Abstract
Large-scale industrial use of chromium(VI) has resulted in widespread contamination with carcinogenic chromium(VI). The abilities of microorganisms to survive in these environments and to detoxify chromate require the presence of specific resistance systems. Here we report identification of the transposon-located (TnOtChr) chromate resistance genes from the highly tolerant strain Ochrobactrum tritici 5bvl1 surviving chromate concentrations of >50 mM. The 7,189-bp-long TnOtChr of the mixed Tn21/Tn3 transposon subfamily contains a group of chrB, chrA, chrC, and chrF genes situated between divergently transcribed resolvase and transposase genes. The chrB and chrA genes, but not chrF or chrC, were essential for establishment of high resistance in chromium-sensitive O. tritici. The chr promoter was strongly induced by chromate or dichromate, but it was completely unresponsive to Cr(III), oxidants, sulfate, or other oxyanions. Plasmid reporter experiments identified ChrB as a chromate-sensing regulator of chr expression. Induction of the chr operon suppressed accumulation of cellular Cr through the activity of a chromate efflux pump encoded by chrA. Expression of chrB, chrC, or chrF in an Escherichia coli sodA sodB double mutant restored its aerobic growth in minimal medium and conferred resistance to superoxide-generating agents menadione and paraquat. Nitroblue tetrazolium staining on native gels showed that ChrC protein had superoxide dismutase activity. TnOtChr appears to represent a mobile genetic system for the distribution of the chromate-regulated resistance operon. The presence of three genes protecting against superoxide toxicity should provide an additional survival advantage to TnOtChr-containing cells in the environments with multiple redox-active contaminants.
Chromium(VI) is one of the major environmental contaminants, which reflects its numerous high-volume industrial applications and poor environmental practices in the disposal of chromium-containing waste products (42). High solubility and tetrahedral conformation of the chromate anion promote its rapid transport across biological membranes (11), and once internalized by cells, Cr(VI) exhibits a variety of toxic, mutagenic, and carcinogenic effects (43). Formation of DNA damage is a major cause of toxic and mutagenic responses in both human and bacterial cells, as evidenced by their increased sensitivity to chromate in the absence of DNA repair (16, 36). Human and other mammalian cells lack any detectable extrusion of chromate, and DNA repair is their main cellular defense mechanism against chromate toxicity. Because bacterial cells are less proficient in repair of chromium-DNA adducts compared to human cells (35), their ability to survive in the environment with heavy chromate contamination required selection of alternative resistance mechanisms. Genes conferring resistance to chromate have been found in Pseudomonas spp. (7, 27), Streptococcus lactis (13), and Cupriavidus metallidurans (28). Unlike other metal resistance systems that allow survival at high millimolar concentrations, currently known chr genes provided protection only in the submillimolar range, and the chromate-inducible systems were not very selective (32). One of the reasons for a limited protective ability of the characterized chr systems could be related to their responsiveness to sulfate, since a strong activation of the efflux pumps could lead to the coextrusion of sulfate and the resulting metabolic deficiency in sulfur donors. Chromate and sulfate are isostructural anions, which makes it difficult for cells to differentiate between them and is the basis for cellular uptake of chromate by sulfate transporters (43).
In the search for highly selective and efficient chromate defense systems, we focused our efforts on the characterization of the genetic factors responsible for chromate resistance of Ochrobactrum tritici strain 5bvl1. This strain was isolated from chromium-contaminated sludge from a wastewater treatment plant receiving wastewaters from tannery industries (14) and was found to be able to grow in the presence of high concentrations of chromate (8). Different alphaproteobacteria belonging to the genus Ochrobactrum have been isolated from clinical and/or environmental samples; however, little is known about their genetic organization and general resistance abilities. We identified a transposon-based chrBACF operon as the key determinant of high chromate tolerance by strain 5bvl1. The activation of this operon was highly selective and provided resistance principally through efficient extrusion of chromate. We also found that the chrB, chrC, and chrF genes complemented a superoxide-sensitive phenotype of sodA sodB double-null Escherichia coli cells, indicating that expression of chrBACF would confer cross-resistance to other environmental contaminants.
MATERIALS AND METHODS
Bacterial strains, plasmids, and growth conditions.
The bacterial strains and plasmids used in this study are listed in Table 1. Tris-buffered mineral salts medium (24) with 0.5% glucose (Tris-glucose medium) was used as minimal medium for growing O. tritici strain 5bvl1 and O. triticiT (type strain). Lysogeny broth (LB) was used as complex medium for O. tritici strains and Escherichia coli. Analytical-grade salts of CrCl3·6H2O, K2CrO4, K2Cr2O7, Na2WO4·2H2O, Na2MoO4·2H2O, Na3AsO4, and Na3VO4 were used to prepare 0.5 M stock solutions, which were sterilized by filtration.
TABLE 1.
Bacterial strains and plasmids used in this study
| Strain or plasmid | Relevant characteristic(s) | Sourcea or reference |
|---|---|---|
| Bacterial strains | ||
| O. tritici 5bvl1 | Wild type; Cr(VI)r | This study |
| O. triticiT | Type strain; Cr(VI)s | BCCM/LMG |
| O. tritici E117 | Mutant of 5bvl1; Tn5 inserted in chrA | This study |
| E. coli DH5α | φ80dlacZΔM15 endA1 recA1 gyrA96 thi-1 hsdR17(rK− mK+) relA1 supE44 deoR (lacZYA-argF)U169 | Promega |
| E. coli S17-1 λpir | Tpr SmrrecA thi pro hsdR hsdM+ RP4:2-Tc Mu:Km Tn7 λpir | Biomedal, Seville, Spain |
| E. coli AB1157 | F−thr-1 leuB6 proA2 his-4 thi-1 argE2 lacY1 galK2 rspL supE44 ara-14 xyl-15 mtl-1 tsx-33 | 17 |
| E. coli JI132 | As AB1157 but (sodA::Mud PR13)25 (sodB-Kan)1-Δ2 | 17 |
| O. triticiTchrFCAB | Type strain carrying chrF, chrC, chrA, and chrB genes from strain 5bvl1 | This study |
| O. triticiTchrCAB | Type strain carrying chrC, chrA, and chrB genes from strain 5bvl1 | This study |
| O. triticiTchrAB | Type strain carrying chrA and chrB genes from strain 5bvl1 | This study |
| O. triticiTchrB | Type strain carrying chrB gene from strain 5bvl1 | This study |
| O. tritici E117:chrA | Mutant of 5bvl1 complemented with the chrA gene cloned in pBBR1MCS-5 vector | This study |
| Plasmids | ||
| pSUP5011 | Tn5-based transposon | DSMZ |
| pGEM-T Easy | Apr; T-tailed PCR product cloning vector | Promega |
| pBBR1MCS-5 | Gmr; oripBBR1MCS Mob+lacZa, broad-host-range cloning and expression vector | 19 |
| pSJ3 | Apr; vector containing the promotorless lacZ | 4 |
| pUTmini-Tn5Tc | Tcr; vector containing the Tn5 transposon | Biomedal, Seville, Spain |
| chrp::lacZ | pSJ3 vector with chr promoter-lacZ fusion | This study |
| chrBp::lacZ | pSJ3 vector with promoter-chrB-lacZ fusion | This study |
| pTrc99A | Apr; expression vector | Amersham Pharmacia Biotech |
DSMZ, Deutsche Sammlung von Mikroorganismen und Zellkulturen GmbH, Braunschweig, Germany; BCCM/LMG, Laboratorium voor Microbiologie, Universiteit Gent, Belgium.
Transposon mutagenesis and screening.
Transposon insertion mutants were generated by mobilization of the suicide plasmid pSUP5011 from the donor strain E. coli S17-1 to the recipient strain O. tritici 5bvl1. In this procedure, the Tn5::Kanr transposon is transferred from E. coli to strain 5bvl1 (Tcr), using the filter mating method (12), where the transposon randomly inserts into the DNA, thus generating a library of insertion mutants. Selection was done on LB plates with tetracycline (10 μg/ml) and kanamycin (750 μg/ml) to obtain cells in which transposition had occurred. This high concentration of kanamycin was necessary to select the transconjugants. The transconjugants were then plated on LB plates, and clones unable to grow in the presence of 2 mM chromate were recovered and subjected to further analyses.
Inverse PCR.
Templates for inverse PCR were prepared from about 1 μg of total DNA digested with enzymes that cut once inside the Tn5 transposon (SalI and SphI). The digested and purified DNA (about 500 ng) was ligated overnight at 14°C in a total volume of 50 μl with 3 U of T4 DNA ligase (Roche, Mannheim, Germany). DNA flanking the Tn5 insertion was amplified by PCR with Taq DNA polymerase (Invitrogen, Carlsbad, CA) and with specific primers (purchased from Sigma-Genomys, St. Louis, MO), designed from the transposon inverted repeats (IRs) and from a region inside of the Tn5 transposon. The PCR products were gel purified, cloned into vector pGEM-T Easy (Promega, Madison, WI) (37) and sequenced. Database searches and sequence analyses were performed by using the BLAST program (2).
RNA isolation and RT-PCR.
Total RNA was obtained from mid-exponential-phase strain 5bvl1 cells grown for 1 h in the absence or presence of 0.5 mM chromate in Tris-glucose medium. Total RNA was isolated by the RNeasy mini kit (Qiagen, Valencia, CA) and then digested with RQ1 RNase-free DNase (Promega, Madison, WI) to remove the residual DNA. cDNA synthesis was done with Sensiscript reverse transcriptase (RT) (Qiagen, Valencia, CA) according to the manufacturer's instructions. Briefly, the reverse primers from the chrB gene were used to create the cDNAs. Standard PCR procedures were used to generate amplicons from 5 μl of the reverse transcription reaction mixture using the specific primer pairs for the chrB gene. Reverse transcriptase PCR (RT-PCR) products were examined by 1% agarose gel electrophoresis. The potential presence of DNA contamination in the mRNA preparations was tested in the reaction mixtures lacking RT.
Reporter gene constructs.
The putative promoter and the chrB gene of the TnOtChr operon were amplified, and recognition sites for enzymes KpnI and XbaI were incorporated into the PCR primers. The PCR product was ligated into KpnI-XbaI-digested vector pSJ3, immediately upstream of the lacZ gene, to generate chrBp::lacZ. A product containing only the promoter sequence (chrp) was also ligated into vector pSJ3, resulting in the construct chrp::lacZ. These plasmids were then transformed into E. coli DH5α cells. Overnight cultures were diluted 100 times with fresh medium and incubated until they reached exponential growth phase (optical density at 600 nm [OD600] of 0.3 to 0.6). Cultures were then distributed into tubes and induced by different chromate concentrations or by other compounds (sulfate, arsenate, tungstate, vanadate, and molybdate). After incubation for one additional hour, β-galactosidase enzymatic assays were performed as previously described (26). The results are presented as mean values from at least three independent experiments.
Construction of O. triticiT chrFCAB, O. triticiT chrCAB, O. triticiT chrAB, and O. triticiT chrB.
Different fragments of the TnOtChr operon, including the promoter region, were amplified, and recognition sites for enzymes PstI and KpnI were incorporated into the PCR primers. The PCR products were ligated into PstI-KpnI-digested vector pSJ3 upstream of the lacZ gene. The fragments containing the chr genes were isolated by digesting the plasmids with NotI and were cloned into the unique NotI site of pUTmini-Tn5 (Tcr). Transfer of the resulting plasmids, pUT::chrB, pUT::chrAB, pUT::chrCAB, and pUT::chrFCAB to recipient strain O. tritici was performed with the donor E. coli S17-1 λpir strain by biparental conjugation using the filter mating method (12). Selection was done on LB plates with tetracycline (10 μg/ml) and kanamycin (30 μg/ml) to obtain cells in which transposition had occurred. Chromosomal integration of chr genes was confirmed by Southern blotting and PCR amplification.
Complementation of E117 mutant with chrA.
The chrp and chrA sequences were PCR amplified from O. tritici strain 5bvl1 and cloned into pBBR1MCS-5, resulting in the plasmid pBBR1::chrAp. This vector was transformed into competent E. coli S17-1 cells and then mobilized into the E117 mutant by biparental conjugation (12). Selection of plasmid-expressing clones, designated E117:chrA, was performed on LB plates with gentamicin (15 μg/ml) and kanamycin (30 μg/ml).
Chromate resistance assays.
Chromate resistance was measured by the growth rates in Tris-glucose minimum medium. Overnight cultures were diluted 100-fold into 300-ml flasks containing 100 ml of fresh medium supplemented with different chromate concentrations. The bacterial suspensions were incubated at 37°C with shaking at 170 rpm for 15 h, and the OD600 was measured. Chromate resistance was also determined by the clonogenic assay. O. tritici strains were grown to an OD600 of 0.2 to 0.3, and serial dilutions of each culture were plated in triplicate on LB plates supplemented with chromate at the indicated concentrations. Plates were incubated at 37°C, and colonies were counted after 3 days.
Chromate uptake.
Overnight cultures were diluted in 100 ml of the new medium and grown to exponential phase. Uptake assays were initiated by adding different chromate concentrations (0.5, 1, and 3 mM) to the suspensions followed by incubation at 37°C with shaking for 3 h. Control and Cr(VI)-exposed cells were harvested by centrifugation and washed twice with cold phosphate-buffered saline. Cellular Cr extracted by a hot nitric acid procedure (25) was measured by graphite furnace atomic absorption spectroscopy using Zeeman background correction (34). Measurements were done using a model 4100ZL Perkin-Elmer GF-AAS instrument. The detection limit was 0.4 pmol of chromium. Intracellular chromium content was expressed as nanogram of Cr per microgram of total cellular protein.
Complementation of an E. coli sodA sodB mutant.
The chrF, chrC, and chrB genes were cloned by PCR from chromosomal DNA from O. tritici strain 5bvl1. The PCR products were double digested with PstI and NcoI and ligated into PstI-NcoI-digested pTrc99A in the presence of 1 U of T4 DNA ligase (Invitrogen, Carlsbad, CA). E. coli JI132 cells were transformed with empty vector pTrc99A or with indicated expression constructs. Positive clones designated pTrc_chrF, pTrc_chrC, pTrc_chrFC, and pTrc_chrB were grown in LB medium with ampicillin (100 μg/ml) to an OD600 of 0.4 when 1 mM of isopropyl-β-d-1-thiogalactopyranoside (IPTG) was added to induce cultures. To test the sensitivity of the transformants to paraquat or menadione, induced cells were treated with different concentrations of these drugs for 1 h at 37°C. Samples were diluted and placed on LB plates, and colonies were counted after 24 h. The growth of JI132-complemented strains was also examined in M9 mineral medium supplemented with 0.5 mM l-amino acids (6, 17).
Determination of SOD activity in crude extracts.
Cells were harvested by centrifugation, resuspended in 50 mM Tris buffer, pH 7.8, and lysed by sonication. After centrifugation (15 min, 16,000 × g, 4°C), the supernatants were collected. Total protein was determined using the Bradford assay (Bio-Rad Laboratories, Hercules, CA). The superoxide dismutase (SOD) activity was measured using a superoxide dismutase assay kit (Cayman Chemical Co., Ann Arbor, MI). The SOD activity on 10% nondenaturing polyacrylamide gels was visualized by nitroblue tetrazolium negative staining (5).
Nucleotide sequence accession number.
The nucleotide sequence determined in this study has been deposited in the GenBank database under accession number EF469735.
RESULTS
Isolation of chromate-sensitive mutants.
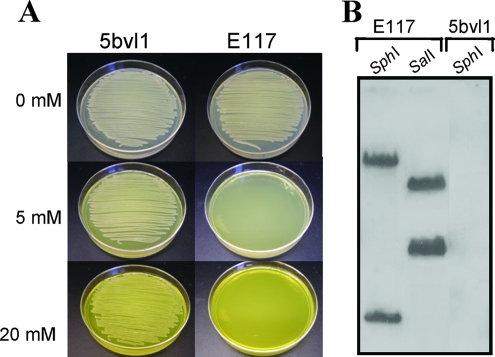
Random mutants were generated by mobilization of the suicide plasmid pSUP5011 from the donor strain E. coli S17-1 into the recipient strain O. tritici 5bvl1. Approximately 4,000 kanamycin-resistant clones were randomly chosen and initially tested for their ability to grow on LB plates containing 2 mM chromate. After 48 h of incubation, chromium-sensitive clones were selected for testing of chromate sensitivity in liquid medium and on plates with a range of high Cr(VI) concentrations. One mutant, designated E117, was particularly sensitive to chromate (Fig. 1A), and it was selected for detailed genetic analysis. Southern blot probing of total genomic DNA digestion products (SphI and SalI) with an internal transposon fragment labeled with dioxigenin-dUTP confirmed the presence of the transposon insertion in the E117 clone (Fig. 1B). Both restriction enzymes generated only two Tn5-positive fragments, indicating that the E117 clone had a single transposon insert.
FIG. 1.
(A) Growth of O. tritici 5bvl1 and E117 mutant on LB plates containing the indicated chromate concentrations. (B) Southern hybridization of total DNA from mutant E117 digested with SphI and SalI and strain 5bvl1 digested with SphI as a control. An internal transposon fragment labeled with dioxigenin-dUTP was used as a probe. Both 5bvl1 and E117 samples were analyzed on the same blot.
Genetic organization of the chromate resistance operon.
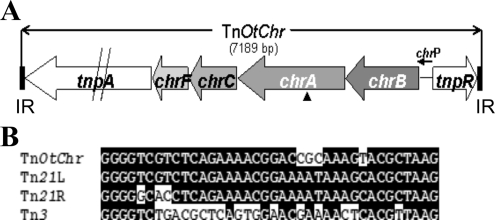
Arbitrary PCR anchored with transposase-specific primers identified the Tn5 insertion site in mutant E117 as being inside an open reading frame (ORF) coding for a protein with a high degree of homology to a chromate transporter (referred to as ChrA) (Fig. 2A). Primer walking experiments revealed a total of six ORFs. Four of the ORFs were located on the same DNA strand between the divergently transcribed tnpR and tnpA genes. Several features typically associated with transposons were identified, and therefore, the name TnOtChr was given. Transposon TnOtChr has a length of 7,189 nucleotides and contains two conserved genes (tnpA and tnpR), which encode transposase and resolvase, respectively. Analysis of these genes revealed that TnOtChr belongs to the large Tn3 family of transposons; however, other sequence elements of TnOtChr were more similar to the Tn21 family. As in Tn3, the tnpA and tnpR genes are divergently transcribed, whereas in the Tn21 subfamily of transposons, the tnpR and tnpA genes are transcribed as a unit (15). The left and right inverted repeats of TnOtChr are 38 bp long and identical to each other (Fig. 2B). They are 33/38 bp identical to the Tn21 left IR and 30/38 bp identical to the Tn21 right IR, but only 19/38 bp identical to the Tn3 IR sequences (left or right). Sequences flanking the transposon were not associated with chromate resistance, which together code for a putative esterase, indicating that the transposon was inserted in the middle of this gene.
FIG. 2.
(A) Physical map of the chr locus in O. tritici 5bvl1 strain. The Tn5 insertion in chrA is indicated by the small black vertical arrowhead. (B) Alignment of the inverted repeat sequences of TnOtChr, Tn21, and Tn3. Identical bases are indicated by dark shading.
The chrA gene encodes a protein showing the highest homology (77% identity and 86% similarity) to a putative chromate transporter from Janthinobacterium sp. strain Marseille (GenBank accession no. YP_001354737). Alignment of amino acid sequences of these proteins with the ChrA from other organisms (C. metallidurans and Pseudomonas aeruginosa) revealed some common structural features. These include the presence of two copies of the motif GGX12VX4WX16PGPX9/8G (X is any residue) (30). Upstream of chrA, primer walking revealed one additional ORF related to chromate resistance, chrB coding for a 312-amino-acid-long protein. ChrB showed 64% identity with a chromate resistance protein of Herminiimonas arsenicoxydans (GenBank accession no. YP_001099204). The alignment of these ChrB proteins with the homologous proteins from C. metallidurans also found a strong similarity in the areas of conserved domains. An ORF (chrC) immediately downstream of chrA encoded a protein product of 202 amino acids, displaying 71% identity to the manganese or iron SOD of the strain C. metallidurans CH34 (GenBank accession no. ABF13060). The next downstream ORF was homologous (83% identity) to a hypothetical protein of Burkholderia xenovorans LB400 (GenBank accession no. ABE37064). This ORF also showed high similarity to both ORFs, referred to as chrF1 and chrF2, present in the plasmid pMOL28 and chromosome of strain C. metallidurans CH34, which code for uncharacterized conserved proteins related to chromate resistance. Consequently, we named this gene and the corresponding protein chrF and ChrF, respectively. The protein ChrF also showed 28% identity and 49% similarity with the C-terminal region of ChrB, and based on BLAST analyses, both proteins showed sequence similarity with SodM-like proteins from other bacterial species. Metal dependence of the activity of these proteins has not yet been established.
The chr operon is functional in type strain O. tritici.
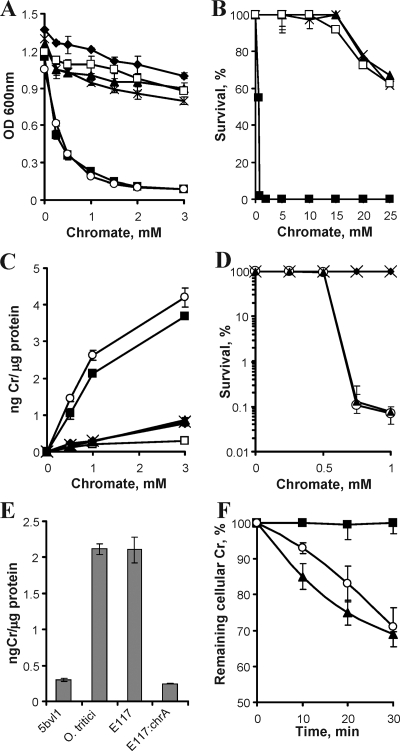
To test whether the discovered genes were functional in a chromate-sensitive Ochrobactrum, E. coli S17-1 λpir cells containing plasmid pUT::chrFCAB, pUT::chrCAB, pUT::chrAB, or pUT::chrB were mated with O. triticiT. The chromate resistance of these constructs was compared to the wild strain 5bvl1 and type strain O. tritici (Fig. 3A). O. triticiT cells containing pUT::chrB (O. triticiT chrB) showed chromate susceptibility that was similar to cells without any construct. Introduction of three other constructs (pUT::chrFCAB, pUT::chrCAB, and pUT::chrAB) made O. triticiT as resistant to Cr(VI) as strain 5bvl1 was. The clonogenic survival studies also found that O. triticiT carrying these different constructs exhibited similar percentages of cells surviving at several chromate concentrations (Fig. 3B). These results indicate that, at least in O. triticiT, chrF and chrC genes did not seem to play a significant role in chromate resistance.
FIG. 3.
(A) Sensitivity of O. tritici strains to chromate. Cultures of O. tritici 5bvl1 (♦), O. triticiT (▪), O. triticiT chrFCAB (▴), O. triticiT chrCAB (×), O. triticiT chrAB (□), and O. triticiT chrB (○) strains were grown in Tris-glucose medium for 15 h at 37°C in the presence of the indicated concentrations of chromate. (B) Clonogenic survival at different chromate concentrations. The cultures were plated on LB plates, and the colonies were counted after 3 days of incubation at 37°C. Strain symbols are as defined above for panel A. (C) Chromate uptake by O. tritici strains. Exponential-phase cells were incubated at 37°C with chromate for 3 h, and cellular chromium concentrations were measured as described in Materials and Methods. (D) Survival of O. tritici 5bvl1 (♦), O. tritici (○), E117 mutant (▴), and E117:chrA (×) strains at different chromate concentrations. The cultures were also plated on LB plates, and the colonies were counted after 3 days of incubation at 37°C. (E) Chromate uptake. Exponential growing cells were exposed to 1 mM chromate for 3 h. Values are means ± standard deviations (error bars) from three independent experiments. (F) Chromate efflux by O. tritici 5bvl1 (○), E117 mutant (▪), and E117:chrA (▴) strains. Cells were first preincubated with 0.1 mM chromate for 1 h to induce the chr operon and then pulse-loaded for 15 min with high chromate concentrations (1 mM chromate for E117 mutant and 5 mM chromate for strains 5bvl1 and E117:chrA), resulting in approximately equal initial levels of cellular chromium. Cells were washed and then incubated in fresh medium for 0 to 30 min at 37°C to assess efflux of cellular chromium. Results are the percentages of remaining cellular concentrations of Cr after normalization for protein content (means ± standard deviations [error bars] from three independent experiments).
To investigate a potential mechanism of resistance, we examined the role of chr genes in chromium accumulation by different bacterial cultures (Fig. 3C). The strains that tolerated high chromate concentrations (O. triticiT 5bvl1, O. triticiT chrFCAB, O. triticiT chrCAB, and O. triticiT chrAB) also accumulated dramatically less chromium, whereas the chromate-sensitive strains (O. tritici type strain and O. triticiT chrB) showed the highest intracellular chromium concentrations. A direct association between chromate sensitivity and increased chromium accumulation was further confirmed by findings that chromate-sensitive type strain O. tritici and E117 mutant contained very high levels of intracellular Cr (Fig. 3D and E). Further studies revealed the critical role of ChrA protein in the chromate detoxification process. Suspensions of Cr(VI)-resistant cells (strains 5bvl1 and E117:chrA) pulse-loaded with chromate showed a clear time-dependent loss of cellular Cr, whereas chromium-sensitive E117 mutant cells exhibited no detectable decrease in Cr levels during 30 min of incubation (Fig. 3F). These findings demonstrated that chromium-resistant strain 5bvl1 cells express a very efficient chromate efflux system that was able to maintain a very low level of cellular Cr, even in media containing millimolar chromate concentrations.
Regulation of the chr promoter.
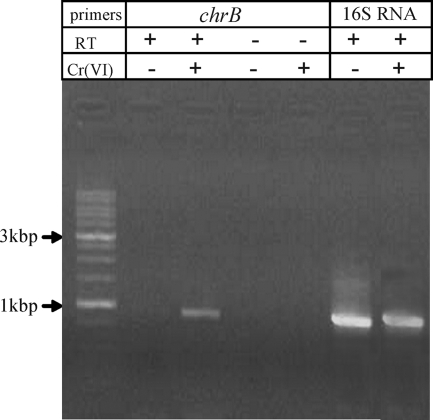
To examine whether the chr operon was inducible by chromate, we utilized a highly sensitive RT-PCR methodology to detect chrB expression in the presence and absence of Cr(VI) (Fig. 4). In Cr(VI)-treated cultures, an RT-PCR product corresponding to the predicted size of 939 bp was readily amplified, while expression of the chrB gene was undetectable in chromium-free cultures. Control RT-PCRs generated 16S rRNA-derived products from all samples, indicating that RNA from untreated cells was amplifiable. A potential PCR amplification from contaminating DNA was ruled out because of the absence of products in the samples lacking reverse transcriptase (Fig. 4, RT lanes).
FIG. 4.
Induction of chrB expression in 5bvl1 cells by Cr(VI). The presence of chrB cDNA was detected by RT-PCR. Total RNA was isolated from control and 0.5 mM chromate-treated cells. The chrB RT-PCR product was amplified from total RNA extracted from mid-log-phase control cells and Cr(VI)-exposed cells. Control PCRs for DNA contamination contained Taq polymerase but lacked reverse transcriptase (RT). The quality of isolated RNA was verified by RT-PCR of 16S rRNA. All samples were analyzed on the same gel.
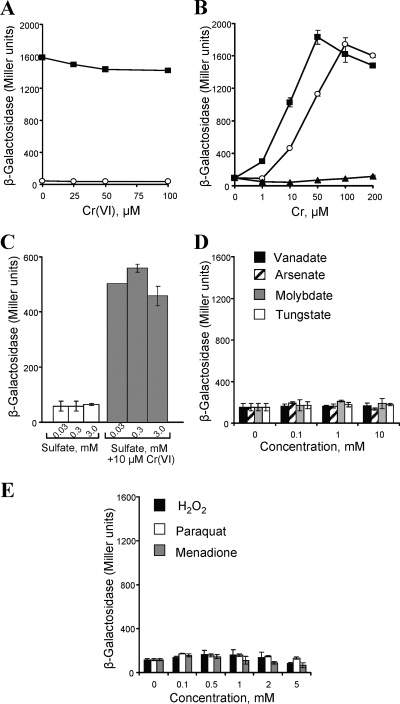
To investigate the mechanisms of chr inducibility, we constructed reporter plasmids by cloning the chrp sequence and chr genes in front of the promoterless lacZ gene in the pSJ3 vector. The chrp::lacZ construct showed a constitutively high β-galactosidase activity which was unaffected by the addition of chromate (Fig. 5A). The role of the ChrB protein in the chromate-mediated induction was tested using a plasmid containing a chrBp::lacZ fusion. This reporter revealed a potent activation by Cr(VI), and at 0.1 mM dose, β-galactosidase activity reached a plateau corresponding to the activity of the chrp::lacZ vector (Fig. 5B). To assess the selectivity of the ChrB-dependent responses, we examined the activity of the chrBp::lacZ reporter in the presence of sulfate, a biological anion that is isostructural with chromate. We determined that the addition of 0.03 to 3 mM sulfate had no effect on the basal activity of the chrBp::lacZ reporter plasmid or its inducibility by Cr(VI) (Fig. 5C). The selectivity of the chromate-sensing ChrB was further evaluated by testing other oxyanions, such as vanadate, arsenate, tungstate, and molybdate, for their ability to act as inducers of the chrBp::lacZ reporter. We found that even at levels more than 1,000 times above the effective chromate concentrations, none of these anions was capable of producing of any detectable responses (Fig. 5D). We also examined whether the chr operon was inducible by oxidative stress by testing hydrogen peroxide and the superoxide-generating reagents paraquat and menadione. We found that none of these oxidants was able to induce significant increases in the reporter expression (Fig. 5E).
FIG. 5.
Regulation of the chr promoter. (A) Reporter activity of the promoterless lacZ-carrying pSJ3 (○) and chrp::lacZ vectors (▪) following treatment of the cells with Cr(VI) for 1 h. (B) Reporter activity using the chrBp::lacZ construct. Cultures at mid-log phase were incubated with or without chromate (○), dichromate (▪), or Cr(III) (▴) for 1 h. (C) Activity of the chrBp::lacZ reporter in cells incubated with different concentrations of sulfate in the presence and absence of 10 μM chromate. (D and E) chrBp::lacZ reporter activity in cells incubated for 1 h in the presence of different concentrations of oxyanions (D) or oxidants (E). Values are means ± standard deviations (error bars) from at least two experiments.
Protective roles of ChrF, ChrC, and ChrB against superoxide toxicity.
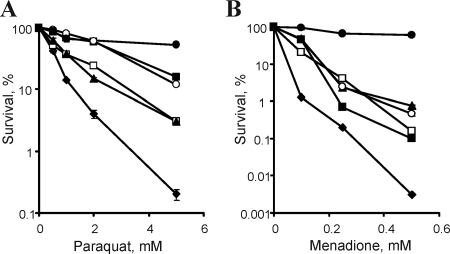
As noted above, ChrC showed some sequence similarity to proteins with SOD activities and ChrB and ChrF showed sequence similarity with putative SOD proteins. This prompted us to conduct additional investigation of the functional roles of these three chr proteins by expressing them in an E. coli sodA sodB double mutant. We found that expression of any of these three proteins strongly increased resistance of sodA sodB cells to the toxicity of superoxide anion-generating reagents paraquat (Fig. 6A) or menadione (Fig. 6B). Although complementation with chrB, chrC, or chrF did not produce the same level of tolerance as that of wild-type E. coli expressing both SodA and SodB, the resistance of the chrB-complemented cells at 0 to 2 mM doses of paraquat was identical to the wild-type strain. The expression of chrB, chrC, or chrF did not increase resistance of E. coli sodA sodB mutant to another organic oxidant, cumene hydroperoxide (not shown), which damages cells primarily via lipid oxidation.
FIG. 6.
Sensitivity of E. coli strains to superoxide-producing paraquat (A) and menadione (B). Cell suspensions were plated on LB plates, and the colonies were counted after 24 h of incubation at 37°C. The wild-type E. coli AB1157 (•) strain and the mutant strain JI132 carrying construct pTrc99A (♦), pTrc_chrF (□), pTrc_chrC (▴), pTrc_chrFC (○), or pTrc_chrB (▪) are shown.
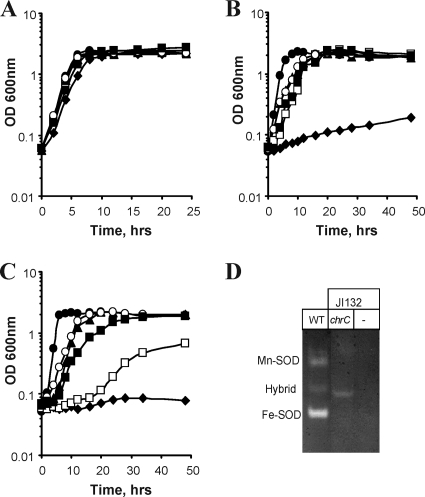
sodA sodB double-deficient E. coli cells are unable to grow under aerobic conditions without sulfur-containing amino acids or aromatic amino acids (6, 17), which is apparently caused by a rapid inactivation of enzymes involved in the biosynthesis of these amino acids by the high level of superoxide in oxygenated cultures. Rescue of the growth defect in sodA sodB double mutant cells in aerobic minimal medium is commonly used as a genetic test for the presence of SOD activity in a protein of interest. We found that the expression of chrF, chrC, and chrB genes restored the ability of SOD-null E. coli cells to grow in media without both aromatic amino acids and sulfur-containing amino acids (Fig. 7B), although cells carrying the pTrc_chrF construct showed a significantly slower growth in medium without methionine and cysteine (Fig. 7C). While the exact reasons for this discrepancy between two conditions are currently unclear, one possibility could be related to a higher content of Cys and Met in the ChrF protein, making it more difficult to synthesize when the availability of these amino acids is limited. Biochemical testing of cellular extracts found no significant changes in SOD activity following expression of chrC and chrF genes in sodA sodB cells (not shown) and a modest increase after complementation with chrB (0.28 ± 0.02 U/mg protein in vector-complemented cells and 1.18 ± 0.22 U/mg protein in chrB-complemented cells, P < 0.001, one-way analysis of variance). Qualitative detection of SOD activity on native protein gels produced consistent positive results only for ChrC-expressing cells (Fig. 7D). Because SOD activity was completely absent in preparations containing phosphate buffer (not shown), the lability of a metal cofactor(s) in expressed chr proteins may explain their low SOD activity and inconsistent results between two enzymatic assays. Further optimization of the in vitro conditions for measurements of SOD activity in Chr proteins is clearly needed. Heterologous expression of Chr proteins may have also led to difficulties in the delivery of metal ions by E. coli metallochaperones to create fully functional SODs. The absence of SOD-null O. triticiT cells and the naturally high resistance of these cells to superoxide-generating agents precluded testing of ChrB, ChrC, and ChrF proteins for SOD activity in their native cellular environment.
FIG. 7.
Growth of E. coli strains in M9 mineral medium under different aerobic conditions. (A to C) The bacteria were grown in the presence of all 20 amino acids (A); all amino acids, except phenylalanine, tryptophan, and tyrosine (B); and all amino acids, except cysteine and methionine (C). Wild-type E. coli AB1157 (•) strain and the mutant strain JI132 carrying construct pTrc99A (♦), pTrc_chrF (□), pTrc_chrC (▴), pTrc_chrFC (○), or pTrc_chrB (▪) are shown. Growth was monitored at 600 nm by examining the OD600. (D) Detection of SOD activity on native gels. Protein extracts (50 μg) were separated on 10% polyacrylamide, and the presence of proteins with SOD activity was visualized by illumination of gels impregnated with a riboflavin-nitroblue tetrazolium mixture. JI132 is a sodA sodB double null E. coli mutant. WT, wild-type E. coli AB1157.
DISCUSSION
O. tritici strain 5bvl1 was isolated from the consortium of bacteria adapted to live in the environment with high concentrations of chromate (14), and we reasoned that this strain should contain genes conferring high tolerance of Cr(VI). This expectation was confirmed by the identification of a set of Cr(VI) resistance genes located on the chromosomally integrated transposable element TnOtChr. The structure of this transposon indicates its ability to distribute the chromate resistance genes. The tnpR and tnpA genes are separated by a region that contains four chromate resistance-related genes. Other transposons carrying mercury resistance genes have been identified (22, 41), and more recently, transposons with arsenic resistance genes were also found (39, 40). A transposon carrying a chromate resistance determinant has already been found in plasmid pB4 from an uncultivatable bacterium (38). However, the pB4-based transposon Tn5719 contained the tnpR (resolvase) and tnpA (transposase) genes physically adjacent to each other as in the majority of the transposons of the Tn21 subfamily (15, 20). More recently, a new plasmid pCNB1 from Comamonas sp. strain CNB-1 which contained the genes for chromate resistance located on a transposon has been described (23). In this case, only two genes, a putative chromate transporter and a regulator, were found on the transposon. Thus, TnOtChr is a unique described transposon in which the chr operon contains SOD-like genes and the tnpR and tnpA genes flanked the chr genes.
Insertion of the Tn5 transposon in the chrA gene (mutant E117) completely abolished chromate resistance, indicating that this gene was essential for growth of strain 5bvl1 in the presence of high Cr(VI). The experiments with the constructs containing the intact and partial chr operons determined that the defect in E117 was caused by the inactivation of ChrA protein, not by Tn5 polar effects on chr expression. Complementation of E117 mutant and type strain O. tritici with chrA conferred the ability to grow in the medium with high chromate concentrations and thus established a direct link between the expression of ChrA and the presence of the chromate-tolerant phenotype. This chromate resistance system operates by maintaining low cellular chromium levels even in the presence of millimolar extracellular chromate concentrations. Decreased accumulation of chromate has been associated with chromate resistance in other microorganisms (3, 29, 31, 33). ChrA proteins from C. metallidurans (29) and Pseudomonas aeruginosa (10) have been functionally characterized as chromate efflux pumps. Direct observations of the cellular loss of chromium via a ChrA-dependent mechanism and the presence of significant amino acid homology among the chrA gene products from different species all indicate that the ChrA protein from strain 5bvl1 also functions as a chromate efflux pump. The chrA gene probably originated by gene duplication followed by gene fusion, since some chr operons contain two adjacent genes encoding the amino- and carboxy-terminal parts of the full-length ChrA protein (30).
Our experiments with the reporter constructs demonstrated that ChrB protein acted as the chromate-sensitive regulator of the chr operon. Unlike ChrB proteins from C. metallidurans that activated the chromate resistance system in the presence of Cr(VI) and Cr(III) (18, 32), ChrB from O. tritici strain 5bvl1 responded exclusively to Cr(VI). Another unique feature of the chr operon from strain 5bvl1 was insensitivity of its induction by chromate on the concentration of sulfate ions. Thus, chromate selectivity of ChrB could be a major factor in the much greater efficiency of the strain 5bvl1 chr system, endowing cells with the ability to grow at >100-fold-higher chromate concentrations relative to the resistance system that exhibits a lower degree of chromate selectivity (32).
The absence of significant effects on chromate sensitivity in strains expressing constructs lacking chrC and chrF genes at first glance appears unusual. From an evolutionary point of view, it is highly unlikely that two genes would be incorporated into the chromate resistance operon while they provide no growth advantage under conditions of chromate exposure. The likely answer probably lies in the fact these genes are part of the transposon-based mobile genetic element. While in O. tritici cells, chrF and chrC apparently do not play a major role in resistance, the expression of the entire chrBACF operon can be important for the survival of other bacteria in the environment with high chromate. Cr(VI) causes cell death via both oxidative and nonoxidative processes, and the relative significance of these mechanisms varies depending on the rate of chromate reduction, nature of intermediate species, and other conditions (43). A common pathway for Cr(VI) reduction to Cr(III) is through Cr(V) and Cr(IV) intermediates which are both capable of redox cycling and can generate large amounts of reactive oxygen species (25). Thus, in bacteria with abnormally high production of toxic superoxide in response to chromate (1), the expression of ChrC, ChrF, and ChrB proteins should provide an important second line of defense against this toxic metal. The water treatment sludge from which the chromate-resistant O. tritici strain 5bvl1 was isolated (14) contained many other toxicants, which is generally true for the majority of chromate-contaminated environments. Thus, the expression of the chr operon can confer cross-resistance to the redox-active contaminants that can either generate superoxide directly or deplete cellular antioxidants, which would potentiate the formation of reactive oxygen species by chromate (25).
Acknowledgments
This work was funded by a fellowship from the Fundação para a Ciência e Tecnologia, Portugal (R.B.) and research grant ES013660 from the National Institute of Environmental Health Sciences (A.Z.).
We thank Jim Imlay for his invaluable advice regarding SOD complementation experiments and for providing us with E. coli sodA sodB double mutant cells.
Footnotes
Published ahead of print on 5 September 2008.
REFERENCES
- 1.Ackerley, D. F., Y. Barak, S. V. Lynch, J. Curtin, and A. Martin. 2006. Effect of chromate stress on Escherichia coli K-12. J. Bacteriol. 1883371-3381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Altschul, S. F., T. L. Madden, A. A. Schaffer, J. Zhang, W. Miller, and D. J. Lipman. 1997. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 253389-3402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Alvarez, A. H., R. Moreno-Sánchez, and C. Cervantes. 1999. Chromate efflux by means of the ChrA chromate resistance protein from Pseudomonas aeruginosa. J. Bacteriol. 1817398-7400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Barragán, M. J. L., M. Carmona, M. T. Zamarro, B. Thiele, M. Boll, G. Fuchs, J. L. García, and E. Díaz. 2004. The bzd gene cluster, coding for anaerobic benzoate catabolism, in Azoarcus sp. strain CIB. J. Bacteriol. 1865762-5774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Beauchamp, C., and I. Fridovich. 1971. Superoxide dismutase: improved assays and an assay applicable to acrylamide gels. Anal. Biochem. 44276-287. [DOI] [PubMed] [Google Scholar]
- 6.Benov, L., and I. Fridovich. 1999. Why superoxide imposes an aromatic amino acid auxotrophy on Escherichia coli. The transketolase connection. J. Biol. Chem. 2744202-4206. [DOI] [PubMed] [Google Scholar]
- 7.Bopp, L. H., A. M. Chakrabarty, and H. L. Ehrlich. 1983. Chromate resistance plasmid in Pseudomonas fluorescens. J. Bacteriol. 1551105-1109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Branco, R., M. C. Alpoim, and P. V. Morais. 2004. Ochrobactrum tritici strain 5bvl1—characterization of a Cr(VI)-resistant and Cr(VI)-reducing strain. Can. J. Microbiol. 50697-703. [DOI] [PubMed] [Google Scholar]
- 9.Cervantes, C., and H. Ohtake. 1988. Plasmid-determined resistance to chromate in Pseudomonas aeruginosa. FEMS Microbiol. Lett. 56173-176. [Google Scholar]
- 10.Cervantes, C., H. Ohtake, L. Chu, T. K. Misra, and S. Silver. 1990. Cloning, nucleotide sequence, and expression of the chromate resistance determinant of Pseudomonas aeruginosa plasmid pUM505. J. Bacteriol. 172287-291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Cervantes, C., J. Campos-Garcia, S. Devars, F. Gutierrez-Corona, H. Loza-Tavera, J. C. Torres-Guzman, and R. Moreno-Sanchez. 2001. Interactions of chromium with microorganisms and plants. FEMS Microbiol. Rev. 25335-347. [DOI] [PubMed] [Google Scholar]
- 12.de Lorenzo, V., and K. N. Timmis. 1994. Analysis and construction of stable phenotypes in gram-negative bacteria with Tn5- and Tn10-derived minitransposons. Methods Enzymol. 235386-405. [DOI] [PubMed] [Google Scholar]
- 13.Efstathiou, J. D., and L. L. McKay. 1977. Inorganic salts resistance associated with a lactose-fermenting plasmid in Streptococcus lactis. J. Bacteriol. 13257-265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Francisco, R., M. C. Alpoim, and P. V. Morais. 2002. Diversity of chromium-resistant and -reducing bacteria in a chromium-contaminated activated sludge. J. Appl. Microbiol. 92837-843. [DOI] [PubMed] [Google Scholar]
- 15.Grinsted, J., F. de la Cruz, and R. Schmitt. 1990. The Tn21 subgroup of bacterial transposable elements. Plasmid 24163-189. [DOI] [PubMed] [Google Scholar]
- 16.Hailer, M. K., P. G. Slade, B. D. Martin, and K. D. Sugden. 2005. Nei deficient Escherichia coli are sensitive to chromate and accumulate the oxidized guanine lesion spiroiminodihydantoin. Chem. Res. Toxicol. 181378-1383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Imlay, J. A., and I. Fridovich. 1992. Suppression of oxidative envelope damage by pseudoreversion of a superoxide dismutase-deficient mutant of Escherichia coli. J. Bacteriol. 174953-961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Juhnke, S., N. Peitzsch, N. Hübener, C. Groβe, and D. H. Nies. 2002. New genes involved in chromate resistance in Ralstonia metallidurans strain CH34. Arch. Microbiol. 17915-25. [DOI] [PubMed] [Google Scholar]
- 19.Kovach, M. E., P. H. Elzer, D. S. Hill, G. T. Robertson, M. A. Farris, R. M. Roop II, and K. M. Peterson. 1995. Four new derivatives of the broad-host-range cloning vector pBBR1MCS carrying different antibiotic-resistance cassettes. Gene 166175-176. [DOI] [PubMed] [Google Scholar]
- 20.Liebert, C. A., R. M. Hall, and A. O. Summers. 1999. Transposon Tn21, flagship of the floating genome. Microbiol. Mol. Biol. Rev. 63507-522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Reference deleted.
- 22.Lund, P. A., S. J. Ford, and N. L. Brown. 1986. Transcriptional regulation of the mercury-resistance genes of transposon Tn501. J. Gen. Microbiol. 132465-480. [DOI] [PubMed] [Google Scholar]
- 23.Ma, Y.-F., J.-F. Wu, S.-Y. Wang, C.-Y. Jiang, Y. Zhang, S.-W. Qi, L. Liu, G.-P. Zhao, and S.-J. Liu. 2007. Nucleotide sequence of plasmid pCNB1 from Comamonas strain CNB-1 reveals novel genetic organization and evolution for 4-chloronitrobenzene degradation. Appl. Environ. Microbiol. 774477-4483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Mergeay, M., D. Nies, H. G. Schlegel, J. Gerits, P. Charles, and F. Van Gijsegem. 1985. Alcaligenes eutrophus CH34 is a facultative chemolithotroph with plasmid-bound resistance to heavy metals. J. Bacteriol. 162328-334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Messer, J., M. Reynolds, L. Stoddard, and A. Zhitkovich. 2006. Causes of DNA single-strand breaks during reduction of chromate by glutathione in vitro and in cells. Free Radic. Biol. Med. 401981-1992. [DOI] [PubMed] [Google Scholar]
- 26.Miller, J. H. 1992. A short course in bacterial genetics. A laboratory manual and handbook for Escherichia coli and related bacteria. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY.
- 27.Mondaca, M. A., C. L. González, and C. A. Zaror. 1998. Isolation, characterization and expression of a plasmid encoding chromate resistance in Pseudomonas putida KT2441. Lett. Appl. Microbiol. 26367-371. [Google Scholar]
- 28.Nies, A., D. H. Nies, and S. Silver. 1989. Cloning and expression of plasmid genes encoding resistances to chromate and cobalt in Alcaligenes eutrophus. J. Bacteriol. 1715065-5070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Nies, A., D. H. Nies, and S. Silver. 1990. Nucleotide sequence and expression of a plasmid-encoded chromate resistance determinant from Alcaligenes eutrophus. J. Biol. Chem. 2655648-5653. [PubMed] [Google Scholar]
- 30.Nies, D. H., S. Koch, S. Wachi, N. Peitzsch, and M. H. Saier, Jr. 1998. CHR, a novel family of prokaryotic proton motive force-driven transporters probably containing chromate/sulfate antiporters. J. Bacteriol. 1805799-5802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ohtake, H., C. Cervantes, and S. Silver. 1987. Decreased chromate uptake in Pseudomonas fluorescens carrying a chromate resistance plasmid. J. Bacteriol. 1693853-3856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Peitzsch, N., G. Eberz, and D. H. Nies. 1998. Alcaligenes eutrophus as a bacterial chromate sensor. Appl. Environ. Microbiol. 64453-458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Pimentel, B. E., R. Moreno-Sánchez, and C. Cervantes. 2002. Efflux of chromate by Pseudomonas aeruginosa cells expressing the ChrA protein. FEMS Microbiol. Lett. 212249-254. [DOI] [PubMed] [Google Scholar]
- 34.Quievryn, G., J. Messer, and A. Zhitkovich. 2002. Carcinogenic chromium(VI) induces cross-linking of vitamin C to DNA in vitro and in human lung A549 cells. Biochemistry 413156-3167. [DOI] [PubMed] [Google Scholar]
- 35.Quievryn, G., E. Petersen, J. Messer, and A. Zhitkovich. 2003. Genotoxicity and mutagenicity of chromium(VI)/ascorbate-generated DNA adducts in human and bacterial cells. Biochemistry 421062-1070. [DOI] [PubMed] [Google Scholar]
- 36.Reynolds, M., E. Peterson, G. Quievryn, and A. Zhitkovich. 2004. Human nucleotide excision repair efficiently removes chromium-DNA phosphate adducts and protects cells against chromate toxicity. J. Biol. Chem. 27930419-30424. [DOI] [PubMed] [Google Scholar]
- 37.Sambrook, J., E. F. Fritsch, and T. Maniatis. 1989. Molecular cloning: a laboratory manual, 2nd ed. Cold Spring Harbor Laboratory, Cold Spring Harbor, NY.
- 38.Tauch, A., A. Schlüter, N. Bischoff, A. Goesmann, F. Meyer, and A. Pühler. 2003. The 79,370-bp conjugative plasmid pB4 consists of an IncP-1β backbone loaded with a chromate resistance transposon, the strA-strB streptomycin resistance gene pair, the oxacillinase gene blaNPS-1, and a tripartite antibiotic efflux system of the resistance-nodulation-division family. Mol. Gen. Genomics 268570-584. [DOI] [PubMed] [Google Scholar]
- 39.Tuffin, I. M., P. de Groot, S. M. Deane, and D. E. Rawlings. 2005. An unusual Tn21-like transposon containing an ars operon is present in highly arsenic-resistant strains of the biomining bacterium Acidithiobacillus caldus. Microbiology 1513027-3039. [DOI] [PubMed] [Google Scholar]
- 40.Tuffin, I. M., S. B. Hector, S. M. Deane, and D. E. Rawlings. 2006. Resistance determinants of a highly arsenic-resistant strain of Leptospirillum ferriphilum isolated from a commercial biooxidation tank. Appl. Environ. Microbiol. 722247-2253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Yeo, C. C., J. M. Tham, S. M. Kwong, S. Yiin, and C. L. Poh. 1998. Tn5563, a transposon encoding putative mercuric ion transport proteins located on plasmid pRA2 of Pseudomonas alcaligenes. FEMS Microbiol. Lett. 165253-260. [DOI] [PubMed] [Google Scholar]
- 42.Zhitkovich, A. 2002. Chromium: exposure, toxicity, and biomonitoring approaches, p. 267-285. In S. H. Wilson and W. A. Suk (ed.), Biomarkers of environmentally associated disease: technologies, concepts, and perspectives. CRC Press LLC, New York, NY.
- 43.Zhitkovich, A. 2005. Importance of chromium-DNA adducts in mutagenicity and toxicity of chromium(VI). Chem. Res. Toxicol. 183-11. [DOI] [PubMed] [Google Scholar]