Abstract
Kingella kingae is a gram-negative bacterium that colonizes the respiratory tract and is a common cause of septic arthritis and osteomyelitis. Despite the increasing frequency of K. kingae disease, little is known about the mechanism by which this organism adheres to respiratory epithelium and seeds joints and bones. Previous work showed that K. kingae expresses long surface fibers that vary in surface density. In the current study, we found that these fibers are type IV pili and are necessary for efficient adherence to respiratory epithelial and synovial cells and that the number of pili expressed by the bacterium correlates with the level of adherence to synovial cells but not with the level of adherence to respiratory cells. In addition, we established that the major pilin subunit is encoded by a pilA homolog in a conserved region of the chromosome that also contains a second pilin gene and a type IV pilus accessory gene, both of which are dispensable for pilus assembly and pilus-mediated adherence. Upon examination of the K. kingae genome, we identified two genes in physically separate locations on the chromosome that encode homologs of the Neisseria PilC proteins and that have only a low level homology to each other. Examination of mutant strains revealed that both of the K. kingae PilC homologs are essential for a wild-type level of adherence to both respiratory epithelial and synovial cells. Taken together, these results demonstrate that type IV pili and the two PilC homologs play important roles in mediating K. kingae adherence.
Kingella kingae is a gram-negative bacterium and is an emerging pathogen that has been recognized increasingly in recent years as a cause of a variety of pediatric illnesses (6, 15, 24, 29, 32). Previously, K. kingae was dismissed in terms of its pathogenic potential because of its infrequent occurrence as a cause of disease. However, recent advances in culture techniques and molecular diagnostics have resulted in high rates of recovery of K. kingae in cases of septic arthritis and osteomyelitis (4, 6, 15, 24, 30). For example, in one recent study, K. kingae was found to account for nearly one-half of all osteoarticular infections in patients under 18 years of age and for a sizable majority of bone and joint infections in children less than 36 months of age (4).
The pathogenesis of invasive K. kingae disease is presumed to begin with colonization of the posterior pharynx. This presumption is supported by recent work by Yagupsky and colleagues demonstrating that the same strain can be isolated from both the respiratory tract and the blood of patients with K. kingae invasive disease (33). Additional work has shown that approximately 70% of young children are colonized by K. kingae at least once per year and that colonization by the same strain of K. kingae can persist for more than 2 months (22, 29, 31, 32). Following colonization, K. kingae must breach the epithelial barrier, a process potentially mediated by a canonical RTX toxin (12), allowing dissemination via the bloodstream to distal sites, such as bones and joints.
An essential step in both colonization of the respiratory tract and seeding of distal sites is adherence to tissues, including the respiratory epithelium and the synovium. To gain insight into the determinants of K. kingae adherence, we examined the role of the long fibers that have been observed on the surface K. kingae (5, 9). We found that these fibers are type IV pili and are essential for K. kingae adherence. Both high-density piliation and low-density piliation were associated with efficient adherence to respiratory epithelial cells, while high-density piliation was required for maximal adherence to synovial cells. The major pilin subunit was found to be encoded by a region that is conserved across several species and contains two pilinlike genes and a type IV pilus accessory gene. Additional analysis identified two predicted proteins with homology to PilC1 and PilC2 in the pathogenic Neisseria species. These two proteins have a low level of homology to each other and are both essential for wild-type levels of adherence to respiratory epithelial and synovial cell lines.
MATERIALS AND METHODS
Bacterial strains.
The strains used in this study are listed in Table 1. Escherichia coli strains were stored at −80°C in Luria-Bertani (LB) broth with 15% glycerol, and K. kingae strains were stored at −80°C in brain heart infusion broth with 30% glycerol. E. coli was routinely grown at 37°C on LB agar or in LB broth supplemented with 100 μg/ml ampicillin, 50 μg/ml kanamycin, or 20 μg/ml tetracycline, as appropriate. K. kingae was grown at 37°C with 5% CO2 on chocolate agar plates supplemented with 50 μg/ml kanamycin or 2 μg/ml tetracycline, as appropriate.
TABLE 1.
Strains used in this study
| Strain | Description | Reference |
|---|---|---|
| 269-492 | K. kingae clinical isolate from St. Louis Children's Hospital | 12 |
| KK01 | Naturally occurring nonspreading/noncorroding derivative of 269-492 | 12 |
| KK03 | Naturally occurring spreading/corroding derivative of 269-492 | 12 |
| 269-492 pilA1::aphA3 | 269-492 with aphA3 insertion in pilA1 | This study |
| 269-492 pilF:aphA3 | 269-492 with aphA3 insertion in pilF | This study |
| 269-492 recJ::aphA3 | 269-492 with aphA3 insertion in recJ | This study |
| 269-492 pilA2 | 269-492 recJ::aphA3 with Y57stop and Y58stop mutations in pilA2 | This study |
| 269-492 pilA1 | 269-492 recJ::aphA3 with Y57stop and Y58stop mutations in pilA1 | This study |
| 269-492 pilA1/pilA2 | 269-492 recJ::aphA3 containing Y57stop and Y58stop mutations in pilA1 and Y57stop and Y58stop mutations in pilA2 | This study |
| 269-492 fimB::tetM | 269-492 recJ::aphA3 containing a tetM insertion in fimB | This study |
| 269-492 pilC1 | 269-492 with tetM insertion in pilC1 | This study |
| 269-492 pilC2 | 269-492 with aphA3 insertion in pilC2 | This study |
| 269-492 pilC1/pilC2 | 269-492 with tetM insertion in pilC1 and aphA3 insertion in pilC2 | This study |
| DH5α | E. coli F− φ80dlacZΔM15 Δ(lacZYA-argF)U169 deoR recA1 endA1 hsdR17(rk− mk+) phoA supE44 thi-1 gyrA96 relA1 | 20 |
Cell lines.
Cell lines were obtained from either the American Tissue Culture Collection or the Duke Comprehensive Cancer Center. Chang cells (human conjunctiva; ATCC CCL-20.2), A549 cells (human type II pneumocytes; ATCC CCL-185), HEp2 cells (human larynx; ATCC CCL-23), and Hig-82 cells (rabbit synovium; ATCC CRL-1832) were cultivated in media at 37°C with 5% CO2 as previously described (12). SW982 cells (human synovium; ATCC HTB-93) were maintained in Leibovitz's L-15 medium supplemented with 10% fetal calf serum at 37°C without CO2.
Plasmid and strain construction.
To create K. kingae gene disruptions, the relevant gene was first cloned into pUC19 and then interrupted with an antibiotic cassette. The resulting plasmid was introduced into K. kingae by natural transformation as described previously (12), and transformants were recovered by selection with the appropriate antibiotic. Correct localization of K. kingae gene disruptions was confirmed by either Southern blotting or PCR. The primers used to generate constructs and to confirm gene disruptions are listed in Table 2. To disrupt pilA1, a DNA fragment containing the gene and flanking sequence was amplified by PCR from K. kingae strain 269-492 with primers pilA1F and pilA1R. This fragment was then ligated into BamHI/EcoRI-digested pUC19, creating pUC19/pilA1. The aphA3 kanamycin resistance cassette was released from pFalcon2 (8) by MluI digestion and ligated into an MluI site within the pilA1 gene to create pUC19/pilA1::aphA3. To disrupt pilF, fragments corresponding to the 5′ and 3′ regions of the gene were individually amplified by PCR using primers pilF5′F and pilF5′R and primers pilF3′F and pilF3′R, respectively. These fragments were ligated into BamHI/EcoRI-digested pUC19 (introducing an MluI site within pilF), creating pUC19/pilF::MluI. The aphA3 cassette was then ligated into the MluI site of pUC19/pilF::MluI, generating pUC19/pilF::aphA3. To disrupt pilC1, fragments corresponding to the 5′ and 3′ regions of the gene were individually amplified by PCR using primers pilC1Δ5F and pilC1Δ5R and primers pilC1Δ3F and pilC1Δ3R, respectively. These fragments were ligated into BamHI/EcoRI-digested pUC19, generating pUC19/pilC1::ClaI (containing an internal deletion in pilC1 and a ClaI site in place of the deleted region). The tetM cassette from pHSX-Tet4 was obtained from H. Seifert (Northwestern University) and was amplified using primers pHSXtet4 5′ ClaI and pHSXtet4 3′ ClaI and then ligated into pUC19/pilC1::ClaI, creating pUC19/pilC1::tetM. The K. kingae pilC2::aphA3 mutant was isolated in a screen for nonadherent mutants (T. Kehl-Fie and J. W. St. Geme III, unpublished data).
TABLE 2.
Primers used in this study
| Primer | Sequence |
|---|---|
| pilA1F | GATCGAATTCGTTCTATTGCCGTTGCCAAGCGTTGTGC |
| pilA1R | GATCGGATCCCATTTCCTATTTCTATTTTTCTAATTATGTATGTATTAAAACGCATAAAGC |
| pilF5′F | GATCGAATTCCACCAGAAGACTTGGCAGAGCTGTGTGC |
| pilF5′R | ACGTACGCGTCCAGCACCATGCCGTATGGACGATAAATC |
| pilF3′F | GATCGGATCCCTTAAATTACGACCTTCAAACTGGTAGCGATTGCC |
| pilF3′R | ACGTACGCGTTAACAGGGCCTACGGGCTCGGGTAAAAC |
| pilC1Δ5F | ACGTGAATTCCGCCTGTTTCTTCGTTGTCGGG |
| pilC1Δ5R | ACGTATCGATCGCAAAGCTGGACAAGGCAAGTGATACC |
| pilC1Δ3F | ACGTATCGATGGGGAAGCAAGAACCAATGGTGAAGATGG |
| pilC1Δ3R | ACGTGGATCCGGCTCTGCGATAGAAACTCCATACGG |
| pHSXtet4 5′ ClaI | ACGTATCGATAAGATTGTAAAATAACAAATATTGGTACATGATTACAG |
| pHSXtet4 3′ ClaI | ACGTATCGATCCACTTGTAGTTTATAATAACTATCTCCTCCTTTACAC |
| Pilin Region Fwd-2 | ACGTGAATTCGCAACTCGTCTTCGCATCGCC |
| Pilin Region Rev-2 | ACGTGTCGACCCAGCAACACCGTCCAATCCAG |
| RecJ+BamHIFwd | CCGCGTTGATTCGGGATCCTTCACAATAATGCAATCG |
| RecJ+BamHIRev | CGATTGCATTATTGTGAAGGATCCCGAATCAACGCGG |
| pfaclconw/BamHIfwd | GCATGGATCCCATCTAAATCTAGGTACTAAAACAATTCATCCAG |
| pfalconw/BamHIrev | GCATGGATCCGTTTGACAGCTTATCATCGATAAACCCAG |
| PilA1K/oFwd-2 | GTGCTTTGACTGAGTAATAAGCTTCTAACAATGC |
| PilA1K/oRev-2 | GCATTGTTAGAAGCTTATTACTCAGTCAAAGCAC |
| PilA2K/oMutFwd | TGCAAAAACATCGGTGGCTGAATAATAAGCTTCGCATAACCAATTTCC |
| PilA2K/oMutRev | GGAAATTGGTTATGCGAAGCTTATTATTCAGCCACCGATGTT TTTGCA |
| PilA1 RT-left | CAAACGTTCACGCGTATCTG |
| PilA1 RT-right | GGCCATGCATTGTTAGAAGC |
| PilA2 RT left#2 | TAATTACGGCATTCCGATGG |
| PilA2 RT-right#2 | GCCTAGTGTGGCGAAAGAAG |
| fimB RT-left | GGCATAGATTTGTCGCTGGT |
| fimB RT-right | ATCAAACGCCACAAACACAG |
| ftsZ RT-left | CCAGAGCGAACCAAAGTCTC |
| ftsZ RT-right | AAGCTATACTCGCCCTGCTG |
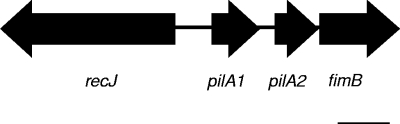
In-frame deletions in pilA1 and pilA2 and a disruption in fimB were produced by first amplifying a fragment containing recJ, pilA1, pilA2, and fimB (Fig. 1) with PCR primers Pilin Region Fwd-2 and Pilin Region Rev-2. This fragment was then ligated into SalI/EcoRI-digested pUC19, creating plasmid pPR. Subsequently, a BamHI site was introduced into recJ in pPR using primers RecJ+BamHIFwd and RecJ+BamHIRev and a QuikChange II XL kit (Stratagene La Jolla, CA), creating pPR::BamHI. The aphA3 cassette was then amplified from pFalcon2 (8) with primers pfalconw/BamHIfwd and pfalconw/BamHIrev and ligated into pPBR::BamHI. Restriction digestion identified a recombinant plasmid with the aphA3 cassette divergently transcribed from the pilin genes, and this plasmid was designated pPRBK. Premature stop codons were introduced at codons encoding Tyr57 and Tyr58 in both pilA1 and pilA2 in pPRBK individually and in combination using primers PilA1K/OFwd-2 and PilA1K/ORev-2 and primers PilA2K/OMutFwd and PilA2K/OMutRev, producing truncation constructs designated pPRBK::pilA1 (pilA1 truncation), pPRBK::pilA2 (pilA2 truncation), and pPRBK::pilA1/2 (pilA1 and pilA2 truncations). Following each step, constructs were sequenced to confirm the presence of the intended mutation and the absence of unintended PCR related mutations. After transformation into K. kingae, integration of the pilA1 and pilA2 truncation mutations into the K. kingae chromosome was confirmed by DNA sequencing. To disrupt fimB, plasmid pPRBK was digested with NruI and ligated to the tetM cassette from pHSX-tet4, creating PRBK::fimBtetM.
FIG. 1.
Diagram of the pilin-encoding region of K. kingae strain 269-492. Scale bar = 500 bp.
Adherence assays.
Bacteria were incubated for 17 to 18 h overnight on chocolate agar or LB agar as appropriate and then resuspended in brain heart infusion broth to an optical density at 600 nm of 0.8. For both qualitative and quantitative assays, approximately 6.5 × 106 CFU was inoculated onto fixed confluent cell monolayers in 24-well plates. Monolayers were fixed by removing the growth medium and then adding 2% glutaraldehyde in 0.1 M sodium phosphate buffer (pH 7.4) and incubating the preparations at 4°C for 2 h with gentle rocking. The monolayers were washed three times with 1× Tris-buffered saline, and then 300 μl of fresh tissue culture medium was added to each well. Following inoculation of bacteria onto monolayers, the 24-well plates were centrifuged for 5 min at 1,000 rpm and then incubated for 25 min at 37°C. Subsequently, monolayers were washed four times with phosphate-buffered saline (PBS) to remove nonadherent organisms. For qualitative assays, monolayers were fixed and stained with Giemsa stain and then examined by light microscopy. For quantitative assays, trypsin-EDTA was added to monolayers, and then the monolayers were incubated for 20 min at 37°C to release adherent bacteria. Appropriate dilutions were prepared and spread on agar plates, and the percent adherence was determined by dividing the number of adherent CFU by the number of CFU in the inoculum. For quantitative assays, each sample was assayed in triplicate.
Transmission electron microscopy.
Bacterial strains were incubated for approximately 17 to 18 h on chocolate agar and then resuspended in PBS. Subsequently, they were pelleted and resuspended in 0.2 M ammonium acetate (pH 7.4). Negative-staining transmission electron microscopy was performed as described previously (12), except that a Philips CM-12 electron microscope (FEI, Hillsboro, OR) was used and the bacteria were not washed after they were added to the grids. Levels of piliation were assessed by examining a minimum of 20 bacteria by electron microscopy and were assigned semiquantitative designations ranging from − to +++. Nonpiliated strains (−) had no visible fibers; KK01-like strains (+) had an average of 2 or fewer visible fibers per bacterium; 269-492-like strains (++) had an average of 3 to 10 visible fibers per bacterium; and KK03-like strains (+++) had an average of more than 10 or more visible fibers per bacterium.
Quantitative real-time PCR.
Bacterial strains were incubated for 17 to 18 h on chocolate agar and then washed once with PBS. To lyse the bacteria, bacterial pellets were resuspended in 1 ml of Tri Reagent (Sigma, St. Louis, MO) prewarmed to 65°C and were then incubated briefly at room temperature. RNA was isolated from the lysed bacteria using an RNeasy Mini kit and the lipid-rich tissue protocol (Qiagen, Valencia CA). To remove residual DNA, the RNA samples were treated with RQ1 DNase (Fisher Scientific, Pittsburgh, PA). The DNase was removed using the Tri Reagent RNA isolation protocol (Sigma, St. Louis, MO). To create cDNA, approximately 2 μg of RNA, random hexamers, and SuperScript II were used according to the manufacturer's instructions (Invitrogen, Carlsbad, CA). Real-time PCR was performed using SYBR green and the following primer sets: pilA1 RT-left and pilA1 RT-right, pilA2 RT left#2 and pilA2 RT-right#2, fimB RT-left and fimB RT-right, and ftsZ RT-left and ftsZ RT-right.
Nucleotide sequence accession numbers.
K. kingae DNA sequences have been deposited in the GenBank database under the following accession numbers: pilin region, EU828772; pilC1, EU828769; pilC2, EU828770; pilFGD region, EU828771; pilTU region, EU828768; and pilMNOPQ region, EU828773.
RESULTS
K. kingae adheres to respiratory epithelial and synovial cells.
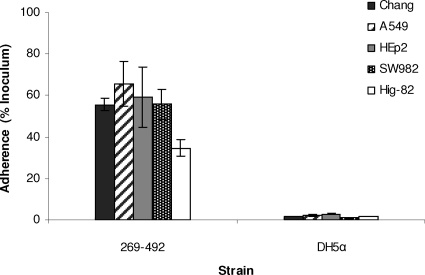
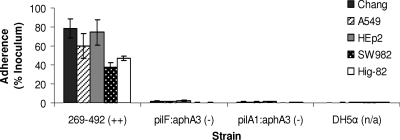
To begin to understand the mechanism of K. kingae colonization of the respiratory tract, we set out to identify cell types to which K. kingae adheres. A clinical isolate from St. Louis Children's Hospital designated 269-492 was examined to determine its adherence to a panel of cultured respiratory cell lines, including Chang cells (human conjunctiva), HEp-2 cells (human larynx), and A549 cells (type II pneumocytes). When it was examined by light microscopy after staining with Giemsa stain, strain 269-492 was found to adhere specifically to Chang, HEp-2, and A549 cells (data not shown). As shown in Fig. 2, quantitative assays confirmed that strain 269-492 adhered at high levels to all of these cell types. Given the role of K. kingae in causing septic arthritis, we also examined strain 269-492 adherence to synovial cell lines, including Hig-82 cells (rabbit synovium) and SW982 cells (human synovium). As shown in Fig. 2, strain 269-492 was capable of efficient adherence to these cells as well.
FIG. 2.
Adherence of K. kingae strain 269-492 to respiratory epithelial cells (Chang, A549, and HEp2 cells) and synovial cells (SW982 and Hig-82 cells). E. coli DH5α was used as a negative control.
Fibers expressed by K. kingae are type IV pili and are required for adherence.
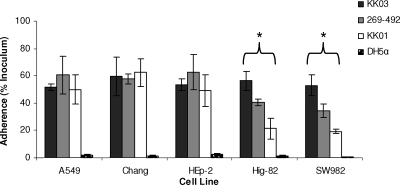
Previous work demonstrated that K. kingae has two colony types, one designated spreading/corroding and correlating with abundant surface fibers and the other designated nonspreading/noncorroding and correlating with sparse surface fibers (5, 9). To investigate the role of these fibers in adherence, colony variants of strain 269-492 designated KK03 (spreading/corroding, expressing abundant surface fibers) and KK01 (nonspreading/noncorroding, expressing sparse fibers) (12) were examined to determine their adherence to respiratory epithelial and synovial cells. As shown in Fig. 3, KK03, KK01, and 269-492 were capable of similar levels of adherence to the Chang, A549, and HEp-2 cell lines. In contrast, the level of adherence to Hig-82 and SW982 cells correlated with the density of surface fibers and was highest for KK03 (∼55% of the inoculum), next highest for 269-492 (∼40% of the inoculum), and lowest for KK01 (∼25% of the inoculum). These observations suggest that the fibers have a role in K. kingae adherence, although it is important to recognize that KK01 and KK03 are colony variants and may have additional phenotypic differences compared with strain 269-492.
FIG. 3.
Adherence of K. kingae wild-type strain 269-492 and spreading/corroding (KK03) and nonspreading/noncorroding (KK01) variants to respiratory epithelial cells (Chang, A549, and HEp2 cells) and synovial cells (SW982 and Hig-82 cells). E. coli DH5α was used as a negative control. *, P ≤ 0.05, as determined by an unpaired t test for KK03 compared to KK01.
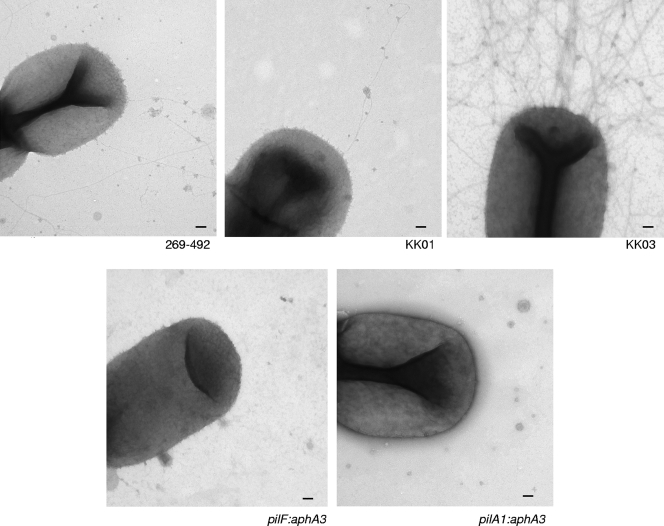
Previous work using antibodies raised against Kingella denitrificans pili suggested that the fibers expressed by K. kingae might be type IV pili (28). Examination of the genome of K. kingae strain 269-492 (Kehl-Fie et al., unpublished data) revealed the presence of several regions encoding proteins with homology to type IV pilus biogenesis components, including regions containing pilFGD-like genes, pilTU-like genes, and pilMNOPQ-like genes. In addition, we identified a region containing two major pilin-like genes that we designated pilA1 and pilA2 and a fimB-like gene (Fig. 1). Overall, the K. kingae PilA1 and PilA2 predicted proteins share approximately 52% identity and 64% similarity. The N-terminal 60 residues are 73% identical and 87% similar, while the C-terminal portions share only 35% identity and 45% similarity. To determine whether the fibers on the surface of K. kingae are type IV pili, we generated disruptions in pilA1 and in pilF (an essential factor in type IV pilus assembly in other organisms [2]) in strain 269-492. As shown in Fig. 4, examination by transmission electron microscopy revealed that both the pilA1 and pilF disruption mutant derivatives of strain 269-492 lacked surface fibers. Further analysis established that the pilA1 and pilF disruption mutants were nonadherent (Fig. 5). Similarly, pilA1 and pilF disruption mutants of the KK01 and KK03 variants of strain 269-492 had undetectable levels of pili and were nonadherent (data not shown). These results demonstrate that the fibers expressed by K. kingae are type IV pili and are necessary for in vitro adherence to respiratory epithelial cells and synovial cells.
FIG. 4.
Transmission electron micrographs of K. kingae strains 269-492, KK01, KK03, 269-492 pilF::aphA3 (pilF:aphA3) and 269-492 pilA1::aphA3 (pilA1:aphA3). Scale bars = 100 nm.
FIG. 5.
Adherence of K. kingae type IV pilus mutants. K. kingae strains 269-492, 269-492 pilF::aphA3 (pilF:aphA3), and 269-492 pilA1::aphA3 (pilA1:aphA3) were assayed to determine their adherence to respiratory epithelial and synovial cell lines. E. coli DH5α was used as a negative control. The level of surface pili on each strain was determined by negative-staining transmission electron microscopy and was scored using a scale ranging from − to +++.
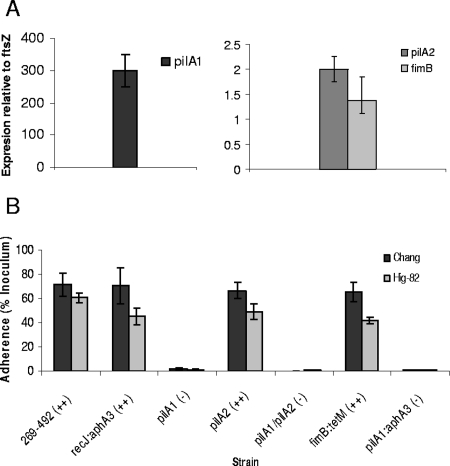
Given the genetic structure of the K. kingae pilA1-pilA2-fimB gene cluster, we examined the levels of transcription of these genes and found that all three were transcribed (Fig. 6). It is noteworthy pilA1 was transcribed at a level that was more than 100-fold greater than the level of transcription of pilA2 and fimB. Given that pilA2 and fimB are transcribed in wild-type organisms, it is possible that the loss of pili observed in the pilA1 disruption mutant was due to a polar effect on pilA2 or fimB rather than to a direct effect on pilA1. To more carefully assess the role of pilA1, pilA2, and fimB in K. kingae type IV pilus expression and adherence, we created in-frame truncations in pilA1 and pilA2 and a disruption of fimB in strain 269-492. Analysis of the pilA1 truncation by real-time PCR revealed no decrease in pilA2 or fimB transcription, and analysis of the pilA2 truncation and the pilA1/pilA2 double truncation revealed no decrease in the level of fimB transcription (data not shown). When they were examined by negative-staining transmission electron microscopy, the pilA2 truncation mutant and the fimB disruption mutant had wild-type levels of surface pili, while the pilA1 truncation mutant and the pilA1/pilA2 double truncation mutant were nonpiliated (data not shown). While the pilA2 truncation mutation and the fimB disruption had no clear effect on pilus expression or colony morphology, we wondered about a potential effect on pilus-mediated adherence. As shown in Fig. 6, the pilA2 truncation mutant and the fimB disruption mutant were capable of wild-type levels of adherence to Chang and Hig-82 cells. Together, these data suggest that PilA1 is the major pilin subunit in K. kingae type IV pili.
FIG. 6.
Analysis of the pilin region. (A) Transcription of pilA1, pilA2, and fimB in K. kingae strain 269-492. The transcription of each gene was measured by quantitative real-time PCR and normalized to the level of the ftsZ transcript. (B) Adherence of K. kingae strains 269-492, 269-492 recJ::aphA3 (recJ:aphA3), 269-492 pilA1 (pilA1), 269-492 pilA2 (pilA2), 269-492 pilA1/pilA2 (pilA1/pilA2), 269-492 fimB::tetM (fimB:tetM), and 269-492 pilA1::aphA3 (pilA1:aphA3) to Chang respiratory epithelial and Hig-82 synovial cells. The level of expressed pili on each strain was determined by negative-staining transmission electron microscopy and was scored using a scale ranging from − to +++.
Both PilC1 and PilC2 contribute to K. kingae adherence.
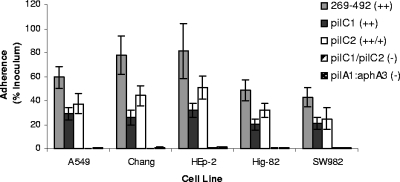
In addition to containing two pilin subunit genes, the genome of K. kingae strain 269-492 contains two genes at separate locations on the chromosome that encode predicted proteins with homology to the Neisseria PilC1 and PilC2 proteins. It is noteworthy that in Neisseria gonorrhoeae and Neisseria meningitidis, the PilC proteins have been shown to contribute to adherence (16, 18, 19). Interestingly, while K. kingae is a member of the Neisseriaceae, K. kingae PilC1 and PilC2 share more similarity with PilY1 from Pseudomonas aeruginosa than with PilC1 and PilC2 from either N. meningitidis or N. gonorrhoeae (Fig. 7). K. kingae PilC1 and PilC2 exhibit a very low level of homology with each other (only 7% identity and 16% similarity overall). To investigate whether K. kingae PilC1 and PilC2 contribute to piliation and adherence, we created disruptions in the pilC1 and pilC2 genes individually and in combination in strain 269-492 and then examined the resulting mutants by using electron microscopy and adherence assays with respiratory epithelial and synovial cells. Both the pilC1 and pilC2 single mutants had visible pili that appeared to have a wild-type structure, while the pilC1/pilC2 double mutant had no detectable pili (data not shown). As shown in Fig. 8, both the pilC1 and pilC2 single mutants were capable of reduced but appreciable adherence to the Chang, A549, HEp-2, Hig-82, and SW982 cell lines, and the level of adherence by the pilC1 mutant to respiratory epithelial cells was slightly lower than that of the pilC2 mutant. In contrast, the pilC1/pilC2 double mutant exhibited negligible adherence comparable to the background adherence. When the pilC1 and pilC2 single mutations were introduced into the KK03 background, the resulting strains had wild-type levels of pili and were capable of reduced but appreciable adherence, mirroring the findings for strain 269-492. Similar to 269-492 pilC1/pilC2, KK03 pilC1/pilC2 had undetectable levels of pili and was nonadherent (data not shown). These results indicate that both PilC1 and PilC2, independent of effects on pilus stability, are necessary for maximal adherence to both respiratory epithelial and synovial cells.
FIG. 7.
Dendrogram showing the relationships between K. kingae 269-492 (KK) PilC1 and PilC2, P. aeruginosa PAO1 (PA) PilY1 (accession number AAG07942.1), N. meningitidis (NM) PilC1 (accession number CAA73469.1) and PilC2 (accession number CAA73470.1), and N. gonorrhoeae (NG) PilC1 (accession number CAA73471.1) and PilC2 (accession number CAA88930.1).
FIG. 8.
Adherence of K. kingae PilC1 and PilC2 mutants. The bars indicate the adherence of K. kingae strains 269-492, 269-492 pilC1 (pilC1), 269-492 pilC2 (pilC2), 269-492 pilC1/pilC2 (pilC1/pilC2), and 269-492 pilA1::aphA3 (pilA1:aphA3) to respiratory epithelial cell lines. The pilC1 mutant and the pilC2 mutant were less adherent than strain 269-492 to all cell lines (P ≤ 0.05, as determined using the unpaired t test). The level of surface-expressed pili on each strain was determined by negative-staining transmission electron microscopy and was scored using a scale ranging from − to +++.
DISCUSSION
K. kingae is a bacterium that is being recognized increasingly as a common cause of septic arthritis and osteomyelitis in young children. However, little is known about the bacterial factors that facilitate colonization of the respiratory tract and promote seeding of deeper tissues. To gain insight into these fundamental processes, we examined the ability of K. kingae to adhere to clinically relevant cell types, including respiratory epithelial cells and synovial cells. We observed efficient adherence that was dependent on the presence of type IV pili, which contain a major pilin subunit encoded by pilA1. Interestingly, the level of adherence to synovial cells correlated with the density of pili, while the levels of adherence to respiratory epithelial cell lines were similar for high-pilus-density and low-pilus-density K. kingae variants. Examination of pilC1 and pilC2 mutants established that both PilC1 and PilC2 are required for wild-type levels of adherence to both respiratory epithelial cells and synovial cells.
Analysis of the K. kingae strain 269-492 genome revealed the presence of a gene cluster containing two major pilin structural genes that we designated pilA1 and pilA2. Truncation mutations in these genes demonstrated that pilA1 encodes the major pilin subunit. The K. kingae pilin gene cluster has several similarities with the pilin gene clusters in Eikenella corrodens (a colonizer of the oral cavity and an important cause of endocarditis) (10, 25, 26), Dichelobacter nodosus (a cause of foot rot in sheep) (13), and K. denitrificans (a less pathogenic relative of K. kingae) (27). Like K. kingae, E. corrodens contains two pilin genes (pilA1 and pilA2) and a pilus accessory factor gene (pilB), which are arranged like pilA1, pilA2, and fimB in K. kingae. In E. corrodens, pilA1 is transcribed at high levels and pilA2 is transcribed at low levels (25). The E. corrodens PilA2 protein is dispensable for pilus formation and function. Interestingly, the E. corrodens PilA2 protein can be the major pilin subunit when the pilA2 gene is moved into the pilA1 position (26). D. nodosus contains a single pilin gene (fimA) followed by a downstream pilus accessory factor gene (fimB), which are transcribed in the same direction (25, 26). K. denitrificans contains pilin genes designated kpdB and kpdD arranged like pilA1 and pilA2 in K. kingae (27). It is not known if the two K. denitrificans pilin genes are followed by a pilus accessory factor gene. While loss of the downstream pilus accessory gene in K. kingae and D. nodosus does not result in any observed phenotypic changes (13), loss of the E. corrodens downstream pilus accessory gene is associated with a loss of twitching motility (26).
While the roles of PilA2 and FimB in K. kingae remain unknown, the similarities in the pilin regions of K. kingae, E. corrodens, D. nodosus, and K. denitrificans suggest that these proteins have a role in pilus function and possibly pathogenesis. Our results indicate that K. kingae PilA2 and FimB are dispensable for adherence and expression of pili. Given the significant differences in amino acid sequence between PilA1 and PilA2, one possible role for PilA2 is to serve as a source of antigenic diversity, replacing PilA1 as the major pilus subunit and possibly facilitating prolonged colonization of the respiratory tract by K. kingae (22, 31). However, this idea does not account for the observed transcription of pilA2, a finding that suggests that PilA2 has a constitutive function. Given the pilinlike N terminus of PilA2, another possibility is that the protein may be incorporated into the pilus as a minor subunit, perhaps providing a scaffold for covalent modification (3, 7, 23) and altering the antigenicity or function of the pilus. Similar to the findings for pilA2, the observed transcription of fimB suggests a role for FimB. Based on results obtained with E. corrodens, it seems reasonable to speculate that FimB may have a role in K. kingae pilus-mediated phenotypes such as twitching motility (26).
In addition to identifying the two pilin genes, analysis of the K. kingae genome revealed the presence of the pilC1 and pilC2 genes, which encode proteins with homology to PilC proteins in other organisms. Unlike their Neisseria counterparts, K. kingae PilC1 and PilC2 have very low levels of homology to each other (1, 18). Loss of either PilC1 or PilC2 from K. kingae resulted in a decrease in adherence, a result that is consistent with findings for pathogenic Neisseria strains, suggesting that the PilC proteins function as adhesins (2, 14, 17, 21). Interestingly, loss of K. kingae PilC1 or PilC2 resulted in only a partial reduction in adherence, in contrast to the phenotype observed for N. meningitidis or N. gonorrhoeae PilC mutants (16, 18, 19). As one might predict based on the work with the Neisseria PilC molecules (11, 16, 19), loss of both K. kingae PilC1 and PilC2 resulted in nonadherent, nonpiliated organisms. Interestingly, the K. kingae PilC1 and PilC2 proteins are more closely related to PilY1 from P. aeruginosa than to the Neisseria PilC molecules, raising the possibility that PilY1 may contribute to adherence, a finding that has not been reported to date. If the K. kingae PilC proteins are adhesins, the differences in adherence observed for the K. kingae PilC1 and PilC2 mutants raise several interesting questions, including what are the host receptors for PilC1 and PilC2 and why do PilC1 and PilC2 mediate different levels of adherence.
To summarize, in this work we report that K. kingae expresses type IV pili that may be involved in multiple steps in the pathogenesis of K. kingae disease, including colonization of the respiratory tract and seeding of joints. Further study of K. kingae type IV pili and the associated functions may provide key insights into the role of type IV pili in K. kingae adherence and colonization, as well as other aspects of K. kingae pathogenicity.
Acknowledgments
We thank Hank Seifert for providing pHSX-Tet4 and for offering insights into the Neisseriaceae and Pablo Yagupsky for contributing general advice about handling K. kingae.
This work was supported by NIH training grant T32-GM07067 to T.K.-F.
Footnotes
Published ahead of print on 29 August 2008.
REFERENCES
- 1.Backman, M., H. Kallstrom, and A. B. Jonsson. 1998. The phase-variable pilus-associated protein PilC is commonly expressed in clinical isolates of Neisseria gonorrhoeae, and shows sequence variability among strains. Microbiology 144149-156. [DOI] [PubMed] [Google Scholar]
- 2.Burrows, L. L. 2005. Weapons of mass retraction. Mol. Microbiol. 57878-888. [DOI] [PubMed] [Google Scholar]
- 3.Castric, P., F. J. Cassels, and R. W. Carlson. 2001. Structural characterization of the Pseudomonas aeruginosa 1244 pilin glycan. J. Biol. Chem. 27626479-26485. [DOI] [PubMed] [Google Scholar]
- 4.Chometon, S., Y. Benito, M. Chaker, S. Boisset, C. Ploton, J. Berard, F. Vandenesch, and A. M. Freydiere. 2007. Specific real-time polymerase chain reaction places Kingella kingae as the most common cause of osteoarticular infections in young children. Pediatr. Infect. Dis. J. 26377-381. [DOI] [PubMed] [Google Scholar]
- 5.Froholm, L. O., and K. Bovre. 1972. Fimbriation associated with the spreading-corroding colony type in Moraxella kingii. Acta Pathol. Microbiol. Scand. Sect. B 80641-648. [PubMed] [Google Scholar]
- 6.Gene, A., J. J. Garcia-Garcia, P. Sala, M. Sierra, and R. Huguet. 2004. Enhanced culture detection of Kingella kingae, a pathogen of increasing clinical importance in pediatrics. Pediatr. Infect. Dis. J. 23886-888. [DOI] [PubMed] [Google Scholar]
- 7.Hegge, F. T., P. G. Hitchen, F. E. Aas, H. Kristiansen, C. Lovold, W. Egge-Jacobsen, M. Panico, W. Y. Leong, V. Bull, M. Virji, H. R. Morris, A. Dell, and M. Koomey. 2004. Unique modifications with phosphocholine and phosphoethanolamine define alternate antigenic forms of Neisseria gonorrhoeae type IV pili. Proc. Natl. Acad. Sci. USA 10110798-10803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hendrixson, D. R., B. J. Akerley, and V. J. DiRita. 2001. Transposon mutagenesis of Campylobacter jejuni identifies a bipartite energy taxis system required for motility. Mol. Microbiol. 40214-224. [DOI] [PubMed] [Google Scholar]
- 9.Henriksen, S. D. 1969. Corroding bacteria from the respiratory tract. 1. Moraxella kingii. Acta Pathol. Microbiol. Scand. 7585-90. [PubMed] [Google Scholar]
- 10.Hood, B. L., and R. Hirschberg. 1995. Purification and characterization of Eikenella corrodens type IV pilin. Infect. Immun. 633693-3696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Jonsson, A. B., G. Nyberg, and S. Normark. 1991. Phase variation of gonococcal pili by frameshift mutation in pilC, a novel gene for pilus assembly. EMBO J. 10477-488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kehl-Fie, T. E., and J. W. St Geme III. 2007. Identification and characterization of an RTX toxin in the emerging pathogen Kingella kingae. J. Bacteriol. 189430-436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kennan, R. M., O. P. Dhungyel, R. J. Whittington, J. R. Egerton, and J. I. Rood. 2001. The type IV fimbrial subunit gene (fimA) of Dichelobacter nodosus is essential for virulence, protease secretion, and natural competence. J. Bacteriol. 1834451-4458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Merz, A. J., and M. So. 2000. Interactions of pathogenic neisseriae with epithelial cell membranes. Annu. Rev. Cell Dev. Biol. 16423-457. [DOI] [PubMed] [Google Scholar]
- 15.Moumile, K., J. Merckx, C. Glorion, P. Berche, and A. Ferroni. 2003. Osteoarticular infections caused by Kingella kingae in children: contribution of polymerase chain reaction to the microbiologic diagnosis. Pediatr. Infect. Dis. J. 22837-839. [DOI] [PubMed] [Google Scholar]
- 16.Nassif, X., J. L. Beretti, J. Lowy, P. Stenberg, P. O'Gaora, J. Pfeifer, S. Normark, and M. So. 1994. Roles of pilin and PilC in adhesion of Neisseria meningitidis to human epithelial and endothelial cells. Proc. Natl. Acad. Sci. USA 913769-3773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Nassif, X., M. Marceau, C. Pujol, B. Pron, J. L. Beretti, and M. K. Taha. 1997. Type-4 pili and meningococcal adhesiveness. Gene 192149-153. [DOI] [PubMed] [Google Scholar]
- 18.Rahman, M., H. Kallstrom, S. Normark, and A. B. Jonsson. 1997. PilC of pathogenic Neisseria is associated with the bacterial cell surface. Mol. Microbiol. 2511-25. [DOI] [PubMed] [Google Scholar]
- 19.Rudel, T., H. J. Boxberger, and T. F. Meyer. 1995. Pilus biogenesis and epithelial cell adherence of Neisseria gonorrhoeae pilC double knock-out mutants. Mol. Microbiol. 171057-1071. [DOI] [PubMed] [Google Scholar]
- 20.Sambrook, J., E. F. Fritsch, and T. Maniatis. 1989. Molecular cloning: a laboratory manual, 2nd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY.
- 21.Scheuerpflug, I., T. Rudel, R. Ryll, J. Pandit, and T. F. Meyer. 1999. Roles of PilC and PilE proteins in pilus-mediated adherence of Neisseria gonorrhoeae and Neisseria meningitidis to human erythrocytes and endothelial and epithelial cells. Infect. Immun. 67834-843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Slonim, A., E. S. Walker, E. Mishori, N. Porat, R. Dagan, and P. Yagupsky. 1998. Person-to-person transmission of Kingella kingae among day care center attendees. J. Infect. Dis. 1781843-1846. [DOI] [PubMed] [Google Scholar]
- 23.Stimson, E., M. Virji, K. Makepeace, A. Dell, H. R. Morris, G. Payne, J. R. Saunders, M. P. Jennings, S. Barker, M. Panico, et al. 1995. Meningococcal pilin: a glycoprotein substituted with digalactosyl 2,4-diacetamido-2,4,6-trideoxyhexose. Mol. Microbiol. 171201-1214. [DOI] [PubMed] [Google Scholar]
- 24.Verdier, I., A. Gayet-Ageron, C. Ploton, P. Taylor, Y. Benito, A. M. Freydiere, F. Chotel, J. Berard, P. Vanhems, and F. Vandenesch. 2005. Contribution of a broad range polymerase chain reaction to the diagnosis of osteoarticular infections caused by Kingella kingae: description of twenty-four recent pediatric diagnoses. Pediatr. Infect. Dis. J. 24692-696. [DOI] [PubMed] [Google Scholar]
- 25.Villar, M. T., J. T. Helber, B. Hood, M. R. Schaefer, and R. L. Hirschberg. 1999. Eikenella corrodens phase variation involves a posttranslational event in pilus formation. J. Bacteriol. 1814154-4160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Villar, M. T., R. L. Hirschberg, and M. R. Schaefer. 2001. Role of the Eikenella corrodens pilA locus in pilus function and phase variation. J. Bacteriol. 18355-62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Weir, S., L. W. Lee, and C. F. Marrs. 1996. Identification of four complete type 4 pilin genes in a single Kingella denitrificans genome. Infect. Immun. 644993-4999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Weir, S., and C. F. Marrs. 1992. Identification of type 4 pili in Kingella denitrificans. Infect. Immun. 603437-3441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Yagupsky, P. 2004. Kingella kingae: from medical rarity to an emerging paediatric pathogen. Lancet Infect. Dis. 4358-367. [DOI] [PubMed] [Google Scholar]
- 30.Yagupsky, P., R. Dagan, C. W. Howard, M. Einhorn, I. Kassis, and A. Simu. 1992. High prevalence of Kingella kingae in joint fluid from children with septic arthritis revealed by the BACTEC blood culture system. J. Clin. Microbiol. 301278-1281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Yagupsky, P., R. Dagan, F. Prajgrod, and M. Merires. 1995. Respiratory carriage of Kingella kingae among healthy children. Pediatr. Infect. Dis. J. 14673-678. [DOI] [PubMed] [Google Scholar]
- 32.Yagupsky, P., N. Peled, and O. Katz. 2002. Epidemiological features of invasive Kingella kingae infections and respiratory carriage of the organism. J. Clin. Microbiol. 404180-4184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Yagupsky, P., N. Porat, and E. Pinco. Pharyngeal colonization by Kingella kingae in children with invasive disease. Pediatr. Infect. Dis. J., in press. [DOI] [PubMed]