Abstract
P58/DNAJc3 defends cells against endoplasmic reticulum (ER) stress. Most P58 molecules are translocated into the ER lumen, and here we report selective and stable binding to misfolded proteins by P58's TPR-containing N-terminal domain. In vitro, too, P58 binds selectively to a model misfolded protein and challenge of that complex with physiological concentrations of the ER lumenal Hsp70-type chaperone BiP encourages disassembly. BiP-induced dissociation of P58 from its substrate depends on the presence of ATP and on interactions with P58's J-domain, which are mediated by invariant residues BiPR197 and P58H422. A functional J-domain also accelerates dissociation of P58 from a model substrate, VSV-Gts045, on the latter's re-folding in vivo. However, J-domain binding can be separated from the ability to promote substrate dissociation by the mutant BiPE201G and a wild-type J-domain fused ectopically to P58H422Q rescues the latter's inability to dissociate from substrate in response to BiP and ATP. These findings are consistent with a model whereby localized activation of the Hsp70-type partner is sufficient to promote substrate handover from the J-domain co-chaperone.
Keywords: chaperones, endoplasmic reticulum, protein folding, translation
Introduction
The high flux of newly synthesized (unfolded) proteins through the endoplasmic reticulum (ER) of secretory cells renders the latter especially dependent on chaperones that function in the ER. For this reason, secretory cells have heightened activity of the ER unfolded protein response (UPR), which couples the stress imposed by the load of unfolded proteins in the lumen of the organelle (ER stress) to activation of genes that encode chaperones that function in the organelle (Schroder and Kaufman, 2005; Bernales et al, 2006).
Surprisingly, one of the genes consistently upregulated by the mammalian UPR was predicted to encode a cytoplasmic protein, P58, which was initially identified as an inhibitor of the cytoplasmically localized kinases PKR (Barber et al, 1994) and PERK (Yan et al, 2002; Van Huizen et al, 2003). PERK mediates an adaptive response to ER stress that limits the influx of unfolded proteins into the lumen of the stressed organelle (Ron and Harding, 2007); however, a modulatory function for P58 in the UPR could not readily explain the phenotype of P58 loss of function. In both worms and mice, loss of P58 sensitizes the animals to the consequences of protein misfolding in the ER lumen (Kapulkin et al, 2005; Ladiges et al, 2005), which is the opposite phenotype of that predicted from the loss of an inhibitor of PERK. Experiments conducted in an effort to explain the P58 knockout phenotype supported a model whereby P58 functions at the cytosolic face of the translocon (Oyadomari et al, 2006). However, new findings (Rutkowski et al, 2007, confirmed here) place P58 predominantly in the lumen of the ER, and therefore suggest a more direct function for the co-chaperone in buffering the load of unfolded proteins in the ER lumen.
Molecular chaperones form transient complexes with hydrophobic residues exposed on extended segments of non-native substrate proteins. P58 (also known as DnaJc3) contains a J-domain found in a large class of co-chaperones that function alongside the Hsp70 family of chaperones. The ability of Hsp70 to bind substrates is tightly regulated by a nucleotide-binding domain that couples ATPase activity to conformational changes that affect the substrate-binding domain (Szabo et al, 1994). In the ATP-bound state, a substrate-binding groove is accessible and substrates associate and dissociate with high ‘on/off' rates, whereas the ADP-bound form stably associates with substrates (Schmid et al, 1994). J-domain co-chaperones (also called Hsp40s) consist of a diverse family of proteins that share a conserved region of ∼70 amino acids, the J-domain, which mediates interactions with partner Hsp70s and markedly stimulates their intrinsically slow rates of ATP hydrolysis (Liberek et al, 1991). A conserved motif consisting of His-Pro-Asp (HPD) found in the J-domain is essential for surface interactions with invariant residues on Hsp70, and mutations of the motif abrogate stimulation of Hsp70 ATPase activity (Tsai and Douglas, 1996; Suh et al, 1998; Jiang et al, 2007).
Several J-domain co-chaperones have been found to bind hydrophobic residue-rich extended segments of unfolded proteins through a separate substrate-interacting domain (Sha et al, 2000; Rudiger et al, 2001; Li et al, 2003) and they are thought to subsequently recruit their Hsp70 partners to that substrate through their J-domain. The current model of sequential binding (first by the co-chaperone and then handover of substrate to Hsp70) is supported by the finding that the J-domain's affinity for Hsp70 is markedly enhanced by the presence of ATP in the latter's nucleotide-binding domain (Wawrzynow and Zylicz, 1995). Substrate transfer from the co-chaperone to the chaperone presumably involves J-domain-induced hydrolysis of ATP, and its conversion to ADP, which remains bound. The transfer of substrate is also favoured by the fact that its engagement at the substrate-binding domain of Hsp70 contributes to ATP hydrolysis at the nucleotide-binding domain (Flynn et al, 1989; Laufen et al, 1999) and the re-cycling of the J-domain co-chaperone is presumably favoured by its disengagement from the ADP-bound Hsp70. However, the function, if any, of the Hsp70 partner in facilitating substrate release from the J-domain co-chaperone has not been addressed. This paper explores the interactions between lumenal P58, its substrates and Hsp70 partner, and exploits the surprising stability of the complex between P58 and its misfolded substrate to address the mechanism of substrate release.
Results
Selective binding of misfolded lumenal proteins by P58
Consistent with the recent publication of Rutkowski et al (2007), we also found that the conserved hydrophobic sequence at P58's N terminus efficiently directs translocation of the protein into the ER lumen and that in vitro translated radiolabelled mouse P58 migrated more slowly on SDS–PAGE than endogenous P58 immunopurified from metabolically labelled ER-stressed mouse fibroblasts or from 293T cells transfected with an expression plasmid for the mouse protein (Supplementary Figure S1). These observations are consistent with the presence of a cleavable N-terminal signal sequence in P58 that directs most of the P58 synthesized in cells into the ER lumen.
P58's localization to the ER lumen suggests that its ability to ameliorate the consequences of ER stress might stem from interactions with misfolded lumenal ER proteins. To explore this possibility, Chinese hamster ovary (CHO) cells were stably transduced with a gene encoding mouse P58 modified to contain a cleavable signal sequence followed by an eight-residue FLAG epitope tag that is recognized by the FLAG–M1 monoclonal antibody only in the context of the protein's N terminus (which is exposed only after cleavage of the signal peptide; Supplementary Figure S2). Immunoprecipitation with the FLAG–M1 antibody thus confines the analysis to lumenal P58 and also overcomes the challenge posed by the fact that none of the antibodies or antisera to P58 in our possession immunoprecipitates the protein under native conditions.
FLAG–P58 recovered by immunopurification from CHO cells was stably associated with a co-expressed misfolding-prone fusion between mutant insulin (INS2C96Y) and green fluorescent protein (GFP) but not with the more abundant co-expressed cytosolic GFP (Supplementary Figure S3). To explore in more detail this suggestion of an interaction of P58 with misfolded proteins, we turned to a well-characterized ER substrate, the folded state of which can be reversibly manipulated by temperature shift of expressing cells. The ts045 mutation in the extracellular/lumenal portion of the vesicular stomatitis virus envelope glycoprotein (VSV-Gts045) causes misfolding and ER retention at the non-permissive temperature of 41°C but is compatible with normal folding and ER exit at the permissive temperature of 32°C (Kreis and Lodish, 1986).
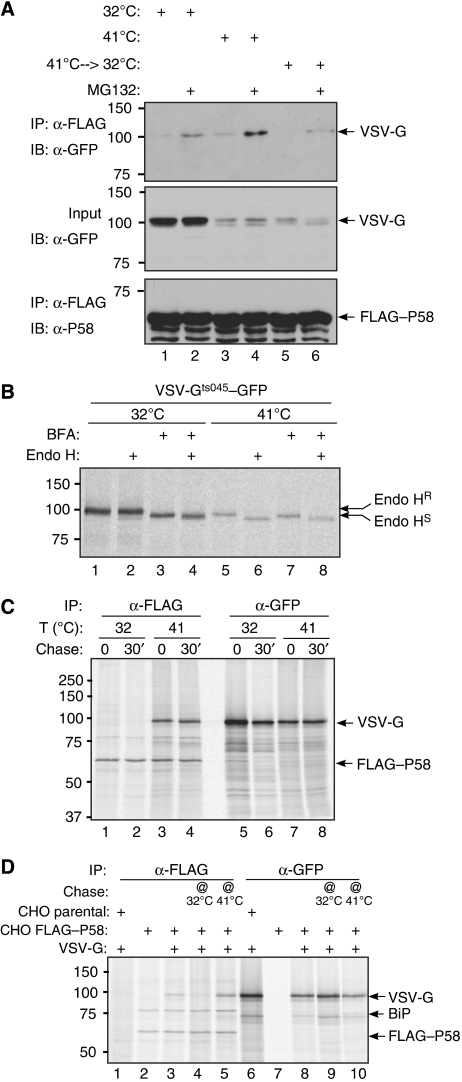
VSV-Gts045, tagged at its cytoplasmic portion by GFP, was expressed transiently in the CHO cells stably expressing FLAG–P58. Immunoblotting of FLAG–M1 immunoprecipitates with a GFP antiserum (that detects VSV-Gts045–GFP) revealed a small amount of VSV-Gts045–GFP in complex with P58 at steady state in all culture conditions (Figure 1A), but this increased by culture at the non-permissive temperature, especially in the presence of the proteasome inhibitor MG132 (Figure 1A, lane 4), which presumably stabilizes VSV-Gts045–GFP (whose half-life in the FLAG–P58 cells is rather short, ∼0.5 h; Supplementary Figure S4). Significantly, shift of cells cultured at the non-permissive temperature back to the permissive temperature led to dissociation of much of the complex within 1 h (Figure 1A, compare lanes 4 and 6).
Figure 1.
P58 selectively associates with misfolded VSV-G in vivo. (A) Immunoblot of GFP-tagged VSV-Gts045 recovered in complex with FLAG–P58 from lysates of FLAG–P58-expressing CHO cells that had been maintained at the permissive temperature (32°C) or the non-permissive temperature (41°C) for 12 h before harvest or switched back to the permissive temperature for the last hour before harvest (41–>32°C). Where indicated, the cells were exposed to the proteasome inhibitor MG132 for 3 h before harvest. The middle panel reports on the amount of VSV-Gts045–GFP in the cell lysates (1.5% of the input) and the lower panel on the recovery of FLAG–P58 in the immunoprecipitate. (B) Autoradiograph of immunopurified VSV-Gts045–GFP from transfected CHO cells metabolically labelled for 30 min at the permissive (32°C) or non-permissive temperature (41°C) in the absence or presence of brefeldin A (BFA). Where indicated, the immunopurified proteins were digested by an endoglycosidase that does not react with Golgi-modified N-linked saccharides (Endo H). The slower migrating Endo H-resistant (Endo HR) and faster migrating Endo H-sensitive (Endo HS) forms of VSV-G–GFP are indicated. (C) Autoradiograph of proteins immunopurified from CHO cells co-expressing FLAG–P58 and VSV-Gts045–GFP. The cells were briefly radiolabelled (30 min) in the presence of BFA at the permissive (32°C) or non-permissive temperature (41°C) and where indicated, chased with unlabelled media for 30 min in the continued presence of BFA, before lysis and immunoprecipitation with anti-FLAG antibody or anti-GFP serum. (D) Same as (C) except that cells were pulse labelled in the presence of BFA at the non-permissive temperature (41°C) and a cold chase was performed for 1 h at the permissive (32°C, lane 4) or non-permissive temperature (41°C, lane 5) in the continued presence of BFA. The lysate of cells lacking FLAG–P58 (CHO parental, lanes 1 and 6) serves as a specificity control for the recovery of VSV-Gts045–GFP in complex with FLAG–P58.
These experiments suggest a dynamic association of P58 and misfolded VSV-G, which could be direct or indirect. Furthermore, as folding (at the permissive temperature) leads to exit from the ER, the above experiment does not tell us whether P58 discriminates between ER-localized folded and misfolded VSV-G. To uncouple folding from ER exit, we used the drug brefeldin A (BFA), which blocks exit of all proteins (folded and unfolded) from the ER (Misumi et al, 1986). To confirm retention of VSV-Gts045–GFP in the ER of cells exposed to BFA, the protein was immunoprecipitated with the anti-GFP serum from cells that were metabolically labelled at the permissive or non-permissive temperature in the absence or presence of BFA. To monitor ER exit, the immunopurified proteins were subjected to endoglycosidase H (Endo H), an enzyme that selectively removes N-linked glycans that had not undergone modification by Golgi enzymes (Trimble et al, 1978). VSV-Gts045–GFP synthesized at the permissive temperature (in the absence of BFA) is mostly Endo H resistant and migrates slowly on SDS–PAGE; the slow mobility presumably reflects further Golgi modifications of N-linked glycans. Inhibition of ER exit by BFA sensitizes the protein to Endo H and enhances its mobility on SDS–PAGE (Figure 1B, lanes 1–4). By contrast, VSV-Gts045–GFP synthesized at the non-permissive temperature is mostly Endo H sensitive and addition of BFA effects a slight (paradoxical) increase in the fraction of the protein that is Endo H resistant; presumably by promoting exposure of misfolded, ER-localized VSV-Gts045–GFP to Golgi-modifying enzymes from the collapsed Golgi apparatus (in the BFA-treated cells) (Figure 1B, lanes 5–8).
Having unlinked VSV-Gts045–GFP folding from ER exit it was then possible to inquire as to the role of VSV-G's misfolding in its association with P58. FLAG–P58 CHO cells transiently expressing VSV-Gts045–GFP were metabolically labelled at the permissive or non-permissive temperature in the presence of BFA, and radiolabelled proteins in complex with P58 (immunopurified with FLAG–M1) were revealed by SDS–PAGE and autoradiography. Labelled VSV-Gts045–GFP was recovered in complex with P58 only in cells labelled at the non-permissive temperature (Figure 1C). The effectiveness of BFA in retaining the correctly folded VSV-Gts045–GFP in the ER (at the permissive temperature) is revealed by the identical mobility of the protein extracted from cells labelled (and chased) at the permissive and non-permissive temperature (lanes 5–8).
To expand on these observations, we metabolically labelled cells at the non-permissive temperature and then either left them at the non-permissive or shifted them to the permissive temperature during a non-labelled (‘cold') chase period. The complex of P58 and VSV-Gts045–GFP, which is stable for 1 h of chase at the non-permissive temperature, dissociated over a similar period of chase at the permissive temperature (Figure 1D, compare lanes 4 and 5). These experiments reveal that P58 selectively associates with misfolded VSV-G and that the latter is released from the complex on folding.
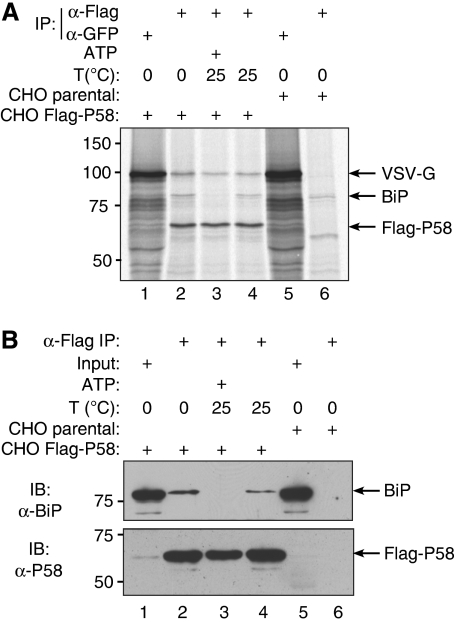
A 78-kDa protein, which is likely to be the abundant ER chaperone BiP, is a reproducible constituent of the P58-containing complexes (Figures 1C and D, and 2; Rutkowski et al, 2007). To determine whether BiP mediates association of VSV-Gts045–GFP with P58, we exposed FLAG–P58 immunopurified complexes (from cells metabolically labelled at the non-permissive temperature) to ATP and Mg2+. As predicted, this led to dissociation of BiP from the complex but had only a minor effect on the content of misfolded VSV-G (Figure 2A and B, compare lanes 3 and 4 in each).
Figure 2.
The complex between P58 and misfolded VSV-G is independent of BiP. (A) Autoradiograph of proteins immunopurified from CHO cells expressing VSV-Gts045–GFP and FLAG–P58 (lanes 1–4) or control, parental CHO cells (lanes 5 and 6), metabolically labelled at the non-permissive temperature. Where indicated, the immunopurified FLAG–P58-containing complex was challenged by incubation at room temperature (25°C) in the presence or absence of ATP (to dissociate bound BiP). The VSV-Gts045–GFP, BiP and FLAG–P58 that remained associated with the immune complex are revealed by autoradiography. (B) BiP and FLAG immunoblot of material prepared as in (A), except that lane 1 contains 1% of the input of the lysate used in lanes 2–4.
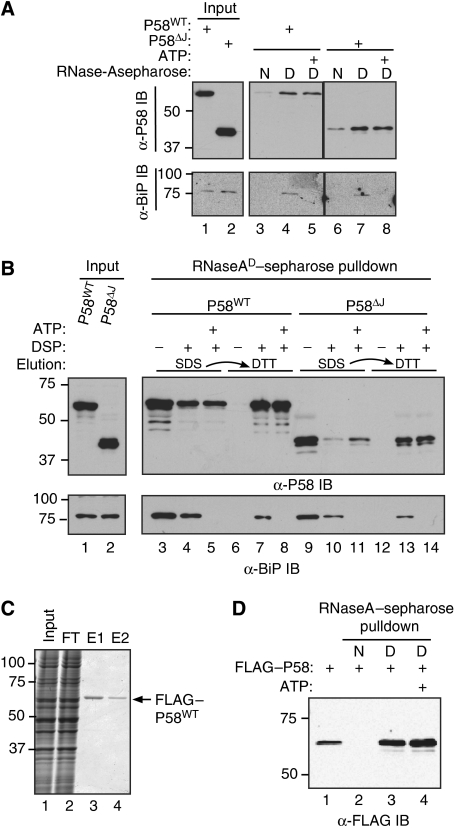
BiP-independent association of P58 with misfolded VSV-Gts045–GFP suggests the possibility of direct binding of misfolded proteins by P58. To explore this possibility, we exposed lysates from mammalian cells expressing P58 to a sepharose resin containing covalently coupled native bovine RNase A or an identical resin on which the RNase A had been reduced and denatured (see Materials and methods section). More P58 was retained on the resin containing denatured RNase A than on the resin containing native RNase A; denaturation of some RNase A molecules during the crosslinking procedure might contribute to the residual binding of P58 to the native resin (Figure 3A, lanes 3 and 6). Endogenous BiP from the cell lysates associated selectively with the denatured bait, but unlike P58, dissociated from the resin in the presence of ATP and Mg2+. This experiment indicates that P58 distinguishes between a folded and misfolded version of the same protein. A C-terminal truncated P58 lacking its J-domain also bound selectively to the denatured protein, indicating that the J-domain is dispensable to selective binding and the TPR motif-containing N-terminus of P58 is sufficient for interaction with substrate (Figure 3A, lanes 6–8).
Figure 3.
Direct binding of P58 to misfolded RNase A in vitro. (A) Immunoblots of full-length FLAG–P58 (P58WT) and C-terminally truncated FLAG–P58 (P58ΔJ) from lysates of transfected 293T cells associated with an affinity resin consisting of native (N) or denatured (D) RNase A coupled covalently to sepharose beads. The resin was reacted with cell lysates, washed, followed by incubation in the presence of ATP where indicated, further washed and the remaining bound material was eluted and revealed by SDS–PAGE. Lanes 1 and 2 report on the input of the binding reactions. The lower panel (a BiP immunoblot) reports on the association of endogenous BiP with the resin. (B) As in (A), except that the crosslinker DSP was added to the assembled complex followed by sequential elution (indicated by the arrow), first with SDS (to recover non-covalently bound proteins) and then with DTT (to reverse the crosslink and recover covalently bound proteins). Where indicated, the pre-formed complex was reacted with ATP and washed extensively before adding cross linker. (C) Coomassie-stained gel of wild-type FLAG–P58 purified from stably expressing CHO cells by FLAG immuno-affinity chromatography and elution by FLAG peptide competition. The crude lysate (input), the column flow-through (FT) and the first and second elution by FLAG peptide competition elution are shown (E1 and E2). (D) Immunoblot of the purified FLAG–P58 (shown in (C)) that associated with the native (N) and denatured (D) RNase A affinity resin. Where indicated, the complex was challenged with ATP. Lane 1 shows 5% of the input used in the other lanes.
To gain further insight into the interaction between P58 and misfolded proteins, we measured the ability of a crosslinking reagent to covalently link P58 and denatured RNase A. A complex between P58 from mammalian cell lysates and the denatured RNase A-containing affinity resin was exposed to DSP, a crosslinker with a 12-Å spacer arm containing a disulphide bond that can be cleaved under reducing conditions. Non-covalently bound P58 was readily eluted by exposing the resin-bound complex to 4% SDS. P58 that remained associated with the resin by the DSP-induced crosslinks was next eluted with 100 mM DTT. Addition of crosslinker reduced the amount of P58 recovered in the first elution (with 4% SDS, which only disrupts non-covalent interactions, Figure 3B, compare lanes 3 and 4) and led to the retention of P58 as a covalently bound complex with denatured RNase A that was recovered in the second elution (with DTT, which cleaves the crosslinker; Figure 3B, compare lanes 6 and 7). Addition of ATP and Mg2+ to the pre-formed complex (and washing the eluted proteins away before adding crosslinker) did not affect the amount of P58 crosslinked to the misfolded RNase A resin, but, as expected, led to BiP's dissociation from the resin and markedly diminished the amount of BiP that was crosslinked (lower panel, compare lanes 4, 5 and 7, 8). P58 lacking the J-domain also bound and crosslinked to the denatured RNase A (lanes 9–14). To determine whether other components in the mammalian cell lysate that were retained on the resin contribute to the specificity of this interaction, we first purified FLAG–P58 from CHO cell lysates by binding it in high salt to a FLAG affinity resin and eluted the bound FLAG–P58 by FLAG peptide competition (Figure 3C). The purified FLAG–P58 also exhibited preference for binding denatured over native RNase A and binding was indifferent to ATP and Mg2+ (Figure 3D).
BiP- and ATP-dependent dissociation of P58 from substrates
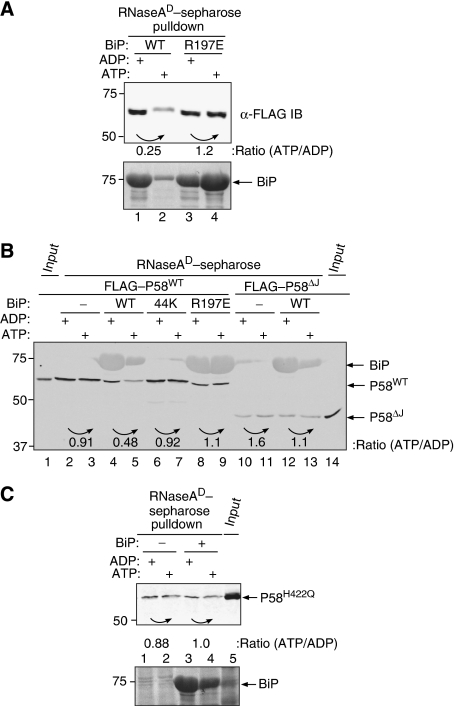
The above observations are consistent with direct binding of P58 to misfolded proteins and suggest that the N-terminal portion of the co-chaperone mediates such binding. However, the presence of a functional J-domain in P58 and physical evidence for its association with BiP (Rutkowski et al, 2007) suggest that the two proteins might cooperate in coping with misfolded proteins. Addition of bacterially expressed P58 to BiP did not enhance the latter's binding to misfolded proteins in vitro (data not shown). However, we noted that challenge of pre-existing complexes of purified FLAG–P58 and denatured RNase A with purified bacterially expressed BiP led to substantial dissociation of P58 from the complex in the presence of ATP and Mg2+ (Figure 4A, lanes 1 and 2). The effect was dependent on a functional interaction with the J-domain and was abolished by a mutation BiPR197E (Awad et al, 2008), in an invariant arginine known to mediate interactions of Hsp70 chaperones with their J-domain co-chaperones (Suh et al, 1998; Jiang et al, 2007) (Figure 4A, lanes 3 and 4). The enhanced apparent mobility of the residual FLAG–P58 (Figure 4A, lanes 1, 3 and 4) reflects a reproducible SDS–PAGE compression artefact caused by large amounts of BiP bound to the resin, whereas the enhanced binding of BiPR197E to the resin in the presence of ATP and Mg2+ (Figure 4A, lane 4) is a reproducible finding explained by the effect of the mutation on the functional coupling of nucleotide and substrate binding by BiP (Awad et al, 2008). BiP-dependent dissociation was similarly observed when the complex between P58 and denatured RNase A was formed in a crude lysate of cells expressing the various proteins, circumventing the need to extensively purify P58 (Figure 4B, lanes 4 and 5). In this simple assay, the effect of ATP and Mg2+ was dependent on the concentration of BiP (Supplementary Figure S5) and was abolished both by truncation of BiP's peptide-binding domain (BiP-44K) and by BiPR197E (Figure 4B, lanes 6–9).
Figure 4.
Addition of BiP and ATP dissociates P58 from misfolded RNase A. (A) Immunoblot of purified FLAG–P58 (shown in Figure 3C and D) associated with denatured RNase A. Where indicated, the pre-formed complex was challenged with 9 μM bacterially expressed wild-type BiP (WT) or mutant BiPR197E in the presence of ADP or ATP. The ratio of P58 retained on the resin was measured by the LI-COR IR imaging system and is indicated for each experimental pair (under the curved arrow). The bound BiP was revealed by staining the blot with Ponceau S (lower panel). (B) As in (A) but using wild-type FLAG–P58 (WT) and a C-terminal truncation mutant FLAG–P58 lacking the J-domain (ΔJ) from mammalian cell lysates associated with denatured RNase A. Where indicated, the pre-formed complex was challenged with 9 μM wild-type BiP (WT), BiP lacking its C-terminal substrate-binding domain (44K) and mutant BiPR197E. The BiP that remained bound to the resin is revealed in this experiment as a residual fluorescent signal on the P58 immunoblot. (C) Same as (B), but using mammalian cell lysates expressing mutant FLAG–P58H422Q.
A role for J-domain interaction with BiP in mediating this ATP/Mg2+-dependent dissociation is further supported by the finding that the association of C-terminally truncated P58 (P58ΔJ) with denatured RNase A was unaffected by the addition of BiP and ATP/Mg2+ (Figure 4B, lanes 10–13). This conclusion is also supported by the finding that a mutation, P58H422Q, in an invariant histidine that mediates J-domain interactions with Hsp70 chaperones (Suh et al, 1998), also abolishes the ability of BiP and ATP/Mg2+ to dissociate the complex between P58 and denatured RNase A (Figure 4C). A role for J-domain-mediated interactions between chaperone and co-chaperone is also supported by the finding that BiP that had bound substrate independently of P58 (i.e. in the presence of ADP) is unable to effect the latter's release when later challenged with ATP (data not shown).
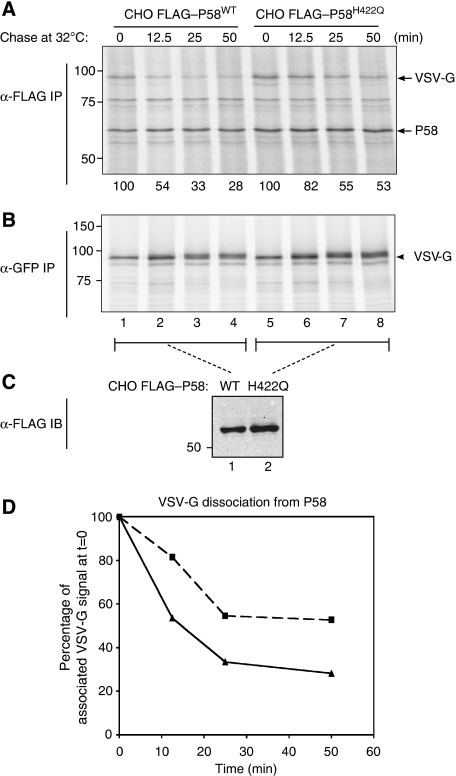
The findings noted above implicate an interaction between BiP and the J-domain of P58 in promoting the latter's dissociation from its substrates in vitro. To determine whether an in vivo correlate of this phenomenon might also exist, we compared the stability of VSV-G binding to wild-type and the J-domain mutant P58H422Q, which is unable to interact with BiP (see below). CHO cells, expressing similar levels of FLAG–P58WT and FLAG–P58H422Q, were transfected with VSV-Gts045–GFP, and radiolabelled proteins associated with FLAG–P58 were recovered by immunoprecipitation at the end of a 30′ labelling pulse conducted at the non-permissive temperature (that promotes VSV-Gts045–GFP misfolding) or following a chase at the permissive temperature (that promotes re-folding). The complex between P58WT and VSV-Gts045–GFP decayed reproducibly faster than the complex with P58H422Q (Figure 5), an observation similar to that reported for ERdj3 (Shen and Hendershot, 2005) and consistent with a role for BiP in promoting the J-domain-dependent dissociation of P58 from its substrate.
Figure 5.
A J-domain mutation that compromises interaction with BiP stabilizes P58's binding to VSV-Gts045–GFP in cells. (A) Autoradiograph of 35S-radiolabelled proteins recovered in an anti-FLAG–M1 immunoprecipitate from lysates of CHO cells stably expressing wild-type P58 or the H422Q J-domain mutant. The cells were transfected with a VSV-Gts045–GFP expression plasmid, pulsed at the non-permissive temperature (41°C) and chased for the indicated time at the permissive temperatures, before isolation of the immune complexes. The location of the radiolabelled P58 and VSV-G is indicated. (B) Autoradiograph of 35S-radiolabelled VSV-G–GFP recovered by immunoprecipitation with anti-GFP serum from the flow through of the reactions shown in (A). (C) Immunoblot comparing the steady-state levels of wild-type and H422Q mutant FLAG–P58 in the CHO cells. (D) Plot of the signal of labelled VSV-G bound to P58 from (A). CHO cells stably expressing FLAG–P58WT (-▴-) and CHO cells stably expressing FLAG–P58H422Q (-▪-).
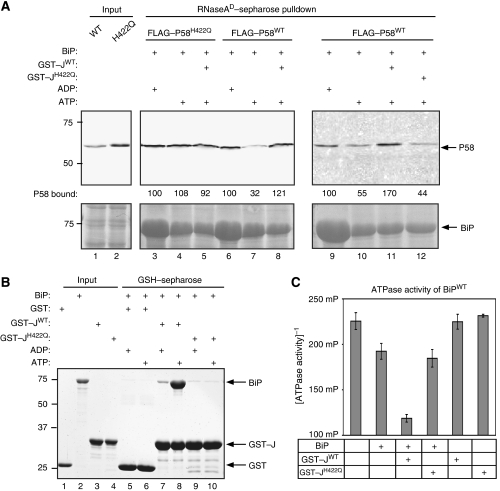
To gain further insight into the molecular mechanisms involved in BiP-induced dissociation of P58 from substrates, we sought to determine whether the isolated J-domain of P58 could compensate (in trans) for BiP's inability to dissociate a J-domain mutant P58H422Q from denatured RNase A. Complexes of unfolded RNase A and the J-domain mutant P58H422Q were assembled in vitro and challenged with BiP and ATP. As expected, the mutation in P58H422Q prevented ATP-mediated complex disassembly (Figure 6A, lanes 3 and 4). Addition of a purified fusion protein of GST with P58's isolated J-domain (residues 384–470) (GST–JWT) had no reproducible effect on the stability of complexes of denatured RNase A and mutant P58H422Q (lane 5). By contrast, when added to complexes of denatured RNase A and P58WT, the isolated J-domain of P58 (GST–JWT) reproducibly attenuated the dissociation by BiP and ATP (lanes 6–8). The ability of GST–JWT to interfere with complex disassembly by BiP was dependent on the functionality of the J-domain, as it was reversed by the H422Q mutation (compare lanes 11 and 12). Functionality of the GST–JWT fusion protein used in these assays was revealed by its ability to bind BiP in an ATP-dependent manner (Figure 6B) and by its ability to promote BiP-dependent ATP hydrolysis (Figure 6C), assays in which the GST–JH422Q mutant proved inert, as expected. These experiments are most consistent with a model whereby the ability of nucleotide-bound BiP to promote dissociation of complexes between misfolded substrates and P58 requires an interaction with the J-domain of the substrate-bound P58 molecule.
Figure 6.
Addition of purified P58 J-domain compromises BiP's ability to promote dissociation of P58 from denatured RNase A. (A) Immunoblot of FLAG–P58WT and a J-domain mutant FLAG–P58H422Q from mammalian cell lysates associated with denatured RNase A. Where indicated, the pre-formed complex was challenged with bacterially expressed BiP (9 μM) in the presence of ADP or ATP and the presence or absence of equimolar wild-type or mutant versions of P58's isolated J-domain (residues 384–470) expressed as a fusion protein with GST (GST–JWT and GST–JH422Q). The amount of P58 retained on the resin following each manipulation was measured by the LI-COR IR imaging system and is indicated (as a percentage of the signal in the unmanipulated sample). BiP's association with the resin is revealed by Ponceau S staining of the blot (lower panel). (B) Coomassie-stained SDS–PAGE of BiP retained on glutathione sepharose beads to which GST, GST–JWT or GST–JH422Q had been pre-bound. Where indicated, the binding buffer was supplemented with ADP or ATP. Lanes 1–4 report on the input proteins used in the experimental lanes 5–10. (C) ATPase activity of BiP alone or in the presence of GST–JWT or GST–JH422Q, measured by the conversion of ATP to ADP. The mean±s.e.m. of the fluorescence polarization signal of AlexaFluor 633-labelled ADP tracer, which is dissociated from an anti-ADP antibody in the presence of ADP generated in the reaction from BiP's ATPase activity, is shown (n=3). The fluorescent polarization signal is inversely proportional to the ATPase activity.
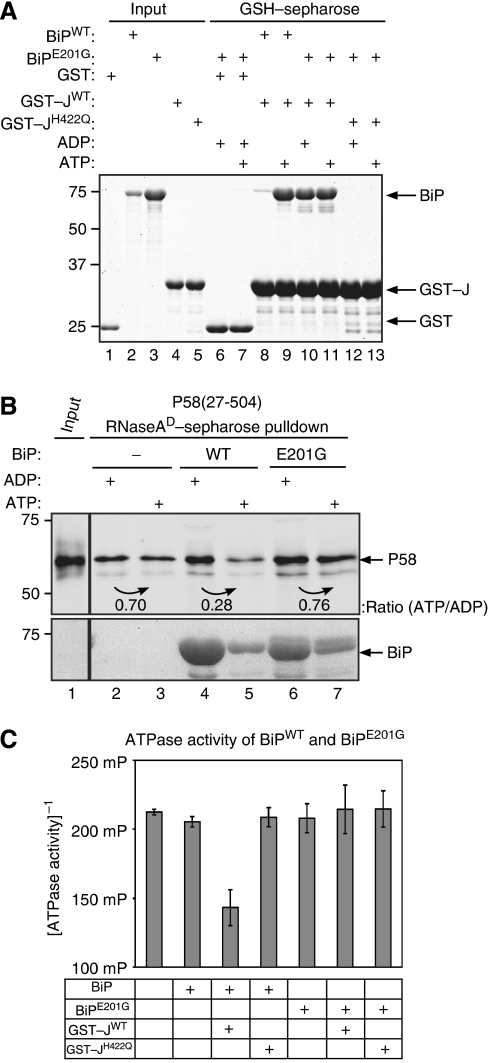
The observations noted above are consistent with both an allosteric model, whereby engagement of P58's J-domain by BiP reduces P58's affinity for substrate, and a kinetic model, whereby the engagement of BiP by P58's J-domain affects BiP's affinity for substrate. To help discriminate between these possibilities, we sought to separate BiP–J-domain interaction from complex disassembly. The mutant BiPE201G bound the isolated J-domain of P58 (GST–JWT) (Figure 7A), but failed to dissociate P58 from substrate (Figure 7B, lanes 4–7), thus separating J-engagement from complex disassembly. The E201G mutation also abolishes BiP's basal (Gaut and Hendershot, 1993) and J-domain-stimulated ATPase activity (Figure 7C) and corrupts the nucleotide dependence of J-binding; unlike wild-type BiP that binds J-domains only in the presence of ATP, J-binding by BiPE201G occurred in the presence of either ADP or ATP (Figure 7A, lanes 8–11). Therefore, the mutation does not discriminate definitively between an anomalous mode of J-domain engagement (that impedes allosteric regulation of P58 by the mutant BiP) from inability of the mutant BiP to complete a cycle of ATP hydrolysis and thereby modulate its affinity for substrate to effect the latter's kinetic partitioning away from P58.
Figure 7.
The BiPE201G mutation separates J-domain binding from dissociation of P58 from denatured RNase A. (A) Coomassie-stained SDS–PAGE of BiP or BiPE201G mutant retained on glutathione sepharose beads to which GST, GST–JWT or GST–JH422Q had been pre-bound. Where indicated, the binding buffer was supplemented with ADP or ATP. Lanes 1–5 report on the input proteins used in the experimental lanes 6–13. (B) Immunoblot of bacterially expressed and purified P58 associated with denatured RNase A. Where indicated, the pre-formed complex was challenged with bacterially expressed BiPWT or BiPE201G (9 μM) in the presence of ADP or ATP. (C) ATPase activity of wild-type BiP and the E201G mutant alone or in the presence of GST–JWT or GST–JH422Q, measured by the conversion of ATP to ADP. The mean±s.e.m. of the fluorescence polarization signal is shown (as in Figure 6C).
To try and de-convolute this issue, we made a V461F mutation in BiP's substrate-binding site (analogous to the V436F mutation in DnaK; Laufen et al, 1999), which compromised both BiP's ability to interact with substrate and to promote dissociation of P58 from substrate (Supplementary Figure S6A). P58's J-domain stimulated ATP hydrolysis by BiPV461F, suggesting that BiPV461F interacts productively with P58-J and supporting a role for substrate binding by BiP in the dissociation reaction (Supplementary Figure S6B). Instability of the complex between BiPV461F and P58-J (Supplementary Figure S6C) seemingly weakens this conclusion, by suggesting that the BiPV461F mutation also affects interactions between BiP's NBD and the P58-J. However, this caveat is mitigated by BiPV461F's inability to bind unfolded substrates and by the known requirement for the immobilized J-protein to serve as a substrate in retaining steady-state binding of Hsp70 (Mayer et al, 1999).
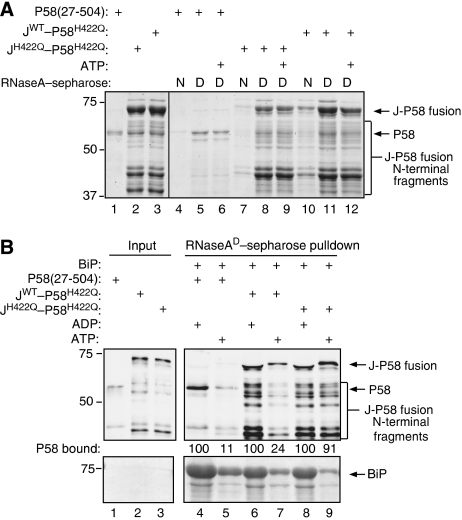
To convey the conformational changes from the J-domain to P58's substrate-binding domain, an allosteric mechanism, if it exists, would likely constrain the relationship between P58's functional domains. Therefore, we tested the ability of an ectopic J-domain fused to the N terminus of a mutant P58H422Q to rescue the latter's inability to undergo BiP/ATP-mediated dissociation from its substrate. The JWT–P58H422Q, JH422Q–P58H422Q fusion proteins and the parental P58WT purified from Escherichia coli exhibited the expected specificity for denatured over native RNase A (Figure 8A). The ectopic wild-type J-domain (but not the H422Q mutant, which serves as a specificity control) rescued the inability of P58H422Q to dissociate from substrate in the presence of BiP/ATP (Figure 8B, lanes 6–9). Interestingly, the presence of an ectopic wild-type J-domain at the N terminus of P58H422Q also enabled BiP/ATP to promote dissociation of C-terminal truncations of JWT–P58H422Q from substrate, arguing that linkage of the J-domain to the substrate-binding domain is sufficient to mediate the effect. These findings reveal the portability of P58's J-domain and argue against a constrained relationship between P58's functional domains that is predicted by an allosteric mechanism for substrate release.
Figure 8.
Ectopic fusion of the J-domain to the N terminus of P58H422Q rescues the protein's inability to dissociate from misfolded RNase A in response to BiP/ATP. (A) Coomassie-stained gel of bacterially expressed proteins associated with the native (N) and denatured (D) RNase A affinity resin. The resin was reacted with purified bacterially expressed P58, or fusion proteins consisting of P58's J-domain (either wild-type or H422Q) fused ectopically to P58H422Q's N terminus, washed and the remaining proteins were eluted in SDS. Lanes 1–3 contain 10% of the input used in the respective binding reactions. The migration of P58, J–P58 fusion proteins and their N-terminal fragments are indicated. (B) Immunoblot of material as in (A). Where indicated, the pre-formed complex with denatured RNase A was challenged with bacterially expressed BiPWT (9 μM) in the presence of ADP or ATP. The amount of P58 retained on the resin following each manipulation was measured by immunoblot with a LI-COR IR imaging system and is indicated (as a percentage of the signal in the unmanipulated sample). Note that the N-terminal fragments of JWT–P58H422Q (but not those of the JH422Q–P58H422Q control) were also dissociated by BiP/ATP.
Discussion
This study confirms that P58, a protein previously believed to reside in the cytosol, possesses a cleavable N-terminal signal peptide that mediates translocation into the ER lumen. Most of the signal detected in immunoprecipitates of radiolabelled endogenous P58 migrates on SDS–PAGE as a single band of higher mobility than the in vitro translation product of the P58 mRNA, suggesting that the signal peptide is cleaved in most P58 molecules in cultured fibroblasts. These observations, together with those reported in a detailed study (Rutkowski et al, 2007), suggest that most of the P58 are located in the ER lumen. Although the new findings do not rule out a role for small amounts of cytoplasmic P58 in modulating the activity of the cytoplasmically located kinase domain of PERK (Yan et al, 2002) or in interacting with proteins that are delayed at the ER translocon (Oyadomari et al, 2006), they behoove us to consider alternative roles for P58. The evidence from this study, of a direct interaction between P58 and misfolded proteins suggests a simple model whereby P58 assists in the handling of unfolded or misfolded lumenal proteins as a lumenal J-domain containing co-chaperone.
ER homoeostasis is dependent on the organelle's ability to rid itself of terminally misfolded/misassembled proteins. Chaperones contribute to this process, presumably by binding to the misfolded proteins and retaining them in a retro-translocation-competent state (Tsai et al, 2002). We have observed no reproducible effect of modulating P58 levels on the degradation of a model misfolded secreted glycoprotein (mutant α1-antitrypsin), a misassembled membrane glycoprotein (T-cell receptor α-subunit) (Oyadomari et al, 2006), a misfolded membrane-associated glycoprotein (VSV-Gts045–GFP; Supplementary Figure S3) or a misfolded secreted non-glycoprotein (a fusion of mutant pro-INS2C96Y and GFP, data not shown). These negative results suggest that P58 does not have a pervasive role in clearing the ER of misfolded proteins by promoting their degradation. Instead, P58 may reduce the burden of unfolded and misfolded proteins by promoting protein folding in the ER lumen. Direct binding of unfolded/misfolded lumenal proteins by P58 explains previous observations that more newly synthesized (metabolically labelled) proteins can be crosslinked to P58 in stressed cells (Oyadomari et al, 2006).
J-domain co-chaperones contribute to protein folding by collaborating with chaperones of the DnaK/Hsp70 class (Craig et al, 2006). J-domain co-chaperone interactions with unfolded substrate proteins are believed to precede binding to the Hsp70 partner. By promoting nucleotide hydrolysis in the latter, J-domain co-chaperones affect an allosteric shift that increases the affinity of the Hsp70 partner for the unfolded protein and promotes stable Hsp70–substrate protein interaction. Most J-domain co-chaperones interact transiently with the unfolded substrate and the handover of the substrate from the J-domain co-chaperone to the Hsp70 partner is envisioned as being driven by mass action (Bukau et al, 2006). P58 seems unusual in that regard, as the complex it forms with unfolded proteins is quite stable (at least in vitro where it survives extensive washing).
We find that physiological concentrations of the abundant ER Hsp70-type chaperone, BiP, can promote dissociation of the P58–substrate complex in vitro. However, this effect requires ATP and is sensitive to mutations in either partner that disrupt J-domain–BiP interactions. Taken together, these observations suggest that a productive interaction between ATP-bound BiP and P58, mediated through the latter's J-domain, is required for the dissociation of P58 from its misfolded substrate. A similar phenomenon is observed in ERdj3's interactions with BiP (Jin et al, 2008), suggesting that reciprocal regulation of co-chaperone binding to substrate by the Hsp70 partner might be a conserved feature of this chaperone network.
Our findings could be explained by simple competition between BiP and P58 for binding sites on the unfolded substrate, leading to P58's displacement by mass action at steady state. This model is disfavoured by the observation that conditions leading to disruption of the P58–substrate complex (i.e. ATP/Mg2+) are associated with less binding of BiP to the denatured RNase A (the substrate) and by the finding that more mutant BiPR197E, which is unable to direct the dissociation of P58, is bound to the substrate than wild-type BiP.
The failure of the isolated J-domain of P58 to complement the BiP/ATP-mediated dissociation of the mutant P58H422Q from its unfolded substrate argues that nucleotide-bound BiP must interact with the J-domain of substrate-bound molecules of P58 for the latter's release to take place. This requirement is consistent with allosteric regulation of P58 by BiP through the J-domain. Several findings argue against this possibility: the three types of J-co-chaperones (I–III) (Cheetham and Caplan, 1998) vary considerably in the arrangement of their J-domain relative to the substrate-binding domain, which seems incompatible with a common allosteric mechanism. The portability of P58's J-domain (Figure 8) also argues against allostery, as the latter is predicted to constrain the relationship of the two domains. BiP's ability to promote dissociation of N-terminal fragments of JWT–P58H422Q provides further evidence against the constraints predicted by the allosteric model; though of course it is impossible to exclude the fortuitous re-creation of a functional machine in the JWT–P58H422Q fusion protein used to test this hypothesis. Finally, the separation of J-domain interaction from substrate release by BiPE201G and BiPV461F argues against allostery. However, it remains formally possible that the mutations in BiP, though permissive for J-domain interactions, selectively corrupt the allosteric mechanism.
More plausible, in our opinion, is an alternative model whereby the J-domain promotes cycles of competitive binding by BiP in proximity to the co-chaperone, followed by dissociation of both partners. The argument for proximity is supported by the observation that the isolated wild-type P58 J-domain (but not the BiP-binding defective mutant JH422Q) interferes (in trans) with BiP/ATP-mediated dissociation of P58 from its substrate. Local competition and kinetic partitioning of the substrate from the J-domain co-chaperone to the Hsp70 partner likely involve ATP binding, hydrolysis and nucleotide exchange by the latter. Although global refolding of the substrate is precluded in our experiments by blocking disulphide-bond formation, local remodelling of the substrate by BiP remains possible. The inability of BiPE201G to properly handle nucleotide would explain its inability to engage a mechanism of kinetic partitioning and substrate remodelling, despite binding the J-domain. Likewise, the BiPV461F mutation's effect on substrate binding also precludes this mechanism. BiP's ability to regulate P58's association with misfolded protein thus fits well with the latter's proposed role as an intermediary that assists protein folding in the ER lumen.
Materials and methods
Mammalian cell culture and in vivo protein expression
CHO.K1 and 293T cells were cultured in standard conditions and transfected with FuGENE 6 transfection reagent (Roche) or by calcium phosphate co-precipitation, respectively. An expression plasmid encoding N-terminally FLAG-tagged mouse P58 (27-504) was constructed in the pFLAG-CMV-1 vector (Sigma) and deletion derivatives and point mutations were introduced by PCR and confirmed by sequencing. The tagged protein-coding sequence was then transferred to the pBABE-puro plasmid and pseudotyped retrovirus produced in 293T cells. These were used to infect CHO.K1 cells and select puromycin-resistant clones that stably express FLAG–P58 (27-504) and its mutant derivatives. Mammalian expression plasmid encoding VSV-Gts045–GFP (Nehls et al, 2000) was a gift of Jennifer Lippincott-Schwartz.
Antisera, immunoblotting and immunoprecipitation
Antisera to mouse P58 and GFP were described earlier (Oyadomari et al, 2006), α-FLAG M2 monoclonal antibody, α-FLAG M1 affinity gel and EZView Red α-FLAG M2 Affinity gel were purchased from Sigma. Immunoblots were developed using HRP-conjugated donkey α-mouse (Zymed) or IRDye 800CW-conjugated goat α-mouse on an Odyssey Infrared Imaging System and analysed by Odyssey software (v2.1) (LI-COR).
To recover proteins associated with P58, CHO.K1 cells stably expressing FLAG–P58 were transfected with VSV-Gts045–GFP plasmid and 48 h later were labelled with 35S-Translabel for 30 min–1 h in the presence of 10 μM MG132 and 5 μg/ml BFA (LC Laboratories) under permissive (32°C) or non-permissive (41°C) conditions followed by cold chase. Cell lysates were prepared in a Ca+2-containing buffer (0.3% Triton X-100, 150 mM NaCl, 20 mM Hepes pH 7.4, 10% glycerol, 10 mM CaCl2, 1 mM DTT, 1 mM PMSF, 4 μg/ml aprotinin and 2 μg/ml pepstatin A) and radiolabelled proteins were immunopurified with α-FLAG M1 affinity gel overnight. The bound proteins were eluted from α-FLAG M1 in 10 mM EDTA, 20 mM Tris pH 7.5, 0.02% Tween-20 or 500 μg/ml FLAG peptide, resolved by SDS–PAGE and detected by the Storm phosphoimager (Molecular Dynamics). To dissociate BiP, immunopurified complexes were exposed to K buffer (20 mM Hepes pH 7.0, 75 mM KCl, 5 mM MgCl2 and 0.01% Tween-20) at room temperature for 30 min in the absence or presence of ATP (5 mM), washed extensively with K buffer, eluted and analysed as above.
Bacterial expression, protein purification and analysis
Construction of plasmids and procedures for protein expression and analysis of BiP's ATPase activity are described in the Supplementary data.
To assay BiP binding, 10 nmol of the GST–P58–J-domain (384–470) wild-type (GST–JWT) or mutant (GST–JH422Q) proteins, or GST, was immobilized on glutathione sepharose and 10 nmol of purified BiP was added to K buffer and allowed to interact in the presence of 5 mM ATP or ADP for 1 h at 4°C. After four washes with K buffer, the remaining proteins were revealed by SDS–PAGE stained with Coomassie.
Chaperone association with denatured RNase A
Generation of a denatured or native RNase A affinity matrix is described in the Supplementary data. Lysates from the aforementioned mammalian cells, or P58 purified from bacteria or CHO cells, were allowed to interact with 10 μl of either the native or denatured RNase A matrix in a total volume of 500 μl by rotation at 30°C for 1 h. The matrices and associated proteins were washed five times with cold buffer K and the associating proteins were eluted with 2 × Laemmli buffer, separated by SDS–PAGE and revealed by Coomassie staining or immunoblotting.
To study chemical crosslinking of P58 to denatured RNase A, lysates from 293T cells expressing FLAG-tagged P58 were prepared in buffer B (0.1% Triton X-100, 20 mM Hepes pH 7.4, 75 mM KCl, 10% glycerol and 1 mM EDTA) and allowed to interact with denatured RNase A as described above. After two washes with buffer K to remove loosely associated proteins, the complexes were incubated at 37°C for 30 min in the presence or absence of 5 mM ATP (Sigma) to release endogenous associated BiP, followed by four more washes with buffer K. Remaining associated proteins were covalently crosslinked to the immobilized denatured RNase A with 50 μM dithiobis(succinimidylpropionate) (DSP) (Pierce) in a total volume of 500 μl at 4°C on rotation for 1 h followed by quenching with 100 mM Tris pH 7.5 for 15 min at 4°C. Non-covalently associated proteins were eluted with 4% SDS, 100 mM Tris pH 6.8 and the matrices were further washed with RIPA three more times. Proteins covalently crosslinked to the denatured RNase A matrix were then recovered by eluting with Laemmli buffer containing 100 mM DTT (which reduces an S–S bond that joins the crosslinker's arms) and analysed by immunoblotting.
Release of P58 from denatured RNaseA
Lysates from CHO cells stably expressing FLAG-tagged P58 or purified bacterially expressed P58 and its mutants were allowed to interact with denatured RNase A matrix in buffer B as described above. After three washes with buffer K, the assembled complexes were challenged with 9 μM of purified bacterially expressed BiP in the presence of 5 mM ATP or ADP in a total volume of 400 μl of buffer K at 37°C for 30 min. Where indicated, bacterially expressed purified GST–P58–J-domain (384–470) (9 μM) was added. Dissociated proteins were removed by three washes with buffer K, the remaining material was eluted with Laemmli buffer and analysed by immunoblotting.
Supplementary Material
Supplementary Figure S1–S6
Acknowledgments
We thank Jennifer Lippincott-Schwartz (NICHD) for the VSV-Gts045–GFP expression plasmid and Peter Arvan (U Michigan) for the INS2C96Y–GPF expression plasmid and Luke Wiseman (NYU) and Matthias Meyer (ZMBH, Heidelberg) for useful suggestions. This study was supported by grants DK047119 and ES08681 to DR and GM54068 to LMH.
References
- Awad W, Estrada I, Shen Y, Hendershot LM (2008) BiP mutants that are unable to interact with endoplasmic reticulum DnaJ proteins provide insights into interdomain interactions in BiP. Proc Natl Acad Sci USA 105: 1164–1169 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barber GN, Thompson S, Lee TG, Strom T, Jagus R, Darveau A, Katze MG (1994) The 58-kilodalton inhibitor of the interferon-induced double-stranded RNA-activated protein kinase is a tetratricopeptide repeat protein with oncogenic properties. Proc Natl Acad Sci USA 91: 4278–4282 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bernales S, Papa FR, Walter P (2006) Intracellular signaling by the unfolded protein response. Annu Rev Cell Dev Biol 22: 487–508 [DOI] [PubMed] [Google Scholar]
- Bukau B, Weissman J, Horwich A (2006) Molecular chaperones and protein quality control. Cell 125: 443–451 [DOI] [PubMed] [Google Scholar]
- Cheetham ME, Caplan AJ (1998) Structure, function and evolution of DnaJ: conservation and adaptation of chaperone function. Cell Stress Chaperones 3: 28–36 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Craig EA, Huang P, Aron R, Andrew A (2006) The diverse roles of J-proteins, the obligate Hsp70 co-chaperone. Rev Physiol Biochem Pharmacol 156: 1–21 [DOI] [PubMed] [Google Scholar]
- Flynn GC, Chappell TG, Rothman JE (1989) Peptide binding and release by proteins implicated as catalysts of protein assembly. Science 245: 385–390 [DOI] [PubMed] [Google Scholar]
- Gaut JR, Hendershot LM (1993) Mutations within the nucleotide binding site of immunoglobulin-binding protein inhibit ATPase activity and interfere with release of immunoglobulin heavy chain. J Biol Chem 268: 7248–7255 [PubMed] [Google Scholar]
- Jiang J, Maes EG, Taylor AB, Wang L, Hinck AP, Lafer EM, Sousa R (2007) Structural basis of J cochaperone binding and regulation of Hsp70. Mol Cell 28: 422–433 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jin Y, Awad W, Hendershot LM (2008) Regulated release of ERdj3 from unfolded proteins by BiP. EMBO J (in press) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kapulkin V, Hiester BG, Link CD (2005) Compensatory regulation among ER chaperones in C. elegans. FEBS Lett 579: 3063–3068 [DOI] [PubMed] [Google Scholar]
- Kreis TE, Lodish HF (1986) Oligomerization is essential for transport of vesicular stomatitis viral glycoprotein to the cell surface. Cell 46: 929–937 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ladiges WC, Knoblaugh SE, Morton JF, Korth MJ, Sopher BL, Baskin CR, MacAuley A, Goodman AG, LeBoeuf RC, Katze MG (2005) Pancreatic beta-cell failure and diabetes in mice with a deletion mutation of the endoplasmic reticulum molecular chaperone gene P58IPK. Diabetes 54: 1074–1081 [DOI] [PubMed] [Google Scholar]
- Laufen T, Mayer MP, Beisel C, Klostermeier D, Mogk A, Reinstein J, Bukau B (1999) Mechanism of regulation of hsp70 chaperones by DnaJ cochaperones. Proc Natl Acad Sci USA 96: 5452–5457 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li J, Qian X, Sha B (2003) The crystal structure of the yeast Hsp40 Ydj1 complexed with its peptide substrate. Structure 11: 1475–1483 [DOI] [PubMed] [Google Scholar]
- Liberek K, Marszalek J, Ang D, Georgopoulos C, Zylicz M (1991) Escherichia coli DnaJ and GrpE heat shock proteins jointly stimulate ATPase activity of DnaK. Proc Natl Acad Sci USA 88: 2874–2878 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mayer MP, Laufen T, Paal K, McCarty JS, Bukau B (1999) Investigation of the interaction between DnaK and DnaJ by surface plasmon resonance spectroscopy. J Mol Biol 289: 1131–1144 [DOI] [PubMed] [Google Scholar]
- Misumi Y, Misumi Y, Miki K, Takatsuki A, Tamura G, Ikehara Y (1986) Novel blockade by brefeldin A of intracellular transport of secretory proteins in cultured rat hepatocytes. J Biol Chem 261: 11398–11403 [PubMed] [Google Scholar]
- Nehls S, Snapp EL, Cole NB, Zaal KJ, Kenworthy AK, Roberts TH, Ellenberg J, Presley JF, Siggia E, Lippincott-Schwartz J (2000) Dynamics and retention of misfolded proteins in native ER membranes. Nat Cell Biol 2: 288–295 [DOI] [PubMed] [Google Scholar]
- Oyadomari S, Yun C, Fisher EA, Kreglinger N, Kreibich G, Oyadomari M, Harding HP, Goodman AG, Harant H, Garrison JL, Taunton J, Katze MG, Ron D (2006) Co-translocational degradation protects the stressed endoplasmic reticulum from protein overload. Cell 126: 727–739 [DOI] [PubMed] [Google Scholar]
- Ron D, Harding H (2007) eIF2a phosphorylation in cellular stress responses and disease. In Translational Control, Sonenberg N, Hershey J, Mathews M (eds) Vol. 13, pp 345–368. Cold Spring Harbor: Cold Spring Harbor Laboratory Press [Google Scholar]
- Rudiger S, Schneider-Mergener J, Bukau B (2001) Its substrate specificity characterizes the DnaJ co-chaperone as a scanning factor for the DnaK chaperone. EMBO J 20: 1042–1050 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rutkowski DT, Kang SW, Goodman AG, Garrison JL, Taunton J, Katze MG, Kaufman RJ, Hegde RS (2007) The role of p58IPK in protecting the stressed endoplasmic reticulum. Mol Biol Cell 18: 3681–3691 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmid D, Baici A, Gehring H, Christen P (1994) Kinetics of molecular chaperone action. Science 263: 971–973 [DOI] [PubMed] [Google Scholar]
- Schroder M, Kaufman RJ (2005) The mammalian unfolded protein response. Annu Rev Biochem 74: 739–789 [DOI] [PubMed] [Google Scholar]
- Sha B, Lee S, Cyr DM (2000) The crystal structure of the peptide-binding fragment from the yeast Hsp40 protein Sis1. Structure 8: 799–807 [DOI] [PubMed] [Google Scholar]
- Shen Y, Hendershot LM (2005) ERdj3, a stress-inducible endoplasmic reticulum DnaJ homologue, serves as a cofactor for BiP's interactions with unfolded substrates. Mol Biol Cell 16: 40–50 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suh WC, Burkholder WF, Lu CZ, Zhao X, Gottesman ME, Gross CA (1998) Interaction of the Hsp70 molecular chaperone, DnaK, with its cochaperone DnaJ. Proc Natl Acad Sci USA 95: 15223–15228 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Szabo A, Langer T, Schroder H, Flanagan J, Bukau B, Hartl FU (1994) The ATP hydrolysis-dependent reaction cycle of the Escherichia coli Hsp70 system DnaK, DnaJ, and GrpE. Proc Natl Acad Sci USA 91: 10345–10349 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trimble RB, Tarentino AL, Plummer TH Jr, Maley F (1978) Asparaginyl glycopeptides with a low mannose content are hydrolyzed by endo-beta-N-acetylglucosaminidase H. J Biol Chem 253: 4508–4511 [PubMed] [Google Scholar]
- Tsai B, Ye Y, Rapoport TA (2002) Retro-translocation of proteins from the endoplasmic reticulum into the cytosol. Nat Rev Mol Cell Biol 3: 246–255 [DOI] [PubMed] [Google Scholar]
- Tsai J, Douglas MG (1996) A conserved HPD sequence of the J-domain is necessary for YDJ1 stimulation of Hsp70 ATPase activity at a site distinct from substrate binding. J Biol Chem 271: 9347–9354 [DOI] [PubMed] [Google Scholar]
- Van Huizen R, Martindale JL, Gorospe M, Holbrook NJ (2003) P58IPK, a novel endoplasmic reticulum stress-inducible protein and potential negative regulator of eif2alpha signaling. J Biol Chem 278: 15558–15564 [DOI] [PubMed] [Google Scholar]
- Wawrzynow A, Zylicz M (1995) Divergent effects of ATP on the binding of the DnaK and DnaJ chaperones to each other, or to their various native and denatured protein substrates. J Biol Chem 270: 19300–19306 [DOI] [PubMed] [Google Scholar]
- Yan W, Frank CL, Korth MJ, Sopher BL, Novoa I, Ron D, Katze MG (2002) Control of PERK eIF2-alpha kinase activity by the endoplasmic reticulum stress-induced molecular chaperone P58IPK. Proc Natl Acad Sci USA 99: 15920–15925 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Figure S1–S6