthracis EsxB functioned as a universal binding and secretion partner for the type VII pathway, the protein might also bind to other WXG100 proteins. To investigate protein-protein interactions for B. anthracis WXG100 factors, we employed the yeast two-hybrid approach. The complete coding sequences of esxB, esxL, esxP, esxQ, esxV, and esxW were cloned into yeast expression vectors pGBKT7 and pGADT7. Resulting plasmids were transformed in pairs into the reporter strain AH109, and protein interactions were screened by plating transformants on selective agar with medium- and high-stringency galactose requirements for growth (Fig. 6). When fused to GAL4BD, EsxW and EsxV displayed trans-activating properties even in the absence of GAL4AD activating partners, precluding their further analysis. As a summary for all other interactions, only the GAL4AD-EsxB and GAL4BD-EsxW pair afforded growth on high-stringency agar, whereas under medium stringency, EsxB homodimer formation (GAL4AD-EsxB and GAL4BD-EsxB) was able to support yeast growth (Fig. 6). All other combinations of plasmid transformants were unable to support yeast growth. Thus, EsxB binds specifically to EsxW or to itself but not to EsxL, EsxP, and EsxV, suggesting that EsxB does not function as a universal binding partner for type VII secretion.
FIG. 6.
Yeast two-hybrid analysis of WXG100 protein interactions. The coding regions of WXG100 proteins were cloned in the bait (AD) and prey (BD) vectors, as indicated on the figure, and the resulting plasmids were transformed into a yeast strain bearing three reporters: ADE2, HIS3, and MEL1 (or lacZ). BD-EsxW and BD-EsxV exhibited intrinsic transactivating properties when fused to the GAL4BD and could not be used in this assay. M and H indicate medium and high stringency of growth, respectively, i.e., autotrophy for leucine-tryptophan (medium) or autotrophy for adenine-histidine-leucine-tryptophan plus the ability to use galactose (high). Combinations in panel A did not yield any productive interactions in these assays; data in panel B suggest positive interactions between EsxB-EsxW and EsxB-EsxB.
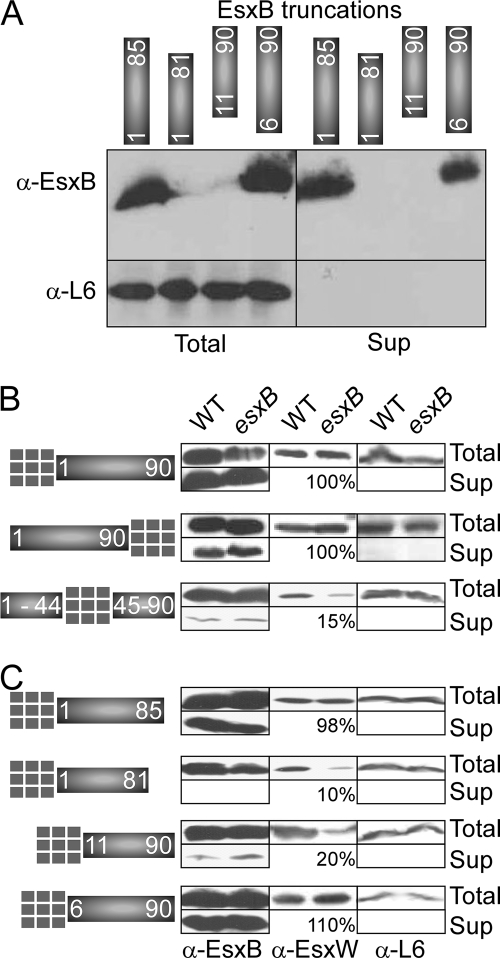
EsxB sequences required for type VII secretion.
To date, most, if not all protein translocation pathways share a common theme: the information required for translocation across biological membranes is contained solely within the peptide sequence itself. To identify sequence elements necessary for secretion, EsxB deletion variants at the N and C termini were generated (Fig. 7A). Deleting five residues at the N or C terminus of EsxB did not affect stability and secretion of EsxB (Fig. 7A). Such variants also supported EsxW secretion in an esxB mutant strain (data not shown). Deletion of more than five residues at either the N or C terminus of EsxB rendered the truncated proteins highly unstable (Fig. 7A) and unable to complement the esxB mutant for EsxW secretion (data not shown).
FIG. 7.
Sequence elements necessary for EsxB secretion. (A) Truncated variants of EsxB lacking either the first 5 and 10 or the last 5 and 9 amino acids at the N and C termini of the protein, respectively, were generated by cloning the corresponding DNA fragments in vector pOS1 carrying the pesxB. These variants are depicted as blocks of protein sequences consisting of residues 6 to 90, 11 to 90, 1 to 85, and 1 to 81. The plasmids were transformed in a mutant strain lacking esxB. Western blot analysis using antibodies against EsxB was used to examine complementation for EsxB production and secretion. The relevant part of the gel is shown. Longer exposure with chemiluminescence reagents did not reveal any truncated or degraded fragments for the EsxB proteins consisting of residues 11 to 90 or 1 to 81. (B and C) Secretion of protein hybrids. Ubiquitin was fused to full-length EsxB either at the N or C terminus or in the middle of the protein (B) or at the N terminus of the truncated variants shown in panel A. All clones were constructed using the same template and vector (pOS1 carrying pesxB; all cloning used the same restriction sites). Plasmids were transformed in wild-type strain Sterne (WT) or an isogenic mutant lacking esxB. Secretion profiles of hybrid proteins, EsxW, and ribosomal protein L6 (cytoplasmic control) were examined by Western blotting using appropriate antibodies. The amount of EsxW protein was evaluated by scanning autoradiogram. A value of 100% indicates no change in EsxW stability. Total, cell culture; Sup, supernatant; α, anti.
As the N and C termini of EsxB appear to be dispensable for secretion, we wondered whether the secretion substrate properties of EsxB tolerate fusions to reporter proteins such as ubiquitin. Both N- and C-terminal ubiquitin hybrids were generated, and designated Ub-EsxB and EsxB-Ub, respectively (Fig. 7B). Secretion of hybrid proteins was measured by immunoblot analysis of fractionated B. anthracis cultures using specific antibodies. Both ubiquitin and EsxB antibodies identified the same protein species and generated similar data; only data with anti-EsxB are shown in Fig. 7B. B. anthracis secreted both Ub-EsxB and EsxB-Ub (Fig. 7B). Further, ubiquitin fusion to the N or C terminus of EsxB truncation variants revealed that the terminal five residues were each dispensable for secretion. However, any further trimming resulted in the accumulation of Ub-EsxB and EsxB-Ub hybrids in the cytoplasm and in loss of secretion (Fig. 7C). Truncated fragments of EsxB encompassing amino acids 10 to 90 or 1 to 81, which are otherwise unstable, could be stabilized by ubiquitin fusion to the N termini of these variants (Fig. 7A and B, compare truncated and hybrid proteins). Hybrids that were unable to be secreted also failed to stabilize EsxW (Fig. 7C, middle panels). Thus, EsxB variants that cannot stably interact with EsxB or EsxW also fail to promote type VII secretion. However, the analysis of truncated EsxB variants did not point to a specific amino acid sequence that is necessary and sufficient for secretion. Rather, with the exception of short segments of N- and C-terminal sequences, the entire backbone of EsxB appeared to be important both for stability and secretion. In an attempt to disrupt EsxB folding, ubiquitin was inserted in the middle of EsxB (Fig. 7). The EsxB-Ub-EsxB hybrid accumulated in the cytoplasm and was not a substrate for secretion (Fig. 7B), thereby corroborating the hypothesis that the folded structure of EsxB is necessary for substrate recognition in the type VII secretion pathway.
DISCUSSION
Bacteria employ specialized secretion pathways to export virulence factors for the subversion of host defense functions. Recent studies have provided evidence that such an alternative protein secretion system, the T7SS (1), may fulfill similar properties for mycobacteria and staphylococci. WXG100-like proteins are proposed to be the secretion substrates for the T7SS although recent evidence suggest that these may not be the only substrates (2, 6, 9, 16). Bioinformatics analysis can be used to identify putative WXG100 proteins in the genome of various bacteria. B. anthracis encodes six putative WXG100 proteins. Five of these proteins harbor long C-terminal extensions, a feature observed only in the genomes of bacilli from the B. cereus group.
Using immunoblot analysis, we have been able to identify conditions that lead to production and secretion of two WXG100 proteins, EsxB and EsxW. By interrogating serum of animals infected with B. anthracis Ames, we determined that a third protein, EsxP, in addition to EsxB and EsxW, is produced during infection and perceived by the immune system for the development of specific antibodies. B. anthracis spores represent the infectious agents of anthrax. Spores are taken up by macrophages, germinate, and replicate intracellularly first (18). After initial intracellular replication, vegetative bacilli are released from macrophages and then replicate extracellularly, avoiding phagocytic killing by virtue of their encapsulation and toxin secretion (18). The genes that enable initial germination, phagosomal escape, and replication of bacilli in macrophages remain largely unknown. Presumably, newly germinating bacilli must use a gene expression program that is different at this stage of the infection and allows for escape from the initial replication niche. If so, factors like EsxL/Q/V, whose synthesis and secretion is not observed when bacilli are grown in vitro, may be produced at early stages of infection. All other organisms examined for a T7SS thus far (M. tuberculosis, Mycobacterium marinum, Mycobacterium bovis, Mycobacterium leprae, and S. aureus) appear to constitutively secrete WXG100 proteins in the culture medium. Hence, B. anthracis is unique in that WXG100 protein production appears to be induced.
Nuclear magnetic resonance solution structure of the ESAT-6-CFP-10 protein complex revealed a four-helix bundle made of two similar and antiparallel helix-turn-helix hairpin structures (24). The interaction between the two proteins is extensive along both helices in each monomer and is thought to take place in the cytosol prior to protein export. Long flexible arms at both ends of the complex could be observed by nuclear magnetic resonance. In particular, the C-terminal amino acids of CFP-10 (residues 85 to 100) were disordered and did not contribute to dimer interaction. Interestingly, the last seven residues of CFP-10 have been shown to interact with one of the ATPases of the T7SS, namely, Rv3871, and serve as a secretion signal (8). Presumably, ESAT-6 does not carry its own secretion signal but piggybacks with CFP-10 for translocation across the mycobacterial envelope. B. anthracis EsxB is most homologous to CFP-10. In B. anthracis, we find that EsxB can be secreted in the absence of any other WXG100 protein. Yeast two-hybrid studies suggest that EsxB may interact with itself and with EsxW. EsxW is very unstable in the absence of EsxB. Unfortunately, the machinery(ies) responsible for secretion of these proteins has not been identified yet. It is not possible to evaluate whether both proteins use the same portal for secretion. Clearly, growth conditions that are optimal for EsxB secretion differ from those that favor EsxW secretion although secretion in the case of EsxW is not very effective in vitro, suggesting additional gating controls in vivo. Truncating the first or last five amino acids of EsxB did not affect secretion of EsxB or EsxW. Hence, the C-terminal arm of B. anthracis EsxB does not appear to bear a secretion signal. Appending ubiquitin to the N or C terminus of EsxB was in either case tolerated for secretion. It seems that the ability of EsxB to be secreted or to interact with EsxW in a manner that stabilizes EsxW could be compromised only by deleting enough amino acids as to disrupt its fold. Indeed, the intervening ubiquitin sequence introduced in the middle of EsxB prevented the hybrid protein from crossing the membrane. We hypothesize that this hybrid was unfolded as it did not support interaction with EsxW in an esxB mutant (EsxW was very unstable).
Clearly, the WXG100 elements of the T7SS are conserved and represent Sec-independent secretion substrates. Yet their regulation, secretion, and destiny during infection remain to be elucidated. The ability to fuse large reporter hybrids to B. anthracis EsxB will be exploited in the future to gain insights into EsxB function during infection. In particular, the possibility that EsxB is a machinery component as opposed to an effector protein of the T7SS remains to be established. If so, the true effector functions of B. anthracis T7SS may be provided by other WXG100 proteins, perhaps those factors that harbor large C-terminal domains.
Supplementary Material
Acknowledgments
We thank Ann Elmer and Kjresti Knox for technical assistance and Bill Blaylock and Olaf Schneewind for critical reading of the manuscript.
E.B. was supported by the Biodefense Training Grant in Host-Pathogen Interactions 5T32 AI065382. We acknowledge membership within and support from the Region V Great Lakes Regional Center of Excellence in Biodefense and Emerging Infectious Diseases Consortium (NIH-NIAID Award 1-U54-AI-057153).
Footnotes
Published ahead of print on 22 August 2008.
Supplemental material for this article may be found at http://jb.asm.org/.
REFERENCES
- 1.Abdallah, A. M., N. C. Gey van Pittius, P. A. Champion, J. Cox, J. Luirink, C. M. Vandenbroucke-Grauls, B. J. Appelmelk, and W. Bitter. 2007. Type VII secretion—mycobacteria show the way. Nat. Rev. Microbiol. 5883-891. [DOI] [PubMed] [Google Scholar]
- 2.Abdallah, A. M., T. Verboom, F. Hannes, M. Safi, M. Strong, D. Eisenberg, R. J. Musters, C. M. Vandenbroucke-Grauls, B. J. Appelmelk, J. Luirink, and W. Bitter. 2006. A specific secretion system mediates PPE41 transport in pathogenic mycobacteria. Mol. Microbiol. 62667-679. [DOI] [PubMed] [Google Scholar]
- 3.Andersen, P., A. B. Andersen, A. L. Sorensen, and S. Nagai. 1995. Recall of long-lived immunity to Mycobacterium tuberculosis infection in mice. J. Immunol. 1543359-3372. [PubMed] [Google Scholar]
- 4.Bachmair, A., D. Finley, and A. Varshavsky. 1986. In vivo half-life of a protein is a function of its amino-terminal residue. Science 234179-186. [DOI] [PubMed] [Google Scholar]
- 5.Berthet, F. X., P. B. Rasmussen, I. Rosenkrands, P. Andersen, and B. Gicquel. 1998. A Mycobacterium tuberculosis operon encoding ESAT-6 and a novel low-molecular-mass culture filtrate protein (CFP-10). Microbiology 1443195-3203. [DOI] [PubMed] [Google Scholar]
- 6.Burts, M. L., and D. Missiakas. 2008. EsaC substrate for the ESAT-6 secretion pathway and its role in persistent infections of Staphylococcus aureus. Mol. Microbiol. 69736-746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Burts, M. L., W. A. Williams, K. DeBord, and D. M. Missiakas. 2005. EsxA and EsxB are secreted by an ESAT-6-like system that is required for the pathogenesis of Staphylococcus aureus infections. Proc. Natl. Acad. Sci. USA 1021169-1174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Champion, P. A., S. A. Stanley, M. M. Champion, E. J. Brown, and J. S. Cox. 2006. C-terminal signal sequence promotes virulence factor secretion in Mycobacterium tuberculosis. Science 3131632-1636. [DOI] [PubMed] [Google Scholar]
- 9.Fortune, S. M., A. Jaeger, D. A. Sarracino, M. R. Chase, C. M. Sassetti, D. R. Sherman, B. R. Bloom, and E. J. Rubin. 2005. Mutually dependent secretion of proteins required for mycobacterial virulence. Proc. Natl. Acad. Sci. USA 10210676-10681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Gaspar, A. H., L. A. Marraffini, E. M. Glass, K. L. Debord, H. Ton-That, and O. Schneewind. 2005. Bacillus anthracis sortase A (SrtA) anchors LPXTG motif-containing surface proteins to the cell wall envelope. J. Bacteriol. 1874646-4655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Gey Van Pittius, N. C., J. Gamieldien, W. Hide, G. D. Brown, R. J. Siezen, and A. D. Beyers. 2001. The ESAT-6 gene cluster of Mycobacterium tuberculosis and other high G+C gram-positive bacteria. Genome Biol. 2research0044.1-0044.18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hsu, T., S. M. Hingley-Wilson, B. Chen, M. Chen, A. Z. Dai, P. M. Morin, C. B. Marks, J. Padiyar, C. Goulding, M. Gingery, D. Eisenberg, R. G. Russell, S. C. Derrick, F. M. Collins, S. L. Morris, C. H. King, and W. R. Jacobs, Jr. 2003. The primary mechanism of attenuation of bacillus Calmette-Guerin is a loss of secreted lytic function required for invasion of lung interstitial tissue. Proc. Natl. Acad. Sci. USA 10012420-12425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Klebe, R. J., J. V. Harriss, Z. D. Sharp, and M. G. Douglas. 1983. A general method for polyethylene-glycol-induced genetic transformation of bacteria and yeast. Gene 25333-341. [DOI] [PubMed] [Google Scholar]
- 14.Lee, V. T., and O. Schneewind. 2001. Protein secretion and the pathogenesis of bacterial infections. Genes Dev. 151725-1752. [DOI] [PubMed] [Google Scholar]
- 15.McDevitt, D., P. Francois, P. Vaudaux, and T. J. Foster. 1994. Molecular characterization of the clumping factor (fibrinogen receptor) of Staphylococcus aureus. Mol. Microbiol. 11237-248. [DOI] [PubMed] [Google Scholar]
- 16.McLaughlin, B., J. S. Chon, J. A. MacGurn, F. Carlsson, T. L. Cheng, J. S. Cox, and E. J. Brown. 2007. A mycobacterium ESX-1-secreted virulence factor with unique requirements for export. PLoS Pathog. 3e105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Missiakas, D., and O. Schneewind. 2007. What has genomics taught us about gram-positive protein secretion and targeting, p. 301-326. In M. J. Pallen, K. E. Nelson, and G. M. Preston (ed.), Bacterial pathogenomics. ASM Press, Washington, DC.
- 18.Mock, M., and A. Fouet. 2001. Anthrax. Annu. Rev. Microbiol. 55647-671. [DOI] [PubMed] [Google Scholar]
- 19.Pallen, M. J. 2002. The ESAT-6/WXG100 superfamily—and a new gram-positive secretion system? Trends Microbiol. 10209-212. [DOI] [PubMed] [Google Scholar]
- 20.Pallen, M. J., R. R. Chaudhuri, and I. R. Henderson. 2003. Genomic analysis of secretion systems. Curr. Opin. Microbiol. 6519-527. [DOI] [PubMed] [Google Scholar]
- 21.Pugsley, A. P. 1993. The complete general secretory pathway in gram-negative bacteria. Microbiol. Rev. 5750-108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Pym, A. S., P. Brodin, R. Brosch, M. Huerre, and S. T. Cole. 2002. Loss of RD1 contributed to the attenuation of the live tuberculosis vaccines Mycobacterium bovis BCG and Mycobacterium microti. Mol. Microbiol. 46709-717. [DOI] [PubMed] [Google Scholar]
- 23.Pym, A. S., P. Brodin, L. Majlessi, R. Brosch, C. Demangel, A. Williams, K. E. Griffiths, G. Marchal, C. Leclerc, and S. T. Cole. 2003. Recombinant BCG exporting ESAT-6 confers enhanced protection against tuberculosis. Nat. Med. 9533-539. [DOI] [PubMed] [Google Scholar]
- 24.Renshaw, P. S., K. L. Lightbody, V. Veverka, F. W. Muskett, G. Kelly, T. A. Frenkiel, S. V. Gordon, R. G. Hewinson, B. Burke, J. Norman, R. A. Williamson, and M. D. Carr. 2005. Structure and function of the complex formed by the tuberculosis virulence factors CFP-10 and ESAT-6. EMBO J. 242491-2498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Renshaw, P. S., P. Panagiotidou, A. Whelan, S. V. Gordon, R. G. Hewinson, R. A. Williamson, and M. D. Carr. 2002. Conclusive evidence that the major T-cell antigens of the Mycobacterium tuberculosis complex ESAT-6 and CFP-10 form a tight, 1:1 complex and characterization of the structural properties of ESAT-6, CFP-10, and the ESAT-6*CFP-10 complex. Implications for pathogenesis and virulence. J. Biol. Chem. 27721598-21603. [DOI] [PubMed] [Google Scholar]
- 26.Schneewind, O., P. Model, and V. A. Fischetti. 1992. Sorting of protein A to the staphylococcal cell wall. Cell 70267-281. [DOI] [PubMed] [Google Scholar]
- 27.Schurter, W., M. Geiser, and D. Mathe. 1989. Efficient transformation of Bacillus thuringiensis and B. cereus via electroporation: transformation of acrystalliferous strains with a cloned delta-endotoxin gene. Mol. Gen. Genet. 218177-181. [DOI] [PubMed] [Google Scholar]
- 28.Sørensen, A. L., S. Nagai, G. Houen, P. Andersen, and A. B. Andersen. 1995. Purification and characterization of a low-molecular-mass T-cell antigen secreted by Mycobacterium tuberculosis. Infect. Immun. 631710-1717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Stanley, S. A., S. Raghavan, W. W. Hwang, and J. S. Cox. 2003. Acute infection and macrophage subversion by Mycobacterium tuberculosis require a specialized secretion system. Proc. Natl. Acad. Sci. USA 10013001-13006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Sterne, M. 1937. Avirulent anthrax vaccine. Onderstepoort J. Vet. Sci. Anim. Ind. 2141-43. [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.