Abstract
The synthesis of “typical” hexa-acylated lipid A occurs via a nine-step enzymatic pathway, which is generally well conserved throughout all gram-negative bacteria. One exception to the rule is Helicobacter pylori, which has only eight homologs to the nine lipid A biosynthetic enzymes. The discrepancy occurs toward the end of the pathway, with H. pylori containing only a single putative secondary acyltransferase encoded by jhp0265. In Escherichia coli K-12, two late acyltransferases, termed LpxL and LpxM, are required for the biosynthesis of hexa-acylated lipid A. Detailed biochemical and genetic analyses reveal that H. pylori Jhp0265 (the protein encoded by jhp0265) is in fact an LpxL homolog, capable of transferring a stearoyl group to the hydroxyl group of the 2′ linked fatty acyl chain of lipid A. Despite the lack of a homolog to LpxM in the H. pylori genome, the organism synthesizes a hexa-acylated lipid A species, suggesting that an equivalent enzyme exists. Using radiolabeled lipid A substrates and acyl-acyl carrier protein as the fatty acyl donor, we were able to confirm the presence of a second H. pylori late acyl transferase by biochemical assays. After synthesis of the hexa-acylated lipid A species, several modification enzymes then function to produce the major lipid A species of H. pylori that is tetra-acylated. Jhp0634 was identified as an outer membrane deacylase that removes the 3′-linked acyl chains of H. pylori lipid A. Together, this work elucidates the biochemical machinery required for the acylation and deacylation of the lipid A domain of H. pylori lipopolysaccharide.
Helicobacter pylori is the only gram-negative bacterium capable of colonizing the human stomach. Such a feat required a great deal of adaptation and, therefore, H. pylori exclusively exists in the human stomach, with no other known reservoir. Such unique host specificity is made possible by the chronic nature of colonization by H. pylori. Unlike most bacterial infections, which are transient, H. pylori can persist in the human stomach for many years allowing transmission between hosts (5, 6). Colonization by H. pylori is usually asymptomatic; however, disease states such as gastritis, peptic ulcer disease, mucosa-associated lymphoid tissue lymphoma, and adenocarcinoma can manifest over time (1, 4, 22). Like other gram-negative bacteria, the major surface component of H. pylori is lipopolysaccharide (LPS). Typically, bacterial LPS is a potent stimulator of the innate immune response; however, H. pylori LPS shows up to 1,000 times lower immunological activity than the LPS of Escherichia coli (17, 20, 21). Therefore, it is thought that the LPS structure of H. pylori has evolved to aid the bacterium in evading the host innate immune system, thereby contributing to chronic infection.
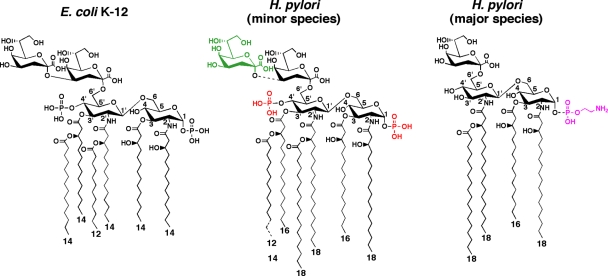
LPS consists of three domains known as lipid A, core, and O antigen (23, 24). For the majority of gram-negative bacteria, production of LPS is necessary for cell growth and viability. The synthesis of “typical” enterobacterial lipid A has been well characterized in E. coli and occurs by a nine-step enzymatic pathway. For the most part, the nine lipid A biosynthetic enzymes are conserved throughout all gram-negative bacteria. Therefore, the majority of gram-negative bacteria are capable of synthesizing a lipid A species closely resembling that of E. coli (Fig. 1) (23, 24).
FIG. 1.
Comparison of E. coli K-12 lipid A to H. pylori minor and major lipid A species. The lipid A of E. coli and the minor lipid A species of H. pylori have very similar structures. The minor lipid A species of H. pylori is then modified by several enzymes, resulting in a reduction in the number of Kdo sugars, phosphate groups, and acyl chains producing the major lipid A species.
Despite the well-conserved lipid A biosynthetic pathway, several lipid A species from a wide range of gram-negative bacteria display considerable heterogeneity compared to E. coli K-12. The heterogeneity can be attributed to the presence of lipid A modification enzymes that either decorate the lipid A species with diverse substituents or remove various functional groups from the lipid A species itself (23, 36). H. pylori is an excellent example of a gram-negative bacterium that modifies its lipid A. The minor lipid A species of H. pylori is similar to that of E. coli and was first identified in H. pylori strain NCTC 11637 and later shown to be synthesized by H. pylori strain 26695 under certain conditions (19, 34). However, the major species is very different, showing changes to the phosphate groups, acyl chains, and the 3-deoxy-d-manno-octulosonic acid (Kdo) sugars (Fig. 1) (19, 33). In some organisms, including H. pylori, modification of the lipid A structure has been shown to promote resistance to cationic antimicrobial peptides and to reduce the endotoxic principle of LPS. The following article clarifies the final steps in the production of the H. pylori minor lipid A species, which involve the addition of the secondary acyl chains to form the hexa-acylated species. The enzyme responsible for the consequent deacylation of the H. pylori lipid A minor species is also identified as Jhp0634.
MATERIALS AND METHODS
Chemicals and other materials.
[γ-32P]ATP and 32Pi were obtained from GE Healthcare. Silica Gel 60 (0.25 mm) thin-layer plates were purchased from EM Separation Technology (Merck). Luria-Bertani (LB) agar and LB broth were from EMD Chemicals. Brucella broth was from Becton Dickinson, and M9 minimal salts were from Difco. Triton X-100 and bicinchoninic acid were from Pierce. All other chemicals were reagent-grade and were purchased from either Sigma or Mallinckrodt.
Bacterial strains and growth conditions.
The bacterial strains and plasmids used in this study are summarized in Table 1. H. pylori strains J99 and 26695 were obtained from the American Type Culture Collection. The H. pylori clinical isolate Hp7-91 was obtained from a human gastric biopsy specimen, as previously described (32). Primary plate cultures of H. pylori were grown from methyl cellulose stock on blood agar medium at 37°C for 36 to 60 h in a microaerobic atmosphere (5% O2, 10% CO2, and 85% N2). The resultant colonies were inoculated into brucella broth supplemented with 7% fetal bovine serum (HyClone) and vancomycin (10 μg/ml). Cells were grown to an A600 of ∼1.0 at 37°C under microaerobic conditions for 24 to 36 h. Prior to every experiment, confirmation of H. pylori was performed by both Gram's staining and a urease test (14). E. coli was typically grown at 37°C in LB broth (18). The E. coli late acyl transferase mutants MKV15, MKV13, and MLK1067 were grown as previously described (38) in M9 minimal medium at 30°C. When required for selection of plasmids, cells were grown in the presence of ampicillin (100 μg/ml).
TABLE 1.
Bacterial strains and plasmids used in this study
| Strain or plasmid | Genotype or description | Source or reference |
|---|---|---|
| Strains | ||
| H. pylori | ||
| J99 | Wild type | ATCC 700824 |
| 26695 | Wild type | ATCC 700392 |
| Hp7-91 | Clinical isolate | |
| Hp7-91/hp0021::cam | H. pylori clinical isolate with chloramphenicol resistance cassette in hp0021 (lpxE) | |
| J99/jhp0265::kan | J99 with kanamycin resistance cassette in jhp0265 | This work |
| J99/jhp0634::cam | J99 with chloramphenicol resistance cassette in jhp0634 | This work |
| J99/jhp0634::cam jhp0634+ | J99/jhp0634::cam rdxA::jhp0634 | This work |
| J99/jhp0265::kan jhp0634::cam | J99 with kanamycin resistance cassette in jhp0265 and chloramphenicol resistance cassette in jhp0634 | This work |
| 26695/hp0280::kan | 26695 with kanamycin resistance cassette in jhp0280 | This work |
| 26695/hp0694::cam | 26695 with chloramphenicol resistance cassette in hp0694 | This work |
| E. coli | This work | |
| XL-1 Blue | recA1 endA1 gyrA96 thi-1 hsdR17 supE44 relA1 lac [F′ proAB lacIqZΔM15::Tn10 (Tetr)] | Stratagene |
| MKV15 | MLK986 lpxP::kan | |
| MKV13 | MLK53 lpxP::kan | |
| MLK1067 | W3110 lpxM::Ωcam | |
| MKV15 (DE3) | MLK986 lpxP::kan (DE3) | This work |
| Plasmids | ||
| pBluescript II SK(+) | Cloning vector; Ampr, lac promoter (lacZ), f1, ColE1 | Stratagene |
| pET21a | Vector containing a T7 promoter; Ampr | Novagen |
| pET21jhp0265 | pET21a containing jhp0265 | This work |
| pET21hp0280 | pET21a containing hp0280 | This work |
| pBLUEjhp0265 | pBluescript II SK(+) containing jhp0265 | This work |
| pBLUEhp0280 | pBluescript II SK(+) containing hp0280 | This work |
| pBSjhp0265 | pBluescript II SK(+) containing jhp0265 ± 1-kb flanking regions | This work |
| pBSjhp0265B | pBSjhp0265 engineered with a BglII restriction site | This work |
| pBSjhp0265::kan | pBSjhp0265B containing a kanamycin resistance cassette in jhp0265 | This work |
| pBShp0694 | pBluescript II SK(+) containing hp0694 ± 1-kb flanking regions | This work |
| pBShp0694N | pBShp0694 engineered with a NdeI restriction site | This work |
| pBShp0694::cam | pBShp0694N containing a chloramphenicol resistance cassette in hp0694 | This work |
| pEThp0954 | pET21a containing hp0954 | This work |
| pET634comp | pEThp0954 with jhp0634 inserted in hp0954 | This work |
Recombinant DNA techniques.
Plasmids were isolated using a QIAprep Spin Minirep Kit (Qiagen). Custom primers were obtained from Integrated DNA Technologies. PCR reagents were purchased from Stratagene. PCR clean up was performed using a Qiaquick PCR Purification Kit (Qiagen). DNA fragments were isolated from agarose gels using a Qiaquick Gel Extraction Kit (Qiagen). Restriction endonucleases, T4 DNA ligase, and Antarctic phosphatase were purchased from New England Biolabs. All modifying enzymes were used according to the manufacturers' instructions.
Overexpression of the late acyl transferases (Jhp0265 and Hp0280) behind a T7lac promoter.
The late acyl transferase genes of H. pylori J99 (jhp0265) and H. pylori 26695 (hp0280) were separately subcloned into pET21a (Novagen) behind the T7lac promoter. The genes were PCR amplified using genomic DNAs as templates. Sequences of primers are shown in Table S1 in the supplemental material. The PCR products and the vector were digested with NdeI and BamHI and ligated overnight at 16°C using T4 DNA ligase (New England BioLabs) to give pET21jhp0265 and pET21hp0280. pET21jhp0265 and pET21hp0280 were transformed into XL-1 Blue (Stratagene) for propagation of the plasmids. pET21jhp0265 and pET21hp0280 were then transformed into MKV15 (DE3) (Table 1) for overexpression of the protein. MKV15 was made DE3-lysogenic using a DE3 Lysogenization Kit (Novagen), according to the manufacturer's instructions.
Cloning of Jhp0265 and Hp0280 into the high-copy-number vector pBluescript SK II(+).
The jhp0265 and hp0280 coding regions, along with a ribosomal binding site, were excised from the plasmids pET21jhp0265 and pET21hp0280 using XbaI and XhoI. The gene fragments were ligated overnight with pBluescript SK II(+), at 16°C using T4 DNA Ligase (New England BioLabs) to give pBLUEjhp0265 and pBLUEhp0280. pBLUEjhp0265 and pBLUEhp0280 were transformed into XL-1 Blue (Stratagene) for propagation of the plasmids. pBLUEjhp0265 and pBLUEhp0280 were then transformed into MKV13, MKV15, and MLK1067 to be used in radiolabeling experiments.
Preparation of cell-free extracts, double-spun cytosol, and washed membrane.
Typically, 500 ml of H. pylori or 200 ml of E. coli cultures was grown to an A600 of ∼1.0 at 37°C and harvested by centrifugation at 6,000 × g for 30 min. All samples were prepared at 4°C. Cell extract, membrane-free cytosol, and washed membranes were prepared as previously described (35) and were stored in aliquots at −20°C. Protein concentration was determined by the bicinchoninic acid method (27), using bovine serum albumin as the standard.
Construction of H. pylori 26695 and J99 late acyl transferase (hp0280 and jhp0265) defective mutants.
The jhp0265 gene and its flanking sequences, including 997 bp upstream and 1,016 bp downstream, were amplified by PCR (primers 1 and 2) from H. pylori J99 genomic DNA using Pfu Turbo (Stratagene) according to the manufacturer's instructions. The amplimer was digested with BamHI and EcoRI, gel purified, and subsequently cloned into the high-copy-number phagemid vector, pBluescript II SK(+) (Stratagene), resulting in the plasmid pBSjhp0265. The vector pBSjhp0265 was then subjected to site-directed mutagenesis, using a QuikChange XL Site-Directed Mutagenesis Kit (Stratagene), to create a BglII (primers 3 and 4) restriction site (pBSjhp0265B). In order to disrupt the jhp0265 gene, a kanamycin resistance cassette (kan) obtained by PCR (primers 5 and 6) from an E. coli-H. pylori shuttle vector (pHel3) (13) was inserted into the BglII and NarI sites of pBSjhp0265B. The resulting plasmid, pBSjhp0265::kan, containing an interrupted jhp0265 gene, was transformed into H. pylori 26695 and J99 by natural transformation (11), and resistant colonies were selected on blood agar plates containing 8 μg/ml of kanamycin. Resistant colonies were repurified on kanamycin-containing plates, and the successful insertion of the resistance cassette was verified by PCR of genomic DNA.
Construction of H. pylori 26695 and J99 3′-O-deacylase (hp0694 and jhp0634) defective mutants.
In a comparison of the gene sequences of H. pylori lpxR homologs found in the microbial database, the 26695 homolog (hp0694) was determined to have a truncation of 237 nucleotides. However, sequencing data using genomic DNA from strain 26695 showed that the reported sequence was missing a thymine between nucleotides 770 and 771, predicting a false stop codon. Using the corrected sequence, we found that hp0694 encodes a full-length homolog of lpxR similar to that of jhp0634 from H. pylori strain J99. The hp0694 gene and its flanking sequences, including 734 bp upstream and 579 bp downstream, were amplified by PCR (primers 7 and 8) from H. pylori 26695 genomic DNA using Pfu Turbo (Stratagene) according to the manufacturer's instructions. The amplimer was digested with XbaI and EcoRI, gel purified, and subsequently cloned into the high-copy-number phagemid vector, pBluescript II SK(+) (Stratagene), resulting in the plasmid pBShp0694. The vector pBShp0694 was then subjected to site-directed mutagenesis, using a QuikChange XL Site-Directed Mutagenesis Kit (Stratagene), to create an NdeI (primers 9 and 10) restriction site (pBShp0694N). In order to disrupt the hp0694 gene, a chloramphenicol resistance cassette (cam) obtained by PCR (primers 11 and 12) from an E. coli-H. pylori shuttle vector (pHel2) (13) was inserted into the NdeI and AvrII sites of pBShp0694N. The resulting plasmid, pBShp0694::cam, containing an interrupted hp0694 gene was transformed into H. pylori 26695 and J99 by natural transformation (11), and resistant colonies were selected on blood agar plates containing 8 μg/ml of chloramphenicol. Resistant colonies were repurified on chloramphenicol-containing plates, and the successful insertion of the resistance cassette was verified by PCR of genomic DNA.
Chromosomal complementation of J99/jhp0634.
rdxA (hp0954) was chosen as the site for chromosomal complementation. RdxA is a nitroreductase that is capable of converting metronidazole from an inactive prodrug into its active form (26). The disruption and consequent inactivation of rdxA by insertion of jhp0634 renders the bacteria resistant to metronidazole because metronidazole can no longer be converted to its active form. hp0954 and its flanking sequences, including 435 bp upstream and 491 bp downstream, were amplified by PCR (primers 13 and 14) from H. pylori 26695 genomic DNA using Pfu Turbo (Stratagene) according to the manufacturer's instructions. The amplimer was digested with XbaI and XhoI, gel purified, and subsequently cloned into pET21a (Novagen), resulting in the plasmid pEThp0954. The vector pEThp0954 was then subjected to inverse PCR (primers 15 and 16) using Pfu Turbo (Stratagene) according to the manufacturer's instructions. The inverse PCR removes 218 bp from the middle of hp0954 and incorporates restriction sites at the ends of the remaining sequence to allow insertion of jhp0634. The inverse PCR amplimer was digested with BamHI and EcoRI, gel purified, and treated with Antarctic phosphatase. The jhp0634 gene was amplified by PCR (primers 17 and 18) from H. pylori J99 genomic DNA using Pfu Turbo (Stratagene) according to the manufacturer's instructions. The jhp0634 amplimer was digested with BamHI and EcoRI, gel purified, and subsequently ligated with the inverse PCR amplimer, resulting in the plasmid pET634comp. The completed pET634 plasmid pET634comp was transformed into H. pylori J99 by natural transformation (11), and resistant colonies were selected on blood agar plates containing 12 μg/ml of metronidazole. Resistant colonies were repurified on metronidazole-containing plates, and the successful insertion of the complementation cassette was verified by PCR of genomic DNA.
Preparation of radiolabeled substrates.
The substrate [4′-32P]lipid IVA was generated from 125 μCi of [γ-32P]ATP and the tetra-acyl-disaccharide 1-phosphate lipid acceptor, using the overexpressed 4′ kinase present in membranes of E. coli BLR(DE3)/pLysS/pJK2 as previously described (35). Kdo2-[4′-32P]lipid IVA was prepared by adding purified E. coli Kdo transferase (KdtA) immediately after the 4′ kinase, as previously described (2). Kdo2-[4′-32P]lipid A was prepared by adding membranes of E. coli BLR(DE3)/HtrB and E. coli BLR(DE3)/MsbB and C12:0-acyl carrier protein (ACP) immediately after the Kdo transferase reaction, as previously described (35). Kdo2-lauroyl-[4′-32P]lipid IVA was prepared in the same way as Kdo2-[4′-32P]lipid A, except that E. coli BLR(DE3)/MsbB membranes were omitted, and the C12:0-ACP was added at a 1:1 molar ratio.
Assay of Jhp0265 activity.
Jhp0265 activity was assayed under optimized conditions in a 10-μl reaction mixture containing 50 mM HEPES, pH 7.5, 0.1% Triton X-100, 50 mM NaCl, 5 mM MgCl2, 0.1 mg/ml bovine serum albumin, 2.5 μM lipid A substrate ([4′-32P]lipid IVA, Kdo2-[4′-32P]lipid IVA, or Kdo2-[4′-32P]lipid A at ∼5,000 cpm/nmol) and either 5 μM ACPSH, C12:0-ACP, C14:0-ACP, C16:0-ACP, or C18:0-ACP. Washed MKV15 (DE3) pET21jhp0265 membranes at 0.01 mg/ml were employed as the enzyme source. Enzymatic reactions were incubated at 30°C for 30 min and terminated by spotting 4.5-μl portions of the mixtures onto Silica Gel 60 thin-layer chromatography (TLC) plates. The plates were dried under a cool air stream for 20 min.
When [4′-32P]lipid IVA was employed as the substrate, reaction products were separated using the solvent chloroform, pyridine, 88% formic acid, and water (50:50:16:5, vol/vol). The reaction products generated from substrates having the Kdo moiety were separated using the solvent chloroform, pyridine, 88% formic acid, and water (30:70:16:10, vol/vol). TLC plates were exposed overnight to a PhosphorImager screen, and product formation was detected and analyzed using a Bio-Rad Molecular Imager PhosphorImager equipped with Quantity One software. The enzyme activity was calculated by determining the percentage of the substrate converted to product.
Assay of unknown acyl transferase activity.
The unknown acyl transferase activity was assayed under optimized conditions in a 10-μl reaction mixture containing 50 mM HEPES, pH 8, 0.2% Triton X-100, 1 mM EDTA, 1 mM dithiothreitol, 2.5 μM lipid A substrate ([4′-32P]lipid IVA, Kdo2-[4′-32P]lipid IVA, Kdo2-lauroyl-[4′-32P]lipid IVA, or Kdo2-[4′-32P]lipid A at ∼5,000 cpm/nmol) and either 5 μM ACPSH, C12:0-ACP, C14:0-ACP, or C18:0-ACP. Washed membranes from H. pylori strain Hp7-91/hp0021::cam (34) at 1 mg/ml were employed as the enzyme source, as indicated in Fig. 6. Enzymatic reactions were incubated at 30°C for 60 min and terminated by spotting 4.5-μl portions of the mixtures onto Silica Gel 60 TLC plates. The plates were dried under a cool air stream for 20 min.
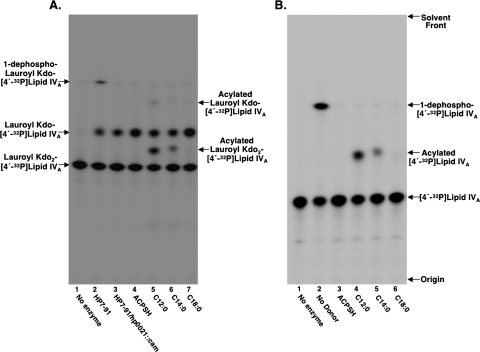
FIG. 6.
Demonstration of a second lipid A late acyl transferase in H. pylori membrane fractions. H. pylori membranes lacking a functional 1-phosphatase (Hp0021) were assayed for late acyl transferase activity with either 2.5 μM lauroyl Kdo2-[4′-32P]lipid IVA-5 μM acyl-ACPs (A) or 2.5 μM [4′-32P]lipid IVA-5 μM acyl-ACPs (B). The lauroyl (C12:0) group is attached at the 2′ position of the substrate used in panel A.
When [4′-32P]lipid IVA was employed as the substrate, reaction products were separated using a solvent consisting of chloroform, pyridine, 88% formic acid, and water (50:50:16:5, vol/vol). The reaction products generated from substrates having the Kdo moiety were separated using solvent consisting of chloroform, pyridine, 88% formic acid, and water (30:70:16:10, vol/vol). TLC plates were exposed overnight to a PhosphorImager Screen and product formation detected and analyzed using a Bio-Rad Molecular Imager PhosphorImager equipped with Quantity One software. The enzyme activity was calculated by determining the percentage of the substrate converted to product.
Isolation and analysis of lipid A species from 32Pi-labeled cells.
MKV13, MKV15, and MLK1067 cells containing pBLUEjhp0265, pBLUEhp0280, and vector controls were grown in G56 minimal medium at 30°C. The cultures were labeled uniformly with 2.5 μCi/ml 32Pi and induced with 0.5 mM isopropyl-β-d-thiogalactopyranoside after 2 h of growth. Bacteria were harvested using a clinical centrifuge and washed with 5 ml of phosphate-buffered saline (pH 7.4). 32P-labeled lipid A was isolated using published protocols (31) and spotted onto a Silica Gel 60 TLC plate (∼10,000 cpm/lane). Lipids were separated using a solvent consisting of chloroform, pyridine, 88% formic acid, and water (50:50:16:5, vol/vol). The TLC plates were exposed overnight to a PhosphorImager screen, and product formation was detected and analyzed using a Bio-Rad Molecular Imager PhosphorImager equipped with Quantity One software.
Large-scale isolation of lipid A for mass spectrometry analysis.
Cultures (500 ml) of each strain were grown overnight at 37°C. Cells were harvested by centrifugation at 6,000 × g for 30 min and washed once with phosphate-buffered saline. The final cell pellets were resuspended in 20 ml of phosphate-buffered saline. Lipid A was released from cells and purified as described previously (34) and stored frozen at −20°C.
Mass spectrometry of lipid A species.
Mass spectra of purified lipids were acquired in the negative ion linear mode using a matrix-assisted laser desorption-ionization-time of flight (MALDI-TOF) mass spectrometer (AXIMA-CFR; Kratos Analytical, Manchester, United Kingdom), equipped with a nitrogen laser (337 nm). The instrument was operated using 20-kV extraction voltage and time-delayed extraction, providing a mass resolution of about ±1 atomic mass units for compounds with an Mr of ∼2,000. Each spectrum represented the average of 100 laser shots, and saturated 6-aza-2-thiothymine in 50% acetonitrile and 10% tribasic ammonium citrate (9:1, vol/vol) served as the matrix. The samples were dissolved in chloroform-methanol (4:1, vol/vol) and deposited on the sample plate, followed by an equal portion of matrix solution (0.3 μl). The sample was dried at 25°C prior to mass analysis.
RESULTS
Jhp0265 functions as a lipid A late acyl transferase.
H. pylori is capable of synthesizing a hexa-acylated lipid A species with two secondary acyl chains although this is only a minor species (Fig. 1) (19, 28, 34). This would suggest that H. pylori has two lipid A secondary acyl chain transferases, homologous to LpxL and LpxM in E. coli. However, the COG (clusters of orthologous groups) database (29) predicts only one late acyl transferase in H. pylori strain J99, designated Jhp0265. This would suggest that either Jhp0265 is dual functional, which is unlikely, or an as yet undiscovered late acyl transferase exists. To characterize the predicted function of jhp0265, jhp0265 was cloned into the pET21a vector and heterologously expressed in E. coli MKV15 (DE3). MKV15 (DE3) is an E. coli strain that lacks the two late acyl transferases LpxL and LpxM and the cold-shock-induced acyl transferase LpxP (38), thereby eliminating interference from endogenous lipid A late acyl transferase enzymes. In vitro assays of overexpressed Jhp0265 membranes using radiolabeled Kdo2-[4′-32P]lipid IVA as the substrate and acyl-ACPs of various chain lengths as the acyl chain donors showed that Jhp0265 is capable of transferring an acyl chain to lipid A (Fig. 2A). To address the dual functionality issue, combinations of acyl-ACPs with different acyl chain lengths were used under the same assay conditions as above. Despite the presence of two different acyl-ACPs, only one product spot was apparent, indicating that Jhp0265 is monofunctional (Fig. 2A). The LpxL/LpxM homologue of H. pylori strain 26695, Hp0280, was also investigated and showed identical results to those of Jhp0265 (data not shown).
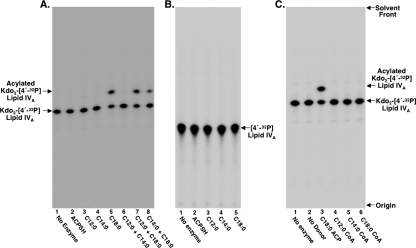
FIG. 2.
Enzymatic assays to investigate Jhp0265 functionality. MKV15 (DE3) pET21jhp0265 membranes were assayed with either 2.5 μM Kdo2-[4′-32P]lipid IVA-5 μM acyl-ACPs (A), 2.5 μM [4′-32P]lipid IVA-5 μM acyl-ACPs (B), or 2.5 μM Kdo2-[4′-32P]lipid IVA-5 μM acyl-CoAs (C). Appearance of a faster migrating, more hydrophobic, reaction product indicates acyl transferase activity.
Jhp0265 displays the same enzymatic characteristics as the E. coli late acyl transferases.
Both LpxL and LpxM have very specific substrate and acyl chain donor specificities. Prior to acylation by LpxL or LpxM, the lipid IVA donor must first be substituted with two Kdo residues at the 6′ position for optimal activity (7, 8). This is also the case with Jhp0265, which cannot use lipid IVA as an acceptor molecule (Fig. 2B) unless the Kdo sugars are present (Fig. 2A). With regard to the acyl chain donor, both LpxL and LpxM utilize acyl-ACPs rather than acyl coenzyme A (acyl-CoA) as their preferred substrates. The length of the acyl chain associated with the ACP is also critical, with LpxL preferring lauroyl (C12:0)-ACP and LpxM preferring myristoyl (C14:0)-ACP. Results shown in Fig. 2C demonstrate that Jhp0265 cannot utilize acyl-CoAs as acyl chain donors. Jhp0265 is also highly specific for stearoyl (C18:0)-ACP compared to lauroyl-ACP and myristoyl-ACP (Fig. 2A). Jhp0265 does display some activity with palmitoyl (C16:0)-ACP (data not shown); however, this is not biologically significant because the H. pylori lipid A does not contain a palmitate in an acyloxyacyl linkage.
In vivo analyses in E. coli show that Jhp0265 is an LpxL homologue.
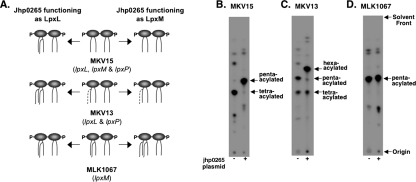
The level of specificity for E. coli LpxL and LpxM, described above, becomes more complex when we take into account the position at which each enzyme adds an acyl chain. LpxL specifically transfers an acyl chain to the hydroxyl group of the 2′ acyl chain of lipid A, and LpxM preferentially transfers an acyl chain to the hydroxyl group of the 3′ acyl chain of lipid A. The order of each addition is also predetermined because LpxM works efficiently only after a secondary acyl chain has been transferred to Kdo2-lipid IVA by LpxL (7, 8, 38). The in vitro data for Jhp0265 presented so far would suggest that Jhp0265 is an LpxL homologue, given that Jhp0265 preferentially transfers a C18:0 acyl-ACP and the published H. pylori lipid A structure shows a C18:0 secondary acyl chain linked to the 2′ acyl chain (Fig. 1). A series of E. coli late acyl transferase mutants were used to confirm this assumption in an in vivo setting. E. coli MKV15 (lpxL lpxP lpxM) has a tetra-acylated lipid A structure (Fig. 3A) (38). E. coli MKV13 (lpxL lpxP) has both tetra- and penta-acylated lipid A structures since some residual LpxM activity is present in vivo (Fig. 3A) (37). Finally, E. coli MLK1067 (lpxM) has a penta-acylated lipid A structure (Fig. 3A) (15). Each strain expressing jhp0265 was radiolabeled with 32Pi, along with a vector control, as described in Materials and Methods. The lipid A was then isolated and visualized using a PhosphorImager. If Jhp0265 adds to the 2′ position, as does E. coli LpxL, the MKV15 mutant expressing jhp0265 would produce a penta-acylated lipid A species, instead of the tetra-acylated species of the vector control (Fig. 3A). The MKV13 mutant overexpressing jhp0265 would be able to make a hexa-acylated lipid A species because addition of an acyl chain by Jhp0265 would allow the endogenous LpxM enzyme to function efficiently (Fig. 3A). MLK1067 overexpressing jhp0265 should show no change in the number of acyl chains because it already has a functional LpxL (Fig. 3A). The results shown in Fig. 3B through D show that all of the above conditions are met, confirming that Jhp0265 adds to the 2′-linked acyl chain and is, therefore, an LpxL homologue.
FIG. 3.
Radiolabeling of E. coli late acyl transferase mutants overexpressing Jhp0265. The center column of panel A shows a cartoon representation of the lipid A species from each of the E. coli lipid A late acyl transferase mutants. The left and right columns demonstrate the expected lipid A structures after expression of Jhp0265. On the left side, it is assumed that Jhp0265 behaves like LpxL, and on the right side, it is assumed that Jhp0265 behaves like LpxM. Each strain was radiolabeled using 2.5 μCi/ml inorganic 32Pi. (B to D) Lipid A was isolated from the indicated strain, separated by TLC, and visualized using a PhosphorImager.
An H. pylori jhp0265 mutant shows no change in lipid A structure.
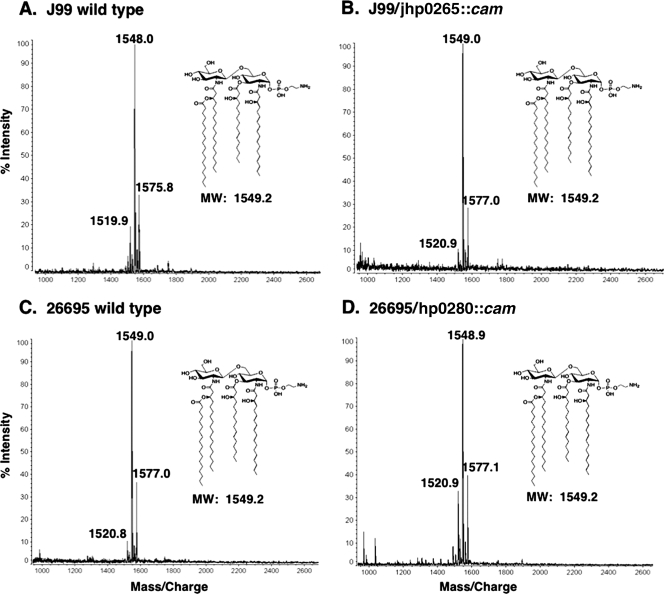
To confirm the in vitro data on Jhp0265, a jhp0265 mutant was constructed in H. pylori strain J99. E. coli lpxL mutants are temperature sensitive for growth at 37°C, whereas the growth rate of the Helicobacter jhp0265 mutant was similar to that of wild-type J99 (data not shown). The J99/jhp0265::kan mutant and J99 wild-type control were grown in liquid medium, and the lipid A was isolated from both strains, followed by mass spectrometric analysis. Surprisingly the mutant showed the same major molecular ion at m/z 1549.0 as wild-type J99 (Fig. 4A and B). Furthermore, tandem mass spectrometry analysis confirmed that the jhp2065 mutant displayed the same acyl chain arrangement as wild-type tetra-acylated H. pylori lipid A (data not shown). Because of this unexpected result, a lipid A late acyl transferase mutant was also constructed and analyzed in strain 26695. The results were the same as for strain J99, as demonstrated in Fig. 4C and D. However, membranes isolated from J99 jhp0265 mutants were unable to catalyze the transfer of a C18:0 fatty acyl chain onto Kdo2-lipid IVA during in vitro assay (data not shown). We speculate that a triacylated lipid A is nonviable in H. pylori, presumably due to a decrease in membrane stability, and that a second site suppressor allowing for C18:0 addition in the absence of functional Jhp0265 permits growth.
FIG. 4.
MALDI-TOF mass spectrometry of H. pylori lipid A late acyl transferase mutants. Panels A and C show spectra for wild-type strains J99 and 26695, respectively. The major ion in panel A is m/z 1,549, and the major ion in panel C is m/z 1,548, which corresponds to the predicted molecular weight (MW) for wild-type H. pylori (1549.2). Panels B and D show spectra for mutant strains J99/jhp0265::cam and 26695/hp0280::cam, respectively. Both mutants have a major ion peak at m/z 1,549, which corresponds to an H. pylori wild-type lipid A structure.
Engineering an H. pylori hexa-acylated lipid A mutant enabled functional studies of Jhp0265.
The tetra-acylated major lipid A species of H. pylori is made by removal of the two 3′ linked acyl chains from the hexa-acylated minor lipid A species (33). Recently an outer membrane enzyme capable of cleaving the 3′ linked acyl chains of lipid A, termed LpxR, was discovered in Salmonella enterica serovar Typhimurium (25), and the H. pylori protein Jhp0634 is a distant homolog (E value of <10−3 using BLASTp). Inactivation of jhp0634 in H. pylori should produce a hexa-acylated major lipid A species. Furthermore, we reasoned that inactivation of H. pylori jhp0265 in the deacylase mutant background would confirm Jhp0265 function because the double mutant would be penta-acylated and, therefore, viable.
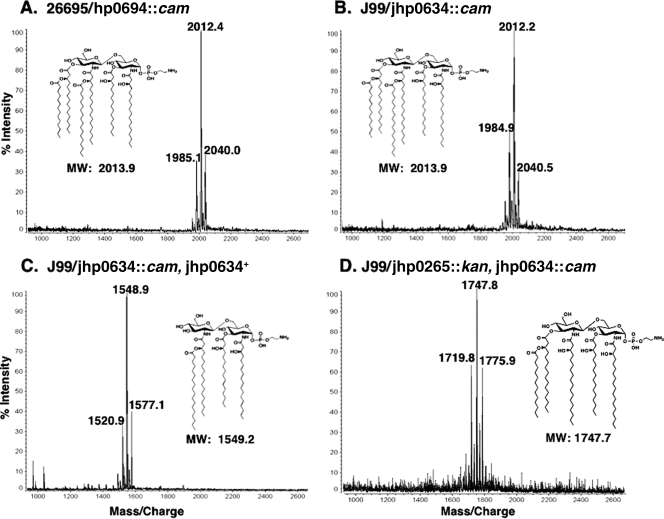
The function of Jhp0634 was investigated in H. pylori J99 by mass spectrometric analysis of the lipid A isolated from a jhp0634 knockout mutant. The jhp0634 mutant synthesized a hexa-acylated lipid A (Fig. 5A), confirming that Jhp0634 was in fact a lipid A 3′-O-deacylase. An identical result was obtained upon deletion of Hp0694, the LpxR homolog in H. pylori strain 26695 (Fig. 5B). Chromosomal complementation of the jhp0634 mutant by insertion of a wild-type copy of jhp0634 into rdxA (jhp0954) (see Materials and Methods) restored the tetra-acylated lipid A phenotype in strain J99 (Fig. 5C), ruling out any polar effects. jhp0265 was then knocked out in the J99/jhp0634::cam background, producing a double mutant. The lipid A isolated from the jhp0634 jhp0265 double mutant was analyzed by mass spectrometry (Fig. 5D). As hypothesized above, the double mutant was penta-acylated and lacked a C18:0 fatty acyl chain at the 2′ position. This is consistent with the in vitro data and confirms that Jhp0265 functions as a lipid A late acyltransferase in whole cells.
FIG. 5.
MALDI-TOF mass spectrometry of H. pylori lipid A acyl chain mutants. Panels A and B show 26695 and J99 3′-O-deacylase mutants, respectively. Both spectra have a major ion peak at m/z 2,012, corresponding to a hexa-acylated lipid A species, showing inactivation of the 3′-O-deacylase. In panel C the complemented J99 3′-O-deacylase mutant is shown. A major ion peak at m/z 1,548.9 indicates that the complemented mutant has reverted back to a wild-type phenotype. Panel D shows the spectrum of a J99 jhp0634 jhp0265 double mutant. The major ion peak at m/z 1,747.8 indicates that the double mutant is penta-acylated. MW, molecular weight.
H. pylori membrane extracts confirm the presence of a second lipid A late acyl transferase.
Because Jhp0265 is not capable of transferring a secondary acyl chain to the 3′ linked acyl chain of H. pylori lipid A, a second acyl transferase must be present in the H. pylori genome that does not share homology to E. coli LpxM. H. pylori/hp0021::cam membranes were assayed using radiolabeled 2′-lauroyl-Kdo2-[4′-32P]lipid IVA as the substrate and acyl-ACPs of various chain lengths as the acyl chain donors in an attempt to demonstrate an activity for a second H. pylori lipid A late acyl transferase. H. pylori/hp0021::cam lacks the 1-phosphatase (31, 34) that is normally responsible for the removal of the 1-phosphate group of H. pylori lipid A, which simplifies interpretation of the assay results by reducing the number of visible products following TLC. However, an H. pylori enzyme that removes the outer Kdo sugar from lauroyl-Kdo2-[4′-32P]lipid IVA is still active under the assay conditions used (Fig. 6A, lane 3). Results shown in Fig. 6A indicate that H. pylori membranes are capable of adding a secondary acyl chain to the 3′ linked acyl chain of lipid A, confirming the presence of a second lipid A late acyl transferase in the H. pylori genome. The experiment shown in Fig. 6A also demonstrates that the unidentified late acyl transferase prefers lauroyl (C12:0)- or myristoyl (C14:0)-ACP, which is consistent with the published H. pylori lipid A structure (Fig. 1). Unlike E. coli LpxM, the H. pylori enzyme was able to transfer an acyl chain to the lipid IVA backbone without prior addition of the Kdo groups or the secondary acyl chain at the 2′ position (Fig. 6B). The enzyme did not utilize acyl-CoA as a substrate (data not shown). Additionally, the enzyme was unable to transfer a fatty acyl chain to hexa-acylated Kdo2-[4′-32P]lipid A. Both the 2′ and 3′ acyl chains of hexa-acylated Kdo2-[4′-32P]lipid A, present on the distal side of the molecule, have secondary acyl chains; therefore, only the acyl chains on the proximal side of the substrate can be substituted. That the unidentified acyl transferase could not transfer an acyl chain to hexa-acylated Kdo2-[4′-32P]lipid A suggests that it acylates lipid A on the distal side of the molecule (data not shown).
DISCUSSION
The constitutive lipid A biosynthetic pathway of E. coli K-12 is a highly ordered process requiring nine enzymatic steps to produce hexa-acylated lipid A. Since lipid A is generally required for viability, the pathway is well conserved in nearly all gram-negative bacteria (23, 36). H. pylori is an exception because it has a single homolog (Jhp0265) to the E. coli late acyl transferases, which we were able to characterize and designate an LpxL homologue. Interestingly, an H. pylori jhp0265 mutant does not produce a triacylated lipid A. Instead, the lipid A species of the jhp0265 mutant is tetra-acylated, just like the wild-type lipid A species, signifying that an additional C18:0 acyltransferase is present in H. pylori. Possible candidates are acyltransferases involved in phospholipid biosynthesis such as PlsB, PlsC, and PlsY (39). H. pylori contains homologs of PlsC and PlsY but no homolog of E. coli PlsB, which is primarily found in gammaproteobacteria. Whether these proteins play a role in acylation of H. pylori lipid A is under investigation. Another possibility would be a PagP-like enzyme that acylates H. pylori lipid A in the outer membrane. In S. enterica serovar Typhimurium and E. coli, PagP catalyzes the addition of a palmitate to one of the primary linked acyl chains of lipid A (3). However, thus far such an activity has not been detected in wild-type H. pylori membranes during in vitro assays (data not shown).
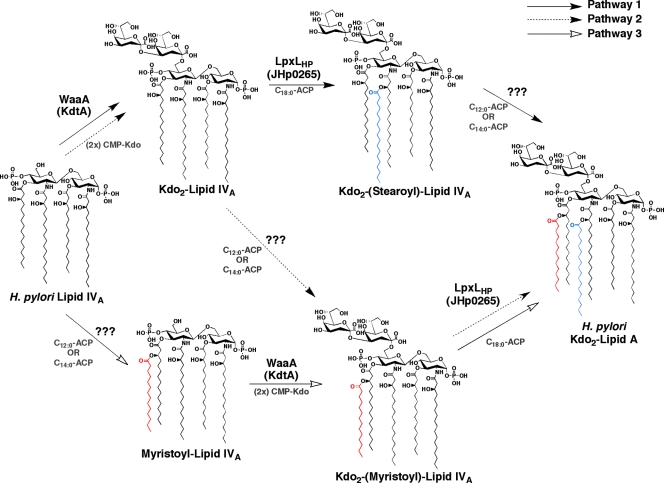
We have also demonstrated that H. pylori expresses a second lipid A late acyl transferase catalyzing the addition of a C12:0 or C14:0 fatty acyl chain; however, its enzymatic properties differ greatly from those of E. coli LpxM. The most striking difference is that the second lipid A late acyl transferase can transfer an acyl chain to lipid IVA in vitro. This means that the order of function for the late acyltransferases cannot be determined (Fig. 7) and opens the question as to how H. pylori ensures that a hexa-acylated lipid A species makes it to the outer membrane. The most likely answer is that the inner membrane flippase, MsbA (10), can efficiently transport only a hexa-acylated lipid A species.
FIG. 7.
Final stages of lipid A synthesis in H. pylori. Because of the less stringent substrate specificity of the unidentified lipid A late acyl transferase of H. pylori, three possible pathways exist. This contradicts the previously established pathway, elucidated in E. coli, which occurs in a very specific and ordered manner.
Even though H. pylori synthesizes a hexa-acylated lipid A, it does not survive long before it is deacylated at the 3′ position to form the tetra-acylated major lipid A species. We confirmed that Jhp0634 is responsible for the 3′-O-deacylase activity by making a knockout mutant. Jhp0634 is a distant homolog of Salmonella LpxR, an outer membrane enzyme that removes the intact acyloxyacyl group at the 3′ position of lipid A (25). Despite the presence of a 3′-O-deacylase in the Salmonella outer membrane, the bacterium does not produce 3′-O-deacylated lipid A species under any growth conditions tested to date. On the other hand, the 3′-O-deacylase of H. pylori is constitutively active, suggesting that in the highly specialized niche that H. pylori occupies, a tetra-acylated lipid A species is advantageous for survival. One explanation for a tetra-acylated lipid A could be evasion of the innate immune system. It is well known that H. pylori lipid A is approximately 1,000-fold less potent at stimulating Toll-like receptor 4 than E. coli lipid A (17, 20, 21). Acyl chain number is a key factor in determining endotoxicity in other bacteria (9, 12, 16, 30). Understanding the biochemical processes required for the synthesis and modification of H. pylori lipid A will facilitate future experiments to define the contribution of its unusual acylation pattern toward evasion of the innate immune response.
Supplementary Material
Acknowledgments
This work was supported by National Institutes of Health grants AI064184 and AI076322 to M.S.T. A.B. was supported by a grant (U54 RR020839 to J. Boeke) from NIH.
Mass spectrometry was carried out at the Middle Atlantic Mass Spectrometry Laboratory at the Johns Hopkins School of Medicine.
Footnotes
Published ahead of print on 29 August 2008.
Supplemental material for this article may be found at http://jb.asm.org/.
REFERENCES
- 1.Bayerdorffer, E., A. Neubauer, B. Rudolph, C. Thiede, N. Lehn, S. Eidt, M. Stolte, et al. 1995. Regression of primary gastric lymphoma of mucosa-associated lymphoid tissue type after cure of Helicobacter pylori infection. Lancet 3451591-1594. [DOI] [PubMed] [Google Scholar]
- 2.Belunis, C. J., and C. R. Raetz. 1992. Biosynthesis of endotoxins. Purification and catalytic properties of 3-deoxy-d-manno-octulosonic acid transferase from Escherichia coli. J. Biol. Chem. 2679988-9997. [PubMed] [Google Scholar]
- 3.Bishop, R. E., H. S. Gibbons, T. Guina, M. S. Trent, S. I. Miller, and C. R. Raetz. 2000. Transfer of palmitate from phospholipids to lipid A in outer membranes of gram-negative bacteria. EMBO J. 195071-5080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Blaser, M. J. 1996. The bacteria behind ulcers. Sci. Am. 274104-107. [DOI] [PubMed] [Google Scholar]
- 5.Blaser, M. J. 1997. The versatility of Helicobacter pylori in the adaptation to the human stomach. J. Physiol. Pharmacol. 48307-314. [PubMed] [Google Scholar]
- 6.Blaser, M. J., and J. C. Atherton. 2004. Helicobacter pylori persistence: biology and disease. J. Clin. Investig. 113321-333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Clementz, T., J. J. Bednarski, and C. R. Raetz. 1996. Function of the htrB high temperature requirement gene of Escherchia coli in the acylation of lipid A: HtrB catalyzed incorporation of laurate. J. Biol. Chem. 27112095-12102. [DOI] [PubMed] [Google Scholar]
- 8.Clementz, T., Z. Zhou, and C. R. Raetz. 1997. Function of the Escherichia coli msbB gene, a multicopy suppressor of htrB knockouts, in the acylation of lipid A. Acylation by MsbB follows laurate incorporation by HtrB. J. Biol. Chem. 27210353-10360. [DOI] [PubMed] [Google Scholar]
- 9.Coats, S. R., T. T. Pham, B. W. Bainbridge, R. A. Reife, and R. P. Darveau. 2005. MD-2 mediates the ability of tetra-acylated and penta-acylated lipopolysaccharides to antagonize Escherichia coli lipopolysaccharide at the TLR4 signaling complex. J. Immunol. 1754490-4498. [DOI] [PubMed] [Google Scholar]
- 10.Doerrler, W. T., M. C. Reedy, and C. R. Raetz. 2001. An Escherichia coli mutant defective in lipid export. J. Biol. Chem. 27611461-11464. [DOI] [PubMed] [Google Scholar]
- 11.Haas, R., T. F. Meyer, and J. P. van Putten. 1993. A flagellated mutants of Helicobacter pylori generated by genetic transformation of naturally competent strains using transposon shuttle mutagenesis. Mol. Microbiol. 8753-760. [DOI] [PubMed] [Google Scholar]
- 12.Hajjar, A. M., R. K. Ernst, J. H. Tsai, C. B. Wilson, and S. I. Miller. 2002. Human Toll-like receptor 4 recognizes host-specific LPS modifications. Nat. Immunol. 3354-359. [DOI] [PubMed] [Google Scholar]
- 13.Heuermann, D., and R. Haas. 1998. A stable shuttle vector system for efficient genetic complementation of Helicobacter pylori strains by transformation and conjugation. Mol. Gen. Genet. 257519-528. [DOI] [PubMed] [Google Scholar]
- 14.Jerris, R. C. 1995. Helicobacter, p. 492-498. In P. R. Murray (ed.), Manual of clinical microbiology, 6th ed., vol. 38. American Society of Microbiology, Washington, DC. [Google Scholar]
- 15.Karow, M., O. Fayet, and C. Georgopoulos. 1992. The lethal phenotype caused by null mutations in the Escherichia coli htrB gene is suppressed by mutations in the accBC operon, encoding two subunits of acetyl coenzyme A carboxylase. J. Bacteriol. 1747407-7418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kawasaki, K., R. K. Ernst, and S. I. Miller. 2004. 3-O-Deacylation of lipid A by PagL, a PhoP/PhoQ-regulated deacylase of Salmonella typhimurium, modulates signaling through Toll-like receptor 4. J. Biol. Chem. 27920044-20048. [DOI] [PubMed] [Google Scholar]
- 17.Mandell, L., A. P. Moran, A. Cocchiarella, J. Houghton, N. Taylor, J. G. Fox, T. C. Wang, and E. A. Kurt-Jones. 2004. Intact gram-negative Helicobacter pylori, Helicobacter felis, and Helicobacter hepaticus bacteria activate innate immunity via Toll-like receptor 2 but not Toll-like receptor 4. Infect. Immun. 726446-6454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Miller, J. R. 1972. Experiments in molecular genetics. Cold Spring Harbor Laboratory Press, Cold Springs Harbor, NY.
- 19.Moran, A. P., B. Lindner, and E. J. Walsh. 1997. Structural characterization of the lipid A component of Helicobacter pylori rough- and smooth-form lipopolysaccharides. J. Bacteriol. 1796453-6463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ogawa, T., Y. Asai, Y. Sakai, M. Oikawa, K. Fukase, Y. Suda, S. Kusumoto, and T. Tamura. 2003. Endotoxic and immunobiological activities of a chemically synthesized lipid A of Helicobacter pylori strain 206-1. FEMS Immunol. Med. Microbiol. 361-7. [DOI] [PubMed] [Google Scholar]
- 21.Ogawa, T., Y. Suda, W. Kashihara, T. Hayashi, T. Shimoyama, S. Kusumoto, and T. Tamura. 1997. Immunobiological activities of chemically defined lipid A from Helicobacter pylori LPS in comparison with Porphyromonas gingivalis lipid A and Escherichia coli-type synthetic lipid A (compound 506). Vaccine 151598-1605. [DOI] [PubMed] [Google Scholar]
- 22.Parsonnet, J., G. D. Friedman, D. P. Vandersteen, Y. Chang, J. H. Vogelman, N. Orentreich, and R. K. Sibley. 1991. Helicobacter pylori infection and the risk of gastric carcinoma. N. Engl. J. Med. 3251127-1131. [DOI] [PubMed] [Google Scholar]
- 23.Raetz, C. R., C. M. Reynolds, M. S. Trent, and R. E. Bishop. 2007. Lipid A modification systems in gram-negative bacteria. Annu. Rev. Biochem. 76295-329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Raetz, C. R., and C. Whitfield. 2002. Lipopolysaccharide endotoxins. Annu. Rev. Biochem. 71635-700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Reynolds, C. M., A. A. Ribeiro, S. C. McGrath, R. J. Cotter, C. R. Raetz, and M. S. Trent. 2006. An outer membrane enzyme encoded by Salmonella typhimurium lpxR that removes the 3′-acyloxyacyl moiety of lipid A. J. Biol. Chem. 28121974-21987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Smeets, L. C., J. J. Bijlsma, S. Y. Boomkens, C. M. Vandenbroucke-Grauls, and J. G. Kusters. 2000. comH, a novel gene essential for natural transformation of Helicobacter pylori. J. Bacteriol. 1823948-3954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Smith, P. K., R. I. Krohn, G. T. Hermanson, A. K. Mallia, F. H. Gartner, M. D. Provenzano, E. K. Fujimoto, N. M. Goeke, B. J. Olson, and D. C. Klenk. 1985. Measurement of protein using bicinchoninic acid. Anal. Biochem. 15076-85. [DOI] [PubMed] [Google Scholar]
- 28.Stead, C., A. Tran, D. Ferguson, Jr., S. McGrath, R. Cotter, and S. Trent. 2005. A novel 3-deoxy-d-manno-octulosonic acid (Kdo) hydrolase that removes the outer Kdo sugar of Helicobacter pylori lipopolysaccharide. J. Bacteriol. 1873374-3383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Tatusov, R. L., M. Y. Galperin, D. A. Natale, and E. V. Koonin. 2000. The COG database: a tool for genome-scale analysis of protein functions and evolution. Nucleic Acids Res. 2833-36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Teghanemt, A., D. Zhang, E. N. Levis, J. P. Weiss, and T. L. Gioannini. 2005. Molecular basis of reduced potency of underacylated endotoxins. J. Immunol. 1754669-4676. [DOI] [PubMed] [Google Scholar]
- 31.Tran, A. X., M. J. Karbarz, X. Wang, C. R. Raetz, S. C. McGrath, R. J. Cotter, and M. S. Trent. 2004. Periplasmic cleavage and modification of the 1-phosphate group of Helicobacter pylori lipid A. J. Biol. Chem. 27955780-55791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Tran, A. X., M. E. Lester, C. M. Stead, C. R. Raetz, D. J. Maskell, S. C. McGrath, R. J. Cotter, and M. S. Trent. 2005. Resistance to the antimicrobial peptide polymyxin requires myristoylation of Escherichia coli and Salmonella typhimurium lipid A. J. Biol. Chem. 28028186-28194. [DOI] [PubMed] [Google Scholar]
- 33.Tran, A. X., C. M. Stead, and M. S. Trent. 2005. Remodeling of Helicobacter pylori lipopolysaccharide. J. Endotoxin Res. 11161-166. [DOI] [PubMed] [Google Scholar]
- 34.Tran, A. X., J. D. Whittimore, P. B. Wyrick, S. C. McGrath, R. J. Cotter, and M. S. Trent. 2006. The lipid A 1-phosphatase of Helicobacter pylori is required for resistance to the antimicrobial peptide polymyxin. J. Bacteriol. 1884531-4541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Trent, M. S., W. Pabich, C. R. Raetz, and S. I. Miller. 2001. A PhoP/PhoQ-induced Lipase (PagL) that catalyzes 3-O-deacylation of lipid A precursors in membranes of Salmonella typhimurium. J. Biol. Chem. 2769083-9092. [DOI] [PubMed] [Google Scholar]
- 36.Trent, M. S., C. M. Stead, A. X. Tran, and J. V. Hankins. 2006. Diversity of endotoxin and its impact on pathogenesis. J. Endotoxin Res. 12205-223. [DOI] [PubMed] [Google Scholar]
- 37.Vorachek-Warren, M. K., S. M. Carty, S. Lin, R. J. Cotter, and C. R. Raetz. 2002. An Escherichia coli mutant lacking the cold shock-induced palmitoleoyltransferase of lipid A biosynthesis: absence of unsaturated acyl chains and antibiotic hypersensitivity at 12 degrees C. J. Biol. Chem. 27714186-14193. [DOI] [PubMed] [Google Scholar]
- 38.Vorachek-Warren, M. K., S. Ramirez, R. J. Cotter, and C. R. Raetz. 2002. A triple mutant of Escherichia coli lacking secondary acyl chains on lipid A. J. Biol. Chem. 27714194-14205. [DOI] [PubMed] [Google Scholar]
- 39.Zhang, Y. M., and C. O. Rock. 27 March 2008. Acyltransferases in bacterial glycerophospholipid synthesis. J. Lipid Res. doi. 10.1194/jlr.R800005-JLR200. [DOI] [PMC free article] [PubMed]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.