Abstract
Ergosterol homeostasis is a critical process for fungal cells. Paralogous zinc cluster transcription factors Upc2p and Ecm22p are major regulators of ergosterol biosynthesis in Saccharomyces cerevisiae. Upc2p and Ecm22p sense and respond to sterol depletion but their mechanism of activation has not been defined. Subcellular localization and functional expression of Upc2p–GFP and Ecm22p-GFP was monitored by fluorescence microscopy and flow cytometry in live yeast cells. Both fusion proteins localized to intracellular membranes and to perinuclear foci. Perinuclear localization of Upc2p-GFP and Ecm22p-GFP was increased when ergosterol biosynthesis was blocked by azole drug treatment. Nuclear localization in response to sterol depletion is consistent with the hypothesis that Upc2p and Ecm22p are trafficked from a membrane to the nucleus as a post-translational mechanism of sterol sensing.
Keywords: Ergosterol, UPC2, ECM22, SREBP, Azole Resistance, GFP, FACS
Introduction
Ergosterol modulates membrane fluidity, stability, permeability and the activities of membrane-bound enzymes in fungi. It is the target of the most commonly used antifungals, the polyenes and azoles [1] and the action of these antifungals is affected by changes in membrane ergosterol levels [2]. Fungal cells are known to regulate biosynthesis of sterols in response to sterol fluxes [3–6] . The transcription factors Upc2p and Ecm22p are major regulators of ergosterol biosynthesis in S.cerevisiae. Upc2p and Ecm22p are Zn[2]-CyS[6] binuclear cluster transcription factors [7, 8] and bind a conserved cis octamer SRE within promoters of ERG genes [4, 9]. When sterol levels are reduced, ERG genes are upregulated by both Upc2p and Ecm22p [10] and both proteins have roles in sterol import and transport within the cell [11]. Upc2p is also a regulator of the hypoxically induced DAN/TIR genes that encode mannoprotein genes responsible for anaerobic cell wall synthesis [12 , 13]. Double deletion of UPC2 and ECM22 is synthetically lethal in the s288C background due to a mutation in HAP1 [14] but a double mutant is viable in the W303 strain background [4, 14]. W303Δecm22Δupc2 is hypersusceptible to the Hmg-CoA reductase inhibitor lovastatin and resistant to the ergosterol binding drug amphotericin B [4].
Upc2p and Ecm22p are highly homologous at in the N-terminal DNA binding domain (DBD) and the C-terminal transmembrane region; the conserved N and C regions are separated by a dissimilar linker domain [4]. Three nuclear localization sequences (NLS) are predicted in the N-terminal domain of Upc2p and four are predicted for Ecm22p [5] . The protein prediction programs TMAP and TMpred predict four transmembrane helices in the C-terminus of Upc2p and Ecm22p, consistent with double pass membrane proteins [4]. Based on promoter occupancy results, Davies et al. have proposed that Upc2p and Ecm22p are constitutively nuclear and are c-terminally bound by an unidentified repressor which inhibits their intrinsic activation capability [10] however the role of the different domains in the function and regulation of Upc2p and Ecm22p have not been experimentally defined.
Upc2p and Ecm22p are analogous in structure to sterol regulatory element-binding proteins (SREBPs) the major regulators of cholesterol homeostasis in mammalian cells. SREBPS are embedded in the endoplasmic reticulum (ER) membrane by two transmembrane α-helices and contain an N-terminal transcription factor domain. The transmembrane domains of SREBP interact with Scap (SREBP cleavage-activating protein). In cholesterol rich conditions SCAP binds the ER membrane protein Insig which retains the SREBP-SCAP precursor complex in the ER. When sterol levels are low, SCAP and Insig do not interact which allows the SCAP-SREBP precursor complex to translocate from the ER to the Golgi where sequential proteolytic cleavages release the N-terminal transcription factor domain which translocates to the nucleus and activates gene transcription,
Recently SREBP and Scap homologues have been identified in the fission yeast Schizosaccharomyces pombe and the pathogenic basidiomycete Cryptococcus neoformans[15–17]. Fungal SREBP homologues have been shown to be proteolytically cleaved in response to sterol and oxygen depletion [16, 17]. SREBP and Scap homologues in the pathogenic fungus Cryptococcus neoformans are essential for transcriptional regulation of sterol genes and required for virulence in mice [16, 18, 19]. These studies demonstrate the importance of fungal SREBPs for anaerobic growth and sterol biosynthesis across fungal phyla.
We propose that Ecm22p and Upc2p are functional analogues to mammalian SREBPs and our data suggests that both proteins may be activated similarly, such that a transmembrane domain anchors full length Upc2p and Ecm22p in the ER membrane. When sterol levels are depleted this membrane localization is destabilized allowing the N-terminal DBD of Upc2p and Ecm22p to translocate to the nucleus via the NLS sequences and activate target genes [5]. Post-translational sterol sensing by transcription factors has been experimentally defined in mammalian cells and other fungal species; our data indicate that sterol sensing may also play a role in the activation of the major regulators of sterol biosynthesis in S.cerevisiae.
S.cerevisiae Upc2p and Ecm22p have been previously studied [14, 20] however, azole induciblity, complete MIC analysis and ergosterol levels have not previously been reported for Upc2p and Ecm22p mutants; nor has previous research addressed the mechanism of Upc2p and Ecm22p activation. In this paper, we characterize the localization of Upc2p and Ecm22p using GFP fusions. We used these fusions to measure azole induction of Upc2p and Ecm22p over 48 hours. We determined that azole susceptibility is a phenotypic indicator of Upc2p and Ecm22p functionality by analyzing FLC susceptibility of UPC2 and ECM22 mutants in two different strain backgrounds.
Materials and Methods
Abbreviations
The following abbreviations are used throughout the text and figures: 5-FC=5-Flucytosine, AMB=Amphotericin B, DBD=DNA binding domain, EBI=ergosterol biosynthesis inhibitor, FACS=Fluorescence activated cell sorting, FLC=Fluconazole, GFP=green fluorescent protein, ITRA=Itraconazole, LOV=Lovastatin, NLS=Nuclear localization sequence, Scap=SREBP cleavage-activating protein, SDS=sodium dodecyl sulfate, SRE=sterol response element, SREBP=Sterol Response Element Binding Protein, TERB=Terbinafine
Strain maintenance and manipulation
Strains utilized in this study are summarized in Table 1. Strains were maintained on the appropriate CSM drop-out media (0.75 g CSM (Bio 101, Inc., Vista, CA), 1.7 g Yeast Nitrogen Base without amino acids or ammonium sulfate, 5 g ammonium sulfate, 20 g dextrose per liter]. Cells were stored at −80° C in selective media supplemented with 10% glycerol. Overnight cultures were inoculated from a single colony from a selective agar plate into selective media and grown overnight at 30°C, 180 rpm. These cultures were then used to inoculate cultures, usually at an OD 600 = 0.1 in CSM media with or without drug, and grown at 30° C, 180 rpm. Media components were obtained from Fisher Scientific (Pittsburgh, PA) or Bio 101, Inc. (Vista, CA). Chemicals were obtained from Fisher, Sigma (St. Louis, MO) or Aldrich (Milwaukee, WI).
Table 1.
MIC of FLC for S.cerevisiae strains used and constructed in this study
| Strain | MIC80 (µg/mL FLC) | Genotype | |
|---|---|---|---|
| 1 |
s288Ca (TW 15627) |
8 | MATa suc2 gal2 mal mel flo1 flo8-1 hap1 |
| 2 |
Ecm22p-GFPb (TW 16704) |
4 | MATa his3Δ1 leu2Δ0 met15Δ0 ura3Δ::ECM22-GFP-HIS3MX6 |
| 3 |
Upc2p-GFPb (TW 15581) |
4 | MATa his3Δ1 leu2Δ0 met15Δ0 ura3Δ0::UPC2-GFP-HIS3MX6 |
| 4 |
Upc2p-GFPΔecm22c (TW 16701) |
4 | MATa his3Δ1 leu2Δ0 met15Δ0 ura3Δ0::UPC2-GFP-HIS3MX6ecm22Δ::URA3 |
| 5 |
Ecm22p-GFPΔupc2c (TW 16711) |
2 | MATa his3Δ1 leu2Δ0 met15Δ0 ura3Δ::ECM22-GFP-HIS3MX6 upc2Δ::URA3 |
| 6 |
BY4741Δupc2b (TW 16723) |
4 | BY4741 MATa his3Δ leu2Δ met15Δura3Δupc2Δ:: KANMX4 |
| 7 |
BY4741Δecm22b (TW 16719) |
4 | BY4741 MATa his3Δ leu2Δ met15Δura3Δ cm22Δ::KANMX4 |
| 8 |
W303-1ad (TW 14967) |
8 | MATa ade2-1 leu2-3,112 his3-1 ura3-52 trp1-100 can1-100 |
| 9 |
W303-1aΔupc2d (TW 14969) |
4 | W303-1a upc2Δ::HIS3 |
| 10 |
W303-1aΔecm22d (TW 14968) |
3 | W303-1a ecm22Δ::TRP1 |
| 11 |
W3031aΔecm22Δupc2d (TW 14970) |
0.032 | W303-1a ecm22:Δ:TRP1 upc2Δ::HIS3 |
| 12 |
W303ΔupcΔecm22::pTEF-UPC2GFPc (TW 15671) |
<0.016 f | W303-1a ecm22:Δ:TRP1 upc2Δ::HIS3 pTEF-UPC2GFP |
| 13 | RFP reference strainse | ND g | EY0987 MATα his3Δ1 leu2Δ0 lys2Δ0 ura3Δ0 |
Deletion of UPC2 and ECM22
The URA3 gene was PCR amplified from the plasmid pJJ242 with oligonucleotides designed with homology upstream and downstream of the endogenous ECM22 locus (5’-agttaagtcgacggtttttttcatttaaggtttatatttcgtttatccatagatctccatctaacataac cgccagggtttcccagtcacgac-3’ and 5’-tattcatcgatgaggttcacattctgtagcactgacccatcgaaagaaaagagtttatattttgataaa agcggataacaatttcacacagga-3’). The PCR product was transformed into the Upc2p-GFP strain by the modified lithium acetate transformation method [21]. Transformants were selected on CSM-URA. UPC2 was deleted in the Ecm22p-GFP background by the same method using oligonucleotides 5’-aaacaaaatgagctttccagaaatagtgaatcaaaaaaagttaagtacaaatatttacagttcagcagtcgcca gggtttcccagtcacgac-3’ and 5’-ggtaacacaatatccctttttttttttttgggaatctatttcgaatattctgcacttttaaattttctaagcggataacaatttc acacagga-3’ to amplify the URA deletion cassette.
PCR and Southern blot analysis
Genomic DNA was prepared as previously described [21]. Transformants were first screened by PCR to determine correct gene integrations. PCR positive transformants were further characterized by Southern blot analysis. 5 µg of genomic DNA was digested separately with PstI or EcoRV, (Promega, Madison, WI) and resolved by electrophoresis in 0.8% agarose gels. A 32P kinase-labeled oligonucleotide was hybridized to the Southern blots to determine the location and copy number of the inserted construct. Standard techniques were used for the Southern blot analyses [22].
Susceptibility testing
Drug susceptibility was determined using the standard CLSI broth microdilution protocol [23] that determines the minimum inhibitory concentration of drug needed to inhibit 80% of cell growth (MIC80). E test strips (AB Biodisk, N.J.), plastic strips with gradients of drug, were used per the manufacturer's instructions. Each strip is placed onto an agar plate containing a lawn of cells, and growth inhibition is measured directly off the plate after 48 h. E-tests were done in triplicate with identical results.
Cell Staining and Fluorescence Microscopy
Cells were treated with increasing concentrations of FLC and visualized microscopically. Cells were grown overnight in CSM, then diluted to approximately 2 × 106 cells/ ml in CSM +/− 10 µg/ml FLC. To stain nuclei the DNA-specific fluorescent dye Hoechst 33258 (1 mg/ml stock in DMSO) was added to a final concentration of 1 µg/mL and cells were incubated in the dark at 30° C, 180 rpm for 5 minutes. Cells were washed twice in PBS pH 7.4. Live cells were immediately observed using a DeltaVision RT optical/digital-sectioning microscope equipped with DIC optics for visible imaging (Applied Precision, Issaquah, WA). An exposure time of 1s in the FITC channel was used for cells grown in the presence of FLC, while an exposure time of 2 s was necessary for cells grown in the absence of FLC to account for differences in intensity due to drug treatment. Images were deconvolved and compiled as projections of digital sections using softWoRx® Explorer 1.2 software. All images were processed identically. For counting experiments at least three fields of view were counted from three independent experiments. For intensity profiles a single image was analyzed following deconvolution using the arbitrary line intensity tool from softWoRx® Explorer 1.2 software (Applied Precision, Issaquah, Washington).
FACS analysis
Overnight cultures grown in CSM were diluted to approximately 2 × 106 cells/ ml in CSM containing 0, 1, 10 or 100 µg/ml FLC. To determine induction for the other EBIs, cells were grown as described above in 10 µg/mL lovastatin, 0.1 µg/mL itraconazole, 10 µg/mL Terbinafine. Cells grown in 2 µg/mL AMB, 0.1µg/mL SDS, and 1 µg/mL 5-FC were analyzed as negative controls. Drug concentrations were chosen based on previous MIC determinations in S.cerevisiae. At specified time points, an aliquot of cells was removed and washed three times in PBS pH 7.4. Cells were diluted 1:40 in ice cold PBS immediately prior to analysis using a Beckman Coulter Epics XL-MCL flow cytometer. 100,000 cells per sample were analyzed. Three independent cultures of each strain/FLC combination were analyzed. The geometric mean of FL1 channel fluorescence for each sample was calculated using FlowJo software 7.2.2 (Tree star, Ashland, Oregon). Geometric means from three independent replicates were averaged and graphed.
Ergosterol levels
Ergosterol levels were measured as described previously [24] with the following modifications. All strains were grown in CSM medium. Strains were grown for 6, 24 and 48 h in CSM containing 0, 1 or 10 µg/ml FLC. Approximately 2 × 109 cells were used for each data point.
Generation of Diploids
RFP reference library stains were mated with both UPC2-GFPΔecm22 and ECM22-GFPΔupc2 strains on solid media as previously described [25]. Diploids were selected on CSM-LYS-MET + 100 µg/mL G418 sulfate (Sigma). Cells were grown overnight in CSM-LYS-MET + 100 µg/mL G418 sulfate. Cells were then diluted to approximately 2 × 106 cells/ ml in CSM- LYS-MET + 100 µg/mL G418 sulfate +/− 10 µg/ml FLC. To stain nuclei the DNA-specific fluorescent dye Hoechst 33258 (1 mg/ml stock in DMSO) was added to a final concentration of 1 µg/mL and cells were incubated in the dark at 30° C, 180 rpm for 5 m. Cells were washed twice in PBS pH 7.4. Live cells were immediately observed using a DeltaVision RT optical/digital-sectioning microscope equipped with DIC optics for visible imaging. Images were deconvolved and compiled as projections of digital sections using softWoRx® Explorer 1.2 software. All images were processed identically.
Results
Localization of Upc2p-GFP and Ecm22p-GFP
Localization of Upc2p and Ecm22p within the nucleus would support the model of activation put forth by Davies et al, while localization outside the nucleus would support an SREBP-like activation model. The two major protein localization projects completed in S.cerevisiae reported conflicting localization of Upc2p-GFP. Huh et al. report Upc2p-GFP to be localized to the nucleus [26, 27] while Habeler et al. report Upc2p C-terminal GFP fusion to be localized to the cytoplasm [26, 27]. Huh et al. reported cytoplasmic and nuclear localization of Ecm22p [27] and Habeler et al. did not report Ecm22p localization. This difference is not unexpected as both studies were done on a large scale and each group used a different methodology.
We obtained strains from both groups to determine the definitive subcellular localization of Upc2p-GFP and Ecm22p-GFP. Habeler’s group constructed an episomal Upc2p-GFP driven by a tef promoter. The plasmid bearing strain expressed high levels of Upc2p-GFP which localized to intracellular membranes and to the cytoplasm (data not shown). To access the functionality of constitutively expressed Upc2p-GFP we transformed the W303Δecm22Δ upc2 mutant (Table 1, row 12) with the tef-Upc2p-GFP plasmid. The Upc2p-GFP plasmid did not complement the azole susceptibility of the W303Δecm22Δ upc2 mutant indicating that this construct was non-functional. Huh et al. used an alternative approach of C-terminal GFP fusions integrated at the endogenous gene locus and driven by the native promoter. Upc2p-GFP and Ecm22p-GFP fusions (Table 1, rows 2 and 3) constructed by Huh’s group had lower expression relative to the episomal Upc2p-GFP as determined by FACS (data not shown) but had WT azole susceptibility (Table 1, rows 2–3). Azole susceptibility indicated that these fusions were functional thus these strains were used for our localization studies.
We hypothesized that intracellular sterol concentration would affect localization of Upc2p and Ecm22p. To test this hypothesis cells were treated with FLC to block sterol synthesis prior to imaging. Upc2p-GFP and Ecm22p-GFP fluorescence was dynamically localized to intracellular membranes and to punctuate perinuclear structures (Figure 1). Some cells displayed a single localization pattern and in some cells both patterns were discernable. The perinuclear structures did not precisely colocalize with the nucleus as shown by co-staining with DAPI (data not shown) or Hoechst (Figure 1). Perinuclear localization was predominant in cells treated with FLC while untreated cells had more membranous localization. To quantify this effect, cells were counted and scored for perinuclear, membranous or combination localization.
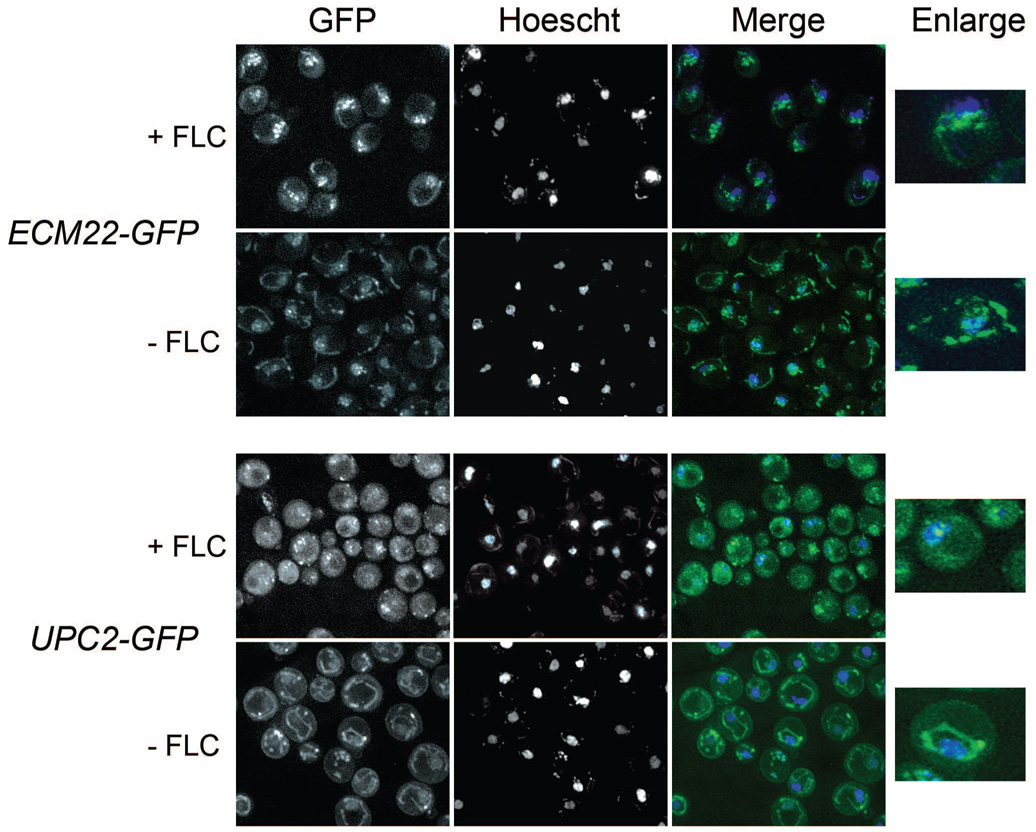
Figure 1. Upc2p-GFP and Ecm22p-GFP are increasingly localized to perinuclear foci in cells treated with FLC.
Cells were grown +/−10 µg/mL FLC for 6 hours prior to visualization by fluorescence microscopy. The GFP channel is shown in the left panel, Hoechst nuclear stain is shown in the middle panel and a merged image is shown on the right.
A. Ecm22p-GFP localization in the presence of untagged UPC2 (Table 1 row 2) is punctuate and nuclear associated in cells treated with FLC and punctuate and membranous in untreated cells.
B. Upc2p-GFP in the presence of untagged ECM22 (Table 1, row 3) is punctuate and nuclear associated in cells treated with FLC and membranous in untreated cells.
Upc2p-GFP was localized to perinuclear foci in 95% of FLC treated cells compared with 16% of non-FLC treated cells. Likewise, Ecm22p-GFP was localized to perinuclear foci in 88% of FLC treated cells compared to 38% of non-FLC treated cells (Table 2). Both GFP fusion proteins exhibited greater association with the nuclear periphery in FLC treated cells and a greater association with membranes in cells grown under non-sterol depleting conditions (Table 2). We observed similar localization by immunofluorescence with an anti-FLAG antibody and a strain expressing a C-terminal Upc2p-6xHIS-FLAG (data not shown) but chose to use GFP as it enabled the observation of live cells.
Table 2.
Perinuclear localization of Upc2p-GFP and Ecm22p-GFP is increased in FLC treated cells
| ECM22-GFP | UPC2-GFP | ECM22-GFP Δupc2 | UPC2-GFP Δecm22 | |||||
|---|---|---|---|---|---|---|---|---|
| FLC | − | + | − | + | − | + | − | + |
| Perinuclear | ||||||||
| 39% | 88% | 16% | 95% | 19% | 79% | 10% | 100% | |
| (31) | (67) | (15) | (56) | (16) | (68) | (6) | (66) | |
| Membrane | ||||||||
| 14% | 1% | 70% | 2% | 24% | 7% | 47% | 0% | |
| (11) | (1) | (68) | (1) | (21) | (6) | (29) | (0) | |
| Combination | ||||||||
| 47% | 11% | 14% | 3% | 57% | 14% | 43% | 0% | |
| (38) | (8) | (14) | (2) | (49) | (12) | (26) | (0) | |
( ) number of cells with observed localization
Localization of Upc2p-GFP and Ecm22p-GFP in UPC2-GFPΔecm22 and ECM22-GFPΔupc2
To further investigate the dynamic localization of Upc2p-GFP and Ecm22p-GFP outside the nucleus we created UPC2-GFPΔecm22 and ECM22-GFPΔupc2 by deleting the paralogous transcription factor in both GFP fusion strains (Table 1, rows 4 and 5). Upc2p and Ecm22p are normally expressed at low levels and to some extent are functionally redundant [4, 10]. Deletion of the paralogous untagged transcription factor improved expression of each GFP fusion protein as detected by FACS analysis and allowed us to examine the localization and expression of each fusion independently of the untagged paralog. The successful deletion of the untagged paralogs in the GFP fusion strains also indicated that both Upc2p-GFP and Ecm22p-GFP are functional fusion proteins as deletion of both UPC2 and ECM22 is synthetically lethal in this strain [14]. Azole susceptibility (Table 1, row 4 and 5) also indicated that GFP did not alter the function of Ecm22p and Upc2p.
The overall localization pattern of Ecm22p-GFP and Upc2p-GFP was similar regardless of whether or not the untagged paralog was deleted. Fluorescence was dynamically localized to intracellular membranes and to punctuate perinuclear structures in both ECM22-GFPΔupc2 (Figure 2A) and UPC2-GFPΔecm22 (Figure 2B). Ecm22p-GFP fluorescence was localized to more diffuse punctuate structures, particularly in the presence of FLC in the ECM22-GFPΔupc2 background (Figure 2A). The FLC induced differential localization we initially observed (Figure 1) was more notable in both ECM22-GFPΔupc2 and UPC2-GFPΔecm22. Ecm22p-GFP was localized to perinuclear foci in 19% of non-FLC treated cells and to 79% of FLC treated cells in ECM22-GFPΔupc2 background (Table 2). Differential localization of Upc2p-GFP in the UPC2Δecm22 background was most remarkable (Figure 2B), as evidenced by perinuclear localization of Upc2p-GFP in 10% of cells in the absence of FLC and pronounced perinuclear localization of GFP in 100% of FLC treated cells (Table 2). Perinuclear GFP was distributed in foci closely associated with the nuclear periphery in UPC2-GFPΔecm22 (Figure 2A). The fluorescence intensity of Hoechst and GFP was analyzed along a line bisecting the nucleus in 10 cells displaying close association of GFP and Hoechst. GFP had peak intensity on one or both sides of the Hoechst nuclear stain for all cells analyzed indicating that the GFP portion of the fusion proteins are not directly associated with DNA. A typical plot is shown in Figure 2C.
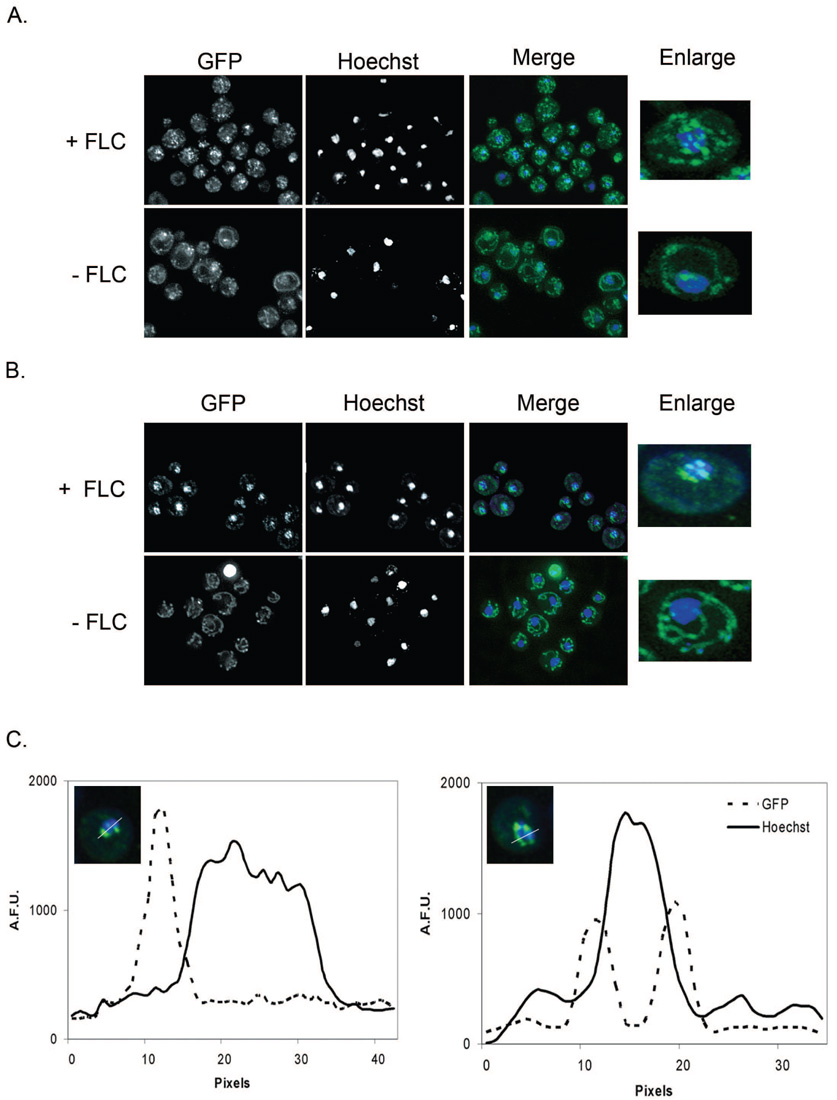
Figure 2. Upc2p-GFPΔecm22 and Ecm22p-GFPΔupc2 localization shifts from membranes to perinuclear foci when cells are exposed to FLC.
Cells were grown +/−10 µg/mL FLC for 6 hours prior to visualization by fluorescence microscopy. GFP is shown in the left panel, Hoechst nuclear stain is shown in the middle panel and a merged image is shown on the right.
A. Ecm22p-GFP localization in the strain ECM22-GFPΔupc2 (Table 1, row 5) is diffusely punctate and nuclear associated in cells treated with FLC and membranous with some cells showing single punctate foci in untreated cells.
B. Upc2p-GFP localization is punctuate and nuclear associated in cells treated with FLC and membranous with some cells showing single punctate foci in the absence of FLC treatment in the strain UPC2-GFPΔecm22 (Table 1, row 4).
C. Punctate perinuclear localization is not within the nucleus. A representative plot of Upc2p-GFP in the strain UPC2-GFPΔecm22 grown in the presence of FLC demonstrates that peak GFP fluorescence is immediately adjacent to, but not within the nucleus. Nuclear associated fluorescence was analyzed by fluorescence intensity line profile. The fluorescence intensities of the GFP (protein fusion) and Hoechst (DNA stain) channels are measured along a line bisecting the nuclei. Fluorescence intensity is plotted on the Y-axis in arbitrary fluorescence units (A.F.U.s) and pixels are plotted on the X-axis.
Colocalization of Upc2p-GFP and Ecm22p-GFP
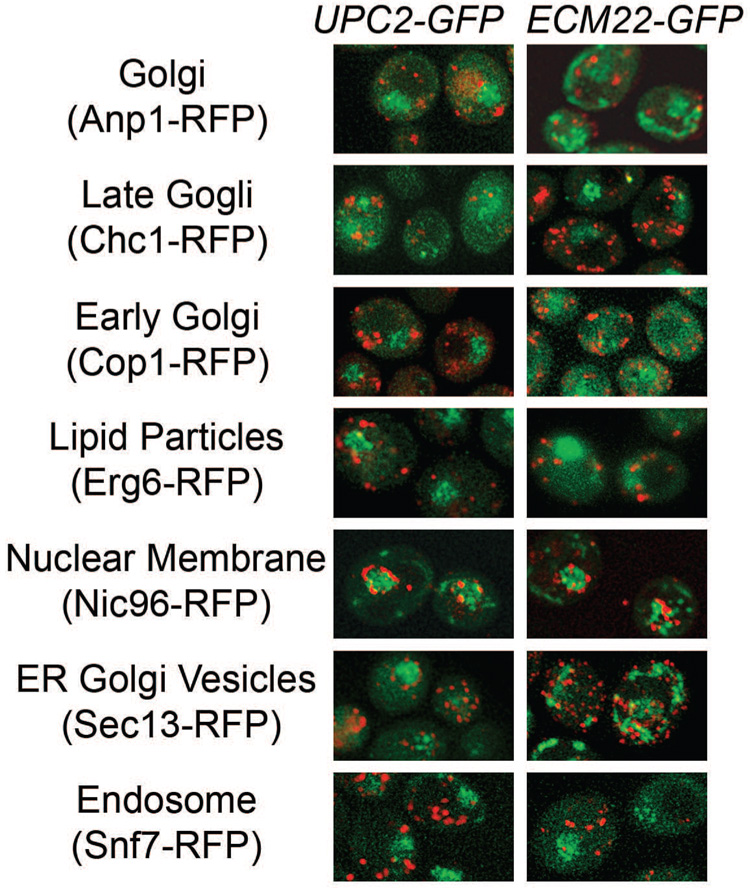
To determine the cellular location of Upc2p-GFP and Ecm22p-GFP cells were stained with vital dyes for mitochondria, vacuole, ER or lipid droplets. No colocalization was observed with any stain for either the membranous or perinuclear localization pattern regardless of the presence of the untagged paralog or FLC exposure (data not shown). To further explore colocalization, ECM22-GFPΔupc2 and UPC2-GFPΔecm22 were mated to a collection of strains expressing RFP-tagged reference proteins. Diploids expressing Upc2p-GFP or Ecm22p-GFP concurrently with one of eleven different RFP-tagged reference proteins were examined using fluorescence microscopy in the presence and absence of FLC. Diploids expressed lower GFP signal most likely due to the introduction of endogenous UPC2 and ECM22 genes from the RFP parent. Some diploid cells displayed differential localization of GFP but in the majority of cells the perinuclear pattern predominated. Perinuclear GFP did not colocalize with the nuclear membrane marker Nic96p-RFP (Figure 3) or the nucleolar marker Sik13p-RFP (data not shown). The membranous localization pattern we observed is consistent with published reference patterns for ER proteins however this localization pattern was not observed in all of the diploid strains. Dynamic localization and lower expression of GFP in diploid cells confounded colocalization; preventing definitive colocalization. Colocalization with the organelles shown in Figure 3 could not be shown but is not be ruled out.
Figure 3. Upc2p-GFP and Ecm22p-GFP do not colocalize with RFP tagged reference proteins.
Diploid cells expressing Upc2p-GFP (right) or Ecm22p-GFP (left) and RFP reference proteins were visualized by fluorescence microscopy. Merged images of RFP and GFP channels are shown. The Pearson coefficient of correlation [33] was calculated for RFP and GFP and none of the reference proteins showed colocalization with Upc2p-GFP or Ecm22p-GFP (data not shown). The genotype of the RFP-tagged cells is found in Table 1.
Azole Induction of Upc2p-GFP and Ecm22p-GFP
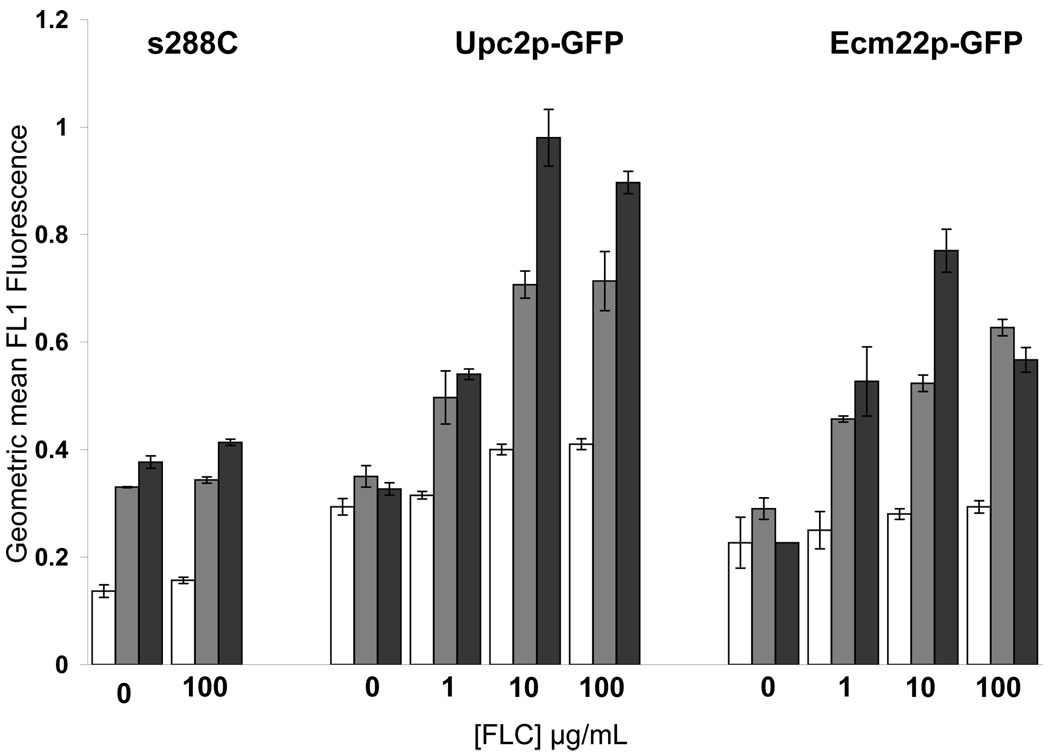
During the imaging experiments shown in Figure 1 and Figure 2 cells treated with FLC displayed a higher GFP signal relative to untreated cells presumably due to increased Upc2p-GFP and Ecm22p-GFP expression in response to sterol depletion. Therefore different exposure times were used to optimize the signal to noise ratio and capture optimal images for localization. To quantify Upc2p-GFP and Ecm22p-GFP induction in response to FLC treatment we measured GFP expression by fluorescence activated cell sorting (FACS). GFP expression was measured by FACS in strains ECM22-GFPΔupc2 and UPC2-GFPΔecm22 with and without FLC treatment over 48 hours. GFP induction correlated with FLC dose and both fusions were maximally induced after 48 hours of treatment with 10 µg/ml FLC. After 24 hours, both fusions had similar levels of GFP induction in 10 and 100 µg/ml FLC but Upc2p-GFP had a slightly higher basal level and a significantly higher level of FLC-responsive induction compared with Ecm22p-GFP. Treatment with 100 µg/ml FLC resulted in less GFP induction than treatment with 10 µg/ml FLC after 48 hours possibly due to cell death at supra-MIC FLC concentrations or decreased protein turnover. The parental stain s288C was used as a non-GFP control and had higher autofluorescence at 24 and 48 hour time points which may indicate that autofluorescence increases as cells grow.
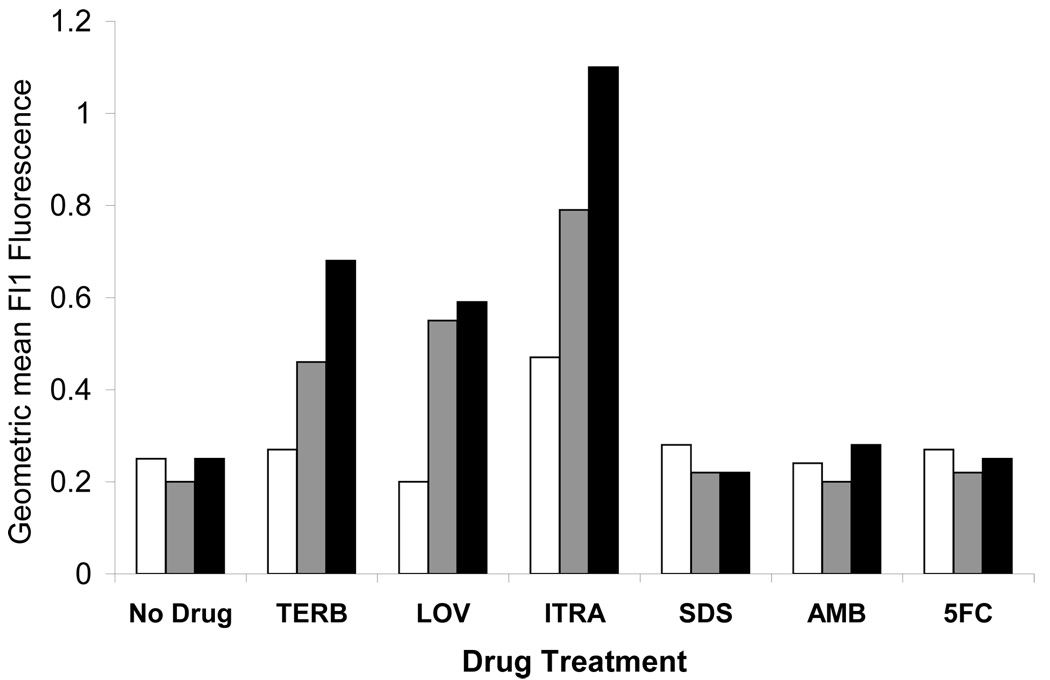
The effect of three other ergosterol biosynthesis inhibitors on Upc2p-GFP and Ecm22p-GFP induction was also measured by FACS. Lovastatin and Terbinafine target enzymes in early ergosterol biosynthesis and Itraconazole is a broad spectrum azole and with the same target as FLC. Agents with targets other than ergosterol biosynthesis were also tested to ensure that the effect was specific to inhibition of ergosterol biosynthesis. Upc2p-GFP and Ecm22p-GFP induction after 24 h of exposure to various agents was measured by FACS. The denaturing detergent SDS, the sterol binding drug amphotericin B and the nucleoside analog 5-flucytosine had no effect on expression of Ecm22p-GFP or Upc2p-GFP after 24 hours, while the drugs with targets in the ergosterol biosynthetic pathway caused Upc2p and Ecm22p induction (Figure 3B). Upc2p and Ecm22p were induced to similar levels after Lovastatin exposure but Upc2p was more highly induced by Terbinafine and Itraconazole. s288C cells had higher autofluorescence when exposed to Itraconazole as compared to the other agents tested in this experiment.
Azole Susceptibility of Upc2p-GFP and Ecm22p-GFP
To validate the functionality of Upc2p-GPF and Ecm22p-GFP both strains were compared to the parental lab strain (s288C). UPC2-GFP, ECM22-GFP, UPC2-GFPΔecm22 and ECM22-GFPΔupc2 had identical growth rates in complete synthetic media in the presence and absence of FLC(data not shown). All the strains used in this study were tested for FLC susceptibility by standard CLSI broth microdilution MIC testing [23] and commercially available FLC E-test. E-test results correlated with MIC80 results for all strains and are not shown. UPC2-GFPΔecm22 and ECM22-GFPΔupc2 were more susceptible to azoles at levels similar to the deletion of either UPC2 or ECM22 in the W303 and BY4741 backgrounds (Table 1 rows 2–10). While single deletion of either UPC2 or ECM22 causes a small increase in azole susceptibility, deletion of both genes is either lethal (s288C) or results in dramatic azole hypersusceptibility (W303) (Table 1). This is the first time the role of both transcription factors in azole susceptibility has been studied. Based on these results we concluded that azole susceptibility depends on Ecm22p and Upc2p function.
Ergosterol Content of Upc2p-GFP and Ecm22p-GFP
Total cellular ergosterol content was also measured to determine the contribution of each transcription factor to sterol synthesis in response to azole treatment. The GFP fusions used for localization studies were analyzed for ergosterol content after growth in the presence of increasing amounts of FLC (Table 3). Ergosterol levels were reduced in a FLC dose-dependent manner. Deletion of UPC2 or ECM22 in both GFP fusion backgrounds caused a significant decrease in ergosterol levels at all time points compared to the parental s288C strain. To ascertain that this decrease was due to the gene deletion and not a dysfunctional GFP fusion, the ergosterol content of the deletions in the absence of the GFP fusion were measured and shown to be equivalent at 48 hours in 0 µg/ml FLC (Table 3) This data demonstrates the importance of both UPC2 and ECM22 transcriptional activity for sterol biosynthesis. This data also validate the functionality of the GFP fusions, as ergosterol levels are depleted in response to FLC similarly to s288C.
Table 3.
Total cellular ergosterol is decreased by deletion of Upc2p or Ecm22p and by FLC treatment
| Time | 6 H | 24H | 48 H | ||||||
|---|---|---|---|---|---|---|---|---|---|
| FLC (µg/ml) | 0 | 1 | 10 | 0 | 1 | 10 | 0 | 1 | 10 |
| s288C | 100.00 | 63.84 | 49.88 | 100.00 | 66.97 | 26.12 | 100.00 | 75.05 | 39.26 |
| ECM22-GFP Δupc2 | 68.22 | 44.44 | 19.57 | 36.48 | 25.87 | 16.34 | 43.41 | 31.56 | 22.65 |
| UPC2-GFP Δecm22 | 79.02 | 70.16 | 31.32 | 45.35 | 38.42 | 8.70 | 40.46 | 32.48 | 13.85 |
| BY4741 Δecm22 | - | - | - | - | - | - | 42.86 a | - | - |
| BY4741 Δupc2 | - | - | - | - | - | - | 41.57a | - | - |
Sterol levels were determined for 48 H in the absence of drug
Discussion
Diverse cellular functions including membrane integrity, sparking of cell division and drug resistance are critically dependent on ergosterol levels. Ergosterol biosynthesis is transcriptionally regulated in response to ergosterol depletion [28] but the mechanism by which this occurs is not yet understood in the model organism S.cerevisiae. The transcription factors Upc2p and Ecm22p regulate ergosterol biosynthesis but the signals that control Upc2p and Ecm22p activation are not known. Our results argue that Upc2p and Ecm22p are post-translationally regulated in a sterol dependent manner.
The key observation that supports this hypothesis is that Upc2p-GFP and Ecm22p-GFP localization shifts from intracellular membranes to perinuculear foci upon sterol depletion (Figure 1 and 2, Table 2). This observation is consistent with the hypothesis that inactive Upc2p and Ecm22p precursors reside in intracellular membranes and sense sterol levels in the membranes directly. In this model, transmembrane localization of Upc2p and Ecm22p is perturbed by low sterol levels allowing the N-terminal transcription factor domain of the proteins to be imported to the nucleus to upregulate ergosterol biosynthesis.
Interestingly, a constitutively active allele of UPC2 designated UPC2-1 is due to glycine to aspartic acid mutation in the c-terminus [11, 29] but the mechanism by which this mutation causes constitutive activation has not been defined. The same mutation also causes hyperactivity of ECM22 [11]. For both Upc2-1p and Ecm22-1p the mutation is found directly adjacent to a predicted transmembrane domain (our unpublished observation). A possible mechanism for hyperactivity is that an aspartic acid in this position interferes with transmembrane localization causing Upc2-1p and Ecm22-1p to be continually processed and targeted to the nucleus in a sterol independent manner.
Our localization data suggest that the C-terminal transmembrane portion of Upc2p and Ecm22p remains extra nuclear (Figure 2C) while the N-terminal DNA binding domain of Upc2p and Ecm22p enters the nucleus via a NLS sequence and upregulates ERG genes by binding to the SRE. The N-terminal transcription factor domain must enter the nucleus for the GFP-fusion strains to have normal azole susceptibility (Table 1). We believe the N-terminal transcription factor domain is separated from the C-terminal transmembrane domain by proteolytic cleavage prior to nuclear import; an analogous model to SREBP regulation of cholesterol biosynthesis. If this if the case then the N-terminus of Upc2p and Ecm22p should localize to the nucleus. Preliminary attempts to create functional N-terminal GFP fusions to Upc2p and Ecm22p have not been successful, although current work in our lab suggests that Upc2p and Ecm22p are proteolytically processed.
To further test the SREBP-like activation model of Upc2p and Ecm22p, we correlated induction of Upc2p-GFP and Ecm22p-GFP with ergosterol levels in FLC treated cells. FLC treatment depletes cellular ergosterol levels within 6 hours (Table 3). FACS analysis shows that Upc2p-GFP and Ecm22p-GFP are upregulated in this time; possibly due to self regulation as both promoters contain an SRE [12, 30]. Exposure to azoles results in a decrease in cellular ergosterol levels at all time points in all strains tested which is linked to induction of Upc2p-GFP and Ecm22p-GFP as measured by FACS analysis. Deletion of Upc2p or Ecm22p also results lower ergosterol levels which is compounded by FLC treatment (Table 3). These data show that Upc2p and Ecm22p localization and induction are influenced by FLC treatment which decreases ergosterol levels of yeast cells. Additionally, azole susceptibility testing (Table 1) demonstrates that fusion of a C-terminal GFP to Upc2p and Ecm22p does not inhibit protein function.
Analysis of UPC2-GFPΔecm22 and ECM22-GFPΔupc2 showed that both Upc2p and Ecm22p are important for the cellular response to azoles, and activation of ergosterol biosynthesis. Deletion of Upc2p or Ecm22p results in 41.57% and 42.86% (respectively) less ergosterol relative to the parental strain after 48 hours of normal growth (Table 3) while deletion of both transcription factors results in much lower total ergosterol levels (unpublished observation). Deletion of either Upc2p or Ecm22p causes a 2-fold increase in azole susceptibility in both the strain backgrounds tested (Table 1) while deletion of both transcription factors is either lethal (s288C strain) or causes extreme azole hypersusceptibility (Table 1, row 11). The difference in ergosterol content, viability and azole susceptibility in single deletions relative to double deletions of Upc2p and Ecm22p suggests that these proteins are able to compensate for each other. This is further supported by data showing that Upc2p and Ecm22p bind the same conserved promoter element [4]. It will be interesting to investigate the differences in function that have selected for the genetic preservation of these highly similar proteins.
Rapid induction of sterol biosynthetic gene expression via nuclear localization of sterol regulatory transcription factors is fundamental for controlled sterol synthesis in mammals and other fungi. We show here that Upc2p and Ecm22p are clearly localized outside of the nucleus (Figure 1 and Figure 2) and in conditions of sterol depletion localization shifts toward the nucleus. Regulated nuclear import of transcriptional regulators is vital for other cellular signaling pathways such as NF-AT in mammalian cells [31] and Crz1p a zinc cluster transcription factor of S.cerevisiae [32]. Similar mechanisms of activation have been suggested for other zinc cluster transcription factors in S.cerevisiae and several other zinc cluster transcription factors are localized in the cytoplasm [7].
Upc2p and Ecm22p do not have direct sequence homology to SREBPs but are functionally related to SREBPs with an N-terminal transcription factor domain and a C-terminal transmembrane domain. The S.pombe SREBP also has relatively low sequence homology to mammalian SREBPs but has been shown to be a functional homolog. Functional analogy is not surprising as it has long been known that ergosterol biosynthetic genes of yeast are transcriptionally regulated in response to ergosterol depletion [28]. Fungal and mammalian SREBPs are a possible case of convergent evolution because regulation of sterol biosynthesis in response to sterol levels is important for cellular integrity and growth. Studies to determine how Upc2p and Ecm22p are proteolytically processed or otherwise modified are currently underway. The C.neoformans SREBP is cleaved by a site 2 protease but a site 1 protease or Scap homologue have not been identified [16, 18]. Scap and Insig homologues regulate sterol sensing in S.pombe [20] but homology searches do not reveal candidate homologues of Scap, Insig or Site-1 and 2 proteases in S.cerevisiae. Unique functional homologues in the sterol regulatory pathway may exist in S.cerevisiae and other fungi that remain to be identified. We are currently using affinity purification to identify other proteins that interact with Upc2p and Ecm22p to regulate ergosterol homeostasis.
Figure 4. Upc2p-GFP and Ecm22p are induced in response to ergosterol biosynthesis inhibitors.
A. Upc2p-GFP and Ecm22p-GFP expression was measured by FACS after 6 H (white bars), 24 H (grey bars) and 48 H (black bars) of growth in increasing FLC concentrations. A wild type strain (s288C) was used as a control. Upc2p and Ecm22p expression was induced by FLC and increased over time.
B. Upc2p-GFP (black bars) and Ecm22p-GFP (gray bars) levels were measured by FACS after cells were treated with ergosterol biosynthesis inhibitors for 24 hours. s288C (white bars) was used as a no GFP control. Drugs that target ergosterol biosynthesis (terbinafine, lovastatin and itraconazole) increased expression of GFP fusion proteins, while drugs with targets outside the ergosterol biosynthetic pathway (SDS, amphotercin B and 5-FC) did not alter Upc2p-GFP and Ecm22p expression.
Acknowledgements
This research was funded by NIH NIDCR RO1 grants DE-11367, DE 14161 and DE17078. We thank Amy DeRocher and Tracy Mitchell (Seattle Biomedical Research Institute, Seattle, Washington) for assistance with microscopy and image analysis. We thank Klaus Natter (University of Graz, Austria) for providing the episomal Upc2p-GFP construct. We thank Felix Lam and the Erin O’Shea Lab (Harvard University, Cambridge, Massachusetts) for the RFP-tagged reference proteins. We thank members of the White laboratory and Amy DeRocher for their valuable comments and support.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Fluckiger U, et al. Treatment options of invasive fungal infections in adults. Swiss Med Wkly. 2006;136(29–30):447–463. doi: 10.4414/smw.2006.11392. [DOI] [PubMed] [Google Scholar]
- 2.Parks LW, Casey WM. Physiological implications of sterol biosynthesis in yeast. Annu Rev Microbiol. 1995;49:95–116. doi: 10.1146/annurev.mi.49.100195.000523. [DOI] [PubMed] [Google Scholar]
- 3.Henneberry AL, Sturley SL. Sterol homeostasis in the budding yeast, Saccharomyces cerevisiae. Semin Cell Dev Biol. 2005;16(2):155–161. doi: 10.1016/j.semcdb.2005.01.006. [DOI] [PubMed] [Google Scholar]
- 4.Vik A, Rine J. Upc2p and Ecm22p, dual regulators of sterol biosynthesis in Saccharomyces cerevisiae. Mol Cell Biol. 2001;21(19):6395–6405. doi: 10.1128/MCB.21.19.6395-6405.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.White TC, Silver PM. Regulation of sterol metabolism in Candida albicans by the UPC2 gene. Biochem Soc Trans. 2005;33(Pt 5):1215–1218. doi: 10.1042/BST20051215. [DOI] [PubMed] [Google Scholar]
- 6.Song JL, et al. The Candida albicans lanosterol 14-alpha-demethylase (ERG11) gene promoter is maximally induced after prolonged growth with antifungal drugs. Antimicrob Agents Chemother. 2004;48(4):1136–1144. doi: 10.1128/AAC.48.4.1136-1144.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.MacPherson S, Larochelle M, Turcotte B. A fungal family of transcriptional regulators: the zinc cluster proteins. Microbiol Mol Biol Rev. 2006;70(3):583–604. doi: 10.1128/MMBR.00015-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Todd RB, Andrianopoulos A. Evolution of a fungal regulatory gene family: the Zn(II)2Cys6 binuclear cluster DNA binding motif. Fungal Genet Biol. 1997;21(3):388–405. doi: 10.1006/fgbi.1997.0993. [DOI] [PubMed] [Google Scholar]
- 9.Germann M, et al. Characterizing sterol defect suppressors uncovers a novel transcriptional signaling pathway regulating zymosterol biosynthesis. J Biol Chem. 2005;280(43):35904–35913. doi: 10.1074/jbc.M504978200. [DOI] [PubMed] [Google Scholar]
- 10.Davies BS, Wang HS, Rine J. Dual activators of the sterol biosynthetic pathway of Saccharomyces cerevisiae: similar activation/regulatory domains but different response mechanisms. Mol Cell Biol. 2005;25(16):7375–7385. doi: 10.1128/MCB.25.16.7375-7385.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Shianna KV, et al. Identification of a UPC2 homolog in Saccharomyces cerevisiae and its involvement in aerobic sterol uptake. J Bacteriol. 2001;183(3):830–834. doi: 10.1128/JB.183.3.830-834.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Davies BS, Rine J. A role for sterol levels in oxygen sensing in Saccharomyces cerevisiae. Genetics. 2006;174(1):191–201. doi: 10.1534/genetics.106.059964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kwast KE, Burke PV, Poyton RO. Oxygen sensing and the transcriptional regulation of oxygen-responsive genes in yeast. J Exp Biol. 1998;201(Pt 8):1177–1195. doi: 10.1242/jeb.201.8.1177. [DOI] [PubMed] [Google Scholar]
- 14.Valachovic M, et al. Cumulative mutations affecting sterol biosynthesis in the yeast Saccharomyces cerevisiae result in synthetic lethality that is suppressed by alterations in sphingolipid profiles. Genetics. 2006;173(4):1893–1908. doi: 10.1534/genetics.105.053025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Chan CK, et al. Mutual exclusivity of DNA binding and nuclear localization signal recognition by the yeast transcription factor GAL4: implications for nonviral DNA delivery. Gene Ther. 1998;5(9):1204–1212. doi: 10.1038/sj.gt.3300708. [DOI] [PubMed] [Google Scholar]
- 16.Chang YC, et al. Sre1p, a regulator of oxygen sensing and sterol homeostasis, is required for virulence in Cryptococcus neoformans. Mol Microbiol. 2007;64(3):614–629. doi: 10.1111/j.1365-2958.2007.05676.x. [DOI] [PubMed] [Google Scholar]
- 17.Hughes AL, Todd BL, Espenshade PJ. SREBP pathway responds to sterols and functions as an oxygen sensor in fission yeast. Cell. 2005;120(6):831–842. doi: 10.1016/j.cell.2005.01.012. [DOI] [PubMed] [Google Scholar]
- 18.Chun CD, Liu OW, Madhani HD. A link between virulence and homeostatic responses to hypoxia during infection by the human fungal pathogen Cryptococcus neoformans. PLoS Pathog. 2007;3(2):e22. doi: 10.1371/journal.ppat.0030022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lee H, et al. Cobalt chloride, a hypoxia-mimicking agent, targets sterol synthesis in the pathogenic fungus Cryptococcus neoformans. Mol Microbiol. 2007;65(4):1018–1033. doi: 10.1111/j.1365-2958.2007.05844.x. [DOI] [PubMed] [Google Scholar]
- 20.Espenshade PJ, Hughes AL. Regulation of sterol synthesis in eukaryotes. Annu Rev Genet. 2007;41:401–427. doi: 10.1146/annurev.genet.41.110306.130315. [DOI] [PubMed] [Google Scholar]
- 21.Adams AKC, Gottschling D. Methods in Yeast Genetics. Cold Springs Harbor: Cold Springs Harbor Laboratory Press; 1995. [Google Scholar]
- 22.Maniatis T, Fritsch EF, Sambrook J. Molecular Cloning: A Laboratory Manual. Cold Spring Harbor, N.Y.: Cold Spring Harbor Laboratory Press; 1982. [Google Scholar]
- 23.Standards NCfCL. Reference method for broth dilution antifungal susceptibliityy testing of yeasts: approved standard M27-A2. 2nd edition. Wayne, PA: Clincial Laboratory Standards Institute; 2002. [Google Scholar]
- 24.Arthington-Skaggs BA, et al. Quantitation of ergosterol content: novel method for determination of fluconazole susceptibility of Candida albicans. J Clin Microbiol. 1999;37(10):3332–3337. doi: 10.1128/jcm.37.10.3332-3337.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Guide to Yeast Genetics and Molecular Biology. In: Guthrie CaF, GR, editors; Abelson JaSM., editor. MEthods in Enzymology. Vol. 194. San Diego: Academic Press, Inc.; 1991. p. 933. [Google Scholar]
- 26.Habeler G, et al. YPL.db: the Yeast Protein Localization database. Nucleic Acids Res. 2002;30(1):80–83. doi: 10.1093/nar/30.1.80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Huh WK, et al. Global analysis of protein localization in budding yeast. Nature. 2003;425(6959):686–691. doi: 10.1038/nature02026. [DOI] [PubMed] [Google Scholar]
- 28.Smith SJ, Crowley JH, Parks LW. Transcriptional regulation by ergosterol in the yeast Saccharomyces cerevisiae. Mol Cell Biol. 1996;16(10):5427–5432. doi: 10.1128/mcb.16.10.5427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Crowley JH, et al. A mutation in a purported regulatory gene affects control of sterol uptake in Saccharomyces cerevisiae. J Bacteriol. 1998;180(16):4177–4183. doi: 10.1128/jb.180.16.4177-4183.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Abramova NE, et al. Regulatory mechanisms controlling expression of the DAN/TIR mannoprotein genes during anaerobic remodeling of the cell wall in Saccharomyces cerevisiae. Genetics. 2001;157(3):1169–1177. doi: 10.1093/genetics/157.3.1169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Beals CR, et al. Nuclear localization of NF-ATc by a calcineurin-dependent, cyclosporin-sensitive intramolecular interaction. Genes Dev. 1997;11(7):824–834. doi: 10.1101/gad.11.7.824. [DOI] [PubMed] [Google Scholar]
- 32.Stathopoulos-Gerontides A, Guo JJ, Cyert MS. Yeast calcineurin regulates nuclear localization of the Crz1p transcription factor through dephosphorylation. Genes Dev. 1999;13(7):798–803. doi: 10.1101/gad.13.7.798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Bolte S, Cordelieres FP. A guided tour into subcellular colocalization analysis in light microscopy. J Microsc. 2006;224(Pt 3):213–232. doi: 10.1111/j.1365-2818.2006.01706.x. [DOI] [PubMed] [Google Scholar]