Abstract
The Myc-associated zinc-finger protein, Miz1, activates transcription of the p21cip1 gene in response to UV irradiation. Miz1 associates with topoisomerase II binding protein1 (TopBP1), an essential activator of the Atr kinase. We show here that Miz1 is required for the recruitment of a fraction of TopBP1 to chromatin, for the protection of TopBP1 from proteasomal degradation and for Atr-dependent signal transduction. TopBP1 that is not bound to chromatin is degraded by the HectH9 (Mule, ARF-BP1 and HUWE1) ubiquitin ligase. Myc antagonizes the binding of TopBP1 to Miz1; as a result, expression of Myc leads to dissociation of TopBP1 from chromatin, reduces the amount of total TopBP1 and attenuates Atr-dependent signal transduction. Our data show that Miz1 and Myc affect the activity of the Atr checkpoint through their effect on TopBP1 chromatin association and stability.
Keywords: Atr, Miz1, Myc, TopBP1
Introduction
Miz1 was initially described as a protein that interacts with the carboxy terminus of the Myc oncoprotein; it is a zinc-finger protein that contains an amino-terminal POZ domain. Miz1 activates transcription of genes encoding the cell cycle inhibitors p15Ink4b and p21Cip1, leading to cell cycle arrest (Adhikary and Eilers, 2005). For example, upregulation of p15ink4b expression in response to TGF-β in keratinocytes requires Miz1, and the skin of Miz1 conditional knockout animals shows phenotypes indicative of defective TGF-β signalling (Gebhardt et al, 2007). Similarly, Miz1 activates multiple genes involved in cell adhesion, metabolism and apoptosis (e.g. Patel and McMahon, 2007).
Miz1 can also repress transcription when it forms complexes with other transcription factors. For instance, Miz1 heterodimerizes with the POZ domain transcription factors Zbtb4 to repress transcription of p21cip1 in peripheral neurons (Weber et al, 2008). Similar to other POZ domain repressor complexes, the Miz1/Zbtb4 complex recruits histone deacetylases through an interaction with the Sin3 adaptor protein (Weber et al, 2008). Miz1 also represses transcription of the p15ink4b and p21cip1 genes in a complex with the Myc oncoprotein. As p15Ink4b can mediate cell cycle arrest in response to TGF-β, repression of p15ink4b is one mechanism through which deregulation of MYC renders cells resistant to the antimitogenic functions of TGF-β. Similarly, Myc represses transcription of the p21cip1 gene through binding to Miz1; as a result, high levels of Myc suppress cell cycle arrest and favour apoptosis in response to DNA damage (Herold et al, 2002; Seoane et al, 2002).
Miz1 also associates with the topoisomerase II binding protein1 (TopBP1) in unstressed cells; TopBP1 dissociates from Miz1 after exposure to UV irradiation (Herold et al, 2002). As association of both Miz1 and E2F1 with TopBP1 leads to transcriptional repression (Liu et al, 2006), dissociation of TopBP1 from Miz1 may facilitate the induction of p21cip1. This suggestion is consistent with the observation that depletion of TopBP1 leads to a p53-independent transcriptional induction of p21cip1 expression and inhibition of cyclin E/Cdk2 kinase (Jeon et al, 2007). Nevertheless, the finding is surprising, as TopBP1 is not a dedicated transcriptional co-repressor but instead is required for the loading of Cdc45 to origins of replication during DNA replication (Van Hatten et al, 2002) and for the activation of the Atr (Atm (ataxia-telangiectasia-mutated)-Rad3-related) kinase in response to stalling of the replicative DNA polymerases δ and ɛ (Kumagai et al, 2006; Delacroix et al, 2007). This stalling (e.g. upon UV irradiation) leads to the accumulation of stretches of single-stranded DNA, which are covered by the single-stranded DNA-binding protein Rpa. Atr is recruited to single-stranded DNA through binding of the Atr-associated protein, Atrip, to Rpa (Zou and Elledge, 2003). Stalled DNA polymerases also recruit a clamp loader (Rad17 replication factor C-RfC), which in turn recruits a clamp complex consisting of the Rad9, Rad1 and Hus1 proteins (the ‘9-1-1' complex) (Byun et al, 2005). The carboxyl-terminal tail of Rad9 does not participate in clamp formation but binds to TopBP1 and, once recruited, TopBP1 activates Atr through a direct protein–protein interaction (Kumagai et al, 2006; Delacroix et al, 2007).
We show here that Miz1 is required for the binding of a fraction of TopBP1 to chromatin and to protect TopBP1 from proteasomal degradation; consequently, Miz1 is required for Atr-dependent signal transduction. Furthermore, we show that TopBP1 that is not bound to chromatin is degraded by the HectH9 (ARF-BP1, Mule and HUWE1) ubiquitin ligase (Adhikary et al, 2005; Chen et al, 2005). Miz1 has been shown earlier to inhibit HectH9-dependent ubiquitination (Adhikary et al, 2005). Finally, we show that Myc displaces TopBP1 from Miz1 and leads to a reduction in chromatin-bound TopBP1 and Atr-dependent signal transduction. Taken together, our data suggest that the relative levels of Myc and Miz1 affect the function of the Atr-dependent DNA checkpoint through the regulation of TopBP1 stability.
Results
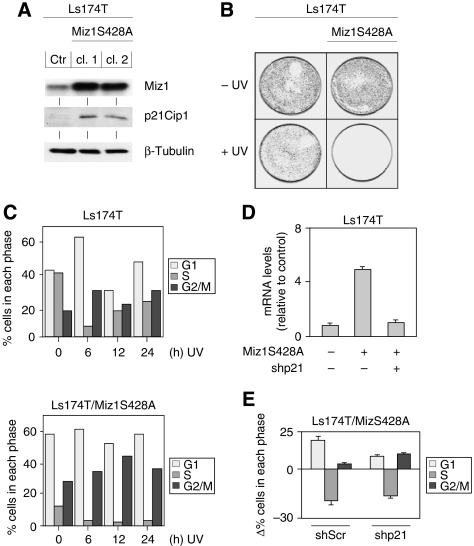
Previous work had shown that Miz1 is phosphorylated by the protein kinase Akt at serine 428 and that expression of a mutant version of Miz1, Miz1S428A, that cannot be phosphorylated by Akt strongly delays the resumption of proliferation in NIH3T3 cells after exposure to low doses of UV irradiation (Wanzel et al, 2005). To better understand the role of Miz1 in this pathway, we expressed Miz1S428A in the human colon carcinoma cell line, Ls174T, which has wild-type p53 (van de Wetering et al, 2002). Several clones and pools of Ls174T cells infected with retroviruses encoding Miz1S428A expressed elevated levels of Miz1 protein. As expected from previous work, these cells displayed elevated levels of p21Cip1 protein and mRNA (Figure 1A and D). Consistent with the presence of elevated levels of p21Cip1, the rate of proliferation of Ls174T/Miz1 and Ls174T/Miz1S428A cells was reduced relative to control cells in the absence of UV irradiation and FACS analysis revealed a decreased percentage of cells in the S phase (Figure 1B, C and E). Furthermore, similar to our previous observations, Ls174T/Miz1S428A cells did not recover in clonogenic assays in response to UVB irradiation, in contrast to control cells (Figure 1B). FACS analysis revealed that the dose of UVB used caused a transient decrease in the percentage of control cells that incorporated BrdU (Figure 1C). In contrast, UVB caused a persistent decrease in the percentage of Ls174T/Miz1S428A cells incorporating BrdU.
Figure 1.
Role of P21CIP1 in Miz1-induced cell cycle arrest. (A) Expression of Miz1S428A enhances the expression of p21Cip1 in human colon carcinoma cells. The panels show immunoblots documenting the expression of Miz1, p21Cip1 and β-tubulin as control in vector (pWZL)-infected (ctr) Ls174T colon carcinoma cells and in two clones of Ls174T cells infected with a retrovirus expressing Miz1S428A. (B) Ls174T/Miz1S428A show an enhanced sensitivity to UV irradiation. The panels show colony assays of control Ls174T and of Ls174T/Miz1S428A cells. Cells were either exposed to 250 J/m2 of UVB irradiation or left untreated as indicated. In total, 50 000 cells were plated for each assay; plates were stained 8 days after irradiation. (C) Ls174T/Miz1S428A cells arrest in both G1 and G2 phases after UV irradiation. Ls174T and Ls174T/Miz1S428A cells were exposed to UVB irradiation; cells were harvested at the indicated times after irradiation. In each case, BrdU was added to the culture 1 h before harvesting. Harvested cells were fixed, stained with α-BrdU antibodies and propidium iodide and subjected to FACS analysis. (D) shRNA-mediated depletion of P21CIP1 mRNA. RNA was isolated from control cells and from Ls174T/Miz1S428A cells infected with retroviruses expressing either control shRNA or shRNA targeting P21CIP1. Expression of P21CIP1 was determined by RQ-PCR using mRNA encoding ribosomal protein S14 as a control. (E) Depletion of P21CIP1 does not alleviate Miz1-induced reduction in S-phase cells. The panel shows the results of a FACS analysis of the cell cycle distribution of Ls174T/Miz1S428A cells that were infected with vectors expressing either control shRNA or shRNA targeting P21CIP1. Each bar represents the difference to control cells.
To rule out the possibility that these phenotypes reflected a gain of function of Miz1S428A, we also expressed wild-type Miz1 in Ls174T cells (Supplementary Figure 1). Expression of Miz1 led to increased levels of p21 (Supplementary Figures 1a and c), reduced the percentage of cells incorporating BrdU in exponentially growing cells (Supplementary Figure 1d) and delayed the resumption of proliferation and DNA synthesis after UV irradiation (Supplementary Figure 1b and d), similar to that observed for Miz1S428A. However, the phenotypes of cells expressing Miz1 were less pronounced than those of cells expressing Miz1S428A, supporting our previous conclusion that phosphorylation by Akt attenuates the cell cycle inhibitory functions of Miz1 (Wanzel et al, 2005).
Ls174T cells expressing Miz1S428A showed elevated levels of cells in both G1 and G2 phases of the cell cycle and arrested in both phases of the cell cycle in response to UVB (Figure 1C). To test whether the elevated levels of p21Cip1 are responsible for both the enhanced G1 and G2 arrest, we expressed an shRNA targeting P21CIP1 in Ls174T/Miz1S428A cells (Figure 1D and E). Expression of the shRNA reduced the levels of P21CIP1 mRNA to the levels seen in control cells and diminished the accumulation of cells in G1 phase. In contrast, depletion of P21CIP1 had no effect on the Miz1-induced reduction of cells in S phase, as cells expressing both Miz1S428A and shP21CIP1 are accumulated in the G2 phase of the cell cycle. Furthermore, depletion of P21CIP1 did not accelerate the resumption of DNA proliferation in Miz1S428A cells upon UV irradiation (not shown). The data show that the induction of P21CIP1 by Miz1 accounts for the arrest of cells expressing Miz1S428A in the G1 phase of the cell cycle, but that these cells arrest in the G2 phase in a P21CIP1-independent manner.
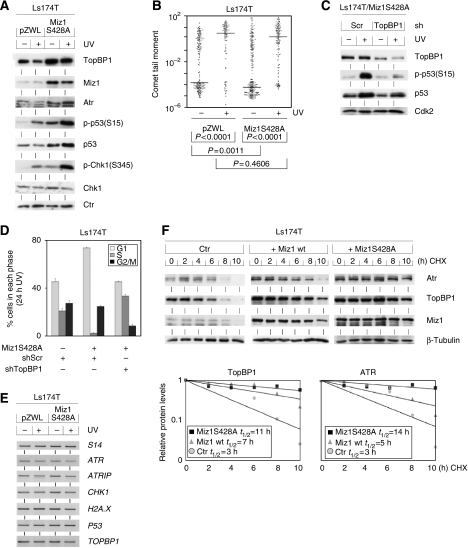
Activation of the Atr kinase results in cell cycle arrest in the S and G2 phases of the cell cycle (Abraham, 2001). As Miz1 associates with TopBP1, which activates Atr, we speculated that Miz1 might affect the level or function of TopBP1 and thereby affect the function of the Atr pathway (Herold et al, 2002). To explore this possibility, we measured the protein levels of TopBP1, Atr, p53, Chk1 and of phosphorylated p53(Ser15) and Chk1(Ser345), downstream substrates of Atr, in control cells and in Ls174T/Miz1S428A cells before and after exposure to UVB (Figure 2A). The steady-state levels of TopBP1 were enhanced in Ls174T/Miz1S428A relative to control cells, both before and after UV irradiation; we also observed a moderate increase in Atr levels. Correlating with these alterations, the levels of phosphorylated p53 and Chk1 were increased in Ls174T/Miz1S428A relative to control cells (Figure 2A); identical results were seen for phosphorylated Atm (not shown). Significant levels of phosphorylated p53 and Chk1 were observed in Ls174T/Miz1S428A cells even in the absence of UV irradiation. The levels of Rad1, Hus1 and Rad9 were unaltered in Ls174T/Miz1S428A relative to control cells (Supplementary Figure 1e and f). These changes might either reflect alterations in the cellular response to DNA damage or might indicate that Miz1 affects the level of DNA damage that is present in cells (e.g. by inducing reactive oxygen species or by inhibiting DNA repair). Comet assays revealed that expression of Miz1S428A did not induce DNA damage by itself or enhance the extent of comet formation in response to UV irradiation, arguing that Miz1 does not induce DNA damage but enhances the signalling activity of the Atr pathway (Figure 2B).
Figure 2.
Miz1S428A stabilizes TopBP1 and Atr and enhances signal transduction by Atr. (A) Expression of Miz1S428A enhances the steady-state levels of TopBP1 and Atr. The panels show immunoblots documenting the expression of the indicated proteins in control and Ls174T/Miz1S428A cells. Cells were either mock-treated (−) or exposed to UVB irradiation (+) 2 h before harvesting. A nonspecific band was used as a loading control (‘Ctr'). (B) Comet assays documenting the extent of DNA damage in control and Ls174T/Miz1S428A cells before and after exposure to UVB. Cells were harvested 12 h after irradiation. The horizontal bar indicates the median comet tail moment for each cell population. (C) Reduction of TopBP1 levels in Ls174T/Miz1S428A cells reduces the phosphorylation of p53. Ls174T/Miz1S428A cells were transfected with vectors expressing shRNA targeting TOPBP1 or control shRNA. Resistant cell pools were selected and exposed to UVB as before; cells were harvested 2 h after UVB irradiation. The panels show immunoblots probed with the indicated antibodies. (D) Reduction in TopBP1 levels facilitates the resumption of DNA synthesis of Ls174T/Miz1S428A after UV irradiation. 24 h after irradiation, the cell pools described in (C) were harvested and the cell cycle distribution was determined by FACS analysis. The cell cycle distribution of Ls174T cells was determined as a control. (E) Miz1 does not regulate the levels of mRNAs that encode Atr checkpoint proteins. The panels show RT–PCR assays that document the mRNA levels of the indicated genes. The cells are identical to those used in (A); RNA was isolated from either mock-treated cells (−) or 2 h after UV irradiation (+). Expression of S14 was used as a control. (F) Regulation of TopBP1 and Atr protein stability by Miz1. At the start of the experiment, 50 μg/ml cycloheximide (CHX) was added to control Ls174T cells and to cells expressing either wild-type Miz1 or Miz1S428A. Cells were harvested and lysates were prepared at the indicated times after the addition of cycloheximide. The lower panels show a quantitation of immunoblots measuring TopBP1 and Atr stability in the indicated cell types. The values are normalized to the expression of β-tubulin.
To test whether the enhanced levels of TopBP1 are required for the prolonged cell cycle arrest of Ls174T/Miz1S428A cells on UV irradiation, we infected these cells with retroviruses that express shRNA targeting TOPBP1 (Figure 2C). Infected cells showed a 2- to 3-fold reduction in TopBP1 levels relative to cells infected with viruses expressing scrambled shRNA and showed reduced levels of p53 phosphorylation. Such cells resumed DNA replication as rapidly as control cells that do not express Miz1S428A after UV irradiation (Figure 2D). Similarly, treatment of Ls174T/Miz1S428A cells with caffeine, an inhibitor of the Atr and Atm kinases, accelerated the resumption of DNA replication after UV irradiation to a similar extent (not shown). We concluded that Miz1 enhances the expression levels of TopBP1 and to a lesser extent Atr and enhances the activity of the Atr signalling pathway towards p53, Chk1 and Atm.
Ls174T/Miz1S428A cells expressed unaltered mRNA levels of ATR, ATRIP, TOPBP1, P53, H2A.X and CHK1 relative to control cells, demonstrating that the alterations in protein levels that are induced by Miz1S428A occur at a post-transcriptional level (Figure 2E). Furthermore, expression of Miz1S428A did not affect mRNA levels of HUS1, RAD1 and RAD9 (Supplementary Figure 1e). To determine whether enhanced protein stability accounts for the enhanced levels of TopBP1 and Atr found in cells expressing Miz1S428A, we treated control and Ls174T cells expressing either wild-type Miz1 or Miz1S428A with cycloheximide and prepared extracts after different times (Figure 2F). Protein levels of both TopBP1 and Atr decreased in control cells with a half-life of approximately 3 h for each protein, demonstrating that both proteins are subject to proteolysis. In contrast, both proteins were essentially stable over the entire time period in cells expressing Miz1S428A (estimated half-life: TopBP1: 11 h; Atr: 14 h). Cells expressing wild-type Miz1 showed an intermediate phenotype (TopBP1: 7 h; Atr: 5 h). Expression of Miz1S428A also stabilized both TopBP1 and Atr in cells exposed to UV irradiation, albeit to a lower extent (Supplementary Figure 2a). Therefore, the increased stability of TopBP1 and Atr seen in cells expressing Miz1S428A is not an indirect consequence of the enhanced activity of the Atr pathway in these cells. Furthermore, enhanced stability of TopBP1 after UV irradiation was also seen in cells treated with the PI3 kinase inhibitor, LY294002, consistent with a role for Akt in regulating Miz1-dependent arrest (Supplementary Figure 2b). The stability of Chk1 and Cdk2 was unaltered in cells expressing Miz1S428A relative to control cells, demonstrating that Miz1 specifically affects TopBP1 and Atr stability (not shown). Furthermore, treatment of cells with the proteasome inhibitor MG132 largely reverted the decrease in TopBP1 and Atr levels that was observed in control cells, demonstrating that both proteins are degraded by the proteasome (not shown).
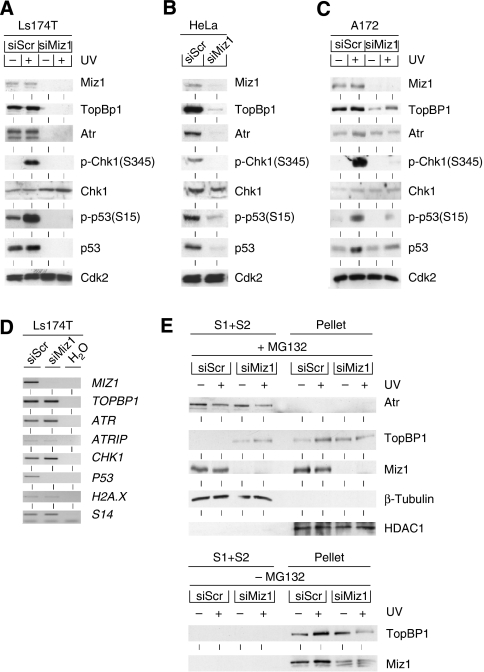
To exclude that these effects solely reflect a function of ectopically expressed Miz1, we depleted endogenous Miz1 in Ls174T cells using retroviral shRNA vectors. Surviving colonies had a 3- to 4-fold reduced expression level of Miz1 and showed a strongly reduced level of Chk1 phosphorylation upon UV irradiation, pointing to a requirement for Miz1 in Atr signalling (Supplementary Figure 3). As TopBP1 is required for normal DNA replication (Kim et al, 2005), the moderate reduction in Miz1 levels observed in surviving cells may reflect a positive selection for the retention of functional Miz1. Therefore, we depleted Miz1 using transient transfection of siRNA (Figure 3A). Depletion of Miz1 in Ls174T cells led to strongly reduced protein levels of TopBP1, Atr and p53. In contrast, the levels of Chk1 and Cdk2 remained unaltered. Neither Chk1 nor p53 was phosphorylated in response to UV irradiation in Miz1-depleted cells, demonstrating that Miz1 is essential for Atr-dependent signal transduction. Identical results were obtained in HeLa cells, excluding the possibility that the effects were restricted to a single cell line (Figure 3B). Furthermore, depletion of Miz1 strongly decreased the levels of TopBP1 and UV-induced Chk1 phosphorylation in A172 glioblastoma and in HCT116 colon carcinoma cells, whereas there was a smaller decrease in Atr levels in these cell lines (Figure 3C and data not shown).
Figure 3.
Miz1 is required for Atr-dependent signal transduction. (A) Depletion of Miz1 reduces the steady-state levels of TopBP1 and Atr and inhibits Atr-dependent signalling. Ls174T colon carcinoma cells were transfected with either scrambled siRNA or siRNA targeting MIZ1; 56 h after transfection, cells were either left untreated (–) or exposed to UVB (+) as indicated. Cells were harvested 2 h after irradiation. The panels show immunoblots documenting the expression and/or phosphorylation status of the indicated proteins. (B) Depletion of Miz1 reduces the steady-state levels of TopBP1 and Atr in HeLa cells. HeLa cells were transfected with either scrambled siRNAs or siRNAs targeting MIZ1 and harvested 56 h later. The panels show immunoblots documenting the expression and/or phosphorylation status of the indicated proteins. (C) Depletion of Miz1 reduces the steady-state levels of TopBP1 and inhibits Atr-dependent signalling in A172 glioblastoma cells. The experiment was performed exactly as described in (A). (D) Miz1 is essential for the expression of P53 mRNA, but not of other mRNAs encoding checkpoint proteins. Ls174T cells were transfected with scrambled siRNA or with siRNA targeting MIZ1; RNA was isolated 36 h after transfection and RT–PCR was used to measure the levels of the indicated mRNAs. (E) A fraction of TopBP1 is released from chromatin in cells depleted of Miz1. Ls174T cells were transfected with scrambled siRNA or with siRNA targeting MIZ1. Where indicated, MG132 was added to cells 50 h after transfection. After 6 h, cells were irradiated with UV as shown; after another 2 h, cells were fractionated into cytosolic (S1), soluble nuclear (S2) and insoluble fractions (pellet). Cytosolic and soluble nuclear fractions were pooled for the immunoblot shown here; HDAC1 and β-tubulin were used as marker proteins for chromatin and soluble/cytosolic fractions, respectively. Control experiments (not shown) had confirmed previous observations (Smits et al, 2006) that treatment with micrococcal nuclease releases TopBP1 from the insoluble pellet.
To determine whether loss of expression of TopBP1, Atr and p53 reflects an essential role of Miz1 in transcription of the corresponding genes, we isolated RNA from both Miz1-depleted Ls174T and HeLa cells and measured the expression of mRNAs encoding multiple checkpoint proteins using RT–PCR. Loss of Miz1 had no significant effects on the mRNA levels of ATR, ATRIP, TOPBP1, CHK1 and H2A.X mRNAs in either Ls174T or HeLa cells (Figure 3D and data not shown); this is consistent with the finding that expression of Miz1S428A did not affect the expression of these mRNAs. In contrast, P53 mRNA levels were significantly lower in cells depleted of Miz1; therefore, the decrease in p53 protein levels observed in Miz1-depleted cells may reflect a role of Miz1 in P53 transcription, stabilization of the p53 protein or both (see below).
Treatment of cells with the proteasome inhibitor MG132 abolished the decrease in TopBP1 and Atr levels observed in Miz1-depleted Ls174T cells, arguing that both proteins are degraded through the proteasome (not shown). To better understand why TopBP1 is degraded in Miz1-depleted cells, we fractionated extracts of Miz1-depleted cells that had been treated with MG132 into soluble and chromatin-bound proteins (Figure 3E). TopBP1 was exclusively recovered in the chromatin-bound pellet in control cells, consistent with previous observations that it is bound to chromatin (Van Hatten et al, 2002). In contrast, a significant fraction of TopBP1 accumulated in the soluble fraction in Miz1-depleted cells in the presence of a proteasome inhibitor; no TopBP1 was recovered in the soluble fraction of Miz1-depleted cells in the absence of MG132, demonstrating that soluble TopBP1 underlies proteasomal turnover (Figure 3E). The bulk of Atr was not stably associated with chromatin independently of the level of Miz1. Therefore, Miz1 is required for chromatin association of a significant fraction of TopBP1 and may stabilize TopBP1 by recruiting it to chromatin.
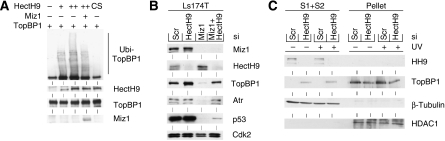
We have shown earlier that Miz1 binds to the HectH9 ubiquitin ligase and that it can inhibit ubiquitination by HectH9, suggesting that HectH9 may also be responsible for the degradation of TopBP1 and/or Atr in the absence of Miz1 (Adhikary et al, 2005). Consistent with this hypothesis, ubiquitination of TopBP1 was virtually undetectable in the absence of HectH9 and ectopic expression of HectH9 led to a substantial ubiquitination of TopBP1 (Figure 4A and Supplementary Figure 4a). In contrast, expression of a point mutant of HectH9, in which the catalytic cysteine is replaced by serine (HectH9CS), did not induce ubiquitination of TopBP1, suggesting that HectH9 itself is a catalytically active E3 ligase towards TopBP1 (Figure 4A). Co-expression of Miz1 reduced ubiquitination of TopBP1 by HectH9. In parallel experiments, HectH9 did not ubiquitinate Atr; furthermore, ectopic expression of HectH9 had no effect on turnover of Atr under any circumstances we tested. We concluded that HectH9 does not directly target Atr.
Figure 4.
Miz1 stabilizes TopBP1 through inhibition of the ubiquitin ligase HectH9. (A) Miz1 inhibits HectH9-mediated ubiquitination of TopBP1. Hek293 cells were transfected with expression plasmids encoding TopBP1, Miz1, HA-tagged ubiquitin and either wild-type ΔNHectH9 or ΔNHectH9CS, in which the catalytic cysteine has been replaced by serine as indicated. ΔNHectH9 comprises amino acids 2473–4374 of full-length HectH9; it is used here as we are unable to efficiently express the full-length, 500 kDa HectH9 protein (Adhikary et al, 2005). MG132 was included in all experimental conditions to block proteasomal degradation. Cell lysates were precipitated with α-HA antibodies bound to agarose and the precipitates were probed with antibodies against TopBP1 (upper panel). The lower panels show input blots for TopBP1, HectH9 and Miz1. (B) Depletion of HectH9 reverts destabilization of TopBP1 in Miz1-depleted cells. Ls174T cells were transfected with siRNA oligonucleotides targeting MIZ1, HECTH9 or a combination of both. Cells were harvested 56 h after transfection. The panels show immunoblots documenting the expression of the indicated proteins. (C) Non-chromatin-bound TopBP1 accumulates on depletion of HectH9. Ls174T cells were transfected with either control siRNA oligonucleotides or siRNA targeting HectH9; 50 h after transfection, cells were fractionated as described in the legend of Figure 3E.
Co-depletion of HectH9 together with Miz1 reverted the decrease in steady-state levels of TopBP1 and Atr, demonstrating that HectH9 is required for the degradation of both proteins in Miz1-depleted cells (Figure 4B). In contrast, depletion of HectH9 only partially restored the expression of p53, demonstrating that the decrease in p53 steady-state levels that is observed in Miz1-depleted cells is, in part, due to HectH9-mediated degradation and, in part, due to changes in P53 transcription. Fractionation of cell extracts showed that HectH9 is not bound to chromatin and TopBP1 is accumulated in the non-chromatin-bound fraction after the depletion of HectH9, similar to that observed after the depletion of Miz1 in the presence of proteasome inhibitors (Figure 4C). Taken together, the data show that HectH9 can ubiquitinate TopBP1 in vivo and that Miz1 protects TopBP1 from HectH9-mediated degradation.
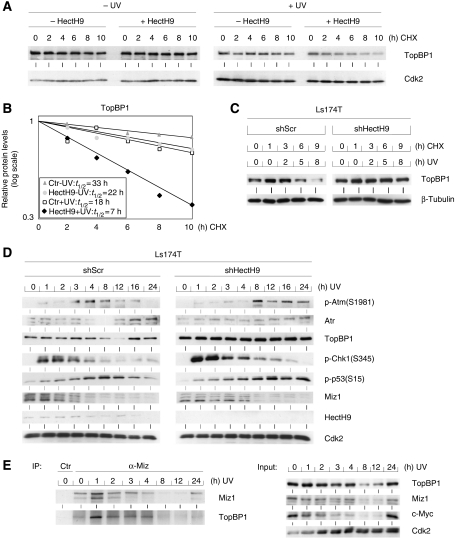
To determine under which physiological circumstances HectH9 has a role in TopBP1 turnover, we transiently transfected expression plasmids encoding TopBP1 and HectH9 into HeLa cells and determined the stability of TopBP1 on treatment with cycloheximide (Figure 5A). Ectopically expressed TopBP1 was stable during the time course of the experiment in unstressed cells, both in the presence and absence of HectH9. We reasoned that this might be due to binding to endogenous Miz1 and used two experiments to test this hypothesis: first, we found that HectH9 induced the turnover of TopBP1S1159A, a point mutant that fails to associate with Miz1 (Supplementary Figure 4b) (Liu et al, 2006). Second, we have shown earlier that TopBP1 dissociates from Miz1 upon UV irradiation (Herold et al, 2002). Consistently, HectH9 destabilized TopBP1 when cells were treated with UV before the addition of cycloheximide (Figure 5A). Quantification of the result showed that the half-life of ectopically expressed TopBP1 after exposure to UV decreased from 18 to 7 h on expression of HectH9 (Figure 5B).
Figure 5.
Regulation of TopBP1 and Atr stability and Atr-dependent signal transduction by HectH9. (A) Expression of HectH9 induces the degradation of TopBP1 on UV irradiation. HeLa cells were transfected with expression vectors encoding HectH9 and TopBP1. Cells were either left untreated (−) or exposed to UVB (+) as shown. After 1 h, cycloheximide was added and lysates were harvested at the indicated times after the addition of cycloheximide. (B) Quantification of the experiment. The values shown are obtained from the experiment shown in (A) and are normalized to the Cdk2 control. For each experimental condition, the signal at the start of the experiment was set to one. (C) Depletion of HectH9 stabilizes TopBP1 after UV irradiation. Ls174T cells were infected with retroviruses expressing either scrambled shRNA or shRNA targeting HectH9. After selection, cells were UV irradiated. 1 h before UV irradiation, cycloheximide was added. Cells were harvested at the indicated times afterwards. The panels show immunoblots probed with the indicated antibodies. (D) HectH9 is required to end Atr-dependent signalling after UV irradiation. The same cells as described in (C) were exposed to irradiation as before. At the indicated times after irradiation, cells were harvested and lysates were prepared. The panels show immunoblots probed with the indicated antibodies. (E) Levels of Myc, Miz1, TopBP1 and of the Miz1/TopBP1 complex after UV irradiation. The experiment was performed as in (D).
To determine whether endogenous HectH9 has a role in regulating TopBP1 stability upon UV irradiation, we depleted HectH9 using retroviruses expressing shRNA that targets HECTH9. Similar to the effects observed using siRNA in Miz1-depleted cells, depletion of HectH9 by shRNA stabilized TopBP1 after UV irradiation, demonstrating a role of endogenous HectH9 in TopBP1 turnover in cells expressing normal levels of Miz1 (Figure 5C). In contrast, depletion of either hHYD (human hyperplastic disc), an E3 ligase implicated in the ubiquitination of TopBP1 (Honda et al, 2002), BARD1 or BRCA1 had no effect on protein levels of TopBP1 (not shown).
To better understand the physiological role of HectH9-mediated turnover of TopBP1, we performed time course experiments in Ls174T cells expressing either shRNA targeting HECTH9 or a scrambled control and harvested extracts at different time points after UV irradiation (Figure 5D and E; for a quantitation of the results, see Supplementary Figure 4c). In control cells, levels of TopBP1 and Miz1 declined around 8 h after UV irradiation; they recovered after 24 h. Consequently, the Miz1/TopBP1 complex was detectable in unstressed cells and for 4 h after irradiation, but was not detectable at 8 and 12 h after irradiation (Figure 5E). We note that the decrease in TopBP1 protein level is preceded by a decrease in TOPBP1 mRNA expression (Supplementary Figure 4c).
In these experiments, phosphorylation of Chk1 rapidly increased after UV irradiation and began to decline after 3–4 h. In contrast, phosphorylation of p53 and Atm increased with a delayed kinetics and began to decline at 12–16 h in control cells. Depletion of HectH9 led to moderately increased levels of Chk1 phosphorylation (similar to that observed in cells expressing Miz1S428A), but had no effect on the kinetics of Chk1 phosphorylation; this may be due to the degradation of phosphorylated Chk1 by Cul1- or Cul4-dependent ubiquitin ligases (Zhang et al, 2005). In contrast, phosphorylation of p53 steadily increased in HectH9-depleted cells up to 24 h after irradiation; furthermore, depletion of HectH9 had no effect on the levels of Miz1, but abrogated the transient decrease in TopBP1 and Atr levels that is observed in control cells. Finally, phosphorylation of Atm (which is mediated by Atr upon UV irradiation; Stiff et al, 2006) was extended in HectH9-depleted cells. Our data show that HectH9 has an essential function in ending Atr-dependent signal transduction and this function correlates with a HectH9-dependent decrease in TopBP1 and Atr steady-state levels before the end of the Atr-dependent signal.
To determine whether HectH9 also has a role in responses to double-strand breaks, we exposed control and HectH9-depleted cells to gamma-irradiation (Supplementary Figure 4d). Depletion of HectH9 had no effect on the time course of Atm and Chk2 phosphorylation, arguing that HectH9 has no role in terminating the checkpoint response to double-strand DNA breaks. In contrast, depletion of HectH9 led to an accumulation of phosphorylated p53, consistent with its role in the degradation of p53 (Chen et al, 2005).
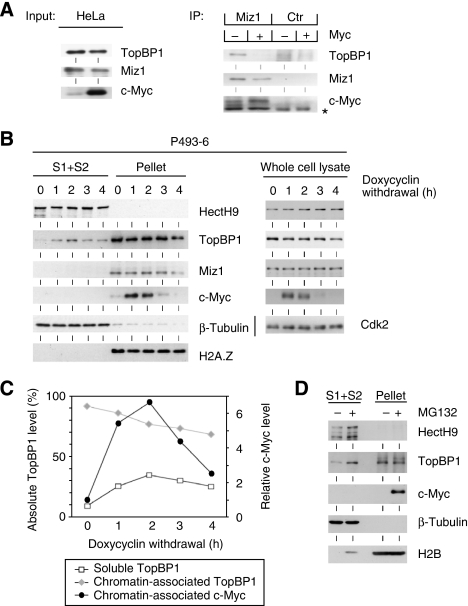
In unstressed cells, a significant fraction of Miz1 is associated with TopBP1 and Myc. We have been unable to co-precipitate TopBP1 with Myc, suggesting that Miz1 forms mutually exclusive complexes with Myc and TopBP1. Consistent with this notion, endogenous TopBP1 was recovered in α-Miz1 immunoprecipitates from control cells, but not from cells that had been transfected with CMV-driven expression plasmids that express Myc (Figure 6A). To demonstrate directly that Myc displaces TopBP1 from chromatin, we used P493-6 B cells, which express Myc under the control of a tetracyclin-repressed promoter (Schuhmacher et al, 1999). Withdrawal of doxycyclin rapidly induces the expression of Myc in these cells (Figure 6B and C). Fractionation experiments showed that virtually all TopBP1 was associated with chromatin in cells before the induction of Myc expression. In contrast, induction of Myc led to a rapid and transient accumulation of TopBP1 in the soluble fraction, which preceded a decrease in the total levels of TopBP1; controls confirmed that this was not due to a cross-contamination of soluble fractions with chromatin proteins, as histone H2A.Z remained bound to the chromatin on induction of Myc. As observed for Ls174T cells, treatment of P493-6 cells with the proteasome inhibitor MG132 further increased the amount of soluble TopBP1 but had little effect on chromatin-bound TopBP1, demonstrating that soluble TopBP1 undergoes rapid turnover (Figure 6D).
Figure 6.
Role of Myc in the regulation of TopBP1 protein levels by Miz1. (A) Myc antagonizes the binding of TopBP1 to Miz1. HeLa cells were transfected with either control vectors or vectors expressing Myc as indicated; at the time of transfection, MG132 was added to inhibit proteasomal turnover of TopBP1. 48 h after transfection, cells were harvested and lysates were immunoprecipitated with α-Miz1 antibodies; immunoprecipitates were probed with α-TopBP1 antibodies to visualize the endogenous Miz1/TopBP1 complex. The asterisk depicts a nonspecific band. (B) Induction of Myc leads to a release of TopBP1 from chromatin. P493-6 B cells were cultured in the presence of doxycyclin and transferred to a medium lacking doxycyclin at the start of the experiment. Cells were harvested at the indicated times after transfer and soluble and chromatin-associated fractions were prepared as described above. The panels document immunoblots probed with the indicated antibodies. (C) Quantitation of the fractionation experiments. The panel shows the changes in the levels of the indicated proteins in each fraction. Average values obtained from two independent experiments are shown. (D) Proteasome inhibition increases the soluble pool of TopBP1. P493-6 cells were treated for 10 h with MG132 and subsequently fractionated as describe before. The panels document immunoblots probed with the indicated antibodies.
Continuous expression of Myc should therefore lower the steady-state levels of TopBP1 by dissociating the protein from Miz1 and displacing it from chromatin. To test this prediction, we infected Ls174T cells with retroviruses expressing human Myc. Immunoblotting showed that infected cells expressed 2- to 3-fold elevated levels of Myc (Figure 7A). Expression of Myc levels led to a decrease in the levels of TopBP1 and reduced phosphorylation of p53 and Chk1 in response to UV irradiation. Similar results were obtained in several independent clones of infected cells, excluding the possibility that the reduction in TopBP1 levels is due to clonal variation (not shown). Ectopic expression of Myc had no effect on the levels of TOPBP1 mRNA, demonstrating that it regulates the steady-state levels of TopBP1 at a post-transcriptional level (Figure 7B). Several clones of Ls174T cells expressing MycV394D, a mutant of Myc that is unable to bind to Miz1 (Herold et al, 2002), showed unaltered levels of TopBP1 and an unaltered phosphorylation of Chk1 upon UV irradiation, suggesting that Myc regulates the levels of TopBP1 through binding to Miz1 (Supplementary Figure 5).
Figure 7.
Myc affects TopBP1 levels and chromatin association. (A) Ectopic expression of Myc reduces the steady-state levels of TopBP1 and Atr. Ls174T cells were infected with retroviruses expressing Myc. After selection, they were either mock treated (−) or exposed to UV irradiation (+). Cells were harvested 2 h after UV irradiation. The panels show immunoblots probed with the indicated antibodies. (B) Ectopic expression of Myc does not affect mRNA levels of checkpoint proteins. Cells were infected and treated as described in (A). Cells were harvested 2 h after UV irradiation and RNA was prepared. ‘∅' designates a control RT–PCR reaction lacking cDNA. (C) Depletion of Myc enhances the steady-state levels of TopBP1. Ls174T cells were transfected with either control siRNA or siRNA targeting MYC. The panels show immunoblots of the indicated proteins. (D) Depletion of Myc depletes soluble and elevates chromatin-bound pools of TopBP1. Ls174T cells were transfected with two different siRNAs targeting MYC; MG132 was added 4 h before fractionation. The fractionation was performed as described before and immunoblots were probed with the indicated antibodies. (E) Depletion of Myc has no effect on the levels of TOPBP1 and ATR mRNAs. Ls174T cells were transfected as described in (C). RNA was isolated 36 h after transfection. RT–PCR assays documenting the expression of the indicated genes are shown.
To test whether endogenous Myc regulates chromatin association of TopBP1, we depleted Myc using soluble siRNA. As expected, this led to an increase in the total levels of TopBP1 and also Atr (Figure 7C). Fractionation experiments performed in the presence of MG132 to block proteasomal degradation showed that reduction of endogenous Myc levels depleted the soluble but enhanced the chromatin-bound pool of TopBP1 (Figure 7D). Parallel RT–PCR experiments showed that the levels of TOPBP1 and ATR mRNAs were unaffected by the depletion of Myc (Figure 7E). Identical results were obtained in HeLa cells (not shown). Time course experiments showed that depletion of Myc had no effect on the initiation of Atr signalling after UV irradiation; instead, Myc-depleted cells showed extended phosphorylation of Atm and p53, but not of Chk1 (Supplementary Figure 6). These data support a model in which Myc antagonizes the binding of TopBP1 to Miz1, leading to a reduction in the levels of chromatin-associated TopBP1 and an attenuation of Atr-dependent signal transduction and suggest that endogenous Myc facilitates the end of checkpoint signalling through this mechanism.
Discussion
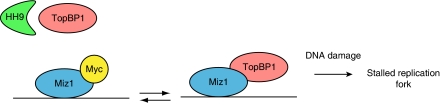
Previous work had shown that TopBP1 associates with Miz1 (Herold et al, 2002; Liu et al, 2006). We have shown here that Miz1 is required for the binding of a significant fraction of TopBP1 to chromatin and for the protection of TopBP1 from proteasomal degradation. This correlates with a requirement for Miz1 in Atr-dependent signal transduction. Time course experiments reveal that Miz1 and TopBP1 form a complex in unstressed cells and early after UV irradiation. The loss of the Miz1/TopBP1 complex at later time points correlates with the end of Atr-dependent signal transduction. Our data raise the possibility that Miz1 itself is a direct part of the Atr-dependent checkpoint; however, we have been unable to detect any association between Miz1 and Atr. We therefore suggest that the Miz1/TopBP1 complex functions as a ‘reservoir', from which TopBP1 is recruited to stalled replication forks. A model describing our findings is shown in Figure 8. How exactly the Miz1-bound chromatin pool of TopBP1 relates to the pool of TopBP1 that is bound to origins of DNA replication remains open at present (Van Hatten et al, 2002).
Figure 8.
Myc and Miz1 regulate TopBP1 stability and Atr/Chk1 function. The figure shows a model summarizing our findings; details are described in the text.
In the absence of Miz1, TopBP1 is released from chromatin and degraded by the HectH9 ubiquitin ligase. HectH9 is not bound to chromatin, therefore Miz1 may sequester TopBP1 from HectH9-mediated degradation. Furthermore, Miz1 binds to a specific domain of HectH9 and may therefore interfere with the binding of TopBP1 to HectH9 (Adhikary et al, 2005); both possibilities are not mutually exclusive. These results extend several previous reports that have implicated HectH9 in the cellular response to DNA damage. Notably, HectH9 itself is a substrate for the Atm/Atr kinases; it is also required for responses to DNA damage in the G1, S and G2 phases of the cell cycle (Mu et al, 2007). For example, depletion of HectH9 leads to radio-resistant DNA synthesis (Mu et al, 2007). HectH9 binds to Cdc6 and degrades it in response to DNA damage, suggesting that degradation of Cdc6 is one of the mechanisms through which HectH9 contributes to the arrest of DNA replication in response to DNA damage (Hall et al, 2007). Similar to what we find for TopBP1, HectH9 degrades a pool of Cdc6 that is released from chromatin (Hall et al, 2007). Notably, however, the time points at which Cdc6 and TopBP1 are degraded appear to be different, as Cdc6 is degraded rapidly, but TopBP1 is degraded with delayed kinetics after UV irradiation, suggesting that HectH9 may have a dual function in arresting DNA replication and in ending checkpoint signalling after DNA damage. TopBP1 contains multiple BRCT domains, which bind to phosphorylated serine and threonine residues: possibly, therefore, phosphorylation of HectH9 by Atm or Atr is required for the degradation of TopBP1, providing a negative feedback signal. Such a scenario could also explain why we have so far failed to reconstitute the reaction in vitro, as phosphorylation of HectH9 by Atr would not occur in an in vitro system.
TopBP1 is also released rapidly from chromatin in response to the induction of Myc. The easiest explanation for this observation is to suggest that Myc competes with TopBP1 for binding to Miz1. Currently, we have no proof that the release of TopBP1 occurs through direct competition for Miz1; however, the rapidity of the release and the fact that Myc needs to bind to Miz1 to reduce the steady-state levels of TopBP1 make this explanation likely. Notably, recent work has demonstrated a transcription-independent function of Myc in DNA replication both in Xenopus and mammalian cells (Dominguez-Sola et al, 2007). Atr activity limits origin usage also during unperturbed DNA replication in Xenopus oocytes (Shechter et al, 2004). Similarly, Chk1-deficient mammalian cells show an increased use of replication origins (Maya-Mendoza et al, 2007). We suggest, therefore, that the effects of Myc on Atr activity that are described here can account for the non-transcriptional functions that have been ascribed to Myc.
The mechanisms described here may provide insight into two further phenomena closely associated with transformation by Myc: first, cells that express deregulated Myc show increased H2A.X phosphorylation and activation of Atm and Chk2, indicative of the accumulation of DNA double-strand breaks (Vafa et al, 2002). This is also observed in animal models such as Myc-induced lymphomas, demonstrating that it does not reflect a response to the artificially stressful conditions in tissue culture (e.g. Gorrini et al, 2007). We show here that the deregulated expression of Myc in human tumour cells reduces the steady-state levels of TopBP1 and Atr and compromises Atr-dependent signal transduction. Inhibition of Chk1 function in mammalian cells leads to double-strand breaks and phosphorylation of Atm/Atr target proteins, such as H2A.X (Syljuasen et al, 2005). Similarly, TopBP1-deficient cells show increased H2A.X phosphorylation and activation of Atm and Chk2, indicative of the accumulation of DNA double-strand breaks in the absence of TopBP1 (Kim et al, 2005). Potentially, therefore, the decrease in Atr function and TopBP1 stability that is induced by Myc can account for the Myc-dependent induction of genomic instability.
Second, activation of a conditional allele of TopBP1 has recently been shown to induce cellular senescence even in the absence of DNA damage (Toledo et al, 2008). Senescence is considered a tumour-protective mechanism that limits oncogenic transformation by Ras; furthermore, Ras-induced senescence is closely related to and involves components of the DNA damage response pathway (Di Micco et al, 2006). Ectopic expression of Myc facilitates oncogenic transformation by Ras; we suggest, therefore, that regulating chromatin association and steady-state levels of TopBP1 may be one pathway through which Myc allows Ras-transformed cells to escape from senescence.
Materials and methods
Cell culture and comet assays
Ls174T cells were stably transfected with an ecotropic receptor and subsequently infected with pZWL-neo-based retroviruses expressing Miz1S428A (Wanzel et al, 2005). A clone expressing elevated levels of Miz1 was selected for further experiments. For FACS analyses, cells were labelled for 1 h with BrdU (20 μM) and stained with α-BrdU antibodies (Becton-Dickinson) and propidium iodide (70 μM). UVB irradiation was carried out using 250 J/m2. Alkaline comet assays were performed with the Comet Assay kit (Trevigen). Data were evaluated using the Wilcoxon rank-sum test.
siRNA
siRNA oligonucleotides were transfected using oligofectamin (Invitrogen). siRNA targeting MIZ1 Myc and HectH9 was obtained from Dharmacon (ON-TARGETplus). shRNA sequences are available upon request.
Ubiquitination assays
293T cells were transfected with CMV-driven expression plasmids expressing HA-Ub, TopBP1, HectH9ΔN and Miz1 as indicated. 36 h after transfection, cells were treated with proteasome inhibitor MG132 (40 μM) for 9 h. Cells were collected, lysed in 200 μl of lysis buffer (50 mM Tris–HCl, pH 7.5, 0.5 mM EDTA, 1% SDS) and boiled for 10 min. Lysates were clarified by centrifugation. Supernatant was diluted 10 times with 0.5% NP-40 buffer and immunoprecipitated with α-HA matrix (Covance; AFC-101P). Immunoprecipitates were washed four times and resolved by 5% SDS–PAGE, followed by immunoblotting with α-TopBP1 antibodies.
Chromatin fractionation
In total, 3 × 106 cells were washed in PBS and re-suspended in 200 μl of solution A (10 mM Hepes-KOH, pH 7.9, 10 mM KCl, 1.5 mM MgCl2, 0.34 M sucrose, 10% glycerol, 1 mM DTT, protease and phosphatase inhibitors, 0.1% Triton X-100). Cells were incubated on ice for 5 min, and cytoplasmic (S1) and nuclear fractions were harvested by centrifugation at 1300 g for 4 min. Isolated nuclei were then washed in solution A, lysed in 150 μl solution B (3 mM EDTA, 0.2 mM EGTA, 1 mM DTT, protease and phosphatase inhibitors), and incubated on ice for 10 min. The soluble nuclear (S2) and chromatin fractions were harvested by centrifugation at 1700 g for 4 min. Isolated chromatin (pellet) was then washed in solution B, spun down at 10 000 g, and resuspended in sample buffer. Fractions S1 and S2 were pooled to one soluble fraction.
Antibodies and immunoblotting
Cell extracts were prepared in either NP-40 buffer (50 mM Tris–HCl, pH 8.0, 150 mM NaCl, 1% NP-40) or TNN buffer (50 mM Tris–HCl, pH 8.0, 120 mM NaCl, 5 mM EDTA, 0.5% NP-40, protease and phosphatase inhibitor cocktail (Roche)). Proteins were resolved by SDS–PAGE and transferred to PVDF membrane (Millipore). Antibodies were purchased from Santa Cruz (Atr, Cdk2, Chk1, p-p53(S15), p53, p21), Novus (TopBP1), Cell Signaling (p-Chk1(S345)), Upstate (p-H2A.X(S139)) and ProSci (HectH9). Antibodies for Miz1 (10E2), Myc (9E10) and HectH9 have been described earlier (Herold et al, 2002; Adhikary et al, 2005). Immunoblots were quantitated using ImageJ software.
RT–PCR
Primer sequences used for RT–PCR are available upon request.
Supplementary Material
Supplementary Figure 1
Supplementary Figure 2
Supplementary Figure 3
Supplementary Figure 4
Supplementary Figure 5
Supplementary Figure 6
Supplementary Information
Acknowledgments
This study was supported by grants from the Deutsche Forschungsgemeinschaft (FOG 551 to ME) and the Thyssen Stiftung (ME), the FP6 Program of the European Union (INTACT; ME and RB), the Dutch Cancer Society (RB) and from the Netherlands Organization for Scientific Research (NWO) (RB). We thank Bianca Jebavy and Michael Neuhaus for excellent technical support.
References
- Abraham RT (2001) Cell cycle checkpoint signaling through the ATM and ATR kinases. Genes Dev 15: 2177–2196 [DOI] [PubMed] [Google Scholar]
- Adhikary S, Eilers M (2005) Transcriptional regulation and transformation by Myc proteins. Nat Rev Mol Cell Biol 6: 635–645 [DOI] [PubMed] [Google Scholar]
- Adhikary S, Marinoni F, Hock A, Hulleman E, Popov N, Beier R, Bernard S, Quarto M, Capra M, Goettig S, Kogel U, Scheffner M, Helin K, Eilers M (2005) The ubiquitin ligase HectH9 regulates transcriptional activation by Myc and is essential for tumor cell proliferation. Cell 123: 409–421 [DOI] [PubMed] [Google Scholar]
- Byun TS, Pacek M, Yee MC, Walter JC, Cimprich KA (2005) Functional uncoupling of MCM helicase and DNA polymerase activities activates the ATR-dependent checkpoint. Genes Dev 19: 1040–1052 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen D, Kon N, Li M, Zhang W, Qin J, Gu W (2005) ARF-BP1/Mule is a critical mediator of the ARF tumor suppressor. Cell 121: 1071–1083 [DOI] [PubMed] [Google Scholar]
- Delacroix S, Wagner JM, Kobayashi M, Yamamoto K, Karnitz LM (2007) The Rad9–Hus1–Rad1 (9-1-1) clamp activates checkpoint signaling via TopBP1. Genes Dev 21: 1472–1477 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Di Micco R, Fumagalli M, Cicalese A, Piccinin S, Gasparini P, Luise C, Schurra C, Garre M, Nuciforo PG, Bensimon A, Maestro R, Pelicci PG, d'Adda di Fagagna F (2006) Oncogene-induced senescence is a DNA damage response triggered by DNA hyper-replication. Nature 444: 638–642 [DOI] [PubMed] [Google Scholar]
- Dominguez-Sola D, Ying CY, Grandori C, Ruggiero L, Chen B, Li M, Galloway DA, Gu W, Gautier J, Dalla-Favera R (2007) Non-transcriptional control of DNA replication by c-Myc. Nature 448: 445–451 [DOI] [PubMed] [Google Scholar]
- Gebhardt A, Kosan C, Herkert B, Moroy T, Lutz W, Eilers M, Elsasser HP (2007) Miz1 is required for hair follicle structure and hair morphogenesis. J Cell Sci 120: 2586–2593 [DOI] [PubMed] [Google Scholar]
- Gorrini C, Squatrito M, Luise C, Syed N, Perna D, Wark L, Martinato F, Sardella D, Verrecchia A, Bennett S, Confalonieri S, Cesaroni M, Marchesi F, Gasco M, Scanziani E, Capra M, Mai S, Nuciforo P, Crook T, Lough J, Amati B (2007) Tip60 is a haplo-insufficient tumour suppressor required for an oncogene-induced DNA damage response. Nature 448: 1063–1067 [DOI] [PubMed] [Google Scholar]
- Hall JR, Kow E, Nevis KR, Lu CK, Luce KS, Zhong Q, Cook JG (2007) Cdc6 stability is regulated by the Huwe1 ubiquitin ligase after DNA damage. Mol Biol Cell 18: 3340–3350 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herold S, Wanzel M, Beuger V, Frohme C, Beul D, Hillukkala T, Syvaoja J, Saluz HP, Hänel F, Eilers M (2002) Negative regulation of the mammalian UV response by Myc through association with Miz-1. Mol Cell 10: 509–521 [DOI] [PubMed] [Google Scholar]
- Honda Y, Tojo M, Matsuzaki K, Anan T, Matsumoto M, Ando M, Saya H, Nakao M (2002) Cooperation of HECT-domain ubiquitin ligase hHYD and DNA topoisomerase II-binding protein for DNA damage response. J Biol Chem 277: 3599–3605 [DOI] [PubMed] [Google Scholar]
- Jeon Y, Lee KY, Ko MJ, Lee YS, Kang S, Hwang DS (2007) Human TopBP1 participates in cyclin E/CDK2 activation and preinitiation complex assembly during G1/S transition. J Biol Chem 282: 14882–14890 [DOI] [PubMed] [Google Scholar]
- Kim JE, McAvoy SA, Smith DI, Chen J (2005) Human TopBP1 ensures genome integrity during normal S phase. Mol Cell Biol 25: 10907–10915 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumagai A, Lee J, Yoo HY, Dunphy WG (2006) TopBP1 activates the ATR–ATRIP complex. Cell 124: 943–955 [DOI] [PubMed] [Google Scholar]
- Liu K, Paik JC, Wang B, Lin FT, Lin WC (2006) Regulation of TopBP1 oligomerization by Akt/PKB for cell survival. EMBO J 25: 4795–4807 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maya-Mendoza A, Petermann E, Gillespie DA, Caldecott KW, Jackson DA (2007) Chk1 regulates the density of active replication origins during the vertebrate S phase. EMBO J 26: 2719–2731 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mu JJ, Wang Y, Luo H, Leng M, Zhang J, Yang T, Besusso D, Jung SY, Qin J (2007) A proteomic analysis of ataxia telangiectasia-mutated (ATM)/ATM-Rad3-related (ATR) substrates identifies the ubiquitin–proteasome system as a regulator for DNA damage checkpoints. J Biol Chem 282: 17330–17334 [DOI] [PubMed] [Google Scholar]
- Patel JH, McMahon SB (2007) BCL2 is a downstream effector of MIZ-1 essential for blocking c-MYC-induced apoptosis. J Biol Chem 282: 5–13 [DOI] [PubMed] [Google Scholar]
- Schuhmacher M, Staege MS, Pajic A, Polack A, Weidle UH, Bornkamm GW, Eick D, Kohlhuber F (1999) Control of cell growth by c-Myc in the absence of cell division. Curr Biol 9: 1255–1258 [DOI] [PubMed] [Google Scholar]
- Seoane J, Le HV, Massague J (2002) Myc suppression of the p21(Cip1) Cdk inhibitor influences the outcome of the p53 response to DNA damage. Nature 419: 729–734 [DOI] [PubMed] [Google Scholar]
- Shechter D, Costanzo V, Gautier J (2004) ATR and ATM regulate the timing of DNA replication origin firing. Nat Cell Biol 6: 648–655 [DOI] [PubMed] [Google Scholar]
- Smits VA, Reaper PM, Jackson SP (2006) Rapid PIKK-dependent release of Chk1 from chromatin promotes the DNA-damage checkpoint response. Curr Biol 16: 150–159 [DOI] [PubMed] [Google Scholar]
- Stiff T, Walker SA, Cerosaletti K, Goodarzi AA, Petermann E, Concannon P, O'Driscoll M, Jeggo PA (2006) ATR-dependent phosphorylation and activation of ATM in response to UV treatment or replication fork stalling. EMBO J 25: 5775–5782 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Syljuasen RG, Sorensen CS, Hansen LT, Fugger K, Lundin C, Johansson F, Helleday T, Sehested M, Lukas J, Bartek J (2005) Inhibition of human Chk1 causes increased initiation of DNA replication, phosphorylation of ATR targets, and DNA breakage. Mol Cell Biol 25: 3553–3562 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Toledo LI, Murga M, Gutierrez-Martinez P, Soria R, Fernandez-Capetillo O (2008) ATR signaling can drive cells into senescence in the absence of DNA breaks. Genes Dev 22: 297–302 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vafa O, Wade M, Kern S, Beeche M, Pandita TK, Hampton GM, Wahl GM (2002) c-Myc can induce DNA damage, increase reactive oxygen species, and mitigate p53 function. A mechanism for oncogene-induced genetic instability. Mol Cell 9: 1031–1044 [DOI] [PubMed] [Google Scholar]
- van de Wetering M, Sancho E, Verweij C, de Lau W, Oving I, Hurlstone A, van der Horn K, Batlle E, Coudreuse D, Haramis AP, Tjon-Pon-Fong M, Moerer P, van den Born M, Soete G, Pals S, Eilers M, Medema R, Clevers H (2002) The beta-catenin/TCF-4 complex imposes a crypt progenitor phenotype on colorectal cancer cells. Cell 111: 241–250 [DOI] [PubMed] [Google Scholar]
- Van Hatten RA, Tutter AV, Holway AH, Khederian AM, Walter JC, Michael WM (2002) The Xenopus Xmus101 protein is required for the recruitment of Cdc45 to origins of DNA replication. J Cell Biol 159: 541–547 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wanzel M, Kleine-Kohlbrecher D, Herold S, Hock A, Berns K, Park J, Hemmings B, Eilers M (2005) Akt and 14-3-3eta regulate Miz1 to control cell-cycle arrest after DNA damage. Nat Cell Biol 7: 30–41 [DOI] [PubMed] [Google Scholar]
- Weber A, Marquardt J, Elzi D, Forster N, Starke S, Glaum A, Yamada D, Defossez PA, Delrow J, Eisenman RN, Christiansen H, Eilers M (2008) Zbtb4 represses transcription of P21CIP1 and controls the cellular response to p53 activation. EMBO J 27: 1563–1574 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang YW, Otterness DM, Chiang GG, Xie W, Liu YC, Mercurio F, Abraham RT (2005) Genotoxic stress targets human Chk1 for degradation by the ubiquitin–proteasome pathway. Mol Cell 19: 607–618 [DOI] [PubMed] [Google Scholar]
- Zou L, Elledge SJ (2003) Sensing DNA damage through ATRIP recognition of RPA–ssDNA complexes. Science 300: 1542–1548 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Figure 1
Supplementary Figure 2
Supplementary Figure 3
Supplementary Figure 4
Supplementary Figure 5
Supplementary Figure 6
Supplementary Information