Abstract
The human type 1 (placenta, breast tumors) and type 2 (gonads, adrenals) isoforms of 3β-hydroxysteroid dehydrogenase/isomerase (3β-HSD) are key enzymes in biosynthesis of all active steroid hormones. Human 3β-HSD1 is a critical enzyme in the conversion of DHEA to estradiol in breast tumors and may be a major target enzyme for the treatment of breast cancer. 3β-HSD2 participates in the production of cortisol and aldosterone in the human adrenal gland. The goals of this project are to evaluate the role of the 2α-cyano group on trilostane (2α-cyano-4α,5α-epoxy-17β-ol-androstane-3-one) and determine which amino acids may be critical for 3β-HSD1 specificity. Trilostane without the 2α-cyano group, 4α,5α-epoxy-testosterone, was synthesized. Using our structural model of 3β-HSD1, trilostane or 4α,5α-epoxy-testosterone was docked in the active site using Autodock 3.0, and the potentially critical residues (Met187 and Ser124) were identified. The M187T and S124T mutants of 3β-HSD1 were created, expressed and purified. Dixon analyses of the inhibition of wild-type 3β-HSD1, 3β-HSD2, M187T and S124T by trilostane and 4α,5α-epoxy-testosterone suggest that the 2α-cyano group of trilostane is anchored by Ser124 in both isoenzymes. Kinetic analyses of cofactor and substrate utilization as well as the inhibition kinetics of M187T and the wild-type enzymes suggest that the 16-fold higher-affinity inhibition of 3β-HSD1 by trilostane may be related to the presence of Met187 in 3β-HSD1 and Thr187 in 3β-HSD2. This structure/function information may lead to the production of more highly specific inhibitors of 3β-HSD1 to block the hormone-dependent growth of breast tumors.
Keywords: 3β-hydroxysteroid dehydrogenase, short-chain dehydrogenase/reductase, structure-based mutagenesis, structure-function relationship
Introduction
The human type 1 (placenta, mammary gland, breast tumors) and type 2 (gonads, adrenals) isoforms of 3β-hydroxysteroid dehydrogenase (EC 1.1.1.145)/steroid Δ5-Δ4-isomerase (EC 5.3.3.1) (3β-HSD/isomerase) are encoded by two distinct genes which are expressed in a tissue-specific pattern [1]. 3β-HSD/isomerase catalyzes the conversion of 3β-hydroxy-5-ene-steroids (dehydroepiandrosterone, pregnenolone) to 3-oxo-4-ene-steroids (androstenedione, progesterone) on a single, dimeric protein containing both enzyme activities [2]. In addition to placenta and other human peripheral tissues, type 1 3β-HSD/isomerase (3β-HSD1) is selectively expressed in mammary glands and breast tumors [3], where it catalyzes the first step in the conversion of dehydroepiandrosterone (DHEA) to estradiol to promote tumor growth. In human adrenals, type 2 3β-HSD/isomerase (3β-HSD2) is required for the production of cortisol and aldosterone [4]. The selective inhibition of human 3β-HSD1 in breast tumors represents a potential new treatment for hormone-sensitive breast cancer. Our studies of the structure/function of human 3β-HSD took a dramatic turn in 2002 when we discovered a 14- to 16-fold higher affinity of purified human 3β-HSD1 for substrate (DHEA) and inhibitor steroids compared to human 3β-HSD2 [5]. A key difference in the structure of the two isoforms was determined to be His156 in 3β-HSD1 and Tyr156 in 3β-HSD2, which is in the catalytic domain, Y154-X-H156/Y156-X-K158, of the isoenzymes [5]. However, His156/Tyr156 was shown to reside in the α-helical subunit interface of the dimeric enzyme. This residue does not interact directly with bound substrate or inhibitor steroids but apparently influences subunit interactions that modify the topography of the active site [6]. Identifying fingerprint residues that interact with key groups on inhibitors, substrates and cofactors may determine the structural basis of the higher affinity of 3β-HSD1 for ligands relative to 3β-HSD2.
The aims of this study are to identify the residue that interacts with the 2α-cyano group of trilostane (Ser124) and to determine the functional significance of a non-identical amino acid in the active sites of the isoenzymes- Met187 in 3β-HSD1 and Thr187 in 3β-HSD2. Docking studies of trilostane with our structural model of human 3β-HSD1 predicts that the 2α-cyano side chain of trilostane (2α-cyano-4α,5α-epoxy-17β-ol-androstane-3-one) may interact with the Ser124 residue of 3β-HSD1. Because Ser124 was reported to be a critical residue for substrate binding in human 3β-HSD1 [7], the S124T mutation of 3β-HSD has been produced, expressed and purified to determine how steric hindrance by the methyl group of Thr124 affects the kinetics of 3β-HSD inhibition by trilostane. In addition, an analog of trilostane that lacks the 2α-cyano group, 4α,5α-epoxy-testosterone, has been synthesized, docking of this analog with 3β-HSD1 has been performed and the inhibition kinetics of 4α,5α-epoxy-testosterone have been compared to those of trilostane. Our structural model also predicts that the Thr187 of 3β-HSD2 is positioned to interact with the carbonyl group of the nicotinamide moiety of enzyme-bound NAD+, while Met187 of 3β-HSD1 is unlikely to interact with this functional group on cofactor. Met187/Thr187 is the only different amino acid in 3β-HSD1 and 3β-HSD2 in the region of the enzyme active-site that approximates the nicotinamide group of NAD+ and the A-ring of bound steroid [5,6], suggesting that this may be a key structural basis for the high affinity of 3β-HSD1 for the inhibitors and substrates relative to 3β-HSD2. To test this prediction, the M187T mutation of 3β-HSD1 has been created, expressed and purified for kinetic analyses of enzyme inhibition by trilostane and 4α,5α-epoxy-testosterone as well as for analyses of substrate and cofactor kinetics. Correlation of the structure/function relationships of the inhibition of 3β-HSD1 by trilostane characterized by these novel S124T and M187T mutations may lead to the development of new, more specific inhibitors of 3ß-HSD1 that can slow the growth of breast tumors without compromising steroidogenesis by 3β-HSD2 in the adrenal enzyme.
2. Methods and materials
2.1 Chemicals
Dehydroepiandrosterone (DHEA), testosterone and pyridine nucleotides were purchased from Sigma Chemical Co. (St. Louis, MO); 5-androstene-3,17-dione from Steraloids Inc. (Newport, RI); reagent grade salts, chemicals and analytical grade solvents from Fisher Scientific Co. (Pittsburgh, PA). Glass distilled, deionized water was used for all aqueous solutions.
2.1 Bioinformatics/Computational Biochemistry/Graphics
As described previously [8], a three dimensional model of human 3β-HSD1 has been developed based upon X-ray structures of two related enzymes: the ternary complex of E-coli UDP-galactose 4-epimerase (UDPGE) with an NAD cofactor and substrate (PDB AC: 1NAH) [9] and the ternary complex of human 17β-hydroxysteroid dehydrogenase (17β-HSD1) with NADP and androstenedione (PDB AC: 1QYX) [10]. Amino acid sequence alignments were performed using CLUSTAL W (1.81) multiple sequence alignment [11].
Using this PDB file for 3β-HSD1 in Autodock 3.0 (The Scripps Research Institute, http://autodock.scripps.edu) [12], the steroid ligand was removed, leaving the NAD+ co-factor in the binding site. All docking experiments were carried out on Autodock 3.0 using the Genetic Algorithm with Local Searching. Independent runs (256) were carried out and the docking results were then analyzed by a ranked cluster analysis. Compounds were identified that had the lowest overall binding energy. The three-dimensional graphics of the enzyme docked with trilostane were created using DeepView/Swiss-PdbViewer (http://www.exspasy.org/spdbv/).
2.2. Site-directed mutagenesis
Using the Advantage cDNA PCR kit (BD Biosciences Clontech, Palo Alto, CA) and pGEM-3βHSD1 as template [13], double-stranded PCR-based mutagenesis was performed with the primers listed below to create the cDNA encoding the S124T and M187T mutants of 3β-HSD1. The forward and reverse primers (mutated codons underlined) used to produce the S124T mutant cDNA were: 5′-CTACACCAGTACCATAGAGGTAGCC-3′; 5′-CCTCTATGGTACTGGTGTAGATGAAGAC-3′, respectively. The forward and reverse primers used to produce the M187T mutant cDNA were: 5′-ACGACCCACGTATATCTATGGGGAAG-3′; 5′-AGATATACGTGGGTCGTAAGGCACAAG-3′, respectively. The presence of the mutated codon and integrity of the entire mutant 3β-HSD cDNA were verified by automated dideoxynucleotide DNA sequencing using the Big Dye Terminator Cycle Sequencing Ready Reaction kit (PE Applied Biosystems, Foster City, CA). Chou-Fasman and Garnier-Osguthorpe-Robson analysis of each mutant enzyme was used to choose amino acid substitutions that produced no apparent changes in the secondary structure of the protein (Protylze program, Scientific and Educational Software, State Line, PA).
2.3. Expression and purification of the mutant and wild-type enzymes
The mutant 3β-HSD1 cDNA was introduced into baculovirus as previously described [13]. Recombinant baculovirus was added to 1.5 × 109 Sf9 cells (1L) at a multiplicity of infection of 10 for expression of each mutant enzyme. The expressed mutant and wild-type enzymes were separated by SDS-polyacrylamide (12%) gel electrophoresis, probed with our anti-3β-HSD polyclonal antibody and detected using the ECL western blotting system with antigoat, peroxidase-linked secondary antibody (Amersham Pharmacia Biotech, Piscataway, NJ). The goat anti-3β-HSD polyclonal primary antibody and rabbit polyclonal anti-Goat IgG (heavy and light cross adsorbed, conjugated to HRP) were obtained from Novus Biologicals (Littleton, CO). Each expressed enzyme was purified from the 100,000 g pellet of the Sf9 cells (2 L) by our published method [2] using Igepal CO 720 (Rhodia, Inc., Cranbury, NJ) instead of the discontinued Emulgen 913 detergent (Kao Corp, Tokyo). Each expressed, purified mutant and wild-type enzyme produced a single major protein band (42.0 kDa) on SDS-polyacrylamide (12%) gel electrophoresis that co-migrated with the purified human 3β-HSD1 control enzyme. Protein concentrations were determined by the Bradford method using bovine serum albumin as the standard [14].
2.4. Synthesis of the trilostane analog
4α,5α-Epoxy-testosterone (trilostane without the 2α-cyano group) was synthesized by reacting testosterone with hydrogen peroxide in chloroform and methanol. Sodium hydroxide was added to alkalinize the mixture as previously described [15]. NMR verified the loss of the 4,5-double bond on testosterone and the addition of the 4α,5α-epoxide group (data not shown). Trilostane (2α-cyano-4α,5α-epoxy-17β-ol-androstane-3-one) [16] was obtained as gift from Gavin P. Vinson, DSc PhD, School of Biological Sciences, Queen Mary University of London.
2.5. Kinetic studies
Michaelis-Menten kinetic constants for the 3β-HSD substrate were determined for the purified mutant and wild-type enzymes in incubations containing dehydroepiandrosterone (DHEA, 2–100 µM) plus NAD+ (0.2 mM) and purified enzyme (0.03 mg) at 27°C in 0.02 M potassium phosphate, pH 7.4,. The slope of the initial linear increase in absorbance at 340 nm per min (due to NADH production) was used to determine 3β-HSD1 activity. Kinetic constants for the isomerase substrate were determined at 27°C in incubations of 5-androstene-3,17-dione (20–100 µM), with or without NADH (0.05 mM) and purified enzyme (0.02 mg) in 0.02 M potassium phosphate buffer, pH 7.4. Isomerase activity was measured by the initial absorbance increase at 241 nm (due to androstenedione formation) as a function of time. Blank assays (zero-enzyme, zero-substrate) assured that specific isomerase activity was measured as opposed to non-enzymatic, "spontaneous" isomerization [17]. Changes in absorbance were measured with a Varian (Sugar Land, TX) Cary 300 recording spectrophotometer. The Michaelis-Menten constants (Km, Vmax) were calculated from Lineweaver-Burke (1/S vs. 1/V) plots and verified by Hanes-Woolf (S vs. S/V) plots. The kcat values (min−1) were calculated from the Vmax values (nmol/min/mg) and represent the maximal turnover rate (nmol product formed/min/nmol enzyme dimer).
Kinetic constants for the 3β-HSD cofactor were determined for the purified mutant and wild-type enzymes in incubations containing NAD+ (10–200 µM), DHEA (100 µM) and purified enzyme (0.03 mg) in 0.02 M potassium phosphate, pH 7.4, at 27°C using the spectrophotometric assay at 340 nm. Kinetic constants for the isomerase cofactor as an allosteric activator were determined in incubations of NADH (0–50 µM), 5-androstene-3,17-dione (100 µM) and purified enzyme (0.02 mg) in 0.02 M potassium phosphate buffer, pH 7.4 at 27°C using the spectrophotometric assay at 241 nm. Zero-coenzyme blanks were used as described above for the substrate kinetics.
Inhibition constants (Ki) were determined for the inhibition of the 3β-HSD1, 3β-HSD2, M187T and S124T activities by trilostane and 4α,5α-epoxy-testosterone using conditions that were appropriate for each enzyme species based on substrate Km values. For 3β-HSD1, the incubations at 27 °C contained sub-saturating concentrations of DHEA (4.0 µM or 8.0 µM), NAD+ (0.2 mM), purified human type 1 enzyme (0.03–0.04 mg) and trilostane (0–0.75 µM) or 4α,5α-epoxy-testosterone (0–10.0 µM) in 0.02 M potassium phosphate buffer, pH 7.4. For 3β-HSD2, similar incubations contained DHEA (15.0 µM or 40.0 µM) and trilostane (0–7.5 µM) or 4α,5α-epoxy-testosterone (0–10.0 µM). For S124T, similar incubations contained DHEA (15.0 µM or 40.0 µM) and trilostane (0–1.0 µM) or 4α,5α-epoxy-testosterone (0–17.5 µM). For M187T, similar incubations contained DHEA (8.0 µM or 20.0 µM) and trilostane (0–5.0 µM) or 4α,5α-epoxy-testosterone (0–17.5 µM). Dixon analysis (I versus 1/V) was used to determine the type or mode of inhibition (competitive, noncompetitive) and calculate the inhibition constant (Ki) values [18, 19]. The Dixon plot is widely used to characterize enzyme inhibition kinetics and is preferable to direct binding analysis, which only determines a dissociation constant for the inhibitor as a ligand and does not determine the type of inhibition. The Ki value represents the inhibitor concentration that reduces maximal enzyme activity by 50% and is considered a measure of the affinity of the enzyme for the inhibitor. A decrease in Ki indicates an increase in affinity [18].
Our triplicate determinations of the kinetic constants used highly purified mutant and wild-type enzymes to produce very reliable results with little variation between the values. The standard deviations reported for the mean values in the kinetic tables show that the variation between triplicates is 5% – 9%. With such low variation in the kinetic determinations, the observed differences of 2- to 65-fold between the kinetic constants of mutant and wild-type enzymes for substrates, cofactors and inhibitor analogs are interpreted to represent real differences.
3. Results
3.1. Predictions of the functions of the Ser124 and Met187/Thr187 residues by docking analysis
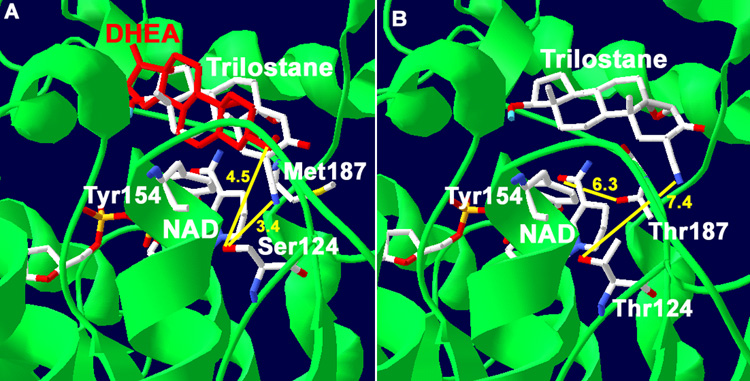
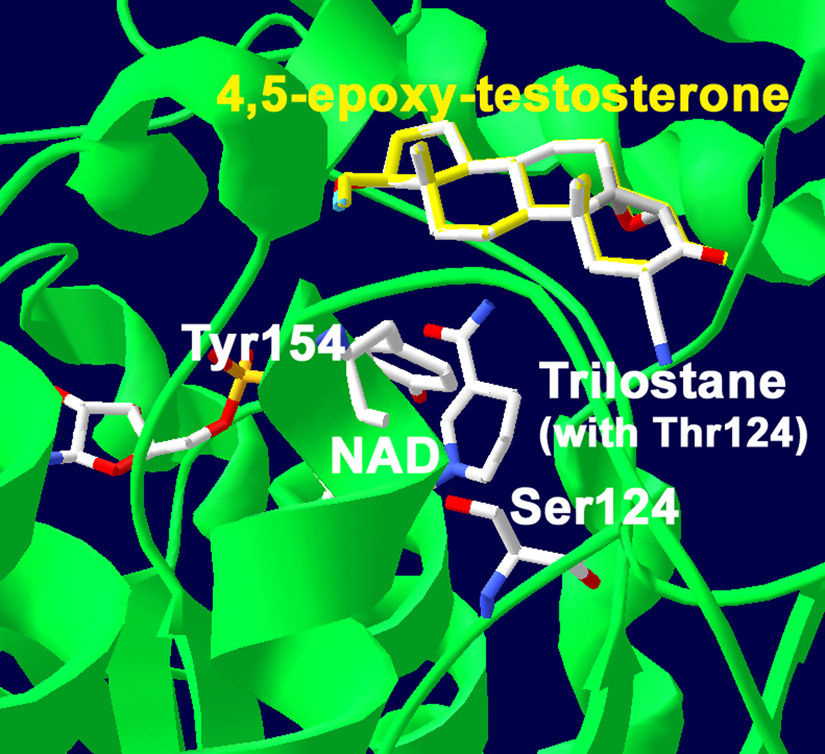
Predictions of the functions of Ser124 and Met187/Thr187 of 3β-HSD1/3β-HSD2 are based on the docking results obtained with our structural model of human 3β-HSD1, which has 51% homology of the Rossmann-fold domain and 40% identity of the key fingerprint residues that interact with bound substrate and cofactor compared to the crystallographic structures of E. coli UDP-galactose-4-epimerase and human 17β-hydroxysteroid dehydrogenase (17β-HSD1) [8]. Site-directed mutagenesis has confirmed the function of the catalytic residues and key substrate and cofactor binding residues as predicted by the structural model [5, 7, 8, 18, 19]. In the current study, trilostane was docked in the active site of our structural model of human 3β-HSD1 using Autodock 3.0. The docking result shown in Fig. 1A had the lowest binding energy of −9.81 kcal/mol and a predicted Ki of 0.072 µM, which is similar to the measured Ki of 0.10 uM for trilostane. As shown in Fig. 1A, the 2α-cyano group of trilostane is positioned to interact with the hydroxyl group of Ser124 (3.4 Å) wild-type 3β-HSD1. Ser124 was previously identified as a key recognition residue of the 3-oxo group of steroid substrate by human 3β-HSD [7]. To test the prediction that Ser124 also functions as a key recognition residue for inhibition by trilostane, the S124T mutant of 3β-HSD1 was created, expressed and purified. In docking studies using the S124T mutant enzyme, the presence of Thr124 produced a substantial shift in the orientation of trilostane (Fig. 1B). In the S124T mutant, the 2α-cyano group of trilostane is at a 2.2-fold greater distance from the hydroxyl group of Thr124 (7.4 Å, Fig. 1B) compared to Ser124 (3.0 Å, Fig. 1A) in wild-type 3β-HSD1. As shown in Fig. 2, when 4α,5α-epoxy-testosterone (trilostane without the 2α-cyano group) is docked with wild-type 3β-HSD1 containing Ser124, this steroid (yellow) superimposes with trilostane docked using the S124T mutant enzyme (with steric hindrance by Thr124 as indicated in Fig. 1B).
Fig. 1.
(A) Docking of trilostane with our structural model of human wild-type 3β-HSD1 shows the predicted interaction between the 2α-cyanogroup of trilostane and Ser124 residue of the wild-type enzyme (3.4 Å in yellow). The interaction of the 3β-hydroxyl group of substrate, DHEA, with Ser124 is also shown (4.5 Å yellow). The overlapping of DHEA (red) and trilostane is consistent with a competitive mode of inhibition. Met187 of 3β-HSD1 is shown to illustrate its lack of interaction with the nicotinamide group of NAD. (B) Docking of trilostane with the S124T mutant of 3β-HSD1 reveals a binding shift so that the 2α-cyanogroup of the inhibitor is 7.4 Å (yellow) from Thr124. In addition, the predicted interaction between the nicotinamide carbonyl of NAD and Thr187 of the M187T mutant enzyme (6.3 Å yellow) is shown. The position of the catalytic Tyr154 is labeled to identify the active site of the enzyme. The protein backbone (green), carbon (white), oxygen (red) and nitrogen (blue) atoms are indicated.
Fig. 2.
Docking of the trilostane analog lacking the 2α-cyano group, 4α,5α-epoxy-testosterone (yellow), with wild-type 3β-HSD1 (containing Ser124) reveals that it is superimposed over trilostane docked with the S124T mutant enzyme (containing Thr124). The position of the catalytic Tyr154 is labeled to identify the active site of the enzyme. The protein backbone (green), carbon (white), oxygen (red) and nitrogen (blue) atoms are indicated.
Also shown in Fig. 1B, the nicotinamide carbonyl of NAD+ is positioned near the hydroxyl group of Thr187 of the M187T 3β-HSD1 mutant (6.3 Å). However, Met187 in wild-type 3β-HSD1 is unlikely to interact with nicotinamide carbonyl on NAD+ according to our docking analysis (Fig. 1A). Because Met187/Thr187 is the only non-identical residue of human 3β-HSD1 and 3β-HSD2 in this enzyme region near the intersection of bound cofactor, inhibitor and substrate (residues 177–194), the M187T mutation of 3β-HSD1 was created, expressed and purified. Kinetic analyses of the expressed, purified mutant enzymes, S124T and M187T, have been performed to test the predictions of these docking results.
3.2. Site-Directed Mutagenesis, Expression and Purification of the Mutant Enzymes
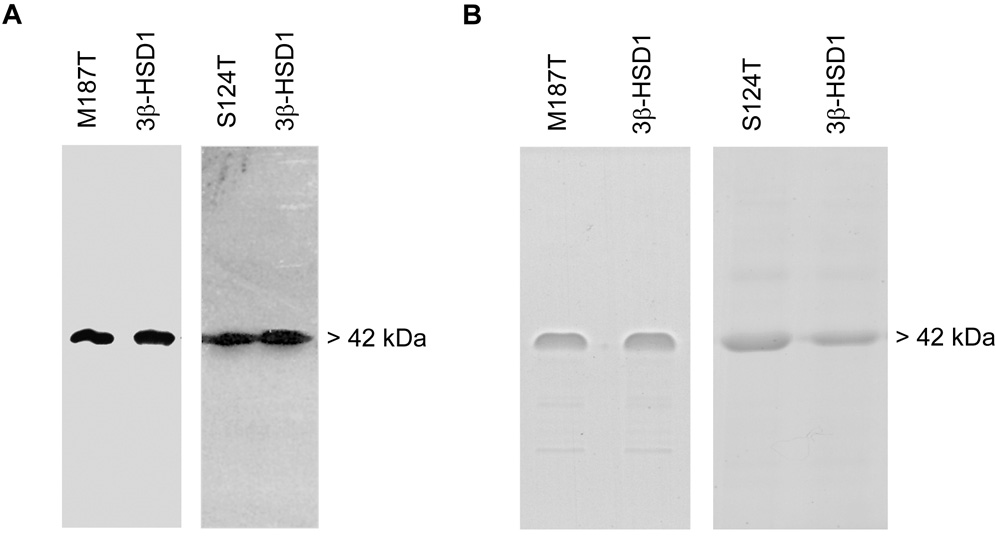
The cDNA encoding the S124T and M187T mutants of 3β-HSD1 were produced by double-stranded, PCR-based mutagenesis and inserted into baculovirus as previously described [13]. As shown by the immunoblots in Fig. 3A, the baculovirus system successfully expressed a single S124T or M187T mutant enzyme protein as well as wild-type human 3β-HSD1, 3β-HSD2 in Sf9 cells. Each expressed 3β-HSD enzyme (42 kDa monomer) was highly purified (90–95%) according to protein bands visible in SDS-PAGE (Fig. 3B) using our published method [2].
Fig. 3.
(A) Western immunoblots of the expressed mutant and wild-type enzymes. The Sf9 cell homogenate (4.0 µg) containing S124T or M187T plus the purified control wild-type 3β-HSD1 (0.05 µg) were separated by SDS-polyacrylamide (7.5%) gel electrophoresis. The 42.0 kDa band of the enzyme monomer was detected using anti-3β-HSD antibody as described in the text. (B) SDS-Polyacrylamide (7.5%) gel electrophoresis of the purified mutant and wild-type enzymes. Each lane was overloaded with 4.0 µg of purified protein, and the bands were visualized by Coomassie Blue staining.
3.3 Kinetic Analyses of the Inhibition of the Wild-type and Mutant Enzymes by the Trilostane Analogs
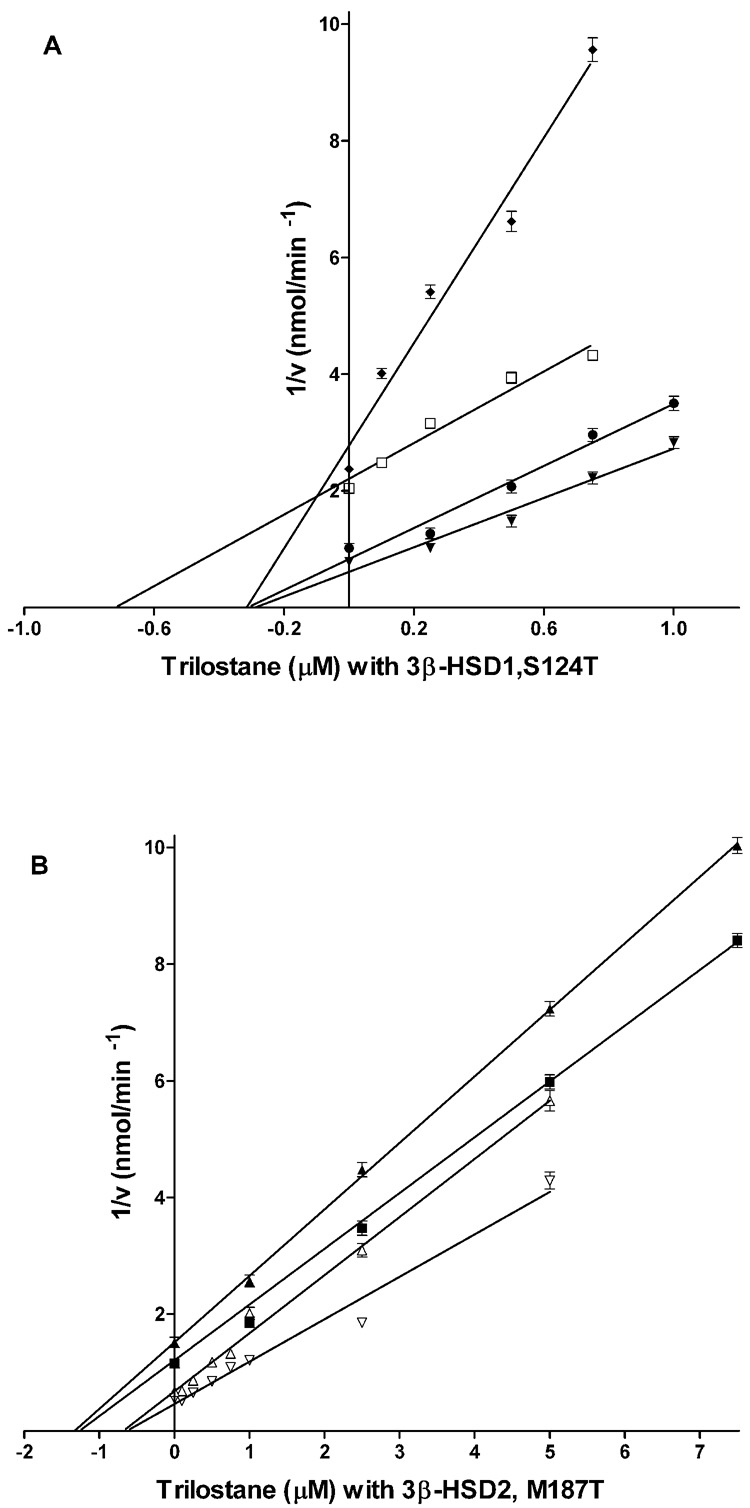
Dixon analyses of the inhibition of wild-type 3β-HSD1, 3β-HSD2, M187T and S124T by trilostane and 4α,5α-epoxy-testosterone produced a profile of Ki values and inhibition modes that clarifies the structural basis for the inhibition of 3β-HSD1 by trilostane with a 16-fold higher-affinity than 3β-HSD2 (Table 1). As illustrated by the Dixon plots, trilostane inhibits 3β-HSD1 (Ki= 0.10 M) in a competitive manner (Fig. 4A) but inhibits 3β-HSD2 (Ki= 1.60 µM) noncompetitively (Fig. 4B).
Table 1.
Comparison of inhibition constants of trilostane and 4α,5α-epoxy-testosterone for purified human 3β-HSD1, 3β-HSD2, M187T and S124T.
| Inhibitor steroid Ki (µM)1 | ||
|---|---|---|
| Enzyme | trilostane | 4α,5α-epoxy-testosterone |
| 3β-HSD1 | 0.10 ± 0.01 (C) | 4.08 ± 0.26 |
| 3β-HSD2 | 1.60 ± 0.10 | 3.75 ± 0.31 (C) |
| M187T | 0.83 ± 0.06 | 2.80 ± 0.19 (C) |
| S124T | 0.41 ± 0.03 | 4.34 ± 0.32 (C) |
For 3β-HSD1, the incubations at 27 °C contained sub-saturating concentrations of DHEA (4.0 µM or 8.0 µM), NAD+ (0.2 mM), purified human type 1 enzyme (0.03–0.04 mg) and trilostane (0–0.75 µM) or 4α,5α-epoxy-testosterone (0–10.0 µM) in 0.02 M potassium phosphate buffer, pH 7.4. For 3β-HSD2, DHEA (15.0 µM or 40.0 µM) and trilostane (0–7.5 µM) or 4α,5α-epoxy-testosterone (0–10.0 µM). For S124T, DHEA (15.0 µM or 40.0 µM) and trilostane (0–1.0 µM) or 4α,5α-epoxy-testosterone (0–17.5 µM). For M187T, DHEA (8.0 µM or 20.0 µM) and trilostane (0–5.0 µM) or 4α,5α-epoxy-testosterone (0–12.5 µM). Dixon analysis (I versus 1/V) was used to determine the type of inhibition and calculate the Ki values. Ki values are means of triplicate determinations ± standard deviations. (C) denotes a competitive mode of inhibition, and no notation indicates a non-competitive mode.
Fig. 4.
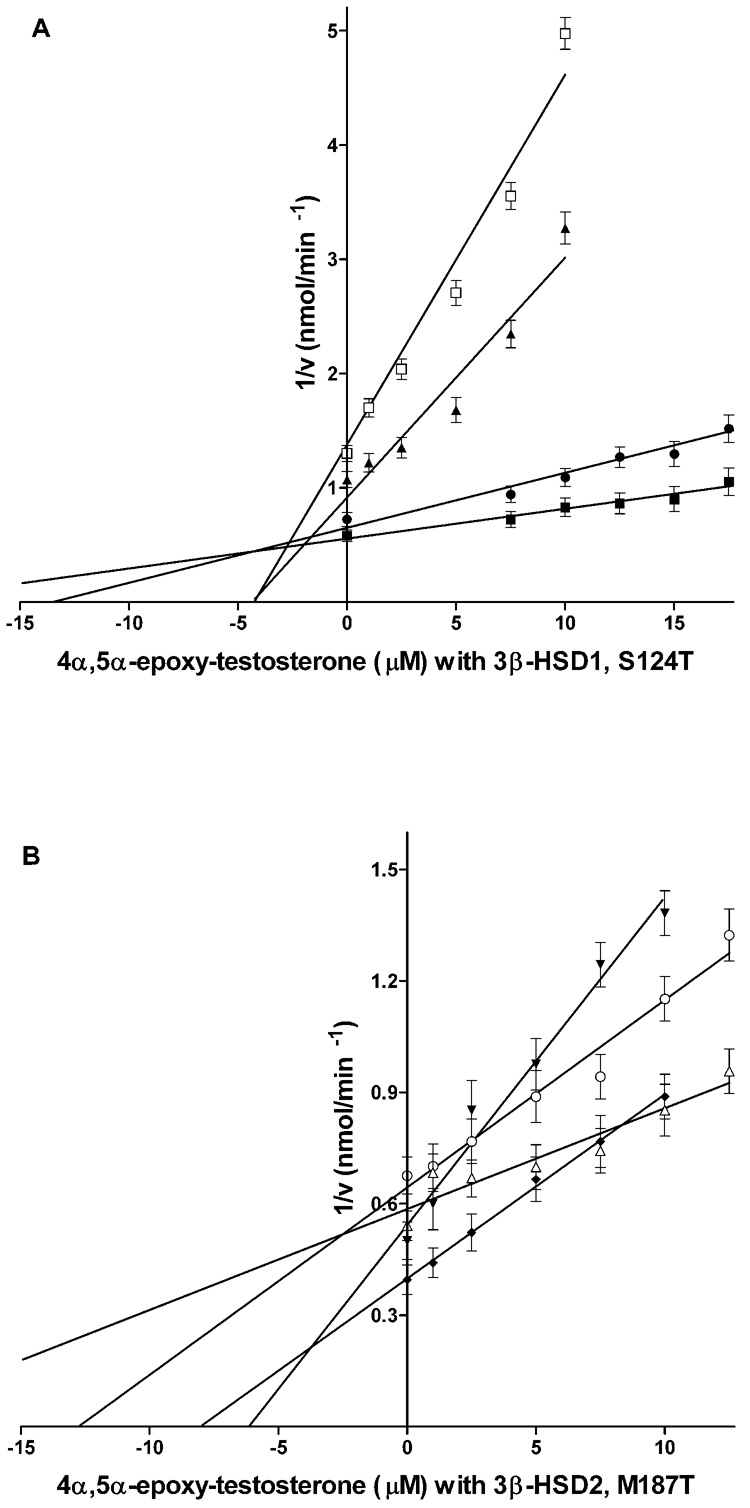
(A) Inhibition of the 3β-HSD activities of the 3β-HSD1 and S124T enzymes by trilostane. For 3β-HSD1, the incubations at 27 °C contained subsaturating concentrations (based on Km values in Table 2) of substrate DHEA, 4.0 µM (◆) or 8.0 µM (□), NAD+ (0.2 mM), purified human 3β-HSD1 enzyme (0.04 mg) and trilostane (0–0.75 µM) in 0.02 M potassium phosphate buffer, pH 7.4. For S124T, similar incubations contained DHEA, 15.0 µM (●) or 40.0 µM (▼) and trilostane (0–1.0 µM). (B) Inhibition of the 3β-HSD activities of the 3β-HSD2 and M187T enzymes by trilostane. For 3β-HSD2, incubations as described above contained DHEA, 15.0 µM (▲) or 40.0 µM (■) and trilostane (0–7.5 µM). For M187T, similar incubations contained DHEA, 8.0 µM (△) or 20.0 µM (▽) and trilostane (0–5.0 µM). Each point on the Dixon plot (1/V vs I) represents the mean of triplicate determinations, and the error bars represent standard deviations. Ki values were calculated from the intersection of the Dixon plots obtained for each enzyme preparation.
The prediction by the docking study that Ser124 may interact with the 2α-cyano group of trilostane is supported by a 4.1-fold increase in Ki value and the shift to a noncompetitive mode of inhibition for the S124T mutant of 3β-HSD1 by trilostane (Fig. 4A, Table 1). The methyl group on Thr124 may sterically hinder the binding of the 2α-cyano group of trilostane (Fig. 1B). In further support, when the inhibitor lacks the 2α-cyano group as in 4α,5α-epoxy-testosterone, wild-type 3β-HSD1 (with Ser124) is inhibited with a 41-fold higher Ki value than trilostane and the mode of inhibition is switched to noncompetitive (Fig. 5A, Table 1, Fig. 2). The effect of the absence of the 2α-cyano group is also seen for the inhibition of 3β-HSD2 by 4α,5α-epoxy-testosterone with an increase in Ki of 2.3-fold with the mode of inhibition being shifted to competitive (Fig. 5B, Table 1).
Fig. 5.
(A) Inhibition of the 3β-HSD activities of the 3β-HSD1 and S124T enzymes by 4α,5α-epoxy-testosterone. For 3β-HSD1, the incubations at 27 °C contained subsaturating concentrations of substrate DHEA, 4.0 µM (□) or 8.0 µM (▲), NAD+ (0.2 mM), purified human 3β-HSD1 enzyme (0.04 mg) and 4α,5α-epoxy-testosterone (0–10.0 µM) in 0.02 M potassium phosphate buffer, pH 7.4. For S124T, similar incubations contained DHEA, 15.0 µM (●) or 40.0 µM (■) and 4α,5α-epoxy-testosterone (0–17.5 µM). (B) Inhibition of the 3β-HSD activities of the 3β-HSD2 and M187T enzymes by 4α,5α-epoxy-testosterone. For 3β-HSD2, similar incubations contained DHEA, 15.0 µM (▼) or 40.0 µM (◆) and 4α,5α-epoxy-testosterone (0–10.0 µM). For M187T, similar incubations contained DHEA, 8.0 µM (○) or 20.0 µM (△) and 4α,5α-epoxy-testosterone (0–17.5 µM). Each point on the Dixon plot (1/V vs. I) represents the mean of triplicate determinations, and the error bars represent standard deviations. Ki values were calculated from the intersection of the Dixon plots obtained for each enzyme preparation.
According to our docking results, Thr187 is positioned to interact with the nicotinamide carbonyl on NAD+ in the M187T mutant of 3β-HSD1 (Figure 1B) and 3β-HSD2 (with Thr187). The low-affinity (high Ki) inhibition profiles of 3β-HSD2 and M187T for both trilostane and 4α,5α-epoxy-testosterone may be related to the effect of Thr187 on cofactor alignment and its influence on inhibitor orientation (Fig. 1B, Table 1), even though direct contact of Met187 or Thr187 with the inhibitors is not predicted by the docking results.. Further analysis of the function of Thr187 is provided by the kinetic studies of substrate and cofactor utilization described below.
3.4 Kinetic Analyses of Substrate and Cofactor Utilization of the Wild-type and Mutant Enzymes
As shown in Table 2, the S124T mutation shifts the high-affinity kinetic profile for the substrate (DHEA) of wild-type 3β-HSD1 (Km= 3.7 µM Kcat= 3.3 min−1) to an 11-fold lower affinity substrate profile (S124T Km= 42.1 µM, Kcat= 7.1 min−1 for DHEA), which is similar to that of wild-type 3β-HSD2 (Km= 47.3 µM, Kcat= 6.9 min−1). The S124T mutant enzyme has 1.6-to 3.2-fold higher Km and Kcat values for isomerase substrates compared to wild-type 3β-HSD1. However, the Km values for the coenzymes of 3β-HSD1 and isomerase measured for the S124T mutant are very similar to those for wild-type 3β-HSD1 (Table 3). These data with S124T support a critical role for Ser124 as a key residue that interacts with the 3-oxo group of the substrate steroids and are consistent with our previous report in which the S124A mutation abolished 3β-HSD activity [7]. Although Thr124 of the S124T mutant can function as a substrate recognition residue for human 3β-HSD1, the presence of the methyl group on Thr124 may sterically hinder the interaction of the amino acid hydroxyl group with the 3-oxo group of the substrate to increase the substrate Km value as well as hinder interaction with the 2α-cyano group of trilostane, as discussed above.
Table 2.
Substrate kinetics for the 3β-HSD and isomerase activities of the purified mutant and wild-type enzymes.
| 3β-HSD1 | Isomerase2 | |||||
|---|---|---|---|---|---|---|
| Purified Enzyme | Km µM | kcat min−1 | kcat/Km min−1 uM−1 | Km µM | kcat min−1 | kcat/Km min−1 uM−1 |
| M187T | 11.0 ± 0.7 | 5.6 ± 0.4 | 0.51 ± 0.04 | 78.1 ± 5.5 | 87.7 ± 5.3 | 1.12 ± 0.07 |
| S124T | 42.1 ± 2.7 | 7.1 ± 0.5 | 0.17 ± 0.01 | 83.7 ± 5.0 | 93.8 ± 9.8 | 1.68 ± 0.09 |
| 3β-HSD1 | 3.7 ± 0.2 | 3.3 ± 0.2 | 0.89 ± 0.04 | 27.9 ± 1.1 | 50.2 ± 2.0 | 1.80 ± 0.08 |
| 3β-HSD2 | 47.3 ± 2.9 | 6.9 ± 0.5 | 0.15 ± 0.01 | 88.4 ± 5.7 | 81.4 ± 6.0 | 0.92 ± 0.06 |
Kinetic constants for the 3β-HSD substrate were determined in incubations containing DHEA (2–100 µM), NAD+ (0.2 mM) and purified enzyme (0.03 mg) in 0.02 M potassium phosphate, pH 7.4.
Kinetic constants for the isomerase substrate were determined in incubations of 5-androstene-3,17-dione (17–150 µM), NADH (0.05 mM) and purified enzyme (0.02 mg) in 0.02 M potassium phosphate buffer, pH 7.4. All values are the means of triplicate determinations ± standard deviations.
Table 3.
Cofactor kinetics for the 3β-HSD and isomerase activities of the purified mutant and wild-type enzymes.
| 3β-HSD1 | Isomerase2 | |||||
|---|---|---|---|---|---|---|
| Purified Enzyme | Km µM | kcat min−1 | kcat/Km min−1 uM−1 | Km µM | kcat min−1 | kcat/Km min−1 uM−1 |
| M187T | 15.3 ± 1.0 | 6.0 ± 0.4 | 0.32 ± 0.02 | 0.5 ± 0.03 | 90.8 ± 5.3 | 181 ± 0.07 |
| S124T | 37.4 ± 2.2 | 5.7 ± 0.3 | 0.15 ± 0.01 | 2.6 ± 0.2 | 86.1 ± 5.8 | 33 ± 0.09 |
| 3β-HSD1 | 34.1 ± 1.7 | 3.5 ± 0.2 | 0.10 ± 0.005 | 4.6 ± 0.2 | 45.0 ± 1.8 | 9.8 ± 0.4 |
| 3β-HSD2 | 86.3 ± 5.6 | 7.1 ± 0.6 | 0.08 ± 0.005 | 12.6 ± 0.9 | 99.1 ± 6.4 | 7.9 ± 0.5 |
Kinetic constants for the 3β-HSD cofactor were determined in incubations containing NAD+ (10–200 µM), dehydroepiandrosterone (100 µM) and purified enzyme (0.03 mg) in 0.02 M potassium phosphate, pH 7.4.
Kinetic constants for the isomerase cofactor were determined in incubations of NADH (0–50 µM), 5-androstene-3,17-dione (100 µM) and purified enzyme (0.02 mg) in 0.02 M potassium phosphate buffer, pH 7.4. All values are the means of triplicate determinations ± standard deviations.
The M187T mutant of 3β-HSD1 produced a 2.2-fold decrease in the Km value for the NAD+ utilization by of 3β-HSD1 and a 9.2-fold decrease in the Km measured for NADH as an allosteric activator of isomerase type 1 with 2- to 3-fold higher Kcat values for both activities (Table 3). These higher affinity kinetic profiles of the M187T mutant for cofactor utilization is predicted by the interaction between Thr187 and the nicotinamide carbonyl of NAD+ shown by the docking results (Fig. 1B). The M187T mutant also produced a 3.0-fold increase in the Km for DHEA and a 2.8-fold increase in the Km for the isomerase substrate, 5-androstene-3,17-dione with parallel shifts in the Kcat and Kcat/Vmax values for both the 3β-HSD and isomerase activities (Table 2). This shift to a low-affinity profile of substrate utilization by the M187T mutation is similar to the inhibition kinetics of trilostane for M187T relative to 3β-HSD1, as discussed above.
4. Discussion
These studies of enzyme-ligand docking, modification of inhibitor structure, enzyme mutagenesis and kinetic analyses of the mutant enzyme strongly support a key interaction of Ser124 of human 3β-HSD with the 2α-cyano group of trilostane. The docking results of our 3β-HSD structural model with trilostane and 4α,5α-epoxy-testosterone correctly predicted this interaction based on kinetic analyses of the S124T mutant enzyme. The competitive mode of inhibition of 3β-HSD1 by trilostane may be due to the overlapping of substrate steroid (DHEA) and trilostane as shown by docking results in Fig. 1A. If that overlap is disrupted by a change in the binding orientation of the inhibitor steroid by the S124T mutation (steric hindrance by the methyl group in Thr124, Fig. 1B), the Ki is increase 4-fold and mode of inhibition is shifted to noncompetitive. When the interaction between Ser124 and the 2α-cyano group is abolished by removal of the 2α-cyano group from trilostane (4α,5α-epoxy-testosterone), the Ki value increases dramatically, and 4α,5α-epoxy-testosterone binds in the same orientation as trilostane that is sterically hindered by Thr124 in the S124T mutant of 3β-HSD1 (Fig. 2). A key role for the 2α-cyano group in the inhibition of 3β-HSD1 and 3β-HSD2 by trilostane is strongly supported by these results.
Met187 in 3β-HSD1 or Thr187 in 3β-HSD2 is the only difference in the amino acid sequences of the two isoenzymes in the area of the protein that interacts with NAD+ and the A-rings of trilostane and the substrate steroid. Thr187 in 3β-HSD2 is a strong hydrogen-bonding donor that is capable of interacting with the carbonyl group on the nicotinamide moiety of enzyme-bound NAD+ and NADH (Fig. 1), while Met187 in 3β-HSD1 is a weak acceptor of hydrogen-bonding that most likely does not interact with the nicotinamide group of the cofactor. The significance of this Met187/Thr187 difference in 3β-HSD1/3β-HSD2 is elucidated by kinetic analyses of the M187T mutation of 3β-HSD1. The M187T mutant may have shifted cofactor binding, which produced the 2- to 9-fold lower Km values for NAD+ and NADH (Table 3). This shift in cofactor binding may have interfered with the utilization of the substrate and the binding of inhibitor steroid to produce 3-fold higher substrate Km values (Table 2) and an 8-fold higher Ki value for trilostane (Table 1). These lower affinity kinetic profiles of M187T for substrate and trilostane are similar to those measured for human 3β-HSD2, which contains Thr187. Because other amino acids participate in the binding of the adenosyl and phosphoribosyl moieties of NAD+ in 3β-HSD and other members of the short-chain dehydrogenase/reductase family of enzymes [20,22], this single mutation (M187T) that affects only the nicotinamide moiety is not sufficient to shift the kinetic profile for overall cofactor utilization of 3β-HSD1 to the lower affinity profile of 3β-HSD2.
Understanding the structure/function relationships of the high-affinity inhibition of 3β-HSD1 by trilostane may lead to the development of new, more specific inhibitors of 3β-HSD1 that may be used to block the production of estradiol from DHEA in breast tumors without compromising steroidogenesis mediated by 3β-HSD2 in the human adrenal enzyme. With the success of the aromatase inhibitors in treating hormone-sensitive breast tumors [21,23], the characterization of a new target enzyme in this biosynthetic pathway enhances our ability to develop new treatments for breast cancer.
Acknowledgments
This research was supported by NIH grant CA114717 (JLT). We thank Gavin P. Vinson, DSc PhD, School of Biological Sciences, Queen Mary University of London, for the providing the trilostane and helpful comments.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Rheaume E, Lachance Y, Zhao H-F, Breton N, Dumont M, de Launoit Y, Trudel C, Luu-The V, Simard J, Labrie F. Structure and expression of a new complementary DNA encoding the almost exclusive 3 beta-hydroxysteroid dehydrogenase/delta 5- delta 4- isomerase in human adrenals and gonads. Mol. Endocrinol. 1991;5:1147–1157. doi: 10.1210/mend-5-8-1147. [DOI] [PubMed] [Google Scholar]
- 2.Thomas JL, Myers RP, Strickler RC. Human placental 3β-hydroxy-5-ene-steroid dehydrogenase and steroid 5-4-ene-isomerase: purification from mitochondria and kinetic profiles, biophysical characterization of the purified mitochondrial and microsomal enzymes. J. Steroid Biochem. 1989;33:209–217. doi: 10.1016/0022-4731(89)90296-3. [DOI] [PubMed] [Google Scholar]
- 3.Gingras S, Moriggl R, Groner B, Simard J. Induction of 3beta-hydroxysteroid dehydrogenase/delta5-delta4 isomerase type 1 gene transcription in human breast cancer cell lines and in normal mammary epithelial cells by interleukin-4 and interleukin-13. Mol. Endocrinol. 1999;13:66–81. doi: 10.1210/mend.13.1.0221. [DOI] [PubMed] [Google Scholar]
- 4.Rainey WE, Carr BR, Sasano H, Suzuki T, Mason JI. Dissecting human adrenal androgen production. Trends Endocrinol. Metab. 2002;13:234–239. doi: 10.1016/s1043-2760(02)00609-4. [DOI] [PubMed] [Google Scholar]
- 5.Thomas JL, Mason JI, Brandt S, Spencer BR, Norris W. Structure/function relationships responsible for the kinetic differences between human type 1 and type 2 3beta-hydroxysteroid dehydrogenase and for the catalysis of the type 1 activity. J. Biol. Chem. 2002;277:42795–42801. doi: 10.1074/jbc.M208537200. [DOI] [PubMed] [Google Scholar]
- 6.Thomas JL, Boswell EL, Scaccia LA, Pletnev V, Umland TC. Identification of key amino acids responsible for the substantially higher affinities of human type 1 3β-hydroxysteroid dehydrogenase/isomerase (3β-HSD1) for substrates, coenzymes and inhibitors relative to human 3β-HSD2. J. Biol. Chem. 2005;280:21321–21328. doi: 10.1074/jbc.M501269200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Thomas JL, Duax WL, Addlagatta A, Scaccia L, Frizzell KA, Carloni SB. Serine 124 completes the Tyr, Lys and Ser triad responsible for the catalysis of human type 1 3β-hydroxysteroid dehydrogenase. J. Mol. Endocrinol. 2004;33:253–261. doi: 10.1677/jme.0.0330253. [DOI] [PubMed] [Google Scholar]
- 8.Pletnev VZ, Thomas JL, Rhaney FL, Holt LS, Scaccia LA, Umland TC, Duax WL. Rational Proteomics V: Structure-based mutagenesis has revealed key residues responsible for substrate recognition and catalysis by the dehydrogenase and isomerase activities in human 3β-hydroxysteroid dehydrogenase/isomerase type 1. J Steroid Biochem Mol Biol. 2006;101:50–60. doi: 10.1016/j.jsbmb.2006.06.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Thoden JB, Frey PA, Holden HM. Crystal structures of the oxidized and reduced forms of UDP-galactose 4-epimerase isolated from Escherichia coli. Biochemistry. 1996;35:2557–2566. doi: 10.1021/bi952715y. [DOI] [PubMed] [Google Scholar]
- 10.Shi R, Lin SX. Cofactor hydrogen bonding onto the protein main chain is conserved in the short chain dehydrogenase/reductase family and contributes to nicotinamide orientation. J. Biol. Chem. 2004;279:16778–16785. doi: 10.1074/jbc.M313156200. [DOI] [PubMed] [Google Scholar]
- 11.Thompson JD, Higgins DG, Gibson TJ. CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position specific gap penalties and weight matrix choice. Nucleic Acids Res. 1994;22:4673–4680. doi: 10.1093/nar/22.22.4673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Morris GM, Goodsell DS, Halliday RS, Huey R, Hart WE, Belew RK, Olson AJ. Automated Docking Using a Lamarckian Genetic Algorithm and and Empirical Binding Free Energy Function. J. Computational Chemistry. 1998;19:1639–1662. [Google Scholar]
- 13.Thomas JL, Evans BW, Blanco G, Mercer RW, Mason JI, Adler S, Nash WE, Isenberg KE, Strickler RC. Site-directed mutagenesis identifies amino acid residues associated with the dehydrogenase and isomerase activities of human type I (placental) 3β-hydroxysteroid dehydrogenase/isomerase. J. Steroid Biochem. Molec. Biol. 1998;66:327–334. doi: 10.1016/s0960-0760(98)00058-2. [DOI] [PubMed] [Google Scholar]
- 14.Bradford MM. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal. Biochem. 1976;72:248–254. doi: 10.1016/0003-2697(76)90527-3. [DOI] [PubMed] [Google Scholar]
- 15.Michne WF, Schroeder JD, Bailey TR, Neumann HC, Cooke D, Young DC, Hughes JV, Kingsley SD, Ryan KA, Putz HS, Shaw LJ, Dutko FJ. Keto/enol epoxy steroids as HIV-1 Tat inhibitors: structure-activity relationships and pharmacophore localization. J. Med. Chem. 1995;17:3197–2206. doi: 10.1021/jm00017a003. [DOI] [PubMed] [Google Scholar]
- 16.Christiansen RG, Neumann HC, Salvador UJ, Bell MR, Schane HP, Jr, Creange JE, Potts GO, Anzalone AJ. Steroidogenesis Inhibitors. 1. Adrenal Inhibitory and Interceptive Activity of Trilostane and Related Compounds. J. Med. Chem. 1984;27:928–931. doi: 10.1021/jm00373a021. [DOI] [PubMed] [Google Scholar]
- 17.Thomas JL, Berko EA, Faustino A, Myers RP, Strickler RC. Human placental 3β-hydroxy-5-ene-steroid dehydrogenase and steroid 5-4-ene-isomerase: purification from microsomes, substrate kinetics, and inhibition by product steroids. J. Steroid Biochem. 1988;31:785–793. doi: 10.1016/0022-4731(88)90287-7. [DOI] [PubMed] [Google Scholar]
- 18.Segel IH. Enzyme Kinetics: Behavior and Analysis of Rapid Equilibrium and Steady-State Enzyme Systems. New York: John Wiley & Sons; 1993. pp. 109–111. [Google Scholar]
- 19.Thomas JL, Duax WL, Addlagatta A, Brandt S, Fuller RR, Norris W. Structure/function relationships responsible for coenzyme specificity and the isomerase activity of human type 1 3β-hydroxysteroid dehydrogenase/isomerase. J. Biol. Chem. 2003;37:35483–35490. doi: 10.1074/jbc.M304752200. [DOI] [PubMed] [Google Scholar]
- 20.Thomas JL, Huether R, Mack VL, Scaccia LA, Stoner RC, Duax WL. Structure/function of human type 1 3β-hydroxysteroid dehydrogenase: an intrasubunit disulfide bond in the Rossmann-fold domain and a Cys residue in the active site are critical for substrate and coenzyme utilization. J Steroid Biochem Mol Biol. 2007;107:80–87. doi: 10.1016/j.jsbmb.2007.02.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Filling C, Berndt KD, Benach J, Knapp T, Prozorovski T, Nordling E, Ladenstein R, Jornvall H, Oppermann U. Critical residues for structure and catalysis in short chain dehydrogenases/reductases. J. Biol. Chem. 2002;277:25677–25684. doi: 10.1074/jbc.M202160200. [DOI] [PubMed] [Google Scholar]
- 22.Santen RJ, Santner SJ, Pauley RJ, Tait L, Kaseta J, Demers LM, Hamilton C, Yue W, Wang JP. Estrogen production via the aromatase enzyme in breast carcinoma- which cell type is responsible. J Steroid Biochem Molec Biol. 1997;61:267–271. [PubMed] [Google Scholar]
- 23.Luo SQ, Martel C, Gauthier S, Merand Y, Belanger A, Labrie C, Labrie F. Long-term inhibitory effects of a novel anti-estrogen on the growth of ZR-75-1 and MCF-7 human breast cancer tumors in nude mice. Intl. J. Cancer. 1997;73:735–739. doi: 10.1002/(sici)1097-0215(19971127)73:5<735::aid-ijc21>3.0.co;2-3. [DOI] [PubMed] [Google Scholar]