Abstract
Genomic analysis shows that the increased complexity of trafficking pathways in mammalian cells involves an expansion of the number of SNARE, Rab and COP proteins. Thus, the human genome encodes four forms of Sec24, the cargo selection subunit of the COPII vesicular coat, and this is proposed to increase the range of cargo accommodated by human COPII-coated vesicles. In this study, we combined X-ray crystallographic and biochemical analysis with functional assays of cargo packaging into COPII vesicles to establish molecular mechanisms for cargo discrimination by human Sec24 subunits. A conserved IxM packaging signal binds in a surface groove of Sec24c and Sec24d, but the groove is occluded in the Sec24a and Sec24b subunits. Conversely, LxxLE class transport signals and the DxE signal of VSV glycoprotein are selectively bound by Sec24a and Sec24b subunits. A comparative analysis of crystal structures of the four human Sec24 isoforms establishes the structural determinants for discrimination among these transport signals, and provides a framework to understand how an expansion of coat subunits extends the range of cargo proteins packaged into COPII-coated vesicles.
Keywords: COPII coat, transport signal, vesicular transport
Introduction
The vesicular trafficking network of eukaryotic cells comprises a dozen or so discrete transport steps, with each step using a conserved set of machinery proteins and common molecular mechanisms for vesicle budding and fusion. Vesicles form in a budding reaction whereby cytoplasmic coat proteins (COPs) assemble on a membrane surface, capture cargo molecules and polymerize into a cage to deform the membrane into a bud (Lee et al, 2004). Cells contain a variety of COPs—including COPI, COPII and several clathrin/adaptin complexes—and specificity in vesicular transport is derived in part through the affinity of each COP for an export site on a particular organelle, and through COP selectivity for transport signal sequences on cargo molecules (Bonifacino and Glick, 2004).
The trafficking network of a specialized mammalian cell is necessarily more complex than that of yeast, and this is associated with a significant increase in the number of trafficking proteins. Thus, Saccharomyces cerevisiae has 31 individual COP subunits (forming 6 COP complexes), 21 SNAREs and 11 Rab GTPases, whereas the human genome encodes 53 COP subunits, 35 SNAREs and 60 Rabs (Bock et al, 2001). An increase in the number of COP subunits in mammalian cells is proposed to expand the repertoire of cargo accommodated by a given class of COP-coated vesicle, as each coat variant is predicted to exhibit differential affinity for cargo transport signals. Support for these ideas comes from a recent study of the COPII coat complex in which the Sec24 isoforms of the coat are shown to have distinct selectivity towards a series of di-hydrophobic signals (Wendeler et al, 2007).
COPII-coated vesicles form on the endoplasmic reticulum (ER) from three subunits—Sar1-GTP, Sec23/24 and Sec13/31—which function in concert to bud ∼60 nm vesicles for transport to the Golgi complex (Lee et al, 2004). Recent studies of S. cerevisiae COPII proteins have shown that Sec24 is the primary cargo selection subunit of the coat (Miller et al, 2002, 2003; Mossessova et al, 2003). Specifically, binding sites have been pinpointed on Sec24 for the YNNSNPF signal of the yeast SNARE Sed5, and for LxxLE and DxE class signals present on certain SNARE and cargo proteins. These transport signals are all short (10–15 residue) sequences nested in the polypeptide chain of the cargo molecule, and in this respect are highly similar to endocytic signals (Bonifacino and Glick, 2004). By contrast, the transport signal on the SNARE protein Sec22 is a conformational epitope that binds to a large surface region of Sec24 and Sec23 (Mancias and Goldberg, 2007).
In this study, we used biochemical and X-ray crystallographic methods to identify mammalian ER exit signals and to define their interactions with human COPII proteins, and we combined this with COPII isoform-specific budding assays to test the relevance of the molecular-level observations. We show that the Sec24 subunits indeed have distinct selectivity towards cargo sorting signals, and we describe molecular mechanisms for this signal discrimination, obtained through X-ray structure determination of the four human Sec24 proteins.
Results and discussion
Syntaxin 5 and membrin are packaged specifically by the mammalian COPII proteins Sec24c and Sec24d
To study the packaging of cargo membrane proteins into mammalian COPII-coated vesicles, we expressed and purified full-length forms of the four human Sec24 isoforms (Sec24a–d), each complexed with Sec23a. These were used in an in vitro budding assay that employed semi-intact cells and pure, recombinant COPII proteins (Mancias and Goldberg, 2007). In an earlier study, we showed that this assay was able to report the isoform-specific packaging of the SNARE protein Sec22 (which is packaged by Sec24a and Sec24b, but not by Sec24c or Sec24d; Mancias and Goldberg, 2007).
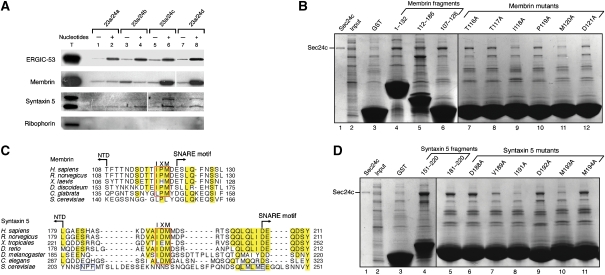
First, we assessed whether any of the four human Sec24 isoforms are capable of packaging the mammalian SNAREs, membrin and syntaxin 5. For this, the semi-intact cells were washed to remove endogenous coat proteins and then incubated with Sec23/24 (comprising Sec23a and Sec24a–d), Sec13a/31a and Sar1a plus GTP (see Materials and methods). The extent of packaging into vesicles of membrin and syntaxin 5 was compared with the packaging of the control cargo protein ERGIC-53 and the control ER-resident ribophorin, by immunoblotting of the vesicle contents (Figure 1A). All four forms of Sec23/24 generated COPII vesicles, as assessed by the packaging of ERGIC-53. Strikingly, the Sec24c and Sec24d isoforms specifically packaged both membrin and syntaxin 5, whereas Sec24a and Sec24b did not enrich the SNAREs above background levels.
Figure 1.
Signal sequences on membrin and syntaxin 5 for binding the Sec24c and d isoforms. (A) An isoform-specific packaging assay is shown. Vesicles were generated from semi-intact HEK-293 or CHO-K1 cells using Sar1a, Sec13a/31a and the four isoforms of human Sec24 (Sec23a/Sec24a–d), in the presence and absence of 0.2 mM GTP and an ATP-regenerating system (see Materials and methods). The presence of ERGIC-53, syntaxin 5, ribophorin (HEK-293 cells) and membrin (CHO-K1 cells) in budded vesicles and total membranes (T) was determined by immunoblotting. Polyclonal antibodies to syntaxin 5 recognize both syntaxin 5 splice variants: a 35-kDa form (lower band) and a 42-kDa form (upper band). (B) Localization of the Sec24c-binding site on membrin. SNARE–GST fusion proteins were incubated with an input mixture comprising Sec24c (0.3 mg/ml) and E. coli lysate (13 mg/ml). Specific binding was assessed by the enrichment of Sec24c from the input (lane 2). Bound proteins were analysed by 4–15% SDS–PAGE and Coomassie blue staining. To pinpoint the transport signal sequence on membrin, residues TTIPMD were mutated in turn to alanine in the context of the membrin (107–126)–GST construct (lanes 7–12). (C) Alignment of membrin and syntaxin 5 sequences. Polypeptide sequences of membrin from six organisms (see Materials and methods) were aligned using ClustalW. Just the linker region (that joins the N-terminal domain to the SNARE motif) is shown here; highly conserved residues are outlined in yellow and the key IxM sequence is boxed in red. The alignment of syntaxin 5 from seven organisms (see Materials and methods) also shows just the linker region; highly conserved residues are outlined in yellow, the predicted IxM sequence is boxed in red, and the YNNSNPF and LxxME motifs within S. cerevisiae syntaxin 5 are boxed in blue. (D) Localization of Sec24c-binding site on the syntaxin 5 linker. Residues of the DVAIDMM sequence were mutated to alanine in the context of the syntaxin 5 (181–200)–GST construct (lanes 6–11).
Thus, the human Sec24 isoforms can be grouped according to their SNARE specificity. Sec24c and Sec24d specifically package membrin and syntaxin 5, whereas Sec24a and Sec24b specifically package Sec22 (Mancias and Goldberg, 2007). This grouping conforms to the sequence similarity of the structural core (that is, excluding the hypervariable N-terminal regions) of the Sec24 isoforms: the sequences of Sec24a and Sec24b are 75% identical, and Sec24c and Sec24d are 66% identical, whereas Sec24a and Sec24c are only 31% identical.
Conserved IxM sequences on membrin and syntaxin 5 bind to Sec24c and Sec24d
To probe for direct interactions between the SNARE proteins and the COPII coat subunits, we expressed portions of the cytosolic domains of membrin and syntaxin 5 fused to the N terminus of glutathione S-transferase (GST), then immobilized the fusion proteins on glutathione Sepharose beads and probed for binding to Sec24c. Purified Sec24c was diluted with Escherichia coli lysate proteins, and the specificity of the interactions was assessed by the enrichment of the coat protein from the input mixture (Figure 1B and D).
Membrin contains an N-terminal folded domain (residues 1–110) upstream of the heptad-repeat SNARE motif and transmembrane domain (Mossessova et al, 2003). To probe the interaction with membrin, we produced two overlapping fragments, one encompassing the N-terminal domain (residues 1–132) and the other the C-terminal SNARE motif (112–186). Somewhat surprisingly, we found that both the N- and C-terminal polypeptides bound to Sec24c (Figure 1B, lanes 4 and 5), implying that the membrin transport signal sequence is situated in the region of overlap of the two constructs. To test this, we produced a membrin fragment that encompasses the linker region, residues 107–126, and we observed an interaction with Sec24c, as predicted (Figure 1B, lane 6); the fragment also binds to Sec24d (data not shown). No binding is observed with the Sec23 subunit (data not shown); thus membrin, similar to almost all other cargo molecules studied to date, binds to the Sec24 subunit of the COPII coat (Lee et al, 2004).
To assess the specificity of the interaction and to pinpoint key residues, we performed an alanine scan through the membrin fragment 107–126. Figure 1B (lanes 7–12) shows the effects of mutations through the central region of the membrin sequence. Mutation of two residues, I118A and M120A significantly impaired binding to Sec24c, whereas mutation of neighbouring residues had no appreciable effect on binding (Figure 1B, lanes 7–12). We refer to this core region of the membrin sequence as the IxM motif; a more definitive test of the function of the IxM motif as a transport signal was possible once we had mapped the interaction site of membrin IxM with Sec24c and Sec24d by X-ray crystallography, and this analysis is described below.
In an earlier study on the S. cerevisiae COPII coat (Mossessova et al, 2003), we detected no interaction between Sec24 and Bos1 (yeast membrin). Consistent with this, the IxM sequence that we have now identified is not present in the yeast Bos1 polypeptide (Figure 1C, upper panel). By contrast, the IxM motif is highly conserved both in sequence and position in membrin proteins of higher eukaryotes; in the sequences that we studied, the methionine residue of IxM is invariant, whereas the isoleucine residue can change conservatively to leucine.
Earlier studies on the S. cerevisiae COPII coat raised a similar puzzle regarding the signals for packaging syntaxin 5 into vesicles (Mossessova et al, 2003). Specifically, a YNNSNPF sequence on yeast Sed5/syntaxin 5 is the primary signal for ER exit, and an LxxME motif may be a secondary signal (Mossessova et al, 2003; Miller et al, 2005); however, as illustrated in Figure 1C (lower panel), the YNNSNPF and LxxME sequences are unique to the S. cerevisiae SNARE protein. These sequences are located in a linker region of the syntaxin 5 polypeptide that separates the N-terminal regulatory domain (NTD) and the C-terminal SNARE motif and transmembrane domain (Bracher and Weissenhorn, 2002). Strikingly, when we studied sequences of the syntaxin 5 linker region from higher eukaryotes, we identified a highly conserved IxM sequence in a central location (Figure 1C, lower panel).
To test whether the IxM sequence on syntaxin 5 binds to Sec24c, we produced an appropriate fragment of syntaxin 5 (fused to GST) containing the putative IxM signal (residues 151–220) and tested this for binding to Sec24c and Sec24d. The ∼70-residue syntaxin 5 peptide does indeed bind to Sec24c and Sec24d (Figure 1D, lane 4 and data not shown). The syntaxin 5 peptide was shortened to just a 20-residue element (residues 181–200), which also binds to the coat proteins (Figure 1D, lane 5). Point mutations to alanine of a core sequence of the 20-residue peptide had the predicted effect: the mutations I191A and M193A impaired binding to Sec24c (Figure 1D, lanes 8 and 10), whereas mutation of neighbouring residues had little or no effect on binding (Figure 1D, lanes 6, 7, 9 and 11).
In summary, we have defined interactions between the COPII coat machinery and mammalian membrin and syntaxin 5, involving a highly conserved IxM motif that we infer is a signal for cargo packaging into COPII-coated vesicles. To confirm the specificity of the interactions, and to establish a structural framework for functional tests of the IxM sequence as a signal for SNARE packaging, we initiated X-ray crystallographic analysis of the coat–signal interactions.
Molecular recognition of the IxM signal by the COPII machinery
For crystallization experiments, we purified truncated versions of the Sec24 proteins that lack the hypervariable, unstructured N-terminal regions (see Materials and methods). These were used in crystallization trials of Sec23/24c–d heterodimers and Sec24c–d monomers, and yielded two new crystal forms for the Sec24c and Sec23a/24d complexes. Synthetic membrin and syntaxin 5 peptides were soaked into fully grown crystals at high concentrations (4 mM), and the co-crystal structures of Sec23a/24d·membrin and Sec23a/24d·syntaxin 5 were determined by standard X-ray crystallographic methods. In both cases, peptide sequences could be identified by difference Fourier analysis and were modelled straightforwardly into residual electron density (see Materials and methods and Table I).
Table 1.
Data collection and refinement statistics
| Complex | Sec23a/24d·syntaxin 5 | Sec23a/24d·membrin | Sec23a/24a·Sec22b·VSVG | Sec23a/24a·Sec22b·Bet1 | Sec24b | Sec24c |
| Peptide soaked | DVAIDMMDP | TTIPMDE | QIYTDIEMNR | GYSACEEEN | — | — |
| Space group | P212121 | P212121 | C2 | C2 | C2 | P212121 |
| Cell parameters a, b, c (Å) | 103.1, 140.8, 152.3 | 103.1, 140.8, 152.4 | 147.8, 97.6, 129.1 | 148.2, 97.2, 129.5 | 161.4, 67.5, 72.4, | 69.7, 183.0, 201.8 |
| β (deg) | 90.1 | 90.0 | 100.4 | |||
| Data processing | ||||||
| Wavelength (Å) | 0.9795 | 0.9795 | 0.9950 | 1.542 | 0.9795 | 0.9795 |
| Resolution (Å) | 50–2.7 | 50–3.0 | 30–2.7 | 25–3.3 | 50–1.8 | 50–2.3 |
| Rmerge (%)a | 8.0 (39.8)b | 9.1 (43.4) | 8.2 (30.9) | 11.1 (39.4) | 5.0 (28.0) | 4.7 (34.6) |
| I/σ | 24.6 (3.2) | 20.2 (3.5) | 15.5 (3.0) | 9.6 (2.2) | 22.5 (3.3) | 27.8 (3.6) |
| Completeness (%) | 99.4 (100.0) | 99.9 (99.8) | 95.1 (97.0) | 92.0 (95.1) | 97.8 (85.0) | 99.4 (98.3) |
| Redundancy | 5.3 (5.1) | 5.2 (5.0) | 3.3 (3.1) | 2.7 (2.6) | 3.1 (2.6) | 3.9 (3.5) |
| Refinement statistics | ||||||
| Data range (Å) | 50–2.7 | 50–3.0 | 30–2.7 | 25–3.3 | 50–1.8 | 50–2.3 |
| Reflections | 58 392 | 43 252 | 46 345 | 24 079 | 65 527 | 104 257 |
| Non-hydrogen atoms | 11 777 | 11 850 | 12 591 | 12 526 | 5815 | 17 569 |
| Water molecules | 150 | 107 | 163 | 0 | 691 | 240 |
| r.m.s. Δ bonds (Å)c | 0.007 | 0.007 | 0.007 | 0.008 | 0.005 | 0.007 |
| r.m.s. Δ angles (deg)c | 1.4 | 1.5 | 1.4 | 1.55 | 1.2 | 1.4 |
| R-factor (%)d | 21.1 | 20.2 | 20.6 | 20.6 | 18.4 | 24.1 |
| Rfree (%)d,e | 26.1 | 25.4 | 27.4 | 29.2 | 21.8 | 28.4 |
| aRmerge=100∑h∑i∣Ii(h)–〈I(h)〉∣/∑h〈I(h)〉, where Ii(h) is the ith measurement and 〈I(h)〉 is the weighted mean of all measurement of I(h) for Miller indices h. | ||||||
| bHighest resolution shell is shown in parentheses. | ||||||
| cRoot-mean-squared deviation (r.m.s. Δ) from target geometries. | ||||||
| dR-factor=100∑∣FP–FP(calc)∣/∑FP. | ||||||
| eRfree was calculated with 5% of the data. | ||||||
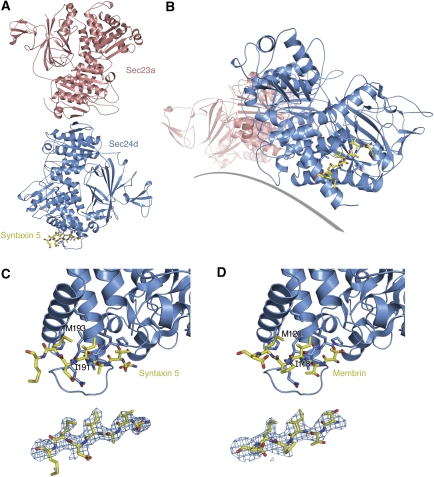
The IxM-containing peptides of syntaxin 5 and membrin bind to a common site on the equatorial surface of Sec24d (Figure 2). The binding site is distal to the interface with Sec23; it is roughly equidistant to the A and B sites described earlier on yeast Sec24 (Mossessova et al, 2003); and we estimate that the binding site is situated ∼10–15 Å from membrane when the COPII coat is bound to the membrane surface.
Figure 2.
Molecular recognition of syntaxin 5 and membrin IxM packaging signals by Sec24d. (A) Crystal structure of the Sec23a/24d complex viewed from the membrane distal surface. Bound to Sec24d is the transport signal of syntaxin 5 (coloured yellow). (B) This alternate view, approximately along the long axis of the Sec23a/24d complex, emphasizes the proximity of the syntaxin 5 signal to the modelled membrane (grey line). (C) Two views of the syntaxin 5 transport signal bound to Sec24d. The upper picture shows a close-up view of the DVAIDMM peptide of syntaxin 5 bound to the COPII protein; the critical Ile191 and Met193 residues are labelled. The lower picture shows the syntaxin 5 peptide together with a difference electron density map (SA omit map; Brünger, 1998), calculated using the Sec23a/24d·syntaxin 5 data at 2.7-Å resolution and contoured at 2.2σ (Table I). (D) Detailed view of the membrin TTIPMD peptide bound to Sec24d. In the upper picture, the Ile118 and Met120 residues on membrin are labelled. The lower picture shows the membrin transport signal and a difference electron density (SA omit) map, calculated using the Sec23a/24d·membrin data at 3.0-Å resolution and contoured at 2.0σ.
The co-crystal structure of syntaxin 5 and Sec23a/24d was determined with the syntaxin 5 peptide 188DVAIDMMDP196 bound, and strong electron density is observed for all residues except the C-terminal Asp195 and Pro196 (Figure 2C). The co-crystal structure comprising membrin peptide bound to Sec23a/24d used the membrin sequence 116TTIPMDE122, and in this case electron density is observed for all residues except C-terminal Glu122 (Figure 2D). The syntaxin 5 and membrin peptides bind in very similar conformations, with the IxM motif in a central position. Specifically, the peptides adopt an extended conformation and bind in a β-strand arrangement to a surface loop connecting helices αL and αM (residues 825–841) in the all-helical domain of Sec24 (Figure 2C and D). The binding site is framed by the most C-terminal α-helix (αS) and the most N-terminal loop-helix in the Sec24d crystal structure. The binding interactions observed between the peptides and Sec24 are consistent with the results of the alanine-scan mutagenesis (Figure 1B and D). Only the isoleucine and methionine side chains of the IxM motif make extensive interactions with Sec24d. Primary hydrophobic contacts are made between the conserved methionine residue and Sec24d residues Leu834 (on the αL–αM element) and His1025 (on helix αS). For both syntaxin 5 and membrin, the isoleucine residue of the IxM motif makes hydrophobic contacts with residues Leu834, Leu836 and Leu1021 of Sec24d. Although the side chains neighbouring the IxM motif do not form contacts with Sec24d, the main chain atoms of these residues likely contribute to binding, specifically through the antiparallel β-strand hydrogen-bonding arrangement involving the αL–αM element (Figures 2C, D and 3C). Finally, the isoleucine and methionine residues of the IxM motif make intra-peptide hydrophobic contacts that may help stabilize the peptide conformation when bound to Sec24d.
Figure 3.
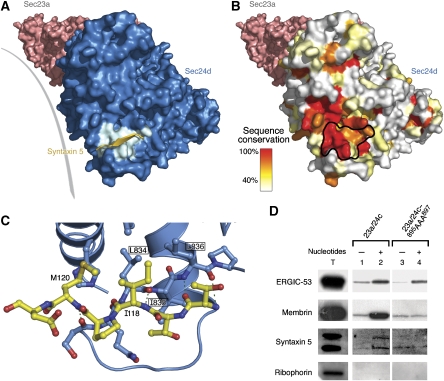
Structural basis for signal-mediated packaging of membrin and syntaxin 5 into COPII vesicles. (A) Molecular surface of the Sec23a/24d complex oriented approximately as in Figure 2B. Sec23a is coloured pink and Sec24d is blue. The IxM-binding site (those residues contacted by the syntaxin 5 transport signal) is highlighted in cyan. The syntaxin 5 peptide is coloured yellow and drawn as an arrow to emphasize its β-strand-type connection to Sec24d. (B) Conservation of the signal-binding site on Sec24d. The molecular surface of Sec24d is coloured according to sequence conservation of the underlying residues in an alignment of Sec24c and Sec24d isoforms from nine organisms (see Materials and methods). The IxM-binding site on Sec24d is outlined in black. (C) Interactions between the IxM signal of membrin and Sec24d. The key residues of membrin, I118 and M120, are labelled. Three central residues of the binding pocket on Sec24d are labelled and boxed: L834, I835 and L836 (the equivalent residues on Sec24c are L895, I896 and L897). (D) In vitro budding assay (performed as in Figure 1A) from semi-intact HEK-293 cells (to monitor packaging of ERGIC-53, syntaxin 5 and ribophorin) and CHO-K1 cells (for membrin). The packaging of cargo was assessed for wild-type Sec24c and the IxM site triple-mutant Sec24c–895AAA897 (This is part of the same experiment as that shown in Figure 1A.). The signal/noise of the blot for syntaxin 5 is limited by the quality of the antibody.
Packaging of syntaxin 5 and membrin by mammalian COPII vesicles
The residues lining the IxM-binding site on Sec24c and Sec24d are highly conserved (Figure 3A and B). Indeed, the key binding-site residues Leu834, Leu836, Leu1021 and His1025 (numbering for Sec24d) are invariant in the Sec24c/d family. Additional residues that may provide stabilization within the αL–αM loop are also conserved, namely Gln833, Ile835 and Pro837.
On the basis of these structural observations, we designed a binding site triple mutant of Sec24c to test the relevance of the co-crystal structure and of the role of the IxM motif in SNARE packaging. Specifically, we mutated the binding site sequence 895LIL897 on Sec24c (equivalent to 834LIL836 on Sec24d) to 895AAA897; the location of these three side chains is shown in Figure 3C. Vesicles generated with the mutant protein contained substantially lower levels (at or near background) of membrin and syntaxin 5 when compared with that obtained with wild-type protein (Figure 3D). This seems not to be due to a general defect in generating COPII vesicles or in cargo packaging, as the mutant coat protein efficiently packaged ERGIC-53.
In summary, we propose that the interaction between the IxM motif and Sec24c/d is the primary mechanism for packaging membrin and syntaxin 5 into COPII-coated vesicles, and that the IxM motif is a highly conserved signal for packaging these SNARE proteins. Although S. cerevisiae Sed5 (syntaxin 5) uses YNNSNPF and LxxME signals (Mossessova et al, 2003; Miller et al, 2005), it seems that IxM is the canonical motif for packaging syntaxin 5 and membrin into COPII-coated vesicles.
Conserved B site for binding Bet1 LxxLE and DxE signal sequences
It was noted in structural studies of the Sec23/24 complex (Bi et al, 2002) that the Sec24 B site—which is now known to bind LxxLE and DxE signals—is well conserved across all isoforms of Sec24 (in particular, the B site is highly similar in S. cerevisiae Sec24 and Lst1, a yeast homologue of Sec24). Nevertheless, in vitro budding experiments show that yeast Sec24 can package Bet1 via its LxxLE motif, whereas Lst1 fails to package Bet1 (Miller et al, 2002). A structural explanation was suggested by X-ray analysis of the Sec24·Bet1 complex; this indicated that subtle residue changes at the B site were responsible for the differential packaging activity of the Sec24 forms. In particular, residue Leu582 of Sec24, which forms van der Waals contacts with Bet1 LxxLE, is changed to aspartic acid (Mossessova et al, 2003).
Interestingly, Leu582 of Sec24, or rather the equivalent residue position in the human proteins, is conserved as a leucine in Sec24a and Sec24b, but is an aspartic acid in Sec24c and Sec24d. On this basis, we tested the prediction that the B site of human Sec24 isoforms has differential selectivity for LxxLE and DxE signal sequences. This was done through pull-down experiments involving the DxE signal of viral VSV-glycoprotein (VSV-G). The DxE sequence of VSV-G was the first ER exit signal to be discovered (Nishimura et al, 1999). More recently, DxE signals have been uncovered in endogenous proteins, in particular yeast Sys1, and this latter protein has been subject to extensive structure–function analysis (Votsmeier and Gallwitz, 2001; Miller et al, 2003; Mossessova et al, 2003).
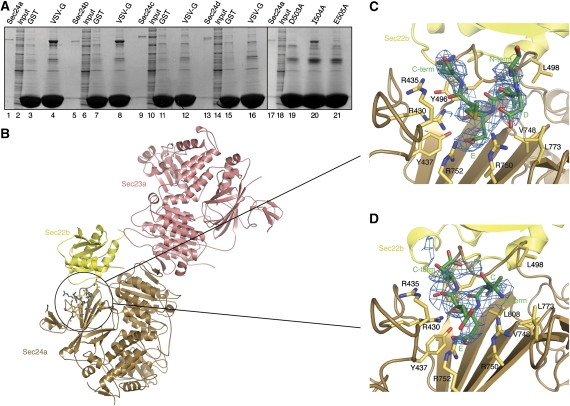
The C-terminal tail of VSV-G was produced as a C-terminal fusion to GST and this was used to test for interactions with the human isoforms Sec24a–d (Figure 4A). VSV-G interacted strongly with Sec24a and Sec24b, but not with Sec24c or Sec24d (Figure 4A, lanes 4, 8, 12 and 16). Alanine mutations of the DxE motif of VSV-G showed a strict requirement for both the aspartate and glutamate residues of the signal (Figure 4A, lanes 19 and 21). By contrast, the central isoleucine residue could be changed to alanine with only minor loss of affinity (Figure 4A, lane 20). In summary, there is indeed selectivity among the human Sec24 isoforms for binding DxE signals.
Figure 4.
Molecular recognition of Bet1 and VSV-G transport signals at the B site of Sec24a and Sec24b. (A) Interactions were tested between the cytoplasmic tail (29 C-terminal residues) of VSV-G and the four human Sec24 isoforms. VSV-G binds to Sec24a and Sec24b (lanes 4 and 8), but not to Sec24c or Sec24d (lanes 12 and 16). Mutation of the aspartate and glutamate residues of the VSV-G DxE motif abrogates binding to Sec24a (lanes 19 and 21); the intervening isoleucine residue can be mutated to alanine without appreciable effect on binding (lane 20). These pull-down experiments were performed as in Figure 1B. (B) Crystal structure of the Sec23a/24a·Sec22b complex, with the membrane-proximal surface facing forward. Circled in the picture is the VSV-G transport signal bound to the B site on Sec24a. Note the proximity of the VSV-G- and Sec22b-binding sites on Sec24a. (C) Interactions between the DxE signal on VSV-G and the B site of Sec24a. Difference electron density for the bound VSV-G peptide (QIYTDIEMNR) was calculated at 2.7-Å resolution and contoured at 1.8σ. (D) Interactions between the Bet1 signal and the B site of Sec24a. Difference electron density for the bound Bet1 peptide (GYSACEEEN) was calculated at 3.3-Å resolution and contoured at 1.7σ.
On the basis of these observations, the minimal VSV-G peptide sequence 499QIYTDIEMNR508 was synthesized. The peptide, which was highly soluble, was soaked into fully grown crystals of the Sec23a/24a·Sec22b complex. The crystal structure of the ∼200 kDa Sec23a/24a·Sec22b complex was determined in an earlier study (Mancias and Goldberg, 2007); importantly, these crystals are grown in a PEG/salt solution that is compatible with peptide binding (see Materials and methods), and the B site is thoroughly exposed to solvent in the crystals and thus available for binding the signal sequence. The crystal structure of Sec23a/24a·Sec22b bound to the VSV-G peptide was refined to 2.7-Å resolution (Figure 4B). In the structure, the B site of Sec24a is very similar, in terms of the conformation of key residue side chains, to the B site of yeast Sec24 when bound to DxE and LxxLE signals (Mossessova et al, 2003). As expected, strong electron density in difference Fourier maps was observed at the B site for a portion of the VSV-G peptide, 502TDIEMN507 (Figure 4C). The binding mode is highly similar to the DxE motif of Sys1 bound to yeast Sec24 (Mossessova et al, 2003). Specifically, the 504IE505 residues of VSV-G adopt a helical configuration and the glutamate side chain makes salt bridge contacts with Arg752 of Sec24a. Importantly, residue Asp503 of VSV-G adopts a left-handed backbone conformation, thereby allowing the carboxylate group of its side chain to form a bivalent interaction with Arg750 of Sec24a (Figure 4C). The same arrangement—involving a left-handed backbone conformation of the aspartate residue—is observed for Sys1, and is the defining feature of the differential binding modes of DxE and LxxLE class signals at the B site (Mossessova et al, 2003).
Finally, we note that the VSV-G sequence, 499QIYTDIEMNR508, has an IxM motif nested in the DxE signal. Despite this, the VSV-G signal does not bind to Sec24c or Sec24d (Figure 4A), suggesting that there are residue restrictions at the sites neighbouring IxM (possibly including a requirement for a short or β-branched amino acid immediately upstream of the isoleucine residue, needed to stabilize the extended β-stranded conformation of the IxM signal). Further studies are required to explore the detailed sequence requirements for IxM binding, and to test the idea that the nesting of a cryptic IxM signal in the DxE motif of VSV-G renders the 499QIYTDIEMNR508 C-terminal tail of the viral protein more resistant functionally to point mutations.
The LxxLE transport signal of yeast Bet1 is believed to be the primary signal for ER exit of this SNARE protein (Miller et al, 2003; Mossessova et al, 2003). Motifs similar to LxxLE are conserved in Bet1 proteins across species, both in terms of sequence and position along the SNARE polypeptide. However, significant sequence variation is observed; thus, human Bet1 bears the motif YxxCE within the sequence 24GYSACEEEN32 at the N terminus of the SNARE motif. Nevertheless, we concluded that human YxxCE is likely to be functionally equivalent to yeast LxxLE, based on an analysis of the yeast Sec24·Bet1 interaction. Specifically, the invariant glutamate residue forms an intricate arrangement of hydrogen-bonding interactions with binding site residues, whereas the role of the leucine residues is merely to form hydrophobic elements that face Sec24 and that help stabilize the peptide helical conformation (Mossessova et al, 2003).
To test whether the YxxCE motif of human Bet1 binds to Sec24a/b in this manner, we co-crystallized the peptide with Sec23a/24a·Sec22b. Initially, crystallization studies proved difficult owing to low solubility of the Bet1 sequence 17–33; similar problems had arisen with the yeast Bet1 peptide (Mossessova et al, 2003). Human Bet1 peptides were sufficiently soluble when N-terminal residues were absent, so the co-crystal structure was determined with the peptide 24GYSACEEEN32. The binding mode of the peptide is highly similar to that of yeast Bet1 LxxLE bound to Sec24. There are four noteworthy observations. First, residue Glu29 of YxxCE forms salt bridge contacts with Arg752 of Sec24a (Figure 4D). Second, Sec24a recognizes the helical configuration of the signal peptide by capping the main chain carbonyl oxygen atoms of 29EEE31 through the guanidinium groups of arginine residues 430 and 435 (Figure 4D). Third, the cysteine residue of the human Bet1 sequence occupies the same binding site as the second leucine of yeast Bet1 LxxLE. Finally, as in the yeast Sec24·Bet1 complex, N-terminal residues of the bound peptide are mobile in the crystal, such that electron density is absent for residues 24GYS26 (see Mossessova et al, for a discussion of the possible role of these upstream residues).
This analysis shows that the B site of Sec24 subunits is conserved from yeast to humans for binding DxE and LxxLE ER exit signals, but only among isoforms of the Sec24a/b class of COPII coat proteins.
Diversity of ER exit signals and binding sites on COPII proteins
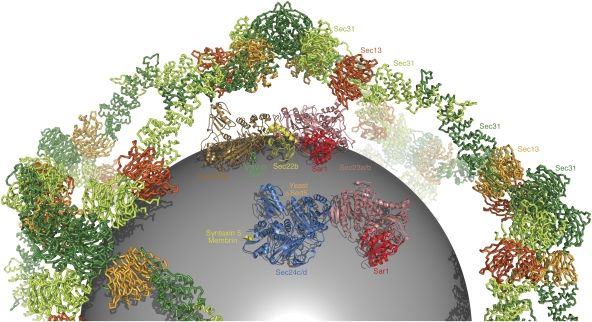
The model in Figure 5 summarizes the results of structural studies on COPII subunit cargo selectivity. The figure includes part of the 60-nm cuboctahedron cage, which is formed from 24 copies of the (Sec13/31)2 assembly unit (Stagg et al, 2006; Fath et al, 2007). Each of the 48 copies of Sec31 in the cage can bind to a Sec23/24·Sar1 complex (Bi et al, 2007), but we have included just two copies of Sec23/24·Sar1 in the figure for clarity.
Figure 5.
Arrangement of cargo-binding sites in the COPII coat. In the figure, the model of the Sec13/31 cage is drawn as a backbone worm and coloured red and green following the convention of Fath et al (2007). Modelled inside the cage is a sphere of diameter 40 nm representing a membrane vesicle. The two copies of the Sec23/24·Sar1 pre-budding complex are composite models based on structural alignments of crystal structures from the current study and from previous studies (Bi et al, 2002; Mossessova et al, 2003). Sec23a is coloured pink, Sar1 (taken from the yeast Sec23/24·Sar1 complex) is red, Sec24a is brown and Sec24d is blue. The bound cargo molecules are coloured as follows: VSV-G/Bet1 is green, Sec22b is yellow, the binding site for yeast Sed5 is labelled with a star in orange and syntaxin 5/membrin is yellow.
Multiple isoforms of Sec24 expand the range of cargo accommodated in mammalian COPII vesicles, but there is no evidence that Sec24 isoforms are segregated into distinct vesicles; instead, mammalian COPII-coated vesicles may include the full complement of Sec24 isoforms. In Figure 5, we have indicated the binding sites for Sec22, Bet1 and VSV-G signals on a model of Sec24a/b, and the binding site of the membrin and syntaxin 5 IxM motif on Sec24c/d, along with the site occupied by the YNNSNPF signal of Sed5/syntaxin 5 in the yeast system. The manner in which Bet1 and Sec22 are packaged by Sec24a and Sec24b is conserved from yeast to humans. By contrast, the YYNSNPF motif seems to be a S. cerevisiae novelty for packaging syntaxin 5 at a site that has (as yet) no known role in the human Sec24 proteins. The conserved mechanism for packaging syntaxin 5 and membrin employs the IxM motif and COPII subunits of the Sec24c/d class.
Structural basis for cargo discrimination by human Sec24 proteins
To complete the structure–function analysis in this study, we sought a structural explanation for the ability of the human Sec24 isoforms to discriminate among membrin, syntaxin 5 and Sec22. To this end, we solved structures of Sec24b and Sec24c, which together with the Sec23a/Sec24d and Sec23a/Sec24a·Sec22 complexes provides a complete set of structures for human Sec24 proteins (see Materials and methods).
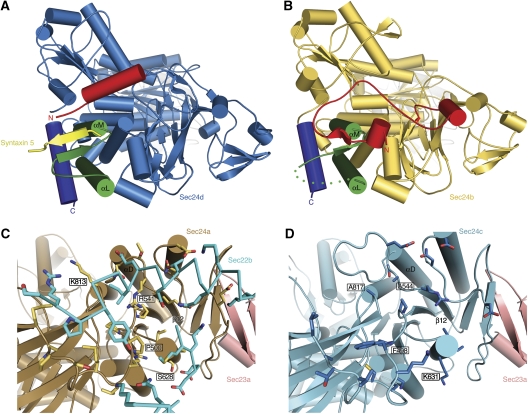
Comparison of the crystal structures provides a clear explanation for the selectivity towards the IxM signal of membrin and syntaxin 5. Figure 6A highlights the αL–αM structural element that forms the core of the IxM-binding site on Sec24c and Sec24d. Strikingly, although the αL–αM conformation is conserved in Sec24a and Sec24b (Figure 6B), it serves as a binding site not for a signal sequence but for a short β-strand in the N terminus of the Sec24 polypeptide (residues 349–353 in Sec24a; 523–527 in Sec24b). This self-interaction involves an antiparallel β-strand arrangement similar to the IxM peptide binding to Sec24c/d. Thus, a peptide element is conserved in Sec24a and Sec24b (coloured red in Figure 6B) to bind across the αL–αM site, whereas this element has a distinct amino-acid sequence in Sec24c and Sec24d and adopts an α-helix that forms part of the IxM-binding site (coloured red in Figure 6A). (The polypeptide chains of the four human Sec24 proteins have three regions: an N-terminal hypervariable region that is probably unstructured in solution; an adjacent short stretch (coloured red in Figure 6A and B) which can affect IxM signal binding and the ‘core' sequence that starts from the β-barrel domain (Bi et al, 2002) and is conserved in all four isoforms.)
Figure 6.
Structural basis for cargo discrimination by human Sec24 isoforms. (A) Crystal structure of the Sec23a/Sec24d·syntaxin 5 complex viewed from the base of Sec24d. The αL–αM loop that forms the binding site for IxM signals is coloured green, the C-terminal helix is deep blue and the variable N-terminal region is red. The syntaxin 5 peptide is yellow. (B) Crystal structure of Sec24b (see Table I). As in (A), the αL–αM loop is coloured green, the C-terminal helix is deep blue and the N-terminal region is red. Note how the N-terminal variable region occupies the IxM-binding site. (C) Structure of Sec24a from the Sec23a/Sec24a·Sec22b complex. Sec24a is coloured brown and Sec23a is pink. Sec24a residues that form the Sec22-binding site are drawn in yellow. Critical interface residues R541 and P500 are labelled. (D) Crystal structure of Sec24c. Sec24c is coloured light blue and Sec23a is modelled in pink (Sec23a is not present in this crystal structure; Table I). Sec24c cannot bind to Sec22 because key binding-site residues are altered: in particular, note that residue 544 is alanine (equivalent to R541 in Sec24a) and residue 498 is phenylalanine (P500 in Sec24a).
Finally, we compared the structures of Sec24c and Sec24d determined in this study with the structure of Sec23a/Sec24a·Sec22b determined earlier (Mancias and Goldberg, 2007) to provide an explanation for Sec24 isoform specificity for the SNARE Sec22. As shown in Figure 6C and D, the conformation of the Sec22-binding site is similar in Sec24a and Sec24c (and also in Sec24b and Sec24d, not shown). However, key residue changes preclude Sec22 binding to Sec24c/d. Several of these are indicated in Figure 6C and D; for example Sec24a residue Pro500 changes to phenylalanine in Sec24c, but the most important change seems to be to Sec24a residue Arg541. This arginine residue was pinpointed originally in a genetic screen for dysfunctional S. cerevisiae Sec24 proteins; it is essential for tight binding of Sec22 to Sec23a/24a, and for packaging into COPII-coated vesicles (Miller et al, 2003; Mancias and Goldberg, 2007). The Sec24a mutant R541A is unable to package Sec22b (as is the Sec24b mutant R715A). Strikingly, the equivalent position in Sec24c harbours an alanine residue (Figure 6D).
Thus, we have described two distinct mechanisms for controlling the specificity of Sec24 proteins for cargo: in the cases of Sec22 and LxxLE/DxE signals, there is a straightforward divergence of binding site residues; in the case of the IxM signal, a more elaborate mechanism entails the occlusion of the IxM-binding site by an N-terminal polypeptide element of the Sec24a/b subunits.
The two classes of Sec24 subunits, Sec24a/b and Sec24c/d, have non-overlapping selectivity for the ER exit signals analysed in this study, and we predict that this trend will continue for the numerous signals that probably remain to be discovered. To date, only one interaction partner is shared by all four Sec24 subunits, namely Sec23 (Fromme et al, 2008).
Materials and methods
Protein production
The expression in insect cells and purification of Sec23/24 complexes was carried out as described (Bi et al, 2002; Mancias and Goldberg, 2007). Briefly, baculoviruses (Bac-to-Bac; Gibco) were prepared encoding the following full-length human proteins: Sec23a, Sec23b, Sec24a, Sec24b, Sec24c, Sec24d and triple-mutant Sec24c (L895A, I896A and L897A); the truncated proteins Sec24a (lacking residues 1–340), Sec24b (lacking residues 1–514), Sec24c (lacking residues 1–299), Sec24c (lacking residues 1–327) and Sec24d (lacking residues 1–266). All proteins contained an N-terminal His6 tag. For protein production, insect cells were harvested 48 h post-infection and protein was purified from cell lysate by Ni2+–IMAC chromatography. The His6 tag was removed with TEV protease (Invitrogen), followed by chromatographic purification using Q-sepharose and Superdex 200 columns. The human Sec13a/31a complex was expressed in baculovirus and purified as described (Kim et al, 2005). Hamster Sar1a was expressed in E. coli and purified as described (Kim et al, 2005). The cytosolic domain of Sec22b was cloned and purified as described (Mancias and Goldberg, 2007).
The cytosolic domains of Homo sapiens syntaxin 5 and membrin were produced by the dual-tagging system of His6–Smt3 and GST (His6–Smt3 fused at the N terminus and GST at the C terminus) and transformed in E. coli Rosetta (DE3) pLysS cells. Protein expression was induced with 1 mM IPTG for 3 h at 30°C. Cells were collected by centrifugation, resuspended in 25 mM Tris–HCl (pH 7.4), 10% (w/v) sucrose and lysed with a single freeze–thaw cycle. DNase I was added to reduce viscosity and, following centrifugation, protein was purified by Ni2+–IMAC chromatography. The His6–Smt3 moiety was removed by Ulp1 protease cleavage (Mossessova and Lima, 2000). All truncated and mutant forms of the SNARE proteins were produced through the same protocol. The C-terminal tail of VSV-G was produced as a C-terminal fusion to GST using the pGEX-4TI vector, transformed in E. coli Rosetta (DE3) pLysS cells, and purified through glutathione Sepharose chromatography. The following peptides (>95% purity) were purchased from Anaspec (San Jose, CA) for crystal soaking experiments: the syntaxin 5 sequence 188DVAIDMMDP196; the membrin sequence 116TTIPMDE122; the VSV-G sequence 499QIYTDIEMNR508 and the Bet1 sequence 24GYSACEEEN32. Finally, E. coli lysate for pull-down assays was prepared according to Mossessova and Lima (2000).
Cell culture, semi-permeabilization and in vitro COPII-budding assay
For the preparation of semi-permeabilized cells to be used in in vitro COPII budding reactions, we used CHO-K1 and HEK-293 cells and followed exactly the procedure described earlier (Mancias and Goldberg, 2007). Likewise, the in vitro COPII budding reactions were carried out as described by Kim et al (2005), incorporating the modifications detailed in Mancias and Goldberg (2007). The reaction has been shown earlier to yield coated vesicles (Kim et al, 2005).
SNARE-binding assays
Assays were carried out according to Mossessova et al (2003). Briefly, a saturating quantity (100–250 μg) of SNARE–GST fusion protein was incubated with 10 μl of a 50% (v/v) slurry of glutathione Sepharose 4B beads (Pharmacia) for 30 min at 4°C. Beads were washed twice with buffer A (150 mM NaCl, 20 mM Tris (pH 7.4), 4 mM DTT, 0.2% (w/v) Triton X-100), leaving 50–100 μg of protein bound to the beads. Beads were then mixed with Sec24a–d (0.3 mg/ml) and E. coli lysate (13 mg/ml) in buffer A (total assay volume 100 μl). The assay mix was incubated for 15 min at 4°C, and beads were washed once with 100 μl buffer A. Proteins were eluted with SDS sample buffer and analysed by 4–15% gradient SDS–PAGE and Coomassie blue staining.
Protein crystallization
Pure Sec23a/24d (truncated 24d lacking residues 1–266) was concentrated to 30 mg/ml in 150 mM NaCl, 20 mM Tris–HCl (pH 7.5), 4 mM DTT and stored at −80°C. Crystals were grown through the hanging-drop method, by adding 1 μl protein solution to 1 μl of well solution comprising 5.5% (w/v) PEG 4000, 0.1 M magnesium sulphate and 100 mM HEPES (pH 7.9). Orthorhombic crystals grew overnight at 22°C (Table I). To form complexes of Sec23a/24d with membrin and syntaxin 5 transport signal sequences, fully grown crystals were soaked with synthetic peptides. For this, crystals were transferred to soaking buffer comprising 6% (w/v) PEG 4000, 0.1 M NaCl, 0.1 M magnesium sulphate, 50 mM HEPES (pH 7.5) plus 4 mM peptide and left overnight at 22°C. These crystals were cryoprotected by transfer to a solution of soaking buffer containing an additional 32% ethylene glycol, and flash frozen in liquid propane. Crystals treated in this manner diffracted synchrotron X-rays beyond 2.7-Å resolution.
The Sec23a/24a·Sec22b complex was crystallized as described (Mancias and Goldberg, 2007). Fully grown crystals were then transferred to soaking buffer comprising 10% (w/v) PEG 4000, 0.1 M NaCl, 0.5 M sodium acetate, 50 mM Tris buffer, pH 7.9, plus 4 mM Bet1 or VSV-G signal peptides, and soaking proceeded overnight at 22°C. Crystals were cryoprotected by transfer to a solution of soaking buffer containing additional 24% ethylene glycol, and flash frozen in liquid propane. Crystals treated in this manner diffracted synchrotron X-rays beyond 2.7-Å resolution.
Pure Sec24b (lacking residues 1–514) was concentrated to 40 mg/ml in 150 mM NaCl, 20 mM Tris pH 7.5, 4 mM DTT and stored at −80°C. Crystals were grown through the hanging-drop method, by adding 1 μl protein solution to 1 μl of well solution comprising 42% ethylene glycol. Crystals (space group: C2; a=161.4, b=67.5, c=72.4 Å, β=100.4°) grew in 1 week at 4°C. Crystals were cryoprotected by transfer to a solution of well buffer and flash frozen in liquid propane. Crystals treated in this manner diffracted synchrotron X-rays beyond 1.8-Å resolution.
Finally, purified Sec24c (lacking residues 1–327) was concentrated to 40 mg/ml in 150 mM NaCl, 20 mM Tris–HCl (pH 7.5), 4 mM DTT and stored at −80°C. Crystals were grown through the hanging-drop method, by adding 1 μl protein solution to 1 μl of well solution comprising 6% (w/v) PEG 4000 and 0.1 M MES at pH 5.8. Crystals (space group P212121, a=69.7, b=183.0, c=201.8 Å) grew overnight at 4°C. Crystals were cryoprotected by transfer to a solution of well buffer containing an additional 32% ethylene glycol and flash frozen in liquid propane. Crystals treated in this manner diffracted X-rays to 2.3-Å resolution.
Crystal structure determination
X-ray diffraction data were collected at beamlines X9A and X-25 of the National Synchrotron Light Source (NSLS), and beamline ID-24 of the Advanced Photon Source. Data were processed with the programs DENZO and SCALEPACK (see Table I). The structure of Sec23a/24d was solved by molecular replacement with the program Phaser (McCoy et al, 2007) using H. sapiens Sec23a and S. cerevisiae Sec24 as the search models (Bi et al, 2002; Mancias and Goldberg, 2007). An initial model of Sec23a/24d was improved by rigid-body and positional refinement, and the full Sec23a/24d structure was built in the resulting electron density map. The crystal structure of the Sec23a/24a·Sec22b complex was described earlier (Mancias and Goldberg, 2007). The structures for Bet1 and VSV-G peptides bound to Sec23a/24a·Sec22b were determined by difference Fourier analysis. Final refinement of the models used the program CNS (Brünger, 1998). The refinement statistics and composition of the final models are summarized in Table I.
The structure of Sec24b was solved by molecular replacement with the program Phaser (McCoy et al, 2007) using S. cerevisiae Sec24 as the search model (Bi et al, 2002). An initial poly-alanine model was improved by rigid-body and positional refinement, and the full Sec24b structure was built in the resulting electron density map. Final refinement with the program CNS (Brünger, 1998) yielded a crystallographic R-factor of 18.4% (Rfree=21.8%) for data between 50.0- and 1.8-Å resolution (Table I).
The structure of Sec24c was also solved by molecular replacement with the program Phaser (McCoy et al, 2007) using S. cerevisiae Sec24 as the search model (Bi et al, 2002). An initial poly-alanine model of Sec24c was improved by rigid-body and positional refinement, and the full Sec24c structure was built in the resulting electron density map. Final refinement with the program CNS (Brünger, 1998) yielded an R-factor of 24.1% (Rfree=28.4%) for data between 50.0- and 2.3-Å resolution (Table I).
Sequence analysis
For preparation of Figure 1C, membrin sequences from six organisms were aligned: H. sapiens (NP_004278), Rattus norvegicus (NP_113873), Xenopus laevis (AAH54249), Dictyostelium discoideum (XP_638899), Candida glabrata (XP_449343) and S. cerevisiae (NP_013179), and syntaxin 5 sequences from seven organisms were aligned: H. sapiens (NP_003155), R. norvegicus (Q08851), X. tropicalis (CAJ83820), Danio rerio (NP_001002333), Drosophila melanogaster (NP_599132), Caenorhabditis elegans (NP_505968) and S. cerevisiae (NP_013126).
For preparation of Figure 3B, Sec24 sequences from the c and d isoform families from nine organisms were aligned: H. sapiens Sec24d (AAH35761), H. sapiens Sec24c (NP_004913), Apis mellifera Sec24c–d (XP_392952), C. elegans Sec24c–d (NP_502178), Aspergillus fumigatus Sec24c–d (XP_754649), Chaetomium globosum Sec24c–d (XP_001227615), Neurospora crassa Sec24 (XP_958077), D. discoideum Sec24 (XP_642481), Schizosaccharomyces pombe Sec24 (CAB58402) and S. cerevisiae Sec24 (NP_014349).
Acknowledgments
We thank Philip Jeffrey and Elena Goldberg for advice on crystallographic analysis and we thank staff at NSLS beamlines X9A and X25 for use of and assistance with synchrotron facilities. This study was supported by grants from the Howard Hughes Medical Institute and the NIH.
Accession numbers The atomic coordinates have been deposited in the Protein Data Bank with the accession codes 3EFO (Sec23a/24d·syntaxin 5), 3EG9 (Sec23a/24d·membrin), 3EGD (Sec23a/24a·Sec22b·VSVG), 3EGX (Sec23a/24a·Sec22b·Bet1), 3EH1 (Sec24b) and 3EH2 (Sec24c).
References
- Bi X, Corpina RA, Goldberg J (2002) Structure of the Sec23/24–Sar1 pre-budding complex of the COPII vesicle coat. Nature 419: 271–277 [DOI] [PubMed] [Google Scholar]
- Bi X, Mancias JD, Goldberg J (2007) Insights into COPII coat nucleation from the structure of Sec23·Sar1 complexed with the active fragment of Sec31. Dev Cell 13: 635–645 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bock JB, Matern HT, Peden AA, Scheller RH (2001) A genomic perspective on membrane compartment organization. Nature 409: 839–841 [DOI] [PubMed] [Google Scholar]
- Bonifacino JS, Glick BS (2004) The mechanisms of vesicle budding and fusion. Cell 116: 153–166 [DOI] [PubMed] [Google Scholar]
- Bracher A, Weissenhorn W (2002) Structural basis for the Golgi membrane recruitment of Sly1p by Sed5p. EMBO J 21: 6114–6124 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brünger AT (1998) Crystallography and NMR system: a new software suite for macromolecular structure determination. Acta Crystallogr D Biol Crystallogr 54: 905–921 [DOI] [PubMed] [Google Scholar]
- Fath S, Mancias JD, Bi X, Goldberg J (2007) Structure and organization of coat proteins in the COPII cage. Cell 129: 1325–1336 [DOI] [PubMed] [Google Scholar]
- Fromme JC, Orci L, Schekman R (2008) Coordination of COPII vesicle trafficking by Sec23. Trends Cell Biol 18: 330–336 [DOI] [PubMed] [Google Scholar]
- Kim J, Hamamoto S, Ravazzola M, Orci L, Schekman R (2005) Uncoupled packaging of amyloid precursor protein and presenilin 1 into coat protein complex II vesicles. J Biol Chem 280: 7758–7768 [DOI] [PubMed] [Google Scholar]
- Lee MC, Miller EA, Goldberg J, Orci L, Schekman R (2004) Bi-directional protein transport between the ER and Golgi. Annu Rev Cell Dev Biol 20: 87–123 [DOI] [PubMed] [Google Scholar]
- Mancias J, Goldberg J (2007) The transport signal on Sec22 or packaging into COPII-coated vesicles is a conformational epitope. Mol Cell 26: 403–411 [DOI] [PubMed] [Google Scholar]
- McCoy AJ, Grosse-Kunstleve RW, Storoni LC, Adams PD, Read RJ (2007) PHASER crystallographic software. J Appl Crystallogr 40: 658–674 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller E, Antonny B, Hamamoto S, Schekman R (2002) Cargo selection into COPII vesicles is driven by the Sec24p subunit. EMBO J 21: 6105–6113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller E, Liu Y, Barlowe C, Schekman R (2005) ER–Golgi transport defects are associated with mutation in the Sed5p-binding domain of the COPII coat subunit Sec24p. Mol Biol Cell 16: 3719–3726 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller EA, Beilharz TH, Malkus PN, Lee MC, Hamamoto S, Orci L, Schekman R (2003) Multiple cargo binding sites on the COPII subunit Sec24p ensure capture of diverse membrane proteins into transport vesicles. Cell 114: 497–509 [DOI] [PubMed] [Google Scholar]
- Mossessova E, Bickford LC, Goldberg J (2003) SNARE selectivity of the COPII coat. Cell 114: 483–495 [DOI] [PubMed] [Google Scholar]
- Mossessova E, Lima CD (2000) Ulp1-SUMO crystal structure and genetic analysis reveal conserved interactions and a regulatory element essential for cell growth in yeast. Mol Cell 5: 865–876 [DOI] [PubMed] [Google Scholar]
- Nishimura N, Bannykh S, Slabough S, Matteson J, Altschuler Y, Hahn K, Balch WE (1999) A di-acidic (DXE) code directs concentration of cargo during export from the endoplasmic reticulum. J Biol Chem 274: 15937–15946 [DOI] [PubMed] [Google Scholar]
- Stagg SM, Gürkan C, Fowler DM, LaPointe P, Foss TR, Potter CS, Carragher B, Balch WE (2006) Structure of the Sec13/31 COPII coat cage. Nature 439: 234–238 [DOI] [PubMed] [Google Scholar]
- Votsmeier C, Gallwitz D (2001) An acidic sequence of a putative yeast Golgi membrane protein binds COPII and facilitates ER export. EMBO J 20: 6742–6750 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wendeler MW, Paccaud JP, Hauri HP (2007) Role of Sec24 isoforms in selective export of membrane proteins from the endoplasmic reticulum. EMBO Rep 8: 258–264 [DOI] [PMC free article] [PubMed] [Google Scholar]