Abstract
Extended Fer-CIP4 homology (EFC)/FCH-BAR (F-BAR) domains generate and bind to tubular membrane structures of defined diameters that are involved in the formation and fission of endocytotic vesicles. Formin-binding protein 17 (FBP17) and Toca-1 contain EFC/F-BAR domains and bind to neural Wiskott–Aldrich syndrome protein (N-WASP), which links phosphatidylinositol (4,5)-bisphosphate (PIP2) and the Rho family GTPase Cdc42 to the Arp2/3 complex. The N-WASP–WASP-interacting protein (WIP) complex, a predominant form of N-WASP in cells, is known to be activated by Toca-1 and Cdc42. Here, we show that N-WASP–WIP complex-mediated actin polymerization is activated by phosphatidylserine-containing membranes depending on membrane curvature in the presence of Toca-1 or FBP17 and in the absence of Cdc42 and PIP2. Cdc42 further promoted the activation of actin polymerization by N-WASP–WIP. Toca-1 or FBP17 recruited N-WASP–WIP to the membrane. Conserved acidic residues near the SH3 domain of Toca-1 and FBP17 positioned the N-WASP–WIP to be spatially close to the membrane for activation of actin polymerization. Therefore, curvature-dependent actin polymerization is stimulated by spatially appropriate interactions of EFC/F-BAR proteins and the N-WASP–WIP complex with the membrane.
Keywords: actin polymerization, EFC domain, F-BAR domain, membrane deformation, N-WASP-WIP complex
Introduction
Dynamic rearrangements of membrane shape occur frequently during the formation of cell protrusions and vesicular trafficking (Farsad and De Camilli, 2003; McMahon and Gallop, 2005; Zimmerberg and Kozlov, 2006). The N-BAR domain proteins induce tubulated membranes when they bind to liposomes in vitro. Analysis of the three-dimensional structure of the N-BAR domain of amphiphysin indicated that it forms a banana-shaped dimer and that the concave surface of this dimer fits into membrane tubules induced by the N-BAR domain (Peter et al, 2004). Therefore, it is postulated that the structure of proteins can sense and/or generate membrane curvature. The concept of formation and sensing of membrane shape by the structure of a protein was further supported by analyses of the extended Fer-CIP4 homology (EFC)/FCH-BAR (F-BAR) and IRSp53-MIM-homology (IMD)/Rac-binding (RCB)/inverse-BAR (I-BAR) domains (Kessels and Qualmann, 2004; Itoh et al, 2005; Suetsugu et al, 2006b;; Mattila et al, 2007; Shimada et al, 2007; Frost et al, 2008). Interestingly, the concave surface of the EFC/F-BAR or N-BAR domains binds to the lipid membranes, whereas the convex surface of the IMD domain binds to the lipid membranes (Kessels and Qualmann, 2004; Suetsugu et al, 2006b; Mattila et al, 2007; Shimada et al, 2007). Thus, the structures of these proteins and their functions appear to be correlated. The EFC/F-BAR domain binds to liposomes with larger diameters than the N-BAR domain, and the diameter of the concave surface of the EFC/F-BAR domain is larger than that of the N-BAR domain (Shimada et al, 2007). The EFC/F-BAR and N-BAR domains are involved in invagination of the plasma membrane, such as during endocytosis, whereas the IMD domain is involved in the formation of protrusive structures, such as filopodia and lamellipodia (Takenawa and Suetsugu, 2007; Scita et al, 2008).
The membrane-binding domain is, in most cases, only a part of a multidomain protein. The obvious hypothesis derived from the organization of the domains of membrane-binding proteins is that the membrane-binding domains recruit or function in concert with membranes with specific curvatures (Takenawa and Suetsugu, 2007; Doherty and McMahon, 2008; Scita et al, 2008). Therefore, the membrane curvature may be a signal directing molecular assembly or catalytic activity. Interestingly, proteins containing BAR, EFC/F-BAR, or IMD/RCB/I-BAR domain also contain a molecule–molecule interaction domain, such as the Src-homology 3 (SH3) or Wiskott–Aldrich syndrome protein (WASP)-homology 2 (WH2) domain, and a catalytic domain, such as the tyrosine kinase, Rho guanine-nucleotide exchange factor (GEF) or Rho GTPase-activating protein (GAP) domains (Fujita et al, 2002; Dawson et al, 2006; Itoh and De Camilli, 2006; Takenawa and Suetsugu, 2007; Scita et al, 2008). However, to our knowledge, there are no reports regarding modulation of the functions of the membrane-binding proteins or their effector or binding proteins by binding of the membrane-binding proteins to membrane of a curvature.
The binding proteins of the SH3 domains of the membrane-binding proteins include neural (N)-WASP, WASP family verprolin homologous protein 2 (WAVE2), and dynamin (Ho et al, 2001, 2004; Itoh et al, 2005; Dawson et al, 2006; Itoh and De Camilli, 2006; Tsujita et al, 2006; Takenawa and Suetsugu, 2007; Yarar et al, 2007; Scita et al, 2008). Dynamin is a GTPase that causes fission of the membrane. N-WASP and WAVE2 are activators of the Arp2/3 complex, which drives the formation of branched actin filaments for cellular motility (Loisel et al, 1999; Pollard and Borisy, 2003; Takenawa and Suetsugu, 2007). Therefore, the activation of N-WASP or WAVE2 is thought to lead to activation of the Arp2/3 complex, which in turn results in rapid actin polymerization.
Induction of actin polymerization can be monitored with an in vitro pyrene-actin assay. The inverse geometry of membrane binding has hindered elucidation of the curvature-selective binding of the IMD/RCB/I-BAR domain. However, proteins with the EFC/F-BAR or N-BAR domains appear to be good models for testing ‘curvature-dependent' actin polymerization. We examined the effect of liposomes with a variety of curvatures on actin polymerization in the presence of membrane-binding proteins. Our finding revealed that actin polymerization by the N-WASP–WASP-interacting protein (WIP) complex is activated in the presence of EFC/F-BAR protein, Toca-1 or formin-binding protein 17 (FBP17), depending on the curvature of liposomes added to reaction mixture. We further showed that the binding of N-WASP, as well as EFC/F-BAR domains, to the liposome was required for actin polymerization.
Results
Full-length FBP17 or Toca-1 binds to membrane
We examined the affinity of the SH3 domains of FBP17, an EFC/F-BAR domain protein, and amphiphysin, an N-BAR domain protein, for N-WASP. The affinity of the FBP17 SH3 domain for N-WASP was higher than that of amphiphysin (Supplementary Figures 1 and 2A). Hence, we further studied the EFC/F-BAR domain proteins to test curvature-dependent signalling.
Most of the liposomes used in this study were prepared from brain total lipids (Folch fraction), which are rich in phosphatidylserine (PS) and contain small amounts (approximately 0.05%) of phosphatidylinositol phosphates (PIPs) and phosphatidylinositol (4,5)-bisphospate (PI(4,5)P2) (Michelsen et al, 1995). We prepared large multilamellar vesicles (LMVs) and small unilamellar vesicles (SUVs) from Folch fraction. LMVs contain liposomes with both large diameters (0.5–1 μm) and smaller diameters, whereas SUVs contain liposomes with small diameters of up to 0.1 μm (Supplementary Figure 3). We prepared full-length EFC/F-BAR domain proteins, FBP17 and Toca-1 from Sf9 cells as glutathione S-transferase (GST) fusion proteins (Supplementary Figure 4). On the basis of the diameters of the resulting tubules, the full-length FBP17 protein deformed large liposomes in LMVs to a similar degree as reported earlier for the isolated EFC domain of FBP17 (Supplementary Figure 5) (Itoh et al, 2005).
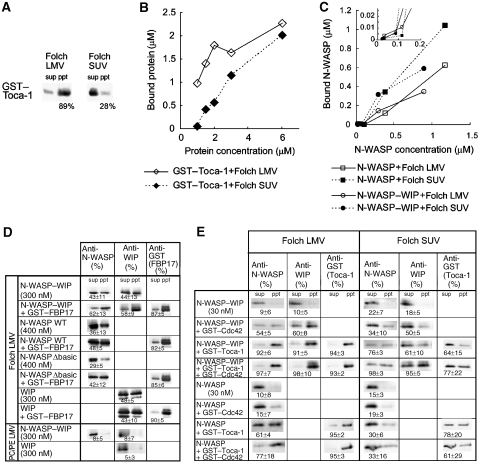
We used a liposome co-sedimentation assay to examine the curvature-selective binding of Toca-1 and FBP17. The sedimentation efficiency of liposomes was dependent on the size of liposomes. Therefore, the percentage of proteins that bound to liposomes was calculated from the percentage of precipitated liposomes determined using fluorescence lipids. The full-length Toca-1 and FBP17 proteins were associated with LMVs and showed weak affinity for SUVs (Figure 1A and B, and data not shown). Approximately 90–100% binding of Toca-1 to LMVs was observed at low protein concentrations, and only a small increase in binding was observed at high protein concentrations. This may result from the heterogeneous nature of the LMVs or the insufficient surface area of liposomes for protein binding. Only a small portion of Toca-1 bound to SUVs at low protein concentrations, but binding of Toca-1 to SUVs increased at higher protein concentrations.
Figure 1.
Recruitment of N-WASP by FBP17 and Toca-1 to the membrane. (A–E) Liposome co-sedimentation assay. (A) Binding of 2 μM GST–Toca-1 protein to 20 μg/ml large multilamellar vesicles (LMVs) and small unilamellar vesicles (SUVs) from the Folch fraction. The percentages of proteins bound to liposomes were calculated from the percentage of liposomes precipitated and are shown with western blots. LMVs contain liposomes of up to 1 μm diameter, whereas SUVs contain liposomes of up to 0.1 μm diameter. sup, supernatant; ppt, precipitate. (B, C) Protein binding to 20 μg/ml Folch liposomes is dependent on protein concentration. The concentration of bound (B) GST–Toca-1, and (C) N-WASP and N-WASP–WIP complex was plotted against the total protein concentration. In (C), the inset shows an enlarged plot for the low concentration range. (D) Recruitment of N-WASP or WIP to liposomes by FBP17. We incubated 300 nM N-WASP–WIP complex, 400 nM wild-type N-WASP, 400 nM Δbasic N-WASP, or 300 nM WIP with 600 nM GST–FBP17 and 50 μg/ml Folch LMVs or PC/PE LMVs. (E) Recruitment of the N-WASP–WIP complex to liposomes by Toca-1 and Cdc42 was examined with a liposome co-sedimentation assay. GST–Toca-1 (600 nM) and GTPγS-loaded GST–Cdc42 (4 μM) were incubated with 30 nM N-WASP–WIP complex or N-WASP and 20 μg/ml Folch LMVs or SUVs, and binding was analysed. The percentages of proteins bound to liposomes were calculated from the percentage of liposome recovery after precipitation (approximately 80–95 and 50% for LMVs and SUVs, see Materials and methods) and are shown with western blots.
N-WASP or the N-WASP–WIP complex binds to the membrane in the presence of FBP17 or Toca-1
One of the most plausible mechanisms for curvature-dependent actin polymerization by full-length FBP17 or Toca-1 is increased local concentration of SH3-binding partners on the membrane of a curvature. N-WASP is known to bind to the membrane (Rohatgi et al, 1999; Prehoda, 2000). Therefore, recruitment of N-WASP to the membrane by SH3 domains and by N-WASP itself should be examined separately. As N-WASP binds to lipids through its basic region (Rohatgi et al, 1999; Prehoda et al, 2000), we prepared N-WASP with a deletion of the basic region (Δbasic N-WASP). In the liposome co-sedimentation assay, N-WASP alone showed preferential affinity for SUVs over LMVs (Figure 1C). The preferential affinity of N-WASP for SUVs may result from the increased surface area of liposomes during the formation of SUVs from LMVs. Although Δbasic N-WASP retained some membrane-binding ability, the co-sedimentation assay with LMVs indicated that FBP17 recruited full-length and Δbasic N-WASP to the membrane to a similar degree (Figure 1D). Therefore, FBP17 could attach N-WASP to the membrane, presumably independently of the membrane binding of N-WASP.
Native N-WASP has been reported to form a protein complex with WIP (Ho et al, 2001, 2004). However, possible lipid-binding activity of the N-WASP–WIP complex has not been reported. We purified recombinant N-WASP–WIP complex (Supplementary Figure 4), and used it in a liposome co-sedimentation assay. Similar amount of N-WASP–WIP bound to Folch liposomes as did N-WASP. The affinity of the N-WASP–WIP complex for SUVs was greater than that for LMVs (Figure 1C). We then examined binding of N-WASP–WIP to various phosphoinositides supplemented in phosphatidylethanolamine (PE) and phosphatidylcholine (PC). Binding of the N-WASP–WIP complex to liposomes containing PS or PIP2 was higher than that to liposomes prepared from PE and PC (Supplementary Figure 6). N-WASP also bound to PS and PI(4,5)P2-containing liposomes (Supplementary Figure 6).
The N-WASP–WIP complex was recruited to Folch LMVs by FBP17 (Figure 1D). WIP has a proline-rich region and may contribute to the association of N-WASP–WIP to FBP17 or Toca-1. The FBP17 SH3 domain bound to WIP alone (Supplementary Figure 2A). Moreover, WIP showed a higher affinity for Folch LMVs than PC/PE LMVs (Figure 1D). However, FBP17 did not efficiently recruit WIP alone to Folch liposome (Figure 1D). Furthermore, the affinity of Toca-1 to the N-WASP–WIP complex was similar to that for N-WASP alone (Supplementary Figure 2B). These data suggest that N-WASP is responsible for the interaction between FBP17 or Toca-1 and the N-WASP–WIP complex at the membrane.
Next, we compared the binding of 30 nM N-WASP–WIP or N-WASP alone to Folch LMVs or SUVs (Figure 1E). Both N-WASP–WIP and N-WASP showed weak binding to Folch liposomes by the co-sedimentation assay (Figure 1C and E). Toca-1 recruited N-WASP and the N-WASP–WIP complex to LMVs (Figure 1E). After the difference in the efficiency of liposome sedimentation was correlated, considerable amounts of N-WASP–WIP complex and Toca-1 were recruited to SUVs in the presence of both N-WASP–WIP and Toca-1 (Figure 1E). The excess concentration of Toca-1 compared with N-WASP–WIP allowed the formation of the N-WASP–WIP-Toca-1 complex with more basic amino acids than N-WASP–WIP complex, which may recruit N-WASP–WIP and Toca-1 to SUVs.
Because Toca-1 but not FBP17 was reported to bind to Cdc42 (Fujita et al, 2002; Ho et al, 2004), we examined the effect of activated GTPγS-loaded Cdc42 on the recruitment of N-WASP–WIP or N-WASP to the membrane in the presence or absence of Toca-1 (Figure 1E). Cdc42 also recruited the N-WASP–WIP complex to LMVs or SUVs (Figure 1E). The membrane recruitment of N-WASP–WIP by Toca-1 and Cdc42 together was not significantly different from that of Toca-1 alone (Figure 1E).
The N-WASP–WIP complex, the Arp2/3 complex, and FBP17 or Toca-1 induce actin polymerization dependent on curvature of liposome, but independent of PIP2
N-WASP and the N-WASP–WIP complex activate actin polymerization through activation of the Arp2/3 complex (Pollard and Borisy, 2003; Takenawa and Suetsugu, 2007). However, the role of curvature-dependent binding of the EFC/F-BAR protein in actin polymerization is unknown. We confirmed that addition of full-length FBP17 or Toca-1 to a reaction mixture containing the N-WASP–WIP complex, Arp2/3, and actin did not activate N-WASP–WIP complex-mediated actin polymerization as reported earlier (Ho et al, 2004) (Figure 2A–C). LMVs or SUVs alone had little effect on N-WASP–WIP complex-mediated actin polymerization (Figure 2A–C). In contrast, simultaneous addition of full-length FBP17 or Toca-1 and LMVs strongly activated N-WASP–WIP-complex-mediated actin polymerization (Figure 2A–C). Despite similar recruitment of N-WASP–WIP to the membrane (Figure 1E), the activation by SUVs was lower than that by LMVs in the presence of FBP17 or Toca-1 (Figure 2A–D). Cleavage of the GST tag of Toca-1 or FBP17 may affect actin polymerization, but cleavage of this tag did not significantly affect actin polymerization induced by Toca-1 and LMVs (Figure 2C).
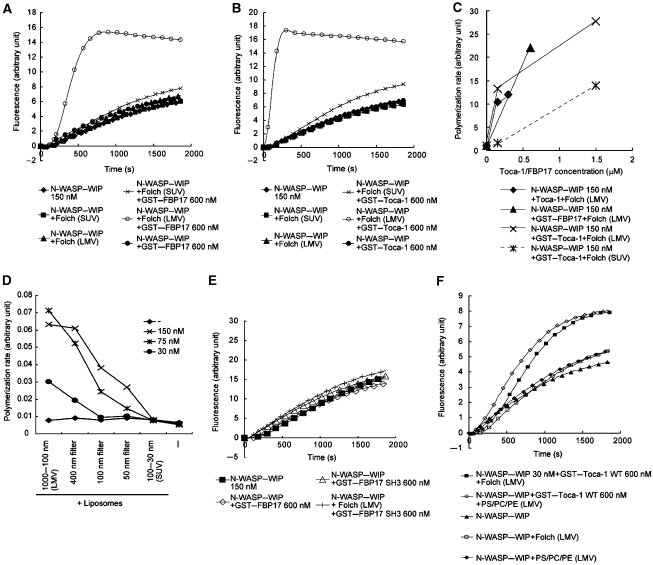
Figure 2.
Induction of membrane curvature-dependent actin polymerization. (A–C) Actin polymerization induced by 150 nM N-WASP–WIP complex, 20 μg/ml Folch liposomes (SUVs of largest diameters 0.1 μm; LMVs of largest diameters 1 μm), and 600 nM (A) GST–FBP17 or (B) GST–Toca-1. (C) Activation of 150 nM N-WASP–WIP complex with increasing concentrations of GST–Toca-1, Toca-1, or GST–FBP17 in the presence of 20 μg/ml SUVs or LMVs from the Folch fraction. (D) Membrane curvature-dependent activation of 30 nM N-WASP–WIP complex in response to various concentrations of GST–Toca-1 and 20 μg/ml SUVs, LMVs, or vesicles made with cyclopore filters. In (C, D), the rate of actin polymerization at 20% polymerization was calculated and plotted against the concentration of Toca-1 or FBP17. (E) Actin polymerization induced by 150 nM N-WASP–WIP complex, 20 μg/ml LMVs, and 600 nM wild-type GST–FBP17 or GST–FBP17 SH3 domain. (F) Actin polymerization induced by 30 nM N-WASP, 600 nM GST–Toca-1, and 20 μg/ml PS/PC/PE LMVs (PE/PC/PS=8/2/3) or Folch LMVs.
The difference in actin polymerization induced by LMVs and SUVs suggests that Toca-1 and FBP17 regulate actin polymerization in a manner dependent on the diameter of the liposomes. We extruded LMVs through cyclopore filters with various pore sizes to form liposomes with the largest defined diameters (Mayer et al, 1986) (Supplementary Figure 3). We then tested the ability of these liposomes to induce actin polymerization in the presence of the N-WASP–WIP complex and various concentrations of Toca-1 (Figure 2D). As these liposome preparations only defined the largest diameter of liposomes, all liposome preparations also contained liposomes of smaller diameters. Within a range of diameters from approximately 30 nm to 1 μm, activation of the N-WASP–WIP complex by Toca-1 was greater when diameters of liposomes were larger (Figure 2D). This indicated that Toca-1-induced actin polymerization was regulated by the curvature or diameter of liposomes.
Proteins containing the SH3 domain or the isolated SH3 domain and liposomes are both known to activate N-WASP-mediated actin polymerization (Takenawa and Suetsugu, 2007). The SH3 domain and lipids may function synergistically to activate N-WASP (Rohatgi et al, 2001). However, in the present study, actin polymerization induced by the SH3 domain, liposomes, and the N-WASP–WIP complex (Figure 2E) was significantly weaker than that induced by full-length FBP17, liposomes, and the N-WASP–WIP complex, indicating that the SH3 domain alone is not sufficient for the activation.
N-WASP was shown earlier to be activated by PIP2 and Cdc42 (Rohatgi et al, 1999). The Folch fraction contains only small amounts of PIPs. To exclude the possibility that PIPs in the Folch fraction are required for activation of actin polymerization, we prepared PIP2-free reconstituted liposomes composed of PS/PC/PE. PS/PC/PE LMVs but not PC/PE LMVs activated N-WASP–WIP complex-mediated actin polymerization in the presence of Toca-1 as effectively as the Folch liposomes (Figure 2F and data not shown). PS/PC/PE SUVs exerted reduced effect on Toca-1 and N-WASP–WIP-mediated activation (data not shown). These results suggest that PIPs are not required for N-WASP–WIP activation in the presence of FBP17 or Toca-1.
N-WASP alone was sufficient for curvature-dependent actin polymerization
N-WASP activation was examined to determine whether curvature-dependent actin polymerization is dependent on intrinsic characteristics of N-WASP or on the formation of N-WASP–WIP complex. The activities of N-WASP alone or the N-WASP–WIP complex alone without liposomes were similar (Supplementary Figure 7A). N-WASP was activated by the addition of liposomes (Supplementary Figure 7A). Consistent with the stronger affinity of N-WASP to Folch SUVs (Figure 1E), Folch SUVs activated N-WASP more strongly than Folch LMVs (Supplementary Figure 7A). The N-WASP–WIP complex was not activated by Folch LMVs or Folch SUVs at this concentration. Actin polymerization induced by N-WASP and FBP17 was enhanced significantly by Folch LMVs (Supplementary Figure 7B). In contrast, polymerization was not enhanced by Folch SUVs (Supplementary Figure 7C), which suggests that FBP17-mediated activation of N-WASP is dependent on membrane curvature but not on N-WASP–WIP complex formation.
The membrane-binding region of N-WASP was essential for actin polymerization induced by FBP17 and the N-WASP–WIP complex in the presence of liposomes
We addressed whether the binding of the basic region of N-WASP to lipids contributed to the synergistic activation observed with FBP17 or Toca-1 and liposomes. We purified the Δbasic N-WASP–WIP complex and examined activation of actin polymerization in the presence of liposomes and FBP17 or Toca-1. Polymerization was considerably weaker with Δbasic N-WASP–WIP complex than with wild-type N-WASP–WIP complex (Figure 3A). Activation of Δbasic N-WASP alone by FBP17 and LMVs was also significantly lower than that of wild-type N-WASP (Supplementary Figure 7B and D). As recruitment to the liposome of the Δbasic N-WASP or Δbasic N-WASP–WIP by FBP17 was similar to the recruitment of wild-type N-WASP or N-WASP–WIP complex by FBP17 in spite of significant difference in actin polymerization (Figure 1D and data not shown, respectively), binding of N-WASP to the membrane through its basic region appears to be important for N-WASP activation by membranes.
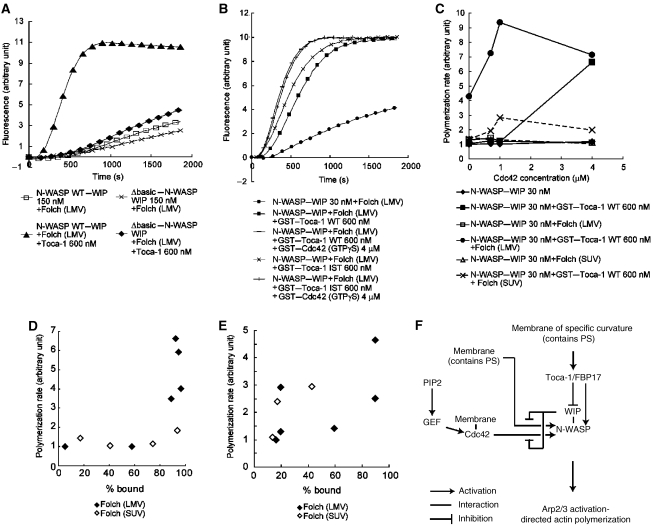
Figure 3.
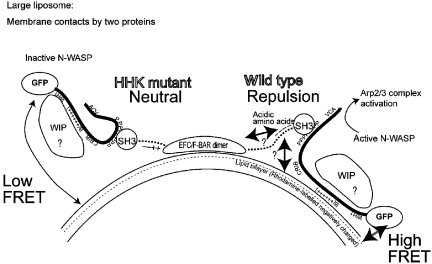
Effects of phosphatidylserine (PS), Cdc42, and the model of N-WASP–WIP-induced actin polymerization. (A) Actin polymerization induced by 150 nM wild-type (WT) or Δbasic N-WASP–WIP complex, 20 μg/ml Folch SUVs or LMVs, and 600 nM Toca-1. (B) Actin polymerization induced by 30 nM N-WASP, 4 μM GST–Cdc42 (GTPγS-loaded), 600 nM wild-type or IST mutant of GST–Toca-1 and 20 μg/ml Folch LMVs. (C) Activation of 30 nM N-WASP–WIP complex with increasing concentrations of GST–Cdc42 (GTPγS-loaded) in the presence of 600 nM wild-type GST–Toca-1 and 20 μg/ml Folch SUVs or LMVs. The rate of actin polymerization at 20% polymerization was calculated and plotted against the Cdc42 concentration. (D) Comparison of the binding of N-WASP–WIP to 20 μg/ml Folch SUVs or LMVs and the rate of actin polymerization. Protein concentrations were the same as those in Figures 1E, 3B and C. (E) Comparison of the binding of N-WASP to 20 μg/ml folch SUVs or LMVs and the rate of actin polymerization. (F) Model of membrane curvature-dependent activation of N-WASP–WIP.
Cdc42 further enhanced the actin polymerization induced by Toca-1 and the N-WASP–WIP complex in the presence of liposomes
The N-WASP–WIP complex is activated by Cdc42 in the presence of Toca-1 (Ho et al, 2004). However, the effect of liposomes on this activation has not been investigated. The N-WASP–WIP complex was not activated by GTPγS-loaded Cdc42 alone in the presence or absence of Folch liposomes, as reported earlier (Ho et al, 2004) (Figure 3C; Supplementary Figure 8A). In contrast, N-WASP was activated by Cdc42 (Supplementary Figure 7E). Cdc42 in the presence of PIP2 produced a higher degree of N-WASP activation as reported earlier (Rohatgi et al, 1999) (Supplementary Figure 7E). Importantly, activation of N-WASP by Folch liposomes or PS/PC/PE liposomes in the presence of Cdc42 was similar to that by Cdc42 and PIP2 (Supplementary Figure 7E and F, and data not shown).
We then examined the effect of Cdc42 on Toca-1-mediated actin polymerization. Addition of Cdc42 to Toca-1 and Folch liposomes increased N-WASP–WIP complex-induced actin polymerization (Figure 3B and C; Supplementary Figure 8A and B). However, as reported earlier, higher concentrations of Cdc42 allowed for N-WASP–WIP activation by Toca-1 in the absence of liposomes (Ho et al, 2004) (Figure 3C; Supplementary Figure 8C).
The contribution of Cdc42–Toca-1 binding to actin polymerization mediated by N-WASP–WIP was further examined by using the IST mutant of Toca-1, which was reported to show reduced affinity for Cdc42 (Ho et al, 2004; Kakimoto et al, 2006). Consistent with this, the IST mutant of Toca-1 had reduced ability to induce N-WASP–WIP activation in the presence of Cdc42 and in the absence of liposomes (Supplementary Figure 8C). However, in the presence of Folch liposomes, activation of N-WASP–WIP by the wild-type and IST mutants of Toca-1 was similar (Figure 3B). The activation of N-WASP–WIP by FBP17, which reportedly shows no affinity to Cdc42 (Kamioka et al, 2004), was also enhanced by Cdc42 (data not shown). Therefore, binding of Cdc42 to Toca-1 appears to have a limited function in N-WASP–WIP-mediated actin polymerization in the presence of membranes.
WIP suppresses activation of N-WASP without membrane binding
We plotted the membrane binding of the N-WASP–WIP complex against the actin polymerization mediated by the N-WASP–WIP complex (Figure 3D). The activation appeared to occur most when N-WASP–WIP protein was bound to LMVs (Figure 3D), which suggest that stable binding of the N-WASP–WIP complex to liposomes is required for N-WASP–WIP activation. In contrast, activation of N-WASP-mediated actin polymerization was less dependent on binding of N-WASP to liposomes (Figure 3E) and occurred in the presence of Cdc42 or Toca-1/FBP17 alone.
These results suggested that WIP suppressed the activation of N-WASP by individual Cdc42, Toca-1/FBP17, or liposomes. The WIP-mediated inhibition of activation was suppressed by Toca-1/FBP17 in combination with large liposomes (Figure 3F). In the presence of liposomes of specific diameters, Toca-1 and FBP17 activated N-WASP–WIP-mediated actin polymerization. In the presence of both liposomes and Toca-1/FBP17, Cdc42 activated N-WASP–WIP complex-mediated actin polymerization. Therefore, Toca-1/FBP17-mediated actin polymerization was dependent on the curvature of the liposomes. As a result, Cdc42-mediated actin polymerization was dependent on the curvature of the liposomes.
However, in contrast to the LMV-bound N-WASP–WIP activation, even when a similar amount of N-WASP–WIP was bound to SUVs, actin polymerization was not activated (Figure 3D). Furthermore, Δbasic N-WASP was not activated by Toca-1 in the presence of LMVs in spite of membrane binding. These observations suggest that the presence of N-WASP or the N-WASP–WIP complex at the membrane is not sufficient to induce actin polymerization. Therefore, we attempted to elucidate the spatial organization of N-WASP in relation to the membrane.
The conserved acidic residues adjacent to the SH3 domain is essential for actin polymerization induced by the N-WASP–WIP complex and FBP17 or Toca-1 in the presence of liposomes
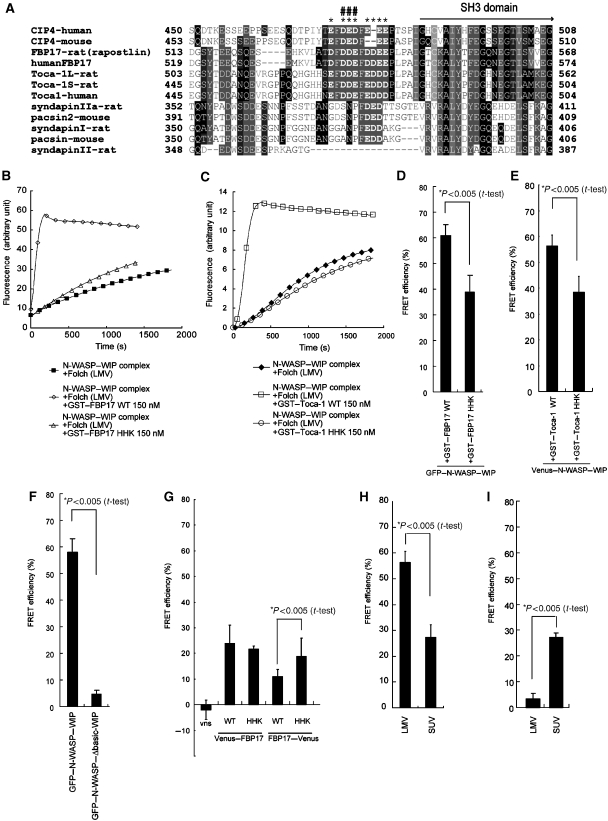
Because N-WASP–WIP was shown to bind to the membrane through the SH3 domain of Toca-1 or FBP17, the position of the SH3 domain in relation to the membrane appears to be important for proper physical positioning of N-WASP. When we aligned the amino-acid sequences adjacent to the SH3 domain of Toca-1, FBP17, CIP4, and syndapin/pacsins, we found conserved acidic amino acids just in front of the SH3 domain among Toca-1, FBP17, and CIP4 (Figure 4A). The inner leaflet of the plasma membrane is negatively charged due to its high PS content. Furthermore, the convex, non-membrane-binding surface of the EFC/F-BAR domain is also negatively charged (Shimada et al, 2007). Therefore, the conserved acidic amino acids would be located away from the membrane (or EFC/F-BAR domain) as a result of electrostatic repulsion. We replaced three of the eight acidic residues in FBP17 and Toca-1 with basic residues (D540H, D541H, and E542K for FBP17, and D528H, D529H, and E530K for Toca-1) to neutralize the charges adjacent to the SH3 domains and referred to the resulting proteins as HHK mutants (Figure 4A).
Figure 4.
Conserved acidic amino acids upstream of the SH3 domain of FBP17 and Toca-1 modulate distance of the protein from the membrane. (A) Alignment of amino-acid sequences of the indicated proteins upstream of the SH3 domains. Alignments were generated with Clustal W software. *Acidic amino acids conserved between FBP17 and Toca-1; #Amino acids mutated in the HHK mutants of FBP17 and Toca-1. (B, C) Actin polymerization induced by 30 nM N-WASP–WIP complex, 20 μg/ml Folch LMVs, and 150 nM wild-type or HHK mutant of (B) GST–FBP17 or (C) GST–Toca-1. (D, E) Fluorescence resonance energy transfer (FRET) between 20 μg/ml rhodamine-labelled Folch LMVs and purified GFP–N-WASP–WIP complex (15 nM) in the presence of wild-type (WT) or HHK mutant of (D) GST–FBP17 (150 nM) and (E) GST–Toca-1 (150 nM). Statistical significance was analysed by Student's t-test. (F) FRET between 20 μg/ml rhodamine-labelled Folch LMVs and wild-type or Δbasic GFP–N-WASP complexed with WIP (15 nM) in the presence or absence of wild-type GST–Toca-1 (150 nM). (G) FRET between 20 μg/ml rhodamine-labelled Folch LMVs and cell lysates expressing N-terminal or C-terminal Venus (a variant of GFP) FBP17 fusion proteins. (H) FRET between 20 μg/ml rhodamine-labelled Folch LMVs or SUVs and purified GFP–N-WASP–WIP complex (15 nM) in the presence of wild-type GST–Toca-1 (150 nM). (I) FRET between 20 μg/ml rhodamine-labelled Folch LMVs or SUVs and purified GFP–N-WASP–WIP complex (100 nM).
The clusters of acidic amino acids in Toca-1 and FBP17 may contribute to the association with N-WASP because N-WASP has basic amino acids that interact with acidic region of N-WASP to form an ‘auto-inhibited structure'. However, recruitment of N-WASP, WIP, or N-WASP–WIP complex to the membrane by full-length HHK mutant Toca-1 or FBP17 was similar to those of wild-type FBP17 (Supplementary Figure 2A, 9, and 10, and data not shown). Further, the affinity of the SH3 domain of the HHK mutant FBP17 to N-WASP was similar to that of wild-type FBP17 (Supplementary Figures 2A, 9, and 10, and data not shown). The affinity of full-length HHK mutant of Toca-1 to N-WASP, WIP, and the N-WASP–WIP complex was similar to that of wild type (Supplementary Figure 2B). The membrane-binding ability and membrane-deforming ability of the HHK mutant of FBP17 were similar to those of wild-type FBP17 (Supplementary Figures 2A and 5). Therefore, the acidic region did not appear to contribute to the interaction of Toca-1 or FBP17 to N-WASP or N-WASP–WIP complex or to the membrane binding of Toca-1 or FBP17.
However, induction of actin polymerization by the HHK mutant of FBP17 or that of Toca-1 in the presence of the N-WASP–WIP complex and LMVs was severely diminished (Figure 4B and C). That N-WASP–WIP itself was associated with the membrane in the presence of HHK mutant (Supplementary Figure 9) suggested that N-WASP associated with the HHK mutant binds to the membrane in a different manner than N-WASP associated with wild-type FBP17 or Toca-1.
The conserved acidic amino acids in Toca-1 and FBP17 affect the distance between the membrane and the N-WASP–WIP complex
We subsequently examined how N-WASP in the N-WASP–WIP complex is associated with the membrane by measuring fluorescence resonance energy transfer (FRET) efficiency between N-terminal green fluorescent protein (GFP)-tagged N-WASP in the N-WASP–WIP complex (Supplementary Figure 4) and Folch liposomes containing rhodamine-labelled PE. Using this strategy, we could measure a decrease or increase in the distance between two molecules.
We measured the fluorescence intensity of GFP in the presence or absence of rhodamine-labelled Folch liposomes and calculated the FRET efficiency. FRET was observed between GFP–N-WASP and rhodamine-labelled liposomes in the presence of FBP17 or Toca-1 (Figure 4D and E). However, in the presence of HHK mutant, the FRET efficiency was significantly decreased (Figure 4D and E), indicating that the distance between N-WASP and the membrane became larger in the presence of the HHK mutant. As recruitment of the N-WASP–WIP complex to liposomes by the wild-type and HHK mutant FBP17 was similar, the larger distance between N-WASP and the membrane in the presence of the HHK mutant may be explained by altered positioning of the SH3 domain relative to the membrane.
We also measured the FRET of wild-type and Δbasic N-WASP to the membrane in the presence of Toca-1. The FRET efficiency of Δbasic N-WASP was significantly weaker than that of wild-type N-WASP, suggesting that the mode of membrane binding of Δbasic N-WASP differed significantly from that of wild-type N-WASP (Figure 4F).
To analyse the distance between the SH3 domain and the membrane, we next examined the FRET efficiency between rhodamine-labelled PE in Folch liposomes and FBP17 tagged N-terminally or C-terminally with the GFP variant Venus (Nagai et al, 2002) (Figure 4G). As expected from the binding of the EFC/F-BAR domain to the membranes, N-terminally Venus-tagged wild-type and HHK mutant of FBP17 showed similar FRET efficiencies (Figure 4G). In contrast, the C-terminally Venus-tagged HHK mutant of FBP17 showed higher FRET efficiency with rhodamine liposomes than did wild-type FBP17 (Figure 4G), which suggests a decrease in the distance between the SH3 domain and the membrane. Therefore, the spatial orientation of the SH3 domain in relation to the membrane was regulated by the charged amino acids located close to the SH3 domain. These data suggest that the position of the SH3 domain of FBP17 or Toca-1 relative to the membrane is essential for the binding of N-WASP to the membrane that triggers actin polymerization (Figure 5).
Figure 5.
Model of curvature-dependent actin polymerization. Both interaction between the membrane and EFC/F-BAR domain proteins and interaction between the membrane and the N-WASP–WIP complex are required for membrane curvature-dependent actin polymerization. The spatial orientation of the SH3 domain appears to be determined by electrostatic repulsion to the membrane or to the convex surface of the EFC/F-BAR domain. This in turn influences the spatial position of the N-WASP–WIP complex and resulting membrane association. The position of WIP relative to the membrane was not determined. The concave surface, as well as side-lying interface (90° rotated surface of the concave surface) of the EFC/F-BAR domain could contact the membrane on induction of actin polymerization.
The distance between the membrane and the N-WASP–WIP complex was altered depending on the size of liposomes
Finally, we addressed the lack of activation of N-WASP–WIP by Toca-1 in the presence of SUVs despite association of N-WASP with membrane. Overall, 76% of N-WASP–WIP molecules were bound to SUVs and 92% of N-WASP–WIP molecules were bound to LMVs in the presence of Toca-1 (Figure 1E). Therefore, we examined the FRET efficiency between GFP–N-WASP–WIP to SUVs in the presence of Toca-1 (Figure 4H). As expected, the SUV-bound N-WASP–WIP had weaker FRET efficiency than did the LMV-bound N-WASP–WIP, indicating that the distance between the N-WASP–WIP complex and SUVs was larger than that between the N-WASP–WIP and LMVs. Consistent with the slightly higher affinity of N-WASP–WIP complex to SUVs than to LMVs (Figure 1C), the FRET efficiency of N-WASP–WIP in the presence of SUVs and in the absence of FBP17 or Toca-1 was higher than that in the presence of LMVs (Figure 4I). Therefore, the distance between N-WASP and the membrane can be affected by the curvature of the liposome in the presence of FBP17 or Toca-1.
Discussion
Actin polymerization has a well-established role in controlling the shape of cellular membranes. The BAR domain superfamily is a sensor and generator of membrane curvature. Among the BAR superfamily proteins, the EFC/F-BAR domain proteins are known to bind to N-WASP and dynamin, which have a function in the fission of endocytosis vesicles through the induction of actin polymerization and mechanical scission of the membrane, respectively. Therefore, it is believed that the BAR superfamily proteins regulate the shape of the membrane. However, whether the shape of the membranes regulates N-WASP- or dynamin-induced actin polymerization or membrane scission was not established. In the present study, we demonstrated that FBP17 and Toca-1 induce activation of N-WASP–WIP complex-mediated actin polymerization in a manner dependent on the curvature of liposomes added to the reaction mixtures.
Activation of signal-transduction cascades such as membrane curvature-dependent actin polymerization may be unique to Toca-1 or FBP17
Because both FBP17 and Toca-1 have curvature-sensing EFC/F-BAR and SH3 domains, recruitment of the effector molecule (the N-WASP–WIP complex) to membranes with a range of curvatures by FBP17 or Toca-1 was expected. However, experiments with HHK mutants of Toca-1 or FBP17 and Δbasic mutants of N-WASP indicated that N-WASP–WIP recruitment to the surface of the membrane was not sufficient to induce actin polymerization. Instead, direct binding of N-WASP–WIP through the basic region to the membrane in the proper spatial organization appeared to be essential for the induction of actin polymerization (Figure 5). Therefore, curvature-dependent actin polymerization requires the association of both EFC/F-BAR proteins and the N-WASP–WIP complex with the membrane in a spatially ordered manner. This finding suggests that most of the BAR domain superfamily protein may recruit their binding partners to the membrane without regulation of the catalytic activities of the binding partners.
Membrane curvature-dependent actin polymerization may occur on the membrane before tubulation or on the tubulated membrane
It is clear that the induction of actin polymerization was dependent on the size of liposome added to the actin polymerization assay system. However, neither the exact diameter of liposome on which actin polymerization occurs nor the orientation of EFC/F-BAR domain on the membrane on induction of actin polymerization was determined. The EFC/F-BAR domain is shown to form tubulated membrane through the formation of laterally contacted spirals of the EFC/F-BAR domain on the surface of tubulated membrane (Shimada et al, 2007; Frost et al, 2008). The concave surface of the EFC/F-BAR domain binds to the tubules. However, at the initial stage of tubule formation, another side-lying interface of EFC/F-BAR domain contacts the membrane (Frost et al, 2008).
Full-length Toca-1 and FBP17 generated tubulated membrane in a manner similar to the EFC/F-BAR domain alone. Therefore, full-length Toca-1 or FBP17 appears to interact with the membrane in a manner similar to the EFC/F-BAR domain proteins. We confirmed that the presence of the N-WASP–WIP complex, the Arp2/3 complex, and actin filaments did not affect the formation of tubule by full-length FBP17 or Toca-1 protein (data not shown).
Actin polymerization had reached maximum within few minutes in our conditions (Figures 2 and 3). The tubulation of the membrane was observed by 2 min of incubation in the presence or absence of the N-WASP–WIP complex, the Arp2/3 complex, and actin (data not shown). Therefore, actin polymerization and the tubulation of the membrane appeared to take place simultaneously.
Therefore, there are two possible modes of actin filament formation. The first involves actin polymerization being induced on the preferential binding of the EFC/F-BAR domain proteins to the relatively large liposomes with diameters of 0.5–1 μm. In this case, the side-lying interface of the EFC/F-BAR domain may bind to the membrane. The second involves actin polymerization occurring on the tubulated liposomes with diameters of ∼150 nm decorated with the Toca-1 or FBP17 proteins (Supplementary Figure 5). In the latter case, the SUVs with a diameter of 150 nm or less are not good substrates for Toca-1 or FBP17, and thus actin polymerization was not induced. Cryo-electron microscopy indicated the existence of membrane surface between spirals of the EFC/F-BAR domain (Frost et al, 2008), which may be utilized for the interaction of the basic region of N-WASP.
As the tubules induced by the EFC/F-BAR domains are quite dynamic (Itoh et al, 2005; Frost et al, 2008), it is difficult to record the diameter of liposome (tubule) at the onset of actin polymerization. However, how the actin polymerization takes place depending on membrane curvature should be examined in future.
The spiral formation of the EFC/F-BAR domains on the tubulated membrane may affect the possible electrostatic repulsion between the SH3 domain and the membrane. As the EFC/F-BAR-domain spiral does not appear to cover the full surface of the tubules, the electrostatic repulsion between the membrane and the acidic residues of FBP17 or Toca-1 appears to be possible (Frost et al, 2008). Instead, the negatively charged convex surface of the EFC/F-BAR domain (Shimada et al, 2007) may contribute to the electrostatic repulsion of the SH3 domain (Figure 5).
Role of WIP in the regulation of N-WASP
WIP has been postulated to suppress the activation of N-WASP. We succeeded in obtaining recombinant N-WASP–WIP complex. The resulting N-WASP–WIP complex was resistant to the liposome- and Cdc42-induced activation of actin polymerization as reported earlier (Figure 3; Supplementary Figure 7) (Martinez-Quiles et al, 2001; Ho et al, 2004). In the presence of liposome and EFC/F-BAR proteins Toca-1 or FBP17, Cdc42 was able to effectively activate the N-WASP–WIP complex (Figure 3).
However, the mechanism of suppression of N-WASP by WIP is still unclear. N-WASP alone was activated by SUV, but the N-WASP–WIP complex was not (Supplementary Figure 7). Interestingly, WIP alone also bound the membrane (Figure 1). The presence of liposome was reported to inhibit the suppression of N-WASP activation by exogenous WIP (Martinez-Quiles et al, 2001). Thus, WIP appears to modulate the binding of N-WASP to membrane presumably by binding of WIP with membrane.
WIP has proline-rich sequence where the SH3 domain binds (Anton et al, 2007). Toca-1 and FBP17 appear to bind to the praline-rich region of WIP. Because Toca-1 and FBP17 form homodimers, it is possible that the two SH3 domains bind to N-WASP and WIP simultaneously, thereby releasing suppression of N-WASP by WIP. However, this is less likely because EFC/F-BAR domain forms a dimer without membrane (Shimada et al, 2007), and both Toca-1 and FBP17 were not able to activate the N-WASP–WIP complex without membrane (Figure 2). As N-WASP alone was activated by FBP17 and the liposome, the activation of the N-WASP–WIP complex is thought to be mainly achieved through conformational change of N-WASP.
Possible physiological significance of the membrane curvature-induced actin polymerization
Toca-1, CIP4, or FBP17 had been shown to be involved in endocytosis (Itoh and De Camilli, 2006; Tsujita et al, 2006; Shimada et al, 2007) and in neuronal morphology (Kakimoto et al, 2006). The structural analysis of the EFC/F-BAR domain demonstrated that the diameter of tubules appears to correspond to the diameter of clathrin-coated pits (Shimada et al, 2007; Frost et al, 2008). Moreover, the F-BAR proteins Toca-1, CIP4, and FBP17 bind to N-WASP and dynamin (Itoh and De Camilli, 2006; Tsujita et al, 2006). The importance of N-WASP-induced actin polymerization in endocytosis has been well established (Merrifield et al, 2005; Takenawa and Suetsugu, 2007). Thus, the most straightforward physiological significance of the actin polymerization reconstituted in this paper is that the membrane curvature generated by clathrin and the EFC/F-BAR proteins induces actin polymerization in endocytosis. Although the actin polymerization may be induced on membrane tubules decorated by EFC/F-BAR proteins, the induction of actin polymerization depending on the size of liposome added to the system may be the first indication that the membrane shape itself could be the intracellular signal-transduction input.
The generation of curvature and the production of phosphoinositides involved in signal transduction appear to be correlated in events such as endocytosis. Therefore, the activation of Cdc42 and the generation of membrane curvature should occur simultaneously, especially at formation of clathrin-coated pits that follows receptor activation (Itoh and De Camilli, 2006). N-WASP is known to bind to the SH3 domains of adaptor proteins such as Grb2, Src, and Nck (reviewed in Takenawa and Suetsugu, 2007). These adaptor proteins also accumulate at the place of receptor activation. However, these adaptor molecules or small G proteins such as Cdc42 have no spatial order of signal transduction, that is, the direction of actin polymerization could be both inwardly or outwardly from the plasma membrane. Thus, the membrane-binding proteins with curvature-sensing modules may control the spatial direction of the results of signal transduction, such as actin polymerization.
Materials and methods
Recombinant proteins
In brief, the cells expressing recombinant proteins were harvested and frozen in liquid nitrogen. The cells were thawed under sonication in Tris-buffered saline without detergents. After clarification of the lysate by centrifugation, proteins were purified using affinity agarose and were eluted with the buffers indicated below after washing on disposable mini columns (CC.07; Assist, Tokyo). GST–FBP17 (human) and wild-type or IST mutant of GST–Toca-1 (rat) were expressed in Sf9 cells using the Bac-to-Bac Baculovirus Expression System (Invitrogen) and were purified with glutathione Sepharose 4B (GE Life Sciences), as described earlier (Suetsugu et al, 2006a). Proteins were eluted in XB (10 mM Hepes (pH 7.9), 100 mM KCl, 2 mM MgCl2, 0.2 mM CaCl2 and 5 mM EGTA) supplemented with glutathione. The GST–SH3 (aa 511–625 of human FBP17 and aa 576–695 of human amphiphysin) domains were expressed in Escherichia coli and purified, as described earlier (Tsujita et al, 2006). Recombinant wild-type and Δbasic (aa 125–194 deleted) N-WASP were expressed in Sf9 cells as His-tagged proteins and were purified with nickel-NTA-agarose (Qiagen). The proteins were eluted with XB supplemented with 100 mM imidazole and 50% glycerol. The N-WASP–WIP complex was expressed in FreeStyle 293 cells, according to the manufacturer's instructions (Invitrogen). N-WASP was expressed in pEF-BOS-myc vector or pEGFP-C1 vector (Clontech). WIP was expressed in pFLAG-CMV-6c vector (Sigma), purified using anti-FLAG-affinity agarose (Sigma), and eluted with 100 μg/ml FLAG peptide in XB supplemented with 50% glycerol. GST–Cdc42 was expressed in Sf9 cells, prepared, and loaded with GTPγS, as described earlier (Suetsugu et al, 2006b).
Liposome preparation
The liposome-binding assay was performed as described earlier (Peter et al, 2004; Tsujita et al, 2006). Liposomes were prepared from total bovine brain lipids (Folch fraction 1; Sigma) (Michelsen et al, 1995). Dried lipids under nitrogen gas were resuspended in XB by mixing with a vortex, followed by hydration at 37°C for 1 h. No sucrose was added to the XB. This preparation yielded the LMVs, which contained liposomes with both small and large diameters (0.1–1 μm). To make liposomes of selected largest diameters, LMVs were subjected to three freeze–thaw cycles followed by extrusion through polycarbonate filter membranes (pore size: 400, 100, or 50 nm) at 65°C (Avanti Polar Lipids). SUVs were made by sonication of LMVs at power level 3 for 5 s with a TIATEC ultra-homogenizer VP-5s, which resulted in liposomes of diameters up to 100 nm. Representative liposome preparations were validated by transmission electron microscopy (Supplementary Figure 3).
Liposome-binding assay
Liposome co-sedimentation assays were performed as follows. To remove aggregated proteins, purified proteins were subjected to centrifugation at 25 000 g for 30 min at 25°C in a TL100 rotor (Beckman). Proteins, at the indicated concentrations, were incubated with liposomes in 50 μl XB for 20 min at room temperature and centrifuged at 25 000 g for 30 min at 25°C in a TL100 rotor. Supernatants and pellets were subjected to SDS–PAGE (polyacrylamide gel electrophoresis), and the gels were stained with Coomassie brilliant blue or subjected to western blot analysis. To calibrate the difference in the amount of the precipitated liposome dependent on liposome preparation, some experiments were performed with liposome supplemented with 1% rhodamine-labelled PE (cat. no. 810158; Avanti Polar Lipids). The fluorescence of each supernatant was measured, and the amount of precipitated liposome was calculated. The percentage of precipitated liposomes was used to calculate the percentage of liposome-bound proteins in the reaction mixtures in Figure 1A and E. Approximately 95% of LMVs and 50% of SUVs were precipitated in the presence of Toca-1. Approximately 80% of LMVs and 50% of SUVs were precipitated in the absence of Toca-1. The increase in LMV sedimentation by the presence of Toca-1 is thought to result from the tubulation that will increase the apparent radius of the liposome. Addition of the N-WASP–WIP complex had no effect on the amount of liposomes precipitated.
For determination of the N-WASP-binding lipids, we used liposomes containing PE/PC/PIPx at a weight ratio of 8/2/1 or PE/PC/PS, phosphatidylinositol (PI), or phosphatidic acid (PA) at a weight ratio of 8/2/3. PE, PC, PI, PA, PS, and PI(4,5)P2 were obtained from Sigma. PI3P, PI4P, PI5P, PI(3,4)P2, PI(3,5)P2, PI(4,5)P2, and PI(3,4,5)P3 were purchased from Echelon Bioscience Inc. The indicated lipid mixtures were resuspended with N-WASP or N-WASP–WIP protein at 1 mg/ml and 400 nM in XB, respectively. Binding was analysed as described above. Representative data from at least three independent experiments are shown.
Negative-staining electron microscopy
Purified proteins were incubated with liposomes, prepared in XB at 25°C for 20 min as described above. This mixture was applied to glow-discharged collodion- and carbon-coated copper grids that were washed in 100 mM Hepes (pH 7.9). The grids were stained with 2% uranyl acetate. At each step, excess solution was removed with filter paper. The dried grids were examined with transmission electron microscopy.
Actin polymerization assay
Actin polymerization assays were performed in XB as described earlier (Suetsugu et al, 2006a). The Arp2/3 complex, actin, and Folch liposomes were used at concentrations of 20 nM, 1.2 μM, and 20 μg/ml, respectively. After the proteins were mixed on ice, pyrene fluorescence was examined at room temperature with a spectrofluorometer (Jasco FP-6500). The rate of actin polymerization was calculated from the slope at 20% polymerization. Representative data from at least three independent experiments are shown.
Antibodies
Anti-N-WASP antibody and anti-WIP antibody were obtained from Cell Signaling Technology and Santa Cruz Biotechnology, respectively.
Statistics
Statistically significant differences were determined using the Student's t-test.
FRET analysis
Rhodamine-labelled or control liposomes were prepared from Folch fraction supplemented with 5% (w/w) rhodamine-labelled PE (cat. no. 810158; Avanti Polar Lipids) or PE. GFP–N-WASP–WIP complex (15 nM), GST–FBP17 (150 nM), and liposomes (20 μg/ml) were incubated in XB for 5 min; the fluorescent intensities were examined by excitation at 488 nm and emission at 507 nm. Alternatively, FreeStyle 293 cells expressing Venus (Nagai et al, 2002)-tagged proteins were lysed by sonication in 20 mM Tris–HCl (pH 7.5), 150 mM NaCl, and 1 mM phenylmethanesulphonyl fluoride. Subsequently, the cell lysate cleared by centrifugation at 20 000 g for 15 min was mixed with the liposomes. The fluorescent intensities were examined by excitation at 488 nm and emission at 528 nm. FRET efficiency was calculated as E = 1 − (F′D/FD, where FD and F′D are the fluorescence intensities of FRET donors (GFP or Venus) in the absence or presence of FRET acceptors (non-labelled or rhodamine-labelled liposomes), respectively. Without donor, no fluorescence was observed at 507 or 528 nm (data not shown). FRET efficiency (E) is described by the equation 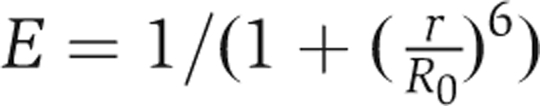 , where r is the distance between the donor (GFP) and the acceptor (rhodamine), and R0 is the Forster distance, which is defined as the distance between the donor and the acceptor when the FRET efficiency is 50% for each pair of donor and acceptor. Thus, the changes in the distance between donor and acceptor could be measured.
, where r is the distance between the donor (GFP) and the acceptor (rhodamine), and R0 is the Forster distance, which is defined as the distance between the donor and the acceptor when the FRET efficiency is 50% for each pair of donor and acceptor. Thus, the changes in the distance between donor and acceptor could be measured.
Supplementary Material
Supplementary Information
Acknowledgments
We thank Chikashi Toyoshima and Haruo Ogawa for help with the electron microscopy experiments; Atsushi Oda for providing the WIP expression vector; Sun Joo Park for actin preparation; Shusaku Kurishu for the Arp2/3 complex; Kazuya Tsujita for FBP17 cDNA; Manabu Negishi for Toca-1 cDNA; and Atsushi Shimada and Tadaomi Takenawa for helpful discussions. SS is supported by grants-in-aid for Scientific Research on Priority Areas from the Ministry of Education, Culture, Sports, Science and Technology of Japan and grants-in-aid from the Japan Science and Technology Corporation (JST), the Kao Foundation for Arts and Sciences, the Cell Science Research Foundation, the Ono Medical Research Foundation, and the Uehara Memorial Foundation.
References
- Anton IM, Jones GE, Wandosell F, Geha R, Ramesh N (2007) WASP-interacting protein (WIP): working in polymerisation and much more. Trends Cell Biol 17: 555–562 [DOI] [PubMed] [Google Scholar]
- Dawson JC, Legg JA, Machesky LM (2006) Bar domain proteins: a role in tubulation, scission and actin assembly in clathrin-mediated endocytosis. Trends Cell Biol 16: 493–498 [DOI] [PubMed] [Google Scholar]
- Doherty GJ, McMahon HT (2008) Mediation, modulation, and consequences of membrane–cytoskeleton interactions. Annu Rev Biophys 37: 65–95 [DOI] [PubMed] [Google Scholar]
- Farsad K, De Camilli P (2003) Mechanisms of membrane deformation. Curr Opin Cell Biol 15: 372–381 [DOI] [PubMed] [Google Scholar]
- Frost A, Perera R, Roux A, Spasov K, Destaing O, Egelman EH, De Camilli P, Unger VM (2008) Structural basis of membrane invagination by F-BAR domains. Cell 132: 807–817 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujita H, Katoh H, Ishikawa Y, Mori K, Negishi M (2002) Rapostlin is a novel effector of Rnd2 GTPase inducing neurite branching. J Biol Chem 277: 45428–45434 [DOI] [PubMed] [Google Scholar]
- Ho HY, Rohatgi R, Lebensohn AM, Le M, Li J, Gygi SP, Kirschner MW (2004) Toca-1 mediates Cdc42-dependent actin nucleation by activating the N-WASP–WIP complex. Cell 118: 203–216 [DOI] [PubMed] [Google Scholar]
- Ho HY, Rohatgi R, Ma L, Kirschner MW (2001) CR16 forms a complex with N-WASP in brain and is a novel member of a conserved proline-rich actin-binding protein family. Proc Natl Acad Sci USA 98: 11306–11311 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Itoh T, De Camilli P (2006) BAR, F-BAR (EFC) and ENTH/ANTH domains in the regulation of membrane–cytosol interfaces and membrane curvature. Biochim Biophys Acta 1761: 897–912 [DOI] [PubMed] [Google Scholar]
- Itoh T, Erdmann KS, Roux A, Habermann B, Werner H, De Camilli P (2005) Dynamin and the actin cytoskeleton cooperatively regulate plasma membrane invagination by BAR and F-BAR proteins. Dev Cell 9: 791–804 [DOI] [PubMed] [Google Scholar]
- Kakimoto T, Katoh H, Negishi M (2006) Regulation of neuronal morphology by Toca-1, an F-BAR/EFC protein that induces plasma membrane invagination. J Biol Chem 281: 29042–29053 [DOI] [PubMed] [Google Scholar]
- Kamioka Y, Fukuhara S, Sawa H, Nagashima K, Masuda M, Matsuda M, Mochizuki N (2004) A novel dynamin-associating molecule, formin-binding protein 17, induces tubular membrane invaginations and participates in endocytosis. J Biol Chem 279: 40091–40099 [DOI] [PubMed] [Google Scholar]
- Kessels MM, Qualmann B (2004) The syndapin protein family: linking membrane trafficking with the cytoskeleton. J Cell Sci 117: 3077–3086 [DOI] [PubMed] [Google Scholar]
- Loisel TP, Boujemaa R, Pantaloni D, Carlier MF (1999) Reconstitution of actin-based motility of Listeria and Shigella using pure proteins. Nature 401: 613–616 [DOI] [PubMed] [Google Scholar]
- Martinez-Quiles N, Rohatgi R, Anton IM, Medina M, Saville SP, Miki H, Yamaguchi H, Takenawa T, Hartwig JH, Geha RS, Ramesh N (2001) WIP regulates N-WASP-mediated actin polymerization and filopodium formation. Nat Cell Biol 3: 484–491 [DOI] [PubMed] [Google Scholar]
- Mattila PK, Pykalainen A, Saarikangas J, Paavilainen VO, Vihinen H, Jokitalo E, Lappalainen P (2007) Missing-in-metastasis and IRSp53 deform PI(4,5)P2-rich membranes by an inverse BAR domain-like mechanism. J Cell Biol 176: 953–964 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mayer LD, Hope MJ, Cullis PR (1986) Vesicles of variable sizes produced by a rapid extrusion procedure. Biochim Biophys Acta 858: 161–168 [DOI] [PubMed] [Google Scholar]
- McMahon HT, Gallop JL (2005) Membrane curvature and mechanisms of dynamic cell membrane remodelling. Nature 438: 590–596 [DOI] [PubMed] [Google Scholar]
- Merrifield CJ, Perrais D, Zenisek D (2005) Coupling between clathrin-coated-pit invagination, cortactin recruitment, and membrane scission observed in live cells. Cell 121: 593–606 [DOI] [PubMed] [Google Scholar]
- Michelsen P, Jergil B, Odham G (1995) Quantification of polyphosphoinositides using selected ion monitoring electrospray mass spectrometry. Rapid Commun Mass Spectrometry 9: 1109–1114 [Google Scholar]
- Nagai T, Ibata K, Park ES, Kubota M, Mikoshiba K, Miyawaki A (2002) A variant of yellow fluorescent protein with fast and efficient maturation for cell-biological applications. Nat Biotechnol 20: 87–90 [DOI] [PubMed] [Google Scholar]
- Peter BJ, Kent HM, Mills IG, Vallis Y, Butler PJ, Evans PR, McMahon HT (2004) BAR domains as sensors of membrane curvature: the amphiphysin BAR structure. Science 303: 495–499 [DOI] [PubMed] [Google Scholar]
- Pollard TD, Borisy GG (2003) Cellular motility driven by assembly and disassembly of actin filaments. Cell 112: 453–465 [DOI] [PubMed] [Google Scholar]
- Prehoda KE, Scott JA, Mullins DR, Lim WA (2000) Integration of multiple signals through cooperative regulation of the N-WASP–Arp2/3 complex. Science 290: 801–806 [DOI] [PubMed] [Google Scholar]
- Rohatgi R, Ma L, Miki H, Lopez M, Kirchhausen T, Takenawa T, Kirschner MW (1999) The interaction between N-WASP and the Arp2/3 complex links Cdc42-dependent signals to actin assembly. Cell 97: 221–231 [DOI] [PubMed] [Google Scholar]
- Rohatgi R, Nollau P, Ho HY, Kirschner MW, Mayer BJ (2001) Nck and phosphatidylinositol 4,5-bisphosphate synergistically activate actin polymerization through the N-WASP–Arp2/3 pathway. J Biol Chem 276: 26448–26452 [DOI] [PubMed] [Google Scholar]
- Scita G, Confalonieri S, Lappalainen P, Suetsugu S (2008) IRSp53: crossing the road of membrane and actin dynamics in the formation of membrane protrusions. Trends Cell Biol 18: 52–60 [DOI] [PubMed] [Google Scholar]
- Shimada A, Niwa H, Tsujita K, Suetsugu S, Nitta K, Hanawa-Suetsugu K, Akasaka R, Nishino Y, Toyama M, Chen L, Liu ZJ, Wang BC, Yamamoto M, Terada T, Miyazawa A, Tanaka A, Sugano S, Shirouzu M, Nagayama K, Takenawa T et al. (2007) Curved EFC/F-BAR-domain dimers are joined end to end into a filament for membrane invagination in endocytosis. Cell 129: 761–772 [DOI] [PubMed] [Google Scholar]
- Suetsugu S, Kurisu S, Oikawa T, Yamazaki D, Oda A, Takenawa T (2006a) Optimization of WAVE2-complex-induced actin polymerization by membrane-bound IRSp53, PIP3, and Rac. J Cell Biol 173: 571–585 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suetsugu S, Murayama K, Sakamoto A, Hanawa-Suetsugu K, Seto A, Oikawa T, Mishima C, Shirouzu M, Takenawa T, Yokoyama S (2006b) The RAC binding domain/IRSp53-MIM homology domain of IRSp53 induces RAC-dependent membrane deformation. J Biol Chem 281: 35347–35358 [DOI] [PubMed] [Google Scholar]
- Takenawa T, Suetsugu S (2007) The WASP-WAVE protein network: connecting the membrane to the cytoskeleton. Nat Rev Mol Cell Biol 8: 37–48 [DOI] [PubMed] [Google Scholar]
- Tsujita K, Suetsugu S, Sasaki N, Furutani M, Oikawa T, Takenawa T (2006) Coordination between the actin cytoskeleton and membrane deformation by a novel membrane tubulation domain of PCH proteins is involved in endocytosis. J Cell Biol 172: 269–279 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yarar D, Waterman-Storer CM, Schmid SL (2007) SNX9 couples actin assembly to phosphoinositide signals and is required for membrane remodeling during endocytosis. Dev Cell 13: 43–56 [DOI] [PubMed] [Google Scholar]
- Zimmerberg J, Kozlov MM (2006) How proteins produce cellular membrane curvature. Nat Rev Mol Cell Biol 7: 9–19 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Information