Abstract
DnaJ proteins often bind to unfolded substrates and recruit their Hsp70 partners. This induces a conformational change in the Hsp70 that stabilizes its binding to substrate. By some unknown mechanism, the DnaJ protein is released. We examined the requirements for the release of ERdj3, a mammalian ER DnaJ, from substrates and found that BiP promoted the release of ERdj3 only in the presence of ATP. Mutations in ERdj3 or BiP that disrupted their interaction interrupted the release of ERdj3. BiP mutants that were defective in any step of the ATPase cycle were also unable to release ERdj3. These results demonstrate that a functional interaction between ERdj3 and BiP, including both a direct interaction and the ability to stimulate BiP's ATPase activity are required to release ERdj3 from substrate and support a model where ERdj3 must recruit BiP and stimulate its high-affinity association with the substrate through activation of ATP hydrolysis to trigger its own release from substrates. On the basis of similarities among DnaJs and Hsp70s, this is likely to be applicable to other Hsp70–DnaJ pairs.
Keywords: BiP, DnaJ-like proteins, endoplasmic reticulum, protein folding
Introduction
The Hsp70 family of molecular chaperones is a highly conserved, widely expressed, and well-studied group of proteins. These chaperones are found in all organisms where they have a function in every cellular organelle and are essential for nearly all cellular processes. The binding of Hsp70 proteins to non-native structures on a vast array of substrate proteins can serve to stabilize folding intermediates, prevent their aggregation, and aid in protein folding and assembly. This is achieved through direct interaction of the C-terminal substrate-binding domain (SBD) of Hsp70 proteins with exposed hydrophobic residues on substrate proteins. Peptide-binding studies suggested a preference for extended chains in which the hydrophobic amino acids would be oriented in a single direction to engage the peptide-binding pocket of the Hsp70 protein, a possibility that is supported by NMR (Landry et al, 1992) and crystallographic (Zhu et al, 1996) studies. The binding and release of substrates to the SBD of Hsp70 proteins are tightly regulated by the highly conserved N-terminal nucleotide-binding domain (NBD) (Liberek et al, 1991b), which can bind either ATP or ADP. When ATP occupies the cleft of the NBD, the SBD is in an open configuration, which has both a high on and high off rate for unfolded proteins. The hydrolysis of ATP to ADP results in a closure of the lid on the SBD, which stabilizes the interaction with bound proteins. Discharge of the unfolded protein occurs when ADP is released and exchanged for ATP. This reopens the lid on the SBD, which allows the bound substrate to be released and provides an opportunity for it to fold. A number of recent studies shed light on the interaction between the two domains, which controls the activity of this group of chaperones (Jiang et al, 2005; Vogel et al, 2006a, 2006b; Liu and Hendrickson, 2007; Awad et al, 2008).
The Hsp70 ATPase cycle, which is essential to the chaperoning process, is controlled by a number of cofactors that regulate either ATP hydrolysis or nucleotide exchange. DnaJ was originally identified along with DnaK (Hsp70) in a genetic screen in Escherichia coli for genes that are required for DNA replication (Saito and Uchida, 1977; Yochem et al, 1978). Later it was shown that DnaK and DnaJ are in the same genetic pathway and that DnaJ stimulates the ATPase activity of DnaK, thereby stabilizing the binding of DnaK to substrates (Liberek et al, 1991a). As is the case with Hsp70s, DnaJ proteins are present in all organisms and all organelles, and the number of DnaJ proteins in an organism often exceeds the number of Hsp70 proteins present (Caplan et al, 1993; Cheetham and Caplan, 1998). All DnaJ proteins possess a highly conserved ∼70 amino-acid ‘J' domain, which contains an invariant tripeptide sequence, His-Pro-Asp, that is required to interact with the ATP-bound form Hsp70 proteins (Mayer et al, 1999). Similar to Hsp70 proteins, at least some DnaJ proteins can bind directly to unfolded substrates (Cheetham and Caplan, 1998; Fan et al, 2003). Peptide-binding studies for E. coli DnaJ revealed significant overlap with the peptides that bound DnaK (Rudiger et al, 2001), arguing that DnaJ was likely to also bind to extended hydrophobic residues on unfolded proteins. This possibility was supported by crystallographic data obtained for a peptide bound to the yeast cytosolic DnaJ protein, Ydj1 (Li et al, 2003). The fact that DnaJs specifically interact with the ATP-bound form of Hsp70s led to a model (Mayer et al, 1999) where DnaJ proteins would bind first to unfolded proteins, recruit the ATP-bound or ‘open' form of Hsp70 to the substrate, and then stimulate its ATPase activity to ‘close' it onto the substrate more stably. This model was supported by data showing that a cytosolic DnaJ protein bound to nascent chains extruding from the ribosome before the Hsp70 protein did (Hendrick et al, 1993), and by in vitro binding studies with DnaK, DnaJ, and denatured luciferase (Szabo et al, 1994) and with HscA and HscB, a specialized Hsp70–DnaJ pair of E. coli (Silberg et al, 2004). However, these studies did not reveal how DnaJ proteins were released from the substrate. Unlike Hsp70, DnaJ proteins do not bind to nucleotide and have not been demonstrated to exist in different conformational states.
The mammalian ER possesses at least six DnaJ family members (Brightman et al, 1995; Meyer et al, 2000; Tyedmers et al, 2000; Yu et al, 2000; Shen et al, 2002; Cunnea et al, 2003; Rutkowski et al, 2007; Petrova et al, 2008). One of these, ERdj3, was shown previously by our group to bind to a number of unfolded proteins in the ER that were BiP substrates (Shen and Hendershot, 2005). When the binding of wild-type and mutant (HPD → QPD) ERdj3 to several different substrates was compared, we consistently found that mutant ERdj3 bound quantitatively better and longer than wild-type ERdj3 (Shen and Hendershot, 2005). The present study was undertaken to better understand the requirements for releasing DnaJ proteins from substrates. Using a series of ERdj3 and BiP mutants, we found that release of ERdj3 was not simply due to a competition with BiP, but that a functional interaction between ERdj3 and BiP was required. We hypothesize that once ERdj3 has recruited BiP to the substrate it must stimulate BiP's ATPase activity to induce high-affinity binding of BiP to the substrate, which produces the signal for ERdj3 release from substrate.
Results
Comparison of the effects of J-domain mutations on ERdj3's ability to associate with substrate both in vivo and in vitro
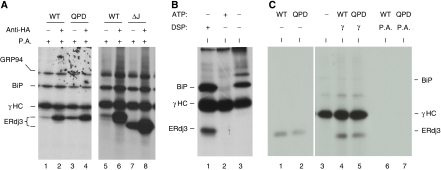
It has been shown that the J domains of DnaJ proteins are important for interactions with their Hsp70 partners, where the signature HPD motif in the J domain has an indispensable function. We demonstrated earlier that a QPD mutation in ERdj3 abrogated its ability to interact with its ER Hsp70 partner, BiP, both physically and functionally (Shen and Hendershot, 2005). To determine whether the interaction with BiP was crucial for ERdj3's ability to bind to unfolded substrates, we examined the ability of wild-type and mutant ERdj3 to bind to immunoglobulin heavy chain (γHC) both in vivo and in vitro. First, we co-expressed γHC along with HA-tagged versions of either wild-type ERdj3 or two different J-domain mutants (both QPD and ΔJ) in COS-1 cells. Co-immunoprecipitation experiments were performed on 3,3′-dithio-bis (propionic acid N-hydroxysuccinimide ester) (DSP)-crosslinked cell lysates. We found that J-domain mutations did not negatively affect the ability of the mutant ERdj3 proteins to bind to γHC in vivo (Figure 1A). In fact, in both cases there was actually more binding of the QPD and ΔJ mutants to the γHC as compared with the binding of wild-type ERdj3 (compare lanes 3 and 7 to lanes 1 and 5). Wild-type and mutant ERdj3 proteins were also expressed in COS-1 cells alone. Protein A Sepharose beads did not precipitate any of the three proteins (Supplementary Figure S1), demonstrating that the binding observed in Figure 1A is dependent on the co-expression of γHC. These data suggested two things; first, as these mutants are unable to interact with BiP, ERdj3 might bind directly to unfolded substrates, and second these ERdj3 mutants might have a higher affinity for substrate or some component of the ER chaperone complex (Meunier et al, 2002). To directly test the first possibility, we developed an in vitro assay to examine the binding of ERdj3 to γHC in the absence of other resident ER chaperones and folding enzymes. It was based on our previous demonstration that BiP can be released from isolated γHC in vitro with ATP leaving the γHC in a conformation that allows them to reassociate with exogenously added BiP (Wei et al, 1995). As shown in Figure 1B (lane 1), ERdj3 can be isolated with γHC only when cells are pretreated with a membrane permeable crosslinker, DSP, whereas BiP's association with γHC is detectable even without crosslinking (Figure 1B, lane 3). However, addition of ATP releases BiP from the γHC (Figure 1B, lane 2). These free γHC were used for binding to in vitro translated ERdj3 proteins (Figure 1C). We found that unlike the in vivo binding assays, the QPD mutant bound to γHC at similar levels as observed for wild-type ERdj3 (Figure 1C, lanes 4 and 5). These data revealed that ERdj3 associates directly with substrates and also argues that the enhanced binding of ERdj3 mutants to γHC in vivo is unlikely to be due to their having a higher affinity for substrate. Instead, it suggested that something else in the cell might be contributing to the difference between wild-type and mutant ERdj3's association with substrate. On the basis of our previous data showing that mutant ERdj3 remains bound to unfolded Ig light chains much longer than wild-type ERdj3 (Shen and Hendershot, 2005), we hypothesized that the J-domain mutations might be affecting release of the ER DnaJ proteins from substrates and that release might be dependent on a functional interaction with BiP.
Figure 1.
ERdj3 binds to γHC directly. (A) COS-1 cells were co-transfected with cDNAs encoding γHC and the indicated HA-tagged ERdj3 constructs. Metabolically labelled, crosslinked cell lysates were immunoprecipitated with anti-HA or Protein A Sepharose alone. Isolated proteins were separated by reducing SDS–PAGE. (B) Ag8.8 cells were metabolically labelled for 16 h with [35S]methionine and cysteine and incubated with (lane 1) or without (lanes 2 and 3) DSP. Cell lysates were prepared with (lane 2) or without (lanes 1 and 3) ATP and immunoprecipitated with Protein A Sepharose. (C) Wild-type (WT) and QPD mutant (Mut) ERdj3 were in vitro translated and run directly (lanes 1 and 2) or incubated with the free γHC immobilized on Protein A Sepharose beads (lanes 4 and 5) prepared as in lane 2 in Figure 1B or with Protein A beads alone (lanes 6 and 7).
Development of an in vitro system to detect binding and release of ERdj3 from substrates
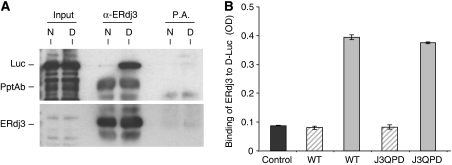
Because the isolation of free γHC was somewhat cumbersome, we wished to develop a simpler in vitro binding assay. We chose denatured firefly luciferase (D-Luc), because it has been widely used as an in vitro substrate for other DnaJ family members, including E. coli DnaJ and two yeast cytosolic DnaJ family members Sis1 and Ydj1 (Szabo et al, 1994; Schumacher et al, 1996; Lu and Cyr, 1998a, 1998b). To determine whether denatured luciferase could serve as an ERdj3 substrate in vitro, we first examined the ability of wild-type ERdj3 to bind to either native or heat-denatured luciferase in solution. For these experiments, luciferase was denatured with heat instead of urea, because we did not want to interfere with the protein–protein interactions required for association and immunoprecipitation. ERdj3 was allowed to interact with native or denatured luciferase and the samples were immunoprecipitated with either a polyclonal anti-ERdj3 antiserum or Protein A Sepharose beads. The association of luciferase was determined by immunoblotting with an anti-luciferase antibody. We found that indeed the binding of ERdj3 to denatured luciferase (D) was readily detectable, whereas its binding to native luciferase (N) was below the level of detection (Figure 2A). This distinction in binding is in keeping with ERdj3 acting as a chaperone and demonstrated that denatured luciferase could be used as an in vitro substrate for ERdj3.
Figure 2.
WT and QPD ERdj3 bind to denatured luciferase similarly in vitro. (A) Temperature denatured (D) or native (N) luciferase (Luc) was directly loaded on a gel (first two lanes) or incubated with bacterially produced recombinant wild-type ERdj3 and then immunoprecipitated with either anti-ERdj3 polyclonal antiserum or with Protein A Sepharose beads alone. Reaction cocktails were subjected to reducing SDS–PAGE and then transferred to a PVDF membrane. The membrane was blotted with either goat anti-luciferase antiserum followed by donkey anti-goat Ig conjugated to HRP or with the polyclonal anti-ERdj3 followed by goat anti-rabbit Ig conjugated to HRP. In both cases, the signal was detected by chemiluminescence. (B) Chemically denatured luciferase (solid grey bars) or binding buffer alone (hatched bars) was used to coat 96-well plates. Recombinant wild-type or the QPD mutant ERdj3 proteins (0.5 μM) were added to the wells and bound ERdj3 was detected with a polyclonal anti-ERdj3 antiserum, followed by donkey anti-rabbit Ig conjugated to alkaline phosphatase. The DNTP substrate was added and after developing, the plates were read on a spectrophotometer and the signal was expressed in OD units. A luciferase-coated well that did not receive ERdj3 protein was treated similarly and serves as a negative control for the antibody (dark grey). All samples were run in triplicate and error bars are indicated.
Next a modified ELISA was developed, which would allow us to readily examine the ability of ERdj3 to bind to luciferase under multiple conditions. For this assay, the luciferase was chemically denatured, as has been done in a number of other studies (Szabo et al, 1994; Lu and Cyr, 1998a, 1998b). First, we tested the binding of wild-type ERdj3 and the QPD mutant to chemically denatured luciferase, which was bound to 96-well plates. Similar to the in vitro binding of these proteins to γHC, we found that both wild-type ERdj3 and the QPD mutant associated equally with chemically denatured luciferase (Figure 2B). Thus, mutation of HPD sequence to QPD did not affect the binding of ERdj3 to chemically denatured luciferase.
ATP does not affect the binding of wild-type ERdj3 or the QPD and HPN mutants to substrate in vitro
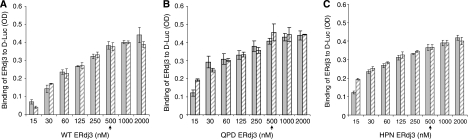
Before testing our hypothesis that ERdj3 release from substrates occurs in response to a functional interaction with BiP, it was necessary to set up an assay to saturate the binding of ERdj3 to luciferase and to ensure that incubation with ATP did not affect this binding. Increasing concentrations of wild-type (Figure 3A), QPD (Figure 3B), or HPN (Figure 3C) recombinant ERdj3 proteins were added to luciferase-coated wells in the absence (Figure 3, solid bar) or presence (Figure 3, hatched bar) of ATP. We found that all three proteins reached saturation binding at concentrations of ∼500 nM and that the inclusion of ATP in the binding buffer did not affect ERdj3's ability to bind to substrate. Thus, in the following experiments 500 nM ERdj3 was used. Quantitation of the amount of luciferase and ERdj3 that were bound to the wells revealed the ERdj3 bound to denatured luciferase at a little under a 1:1 ratio (Supplementary Figure S2).
Figure 3.
WT and mutant (QPD and HPN) ERdj3 bind to D-Luc similarly and ATP does not affect their binding. Chemically denatured luciferase was used to coat the wells and the indicated amounts of WT (A) QPD (B), or HPN (C) ERdj3 were added to the wells with (hatched bars) or without (solid bars) ATP. The plates were developed with anti-ERdj3 as described in Figure 2. The arrow indicates the concentration of ERdj3 that was used in the following experiments.
BiP promoted the release of wild-type ERdj3 from chemically denatured luciferase in the presence of ATP
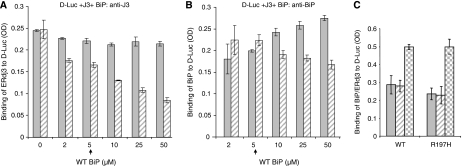
If a functional interaction between BiP and ERdj3 is critical for releasing ERdj3 from substrates, the amount of ERdj3 that is associated with substrate is expected to decrease in the presence of BiP in an ATP-dependent manner. To examine this possibility, increasing amounts of BiP were added to wells containing ERdj3 bound to denatured luciferase in the absence and presence of ATP (Figure 4A). We found that the addition of increasing amounts of BiP in the absence of ATP did not affect ERdj3's association with denatured luciferase (Figure 4A, solid bar), although there was a detectable increase in the binding of BiP to the substrate (Figure 4B, solid bar). This demonstrates that release of ERdj3 does not occur through a simple competition between these two chaperones for substrate and further suggests that BiP- and ERdj3-binding sites are not completely overlapping. However, when ATP was included with BiP, we found that the ability of BiP to release ERdj3 was dependent not only on the concentration of BiP but also required ATP (Figure 4A, hatched bar). The binding of BiP to denatured luciferase did not increase (Figure 4B, hatched bar) when ERdj3 was released (Figure 4A, hatched bar), again suggesting that ERdj3 release does not occur due to a simple competition with BiP. To examine this from the other direction, we first bound either wild-type BiP or a BiP mutant that cannot interact with ER DnaJ proteins (R197H) (Awad et al, 2008) to denatured luciferase and measured the ability of ERdj3 to release them. We found that the addition of ERdj3 did not induce a reduction in the binding of either wild-type or mutant BiP to luciferase-coated well (Figure 4C, solid and hatched bars), even though we could readily measure the binding of ERdj3 to luciferase (Figure 4C, checkered bars). This further argues that the release of ERdj3 does not occur due to a simple competition between BiP and ERdj3 for binding sites on the substrate.
Figure 4.
BiP releases ERdj3 from D-Luc in an ATP-dependent manner. (A) Chemically denatured luciferase was added to the wells followed by ERdj3 binding as described earlier. After washing, the indicated amounts of WT BiP were added to the ERdj3–luciferase complexes with (hatched bar) or without (solid bar) ATP and incubated for an additional 1 h at room temperature. After washing, the amount of ERdj3 that remained bound to luciferase was detected with an anti-ERdj3 antiserum. (B) On a parallel plate, the amount of BiP that was associated with luciferase was determined by incubating with an anti-BiP antiserum. (C) Either wild-type or mutant (R197H) BiP was allowed to bind to luciferase. After washing, ERdj3 was added to half the wells and the amount of BiP that was bound without ERdj3 (solid bars) or with ERdj3 (hatched bars) was measured with an anti-BiP antiserum. In a parallel set of wells, ERdj3 binding was measured with an anti-ERdj3 antibody (checkered bars).
BiP mutants that do not interact with ERdj3 failed to promote the release of ERdj3 from substrate
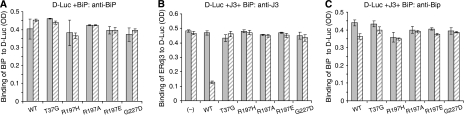
The requirement of ATP for BiP to release ERdj3 from the substrate suggested that a functional interaction between BiP and ERdj3 might be necessary. To test this possibility, we examined the ability of a number of different BiP mutants to release ERdj3 from luciferase. A highly conserved arginine on the ATPase domain of Hsp70 proteins (R197 in BiP) has been shown to be essential for interaction with the HPD motif on DnaJ proteins (Gassler et al, 1998; Suh et al, 1998; Alder et al, 2005). We recently made three substitutions at this site (R197H, R197A, and R197E), all of which have ATPase activity equal to or greater than wild-type BiP, but none of them can bind or be further stimulated by J proteins (Awad et al, 2008). Two other BiP mutants were also tested; a G227D mutant that cannot bind to ATP and a T37G mutant that cannot undergo the ATP-induced conformational change that is required for its chaperoning activity (Wei et al, 1995). Recombinant proteins corresponding to each of these mutants were made and tested both for their ability to bind to luciferase and to release wild-type ERdj3. We found that all five BiP mutants were able to bind equivalently to luciferase when tested at a concentration of 5 μM (Figure 5A), whereas only very low levels of background binding to the wells were observed for all of these proteins when luciferase was not present (Supplementary Figure S3). Although all of the mutants bound to luciferase both alone and in the presence of ERdj3 (Figure 5C), none of them was able to release ERdj3 from this substrate even in the presence of ATP (Figure 5B). As only wild-type BiP was able to release ERdj3 from substrate, it was possible that the loss of ERdj3 was due to refolding of luciferase on the plate. To exclude this possibility, we examined the enzymatic activity of luciferase after interaction with ERdj3 and various BiP mutants and found no evidence of productive refolding, even though our assay could have detected as little as 0.05% renaturation (Supplementary Figure S4).
Figure 5.
Only WT BiP releases ERdj3 from D-Luc. (A) Chemically denatured luciferase was added to the wells, which were then incubated with wild-type or mutant BiP. The binding of BiP to D-Luc was performed in the absence of ERdj3 with (hatched bars) and without ATP (solid bars) and detected with anti-BiP serum. (B, C) Denatured luciferase was added to wells and wild-type ERdj3 was allowed to bind as described. After washing, either wild-type or mutant BiP was added with (hatched bars) or without (solid bars) ATP. The amount of ERdj3 remaining was detected with an anti-ERdj3 antiserum (B) and the binding of BiP was detected with an anti-BiP antiserum (C) and expressed in OD units.
Wild-type BiP does not release two ERdj3 mutants, QPD and HPN, from luciferase
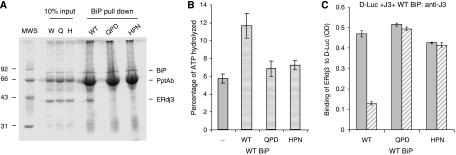
To further explore the possibility that a functional interaction between ERdj3 and BiP was required for ERdj3 release, we produced two ERdj3 proteins in which the HPD motif had been mutated and therefore should not interact functionally with wild-type BiP based on previously defined DnaJ mutants (Wall et al, 1994). The first of these, HPD → QPD, disrupts binding to BiP and stimulation of its ATPase activity without interfering with the ability of this mutant to bind to substrates (Shen and Hendershot, 2005). The second mutant HPD → HPN corresponds to a DnaJ mutant that was defective in interacting with wild-type DnaK (Suh et al, 1998). We first tested the ability of these two mutants to bind to BiP in vitro and to stimulate its ATPase activity. Wild-type recombinant BiP was immobilized on Protein A Sepharose beads by immunoprecipitation and wild-type and mutant ERdj3 proteins were allowed to bind in the presence of ATP (Figure 6A). We found that both mutations interfered with the ability of ERdj3 to bind to BiP, which is in keeping with data from a number of other DnaJ family members (Tsai and Douglas, 1996; Kelley and Georgopoulos, 1997; Wittung-Stafshede et al, 2003). Next the ability of the mutant ERdj3 proteins to stimulate the ATPase activity of BiP was compared with that of wild-type ERdj3. We found that wild-type ERdj3 stimulated BiP's ATPase activity about two-fold, which is in keeping with previous data obtained with only the J domain and glycine/phenylalanine regions of ERdj3 (Shen and Hendershot, 2005), whereas the HPN and QPD mutants were unable to appreciably increase the hydrolysis of ATP (Figure 6B).
Figure 6.
Wild-type BiP can only release wild-type ERdj3 from D-Luc. (A) Recombinant wild-type BiP (5 μg) was immunoprecipitated with a polyclonal anti-BiP antiserum and Protein A Sepharose beads. After washing, 20 μg of the indicated recombinant ERdj3 proteins was added to the BiP beads in ATPase buffer containing ATP. One-tenth of the reaction was removed for direct loading and the remaining nine-tenths were incubated at 4°C for 1 h. After washing, the samples were analysed by reducing SDS–PAGE and the gel was stained with Brilliant Coomassie Blue to visualize proteins. (B) ATPase assays were performed on wild-type BiP alone or with a four-fold molar excess of wild-type, HPN, or QPD ERdj3. ATP hydrolysis was measured by quantitating ADP and expressing it as a percentage of total nucleotide. (C) An experiment similar to that described in the previous figure was performed, except that either wild-type or mutant ERdj3 was bound to luciferase first. After washing, wild-type BiP was added with (hatched bars) or without (solid bars) ATP. The amount of ERdj3 that remained bound was detected with an anti-ERdj3 antiserum and expressed in OD units.
We next performed an experiment similar to those described in Figures 4 and 5, except that this time we were asking whether wild-type BiP was only capable of releasing wild-type ERdj3 or whether it was also able to release the two ERdj3 mutants that did not functionally interact with BiP. If BiP released the ERdj3 mutants, it would argue that a functional BiP–ATP–substrate interaction was required, but that there was no need for a functional BiP–ERdj3 interaction. We found that only wild-type ERdj3 was released by wild-type BiP in the presence of ATP, whereas both the QPD and HPN mutants remained bound to the substrate even in the presence of ATP (Figure 6C). The combination of this experiment and the previous one (Figure 5A) demonstrates that both a BiP–ATP–substrate interaction and a functional BiP–ERdj3 interaction are required to release ERdj3 from substrate. In a search for allele-specific suppressors of DnaJ HPN and QPD mutants, no suppressors were found for the QPD mutant, but three different DnaK mutants were identified that restored growth at temperatures that were non-permissive for the HPN mutant (Suh et al, 1998). Of these, the DnaK R167H mutant (analogous to our R197H mutant) bound better than wild-type DnaK to the DnaJ HPN mutant. Thus, we wished to determine whether our HPN ERdj3 mutant and our R197H BiP mutant would constitute a functional pair that could rescue the inability of HPN ERdj3 to be released from substrate. However, we found that the HPN mutant was as defective in binding to R197H BiP mutant as either wild-type ERdj3 or the QPD mutant and was unable to significantly trigger the release of the HPN mutant (Supplementary Figure S5). Thus, unfortunately the HPN ERdj3 mutant did not appear to re-establish a functional pair with the R197H BiP mutant by this criterion.
BiP and ATP must be added simultaneously to the ERdj3–substrate complex to promote ERdj3 release
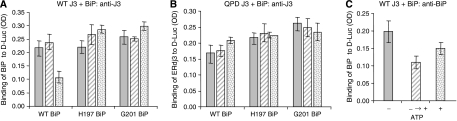
To determine whether recruitment of BiP to the site of ERdj3–substrate interaction was sufficient to release ERdj3 or if it was important that BiP also hydrolyse ATP, we included a BiP mutant (E201G) that was able to interact with DnaJ proteins (Petrova et al, 2008, in press) but that was defective in ATP hydrolysis (Gaut and Hendershot, 1993). We found that even though ERdj3 would be expected to recruit the E201G mutant locally, this BiP mutant was as unable as the R197H BiP mutant, which could not interact with ERdj3 to induce the release of ERdj3 under any condition (Figure 7A and B).
Figure 7.
Both BiP and ATP must be added simultaneously to induce the release of ERdj3 from substrate. Luciferase-binding assays were performed with wild-type ERdj3 or the QPD mutant. The indicated BiP proteins were added with no ATP (solid bars), with ATP as done in the previous experiments (stippled bars) or BiP was allowed to bind in the absence of ATP but after washing unbound BiP away, the D-Luc–ERdj3–BiP complexes were further incubated with ATP (hatched bars). The amount of ERdj3 remaining associated with D-Luc (A, B) and the amount of BiP in the D-Luc–wild-type ERdj3 complexes (C) were determined by ELISA.
We also tested whether ATP needed to be included simultaneously to trigger release. When wild-type BiP was allowed to bind to the ERdj3–substrate complex in the absence of ATP and then washed to remove unbound BiP, challenging this complex with ATP did not lead to the release of ERdj3 (Figure 7A), even though it did induce the release of BiP (Figure 7C). Taken together, these data argue that BiP must be recruited locally to the site of ERdj3–substrate interaction and that ERdj3 must induce ATP hydrolysis in BiP to receive a signal to be released.
Discussion
There is now a significant amount of data to argue that DnaJ proteins bind to unfolded proteins initially and due to their ability to interact specifically with the ATP form of Hsp70s serve to recruit the open form of the Hsp70 to the substrate. Hsp70 proteins must be able to interact with a DnaJ protein as well as to bind and hydrolyse ATP to be efficiently recruited to the substrate (Wawrzynow et al, 1995a). Much less is understood about how DnaJ proteins leave the substrate once Hsp70 has been recruited, although several models have been proposed. First, it is possible that once the DnaJ protein contacts an Hsp70, it releases the unfolded protein and the Hsp70 captures it (Rudiger et al, 2001). The identification of stable DnaJ–Hsp70–substrate complexes (Szabo et al, 1994; Wawrzynow et al, 1995a; Han and Christen, 2003; Shen and Hendershot, 2005) makes this scenario less likely, as do our data showing that the E201G BiP mutant, which does make contact with ERdj3, is unable to induce the release of ERdj3. Second, it is possible that, in addition to the conformational change that DnaJ induces in the Hsp70 protein, upon substrate binding and ATP hydrolysis the Hsp70 protein provides a reciprocal signal to the DnaJ protein that causes its release. It is known that type I and II DnaJ proteins have a second interaction site with the SBD of Hsp70s (Laufen et al, 1999), but it is not clear which domain of DnaJ is involved in this binding (Wall et al, 1995; Wawrzynow and Zylicz, 1995b; Linke et al, 2003; Sahi and Craig, 2007), but if it involved the SBD of DnaJ proteins, this second interaction could induce its release. The fact that similar requirements for release were observed with P58/DNAJ3c, a type III DnaJ protein (Petrova et al, 2008), makes this possibility less likely as the SBD of this group of proteins does not evidently resemble that of type I and II DnaJ proteins and probably does not share the same orientation to the J domain (Gruschus et al, 2004). However, in the absence of a full-length structure for the various types of DnaJ proteins, we do not know the relative orientation of J domains to SBD.
Finally, it has been proposed that once the J domain interacts with the NBD of Hsp70, it reorients the Hsp70 and in some way wrenches the substrate from DnaJ (Landry, 2003). Our demonstration that wild-type BiP, which binds the substrate but cannot release the QPD or HPN mutant, is compatible with this model, as the absence of the BiP–ERdj3 interaction would not allow BiP to wrench the substrate from ERdj3. This wrenching would need to happen before ATP is hydrolysed, as DnaJ domains lose affinity for the ADP-bound form of Hsp70 (Wawrzynow and Zylicz, 1995b). Our finding that ATP must be added simultaneously for the BiP-mediated release to occur argues that the formation of this three-way complex is critical to release, as in the absence of ATP, BiP would not interact with ERdj3 and be concentrated locally. However, the fact that E201G BiP is able to interact with substrate and ERdj3 but cannot induce the release of ERdj3 argues that, although necessary, the formation of the three-way complex is not sufficient to wrench the DnaJ protein from the substrate. Instead, our data argue that in addition to recruiting BiP to the substrate, the DnaJ protein may be able to ‘sense' that ‘it' has stimulated the ATPase activity of BiP or that BiP must assume the capability to wrench the substrate from the DnaJ partner in an ATP-dependent manner.
Our discovery that the release of ERdj3 from substrate is dependent on a functional interaction with BiP is likely to be true of other DnaJ–Hsp70 pairs. Recent data from Petrova et al demonstrate a similar requirement for the release of another ER-localized DnaJ protein, P58, from substrate. Mutations in either P58 or BiP that disrupted the interaction between them blocked the release of P58 from misfolded RNase A. In addition, an earlier in vitro study found that Ydj1 bound to a substrate and prevented its aggregation, but it could not fold the protein unless Hsp70 and ATP were present (Lu and Cyr, 1998a, 1998b; Fan et al, 2005). In view of our data, it is reasonable to suggest that DnaJ remained bound to the substrate in the absence of DnaK, thereby preventing its aggregation, but also preventing it from folding. Only in the presence of ATP would both chaperones be released allowing the substrate to fold. We would speculate, based on our data, that in the absence of ATP that both DnaK and DnaJ might bind, but folding would not occur.
Our previous characterization of the interaction of ERdj3 with substrates in cultured cell lines revealed that wild-type ERdj3 disappeared from BiP–substrate complexes long before folding was complete, whereas a QPD mutant remained associated with the substrate (Shen and Hendershot, 2005). This led us to speculate that the prolonged binding of the QPD mutant to substrates could be due to its inability to recognize that BiP had bound productively to the substrate. The data presented here confirm that the release of ERdj3 from substrate, at least in vitro, required an interaction between the J domain of ERdj3 and NBD of BiP and argue that indeed this is the reason for prolonged association of mutant ERdj3 with substrates in vivo. The in vitro release of ERdj3 from substrate required BiP's ATPase activity (T37G, G227D, and E201G), its ability to interact with ERdj3 (R197H), and presumably its substrate-binding ability (Petrova et al, 2008). These requirements would ensure that once a DnaJ protein engages an unfolded substrate, it would remain bound to prevent aggregation until it had recruited an open form of Hsp70 to the substrate, allowed the Hsp70 to initially associate with the substrate and then hydrolyse ATP to form a more stable interaction. Only when all of these steps had occurred would the DnaJ protein release from the Hsp70 and from the substrate. This scenario is consistent with most published data showing that DnaJ proteins bind first and more transiently than Hsp70, which remain associated until folding is complete.
In summary, our studies reveal that BiP promotes the release of ERdj3 from substrates in the presence of ATP. This is not due to a competition for binding sites on the substrate once BiP is recruited through its association with ERdj3, but rather it requires a functional interaction between ERdj3 and BiP. This includes both the ability to physically interact with each other and the ability of ERdj3 to stimulate the ATPase activity of BiP. The fact that similar data were observed with another ERdj–BiP pair (Petrova et al, 2008, in press) argues that this mechanism is likely to be used for the release of other DnaJ proteins from substrates.
Materials and methods
Cell culture, transfection, and immunoprecipitation
COS-1 monkey kidney fibroblast cells were maintained in DMEM supplemented with 10% fetal bovine serum, 2 mM L-glutamine, and 1% Fungisone in 3% CO2. Cells were transfected with the indicated vectors using the Fugene 6 transfection reagent (Roche Diagnostics), and after 48 h post-transfection, cells were labelled with [35S]Translabel (Amersham Biosciences) for 3 h. Cells were treated with 150 μg/ml DSP, a membrane-permeable crosslinking reagent (Sigma-Aldrich) for 1 h on ice. Cell lysates were prepared using an NP-40 lysing buffer and immunoprecipitated with indicated antisera followed by binding to Protein A Sepharose beads. The immunoprecipitated complexes were analysed by SDS–PAGE under reducing conditions, and the signal was detected using Amplify (Amersham Biosciences) for radiographic visualization.
In vitro translation and heavy chain-binding assay
Ag8.8 murine plasmacytoma cells were maintained in RPMI 1640 supplemented with 10% fetal bovine serum, 2 mM L-glutamine, and 1% Fungizone in 5% CO2. Cells were metabolically labelled for 16 h with [35S]Translabel (Amersham Biosciences) and treated with or without DSP prior to lysing. Cell lysates were prepared and Ig heavy chains were isolated by binding to Protein A as described earlier (Wei et al, 1995). To release BiP from heavy chains, non-crosslinked samples were supplemented with 1 mM MgCl2, 25 mM KCl, and 1 mM ATP. These samples are the source of free γHC used in the in vitro binding assays, except that the beads were washed an additional three times in PBS to reduce detergent in the samples, which interferes with the ability of ERdj3 to remain associated with unfolded substrates (Shen and Hendershot, 2005).
The cDNAs encoding wild-type and mutant (H35Q) ERdj3 were transcribed from the T7 promoter of 3HADSL-ERdj3 (Stratagene) and translated using [35S]methionine (Amersham Biosciences) and the TNT-coupled rabbit reticulocyte lysate (Promega). Equivalent counts for the two protein products were loaded directly on reducing SDS–polyacrylamide gels or incubated with either uncoupled Protein A Sepharose beads (washed three times in PBS), or with the Protein A Sepharose beads to which free γHC were bound. After incubating for 1 h on ice, the beads were washed three times with PBS and bound proteins were subjected to reducing SDS–PAGE.
IP western
Firefly luciferase (Promega) was left untreated (N) or heat denatured (D) at 42°C for 1 h. In both cases, 0.5 μg of protein was incubated with recombinant wild-type ERdj3 protein (2.0 μg) in PBS and immunoprecipitated with either anti-ERdj3 polyclonal antiserum followed by Protein A Sepharose beads or with Protein A Sepharose beads alone. Bound proteins were subjected to reducing SDS–PAGE and then transferred to a PVDF membrane (Bio-Rad), which was blotted with an anti-luciferase antiserum (1:1000) (Promega). Donkey anti-goat Ig conjugated to HRP (1:5000) was used as a secondary antibody and the signal was detected by chemiluminescence (ECL).
Protein purification
Expression of His-tagged wild-type and mutant ERdj3 was induced in E. coli M15 cells with 0.1 mM isopropyl β-D-thiogalactoside (Sigma-Aldrich) followed by growth for 18 h at 18°C. The recombinant proteins were purified on Ni2+-agarose columns under non-denaturing conditions (Qiagen QIAexpress system), dialysed in 25 mM sodium phosphate buffer (pH 7.0), containing 150 mM NaCl, 0.02% Triton X-100, 50% glycerol, and a protease inhibitor cocktail (Roche), and stored at −20°C. His-tagged wild-type and mutant BiP proteins were induced with 1 mM IPTG for 2 h at 37°C, purified on Ni2+-agarose columns, dialysed and stored in 20 mM HEPES buffer (pH 7.2) with 50 mM KCl, 5 mM MgCl2, 0.01% NP-40, 50% glycerol, and a protease inhibitor cocktail.
Measurement of complex formation between ERdj3 proteins and denatured luciferase
Firefly luciferase was denatured in a buffer containing 7 M urea, 25 mM HEPES (pH 7.5), 50 mM KCl, 5 mM EDTA, 5 mM MgCl2, and 5 mM dithiothreitol at room temperature for 40 min and then diluted into PBS containing 0.05% BSA (final concentration 5 μg/ml). A portion of 100 μl of this solution was added into each well of a 96-well microtitre plate purchased from Thermo (Immulon 2HB Flat Bottom Microtiter Plate) and allowed to bind overnight at 4°C. Wells were washed with PBS and blocked with 200 μl PBS containing 1% BSA for 1 h at room temperature. The indicated concentrations of wild-type or mutant ERdj3 in 100 μl of PBS with 0.05% BSA was added to the wells and incubated for 1 h at room temperature, followed by washing with PBS to remove unbounded ERdj3. The amount of ERdj3 that remained bound to denatured luciferase was detected with a polyclonal anti-ERdj3 antiserum (Shen and Hendershot, 2005) followed by donkey anti-rabbit Ig conjugated to alkaline phosphatase (Promega). 4-Nitrophenyl phosphate disodium salt hexahydrate (Sigma-Aldrich) was added, and after stopping the reaction with 0.75 M NaCl, the plates were read on a spectrophotometer at a wavelength 405 nm. Negative controls were set up for each plate in wells that did not contain either denatured luciferase, ERdj3, or each of the antibodies, but which included all the other steps of the reaction.
Release of ERdj3 from luciferase
To detect the amount of BiP binding to denatured luciferase, recombinant hamster BiP was added to wells coated with denatured luciferase instead of ERdj3, and incubated as above, except that a rabbit polyclonal anti-rodent BiP antiserum was used to detect BiP binding. To test the ability of wild-type and mutant BiP to release ERdj3 from luciferase, ERdj3 was first bound to luciferase as above. After washing away unbound ERdj3, the indicated amounts of recombinant BiP proteins were added to the wells in PBS containing 1 mM MgCl2, 25 mM KCl, and either 1 mM ATP or no nucleotide. The plates were incubated for 1 h at room temperature, and the amount of ERdj3 or BiP associated with the denatured luciferase was determined as above.
Luciferase activity assay
Triplicate samples of serial dilutions of native (N) or denatured (D) luciferase in PBS were added to a 96-well microtitre plate. Next, 100 μl of luciferase assay reagent (Promega) was injected into each well and chemiluminescence was read immediately on a 1420 Multilabel Counter (PerkinElmer). To test whether denatured luciferase was able to be refolded in the wells, 96 wells were coated with 0.5 μg D-Luc and reactions were performed as described above. The denatured luciferase was further incubated alone, with ERdj3, or with ERdj3 and BiP in the presence or absence of ATP. The luciferase assay reagent was injected into each well, and luciferase activity was measured on the plates. Each experimental condition was performed in triplicate.
ATPase assay
ATPase assays were performed as described earlier (Chevalier et al, 1998). Briefly, 1 μM of the various recombinant BiP proteins was incubated alone or with 0.5 μM of the indicated full-length ERdj3 proteins at 37°C for 20 min in ATPase buffer containing [γ-32P]ATP (PerkinElmer). After chromatography, the radioactive ATP and free phosphate signals were quantified by phosphoimager analysis (Molecular Dynamics, Sunnyvale, CA) using Image Quant software. The free phosphate signal was expressed as a percentage of the total phosphate signal. Data were deduced from three independent experiments, and the error bars represent standard deviations (s.d.).
Supplementary Material
Supplementary Figures S1
Supplementary Figures S2
Supplementary Figures S3
Supplementary Figures S4
Supplementary Figures S5
Supplementary Data
Acknowledgments
We thank Drs Ying Shen, Jeffrey Brodsky, and Craig Scott for helpful scientific discussions and Melissa Mann for technical help. This study was supported by NIH Grant GM54068 (LMH), the Cancer Center CORE Grant CA21765, and the American Lebanese Syrian Associated Charities of St Jude Children's Research Hospital.
References
- Alder NN, Shen Y, Brodsky JL, Hendershot LM, Johnson AE (2005) The molecular mechanisms underlying BiP-mediated gating of the Sec61 translocon of the endoplasmic reticulum. J Cell Biol 168: 389–399 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Awad W, Estrada I, Shen Y, Hendershot LM (2008) BiP mutants that are unable to interact with endoplasmic reticulum DnaJ proteins provide insights into interdomain interactions in BiP. Proc Natl Acad Sci USA 105: 1164–1169 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brightman SE, Blatch GL, Zetter BR (1995) Isolation of a mouse cDNA encoding MTJ1, a new murine member of the DnaJ family of proteins. Gene 153: 249–254 [DOI] [PubMed] [Google Scholar]
- Caplan AJ, Cyr DM, Douglas MG (1993) Eukaryotic homologues of Escherichia coli dnaJ: a diverse protein family that functions with hsp70 stress proteins. Mol Biol Cell 4: 555–563 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheetham ME, Caplan AJ (1998) Structure, function and evolution of DnaJ: conservation and adaptation of chaperone function. Cell Stress Chaperones 3: 28–36 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chevalier M, King L, Blond S (1998) Purification and properties of BiP. Methods Enzymol 290: 384–409 [DOI] [PubMed] [Google Scholar]
- Cunnea PM, Miranda-Vizuete A, Bertoli G, Simmen T, Damdimopoulos AE, Hermann S, Leinonen S, Huikko MP, Gustafsson JA, Sitia R, Spyrou G (2003) ERdj5, an endoplasmic reticulum (ER)-resident protein containing DnaJ and thioredoxin domains, is expressed in secretory cells or following ER stress. J Biol Chem 278: 1059–1066 [DOI] [PubMed] [Google Scholar]
- Fan CY, Lee S, Cyr DM (2003) Mechanisms for regulation of Hsp70 function by Hsp40. Cell Stress Chaperones 8: 309–316 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fan CY, Ren HY, Lee P, Caplan AJ, Cyr DM (2005) The type I Hsp40 zinc finger-like region is required for Hsp70 to capture non-native polypeptides from Ydj1. J Biol Chem 280: 695–702 [DOI] [PubMed] [Google Scholar]
- Gassler CS, Buchberger A, Laufen T, Mayer MP, Schroder H, Valencia A, Bukau B (1998) Mutations in the DnaK chaperone affecting interaction with the DnaJ cochaperone. Proc Natl Acad Sci USA 95: 15229–15234 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gaut JR, Hendershot LM (1993) Mutations within the nucleotide binding site of immunoglobulin-binding protein inhibit ATPase activity and interfere with release of immunoglobulin heavy chain. J Biol Chem 268: 7248–7255 [PubMed] [Google Scholar]
- Gruschus JM, Han CJ, Greener T, Ferretti JA, Greene LE, Eisenberg E (2004) Structure of the functional fragment of auxilin required for catalytic uncoating of clathrin-coated vesicles. Biochemistry 43: 3111–3119 [DOI] [PubMed] [Google Scholar]
- Han W, Christen P (2003) Mechanism of the targeting action of DnaJ in the DnaK molecular chaperone system. J Biol Chem 278: 19038–19043 [DOI] [PubMed] [Google Scholar]
- Hendrick JP, Langer T, Davis TA, Hartl FU, Wiedmann M (1993) Control of folding and membrane translocation by binding of the chaperone DnaJ to nascent polypeptides. Proc Natl Acad Sci USA 90: 10216–10220 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang J, Prasad K, Lafer EM, Sousa R (2005) Structural basis of interdomain communication in the Hsc70 chaperone. Mol Cell 20: 513–524 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kelley WL, Georgopoulos C (1997) The T/t common exon of simian virus 40, JC, and BK polyomavirus T antigens can functionally replace the J-domain of the Escherichia coli DnaJ molecular chaperone. Proc Natl Acad Sci USA 94: 3679–3684 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Landry SJ (2003) Swivels and stators in the Hsp40–Hsp70 chaperone machine. Structure 11: 1465–1466 [DOI] [PubMed] [Google Scholar]
- Landry SJ, Jordan R, McMacken R, Gierasch LM (1992) Different conformations for the same polypeptide bound to chaperones DnaK and GroEL. Nature 355: 455–457 [DOI] [PubMed] [Google Scholar]
- Laufen T, Mayer MP, Beisel C, Klostermeier D, Mogk A, Reinstein J, Bukau B (1999) Mechanism of regulation of hsp70 chaperones by DnaJ cochaperones. Proc Natl Acad Sci USA 96: 5452–5457 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li J, Qian X, Sha B (2003) The crystal structure of the yeast Hsp40 Ydj1 complexed with its peptide substrate. Structure (Camb) 11: 1475–1483 [DOI] [PubMed] [Google Scholar]
- Liberek K, Marszalek J, Ang D, Georgopoulos C, Zylicz M (1991a) Escherichia coli DnaJ and GrpE heat shock proteins jointly stimulate ATPase activity of DnaK. Proc Natl Acad Sci USA 88: 2874–2878 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liberek K, Skowyra D, Zylicz M, Johnson C, Georgopoulos C (1991b) The Escherichia coli DnaK chaperone, the 70-kDa heat shock protein eukaryotic equivalent, changes conformation upon ATP hydrolysis, thus triggering its dissociation from a bound target protein. J Biol Chem 266: 14491–14496 [PubMed] [Google Scholar]
- Linke K, Wolfram T, Bussemer J, Jakob U (2003) The roles of the two zinc binding sites in DnaJ. J Biol Chem 278: 44457–44466 [DOI] [PubMed] [Google Scholar]
- Liu Q, Hendrickson WA (2007) Insights into hsp70 chaperone activity from a crystal structure of the yeast Hsp110 Sse1. Cell 131: 106–120 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu Z, Cyr DM (1998a) The conserved carboxyl terminus and zinc finger-like domain of the co-chaperone Ydj1 assist Hsp70 in protein folding. J Biol Chem 273: 5970–5978 [DOI] [PubMed] [Google Scholar]
- Lu Z, Cyr DM (1998b) Protein folding activity of Hsp70 is modified differentially by the hsp40 co-chaperones Sis1 and Ydj1. J Biol Chem 273: 27824–27830 [DOI] [PubMed] [Google Scholar]
- Mayer MP, Laufen T, Paal K, McCarty JS, Bukau B (1999) Investigation of the interaction between DnaK and DnaJ by surface plasmon resonance spectroscopy. J Mol Biol 289: 1131–1144 [DOI] [PubMed] [Google Scholar]
- Meunier L, Usherwood YK, Chung KT, Hendershot LM (2002) A subset of chaperones and folding enzymes form multiprotein complexes in endoplasmic reticulum to bind nascent proteins. Mol Biol Cell 13: 4456–4469 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meyer HA, Grau H, Kraft R, Kostka S, Prehn S, Kalies KU, Hartmann E (2000) Mammalian Sec61 is associated with Sec62 and Sec63. J Biol Chem 275: 14550–14557 [DOI] [PubMed] [Google Scholar]
- Petrova K, Oyadomari S, Hendershot L, Ron D (2008) Regulated association of misfolded endoplasmic reticulum lumenal proteins with P58/DNAJc3. EMBO J (in press) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rudiger S, Schneider-Mergener J, Bukau B (2001) Its substrate specificity characterizes the DnaJ co-chaperone as a scanning factor for the DnaK chaperone. EMBO J 20: 1042–1050 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rutkowski DT, Kang SW, Goodman AG, Garrison JL, Taunton J, Katze MG, Kaufman RJ, Hegde RS (2007) The role of p58IPK in protecting the stressed endoplasmic reticulum. Mol Biol Cell 18: 3681–3691 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sahi C, Craig EA (2007) Network of general and specialty J protein chaperones of the yeast cytosol. Proc Natl Acad Sci USA 104: 7163–7168 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saito H, Uchida H (1977) Initiation of the DNA replication of bacteriophage lambda in Escherichia coli K12. J Mol Biol 113: 1–25 [DOI] [PubMed] [Google Scholar]
- Schumacher RJ, Hansen WJ, Freeman BC, Alnemri E, Litwack G, Toft DO (1996) Cooperative action of Hsp70, Hsp90, and DnaJ proteins in protein renaturation. Biochem 35: 14889–14898 [DOI] [PubMed] [Google Scholar]
- Shen Y, Hendershot LM (2005) ERdj3, a stress-inducible endoplasmic reticulum DnaJ homologue, serves as a cofactor for BiP's interactions with unfolded substrates. Mol Biol Cell 16: 40–50 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen Y, Meunier L, Hendershot LM (2002) Identification and characterization of a novel endoplasmic reticulum (ER) DnaJ homologue, which stimulates ATPase activity of BiP in vitro and is induced by ER stress. J Biol Chem 277: 15947–15956 [DOI] [PubMed] [Google Scholar]
- Silberg JJ, Tapley TL, Hoff KG, Vicery LE (2004) Regulation of the HscA ATPase reaction cycle by the co-chaperone HscB and the iron–sulfur cluster assembly protein IscU. J Biol Chem 279: 53924–53931 [DOI] [PubMed] [Google Scholar]
- Suh WC, Burkholder WF, Lu CZ, Zhao X, Gottesman ME, Gross CA (1998) Interaction of the Hsp70 molecular chaperone, DnaK, with its cochaperone DnaJ. Proc Natl Acad Sci USA 95: 15223–15228 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Szabo A, Langer T, Schroder H, Flanagan J, Bukau B, Hartl FU (1994) The ATP hydrolysis-dependent reaction cycle of the Escherichia coli Hsp70 system DnaK, DnaJ, and GrpE. Proc Natl Acad Sci USA 91: 10345–10349 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsai J, Douglas MG (1996) A conserved HPD sequence of the J-domain is necessary for YDJ1 stimulation of Hsp70 ATPase activity at a site distinct from substrate binding. J Biol Chem 271: 9347–9354 [DOI] [PubMed] [Google Scholar]
- Tyedmers J, Lerner M, Bies C, Dudek J, Skowronek MH, Haas IG, Heim N, Nastainczyk W, Volkmer J, Zimmermann R (2000) Homologs of the yeast Sec complex subunits Sec62p and Sec63p are abundant proteins in dog pancreas microsomes. Proc Natl Acad Sci USA 97: 7214–7219 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vogel M, Bukau B, Mayer MP (2006a) Allosteric regulation of Hsp70 chaperones by a proline switch. Mol Cell 21: 359–367 [DOI] [PubMed] [Google Scholar]
- Vogel M, Mayer MP, Bukau B (2006b) Allosteric regulation of Hsp70 chaperones involves a conserved interdomain linker. J Biol Chem 281: 38705–38711 [DOI] [PubMed] [Google Scholar]
- Wall D, Zylicz M, Georgopoulos C (1994) The NH2-terminal 108 amino acids of the Escherichia coli DnaJ protein stimulate the ATPase activity of DnaK and are sufficient for lambda replication. J Biol Chem 269: 5446–5451 [PubMed] [Google Scholar]
- Wall D, Zylicz M, Georgopoulos C (1995) The conserved G/F motif of the DnaJ chaperone is necessary for the activation of the substrate binding properties of the DnaK chaperone. J Biol Chem 270: 2139–2144 [DOI] [PubMed] [Google Scholar]
- Wawrzynow A, Banecki B, Wall D, Liberek K, Georgopoulos C, Zylicz M (1995a) ATP hydrolysis is required for the DnaJ-dependent activation of DnaK chaperone for binding to both native and denatured protein substrates. J Biol Chem 270: 19307–19311 [DOI] [PubMed] [Google Scholar]
- Wawrzynow A, Zylicz M (1995b) Divergent effects of ATP on the binding of the DnaK and DnaJ chaperones to each other, or to their various native and denatured protein substrates. J Biol Chem 270: 19300–19306 [DOI] [PubMed] [Google Scholar]
- Wei J-Y, Gaut JR, Hendershot LM (1995) In vitro dissociation of BiP:peptide complexes requires a conformational change in BiP after ATP binding but does not require ATP hydrolysis. J Biol Chem 270: 26677–26682 [DOI] [PubMed] [Google Scholar]
- Wittung-Stafshede P, Guidry J, Horne BE, Landry SJ (2003) The J-domain of Hsp40 couples ATP hydrolysis to substrate capture in Hsp70. Biochemistry 42: 4937–4944 [DOI] [PubMed] [Google Scholar]
- Yochem J, Uchida H, Sunshine M, Saito H, Georgopoulos CP, Feiss M (1978) Genetic analysis of two genes, dnaJ and dnaK, necessary for Escherichia coli and bacteriophage lambda DNA replication. Mol Gen Genet 164: 9–14 [DOI] [PubMed] [Google Scholar]
- Yu M, Haslam RH, Haslam DB (2000) HEDJ, an Hsp40 co-chaperone localized to the endoplasmic reticulum of human cells. J Biol Chem 275: 24984–24992 [DOI] [PubMed] [Google Scholar]
- Zhu X, Zhao X, Burkholder WG, Gragerov A, Ogata CM, Gottesman ME, Hendrickson WA (1996) Structural analysis of substrate binding by the molecular chaperone DnaK. Science 272: 1606–1614 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Figures S1
Supplementary Figures S2
Supplementary Figures S3
Supplementary Figures S4
Supplementary Figures S5
Supplementary Data