Abstract
The effect of nitrogen regulation on the level of transcriptional control has been investigated in a variety of bacteria, such as Bacillus subtilis, Corynebacterium glutamicum, Escherichia coli, and Streptomyces coelicolor; however, until now there have been no data for mycobacteria. In this study, we found that the OmpR-type regulator protein GlnR controls nitrogen-dependent transcription regulation in Mycobacterium smegmatis. Based on RNA hybridization experiments with a wild-type strain and a corresponding mutant strain, real-time reverse transcription-PCR analyses, and DNA binding studies using cell extract and purified protein, the glnA (msmeg_4290) gene, which codes for glutamine synthetase, and the amtB (msmeg_2425) and amt1 (msmeg_6259) genes, which encode ammonium permeases, are controlled by GlnR. Furthermore, since glnK (msmeg_2426), encoding a PII-type signal transduction protein, and glnD (msmeg_2427), coding for a putative uridylyltransferase, are in an operon together with amtB, these genes are part of the GlnR regulon as well. The GlnR protein binds specifically to the corresponding promoter sequences and functions as an activator of transcription when cells are subjected to nitrogen starvation.
Almost all of the macromolecules in a bacterial cell, including proteins, nucleic acids, and cell wall components, contain nitrogen. Consequently, prokaryotes have developed elaborate mechanisms to provide an optimal nitrogen supply for metabolism and to overcome and survive under nitrogen starvation conditions. We are especially interested in nitrogen metabolism and regulation in mycolic acid-containing actinomycetes. In the actinomycete group, the uptake and assimilation of nitrogen sources have been studied most intensively for Corynebacterium glutamicum (for reviews, see references 4, 5, 6, 9) and Streptomyces coelicolor (for a review, see reference 21).
In corynebacteria, like the amino acid-producing strains of C. glutamicum and Corynebacterium efficiens or the pathogen Corynebacterium diphtheriae, the expression of genes coding for proteins involved in uptake and assimilation of nitrogen sources is under the control of a central regulatory protein, the TetR-type regulator AmtR (14, 17, 28). In C. glutamicum AmtR blocks transcription of at least 33 genes (2), including genes which encode ammonium transporters (amtA and amtB), ammonium assimilation enzymes (glnA and gltBD), creatinine (crnT and codA) and urea (urtABCDE and ureABCEFGD) transport and metabolism, a number of biochemically uncharacterized enzymes and transport systems, and signal transduction proteins (glnD and glnK).
For S. coelicolor, it has been shown that various genes encoding nitrogen metabolism-related proteins are under the control of a different regulatory protein, the OmpR-type regulator GlnR (8, 26). GlnR acts as a transcriptional activator for at least 15 genes encoding proteins related to nitrogen uptake, metabolism, and regulation, as well as proteins with unknown functions. The GlnR-regulated genes include amtB encoding a putative ammonium uptake system, glnK and glnD coding for signal transduction proteins, the glutamine synthetase-encoding glnA and glnII genes, ureA encoding urease subunit gamma, and nirB coding for a large subunit of a putative nitrite reductase. Interestingly, a second GlnR regulator is encoded in the S. coelicolor genome; this regulator, GlnRII, also binds to the upstream regions of glnA, glnII, amtB, glnK, and glnD (8). The regulatory function of this protein is still unknown.
Less functional information is available for mycobacteria (10). Previous work on nitrogen uptake and assimilation concentrated mainly on the major pathogenic member of the genus, Mycobacterium tuberculosis. While extensive data are available for glutamine synthetases, especially the physiologically crucial GlnA1 enzyme (12), the associated adenylyltransferase GlnE (7, 18), and the uridylyl transferase GlnD (20), not much information is available for the transcriptional regulation of nitrogen metabolism in mycobacteria (19). In Mycobacterium smegmatis, homologs of all previously characterized genes encoding glutamine synthetases in M. tuberculosis are present in the genome (GlnA1, MSMEG_4290; GlnA2, MSMEG_4294; GlnA3, MSMEG_3561; GlnA4, MSMEG_2595), as are open reading frames encoding additional glutamine synthetase-like proteins with unknown physiological functions (e.g., MSMEG_1116, MSMEG_3827, MSMEG_5374, and MSMEG_6693).
In order to investigate the control of ammonium assimilation in M. smegmatis, we performed an in silico analysis using the previously described genome sequence of M. smegmatis mc2155 encoding homologs of known nitrogen control proteins and their corresponding cis-acting elements. The results of this analysis were verified experimentally.
MATERIALS AND METHODS
Bioinformatic analyses.
To screen genes encoding well-investigated nitrogen transcriptional regulators (amtR of C. glutamicum, as well as glnR and glnRII of S. coelicolor), genome data were obtained from The Institute for Genomic Research (http://www.tigr.org). To find the most representative homologs, we used single-genome protein BLAST available at the following genome server sites for well-characterized bacterial species: TigrBLAST (http://tigrblast.tigr.org/cmr-blast/) for mycobacterial species, CoryneRegNet (http://www.cebitec.uni-bielefeld.de) (1) for corynebacteria, and the S. coelicolor database ScoDB (http://streptomyces.org.uk). Sequence alignment was performed with CLUSTALW by using predefined algorithms of The European Bioinformatics Institute at The European Molecular Biology Laboratory (www.ebi.ac.uk/clustalw).
In a parallel analysis, all mycobacterial genomes available at the NCBI GenBank were screened for conserved cis elements using the previously described binding motifs of corynebacterial AmtR (2, 28) and Streptomyces GlnR (21) as query sequences. The PreDetector program (13) was used to screen the available mycobacterial genomes after successful positive control analyses were performed with the AmtR and GlnR binding motifs in the genomes of C. glutamicum and S. coelicolor, respectively. Screening for putative cis elements was performed by first creating positional weight matrices with different lengths (in bp) for the known cis elements of AmtR and GlnR with the Weight Matrix Creation module of PreDetector and then using these weight matrices in the Regulon Prediction module with the genomes of interest.
Strains and growth conditions.
Bacteria were routinely grown at 37°C in baffled flasks with agitation. Mycobacterial strains were grown in Middlebrook 7H9 liquid medium (Difco Laboratories) (containing [per 900 ml] approximately 0.5 g ammonium sulfate, 0.5 g l-glutamic acid, 0.1 g sodium citrate, 1.0 mg pyridoxine, 0.5 mg biotin, 2.5 g disodium phosphate, 1.0 g monopotassium phosphate, 0.04 g ferric ammonium citrate, 0.05 g magnesium sulfate, 0.5 mg calcium chloride, 1.0 mg zinc sulfate, and 1.0 mg copper sulfate) supplemented with 0.2% glycerol and 0.05% Tween 80 or on Middlebrook 7H9 medium (Difco Laboratories) containing 1.5% agar supplemented with 0.2% glycerol. When appropriate, antibiotics were added at the following concentrations: hygromycin, 200 μg ml−1 for Escherichia coli and 50 μg ml−1 for M. smegmatis; kanamycin, 30 μg ml−1 for E. coli and 10 μg ml−1 for M. smegmatis; and streptomycin, 400 μg ml−1 for M. smegmatis. In order to study the effects of nitrogen starvation, a fresh M. smegmatis culture was used to inoculate Middlebrook 7H9 medium for overnight growth. This culture, which had an optical density at 600 nm (OD600) of approximately 2 to 3 after overnight growth, was used to inoculate fresh Middlebrook 7H9 medium to obtain an OD600 of approximately 0.2, and cells were grown for 10 to 11 h until the exponential growth phase was reached (OD600, approximately 0.6 to 0.8). To induce nitrogen starvation, cells were harvested by centrifugation, washed, and resuspended in prewarmed Middlebrook 7H9 medium without a nitrogen source (Middlebrook 7H9 medium lacking ammonium sulfate and glutamic acid and containing iron citrate instead of iron ammonium citrate). As a control, unstarved cells were harvested, washed, and transferred using Middlebrook 7H9 medium. Alternatively, nitrogen limitation was induced by addition of methionine sulfoximine (MSX) (final concentration, 200 μM) to Middlebrook 7H9 medium. This substrate analog inhibits glutamine synthetase activity and consequently blocks nitrogen metabolism (15, 16).
General molecular biology techniques.
For plasmid isolation, transformation, and cloning, standard techniques were used (22). E. coli strain JM109 (31) was used as the cloning host. Plasmids were subsequently transferred into competent M. smegmatis cells by electroporation. Chromosomal DNA was extracted from 100-ml cultures grown to stationary phase as described previously (3). DNA sequence analyses were carried out using a BigDye Terminator V3.1 cycle sequencing kit (Perkin Elmer).
Construction of a glnR deletion strain.
In order to generate a glnR deletion, two DNA fragments flanking the glnR gene (1 kb up- and downstream of glnR) were PCR amplified using chromosomal DNA from M. smegmatis strain SMR5 as the template. For subsequent cloning in the pML814 vector (ColE1 origin, FRT-hyg-FRT rpsL, Ampr Hygr; 6,220 bp; general deletion vector; kindly provided by M. Niederweis), SwaI and PacI restriction sites were introduced into the primer sequences used for amplification of the upstream sequence of glnR and SpeI and PmeI restriction sites were introduced into the primer sequences used for amplification of the downstream sequence of glnR (see Table S1 in the supplemental material). The resulting plasmid, pML814ΔglnR, carried an FRT-hyg-FRT expression cassette (24) flanked by the sequences upstream and downstream of glnR. M. smegmatis SMR5, a streptomycin-resistant derivative of M. smegmatis mc2155, was transformed with plasmid pML814ΔglnR, and transformants were selected on hygromycin-containing plates to obtain a single crossover (24). After verification of the single-crossover event using PCR, cells were selected on plates containing hygromycin and streptomycin. Clones on these plates should have lost the vector and should have had the FRT-hyg-FRT cassette integrated into the chromosome. After verification of the double-crossover event using PCR, the FLP recombinase was used to specifically remove the hyg gene from the chromosome, generating a marker-free deletion mutant and allowing the hyg gene to be reused as resistance marker. Selection of clones was carried out using plates containing hygromycin and streptomycin. Deletion of glnR in the resulting strain, MH1, was verified by PCR and Southern blotting (data not shown).
Construction of antisense probes.
For generation of antisense probes, internal DNA fragments of the corresponding genes were amplified by PCR (the primers used for the different probes are shown in Table S1 in the supplemental material). The reverse primers encoded the promoter region for T7 polymerase, which allowed in vitro transcription of probes using T7 polymerase.
RNA preparation, hybridization analyses, and real-time reverse transcription-PCR (RT-PCR).
M. smegmatis RNA was prepared from 6-ml culture samples using a NucleoSpin RNA II kit (Macherey Nagel, Düren, Germany). If necessary, a second DNase digestion was performed with Turbo DNase (Ambion) to completely remove the chromosomal DNA. RNA samples were stored at −80°C.
Antisense probes that were between 0.2 and 0.5 kb long and were used for analysis of gene transcription were generated by PCR and subsequent labeling with a digoxigenin (DIG) RNA-labeling mixture (Roche, Mannheim, Germany) and T7 polymerase (NEB, Frankfurt, Germany). RNA (1 μg per time point) was spotted onto nylon membranes using a Schleicher & Schuell (Dassel, Germany) Minifold I dot blotter. Hybridization of DIG-labeled RNA probes was detected with X-ray film (Amersham Hyperfilm MP; GE Healthcare) using alkaline phosphatase-conjugated anti-DIG Fab fragments and chloro-5-substituted adamantyl-1,2-dioxetane phosphate (CSPD) as a light-emitting substrate as recommended by the supplier (Roche, Mannheim, Germany). All experiments were carried out at least in duplicate with independent cultures (biological replicates).
For real-time RT-PCR, an MyiQ single-color real-time PCR detection system (Bio-Rad, Munich, Germany), a QuantiTect SYBR green RT-PCR kit (Qiagen, Hilden, Germany), primers at a concentration of 1 μM, and 100 ng of template RNA were used. RT was carried out at 50°C for 30 min, and the reverse transcriptase was inactivated and the polymerase was activated by 15 min of incubation at 95°C. The PCR was carried out by using 40 cycles of DNA denaturation for 15 s at 94°C, primer annealing for 30 s at 60°C, and DNA polymerization for 15 s at 72°C. The PCR was followed by a melting curve program (55 to 100°C with a heating rate of 1°C per 10 s) and then a cooling program (25°C). No-template controls were included with all reactions. Data were analyzed using the MyiQ single-color real-time PCR detection system software.
Construction of glnR-carrying plasmids.
For complementation assays, the glnR gene was amplified by PCR using chromosomal DNA of M. smegmatis as the template and oligonucleotides glnR-fw-PacI and glnR-rev-SwaI (see Table S1 in the supplemental material). The PCR products were ligated to plasmid pMN016 (psmyc-mspA, ColE1 origin, PAL5000 origin, Hygr; 6,164 bp), (25) using the SwaI and PacI sites, which was introduced by using the oligonucleotide primers. The resulting plasmid, pMN016-glnR, was sequenced and used as a control.
For purification of GlnR for gel retardation assays, the glnR gene was amplified by PCR (for the primers used, see Table S1 in the supplemental material) and ligated to plasmid pQE70 (Qiagen, Hilden, Germany) in order to add a C-terminal His6 tag to the expressed protein, resulting in plasmid pQE-glnR-His. For expression of His-tagged GlnR in M. smegmatis, the corresponding DNA fragment was excised from plasmid pQE-glnR-His by HindIII/SphI restriction and ligated to dephosphorylated and HindIII/SphI-cut plasmid pMN016, resulting in plasmid pMN016-glnR-His.
Purification of GlnR and gel retardation experiments.
For gel shift assays, protein extracts were prepared from M. smegmatis wild-type strain SMR5 carrying plasmid pMN016-glnR-His. Cells were cultivated in Middlebrook 7H9 medium containing antibiotics as described above, harvested by centrifugation (4,000 × g, 15 min, 4°C), and suspended in 300 mM NaCl-50 mM NaH2PO4-20 mM imidazole (pH [NaOH] 8.0) (2 ml g cells−1) containing lysozyme (2 mg ml−1) and Complete protease inhibitor as recommended by the supplier (Roche, Mannheim, Germany). Cells were subsequently disrupted by ultrasonic treatment. Cell debris was removed by centrifugation (14,000 × g, 30 min, 4°C), and the protein extract was loaded on a 5-ml Ni-nitrilotriacetic acid column in a chromatography apparatus (Äkta prime; GE Healthcare, Munich, Germany). Washing and subsequent elution with 300 mM NaCl-50 mM NaH2PO4-500 mM imidazole (pH [NaOH] 8.0) were carried out as recommended by the supplier of the Ni-nitrilotriacetic acid matrix (GE Healthcare, Munich, Germany); the purified protein was stored at −80°C.
Target DNA for the gel shift assays was synthesized by PCR (for the primers used, see Table S1 in the supplemental material) and was purified by agarose gel electrophoresis. To label the DNA and to prepare the reaction mixture for the gel shift assay, a DIG gel shift kit (Roche, Mannheim) was used as recommended by the supplier. Separation by gel electrophoresis was performed in native 6% polyacrylamide gels (Anamed Electrophorese GmbH, Darmstadt, Germany) using 0.5× Tris-borate-EDTA buffer as the running buffer. Subsequently, the labeled DNA was transferred to a nylon membrane (Roche, Mannheim, Germany) by electroblotting as described in the protocol of the DIG gel shift kit (Roche, Mannheim, Germany). For detection of the labeled DNA, X-ray film was used.
DNA affinity purification of GlnR.
For DNA affinity purification with magnetic beads, target DNAs were amplified by PCR using biotinylated oligonucleotides (see Table S1 in the supplemental material). For preparation of DNA-coated magnetic beads, M-280 Dynabeads coated with streptavidin (Dynal, Oslo, Norway) were washed and resuspended in 200 μl of 1× DNA binding buffer (5 mM Tris-HCl [pH 7.5], 0.5 mM EDTA, 1 M NaCl), and biotinylated target DNA was added. After incubation for 30 min at room temperature to allow DNA binding to the streptavidin-coated magnetic beads, the beads were washed three times with 500 μl of 1× DNA binding buffer and subsequently stored at 4°C in Tris-EDTA buffer containing 0.02% NaN3. Before use, the DNA-coated magnetic beads were washed three times in 1 ml ice-cold protein binding buffer (phosphate-buffered saline) (pH 7.4) (22). For preparation of protein, M. smegmatis wild-type strain SMR5 was grown in 500 ml Middlebrook 7H9 medium until the OD600 was approximately 2. Cells were harvested by centrifugation (4,000 × g, 15 min, 4°C) and resuspended in 4 ml ice-cold lysis buffer (50 mM Tris-HCl, 70 mM KCl, 1 mM EDTA, 1 mM dithiothreitol, 10% glycerol, 400 μl Roche Complete protease inhibitor; pH 8.0). The cell suspension was transferred to tubes containing glass beads, and the cells were disrupted by vigorous shaking at 6.5 m s−1 for 1 min using a FastPrep FP120 instrument (Q-BIOgene, Heidelberg, Germany). The cell debris and glass beads were removed by centrifugation (14,000 × g, 4 min, 4°C) before the membranes were separated by ultracentrifugation (267,000 × g, 30 min, 4°C). One-milliliter aliquots of the supernatant containing the cytoplasmic proteins were incubated with the DNA-coated beads for 45 min at 4°C with continuous agitation. The beads were separated from the protein extract and washed twice with 500 μl ice-cold protein binding buffer. This procedure was repeated three times. Finally, the proteins bound to the DNA were eluted. To do this, the beads were resuspended in 20-μl portions of elution buffer (phosphate-buffered saline with the sodium chloride concentration increasing from 200 mM to 1,000 mM in 100 mM steps). The elution fractions were analyzed by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) (23), and proteins were identified by peptide mass fingerprinting.
Mass spectrometry.
Matrix-assisted laser desorption ionization-time of flight mass spectrometry (MALDI-TOF MS) and peptide mass fingerprint analysis were carried out by the bioanalytics service unit at the Center for Molecular Medicine of Cologne.
RESULTS
In silico analysis of transcription regulators and corresponding binding sites.
A bioinformatic analysis was carried out to identify M. smegmatis homologs of known global transcriptional regulators of nitrogen control in actinomycetes. Interestingly, both a homolog of the C. glutamicum regulator AmtR and a protein with a high level of identity to GlnR, the regulator of nitrogen metabolism in S. coelicolor, were found in M. smegmatis. M. smegmatis AmtR shows 42% identity to C. glutamicum AmtR, and GlnR exhibits 55% amino acid identity to S. coelicolor GlnR. However, when the previously described genome sequences of M. tuberculosis, Mycobacterium bovis, and Mycobacterium avium were screened, only a homolog of glnR was found. Obviously, GlnR proteins are highly conserved in actinobacteria (Fig. 1) (26), and based on their wide distribution in different genera, including Actinomyces, Amycolatopsis, Arthrobacter, Bifidobacterium, Frankia, Leifsonia, Mycobacterium, Nocardia, Rhodococcus, Streptomyces, and Thermobifida, they are most likely involved in nitrogen regulation in mycobacteria. While no homologs of the S. coelicolor GlnR protein have been found in the available corynebacterial genomes, the organization of the genome sequences of the genera mentioned above is conserved and the genomes contain an apparently monocistronic operon with no open reading frames close to the corresponding glnR genes.
FIG. 1.
Alignment of GlnR proteins from various actinomycetes. Abbreviations used for GlnR homologs with annotation numbers: MSMEG, Mycobacterium smegmatis; MAP, Mycobacterium avium subsp. paratuberculosis; Mb, Mycobacterium bovis; Rv, Mycobacterium tuberculosis H37rv; SCO, Streptomyces coelicolor; SAV, Streptomyces avermitilis; nfa, Nocardia farcinica; RHA1, Rhodococcus sp. strain RHA1; FRAAL, Frankia alni ACN14a; Francci3, Frankia sp. strain Cci3; Tfu, Thermobifida fusca; BL, Bifidobacterium longum; AAur, Arthrobacter aurescens; Lxx, Leifsonia xylii; ANA, Actinomyces naeslundii; AAN77733, Amycolatopsis mediterranei. Identical amino acid residues are indicated by a black background, and similar amino acids are indicated by a gray background. M. smegmatis GlnR contains a putative phosphorylation pocket, located at amino acid residues 43 to 53, and a C-terminal DNA binding domain (helix turn helix), located at amino acid residues 144 to 218 (indicated by a bar above the sequence). *, identical residues; :, conserved substitutions/residues; ., semiconserved substitutions.
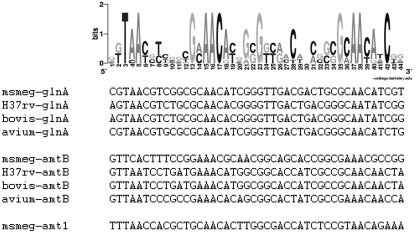
Using a parallel approach, in which the genomes of M. smegmatis, M. tuberculosis, M. bovis, and M. avium were screened for known cis-acting elements of AmtR (2, 28), corresponding binding sites could not be identified. In contrast, using the GlnR motifs of S. coelicolor, Streptomyces avermitilis, and Streptomyces scabies (21) as query sequences, putative binding sites were detected in the available mycobacterial genomes, and these sites included three highly conserved cis elements in M. smegmatis (Fig. 2). These putative binding motifs were located upstream of the glnA gene (msmeg_4290), which codes for glutamine synthetase and is homologous to the M. tuberculosis glnA1 gene coding for the physiologically crucial GS enzyme, as well as upstream of amtB (msmeg_2425) and amt1 (msmeg_6259), which are ammonium permease-encoding genes. No GlnR cis elements were detected upstream of urease subunit-encoding genes (msmeg_1091 and msmeg_3627), glutamate synthase-encoding genes (msmeg_3225, msmeg_5594, and msmeg_6459), or other genes encoding putative glutamine synthetases (msmeg_1116, msmeg_2595, msmeg_3561, msmeg_3827, msmeg_4294, msmeg_5374, and msmeg _6693).
FIG. 2.
GlnR binding motif in mycobacteria. Sequence logos of putative GlnR cis elements were identified upstream of the M. smegmatis (msmeg), M. tuberculosis (H37rv), M. bovis (bovis), and M. avium (avium) glnA, amtB, and amt1 genes (amt1 is present only in the M. smegmatis genome). The standard code of the Weblogo server is shown in gray scale at the top.
The M. smegmatis amtB gene (msmeg_2425) is localized in a gene cluster together with glnK (msmeg_2426), which encodes a PII-type signal transduction protein, and glnD (msmeg_2427), which encodes a putative uridylyltransferase. By using RT-PCR, a common transcript of these three genes was detected (data not shown), adding glnK and glnD to the putative GlnR regulon.
DNA affinity purification of GlnR.
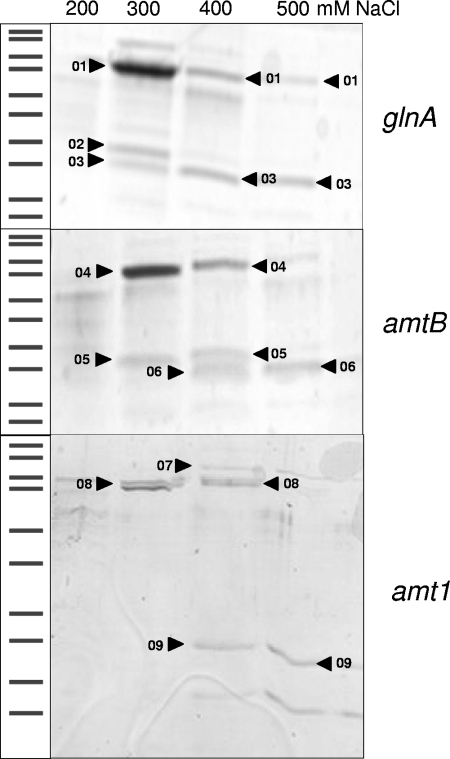
As described above, putative GlnR binding motifs were detected upstream of the glnA gene, as well as upstream of amtB and amt1. The corresponding promoter regions, including the GlnR binding site, were amplified by PCR and bound to magnetic beads. The beads were incubated with M. smegmatis cytoplasmic proteins. Subsequently, bound proteins were eluted, separated by SDS-PAGE (Fig. 3), and identified by MALDI-TOF MS and peptide mass fingerprinting (Table 1). In addition to transcription termination factor Rho, DNA topoisomerase I, and a protein annotated as a hypothetical protein, GlnR was isolated using this approach. In fact, GlnR was purified with all promoter DNA sequences used, supporting the hypothesis that there is transcriptional control of these genes by this regulator. Although binding of GlnR to the promoter regions used seemed to be specific, as indicated by the high NaCl concentrations necessary for elution, in principle, binding of GlnR could also be explained by a general DNA binding property of the protein. Therefore, genetic analyses were carried out to further characterize the GlnR function.
FIG. 3.
DNA affinity purification of GlnR. Magnetic beads coated with glnA, amtB, and amt1 promoter DNA were incubated with M. smegmatis cytoplasmic proteins. After washing steps, proteins bound to the DNA fragments were eluted using buffers containing different NaCl concentrations. Eluted proteins were separated by SDS-PAGE and subjected to tryptic in-gel digestion, MALDI-TOF MS, and peptide mass fingerprint analyses. The numbers indicate proteins identified by this approach, as shown in Table 1.
TABLE 1.
Proteins isolated by DNA affinity purificationa
| Accession no. | Protein | No. | Mol wt | Sequence coverage (%) | Promoter DNA |
|---|---|---|---|---|---|
| MSMEG_4954 | Transcription termination factor Rho | 01 | 71,762 | 60 | glnA |
| MSMEG_3081 | Hypothetical protein | 02 | 34,499 | 71 | glnA |
| MSMEG_5784 | GlnR | 03 | 27,933 | 39-60 | glnA |
| MSMEG_4954 | Transcription termination factor Rho | 04 | 71,762 | 60 | amtB |
| MSMEG_3081 | Hypothetical protein | 05 | 34,499 | 62-71 | amtB |
| MSMEG_5784 | GlnR | 06 | 27,933 | 47-54 | amtB |
| MSMEG_6157 | DNA topoisomerase I | 07 | 102,137 | 46 | amt1 |
| MSMEG_4954 | Transcription termination factor Rho | 08 | 71,762 | 37 | amt1 |
| MSMEG_5784 | GlnR | 09 | 27,933 | 26 | amt1 |
Identification of proteins after DNA affinity purification was carried out by performing tryptic in-gel digestion, MALDI-TOF MS, and peptide mass fingerprint analyses. The numbers are the same as the numbers in Fig. 3.
GlnR-dependent transcriptional response to nitrogen starvation.
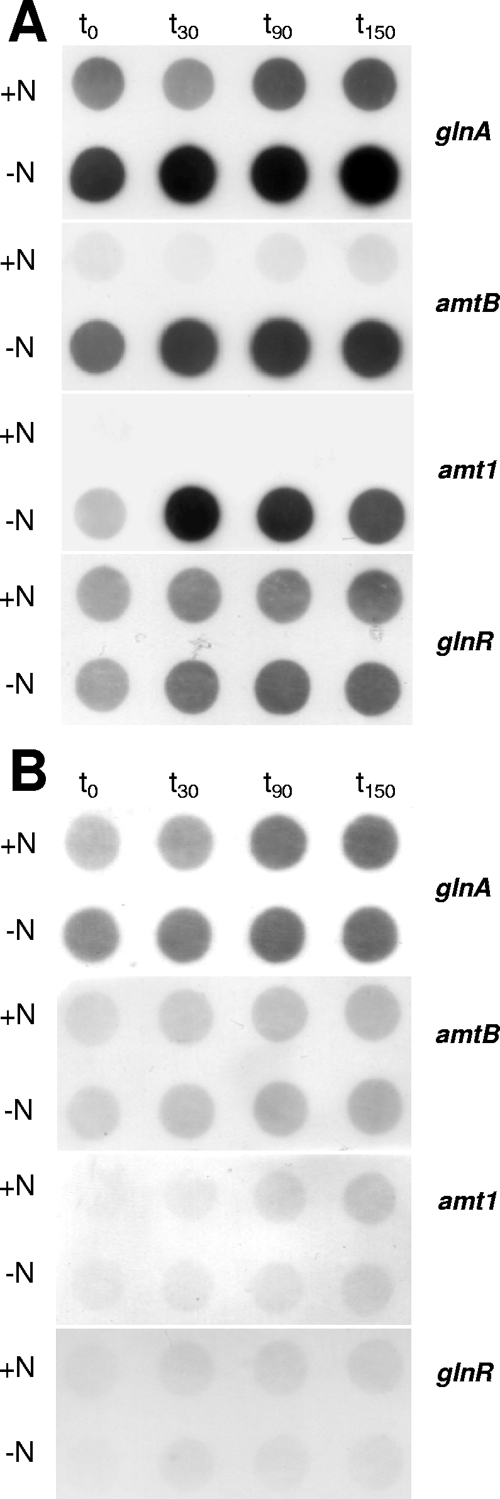
To validate the data obtained by in silico analyses and DNA affinity purification and to elucidate the putative role of GlnR in nitrogen control in M. smegmatis, a glnR deletion was introduced into the M. smegmatis genome. During growth on Middlebrook 7H9 medium, no difference between the wild-type and mutant strains was observed. The doubling times were between 4.5 and 5 h (data not shown). The response to nitrogen deprivation (i.e., the response after transfer of the bacteria to nitrogen-free Middlebrook 7H9 medium) was analyzed by performing RNA hybridization experiments. The transcription profiles of the M. smegmatis wild-type strain and ΔglnR strain MH1 were compared for the glnA, amtB, and amt1 genes. As expected based on the results of the bioinformatic analyses and DNA affinity purification experiments described above, the transcript levels of these genes increased during nitrogen deprivation, while no increase in the level of transcription of glnA, amtB, and amt1 was observed in glnR mutant strain MH1. This suggests that GlnR is in fact a nitrogen-dependent regulator in M. smegmatis and also indicates that GlnR works as an activator of transcription (Fig. 4A). The putative expression regulation of GlnR itself was tested by performing RNA hybridization using a glnR probe (Fig. 4B). Due to deletion of glnR, strain MH1 showed no glnR transcription signal, while the wild-type samples exhibited constitutive expression, which was not significantly altered upon nitrogen deprivation. This result indicates that glnR transcription is not subject to nitrogen control.
FIG. 4.
Transcriptional responses of the M. smegmatis wild-type strain and glnR deletion strain MH1 to nitrogen deprivation: hybridization of RNA isolated from M. smegmatis cells before and after induction of nitrogen starvation by incubation in nitrogen-free medium. (A) Transcription of the glutamine synthetase-encoding glnA gene, amtB and amt1 coding for ammonium uptake systems, and glnR in the wild type. The time points indicated (t0, t30, t90, and t150) are sampling times (in min) after resuspension of centrifuged and washed cells. One microgram of total RNA per spot was applied. (B) Transcription of genes in M. smegmatis glnR deletion strain MH1.
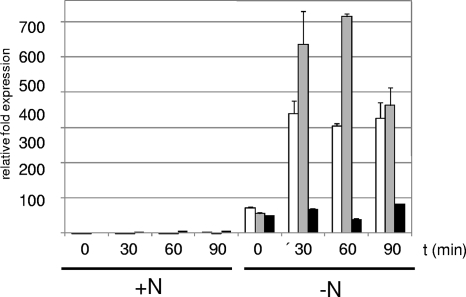
The regulation of mRNA levels in response to nitrogen starvation was also analyzed by performing real-time RT-PCR experiments. For the wild type (Fig. 5), these experiments showed that, compared to the levels after growth in Middlebrook 7H9 medium, the levels of transcripts were significantly increased after the washing step in Middlebrook 7H9 medium without a nitrogen source by factors of 50% ± 1% for glnA, 72% ± 3% for amtB, and 56% ± 3% for amt1. After 30 min the transcript levels were increased by 64% ± 4% for glnA, 340% ± 35% for amtB, and 537% ± 95% for amt1. For all time points, the upregulation factor was significantly higher for the ammonium transporter-encoding genes amtB and amt1 than for glnA. Again, regulation of glnR was not observed (data not shown). For ΔglnR mutant MH1 no increases in mRNA levels in response to nitrogen starvation were observed. In fact, when the steady-state level for the wild type (zero time) was defined as 100%, the levels of transcription in the mutant were 19% ± 2% for glnA, 12% ± 1% for amtB, and 31% ± 1% for amt1, which is consistent with an activator function of GlnR. Moreover, glutamine synthetase activity was reduced in the mutant. While 0.230 ± 0.016 U (mg protein)−1 was observed in the wild type, only 0.148 ± 0.009 U (mg protein)−1 was observed in strain MH1.
FIG. 5.
Real-time RT-PCR of the M. smegmatis wild-type strain. Cells were grown until the exponential growth phase was reached (OD600, approximately 0.6 to 0.8). To induce nitrogen starvation, cells were harvested, washed, and resuspended in prewarmed Middlebrook 7H9 medium without a nitrogen source (−N); prewarmed standard Middlebrook 7H9 medium was used as a control (+N). RNA was isolated at the indicated time points, and transcription of amtB (open bars), amt1 (gray bars), and glnA (black bars) was monitored by real-time RT-PCR. For each gene the control value at zero time was defined as 1.
Induction of GlnR-dependent transcription response by MSX treatment.
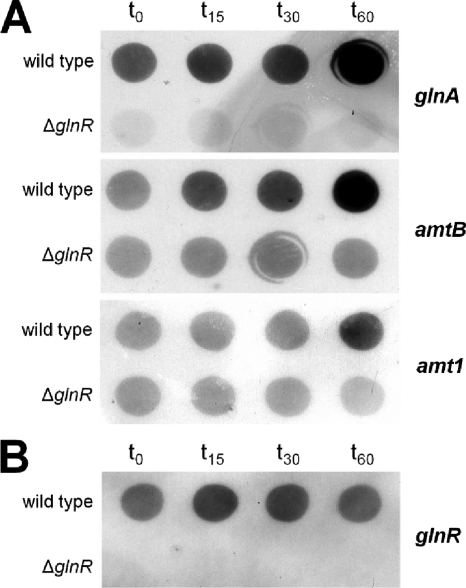
As an alternative approach for inducing nitrogen starvation without changing the medium, the glutamate analog MSX was tested. MSX is known to inactivate glutamine synthetase and impair ammonium assimilation (15, 16). The growth tests carried out showed that addition of MSX to a final concentration of 200 μM did not impair growth of the wild type (doubling time, 4.5 to 5.0 h), while glnR mutant MH1 stopped growing after approximately 4 h of MSX treatment. At the mRNA level, within 15 min after addition of MSX enhanced transcription of glnA and amtB was observed in the wild type, while an increase in amt1 transcription was detected after 60 min (Fig. 6A), indicating that MSX in fact induces the cellular nitrogen starvation response at the level of transcription. As expected, this response at the level of transcription of glnA, amtB, and amt1 was not observed in glnR mutant strain MH1. Expression regulation of GlnR itself was also tested by RNA hybridization using a glnR probe (Fig. 6B). As expected, wild-type samples exhibited constitutive expression of glnR, which was not significantly altered upon addition of MSX, confirming that glnR transcription is not subject to nitrogen control.
FIG. 6.
Induction of the nitrogen starvation response by MSX: hybridization of RNA isolated from M. smegmatis cells before (t0) and after addition of MSX. t15, 15 min after addition of MSX; t30, 30 min after addition of MSX; t60, 60 min after addition of MSX. (A) Transcription of the glutamine synthetase-encoding glnA gene, as well as amtB and amt1 coding for ammonium uptake systems. (B) Transcription of M. smegmatis glnR.
Complementation of strain MH1 by M. smegmatis glnR.
To exclude the possibility of a polar effect of the glnR deletion, a complementation experiment was carried out. To do this, strain MH1 was transformed with plasmid pMN016 (control) and with the glnR-carrying plasmid pMN016-glnR (Fig. 7). Transcription of glnR was tested as a control (Fig. 7A). While the control plasmid without the glnR gene had no influence on transcription of glnA, amtB, and amt1, transcription of these genes in response to nitrogen starvation was increased as it was in the wild type when glnR was supplied in trans by plasmid pMN016-glnR (Fig. 7B). Consequently, the lack of nitrogen control in strain MH1 was caused exclusively by the glnR deletion and could be cured by complementation with a plasmid-encoded glnR gene.
FIG. 7.
Complementation of M. smegmatis glnR deletion strain MH1: hybridization of RNA isolated from M. smegmatis cells carrying control plasmid pMN016 or glnR delivery plasmid pMN016-glnR before (t0) and after induction of nitrogen starvation by addition of MSX. t15, 15 min after addition of MSX; t30, 30 min after addition of MSX; t60, 60 min after addition of MSX. (A) Transcription control for M. smegmatis glnR. (B) Transcription of glnA, amtB, and amt1.
Gel retardation assays.
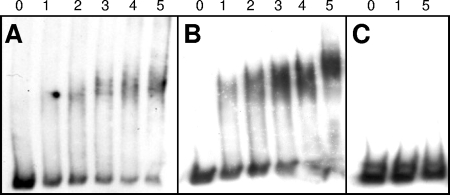
DNA affinity purification revealed that GlnR is able to bind to the promoter regions of glnA, amtB, and amt1. As an independent approach to verify GlnR binding, gel retardation experiments were carried out using corresponding promoter DNAs and cell extracts from the M. smegmatis wild-type strain and glnR deletion mutant MH1. As expected, a promoter DNA fragment of the amtB gene was shifted by the wild-type cell extract, while identical amounts of MH1 proteins had no influence on DNA mobility (data not shown). Nevertheless, these experiments with cell extracts did not exclude the possibility of indirect effects of GlnR on DNA motility. Therefore, GlnR was purified and used as an isolated protein in gel retardation experiments (Fig. 8). Again, the promoter regions of amtB and glnA exhibited decreased motility in the gel when GlnR protein was added (Fig. 8A and B), showing that this transcriptional regulator binds to the corresponding DNA. To exclude the possibility of nonspecific, general DNA binding activity of GlnR, the corresponding glnR promoter DNA was tested. As shown by the RNA hybridization experiments described above (Fig. 4 and 6), glnR expression was not autoregulated, and in fact, no binding of GlnR was observed (Fig. 8C). Furthermore, promoter regions of urease-encoding genes (ureE1 [msmeg_1091] and ureA2 [msmeg_3627]) were tested, and no retardation was observed for these DNA fragments (data not shown). Together, these data indicate that there was specific binding of GlnR to the promoter regions of genes, showing that there was GlnR-dependent upregulation of transcription upon nitrogen deprivation.
FIG. 8.
Gel retardation assay. amtB (220 bp) (A), glnA (219 bp) (B), and glnR (249 bp) (C) apparent promoter fragments were incubated with different amounts of purified GlnR protein. Lanes 0, 1, 2, 3, 4, and 5 contained 0, 1, 2, 3, 4, and 5 μl of a 0.3-μg μl−1 solution, respectively.
DISCUSSION
Previous work on nitrogen metabolism and its regulation in mycobacteria concentrated strongly on glutamine synthetase in M. tuberculosis, which is essential in this bacterium and consequently an important drug target (11, 12, 27). Additionally, studies of the glnE gene product adenylyltransferase, which is crucial for posttranslational modification and regulation of glutamine synthetase, and the glnD-encoded uridylyltransferase, which is putatively involved in nitrogen signal transduction, were carried out (11, 12, 18, 20). In this paper, we describe the first in vivo characterization of GlnR as a regulator of nitrogen control in mycobacteria. In M. smegmatis, GlnR binding sites were identified upstream of the glnA, amtB, and amt1 genes. As in other actinomycetes (14), the M. smegmatis amtB gene forms an operon with downstream glnK and glnD genes. A core ammonium assimilation regulon seems to be controlled by M. smegmatis GlnR and to regulate transcription of glnA, which encodes glutamine synthetase, the ammonium transporter-encoding genes amtB and amt1, and corresponding signal transduction components encoded by glnK and glnD. The signal transduction to M. smegmatis GlnR is unclear, a situation which is similar to that in S. coelicolor. Based on the conserved phosphorylation domain and analogous to the situation in E. coli (32), involvement of a protein kinase is assumed. However, the corresponding protein is unknown.
Based on the data obtained for M. smegmatis in this study and the data obtained in previous studies focusing on S. coelicolor (8, 26, 29, 30) and Amycolatopsis mediterranei (33, 34), GlnR seems to be a major regulator of ammonium assimilation in actinomycetes. This conclusion is supported by the results of electrophoretic mobility shift assays, which showed that S. coelicolor GlnR is able to bind to the glnA promoter regions of Bifidobacterium longum, Frankia sp. strain EAN1, M. tuberculosis, Nocardia farcinica, Nocardioides sp. strain JS614, Propionibacterium acnes, and Rhodococcus sp. strain RHA1 in vitro (26). In contrast to the situation in most other actinomycetes, GlnR does not play a role in nitrogen control in the genus Corynebacterium. In this genus, no GlnR homologs were observed; instead, AmtR is the central nitrogen control protein (28).
Compared to the recently described extended S. coelicolor GlnR regulon (26), M. smegmatis GlnR has a reduced number of target genes. For the NADP-dependent glutamate dehydrogenase gene gdhA (msmeg_5442), the urease operon (msmeg_2627 to msmeg_3622), and the nitrite reductase genes (msmeg_0427 and msmeg_0428), genes that are under the control of GlnR in S. coelicolor, no GlnR cis elements were found in this study. In fact, preliminary data indicate that there may be nitrogen-dependent regulation of the nitrite reductase and urease (T. Bräu, personal communication), and AmtR might be an interesting candidate for a second nitrogen regulator, considering the presence of a gene encoding an AmtR homolog in the genome of M. smegmatis and the fact that for C. glutamicum the urease-encoding genes are under the control of the AmtR repressor (2). Together with N. farcinica, Rhodococcus sp. strain RHA1, and Arthrobacter aurescens, M. smegmatis seems to be one of the few actinomycetes besides the members of the corynebacterium family to have an AmtR homolog. Thus, future studies should focus on the role of AmtR in M. smegmatis using comparisons of transcriptome data for wild-type and deletion mutant strains.
Supplementary Material
Acknowledgments
We thank D. Hillmann (Erlangen, Germany) for providing M. smegmatis strains, M. Niederweis (Birmingham, AL) for providing plasmids, and A. Lüdke (Erlangen, Germany) for help with protein analysis.
A.B., J.A., K.H., and F.T. were supported by the Deutsche Forschungsgemeinschaft (grant SFB 473), and J.S. received a fellowship from the Deutscher Akademischer Austausch Dienst.
Footnotes
Published ahead of print on 8 August 2008.
Supplemental material for this article may be found at http://jb.asm.org/.
REFERENCES
- 1.Baumbach, J. 2007. CoryneRegNet 4.0—a reference database for corynebacterial gene regulatory networks. BMC Bioinformatics 8429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Beckers, G., J. Strösser, U. Hildebrandt, J. Kalinowski, M. Farwick, R. Krämer, and A. Burkovski. 2005. Regulation of AmtR-controlled gene expression in Corynebacterium glutamicum: mechanism and characterization of the AmtR regulon. Mol. Microbiol. 58580-595. [DOI] [PubMed] [Google Scholar]
- 3.Belisle, J. T., and M. G. Sonnenberg. 1998. Isolation of genomic DNA from mycobacteria. Methods Mol. Biol. 10131-44. [DOI] [PubMed] [Google Scholar]
- 4.Burkovski, A. 2003. Ammonium assimilation and nitrogen control in Corynebacterium glutamicum and its relatives: an example for new regulatory mechanisms in actinomycetes. FEMS Microbiol. Rev. 27617-628. [DOI] [PubMed] [Google Scholar]
- 5.Burkovski, A. 2005. Nitrogen metabolism and its regulation, p. 333-349. In M. Bott and L. Eggeling (ed.), Handbook of Corynebacterium glutamicum. CRC Press LLC, Boca Raton, FL.
- 6.Burkovski, A. 2007. Nitrogen control in Corynebacterium glutamicum: proteins, mechanisms, signals. J. Microbiol. Biotechnol. 17187-194. [PubMed] [Google Scholar]
- 7.Carroll, P., C. A. Pashley, and T. Parish. 2008. Functional analysis of GlnE, an essential adenylyl transferase in Mycobacterium tuberculosis. J. Bacteriol. 1904894-4902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Fink, D., N. Weisschuh, J. Reuther, W. Wohlleben, and A. Engels. 2002. Two transcriptional regulators GlnR and GlnRII are involved in regulation of nitrogen metabolism in Streptomyces coelicolor A3(2). Mol. Microbiol. 46331-347. [DOI] [PubMed] [Google Scholar]
- 9.Hänβler, E., and A. Burkovski. 2008. Molecular mechanisms of nitrogen control in corynebacteria, p. 183-201. In A. Burkovski (ed.), Corynebacteria: genomics and molecular biology. Caister Academic Press, Wymondham, United Kingdom.
- 10.Harper, C., D. Hayward, I. Wiid, and P. Van Helden. Regulation of nitrogen metabolism in Mycobacterium tuberculosis: a comparison with mechanisms in Corynebacterium glutamicum and Streptomyces coelicolor. IUBMB Life, in press. [DOI] [PubMed]
- 11.Harth, G., D. L. Clemens, and A. A. Horwitz. 1994. Glutamine synthetase of Mycobacterium tuberculosis: extracellular realease and characterization of its enzymatic activity. Proc. Natl. Acad. Sci. USA 919342-9346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Harth, G., S. Maslesa-Galic, M. V. Tullius, and A. A. Horwitz. 2005. All four Mycobacterium tuberculosis glnA genes encode glutamine synthetase activities but only GlnA1 is abundantly expressed and essential for bacterial homeostasis. Mol. Microbiol. 581157-1172. [DOI] [PubMed] [Google Scholar]
- 13.Hiard, S., R. Maree, S. Colson, P. A. Hoskisson, F. Titgemeyer, G. van Wezel, B. Joris, L. Wehenkel, and S. Rigali. 2007. PREDetector: a new tool to identify regulatory elements in bacterial genomes. Biochem. Biophys. Res. Commun. 357861-864. [DOI] [PubMed] [Google Scholar]
- 14.Jakoby, M., L. Nolden, J. Meier-Wagner, R. Krämer, and A. Burkovski. 2000. AmtR, a global repressor in the nitrogen regulation system of Corynebacterium glutamicum. Mol. Microbiol. 37964-977. [DOI] [PubMed] [Google Scholar]
- 15.Khan, A., S. Akhtar, J. Ahmad, and D. Sarkar. 2008. Presence of a functional nitrate pathway in Mycobacterium smegmatis. Microb. Pathog. 4471-77. [DOI] [PubMed] [Google Scholar]
- 16.Manning, J. M., S. Moore, W. B. Rowe, and A. Meister. 1969. Identification of l-methionine S-sulfoximine as the diastereoisomer of l-methionine SR-sulfoximine that inhibits glutamine synthetase. Biochemistry 82681-2686. [DOI] [PubMed] [Google Scholar]
- 17.Nolden, L., G. Beckers, and A. Burkovski. 2002. Nitrogen assimilation in Corynebacterium diphtheriae: pathways and regulatory cascades. FEMS Microbiol. Lett. 208287-293. [DOI] [PubMed] [Google Scholar]
- 18.Parish, T., and N. G. Stoker. 2000. glnE is an essential gene in Mycobacterium tuberculosis. J. Bacteriol. 1825715-5720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Pashley, C. A., A. C. Brown, D. Robertson, and T. Parish. 2006. Identification of the Mycobacterium tuberculosis glnE promoter and its response to nitrogen availability. Microbiology 1522727-2734. [DOI] [PubMed] [Google Scholar]
- 20.Read, R., C. A. Pashley, D. Smith, and T. Parish. 2007. The role of GlnD in ammonium assimilation in Mycobacterium tuberculosis. Tuberculosis 87384-390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Reuther, J., and W. Wohlleben. 2007. Nitrogen metabolism in Streptomyces coelicolor: transcriptional and post-translational regulation. J. Mol. Microbiol. Biotechnol. 12139-146. [DOI] [PubMed] [Google Scholar]
- 22.Sambrook, J., E. F. Fritsch, and T. Maniatis. 1989. Molecular cloning: a laboratory manual, 2nd ed. Cold Spring Harbor Laboratory, Cold Spring Harbor, NY.
- 23.Schägger, H., and G. von Jagow. 1987. Tricine-sodium dodecyl sulfate-polyacrylamide gel electrophoresis for the separation of proteins in the range from 1 to kDa. Anal. Biochem. 166368-379. [DOI] [PubMed] [Google Scholar]
- 24.Stephan, J., V. Stemmer, and M. Niederweis. 2004. Consecutive gene deletions in Mycobacterium smegmatis using the yeast FLP recombinase. Gene 343181-190. [DOI] [PubMed] [Google Scholar]
- 25.Stephan, J., J. Bender, F. Wolschendorf, C. Hoffmann, E. Roth, C. Mailänder, H. Engelhardt, and M. Niederweis. 2005. The growth rate of Mycobacterium smegmatis depends on sufficient porin-mediated influx of nutrients. Mol. Microbiol. 58714-730. [DOI] [PubMed] [Google Scholar]
- 26.Tiffert, Y., P. Supra, R. Wurm, W. Wohlleben, R. Wagner, and J. Reuther. 2008. The Streptomyces coelicolor GlnR regulon: identification of new GlnR targets and evidence for a central role of GlnR in nitrogen metabolism in actinomycetes. Mol. Microbiol. 67436-446. [DOI] [PubMed] [Google Scholar]
- 27.Tullius, M. V., G. Harth, and A. A. Horwitz. 2003. Glutamine synthetase GlnA1 is essential for growth of Mycobacterium tuberculosis in human THP-1 macrophages and guinea pigs. Infect. Immun. 713927-3936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Walter, B., E. Hänβler, J. Kalinowski, and A. Burkovski. 2007. Nitrogen metabolism and nitrogen control in corynebacteria: variations of a common theme. J. Mol. Microbiol. Biotechnol. 12131-138. [DOI] [PubMed] [Google Scholar]
- 29.Wray, L. V., and S. H. Fisher. 1993. The Streptomyces coelicolor glnR gene encodes a protein similar to other bacterial response regulators. Gene 130145-150. [DOI] [PubMed] [Google Scholar]
- 30.Wray, L. V., M. R. Atkinson, and S. H. Fisher. 1991. Identification and cloning of the glnR locus, which is required for transcription of the glnA gene in Streptomyces coelicolor A3(2). J. Bacteriol. 1737351-7360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Yanisch-Perron, C., L. Vieira, and J. Messing. 1985. Improved M13 phage cloning vectors and host strains: nucleotide sequences of M13mp18 and pUC19 vectors. Gene 33103-119. [DOI] [PubMed] [Google Scholar]
- 32.Yoshida, T., L. Qin, L. A. Egger, and M. Inouye. 2006. Transcription regulation of ompF and ompC by a single transcription factor, OmpR. J. Biol. Chem. 28117114-17123. [DOI] [PubMed] [Google Scholar]
- 33.Yu, H., W. Peng, Y. Liu, T. Wu, Y. Yao, M. Cui, W. Jiang, and G.-P. Zhao. 2006. Identification and characterization of glnA promoter and its corresponding trans-regulating protein GlnR in the rifamycin SV producing actinomycete, Amycolatopsis mediterranei U32. Acta Biochim. Biophys. Sin. 38831-843. [DOI] [PubMed] [Google Scholar]
- 34.Yu, H., Y. Yao, Y. Liu, R. Jiao, W. Jiang, and G.-P. Zhao. 2007. A complex role of Amycolatopsis mediterranei GlnR in nitrogen metabolism and related antibiotics production. Arch. Microbiol. 18889-96. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.