Abstract
Enteropathogenic Escherichia coli (EPEC) and enterohemorrhagic E. coli (EHEC) are diarrheagenic pathogens that colonize the intestinal tract through the formation of attaching and effacing lesions, induced by effectors translocated via a type III secretion system (T3SS) encoded on the locus of enterocyte effacement (LEE). In EHEC O157, numerous virulence factors, including around 40 T3SS effectors, have been identified. Most of them are encoded on genomic islands (GEIs) such as prophages and integrative elements. For EPEC, however, no systematic search of GEIs and virulence-related genes carried therein has been done, and only a limited number of virulence factors have been identified so far. In this study, we performed a systemic and genome-wide survey of the GEIs in strain B171-8, one of the prototype strains of EPEC, by the combined use of whole-genome PCR scanning and fosmid mapping and identified 22 large GEIs, including nine lambda-like prophages, three P2-like prophages, the LEE, and three additional integrative elements. On these prophages and integrative elements, we found genes for a set of T3SS proteins, a total of 33 T3SS effectors or effector homologues, and 12 other virulence factors which include five nonfimbrial adhesins. Most of the T3SS effector families identified are also present in EHEC O157, but B171-8 possesses a significantly smaller number of effectors. Not only the presence or absence of Shiga toxin genes but also the difference in the T3SS effector repertoire should be considered in analyzing the pathogenicity of EPEC and EHEC strains.
Rapidly increasing numbers of bacterial genome sequences are being determined, and their comparison has revealed that bacterial genomes consist of a conserved “core gene pool” and strain-specific “flexible gene pools” (22). The “flexible gene pools” are often carried on so-called “genomic islands” (GEIs), which have been acquired by lateral gene transfer and are mostly related to mobile genomic elements, such as bacteriophages, integrative conjugative elements, and transposons (7, 22). GEIs or the genes on the GEIs that are retained on a bacterial genome confer various advantages to the host bacterium, such as increased fitness in a specific host environment and increased pathogenicity. Thus, the identification and functional analysis of GEIs are pivotal steps toward a proper understanding of important strain-specific features.
Escherichia coli comprises genotypically and phenotypically divergent strains. A fraction of the strains cause diverse intestinal and extraintestinal diseases in humans by means of individually acquired virulence factors (15). So far, the whole genome sequences of nine E. coli strains, including three benign laboratory strains (K-12 strains MG1655, W3110, and DH10B) and six pathogenic strains (two enterohemorrhagic E. coli [EHEC] O157, three uropathogenic E. coli, and one avian pathogenic E. coli strain), have been published (5, 6, 10, 16, 23-25, 41, 53). Comparisons of these genome sequences have revealed that each genome contains a large amount of strain-specific sequences. For instance, a comparison of EHEC O157 strain RIMD 0509952 (referred to as O157 Sakai) and K-12 MG1655 has revealed that they share a total of 4.1 Mb of sequences but that O157 Sakai and K-12 each contains 1.4 Mb and 0.5 Mb of strain-specific sequences (referred to as S loops and K loops, respectively) (24). Importantly, most of the large S loops and K loops are prophages or integrative elements (genetic elements that contain integrase genes but no other genes related to phages and conjugal transfer functions). In O157 Sakai, 18 prophages and 6 integrative elements were identified, and they carry most of the virulence-related genes of O157.
Enteropathogenic E. coli (EPEC) is a major cause of infant diarrhea in nonindustrialized countries (9). Typical EPEC strains are defined by the presence of the locus of enterocyte effacement (LEE) and the EPEC adherence factor (EAF) plasmid. The EAF plasmid encodes bundle-forming pili that are required for bacterium-bacterium interaction and microcolony formation (11, 19). The LEE encodes a set of proteins constituting a type III secretion system (T3SS) machinery, several effector proteins secreted by the T3SS, an adhesin called “intimin,” and others (27). LEEs are also possessed by EHEC strains. In O157 Sakai, we recently found that in addition to the seven LEE-encoded effectors, 32 proteins encoded in non-LEE loci are secreted by the LEE-encoded T3SS (referred to as non-LEE-encoded effectors) (48). Almost all of these non-LEE-encoded effectors have been carried over into the O157 genome by prophages and integrative elements. Although the functions of most of the newly identified effectors have yet to be elucidated, they are believed to be required for O157 to express its full virulence. In EPEC, however, a limited number of non-LEE-encoded effectors have been identified so far (43). This is partly because no systematic search of GEIs and the virulence-related genes carried therein has been done in EPEC strains.
EPEC strain B171-8 is one of the prototype strains of typical EPEC. The whole sequence of the EAF plasmid of EPEC B171-8 has already been published (49). In this study, we analyzed the genome of EPEC B171-8 by the combined use of whole-genome PCR scanning (WGPScanning) (40) and fosmid mapping and identified a total of 22 large GEIs (>10 kb). On these GEIs, a set of proteins for the T3SS, 33 T3SS effector homologues, and 12 other potential virulence factors were identified.
MATERIALS AND METHODS
Bacterial strains, growth conditions, and DNA extraction.
EPEC B171-8 (O111:NM) was isolated from an outbreak of infant diarrhea in Washington, DC, in 1983 (21). E. coli EPI300-T1 [F− mcrA Δ(mmr-hsdRMS-mcrBC) φ80dlacZΔM15 ΔlacX74 recA1 endA1 araD139 Δ(ara,leu)7697 galU galK λ− rpsL nupG trfA tonA dhfr] (Epicentre, Madison, WI) was used as the host strain for fosmid library construction.
For genomic DNA extraction, cells were grown to stationary phase at 37°C in Luria-Bertani (LB) medium, and the genomic DNA was purified using a Genomic-tip 100/G and genomic DNA buffer set (Qiagen, Tokyo, Japan) in accordance with the manufacturer's instructions. For fosmid DNA extraction, cells were grown for 5 h to early stationary phase at 37°C in LB medium containing chloramphenicol (12.5 μg/ml) and CopyControl induction solution (1 μl in 1 ml LB medium; Epicentre). The fosmid DNA was isolated using a Kurabo PI-1100 automatic DNA isolation system (Kurabo Industries, Ltd., Japan).
WGPScanning analysis.
WGPScanning analysis of EPEC B171-8 was performed as previously described (40), with some modifications. In brief, we used 453 pairs of primers that cover the chromosome backbone shared by K-12 and O157 Sakai. All of the primer sequences are available at our website (http://genome.naist.jp/bacteria/o157/pcrscan.html). Long PCR was performed using an LA Taq PCR kit (Takara Shuzo, Kyoto, Japan), with 1 ng of genomic DNA as the template, with 30 cycles of a two-step amplification program of 20 s at 98°C and 16 min at 69°C. The PCR products were separated by field inversion gel electrophoresis, and the product sizes were estimated by a lane analyzer (Atto Corp., Tokyo, Japan).
Construction of fosmid library.
A fosmid library for EPEC B171-8 was constructed using a CopyControl fosmid library production kit (Epicentre) in accordance with the manufacturer's instructions, with some modifications. The genomic DNA was sheared using a 26-gauge syringe. After blunting and phosphorylation of the DNA fragments, the fragments were separated by pulsed-field gel electrophoresis with 1% certified low-melt agarose (Bio-Rad Laboratories, Inc., Tokyo, Japan). Fragments of around 40 kb were recovered from the gel by using GELase (Epicentre). After the DNA solution was concentrated by means of a Microcon YM-100 filter (Millipore), the fragments were ligated with the pCC1FOS vector, packaged into phage particles, and transfected into E. coli EPI300-T1 cells. We collected 2,880 chloramphenicol-resistant clones and used them for fosmid mapping analysis.
End sequencing of fosmid clones.
The end sequences of the fosmid clones were determined using the Fos-F (5′-TCCCAGTCACGACGTTG-3′) and Fos-R (5′-ACCATGATTACGCCAAGC-3′) primers, with approximately 50 ng of fosmid DNA as the template. All base calls with a quality value of <20 in the phred program (18) were removed from each read. We used only reads longer than 200 bp for further analysis.
The sequences of the fosmids or PCR products were determined by the standard random shotgun strategy. To selectively sequence the passenger regions of the lambda-like phages, each region was amplified by long PCR, using a relevant fosmid clone as the template and primers separately targeting the phage tail region and the chromosomal backbone. The fosmid clones and primers used are listed in Table S1 in the supplemental material. An ABI3730 automated sequencer (Applied Biosystems, Foster City, CA) was used for the sequence collection. Sequencher software (Gene Cord Corp., Ann Arbor, MI) was used for the assembly of the shotgun reads.
Translocation assay.
For the translocation assay of T3SS effector candidates, we employed the TEM-1 β-lactamase system (8). The first 300 nucleotides of each gene were amplified by PCR. N-terminal translational fusion with the TEM-1 part of each PCR product was done using the pKTEM vector (kindly provided by Eric Oswald, INRA-ENVT, Toulouse, France), a pBBR1MCS2 derivative containing the TEM-1 gene at the XhoI-XbaI site (30). The recombinant plasmids were introduced into B171-8 cells and used for the translocation assay. All primers used to construct the fusion genes are listed in Table S2 in the supplemental material.
A translocation assay was performed as described by Charpentier and Oswald (8). HeLa cells grown on EZView glass-bottomed culture plates (LB 24-well plates; Iwaki, Tokyo, Japan) were infected with B171-8 cells containing each recombinant plasmid. After incubation at 37°C in 5% CO2 for 1.5 h, the cell cultures were washed three times with phosphate-buffered saline (PBS) and covered with 200 μl of CCF2/AM solution (Invitrogen). After a 1.5-h incubation in the dark at room temperature, the cells were washed three times with PBS. Each well was soaked in 500 μl of PBS and inspected with a Radiance2100 confocal laser scanning microscope (Bio-Rad Laboratories, Inc.). The fluorescence intensity (emission wavelengths, 460 and 530 nm) of each well was also scanned by a SpectraMax GeminiXS fluorescence microplate reader (Molecular Devices Corporation, Sunnyvale, CA).
Nucleotide sequence accession numbers.
All of the DNA sequences determined in this study have been submitted to the DDBJ/GenBank/EMBL database (accession numbers AB426048 to AB426064).
RESULTS
Comparison of the chromosome structures of EPEC B171-8 and K-12 by WGPScanning.
In this analysis, we used 453 primer pairs targeted to the backbone sequences shared by K-12 and O157 Sakai. Nine K-12 chromosome segments that contain large K loops were not amplified, as anticipated (see Table S3 in the supplemental material), but all other segments were successfully amplified from K-12. In the initial scanning of EPEC B171-8, 60 segments yielded no PCR products (see the initial data in Fig. 1 and Table S3 in the supplemental material for more details). Since the failure in PCR amplification could have been due to base changes or deletions in the primer target sequences, these 60 segments were further analyzed by using different combinations of primers (see the final data in Fig. 1 and Table S3 in the supplemental material).
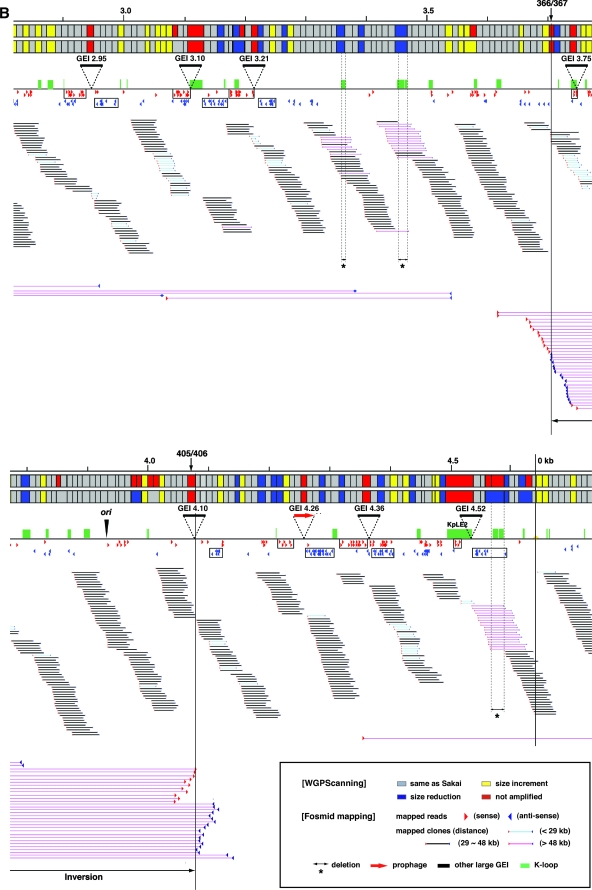
FIG. 1.
Summary of WGPScanning and fosmid mapping analyses of EPEC B171-8. The results of the WGPScanning analysis of EPEC B171-8 are shown in the upper part of each segment. The initial data from the WGPScanning analysis are presented on the upper line, and the final results incorporating the data obtained by additional PCR analyses are shown on the lower line. Those segments which yielded PCR products with the same sizes as those from the reference strain (K-12) are indicated in gray, and those that were not amplified are shown in red. Those segments showing size reductions and size increments are indicated in dark blue and yellow, respectively. The results of fosmid mapping are shown in the bottom part of each panel. The positions of the end sequences of each fosmid clone on the K-12 chromosome are indicated by red (sense) and blue (antisense) arrowheads. The fosmid clones, whose end sequences were mapped on the K-12 chromosome, were classified into three groups (light blue bars, <29 kb; black bars, 29 to 48 kb; and pink bars, >48 kb). The positions of the K loops, regions deleted in B171-8, prophages, and other large GEIs integrated in the B171-8 chromosome are also indicated. The data from the first half and second half of the chromosome are shown in panels A and B, respectively.
In this WGPScanning analysis, we identified 59 chromosome segments of B171-8 which showed size reduction compared with K-12 and 57 segments which showed size increments. Of the 59 segments with size reduction, 14 were segments containing large K loops in K-12, suggesting that whole or parts of these K loops are missing in EPEC B171-8. Among the 57 segments with size increments, one segment was >10 kb larger (15.2-kb increment) than that in K-12, but the size differences observed in other segments were 5 to 10 kb (10 segments) or <5 kb (46 segments). Finally, 31 segments remained unampli-fied. The insertion of large GEIs or inversions/translocations is likely to have occurred in these chromosomal loci.
Fosmid mapping analysis.
We constructed a fosmid library of the EPEC B171-8 genomic DNA, collected 2,880 clones, and used them for mapping analysis of the K-12 chromosome. Our analysis of 95 clones randomly selected from the 2,880 clones indicated that their insert sizes ranged from 31.8 to 49.0 kb (40.5 kb on average) (see Fig. S1 in the supplemental material). The end sequences of the fosmid clones were determined, and 2,410 clones, from which high-quality sequences (>200 bp) were obtained from both ends, were used for the mapping analysis. Since the chromosome size of EPEC B171-8 was estimated to be about 5,250 kb by pulsed-field gel electrophoresis analysis of I-CeuI-digested genomic DNA (data not shown), the final coverage was 18.6 times (Table 1).
TABLE 1.
Summary of B171-8 fosmid library
| Parameter | Value |
|---|---|
| Total no. of clones | 2,880 |
| Average size (kb) of insert DNAa | 40.5 |
| No. of clones used for mapping analysisb | 2,410 |
| Estimated total length (kb) of insert DNA | 97,605 |
| Estimated size (kb) of B171-8 chromosome | 5,250 |
| Coverage (times) of the chromosome by fosmid clones | 18.6 |
Estimated from the results of field inversion gel electrophoresis analysis of 95 randomly selected clones.
High-quality end sequences (>200 bp) were obtained from both ends for these clones.
A FASTA search was performed for all 4,820 end sequences against the chromosome sequence of K-12 MG1655. About 80% (1,996,680 bp) of the total read length of the end sequences (2,474,123 bp) showed ≥90% identity to the K-12 sequence, implying that about 20% of the B171-8 genome sequence (approximately 1 Mb) is absent in K-12. Of the 3,923 end sequences that were mapped to the K-12 chromosome (threshold, ≥90% identity and ≥30% coverage), 3,791 had single hits to the K-12 chromosome and the others hit multiple loci (Fig. 1).
For 1,507 clones (62.5% of the 2,410 clones), both end sequences showed single hits (Table 2; see Table S4 in the supplemental material). Using the distances between their end sequences on the K-12 chromosome, we classified these clones into the following four groups: “short” clones (the distance was <29 kb), “normal” clones (29 to 48 kb), “long” clones (48 to 100 kb), and “superlong” clones (>100 kb) (Fig. 1; see Fig. S2 in the supplemental material). By examining the locations of the “long” clones, we identified 16 regions where large deletions have taken place (Fig. 1). All of these regions exhibited a large size reduction in the WGPScanning analysis.
TABLE 2.
Summary of FASTA search of fosmid end sequences against the K-12 chromosome
| FASTA search result for end sequences against K-12 chromosome sequencea | No. of clones |
|---|---|
| Single hit/single hit | 1,507 |
| Single hit/multiple hits | 101 |
| Single hit/no hit | 676 |
| Multiple hits/multiple hits | 1 |
| Multiple hits/no hit | 29 |
| No hit/no hit | 96 |
| Total | 2,410 |
Threshold, >90% identity with >30% alignment.
The end sequences of most “superlong” clones were mapped to two chromosome loci located in segments 366/367 and 405/406 (Fig. 1), suggesting that a large inversion occurred between these loci in B171-8. The origin of replication is located between these two loci. Other “superlong” clones that were dispersed on the chromosome were probably generated by mispositioning of the end sequences by cross-matching to homologous sequences or translocations of short fragments.
By searching for genomic regions that were covered by “short” clones or clones for which only one end sequence was mapped (referred to as “single-end-mapped” clones), we identified 22 regions which contained large insertions (Fig. 1). These regions corresponded to the unamplified segments or the segment with a 15.2-kb increment identified in the WGPScanning analysis. Among the 31 unamplified segments, 27 were associated with the insertion of 21 large GEIs and 2 were associated with the large inversion described above (Fig. 1). The remaining two unamplified segments were covered by multiple “normal” clones, which made it unlikely that these segments would contain large GEIs. These results indicate that we most probably identified all of the large (>10 kb) GEIs of EPEC B171-8 by the combination of WGPScanning and fosmid mapping analyses.
The results of the BlastX search of no-hit end sequences of “single-end-mapped” clones against the GenBank nr database indicate that 13 of the 22 GEIs are prophages (nine lambda-like phages, three P2-like phages, and one phage of an unknown phage group). The others are the LEE, two fimbrial biosynthesis operons, a lipopolysaccharide biosynthesis operon, the IAHP cluster (13), and ETT2 (E. coli type III secretion system 2), carrying the second set of T3SS genes that are widely distributed in E. coli (24). The origins of the remaining three GEIs were not predicted (Table 3; see Table S5 in the supplemental material).
TABLE 3.
Large GEIs identified in EPEC B171-8a
| GEI | Size (bp) | Description | Integration site |
|---|---|---|---|
| GEI 0.24 | ND | IAHP cluster | ND |
| GEI 0.58 | ND | Prophage (lambda-like) | tRNA-argU |
| GEI 0.81 | ND | Prophage (lambda-like) | ybhC-ybhB |
| GEI 1.03 | ND | Prophage (lambda-like) | tRNA-serT |
| GEI 1.18 | ND | Prophage (lambda-like) | potB |
| GEI 1.31 | 13,715 | Prophage (unknown) | tonB |
| GEI 1.79 | 39,958 | Prophage (P2-like) | btuC-infA |
| GEI 1.94 | ND | Prophage (lambda-like) | yecE |
| GEI 1.98 | 38,742 | Prophage (P2-like) | tRNA-ileZ |
| GEI 2.04 | ND | Prophage (lambda-like) | tRNA-serU |
| GEI 2.06 | ND | Lipopolysaccharide biosynthesis operon | ND |
| GEI 2.21 | ND | Prophage (lambda-like) | mlrA |
| GEI 2.43 | 38,427 | Prophage (P2-like) | pdxB-flk |
| GEI 2.69 | ND | Prophage (lambda-like) | yfhL |
| GEI 2.95 | ND | ETT2 | ND |
| GEI 3.10 | 58,437 | Integrative element (LEE) | tRNA-pheV |
| GEI 3.21 | 27,511 | Integrative element | tRNA-ileX |
| GEI 3.75 | ND | Fimbrial biosynthesis operon | ND |
| GEI 4.10 | ND | Fimbrial biosynthesis operon | ND |
| GEI 4.26 | ND | Prophage (lambda-like) | dusA |
| GEI 4.36 | 59,233 | Integrative element | tRNA-pheU |
| GEI 4.52 | ND | Integrative element | tRNA-leuX |
ND, not determined.
We next determined the DNA sequences of the prophage regions downstream of the tail genes in all nine lambda-like phages because it is known that, in EHEC O157, these regions of lambda-like phages (called “exchangeable effector loci” [EELs]) frequently carry virulence-related genes, especially those for non-LEE-encoded T3SS effectors (24, 48). In addition, we determined the whole sequences of the LEE, four other prophages, two GEIs of unknown origin, and part of the sequence of one GEI of unknown origin. This sequencing analysis revealed that the three undefined GEIs are integrative elements and that the undefined prophage is a prophage remnant of unknown origin.
Comparison of the LEE of EPEC B171-8 with other sequenced LEEs.
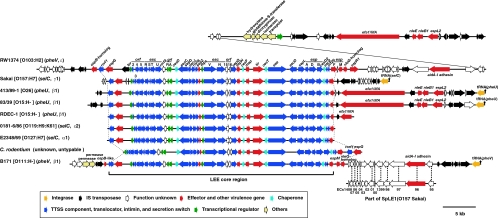
The LEE of B171-8 is 58 kb long and is integrated into the 3′ end of the pheV tRNA gene. The gene organization of the LEE core region of B171-8 is almost identical to those of other sequenced LEEs (14, 17, 20, 24, 26, 47, 55) (Fig. 2). The LEE core region of B171-8 is associated with additional flanking sequences, as in some of the other LEEs.
FIG. 2.
Comparison of the LEE of B171-8 with other sequenced LEEs. The gene organizations of the LEE of EPEC B171-8 and eight other sequenced LEEs are shown. A part of SpLE1, a large integrative element of O157 Sakai, is very similar to a part of the right flanking region of the B171-8 LEE. The genes on the two elements exhibiting high similarity (≥90% identity with ≥90% alignment) are indicated by dotted lines.
The sequences of the LEE-flanking regions are available only for EHEC O157 Sakai, EPEC O103 strain RW1374, EPEC O26 strain 413/89-1, and EPEC O15 strain 83/39 (24, 26, 47). Although the LEE-flanking regions of these strains are highly variable, all encode homologues of adhesins (LifA/Efa1 or AidA-I) and several T3SS effectors. In the LEE-flanking regions of EPEC B171-8, an Aid-I homologue and three T3SS effectors, belonging to the OspB, EspM, and NleG families, are encoded (the OspB homologue is truncated by an insertion element [IS element]). In the left flanking region, two different permeases are encoded. These permease genes are absent in K-12 and O157 Sakai but are found at the same locus in some other sequenced E. coli strains (e.g., E. coli strains CFT073, E24377A, and HS). This suggests that the two permease genes are part of the backbone gene set of E. coli and have been deleted in O157 and K-12. We could not define the att sequences of the LEE in EPEC B171-8, but an IS element is present between the two permease genes and the ospB homologue. This suggests that some IS element-mediated deletion has taken place in the left junction, which probably eliminated the attL sequence. It is also noteworthy that a part of the right flanking region is almost identical to a part of SpLE1, a large integrative element of O157 Sakai (Fig. 2), indicating that the LEE of B171-8 has a complicated evolutionary history.
EELs of lambda-like prophages.
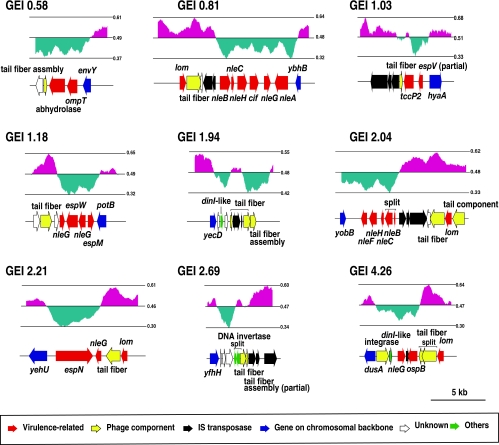
We found that seven of the nine lambda-like prophages carry virulence-related genes in their EELs. These include genes for 20 T3SS effectors or effector homologues (five of them were pseudogenes), OmpT, and alpha/beta hydrolase (Fig. 3 and Table 4). In addition, at least four lambda-like prophages encode Lom homologues, which may be involved in adherence to human epithelial cells or survival in macrophages (1, 2, 51). It is noteworthy that most of these genes or their close homologues are also present in O157 Sakai, with a few exceptions, such as the Cif and OspB homologues.
FIG. 3.
Gene maps of the EELs of nine lambda-like prophages identified in B171-8. The gene organizations and GC contents of the EELs of the nine lambda-like prophages identified in B171-8 are shown.
TABLE 4.
Potential virulence factors identified in EPEC B171-8
| Locus | Productb | Length (aa) | Homology (% identity [length of alignment {aa}])
|
|
|---|---|---|---|---|
| BLAST search, O157 Sakai proteins | BLAST search, nr databasea | |||
| GEI 0.58 | Alpha/beta hydrolase | 494 | 99 (494) to ECs1662 | |
| GEI 0.58 | Outer membrane protease | 317 | 99 (317) to ECs1663 (OmpT) | |
| GEI 0.81 | Lom | 199 | 97 (199) to ECs1122 (Lom) | |
| GEI 0.81 | T3SS effector NleB homolog | 326 | 98 (326) to ECs0846 (NleB) | |
| GEI 0.81 | T3SS effector NleC homologψ | 94 (99) to ECs0847 (NleC) | ||
| GEI 0.81 | T3SS effector NleH homolog | 293 | 92 (293) to ECs0848 (NleH) | |
| GEI 0.81 | T3SS effector Cif | 282 | No hit | 99 (282) to Cif (REPEC E22) |
| GEI 0.81 | T3SS effector NleG homolog | 191 | 78 (189) to ECs1994 (NleG) | |
| GEI 0.81 | T3SS effector NleA homolog | 412 | 75 (441) to ECs1812 (NleA) | |
| GEI 1.03 | T3SS effector TccP2 | 297 | 97 (189) to ECs1126 (TccP2) | |
| GEI 1.03 | T3SS effector EspV homologψ | 100 (50) to ECs1127 (EspV) | ||
| GEI 1.18 | T3SS effector NleG homolog | 159 | 99 (140) to ECs3488 (NleG) | |
| GEI 1.18 | T3SS effector EspW homolog | 352 | 98 (352) to ECs3487 (EspW) | |
| GEI 1.18 | T3SS effector NleG homolog | 216 | 99 (216) to ECs3486 (NleG) | |
| GEI 1.18 | T3SS effector EspM homologc | 196 | 86 (195) to ECs3485 (EspM) | |
| GEI 2.04 | T3SS effector NleB homologψ | 99 (326) to ECs0846 (NleB) | ||
| GEI 2.04 | T3SS effector NleC homologψ | 97 (99) to ECs0847 (NleC) | ||
| GEI 2.04 | T3SS effector NleH homologψ | 86 (214) to ECs1814 (NleH) | ||
| GEI 2.04 | T3SS effector NleF homolog | 189 | 92 (189) to ECs1815 (NleF) | |
| GEI 2.04 | Lom | 199 | 47 (207) to ECs1122 (Lom) | 98 (198) to Lom (E. coli E110019) |
| GEI 2.21 | T3SS effector EspN homolog | 1,135 | 38 (1,146) to ECs1561 (EspN) | 34 (173) to CNF3 (E. coli C48a), 32% [181] to CNF1 (E. coli A70.1) |
| GEI 2.21 | T3SS effector NleG homolog | 189 | 88 (189) to ECs1994 (NleG) | |
| GEI 2.21 | Lom | 199 | 98 (199) to ECs1649 (Lom) | |
| GEI 3.10 | T3SS effector OspB homologψ | No hit | 35 (137) to OspB (S. sonnei Ss046) | |
| GEI 3.10 | T3SS effector EspG | 398 | 83 (397) to ECs4590 (EspG) | |
| GEI 3.10 | T3SS effector EspZ | 100 | 67 (100) to ECs4571 (EspZ) | |
| GEI 3.10 | T3SS effector EspH | 171 | 69 (168) to ECs4564 (EspH) | |
| GEI 3.10 | T3SS effector Map | 203 | 80 (203) to ECs4562 (Map) | |
| GEI 3.10 | T3SS effector Tir | 538 | 66 (559) to ECs4561 (Tir) | |
| GEI 3.10 | T3SS effector EspB | 314 | 70 (317) to ECs4554 (EspB) | |
| GEI 3.10 | T3SS effector EspF | 175 | 78 (173) to ECs4550 (EspF) | |
| GEI 3.10 | T3SS effector EspM homolog | 194 | 54 (191) to ECs1825 (EspM) | |
| GEI 3.10 | T3SS effector NleG homolog | 212 | 62 (211) to ECs1824 (NleG) | |
| GEI 3.10 | AidA-I adhesin-like protein | 949 | 96 (949) to ECs1396 | |
| GEI 3.21 | LifA/Efa1 homolog | 2981 | 28 (1,891) to pO157_059 | |
| GEI 4.26 | T3SS effector NleG homolog | 306 | N terminus, 100 to ECs2226 (NleG) (123); C terminus, 93 to ECs2227 (NleG) (94) | |
| GEI 4.26 | T3SS effector OspB homolog | 291 | No hit | 34 (252) to OspB (S. sonnei Ss046) |
| GEI 4.26 | Lom | 199 | 96 (199) to ECs1649 (Lom) | |
| GEI 4.36 | PagC-like protein | 182 | 98 (182) to ECs3850 | |
| GEI 4.36 | T3SS effector EspL homolog | 549 | 99 (549) to ECs3855 (EspL) | |
| GEI 4.36 | T3SS effector NleB homologψ | 98 (329) to ECs3857 (NleB) | ||
| GEI 4.36 | T3SS effector NleE homolog | 224 | 99 (224) to ECs3858 (NleE) | |
| GEI 4.36 | LifA/Efa1 homologψ | 21 (2,164) to pO157_059 | 100 (2,151) to Efa1 (E. coli O157 strain E45035) | |
| GEI 4.36 | AidA-I adhesin-like protein | 948 | 88 (949) to ECs1396 | |
| GEI 4.52 | AidA-I adhesin-like protein | 948 | 88 (949) to ECs1396 | |
Only the data for proteins that showed <50% identity to O157 Sakai proteins are given.
ψ, pseudogenes (the BlastX program was used for the homology search of pseudogenes).
This protein was originally named “TrcA” (50).
P2-like prophages.
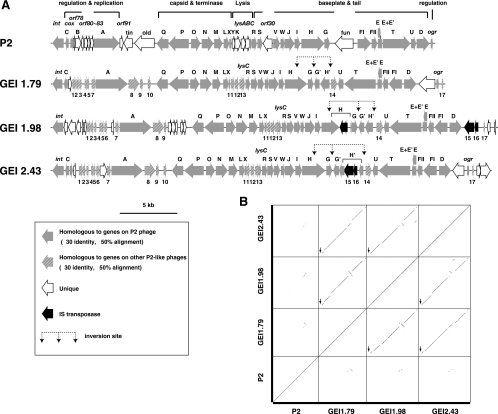
The three P2-like prophages are significantly divergent from phage P2 in DNA sequence (4), but their gene organizations are almost the same as that of the P2 phage, and most genes exhibit >30% amino acid sequence identity to the P2 phage genes. Interestingly, the three P2-like prophages are very similar to each other, even at the DNA sequence level (Fig. 4), although each prophage genome contains several unique regions, such as those encoding integrases and immunity proteins (C and Cox). Another notable feature of the three P2-like prophages is the presence of an inversion system in the tail fiber region. We detected inversion by PCR in the prophage on GEI 1.79 but not in others (data not shown). IS insertion probably inactivated the inversion systems of the two phages on GEI 1.98 and GEI 2.43. The prophage on GEI 1.98 is probably defective because an IS insertion disrupted the H gene for the tail fiber protein and the ogr gene is missing. In contrast, although the inversion system of the prophage on GEI 2.43 is inactive, this prophage may be intact because no other genetic defect was found in the prophage genome, similar to that on GEI 1.79.
FIG. 4.
Comparison of the three P2-like prophages identified in B171-8. (A) The gene organizations of the P2 phage and three P2-like prophages of B171-8 are shown. Seventeen gene families that are not present in phage P2 but are present in other P2 family members are indicated by numbers. Gene family 14 is comprised of invertase genes involved in the inversion of tail fiber regions. (B) Dot plot matrices of the nucleotide sequences of the P2 phage and the three P2-like prophages of EPEC B171-8 are shown. The vertical arrows indicate prophage regions where genes for immunity-related proteins and integrases reside.
The three P2-like prophages all appear to carry many morons in several regions, such as those between the A and Q genes, between the H and U genes, and downstream of the D gene. However, most are of unknown function and include no homologues of known virulence factors.
Other integrative elements and a phage remnant.
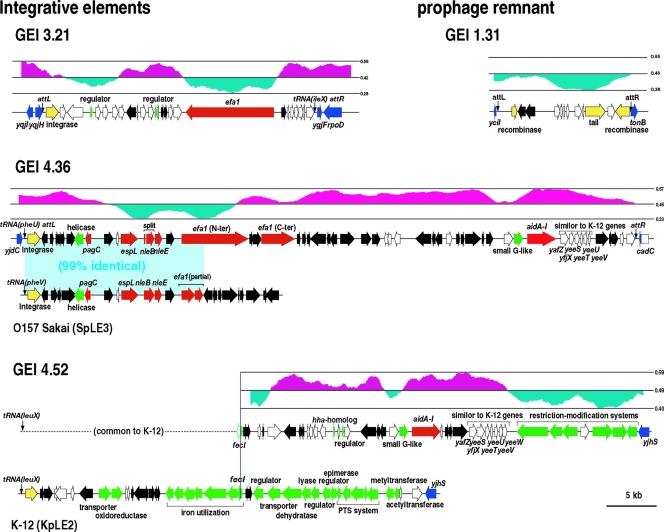
GEI 3.21 is an integrative element integrated in the 3′ end of the ileX tRNA gene. It contains three IS elements and genes for two AlpA-type transcriptional regulators and a LifA/Efa1 adhesin-like protein (Fig. 3). Most of the other genes are also conserved in some pathogenic E. coli strains, but their functions are unknown.
GEI 4.36 is an integrative element integrated into the pheU tRNA gene. The integrase gene of the GEI is identical to that of SpLE3 of O157 Sakai, which is integrated into the pheV tRNA gene on the O157 Sakai chromosome. Furthermore, a part of the GEI is almost identical to a part of SpLE3 (Fig. 3). Both elements contain numerous IS elements and encode three T3SS effectors (EspL2, NleB, and NleE; NleB is split in EPEC B171-8 but intact in O157 Sakai), a PagC-like protein, and an interrupted LifA/Efa1 adhesin-like protein. The 26-kb region of GEI 4.36 that is not present in SpLE3 encodes a small GTP-binding protein and an AidA-I adhesin-like protein.
GEI 4.52 also seems to be an integrative element integrated into the leuX tRNA gene, where an integrative element (KpLE2) is integrated in K-12. Because the left part of GEI 4.52 showed no structural difference from KpLE2 in the WGPScanning and fosmid mapping analyses, we determined the sequence of the right part of GEI 4.52, a 39-kb region between the fecI and yjhS genes. While KpLE2 contains 18 genes for various enzymes, transporters, and regulators in the fecI to yjhS region, the integrative element GEI 4.52 encodes restriction-modification systems, a small GTP-binding protein of unknown function, an AidA-I-like adhesin-like protein, an Hha homologue, and an AlpA-type transcriptional regulator. This region also contains many IS elements and a set of genes for hypothetical proteins. Similar sets of hypothetical genes are present on GEI 4.36 (Fig. 3) and also on three prophage-like elements of K-12 (Cp4-6, Cp4-44, and Cp4-57).
GEI 1.31 is a highly degraded prophage remnant. This prophage contains no virulence-related genes.
Translocation assay of T3SS effector homologues and GEI-encoded proteins of unknown function.
Among the genes for 26 T3SS effectors or effector homologues identified in non-LEE loci (including the regions flanking the LEE core), 19 appear to encode intact proteins, while 7 are apparently pseudogenes (Table 4). In addition, EPEC B171-8 contains one gene encoding an EspM homologue (originally annotated as trcP) which resides on plasmid pB171 (49), but this gene has also been inactivated by an IS element inserted into the 5′-end region.
Of the 19 intact effector homologues encoded by the non-LEE loci, 17 are homologous to the effector proteins of O157 (48): 7 are nearly identical to their O157 counterparts, but 10 show lower amino acid sequence identities (38 to 92%). Of the two that are not present in O157, the Cif protein of B171-8 is 99% identical in amino acid sequence to that of the rabbit EPEC strain E22 (31), but the sequence of the OspB homologue is only 34% identical to the OspB protein of Shigella sonnei strain Ss046. In addition, many genes of unknown function, identified in the EELs of the lambda-like prophages and on the other prophages and integrative elements, could include new effector genes.
We therefore examined whether or not the 10 divergent effector homologues are translocated into host cells by using the TEM-1 assay system as described in Materials and Methods. TccP2 was used as a positive control in this assay (54). Forty-seven proteins of unknown function, encoded by the lambda-like prophages, P2-like prophages, and other sequenced GEIs, were also examined. Among the 10 effector homologues examined, 7 (NleH and NleG from GEI 0.81, EspM from GEI 1.18, NleG from GEI 2.21, NleG and EspM from GEI 3.10, and OspB from GEI 4.26) were clearly translocated into HeLa cells from B171-8 (see Table S6 in the supplemental material). We did not observe a clear translocation of NleA from GEI 0.81, NleF from GEI 2.04, and EspN from GEI 2.21, but these proteins will need to be examined by means of different assay systems. In contrast, none of the 47 proteins of unknown function were translocated in this assay.
DISCUSSION
Microarray- and PCR-based techniques have frequently been used for comparative genomic studies of various bacterial pathogens, and such studies have successfully revealed variations in the gene repertoire and genomic diversity among closely related strains. For instance, we previously found, by WGPScanning and comparative genomic hybridization analyses, that a remarkable variation in genomic structure and gene repertoire is present among O157 EHEC isolates and that there are much larger differences between O157 and non-O157 EHEC strains (37, 38). These approaches, however, provide only limited information on strain-specific GEIs, because genomic regions longer than 20 kb are usually difficult to amplify by PCR and because comparative genomic hybridization analyses provide no information on the genes that are absent in the reference genomes. Therefore, in this study, we performed a systematic genome-wide survey of GEIs on the EPEC B171-8 genome by employing two methods, WGPScanning and fosmid mapping, in combination (Fig. 1). This approach allowed us to identify all of the GEIs larger than 10 kb, which comprise 13 prophages, four integrative elements, and five other GEIs (Table 3).
Nine of the 13 prophages identified on the B171-8 chromosome are lambda-like phages, and 3 are P2-like phages. The three P2-like phages exhibit a surprisingly high level of similarity to each other (Fig. 4). Multiple lysogenization of very similar bacteriophages has been described for the lambda-like phages of O157 (24, 39). Our present data indicate that multiple P2-like phages that contain nearly identical genome sequences can also be lysogenized and stably maintained in some E. coli strains. The replacement or sequence diversification of genes for phage immunity and integrase may allow this in B171-8 (Fig. 4). The sequence similarity of the lambda-like prophages of B171-8 is unknown. However, like the lambda-like phages of O157 (24, 48), they carry numerous virulence-related genes, especially those for non-LEE-encoded T3SS effectors, in the EELs (Fig. 3 and Table 4). One of the integrative elements identified is the LEE of this strain (Fig. 2), and other integrative elements also carry many virulence-related genes, including those for non-LEE-encoded T3SS effectors and nonfimbrial adhesins (Fig. 5 and Table 4).
FIG. 5.
Gene maps for three integrative elements and a prophage remnant identified in B171-8. The gene organizations and GC contents of three integrative elements and a prophage remnant are shown. The gene organizations of SpLE3 of O157 Sakai and KpLE2 of K-12 are also shown. The genes are colored according to their categories, as indicated in the box in Fig. 3.
Among the virulence-related genes found in B171-8, of particular interest are those for the two LifA/Efa1 homologues and the three AidA-I homologues. The LifA/Efa1 family was first identified in EPEC strain E2348/69 as lymphostatin, which induces a profound suppression of lymphocyte activity (29). This protein family was also identified as an adhesin in EHEC and is a factor essential for colonic colonization in Citrobacter rodentium (28, 35, 45). The lifA/efa1 homologue on GEI 4.36 has been interrupted by an IS insertion. However, the gene product may be functional, because it has been shown that the disruption of the N-terminal part of the lifA/efa1 homologue on SpLE3 of O157, which has a sequence almost identical to that on GEI 4.36 but is much shorter, resulted in reduced expression and secretion of LEE-encoded proteins and reduced adherence to cultured cells (44). Also of note is that the two LifA/Efa1 homologues of B171-8 show only 30% amino acid sequence identity, suggesting that they may have somewhat different functions. In contrast, the three AidA-I homologues are very similar to each other and also to that of O157 (88 to 96% identity to ECs1396). Among these, two homologues, on GEI 4.36 and GEI 4.52, appear to have very recently been duplicated in EPEC B171-8, because the 4.5-kb fragments on these two GEIs, which encode the AidA-I adhesin homologues as well as small GTP-binding proteins of unknown function, have identical nucleotide sequences. AidA-I is an autotransporter protein which has been shown to mediate adhesion to human cells in diffusely adherent intestinal pathogenic E. coli strains (3). The acquisition of multiple genes for AidA-I-like proteins may have conferred on EPEC B171-8 an increased ability to colonize the host intestine.
The identification of 34 genes encoding T3SS effectors or effector homologues may be important for future studies of B171-8 pathogenicity. T3SS-expressing pathogens export a cocktail of effectors into the host cell, and various responses are induced in the host cell by these effectors. The 34 T3SS effectors or effector homologues of B171-8 are classified into 22 effector protein families. Intriguingly, all but Cif and OspB are also present in EHEC O157. We explored the new effectors by examining the genes of unknown function identified on the GEIs, but none were translocated into the host cells. In a homology search of the draft sequence of B171-8 now available in the GenBank database (accession no. AAJX00000000), we detected no effector homologue other than those identified in this study and some effector-like proteins that are also present in LEE-negative E. coli strains (46). Thus, it is very likely that EPEC B171-8 contains a significantly smaller number of T3SS effectors than O157 does. Intact genes for the effector proteins belonging to the NleC, NleD, EspK, EspX, EspN, EspO, EspR, and EspJ families identified in O157 (46) are not present in EPEC B171-8, although little is known about their roles in the pathogenesis of O157 (12, 34, 48, 52). On the other hand, Cif and an OspB homologue were found in EPEC B171-8 but not in O157. Cif triggers an irreversible cytopathic effect characterized by the inhibition of the cell cycle G2/M-phase transition and the progressive recruitment of focal adhesion plaques leading to the assembly of stress fibers (33, 36, 46). The function of OspB is still unclear, and the OspB homologue found in B171-8 is highly divergent from that of Shigella flexneri (42), but the B171-8 OspB homologue was translocated into HeLa cells from B171-8 (see Table S6 in the supplemental material). These findings indicate that not only the presence or absence of Shiga toxin genes but also the difference in the repertoire of T3SS effectors should be considered in examining the pathogenicity of O157 and B171-8, representing the EHEC and EPEC pathotypes, respectively.
Finally, we briefly mention a methodological aspect of this study. New sequencing technologies are currently becoming widely available, although they were not available when we planned and performed this study. These technologies are very powerful for resequencing bacterial genomes but produce only short sequence data. Therefore, resequencing of relatively large bacterial genomes (E. coli genomes, for instance), which usually contain substantial amounts of repeat sequences, by these technologies yields highly fragmented draft sequences. These data are very useful for identifying single nucleotide polymorphisms in the conserved chromosome backbone, but it is not easy to directly use systematic analysis of accessory chromosomal elements, which are strain specific (no reference sequence available) and often contain many repeated sequences. For example, Manning et al. recently pyrosequenced an O157:H7 strain isolated in the 2006 spinach outbreak and obtained 201 large and 680 small contigs (32). Thus, we believe that the approach we employed in this study is still of significant usefulness to systematically identify strain-specific large genomic regions and that combining one of the new sequencing technologies with WGPScanning and/or fosmid mapping would be the most powerful approach.
Supplementary Material
Acknowledgments
This work was supported by a Grant-in-Aid for Scientific Research on Priority Area Applied Genomics from the Ministry of Education, Science, and Technology of Japan, a grant-in-aid from the Ministry of Health, Labor and Welfare of Japan (H17-Sinkou-ippan-019), and a grant from the Yakult Foundation.
We thank Akemi Yoshida, Yumiko Takeshita, and Noriko Kanemaru for their technical assistance.
Footnotes
Published ahead of print on 29 August 2008.
Supplemental material for this article may be found at http://jb.asm.org/.
REFERENCES
- 1.Barondess, J., and J. Beckwith. 1990. A bacterial virulence determinant encoded by lysogenic coliphage lambda. Nature 346871-874. [DOI] [PubMed] [Google Scholar]
- 2.Barondess, J., and J. Beckwith. 1995. bor gene of phage lambda, involved in serum resistance, encodes a widely conserved outer membrane lipoprotein. J. Bacteriol. 1771247-1253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Benz, I., and M. Schmidt. 1989. Cloning and expression of an adhesin (AIDA-I) involved in diffuse adherence of enteropathogenic Escherichia coli. Infect. Immun. 571506-1511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bertani, L., and E. Six. 1988. The P2-like phages and their parasite, P4. p. 73-143. In R. Calendar (ed.), The bacteriophages. Oxford University Press, Oxford, United Kingdom.
- 5.Blattner, F., G. R. Plunkett, C. Bloch, N. Perna, V. Burland, M. Riley, J. Collado-Vides, J. Glasner, C. Rode, G. Mayhew, J. Gregor, N. Davis, H. Kirkpatrick, M. Goeden, D. Rose, B. Mau, and Y. Shao. 1997. The complete genome sequence of Escherichia coli K-12. Science 2771453-1474. [DOI] [PubMed] [Google Scholar]
- 6.Brzuszkiewicz, E., H. Brüggemann, H. Liesegang, M. Emmerth, T. Olschläger, G. Nagy, K. Albermann, C. Wagner, C. Buchrieser, L. Emody, G. Gottschalk, J. Hacker, and U. Dobrindt. 2006. How to become a uropathogen: comparative genomic analysis of extraintestinal pathogenic Escherichia coli strains. Proc. Natl. Acad. Sci. USA 10312879-12884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Burrus, V., and M. Waldor. 2004. Shaping bacterial genomes with integrative and conjugative elements. Res. Microbiol. 155376-386. [DOI] [PubMed] [Google Scholar]
- 8.Charpentier, X., and E. Oswald. 2004. Identification of the secretion and translocation domain of the enteropathogenic and enterohemorrhagic Escherichia coli effector Cif, using TEM-1 beta-lactamase as a new fluorescence-based reporter. J. Bacteriol. 1865486-5495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Chen, H., and G. Frankel. 2005. Enteropathogenic Escherichia coli: unravelling pathogenesis. FEMS Microbiol. Rev. 2983-98. [DOI] [PubMed] [Google Scholar]
- 10.Chen, S., C. Hung, J. Xu, C. Reigstad, V. Magrini, A. Sabo, D. Blasiar, T. Bieri, R. Meyer, P. Ozersky, J. Armstrong, R. Fulton, J. Latreille, J. Spieth, T. Hooton, E. Mardis, S. Hultgren, and J. Gordon. 2006. Identification of genes subject to positive selection in uropathogenic strains of Escherichia coli: a comparative genomics approach. Proc. Natl. Acad. Sci. USA 1035977-5982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Clarke, S., R. Haigh, P. Freestone, and P. Williams. 2003. Virulence of enteropathogenic Escherichia coli, a global pathogen. Clin. Microbiol. Rev. 16365-378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Dahan, S., S. Wiles, R. La Ragione, A. Best, M. Woodward, M. Stevens, R. Shaw, Y. Chong, S. Knutton, A. Phillips, and G. Frankel. 2005. EspJ is a prophage-carried type III effector protein of attaching and effacing pathogens that modulates infection dynamics. Infect. Immun. 73679-686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Das, S., and K. Chaudhuri. 2003. Identification of a unique IAHP (IcmF associated homologous proteins) cluster in Vibrio cholerae and other proteobacteria through in silico analysis. In Silico Biol. 3287-300. [PubMed] [Google Scholar]
- 14.Deng, W., Y. Li, B. Vallance, and B. Finlay. 2001. Locus of enterocyte effacement from Citrobacter rodentium: sequence analysis and evidence for horizontal transfer among attaching and effacing pathogens. Infect. Immun. 696323-6335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Dobrindt, U. 2005. (Patho-)genomics of Escherichia coli. Int. J. Med. Microbiol. 295357-371. [DOI] [PubMed] [Google Scholar]
- 16.Durfee, T., R. Nelson, S. Baldwin, G. R. Plunkett, V. Burland, B. Mau, J. Petrosino, X. Qin, D. Muzny, M. Ayele, R. Gibbs, B. Csörgo, G. Pósfai, G. Weinstock, and F. Blattner. 2008. The complete genome sequence of Escherichia coli DH10B: insights into the biology of a laboratory workhorse. J. Bacteriol. 1902597-2606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Elliott, S., L. Wainwright, T. McDaniel, K. Jarvis, Y. Deng, L. Lai, B. McNamara, M. Donnenberg, and J. Kaper. 1998. The complete sequence of the locus of enterocyte effacement (LEE) from enteropathogenic Escherichia coli E2348/69. Mol. Microbiol. 281-4. [DOI] [PubMed] [Google Scholar]
- 18.Ewing, B., L. Hillier, M. Wendl, and P. Green. 1998. Base-calling of automated sequencer traces using phred. I. Accuracy assessment. Genome Res. 8175-185. [DOI] [PubMed] [Google Scholar]
- 19.Frankel, G., A. Phillips, I. Rosenshine, G. Dougan, J. Kaper, and S. Knutton. 1998. Enteropathogenic and enterohaemorrhagic Escherichia coli: more subversive elements. Mol. Microbiol. 30911-921. [DOI] [PubMed] [Google Scholar]
- 20.Gärtner, J., and M. Schmidt. 2004. Comparative analysis of locus of enterocyte effacement pathogenicity islands of atypical enteropathogenic Escherichia coli. Infect. Immun. 726722-6728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Girón, J., A. Ho, and G. Schoolnik. 1991. An inducible bundle-forming pilus of enteropathogenic Escherichia coli. Science 254710-713. [DOI] [PubMed] [Google Scholar]
- 22.Hacker, J., B. Hochhut, B. Middendorf, G. Schneider, C. Buchrieser, G. Gottschalk, and U. Dobrindt. 2004. Pathogenomics of mobile genetic elements of toxigenic bacteria. Int. J. Med. Microbiol. 293453-461. [DOI] [PubMed] [Google Scholar]
- 23.Hayashi, K., N. Morooka, Y. Yamamoto, K. Fujita, K. Isono, S. Choi, E. Ohtsubo, T. Baba, B. Wanner, H. Mori, and T. Horiuchi. 2006. Highly accurate genome sequences of Escherichia coli K-12 strains MG1655 and W3110. Mol. Syst. Biol. 22006.0007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hayashi, T., K. Makino, M. Ohnishi, K. Kurokawa, K. Ishii, K. Yokoyama, C. Han, E. Ohtsubo, K. Nakayama, T. Murata, M. Tanaka, T. Tobe, T. Iida, H. Takami, T. Honda, C. Sasakawa, N. Ogasawara, T. Yasunaga, S. Kuhara, T. Shiba, M. Hattori, and H. Shinagawa. 2001. Complete genome sequence of enterohemorrhagic Escherichia coli O157:H7 and genomic comparison with a laboratory strain K-12. DNA Res. 811-22. [DOI] [PubMed] [Google Scholar]
- 25.Johnson, T., S. Kariyawasam, Y. Wannemuehler, P. Mangiamele, S. Johnson, C. Doetkott, J. Skyberg, A. Lynne, J. Johnson, and L. Nolan. 2007. The genome sequence of avian pathogenic Escherichia coli strain O1:K1:H7 shares strong similarities with human extraintestinal pathogenic E. coli genomes. J. Bacteriol. 1893228-3236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Jores, J., S. Wagner, L. Rumer, J. Eichberg, C. Laturnus, P. Kirsch, P. Schierack, H. Tschäpe, and L. Wieler. 2005. Description of a 111-kb pathogenicity island (PAI) encoding various virulence features in the enterohemorrhagic E. coli (EHEC) strain RW1374 (O103:H2) and detection of a similar PAI in other EHEC strains of serotype O103:H2. Int. J. Med. Microbiol. 294417-425. [DOI] [PubMed] [Google Scholar]
- 27.Kaper, J., J. Nataro, and H. Mobley. 2004. Pathogenic Escherichia coli. Nat. Rev. Microbiol. 2123-140. [DOI] [PubMed] [Google Scholar]
- 28.Klapproth, J., M. Sasaki, M. Sherman, B. Babbin, M. Donnenberg, P. Fernandes, I. Scaletsky, D. Kalman, A. Nusrat, and I. Williams. 2005. Citrobacter rodentium lifA/efa1 is essential for colonic colonization and crypt cell hyperplasia in vivo. Infect. Immun. 731441-1451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Klapproth, J., I. Scaletsky, B. McNamara, L. Lai, C. Malstrom, S. James, and M. Donnenberg. 2000. A large toxin from pathogenic Escherichia coli strains that inhibits lymphocyte activation. Infect. Immun. 682148-2155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kovach, M., P. Elzer, D. Hill, G. Robertson, M. Farris, R. N. Roop, and K. Peterson. 1995. Four new derivatives of the broad-host-range cloning vector pBBR1MCS, carrying different antibiotic-resistance cassettes. Gene 166175-176. [DOI] [PubMed] [Google Scholar]
- 31.Loukiadis, E., R. Nobe, S. Herold, C. Tramuta, Y. Ogura, T. Ooka, S. Morabito, M. Kérourédan, H. Brugère, H. Schmidt, T. Hayashi, and E. Oswald. 2008. Distribution, functional expression, and genetic organization of Cif, a phage-encoded type III-secreted effector from enteropathogenic and enterohemorrhagic Escherichia coli. J. Bacteriol. 190275-285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Manning, S. D., A. S. Motiwal, A. C. Springman, W. Qi, D. W. Lacher, L. M. Ouellette, J. M. Mladonicky, P. Somsel, J. T. Rudrik, S. E. Dietrich, W. Zhang, B. Swaminathan, D. Alland, and T. S. Whittam. 2008. Variation in virulence among clades of Escherichia coli O157:H7 associated with disease outbreaks. Proc. Natl. Acad. Sci. USA 1054868-4873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Marchès, O., T. Ledger, M. Boury, M. Ohara, X. Tu, F. Goffaux, J. Mainil, I. Rosenshine, M. Sugai, J. De Rycke, and E. Oswald. 2003. Enteropathogenic and enterohaemorrhagic Escherichia coli deliver a novel effector called Cif, which blocks cell cycle G2/M transition. Mol. Microbiol. 501553-1567. [DOI] [PubMed] [Google Scholar]
- 34.Marchés, O., S. Wiles, F. Dziva, R. La Ragione, S. Schüller, A. Best, A. Phillips, E. Hartland, M. Woodward, M. Stevens, and G. Frankel. 2005. Characterization of two non-locus of enterocyte effacement-encoded type III-translocated effectors, NleC and NleD, in attaching and effacing pathogens. Infect. Immun. 738411-8417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Nicholls, L., T. Grant, and R. Robins-Browne. 2000. Identification of a novel genetic locus that is required for in vitro adhesion of a clinical isolate of enterohaemorrhagic Escherichia coli to epithelial cells. Mol. Microbiol. 35275-288. [DOI] [PubMed] [Google Scholar]
- 36.Nougayrède, J., F. Taieb, J. De Rycke, and E. Oswald. 2005. Cyclomodulins: bacterial effectors that modulate the eukaryotic cell cycle. Trends Microbiol. 13103-110. [DOI] [PubMed] [Google Scholar]
- 37.Ogura, Y., K. Kurokawa, T. Ooka, K. Tashiro, T. Tobe, M. Ohnishi, K. Nakayama, T. Morimoto, J. Terajima, H. Watanabe, S. Kuhara, and T. Hayashi. 2006. Complexity of the genomic diversity in enterohemorrhagic Escherichia coli O157 revealed by the combinational use of the O157 Sakai oligoDNA microarray and the whole genome PCR scanning. DNA Res. 133-14. [DOI] [PubMed] [Google Scholar]
- 38.Ogura, Y., T. Ooka, Asadulghani, J. Terajima, J. P. Nougayrède, K. Kurokawa, K. Tashiro, T. Tobe, K. Nakayama, S. Kuhara, E. Oswald, H. Watanabe, and T. Hayashi. 2007. Extensive genomic diversity and selective conservation of virulence-determinants in enterohemorrhagic Escherichia coli strains of O157 and non-O157 serotypes. Genome Biol. 8R138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Ohnishi, M., K. Kurokawa, and T. Hayashi. 2001. Diversification of Escherichia coli genomes: are bacteriophages the major contributors? Trends Microbiol. 9481-485. [DOI] [PubMed] [Google Scholar]
- 40.Ohnishi, M., J. Terajima, K. Kurokawa, K. Nakayama, T. Murata, K. Tamura, Y. Ogura, H. Watanabe, and T. Hayashi. 2002. Genomic diversity of enterohemorrhagic Escherichia coli O157 revealed by whole genome PCR scanning. Proc. Natl. Acad. Sci. USA 9917043-17048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Perna, N., G. R. Plunkett, V. Burland, B. Mau, J. Glasner, D. Rose, G. Mayhew, P. Evans, J. Gregor, H. Kirkpatrick, G. Pósfai, J. Hackett, S. Klink, A. Boutin, Y. Shao, L. Miller, E. Grotbeck, N. Davis, A. Lim, E. Dimalanta, K. Potamousis, J. Apodaca, T. Anantharaman, J. Lin, G. Yen, D. Schwartz, R. Welch, and F. Blattner. 2001. Genome sequence of enterohaemorrhagic Escherichia coli O157:H7. Nature 409529-533. [DOI] [PubMed] [Google Scholar]
- 42.Santapaola, D., M. Casalino, A. Petrucca, C. Presutti, C. Zagaglia, F. Berlutti, B. Colonna, and M. Nicoletti. 2002. Enteroinvasive Escherichia coli virulence-plasmid-carried apyrase (apy) and ospB genes are organized as a bicistronic operon and are subject to differential expression. Microbiology 1482519-2529. [DOI] [PubMed] [Google Scholar]
- 43.Spears, K., A. Roe, and D. Gally. 2006. A comparison of enteropathogenic and enterohaemorrhagic Escherichia coli pathogenesis. FEMS Microbiol. Lett. 255187-202. [DOI] [PubMed] [Google Scholar]
- 44.Stevens, M., A. Roe, I. Vlisidou, P. van Diemen, R. La Ragione, A. Best, M. Woodward, D. Gally, and T. Wallis. 2004. Mutation of toxB and a truncated version of the efa-1 gene in Escherichia coli O157:H7 influences the expression and secretion of locus of enterocyte effacement-encoded proteins but not intestinal colonization in calves or sheep. Infect. Immun. 725402-5411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Stevens, M., P. van Diemen, G. Frankel, A. Phillips, and T. Wallis. 2002. Efa1 influences colonization of the bovine intestine by Shiga toxin-producing Escherichia coli serotypes O5 and O111. Infect. Immun. 705158-5166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Taieb, F., J. Nougayrède, C. Watrin, A. Samba-Louaka, and E. Oswald. 2006. Escherichia coli cyclomodulin Cif induces G2 arrest of the host cell cycle without activation of the DNA-damage checkpoint-signalling pathway. Cell. Microbiol. 81910-1921. [DOI] [PubMed] [Google Scholar]
- 47.Tauschek, M., R. Strugnell, and R. Robins-Browne. 2002. Characterization and evidence of mobilization of the LEE pathogenicity island of rabbit-specific strains of enteropathogenic Escherichia coli. Mol. Microbiol. 441533-1550. [DOI] [PubMed] [Google Scholar]
- 48.Tobe, T., S. Beatson, H. Taniguchi, H. Abe, C. Bailey, A. Fivian, R. Younis, S. Matthews, O. Marches, G. Frankel, T. Hayashi, and M. Pallen. 2006. An extensive repertoire of type III secretion effectors in Escherichia coli O157 and the role of lambdoid phages in their dissemination. Proc. Natl. Acad. Sci. USA 10314941-14946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Tobe, T., T. Hayashi, C. Han, G. Schoolnik, E. Ohtsubo, and C. Sasakawa. 1999. Complete DNA sequence and structural analysis of the enteropathogenic Escherichia coli adherence factor plasmid. Infect. Immun. 675455-5462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Tobe, T., I. Tatsuno, E. Katayama, C. Wu, G. Schoolnik, and C. Sasakawa. 1999. A novel chromosomal locus of enteropathogenic Escherichia coli (EPEC), which encodes a bfpT-regulated chaperone-like protein, TrcA, involved in microcolony formation by EPEC. Mol. Microbiol. 33741-752. [DOI] [PubMed] [Google Scholar]
- 51.Vica Pacheco, S., O. García González, and G. Paniagua Contreras. 1997. The lom gene of bacteriophage lambda is involved in Escherichia coli K12 adhesion to human buccal epithelial cells. FEMS Microbiol. Lett. 156129-132. [DOI] [PubMed] [Google Scholar]
- 52.Vlisidou, I., O. Marchés, F. Dziva, R. Mundy, G. Frankel, and M. Stevens. 2006. Identification and characterization of EspK, a type III secreted effector protein of enterohaemorrhagic Escherichia coli O157:H7. FEMS Microbiol. Lett. 26332-40. [DOI] [PubMed] [Google Scholar]
- 53.Welch, R., V. Burland, G. R. Plunkett, P. Redford, P. Roesch, D. Rasko, E. Buckles, S. Liou, A. Boutin, J. Hackett, D. Stroud, G. Mayhew, D. Rose, S. Zhou, D. Schwartz, N. Perna, H. Mobley, M. Donnenberg, and F. Blattner. 2002. Extensive mosaic structure revealed by the complete genome sequence of uropathogenic Escherichia coli. Proc. Natl. Acad. Sci. USA 9917020-17024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Whale, A., R. Hernandes, T. Ooka, L. Beutin, S. Schüller, J. Garmendia, L. Crowther, M. Vieira, Y. Ogura, G. Krause, A. Phillips, T. Gomes, T. Hayashi, and G. Frankel. 2007. TccP2-mediated subversion of actin dynamics by EPEC 2—a distinct evolutionary lineage of enteropathogenic Escherichia coli. Microbiology 1531743-1755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Zhu, C., T. Agin, S. Elliott, L. Johnson, T. Thate, J. Kaper, and E. Boedeker. 2001. Complete nucleotide sequence and analysis of the locus of enterocyte effacement from rabbit diarrheagenic Escherichia coli RDEC-1. Infect. Immun. 692107-2115. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.