Abstract
The enteroviruses poliovirus (PV), Coxsackie B virus (CVB) and rhinovirus (HRV) are members of Picornaviridae that inhibit host cell translation early in infection. Enterovirus translation soon predominates in infected cells, but eventually also shuts off. This complex pattern of modulation of translation suggests regulation by a multifactorial mechanism. We report here that eIF5B is proteolytically cleaved during PV and CVB infection of cultured cells, beginning at 3 hours post-infection and increasing thereafter. Recombinant PV, CVB and HRV 3Cpro cleaved purified native rabbit eukaryotic initiation factor (eIF) 5B in vitro at a single site (VVEQ↓G, equivalent to VMEQ↓G479in human eIF5B) that is consistent with the cleavage specificity of enterovirus 3C proteases. Cleavage separates the N-terminal domain of eIF5B from its essential conserved central GTPase and C-terminal domains. 3Cpro-mediated cleavage of eIF5B may thus play an accessory role in the shut-off of translation that occurs in enterovirus-infected cells.
Keywords: coxsackievirus, eIF5B, poliovirus, protease, rhinovirus, translation shutoff
Introduction
Infection by enteroviruses such as poliovirus (PV), Coxsackie B virus (CVB) and human rhinovirus (HRV), members of the family Picornaviridae, leads to inhibition of host cell protein synthesis. During PV infection, shutoff begins within 2h, is complete with 4h, and is followed by a phase during which viral translation is also progressively inhibited (Etchison et al., 1982). The biphasic nature of these changes suggests that shutoff occurs by a multifactorial mechanism, so that factors required for translation of cellular mRNAs (predominantly following cap-dependent initiation) are targeted during the first phase of shutoff and factors that are also required for translation of viral mRNAs may be inactivated later. Enterovirus mRNAs all contain an internal ribosomal entry site (IRES) that promotes initiation by cap-independent ribosomal binding to the mRNA (Hellen and Sarnow, 2001), unlike most cellular mRNAs, which are translated following cap-dependent initiation
Whereas the mechanism of initiation on enterovirus IRESs is incompletely characterized, many details of the cap-dependent initiation mechanism have been elucidated (Pestova et al., 2007). First, eIF1, eIF1A and eIF3 promote binding of eIF2•GTP•Met-tRNAMetito a 40S ribosomal subunit to form a 43S preinitiation complex. Its attachment to the capped 5′ end of mRNA is mediated by eIF4F (which consists of the eIF4E cap-binding subunit, the eIF4A RNA helicase and eIF4G), and is enhanced by eIF4B and the poly(A) binding protein (PABP). eIF4G coordinates ribosomal recruitment to mRNAs by binding PABP, mRNA and eIFs 3, 4E and 4A. 43S complexes scan to the initiation codon, forming 48S complexes in which the Met-tRNAMeti anticodon is base-paired to the initiation codon in the ribosomal peptidyl (P) site. In the final subunit-joining stage, eIF5 induces hydrolysis of eIF2-bound GTP, releasing Met-tRNAMeti into the P site, and eIF5B mediates displacement of eIF2•GDP and other factors from the 48S complex and joining of a 60S subunit to yield an 80S ribosome.
The earliest changes in the translation apparatus during enterovirus infection involve cleavage by the virus-encoded 2A protease (2Apro) of eIF4GI (which precedes the shutoff of host protein synthesis), and with slightly slower kinetics, of the less abundant eIF4GII (Etchison et al., 1982; Gradi et al., 1998). eIF4GI/eIF4GII are cleaved at a single site, splitting domains that bind PABP and eIF4E from those that bind eIF3 and eIF4A (Lamphear et al., 1995), thereby abrogating eIF4G's function in bridging factors that bind capped mRNA, that unwind mRNA and that bind the ribosome. PABP is also cleaved in enterovirus-infected cells, predominantly by the 3C protease (3Cpro), which separates PABP into an N-terminal fragment that binds eIF4G and the 3′ poly(A) tail of mRNA, and a C-terminal fragment that binds eIF4B and the termination factor eRF3 (Joachims et al., 1999; Kerekatte et al., 1999; Kuyumcu-Martinez et al., 2002). In addition, the translation regulator 4E-BP1 is dephosphorylated during infection. This event sequesters eIF4E and abrogates eIF4F's cap-binding function, which contributes to the shutoff of host protein synthesis and activation of IRES-mediated translation (Gingras et al., 1996; Svitkin et al., 2005), but cannot fully account for shutoff because it lags behind this process.
To obtain a more complete overview of the shut-off process, the status in PV and CVB type 3 (CVB3)-infected cells of factors involved in subunit joining was analyzed. eIF5B was found to be cleaved at a single site during enterovirus infection. Cleavage in vitro was mediated by 3Cpro alone and was specific, because eIF1, eIF1A, eIF4B, eIF4F and eIF5 were not substrates for 3Cpro. The time course of eIF5B proteolysis suggest that its cleavage could contribute to the shutoff of host and viral translation observed in enterovirus-infected cells.
Results and Discussion
Cleavage of eIF5B in enterovirus-infected cells
To determine whether the abundance or integrity of eIF5B was altered during enterovirus infection, cytoplasmic lysates from poliovirus type 1 (PV1)(Mahoney)- and Coxsackievirus B3 (CVB3)-infected 293T cells harvested at various times p.i. (Fig. 1A, 1B) were immunoblotted using polyclonal antibodies directed against the carboxy-terminal amino acid residues 921-1220 of human eIF5B (Wilson et al., 1999). A ∼ 70 kD protein present in mock-infected cells that cross-reacted with these antibodies is indicated by asterisks (Figs. 1C, 1D) and served as an internal loading control. The origin of this and another cross-reactive protein of ∼110 kD (Fig. 1C) have not been established. A ∼83 kD putative eIF5B cleavage product first appeared at 3h p.i., a time point that coincided with the initial appearance of viral proteins (Fig. 1A, 1B), and subsequently increased in prominence. Cleavage of eIF5B therefore begins early in enterovirus infection, at a similar time to cleavage of eIF4G and PABP (Etchison et al., 1982; Gradi et al., 1998; Kerekatte et al., 1999). Full length eIF5B with an apparent molecular weight in these gel/buffer conditions of 175 kD (Pestova et al., 2000) was readily detected by immunoblotting in lysates of 293T and HeLa cells (Fig. 1 and data not shown), but its abundance during the course of infection suggested that only partial cleavage of eIF5B had occurred. The level of cleavage varied between experiments done with PV1 and CVB3-infected 293T cells, in some instances being lower than shown in Fig. 1, and did not correlate with the multiplicity of infection (data not shown). The transient and variable appearance of additional smaller immunoreactive bands in some infections (data not shown), suggested that further cleavage or degradation of cleavage products occurred in infected cells, possibly due to proteasome or caspase activation, a characteristic of PV-infected cells (Belov et al., 2003; Barral et al., 2007).
Figure 1.
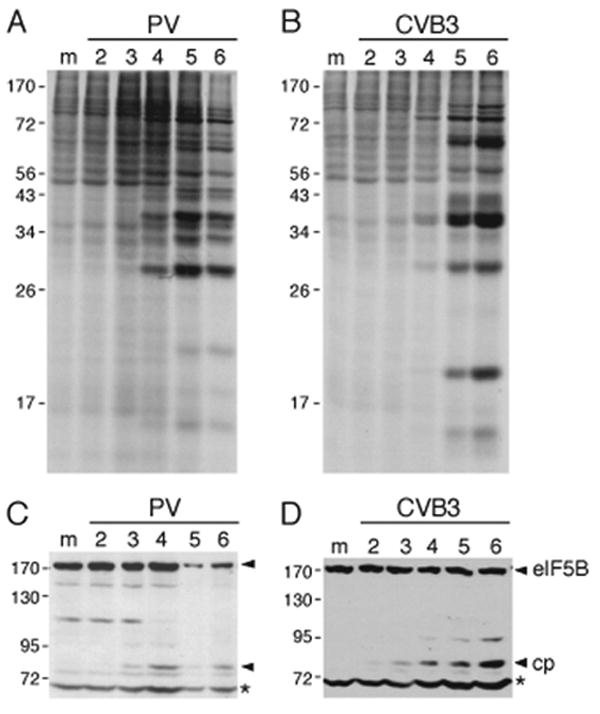
Cleavage of eIF5B during enterovirus infections. (A, B) 293 T cells were mock infected (M) or infected with (A) poliovirus or (B) Coxsackievirus B3 (CVB3) and pulse labeled with [35S]Methionine. Samples from mock infected cells were pulse-labeled at 1 hour p.i. and harvested at 2 hours p.i.; samples for infected cells were harvested at the indicated hours p.i. after pulse-labeling for 1 hour. Aliquots were then analyzed by SDS-PAGE. (C, D) Aliquots of samples of mock-infected 293T (m) or 293T cells infected with (C) poliovirus and (D) Coxsackievirus B3 and harvested at the indicated times (hours) p.i. were subjected to SDS-PAGE and immunoblotted with a polyclonal antibodies to eIF5B. A ∼70 kD host protein present in mock-infected cells that cross-reacted with these antibodies is indicated by asterisks (*) and served as an internal loading control. The positions of molecular weight markers, of intact eIF5B and of a cleavage product (cp) are indicated.
The observations that the level of eIF5B cleavage in enterovirus-infected cells was variable suggested that eIF5B might under some circumstances be protected from 3Cpro-mediated cleavage that we reasoned might result either from post-transcriptional modification, such as phosphorylation (e.g. Beausoleil et al., 2004) or from interaction with other proteins. The kinases that phosphorylate eIF5B are not known and phospho-specific eIF5B antibodies are not available, so the influence of phosphorylation on eIF5B's susceptibility to cleavage could not be characterized. eIF5B's interacting partners include eIF1A, 40S and 60S ribosomal subunits (Marintchev et al., 2003; Olsen et al., 2003; Unbehaun et al., 2007), but their presence did not affect cleavage of eIF5B by HRV or PV 3Cpro in vitro (data not shown). The reason for the variable level of cleavage of eIF5B in infected cells therefore remains to be determined.
Direct cleavage of eIF5B by enterovirus 3C protease
The analysis of virus-infected cell lysates suggested that enteroviruses encode a protease that cleaved eIF5B, yielding at least one fragment detectable by immunoblotting with antibodies directed against its C-terminal region. Incubation with purified recombinant HRV type 14 (HRV14) 3Cpro resulted in partial cleavage of purified native rabbit eIF5B, yielding fragments (cp1 and cp2) with apparent molecular weights of ∼85 kDa and ∼68 kDa (Fig. 2A, lane 2). cp1 was recognized by mouse polyclonal antibodies directed against eIF5B1121-1219(data not shown). In similar experiments carried out with purified CVB3 3Cpro, purified native eIF5B was also partially cleaved (Fig. 2C, lane 2), whereas purified PV 3Cpro cleaved purified eIF5B to near-completion, in both instances yielding cp1 and cp2 cleavage fragments with electrophoretic mobilities like those of the HRV 3Cpro cleavage products (Fig. 2A, lanes 2, 3). These cleavage reactions were performed overnight at 4°C, but the specificity of cleavage was not changed on incubation for shorter periods at 37°C (data not shown). We cannot rule out the possibility that 3CDpro, the major form of 3Cpro in infected cells (Ypma-Wong et al., 1988) cleaves eIF5B with different kinetics than 3Cpro.
Figure 2.

Specific cleavage of eIF5B by CVB3, HRV14 and PV1 3C proteases. (A) Purified native rabbit eIF5B (lanes 1-3) and purified recombinant human eIF5 (lanes 4-6), (B) purified recombinant human eIF1 (lanes 1-3) and purified recombinant human eIF1A (lanes 4-6), (C) purified native rabbit eIF5B (lanes 1-3), (D) control protease substrate (lanes 1, 2) and purified native rabbit eIF4F, and (E) purified recombinant human eIF5B587-1220(eIFΔ5B) (lanes 1, 2) and purified recombinant human eIF4B (lanes 3, 4) were incubated alone, with purified recombinant CVB3 3Cpro, HRV 14 3Cpro or PV1 3Cpro, as indicated. Reaction mixtures were then analyzed by SDS-polyacrylamide gel electrophoresis and Coomassie staining. The positions of 3C proteases, of initiation factors and, in the case of eIF5B, of their cp1 and cp2 cleavage products on gels are indicated to the right of each panel. The asterisk * indicates a contaminant co-purifying with recombinant eIF4B (c.f. Pisarev et al., 2007).
Cleavage of eIF5B by HRV14 and PV 3Cpro was specific, because whereas a control substrate was cleaved to completion after incubation with HRV14 3Cpro (Fig. 2D, lane 2), the integrity and abundance of purified recombinant eIF1, eIF1A, eIF4B and eIF5, and of purified native eIF4F were unaltered after incubation with HRV14 and PV 3Cpro in identical conditions (Fig. 2A, lanes 5, 6; Fig. 2B, lanes 2, 3, 5, 6; Fig. 2D, lane 4; Fig. 2E, lane 4). A recombinant fragment of eIF5B corresponding to amino acid residues 587-1220 (“eIFΔ5B”) was not cleaved by HRV14 3Cpro in these conditions (Fig. 2E, lanes 1, 2), indicating that cleavage likely occurred upstream of a.a. 587. These observations extend earlier reports that eIF2, eIF3, eIF4A, eIF4B and eIF4E are not cleaved during enterovirus infections (Duncan et al., 1983; Etchison et al., 1984; Lee et al., 1985; O'Neill and Racaniello, 1989).
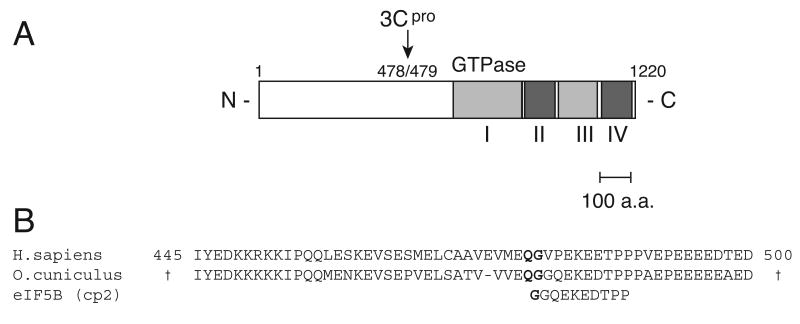
To map the cleavage site precisely, the N-terminal sequence of the cp1 fragment of native rabbit eIF5B was determined by Edman sequencing to be [G/S]GQEK[E/S]DTPP. This peptide sequence aligns precisely with the deduced sequence of a fragment of rabbit genomic DNA (Fig. 3B) and aligns well with a single conserved region of human eIF5B, which is consistent with cleavage of human eIF5B at Q478-G479 (Fig. 3B). The molecular weight of the C-terminal cp1 cleavage fragment of human eIF5B is 83.8 kD, which is consistent with the molecular weight of the cleavage fragment identified in enterovirus-infected 293T cells and in in vitro reactions done using purified eIF5B and CVB3, HRV14 and PV1 3C proteases (e.g. Figs. 1, 2A, 2C). The highly charged amino-terminal domain of eIF5B is responsible for its anomalous mobility in SDS-PAGE (Wilson et al., 1999; Pestova et al., 2000), and accordingly, the molecular weight of the N-terminal cp2 cleavage fragment of human eIF5B (55.1 kD) is less than that predicted on the basis of its mobility in SDS-PAGE (∼68kDa). Cleavage at VVEQ↓G in rabbit eIF5B and likely at VMEQ↓G479in human eIF5B is consistent with the cleavage specificity of enterovirus 3C proteases, for which the principal determinants for cleavage are Gln-Gly (Q-G) amino acid pairs flanking the scissile bond, and any of a small subset of amino acids (usually Ala or Val, but also other amino acids with small, aliphatic side chains) at the P4 position (Cordingley et al., 1990). It is also consistent with the cleavage specificity of enterovirus 3C proteases for cellular substrates such as TATA-binding protein, PABP, and NF-κB (p65-RelA) in which the principal 3C cleavage sites are ASPQ↓G (Das and Dasgupta, 1993), VHVQ↓G (Kuyumci-Martinez et al., 2002), and LLNQ↓G (Neznanov et al., 2005), respectively. Despite the structural homology of enteroviral 3C proteinases, sequence variation between the three proteinases tested here (42% - 59% pairwise amino acid identity) includes residues that determine substrate specificity at P2 and P2′ positions relative to the scissile bond (Matthews et al., 1999; Binford et al., 2005; Lee et al., 2007). These differences may account for the different extents of cleavage observed here.
Figure 3.
(A) Diagrammatic representation of human eIF5B. The site of cleavage by HRV14 3Cpro is shown relative to the positively charged N-terminal domain and the four conserved structural domains (I–IV) (54). (B) CLUSTAL-W alignment of the N-terminal sequence of the 3Cpro cleavage product cp1 of rabbit (Oryctolagus cuniculus) eIF5B, the corresponding eIF5B coding sequence, deduced from an O. cuniculus genomic fragment (Genbank: AAGW01235144.1), and the sequence of human eIF5B (Genbank: NM_015904).
The large amino-terminal domain (NTD) that is cleaved from mammalian eIF5B by 3Cpro does not significantly influence nucleotide binding by this factor (Pisareva et al., 2007), but the kinetics of other individual steps in the working cycle of mammalian eIF5B have not yet been characterized. eIF5B is one of the least abundant initiation factors (Pisarev et al., 2007), so that any change in its activity caused by proteolytic cleavage might therefore contribute to enterovirus-induced shut-off of translation. There are several indications how eIF5B's activity might be altered by 3Cpro-mediated cleavage, which separates the N-terminal domain of eIF5B from its essential conserved central GTPase and C-terminal domains. First, the GTP-dependency of the activity of eIF5B in ribosomal subunit joining is altered in some circumstances by deletion of an N-terminal fragment from eIF5B that is similar in size to that cleaved off by 3Cpro (Pestova et al., 2000). Second, the NTD of yeast eIF5B binds directly to eIF1A (Olsen et al., 2003), a component of 43S complexes that binds directly to the 40S subunit, so that deletion of the eIF5B NTD might affect the kinetics of binding of eIF5B to 48S complexes or of its release from assembled 80S ribosomes. Third, by analogy with the corresponding domain in eIF5B's prokaryotic homologue IF2, which is required for maximal growth, the NTD could interact with or induce conformational changes in the small ribosomal subunit (Moreno et al., 1999; Caserta et al., 2006).
In summary, these studies have identified eIF5B as a novel enterovirus 3Cpro substrate, and the time course of eIF5B proteolysis in infected cells suggest that it could contribute to the virus-induced shutoff of host and viral translation. 3Cpro impairs cap-mediated initiation (Kuyumcu-Martinez et al., 2004) and initiation mediated by PV1 and CVB3 IRESs (our unpublished data). Future studies will be directed to identifying the contribution of eIF5B cleavage to shutoff relative to the effects of 3Cpro-mediated cleavage of other factors involved in enterovirus translation, such as PABP and the IRES trans-acting factors PTB and PCBP2 (Back et al., 2002; Perera et al., 2007).
Methods
Cells and virus infection
293T cells were cultured in Dulbecco's Modified Eagle Medium (DMEM) supplemented with glutamine and 10% fetal bovine serum (FBS). Poliovirus (Type 1, Mahoney) and Coxsackievirus B3 were grown in HeLa cells and purified on cesium chloride gradients (Jones and Ehrenfeld, 1983) and from infected HeLa supernatants by polyethylene glycol precipitation (Kuyumcu-Martinez et al., 2002), respectively. 293T cells or HeLa cells were infected with PV1 at MOI=30 or CVB3 at MOI=10 in serum-free DMEM. At one hour post-infection, media was removed and replaced with DMEM containing 2% FBS. Metabolic labeling was done by incubating cells for one hour in (−met, −cys) DMEM containing 2% dialyzed FBS and 35S-Trans label (MP Biomedicals) at 30 μCi/mL.
Purification of ribosomal subunits and eukaryotic initiation factors
Native eIF4F and eIF5B were purified from rabbit reticulocyte lysate (Pisarev et al., 2007). Recombinant eIF1, eIF1A, eIF4B, eIF5 and eIF5B587-1220(eIFΔ5B) were overexpressed in Escherichia coli and purified (Pisarev et al., 2007). The integrity and activity of these factors and ribosomal subunits were assessed by polyacrylamide gel electrophoresis and using functional assays dependent on each of them (Pisarev et al., 2007).
eIF5B cleavage assays, sequencing, and blotting
Recombinant PV 3Cpro and CVB3 3Cpro were expressed in E. coli and purified as described (Joachims et al., 1999; Zell et al., 2002); recombinant HRV14 3Cpro was from Novagen (Madison, WI). To assay cleavage of individual initiation factors, 2μg eIF5B, eIFΔ5B, eIF1, eIF1A, eIF5, eIF4B or eIF4F was incubated overnight at 4°C in cleavage buffer (150 mM NaCl, 50 mM Tris HCl pH 7.5) and with 1 μg HRV 3Cpro, 1 μg CVB3 3Cpro A, 0.5 μg PV 3Cpro or without added protease. Cleavage products were analyzed by SDS-polyacrylamide gel electrophoresis (SDS-PAGE); gels were stained with coomassie blue. Immunoblotting of eIF5B and its cleavage products was done using polyclonal rabbit antibodies directed against amino acid residues 921-1220 of human eIF5B (Wilson et al., 1999) or polyclonal mouse antibodies directed against amino acid residues 1121-1219 of human eIF5B (Abnova). The eIF5B cp1 cleavage product was resolved by SDS-PAGE and transferred to PVDF membrane for amino-terminal sequencing.
Acknowledgments
This work was supported by PHS grant AI-51340 and by award 0755900T from the American Heart Association (Heritage Affiliate) (to C.U.T.H.) and by PHS grant AI-50237 (to R.E.L). We thank Tatyana Pestova for the gift of native eIF5B, Stuart Wilson for the anti-eIF5B antibodies, Roland Zell for the CVB3 3Cpro expression vector and Irina Abaeva for purifying recombinant CVB3 3Cpro. Protein sequence analysis was provided by The Rockefeller University Protein/DNA Technology Center, which is supported in part by NIH shared instrumentation grants and by funds provided by the U.S. Army and Navy for purchase of equipment.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Back SH, Kim YK, Kim WJ, Cho S, Oh HR, Kim JE, Jang SK. Translation of polioviral mRNA is inhibited by cleavage of polypyrimidine tract-biniding proteins executed by polioviral 3C(pro) J Virol. 2002;76:2529–2542. doi: 10.1128/jvi.76.5.2529-2542.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barral PM, Morrison JM, Drahos J, Gupta P, Sarkar D, Fisher PB, Racaniello VR. MDA-5 is cleaved in poliovirus-infected cells. J Virol. 2007;81:3677–3684. doi: 10.1128/JVI.01360-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beausoleil SA, Jedrychowski M, Schwartz D, Elias JE, Villen J, Li J, Cohn MA, Cantley LC, Gygi SP. Large-scale characterization of HeLa cell nuclear phosphoproteins. Proc Natl Acad Sci USA. 2004;101:12130–12135. doi: 10.1073/pnas.0404720101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Belov GA, Romanova LI, Tolskaya EA, Kolesnikova MS, Lazebnik YA, Agol VI. The major apoptotic pathway activated and suppressed by poliovirus. J Virol. 2003;77:45–56. doi: 10.1128/JVI.77.1.45-56.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Binford SL, Maldonado F, Brothers MA, Weady PT, Zalman LS, Meador JW, 3rd, Matthews DA, Patick AK. Conservation of amino acids in human rhinovirus 3C protease correlates with broad-spectrum antiviral activity of rupintrivir, a novel human rhinovirus 3C protease inhibitor. Antimicrob Agents Chemother. 2005;49:619–626. doi: 10.1128/AAC.49.2.619-626.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caserta E, Tomsic J, Spurio R, La Teana A, Pon CL, Gualerzi CO. Translation initiation factor IF2 interacts with the 30S ribosomal subunit via two separate binding sites. J Mol Biol. 2006;362:787–799. doi: 10.1016/j.jmb.2006.07.043. [DOI] [PubMed] [Google Scholar]
- Cordingley MG, Callahan PL, Sardana VV, Garsky VM, Colonno RJ. Substrate requirements of human rhinovirus 3C protease for peptide cleavage in vitro. J Biol Chem. 1990;265:9062–9065. [PubMed] [Google Scholar]
- Das S, Dasgupta A. Identification of the cleavage site and determinants required for poliovirus 3CPro-catalyzed cleavage of human TATA binding transcription factor TBP. J Virol. 1993;67:3326–3331. doi: 10.1128/jvi.67.6.3326-3331.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duncan R, Etchison D, Hershey JWB. Protein synthesis eukaryotic initiation factors 4A and 4B are not altered by poliovirus infection of HeLa cells. J Biol Chem. 1983;258:7236–7239. [PubMed] [Google Scholar]
- Etchison D, Hansen J, Ehrenfeld E, Edery I, Sonenberg N, Milburn S, Hershey JWB. Demonstration in vitro that eucaryotic initiation factor 3 is active but a cap binding protein complex is inactive in poliovirus-infected HeLa cells. J Virol. 1984;51:832–837. doi: 10.1128/jvi.51.3.832-837.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Etchison D, Milburn S, Edery I, Sonenberg N, Hershey JWB. Inhibition of HeLa cell protein synthesis following poliovirus infection correlates with the proteolysis of a 220,000 dalton polypeptide associated with eukaryotic initiation factor 3 and a cap-binding complex. J Biol Chem. 1982;257:14806–14810. [PubMed] [Google Scholar]
- Gingras AC, Svitkin Y, Belsham GJ, Pause A, Sonenberg N. Activation of the translational repressor 4E-BP1 following infection with encephalomyocarditis virus and poliovirus. Proc Natl Acad Sci USA. 1996;93:5578–5583. doi: 10.1073/pnas.93.11.5578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gradi A, Svitkin YV, Imataka H, Sonenberg N. Proteolysis of human eukaryotic translation initiation factor eIF4GII, but not eIF4GI, coincides with the shutoff of host protein synthesis after poliovirus infection. Proc Natl Acad Sci USA. 1998;95:11089–11094. doi: 10.1073/pnas.95.19.11089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hellen CUT, Sarnow P. Internal ribosome entry sites in eukaryotic mRNA molecules. Genes Dev. 2001;15:1593–1612. doi: 10.1101/gad.891101. [DOI] [PubMed] [Google Scholar]
- Joachims M, Van Breugel PC, Lloyd RE. Cleavage of poly(A)-binding protein by enterovirus proteases concurrent with inhibition of translation in vitro. J Virol. 1999;73:718–727. doi: 10.1128/jvi.73.1.718-727.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones C, Ehrenfeld E. The effect of poliovirus infection on the translation in vitro of VSV messenger ribonucleoprotein particles. Virology. 1983;129:415–430. doi: 10.1016/0042-6822(83)90180-0. [DOI] [PubMed] [Google Scholar]
- Kerekatte V, Keiper BD, Badorff C, Cai A, Knowlton KU, Rhoads RE. Cleavage of poly(A)-binding protein by coxsackievirus 2A protease in vitro and in vivo: another mechanism for host protein synthesis shutoff? J Virol. 1999;73:709–717. doi: 10.1128/jvi.73.1.709-717.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuyumcu-Martinez NM, Joachims M, Lloyd RE. Efficient cleavage of ribosome-associated poly(A)-binding protein by enterovirus 3C protease. J Virol. 2002;76:2062–2074. doi: 10.1128/jvi.76.5.2062-2074.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuyumcu-Martinez NM, Van Eden ME, Younan P, Lloyd RE. Cleavage of poly(A)-binding protein by poliovirus 3C protease inhibits host cell translation: a novel mechanism for host translation shutoff. Mol Cell Biol. 2004;24:1779–1790. doi: 10.1128/MCB.24.4.1779-1790.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lamphear BJ, Kirchweger R, Skern T, Rhoads RE. Mapping of functional domains in eukaryotic protein synthesis initiation factor 4G (eIF4G) with picornaviral proteases. Implications for cap-dependent and cap-independent translational initiation. J Biol Chem. 2005;270:21975–21983. doi: 10.1074/jbc.270.37.21975. [DOI] [PubMed] [Google Scholar]
- Lee ES, Lee WG, Yun SH, Rho SH, Im I, Yang ST, Sellamuthu S, Lee YJ, Kwon SJ, Park OK, Jeon ES, Park WJ, Kim YC. Development of potent inhibitors of the coxsackievirus 3C protease. Biochem Biophys Res Commun. 2007;358:7–11. doi: 10.1016/j.bbrc.2007.03.208. [DOI] [PubMed] [Google Scholar]
- Lee KAW, Edery I, Sonenberg N. Isolation and structural characterization of cap-binding proteins from poliovirus-infected HeLa cells. J Virol. 1985;54:515–524. doi: 10.1128/jvi.54.2.515-524.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marintchev A, Kolupaeva VG, Pestova TV, Wagner G. Mapping the binding interface between human eukaryotic initiation factors 1A and 5B: a new interaction between old partners. Proc Natl Acad Sci USA. 2003;100:1535–1540. doi: 10.1073/pnas.0437845100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matthews DA, Dragovich PS, Webber SE, Fuhrman SA, Patick AK, Zalman LS, Hendrickson TF, Love RA, Prins TJ, Marakovits JT, Zhou R, Tikhe J, Ford CE, Meador JW, Ferre RA, Brown EL, Binford SL, Brothers MA, DeLisle DM, Worland ST. Structure-assisted design of mechanism-based irreversible inhibitors of human rhinovirus 3C protease with potent antiviral activity against multiple rhinovirus serotypes. Proc Natl Acad Sci USA. 1999;96:11000–11007. doi: 10.1073/pnas.96.20.11000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moreno JM, Drskjotersen L, Kristensen JE, Mortensen KK, Sperling Petersen HU. Characterization of the domains of E. coli initiation factor IF2 responsible for recognition of the ribosome. FEBS Letts. 1999;455:130–134. doi: 10.1016/s0014-5793(99)00858-3. [DOI] [PubMed] [Google Scholar]
- Neznanov N, Chumakov KM, Neznanova L, Almasan A, Banerjee AK, Gudkov AV. Proteolytic cleavage of the p65-RelA subunit of NF-kappaB during poliovirus infection. J Biol Chem. 2005;280:24153–24158. doi: 10.1074/jbc.M502303200. [DOI] [PubMed] [Google Scholar]
- Olsen DS, Savner EM, Mathew A, Zhang F, Krishnamoorthy T, Phan L, Hinnebusch AG. Domains of eIF1A that mediate binding to eIF2, eIF3 and eIF5B and promote ternary complex recruitment in vivo. EMBO J. 2003;22:193–204. doi: 10.1093/emboj/cdg030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O'Neill RE, Racaniello VR. Inhibition of translation in cells infected with a poliovirus 2Apro mutant correlates with phosphorylation of the alpha subunit of eucaryotic initiation factor 2. J Virol. 1989;63:5069–5075. doi: 10.1128/jvi.63.12.5069-5075.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perera R, Daijogo S, Walter BL, Nguyen JH, Semler BL. Cellular protein modification by poliovirus: the two faces of poly(rC)-binding protein. J Virol. 2007;81:8919–8932. doi: 10.1128/JVI.01013-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pestova TV, Lomakin IB, Lee JH, Choi SK, Dever TE, Hellen CUT. The joining of ribosomal subunits in eukaryotes requires eIF5B. Nature. 2000;403:332–335. doi: 10.1038/35002118. [DOI] [PubMed] [Google Scholar]
- Pestova TV, Lorsch JR, Hellen CUT. The mechanism of translation initiation in eukaryotes. In: Mathews MB, Sonenberg N, Hershey JWB, editors. Translational control in biology and medicine. Cold Spring Harbor Laboratory Press; Cold Spring Harbor, NY: 2007. pp. 87–128. [Google Scholar]
- Pisarev AV, Unbehaun A, Hellen CUT, Pestova TV. Assembly and analysis of eukaryotic translation initiation complexes. Methods Enzymol. 2007;430:147–177. doi: 10.1016/S0076-6879(07)30007-4. [DOI] [PubMed] [Google Scholar]
- Pisareva VG, Hellen CUT, Pestova TV. Kinetic analysis of interaction of guanine nucleotides with eukaryotic translation initiation factor eIF5B. Biochemistry. 2007;46:2622–2629. doi: 10.1021/bi062134g. [DOI] [PubMed] [Google Scholar]
- Svitkin YV, Herdy B, Costa-Mattioli M, Gingras AC, Raught B, Sonenberg N. Eukaryotic translation initiation factor 4E availability controls the switch between cap-dependent and internal ribosomal entry site-mediated translation. Mol Cell Biol. 2005;25:10556–10565. doi: 10.1128/MCB.25.23.10556-10565.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Unbehaun A, Marintchev A, Lomakin IB, Didenko T, Wagner G, Hellen CUT, Pestova TV. Position of eukaryotic initiation factor eIF5B on the 80S ribosome mapped by directed hydroxyl radical probing. EMBO J. 2007;26:3109–3123. doi: 10.1038/sj.emboj.7601751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilson SA, Sieiro-Vazquez C, Edwards NJ, Iourin O, Byles ED, Kotsopoulou E, Adamson CS, Kingsman SM, Kingsman AJ, Martin-Rendon E. Cloning and characterization of hIF2, a human homologue of bacterial translation initiation factor 2, and its interaction with HIV-1 matrix. Biochem J. 1999;342:97–103. [PMC free article] [PubMed] [Google Scholar]
- Ypma-Wong MF, Dewalt PG, Johnson VH, Lamb JG, Semler BL. Protein 3CD is the major poliovirus proteinase responsible for cleavage of the P1 capsid precursor. Virology. 1988;166:265–270. doi: 10.1016/0042-6822(88)90172-9. [DOI] [PubMed] [Google Scholar]
- Zell R, Sidigi K, Bucci E, Stelzner A, Görlach M. Determinants of the recognition of enteroviral cloverleaf RNA by coxsackievirus B3 proteinase 3C. RNA. 2002;8:188–201. doi: 10.1017/s1355838202012785. [DOI] [PMC free article] [PubMed] [Google Scholar]