Abstract
Bloom syndrome (BS) is a rare cancer-predisposing disorder in which the cells of affected persons have a high frequency of somatic mutation and genomic instability. BLM, the protein altered in BS, is a RecQ DNA helicase. This report shows that BLM is found in the nucleus of normal human cells in the nuclear domain 10 or promyelocytic leukemia nuclear bodies. These structures are punctate depots of proteins disrupted upon viral infection and in certain human malignancies. BLM is found primarily in nuclear domain 10 except during S phase when it colocalizes with the Werner syndrome gene product, WRN, in the nucleolus. BLM colocalizes with a select subset of telomeres in normal cells and with large telomeric clusters seen in simian virus 40-transformed normal fibroblasts. During S phase, BS cells expel micronuclei containing sites of DNA synthesis. BLM is likely to be part of a DNA surveillance mechanism operating during S phase.
Keywords: RecQ DNA helicase, nuclear domain 10, promyelocytic leukemia body, telomere, nucleolus
Bloom syndrome (BS) is a rare cancer-predisposing autosomal recessive disorder characterized by genomic instability, immunodeficiency, infertility, and small stature (1, 2). BS cells have a distinctive genomic instability: a high frequency of sister chromatid exchanges (SCEs) and elevated somatic mutations. BLM, the gene mutated in BS, encodes a DNA helicase (BLM) of the RecQ family (3). BLM is most identical to the two yeast proteins Sgs1p and Rqh1p (4, 5), and its expression can partially complement Saccharomyces cerevisiae sgs1 phenotypes (6, 7). The mouse BLM gene is highly expressed in testis and other cycling cells (8) and when disrupted can create an embryonic lethal phenotype (9). All other known mutations in RecQ DNA helicase family members are recessive loss-of-function mutations and are not lethal.
There are four other human genes in the RecQ family: RecQL/RecQ1, WRN, RecQ4, and RecQ5 (10, 11). WRN is the gene mutated in Werner syndrome, a premature aging disorder, and Werner syndrome cells show features of genomic instability (12). Work from the Guarente laboratory has shown the nucleolar localization of WRN and a relationship between aging and destabilization of the nucleolus in sgs1 mutants (13, 14), suggesting that instability of rDNA contributes to aging. Mutations in the RECQ4 gene have been found in persons with Rothmund–Thomson syndrome, a rare premature aging and cancer-prone disorder (15). Previous work from this laboratory (7) demonstrated the DNA helicase activity of BLM and that transfection of the normal BLM cDNA into BS cells reduces the frequency of SCEs and restores BLM to a focal nuclear pattern. In this report, the location of BLM is examined in relationship to other nuclear structures.
Materials and Methods
Cell Lines.
The following cell lines were from the Coriell Institute (Camden, NJ): simian virus 40 (SV40)-transformed normal human fibroblast GM00637 (HG2855), WI38 normal human lung fibroblast AG06814F, normal human skin fibroblast AG01437B (HG3004), SV40-transformed BS fibroblast GM08555 (HG2522), BS fibroblast GM3510, from persons of Ashkenazi Jewish descent, homozygous for the blmAsh (2281 Δ6ins7) mutation (HG3005), BS fibroblast GM2548, blm514X/blmQ572X (HG3002), and BS fibroblast GM03509, homozygous for blmS595X (HG3006). HG2619, a normal fibroblast cell line, and HG2940, a BS fibroblast cell line, were described (7). The BS cell lines used here lack BLM by Western analysis and immunofluorescence (7).
Antibodies.
The BLM antibody was prepared as described (7). Sources of antibodies were: mouse monoclonal anti (α)-BrdUrd/fluorescein isothiocyanate (FITC), Becton-Dickinson; mouse monoclonal α-proliferating cell nuclear antigen (PCNA), Sigma; human autoimmune sera ANA-N (α-nucleolar) and ANA-C (α-centromere), Sigma; mouse monoclonal α-promyelocytic leukemia (PML), Santa Cruz; FITC and Texas Red- (TR) conjugated secondary (2°) antibodies, Jackson ImmunoResearch (donkey α-mouse and α-rabbit; sheep α-human). The rabbit polyclonal α-replication protein A (RPA) (16) was from C. J. Ingles (Banting and Best Institute, Toronto), the α-TRF1 rabbit polyclonal was from T. de Lange (17), and the rabbit polyclonal α-WRN was made as described (13). When cells were stained with two primary (1°) rabbit antibodies, an intermediate blocking step with normal rabbit serum followed by 0.1 mg/ml goat α-rabbit Fab fragment (Jackson ImmunoResearch) was performed after the first 1°/2° antibody incubation.
Cell Culture, Immunofluorescent Staining, and in Situ Hybridization.
Cells were grown in DMEM plus 10% fetal bovine serum at 37°C with 5% CO2 on Fisher Superfrost Plus slides in 150-mm Petri dishes or on 22 × 22 mm glass coverslips in 6-well plates. Slides fixed in methanol/acetone (1:1) were air dried and stored at −70°C. The slides were stained as described (7). Cells were labeled with 0.1 mM BrdUrd for 10 min. For α-BrdUrd staining, the slides were fixed in 2% paraformaldehyde followed by denaturation in 2 M HCl at 37°C for 15 min. Some nuclei were stained in 0.2 μg/ml 4′,6-diamidino-2-phenylindole. At least 200 cells were counted for BrdUrd patterns and at least 500 were counted for micronuclei (MN). The in situ hybridization probe was transcribed from plasmid pTH5 (17) with SP6 RNA polymerase and digoxigenin (DIG)-labeled UTP (Boehringer Mannheim). The cells were fixed and hybridized as described (18). An FITC-labeled α-DIG mouse monoclonal antibody (Sigma) and a donkey α-mouse FITC/2° antibody were used to detect the probe.
Cell Cycle Analysis.
HG2619 cells were grown on slides then washed three times in PBS and grown in 0.02% serum for 2 days. At time 0, 20% serum was added. One slide was labeled with BrdUrd every hour for 24 h. A control mass culture was maintained in 10% serum. Slides were rinsed in PBS, fixed for 2 min in methanol/acetone (1:1) at room temperature, and air dried. Patterns were scored after staining.
Image Analysis.
Slides were examined with a Zeiss Axioskop fluorescence microscope equipped with narrow band-pass filters and a 63 or 100× objective. Images were captured separately with a charge-coupled device camera (Princeton Instruments Micromax 1400) and pseudocolored using IP Lab (Vienna, VA) spectrum 3.1 software. Merged images were created in Adobe Systems (San Jose, CA) photoshop 4.0. Confocal images were obtained at the Memorial Sloan–Kettering Microscope Core Facility by using a Zeiss confocal laser scanning microscope LSM 510.
Gel Transfer-Hybridization Analysis.
Genomic DNA (19) was digested with AluI and HinfI, and Southern analysis and quantitation was performed as described (20).
Results
Localization of BLM and Sites of DNA Synthesis.
BLM is found in the nucleus of normal human fibroblasts in two distinct structures: small numbers of brightly staining spherical foci and larger diffuse patches (Fig. 1 A-C), as well as in an overall diffuse distribution. Previous studies of BS cells document disturbances in DNA replication (21, 22). The localization of BLM was compared with sites of DNA replication by pulse-labeling cells with BrdUrd (23, 24) and by localization of PCNA (25). There was little overlap between sites of BrdUrd incorporation or sites of PCNA localization and BLM (Fig. 1 A and B and data not shown). The BLM-containing foci are partially coincident with DNA replication foci in late S phase (Fig. 1C).
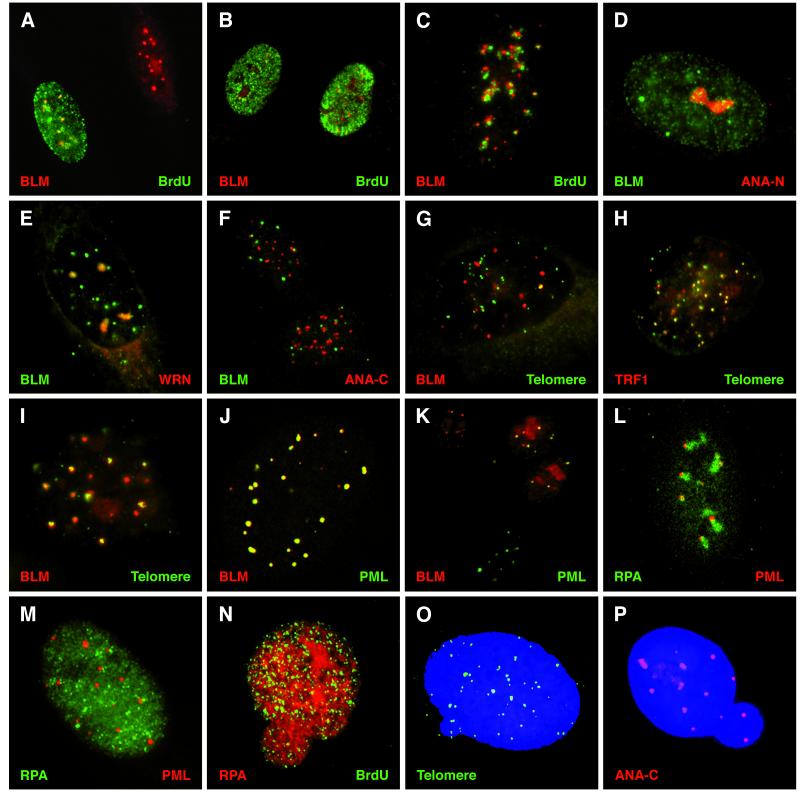
Figure 1.
Localization of α-BLM and sites of DNA replication, repeated sequence elements, and ND10s. (A–C) Normal human diploid fibroblasts (HG2619 or WI38) double-labeled with α-BLM (α-rabbit TR/2°) and α-BrdUrd mouse (α-mouse FITC/2°). Representative BrdUrd patterns are shown: pattern 3, middle to late S phase (A); patterns 1 and 2, early S phase (B); pattern 4, late S phase (C); pattern 5, small numbers of foci (not shown). (D) HG2619 cell stained with α-BLM (α-rabbit FITC/2°) and α-nucleolar human autoimmune sera ANA-N (α-human TR/2°). (E) HG2619 cell stained with α-BLM (α-rabbit FITC/2°) and α-WRN (α-rabbit TR/2°). (F) HG2619 cells stained with α-BLM (α-rabbit FITC/2°) and α-centromere human autoimmune sera ANA-C (α-human TR/2°). (G) WI38 cell stained with α-BLM (α-rabbit TR/2°) and hybridization of a DIG-labeled telomeric sequence probe (α-DIG FITC/2°). (H) SV40-transformed-BS fibroblast cell (HG2522) stained with α-TRF1 rabbit (TR α rabbit/2°) and hybridization of a DIG-labeled telomeric sequence probe (α-DIG FITC/2°). (I) SV40-transformed normal fibroblast cell (HG2855) stained with α-BLM (α-rabbit TR/2°) and hybridization of a DIG-labeled telomeric sequence probe (α-DIG FITC/2°). (J) Confocal image of a normal cell (HG2619) stained with α-BLM (α-rabbit TR/2°) and α-PML (α-mouse FITC/2°). (K) WI38 cells stained with α-BLM (α-rabbit TR/2°), α-PML (α-mouse FITC/2°). (L) Confocal image of HG2619 cell stained with α-RPA (α-rabbit FITC/2°) and α-PML (α-mouse TR/2°). (M) BS cell (HG2940) stained with α-RPA (α-rabbit FITC/2°) and α-PML (α-mouse TR/2°). (N) BS cell (HG3002) stained with α-RPA (α-rabbit TR/2°) and α-BrdUrd (α-mouse FITC/2°). (O) BS cell (HG3005) stained with 4′,6-diamidino-2-phenylindole and hybridization of a DIG-labeled telomeric sequence probe (α-DIG FITC/2°). (P) BS cell (HG2940) stained with 4′,6-diamidino-2-phenylindole and α-ANA-C (α-human TR/2°).
Normal and BS fibroblasts were scored for patterns of BrdUrd incorporation (Table 1). Both normal cell lines surveyed showed previously characterized patterns (23, 24) of BrdUrd incorporation (Fig. 1 A–C). The BS cell lines had nearly normal BrdUrd incorporation patterns but fewer total cells were in S phase (Table 1), staining was less intense, and the cultures contained more cells with a thicker peripheral staining pattern as compared with normal (Fig. 1A, pattern 3).
Table 1.
Patterns of BrdUrd Incorporation sites in normal and BS human fibroblast cell lines
| Fibroblast cell line | Pattern 1 (early S) | Pattern 2 (early-mid S) | Pattern 3 (mid-late S) | Pattern 4 (late S) | Pattern 5 (late S) | % BrdUrd labeled | Percent MN |
|---|---|---|---|---|---|---|---|
| HG3004 | 16 | 42 | 20 | 17 | 2 | 47 | 2 |
| HG2619 | 32 | 39 | 10 | 13 | 5 | 37 | 6 |
| HG3002 (BS) | 7 | 36 | 46 | 7 | 4 | 27 | 21 |
| HG3005 (BS) | 1 | 21 | 59 | 10 | 9 | 15 | 25 |
| HG3006 (BS) | 16 | 36 | 36 | 5 | 12 | 14 | 18 |
BLM Is Found in the Nucleolus of S Phase Cells and Colocalizes with WRN.
During this study, the patches of α-BLM staining (Fig. 1 A and B) were noted to frequently coincide with the excluded nucleolar regions in early S phase. This suggested that the diffuse patches might be the nucleolus. Costaining with α-BLM and the human autoimmune α-nucleolar antigen ANA-N (26) shows that BLM does reside in the nucleolus (Fig. 1D). BLM and WRN (13) also colocalize (Fig. 1E). In normal diploid human fibroblasts, 60–80% of the nuclei have detectable α-BLM staining. Four α-BLM staining patterns are prevalent (Fig. 2): foci only (43%, pattern 1), foci and an overall diffuse staining (36%, pattern 2), foci and patches (20%, pattern 3), and peripheral nuclear clusters (1%, pattern 4). The identification of BLM in the nucleolus in a small percentage of total cells (20%) in a mass culture suggested that the appearance of BLM in the nucleolus might be cell cycle dependent.
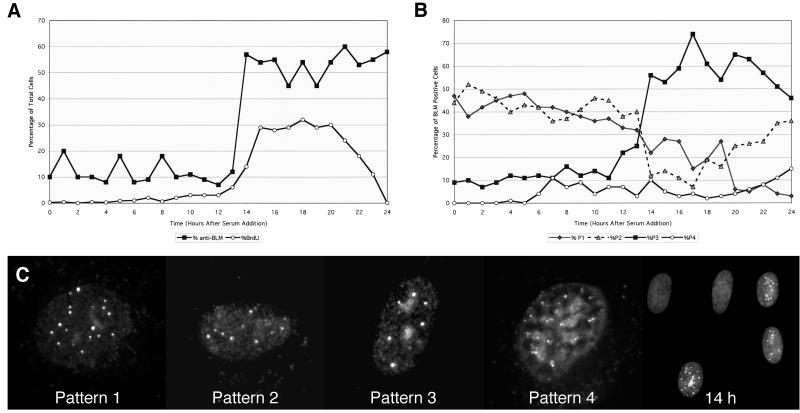
Figure 2.
Patterns of α-BLM staining during the cell cycle. Normal human diploid fibroblasts (HG2619) were serum-starved for 48 h and returned to growth by serum addition. Cells were harvested every hour and stained with α-BLM (α-rabbit TR/2°). Slides were pulse-labeled with BrdUrd and stained with α-BrdUrd (α-mouse FITC/2°). (A) Cells on slides were observed and counted for α-BLM staining and BrdUrd incorporation. (B) Distribution of α-BLM staining patterns as a function of time after serum addition. (C) Representative BLM patterns and a field of cells at 14 h after serum addition stained with α-BLM (α-rabbit TR/2°).
Normal diploid fibroblasts arrested by starvation were induced to grow by serum addition. For the first 12 h, the number of cells labeling with α-BLM and α-BrdUrd remained consistently low (Fig. 2A). Most cells did not label with α-BLM (80% or more) and those that did showed mainly two patterns of BLM localization (Fig. 2C): pattern 1 (foci only) and pattern 2 (foci and an overall diffuse staining). The time course of BrdUrd labeling demonstrated that the cells enter S phase 12–14 h after serum addition (Fig. 2A). Concomitant with the initiation of S phase the percentage of cells labeling with α-BLM increases and the frequency of the patterns changes (Fig. 2 A and B). Pattern 3 becomes the predominant staining pattern in S phase (Fig. 2C, representative field at 14 h). Pattern 4 (fewer smaller foci mostly at the nuclear periphery) becomes most prevalent as S phase ends.
BLM Is Found at a Subset of Telomeres.
Studies on the yeast orthologs of BLM suggest that these helicases are necessary for maintaining the stability of repeated sequence elements in the genome (5, 6, 27–29). Costaining experiments with α-BLM and the human autoimmune α-centromere antigen ANA-C (26) show that α-BLM staining does not greatly overlap but does closely associate with the centromeres (Fig. 1F). In situ hybridization with a human telomeric probe and α-BLM staining was performed. A small subset of telomeres colocalized with α-BLM (Fig. 1G) in normal human diploid fibroblasts. In SV40-transformed normal human fibroblasts, however, there was a strong coincident staining with large clusters (18) of telomeric repeats (Fig. 1I). This coincident staining was confirmed using confocal microscopy (data not shown). BS fibroblasts and SV40-transformed BS cells show normal small punctate telomeric signals (Fig. 1H and data not shown) that colocalize with the telomere repeat binding protein TRF1 (17) and do not have the strong in situ telomere signals seen in SV40-transformed normal human fibroblasts (Fig. 1I).
Telomeres from Cells of Persons with BS Are Not Dramatically Altered from Normal.
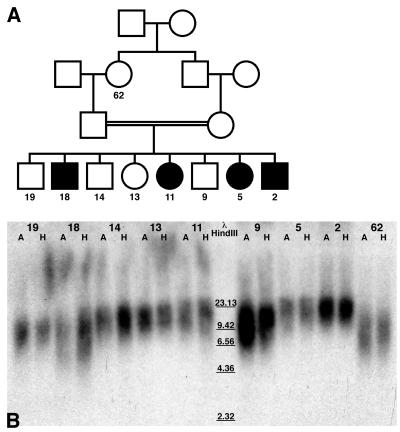
The colocalization of BLM at some telomeres suggested that BLM may play a role in telomere structure or replication. Telomere repeat lengths of four affected and four unaffected children from a consanguineous marriage of two individuals of Ashkenazi Jewish descent were analyzed (Fig. 3). Although this family is a small population, the strong degree of relatedness of the children provides the best comparison between the unaffected and BS populations. These data show that there is no large difference among the samples (Table 2), indicating that BLM is not a major structural or regulatory factor in maintaining telomere length.
Figure 3.
Analysis of telomere repeat lengths of genomic DNA from a consanguineous family of Ashkenazi Jewish decent. (A) Pedigree of family. Affected individuals are indicated by filled symbols. The age of the person when the sample was drawn is indicated. (B) Whole blood DNA from eight sibs ranging in ages from 2 to 19 years and the paternal grandmother (62 years of age) was digested with restriction endonucleases and displayed. A lanes are genomic DNA digests with AluI restriction enzyme. H lanes are genomic DNA digests with restriction enzyme HinfI. Data are shown in Table 2.
Table 2.
Telomere restriction fragment lengths from a consanguineous Ashkenazi Jewish family
| Age | Diagnosis | AluI fragment length, kb | Hinfl fragment length, kb |
|---|---|---|---|
| 62 | Normal | 11.13 | 11.02 |
| 19 | Normal | 12.20 | 10.85 |
| 18 | BS | 12.46 | 11.02 |
| 14 | Normal | 15.78 | 14.07 |
| 13 | Normal | 15.45 | 12.33 |
| 11 | BS | 15.78 | 14.30 |
| 9 | Normal | 14.41 | 13.49 |
| 5 | BS | 17.96 | 14.61 |
| 2 | BS | 18.54 | 15.40 |
BLM Is Located in Nuclear Domain 10 (ND10).
The number and morphology of the BLM-containing nuclear bodies suggested that they might be ND10s (30, 31). Costaining with α-PML and α-BLM shows nearly coincident staining (Fig. 1J). This is confirmed in optical sections by confocal microscopy (data not shown). Foci containing PML and BLM are found in several sizes (Fig. 1 J and K). Based on cell cycle analysis, cells in G0 or G1 have little or no BLM in the ND10s (Fig. 2; Fig. 1K, bottom cell). Therefore BLM is found in three distinct locations in the nucleus of cycling normal human cells: mainly in the ND10s, in the nucleolus at S phase, and at a select subset of telomeres. Analysis of α-PML staining of BS fibroblasts demonstrates a normal morphology and number of ND10s in the nucleus, indicating that BLM is not an essential subunit of these bodies (Fig. 1M). Previous studies on ND10s have reported the close association of these bodies with centromeres (32), as confirmed here (Fig. 1F).
BS Cells Bud Out Micronuclei at S Phase.
BS cells appear to have nearly normal nuclear structures using most of the nuclear antigens surveyed here (Table 1, Fig. 1 H and M–P). BS cells also show normal nucleolar morphology and WRN staining (data not shown). Normal and BS cells were examined for the localization of the single-stranded DNA-binding protein RPA (16) and for sites of DNA synthesis. Recently BLM and RPA have been shown to colocalize in foci on mouse meiotic chromosomes (33). RPA is found primarily in small punctate foci but can be seen in some larger aggregates (34). In normal cells, some larger RPA clusters can be seen in close proximity to ND10s (Fig. 1L). These structures are found in 3% of cells in normal diploid fibroblast cultures (HG2619) but are rarely seen in BS fibroblasts. When they are present, the RPA clusters are much reduced in size (Fig. 1M).
BS fibroblasts were, however, found to bud out MN containing RPA and sites of BrdUrd incorporation (Fig. 1N). Normal fibroblasts (HG2619 and HG3004) expel MN at 2–6%; the BS cell lines used here (HG3002, HG3005, and HG3006) expel MN at 18–25% (Table 1). The MN budding from the BS cells can contain telomeric sequences and centromeres (Fig. 1 O and P). The BrdUrd-incorporating MN are found in the cytoplasm of BS cells and outside the cell (data not shown). Expelling replicating chromatin as MN demonstrates that replicative events in BS cells fail at a high frequency, resulting in cell death and/or chromosome abnormalities.
Discussion
The BS Gene Product Is Found in ND10s and in the Nucleolus of Normal Human Cells.
ND10s are sites of early viral gene expression and DNA replication (32). DNA viruses may target these bodies for the DNA manipulation enzymes they contain and to interfere with cell cycle control and block an apoptotic response pathway (35–38). The dynamics of PML localization and assembly, and dispersion of ND10 foci are critical to the progression and therapy of acute promyelocytic leukemia (APL) (33–35, 39). Recent studies have demonstrated that PML is the critical factor required for ND10 formation and that BS cells have ND10s of normal morphology (38, 39). In APL cells containing the PML-RARα fusion protein, both BLM and PML are dispersed to microspeckles; treatment with retinoic acid restores both proteins to the nuclear bodies (39). BLM is also delocalized in PML−/− cells (38, 39). Both the presence of PML and the displacement activity of BLM are required for proper assembly of BLM into the ND10s (7). In one model, the ND10s may be a deposition site for various proteins such as helicases and topoisomerases which, when unregulated, may be deleterious. These DNA manipulation enzymes can be sequestered in the ND10s until needed.
BLM Is Found in the Nucleolus of Cells in S Phase.
The presence of BLM in the nucleolus of normal human cells during S phase suggests a cell cycle-dependent role. In early G1 of normal human fibroblasts BLM is found only in the ND10s. In contrast, WRN is confined to the nucleolus at all stages of the cell cycle (13). Another study has documented the nucleolar localization of WRN in cycling cells and the loss of the nucleolar signal upon quiescence (40). An immunofluorescence study of WRN localization using monoclonal instead of polyclonal antibodies found the protein in the nucleoplasm rather than in the nucleolus (41). Confluent fibroblasts show little or no BLM or WRN nucleolar staining by immunofluorescent analysis (data not shown); this provides a plausible explanation for the conflicting results obtained for the localization of WRN. A recent report confirms the cell cycle-dependent expression of BLM, but not of WRN (42). Despite colocalization of the two proteins in the nucleolus during S phase, antibodies directed against these two RecQ helicases fail to precipitate each other (42). These reports show that BLM and WRN are differentially regulated during the cell cycle, and that although they colocalize during S phase, they are likely to function in distinct complexes.
BLM Is Found at Some Telomeric Repeats.
The colocalization of BLM with some telomeric sequences is consistent with a study demonstrating that BLM can unwind G4 DNA (43). BLM colocalizes with telomeric clusters (not single foci) from within the ND10s. In SV40-transformed BS fibroblasts, TRF1 colocalizes with telomeres; the large telomere clusters evident in the normal SV40-transformed cells are absent. Some telomerase-negative SV40-transformed human cell lines contain long heterogeneous telomeres maintained by the ALT mechanism and found to be associated with ND10-like structures (44). The number and morphology of these bodies suggests that they are ND10s and that BLM is a component of these foci.
The presence of BLM at a subset of telomeres suggests a role for BLM in maintaining telomere structure or function. Metaphase chromosome spreads from BS cells do not show random end fusions (1, 2). Analyses of telomeric repeat length of a consanguineous family of Ashkenazi descent did not reveal a significant alteration in telomere repeat length among the four affected and four unaffected children in this family. The rate of telomere shortening in the BS individuals is more rapid than in the unaffected, but the population size is too small to be conclusive. BS fibroblast cell lines have normal telomeric in situ hybridization patterns that colocalize with TRF1 (data not shown). These data demonstrate that BLM is not a major component of the telomere complex and are consistent with BLM as part of a DNA surveillance mechanism.
BS Cell Lines Produce MN at S Phase.
Abnormalities in DNA replication have been reported in BS cells (21, 22). This report shows that BS cells bud out MN at an elevated frequency during S phase and have differences in BrdUrd patterns when compared to normal cells. The elevated frequency of MN in BS cells has been shown in primary patient isolates (45). The formation of MN during S phase (46) has been proposed to be part of a p53-dependent process in response to stalled forks caused by a hydroxyurea block. This result is reminiscent of Schizosaccharomyces pombe rqh1+ mutants that have hydroxyurea-dependent checkpoint phenotypes apparently due to the formation of irreversible DNA damage (4). BS cells have been shown to have normal mismatch and nucleotide excision repair pathways and most cell lines are not radiation-sensitive (reviewed in refs. 1 and 2), indicating that most DNA repair functions are within normal ranges. The exact nature of the type of DNA damage accumulating in BS cells is unknown but it may involve whole chromosomes as centromeres are found in the MN. Work on a bifunctional DNA crosslinker (47) demonstrates that crosslinking mammalian chromosomes within GC rich regions creates quadriradials as well as cells with lobular nuclei, cytoplasmic bridges, and MN, all phenotypes seen in BS cells. A block in the DNA template caused by a stalled fork or crosslink between chromosomes can be bypassed by a homologous recombination event leading to SCEs (48). BS cells could use this pathway to complete replication.
The results here support a model in which BLM surveys replicating regions of the genome from ND10s and that it acts on replication forks in late S phase. Recently it has been shown that the RecQ helicase and RecJ nuclease in Escherichia coli act on stalled forks to facilitate recovery of replication after DNA damage (49). This suggests a model for RecQ helicases in the resolution of stalled replication forks, most likely in repeated sequence elements in the human genome such as telomeres and rDNA. A high frequency of aberrant replicative events in cells of persons with BS leads to the clinical features: small size, infertility, and immune system defects. The evolution of cancers in persons with BS arises from the selection of cells with high replicative success, i.e., those progressed toward the transformed state.
Acknowledgments
We thank those who contributed reagents: α-TRF1, plasmid pTH5 and in situ protocol, Titia de Lange, Rockefeller University; and α-RPA, C. James Ingles, Banting and Best Institute, University of Toronto. We thank David Spector for suggesting that the BLM foci are ND10s, Gerd Maul for advice, Wouter Wiegant for suggestions on in situ images, and Jim Noonan for critical review of the manuscript. Confocal images were obtained with the assistance of Katia Manova and John Waka, Memorial Sloan–Kettering Microscopy Core Facility. This work was supported by National Institutes of Health Grant CA50897-08 (James L. German III, principal investigator), American Cancer Society Grant RD-395 (to N.N.), and funds from the New York Blood Center.
Abbreviations
- BS
Bloom syndrome
- SCE
sister chromatid exchange
- RPA
replication protein A
- PCNA
proliferating cell nuclear antigen
- ND10
nuclear domain 10
- PML
promyelocytic leukemia
- MN
micronuclei
- TR
Texas Red
- FITC
fluorescein isothiocyanate
- α-
anti-
- DIG
digoxigenin
- SV40
simian virus 40
Footnotes
This paper was submitted directly (Track II) to the PNAS office.
Article published online before print: Proc. Natl. Acad. Sci. USA, 10.1073/pnas.090525897.
Article and publication date are at www.pnas.org/cgi/doi/10.1073/pnas.090525897
References
- 1.German J. Medicine. 1993;72:393–406. [PubMed] [Google Scholar]
- 2.German J, Ellis N A. In: The Genetic Basis of Human Cancer. Vogelstein B, Kinzler K W, editors. New York: McGraw–Hill; 1998. pp. 301–315. [Google Scholar]
- 3.Ellis N A, Groden J, Ye T-Z, Straughen J, Lennon D, Ciocci S, Proytcheva M, German J. Cell. 1995;83:655–666. doi: 10.1016/0092-8674(95)90105-1. [DOI] [PubMed] [Google Scholar]
- 4.Stewart E, Chapman C R, Al-Khodiary F, Carr A M, Enoch T. EMBO J. 1997;16:2682–2692. doi: 10.1093/emboj/16.10.2682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kusano K, Berres M E, Engels W R. Genetics. 1999;151:1027–1039. doi: 10.1093/genetics/151.3.1027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Yamagata K, Kato J, Shimamoto A, Goto M, Furuichi Y, Ikeda H. Proc Natl Acad Sci USA. 1998;95:8733–8738. doi: 10.1073/pnas.95.15.8733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Neff N F, Ellis N A, Ye T Z, Noonan J, Huang K, Sanz M, Proytcheva M. Mol Biol Cell. 1999;10:665–676. doi: 10.1091/mbc.10.3.665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Seki T, Wang W S, Okumura N, Seki M, Katada T, Enomoto T. Biochim Biophys Acta. 1998;398:377–381. doi: 10.1016/s0167-4781(98)00066-9. [DOI] [PubMed] [Google Scholar]
- 9.Chester N, Kuo F, Kozak C, O'Hara C D, Leder P. Genes Dev. 1998;12:3382–3392. doi: 10.1101/gad.12.21.3382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Puranam K L, Blackshear P J. J Biol Chem. 1994;269:29838–29845. [PubMed] [Google Scholar]
- 11.Kitao S, Ohsugi I, Ichikawa K, Goto M, Furuichi Y, Shimamoto A. Genomics. 1998;54:443–452. doi: 10.1006/geno.1998.5595. [DOI] [PubMed] [Google Scholar]
- 12.Yu C-E, Oshima J, Fu Y-H, Wijsman E M, Hisama F, Alisch R, Matthews S, Nakura J, Miki T, Ouais S, et al. Science. 1996;272:258–262. doi: 10.1126/science.272.5259.258. [DOI] [PubMed] [Google Scholar]
- 13.Marciniak R A, Lombard D B, Johnson F B, Guarente L. Proc Natl Acad Sci USA. 1998;95:6887–6892. doi: 10.1073/pnas.95.12.6887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Sinclair D A, Mills K, Guarente L. Science. 1997;277:1313–1316. doi: 10.1126/science.277.5330.1313. [DOI] [PubMed] [Google Scholar]
- 15.Kitao S, Shimamoto A, Goto M, Miller R A, Smithson W A, Lindor N M, Furuichi Y. Nat Genet. 1999;22:82–84. doi: 10.1038/8788. [DOI] [PubMed] [Google Scholar]
- 16.He Z, Henricksen L A, Wold M S, Ingles C J. Nature (London) 1995;374:566–569. doi: 10.1038/374566a0. [DOI] [PubMed] [Google Scholar]
- 17.Chong L, van Steensel B, Broccoli D, Erdjument-Bromage H, Hanish J, Tempst P, de Lange T. Science. 1995;270:1663–1667. doi: 10.1126/science.270.5242.1663. [DOI] [PubMed] [Google Scholar]
- 18.Henderson S, Allsopp R, Spector D, Wang S-S, Harley C. J Cell Biol. 1996;134:1–12. doi: 10.1083/jcb.134.1.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.German J, Roe A M, Leppert M F, Ellis N A. Proc Natl Acad Sci USA. 1994;91:6669–6673. doi: 10.1073/pnas.91.14.6669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Metcalfe J A, Parkhill J, Campbell L, Stacey M, Biggs P, Byrd P J, Taylor M R. Nat Genet. 1996;13:350–353. doi: 10.1038/ng0796-350. [DOI] [PubMed] [Google Scholar]
- 21.Hand R, German J. Proc Natl Acad Sci USA. 1975;72:758–762. doi: 10.1073/pnas.72.2.758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lonn U, Lonn S, Nylen U, Winblad G, German J. Cancer Res. 1990;50:3141–3145. [PubMed] [Google Scholar]
- 23.Fox M H, Arndt-Jovin D J, Jovin T M, Baumann P H, Robert-Nicoud M. J Cell Sci. 1991;99:247–253. doi: 10.1242/jcs.99.2.247. [DOI] [PubMed] [Google Scholar]
- 24.O'Keefe R T, Henderson S C, Spector D L. J Cell Biol. 1992;116:1095–1110. doi: 10.1083/jcb.116.5.1095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Waseem N, Lane D P. J Cell Sci. 1990;96:121–129. doi: 10.1242/jcs.96.1.121. [DOI] [PubMed] [Google Scholar]
- 26.Leung P S C, Gershwin M E. Curr Opin Immunol. 1989;2:567–575. doi: 10.1016/0952-7915(90)90012-6. [DOI] [PubMed] [Google Scholar]
- 27.Gangloff S, McDonald J P, Bendixen C, Arthur L, Rothstein R. Mol Cell Biol. 1994;14:8391–8398. doi: 10.1128/mcb.14.12.8391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Watt P M, Louis E J, Borts R H, Hickson I H. Cell. 1995;81:253–260. doi: 10.1016/0092-8674(95)90335-6. [DOI] [PubMed] [Google Scholar]
- 29.Lu J, Mullen J R, Brill S J, Kleff S, Romeo A M, Sternglanz R. Nature (London) 1996;383:678–679. doi: 10.1038/383678a0. [DOI] [PubMed] [Google Scholar]
- 30.Ascoli C A, Maul G G. J Cell Biol. 1991;112:785–795. doi: 10.1083/jcb.112.5.785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Dyck J A, Maul G G, Miller W H, Jr, Chen J D, Kakizuka A, Evans R M. Cell. 1994;76:333–343. doi: 10.1016/0092-8674(94)90340-9. [DOI] [PubMed] [Google Scholar]
- 32.Maul G G. BioEssays. 1998;20:660–667. doi: 10.1002/(SICI)1521-1878(199808)20:8<660::AID-BIES9>3.0.CO;2-M. [DOI] [PubMed] [Google Scholar]
- 33.Walpita D, Plug A W, Neff N F, German J, Ashley T. Proc Natl Acad Sci USA. 1999;96:5622–5627. doi: 10.1073/pnas.96.10.5622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Wold M S. Annu Rev Biochem. 1997;66:61–92. doi: 10.1146/annurev.biochem.66.1.61. [DOI] [PubMed] [Google Scholar]
- 35.Hodges M, Tissot C, Howe K, Grimwade D, Freemont P. Am J Hum Genet. 1998;63:297–304. doi: 10.1086/301991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Quignon F, De Bels F, Koken M, Feunteun J, Ameisen J-C, de Thé H. Nat Genet. 1998;20:259–265. doi: 10.1038/3068. [DOI] [PubMed] [Google Scholar]
- 37.Wang Z-G, Ruggero D, Ronchetti S, Zhong S, Gaboli M, Rivi R, Pandolfi P P. Nat Genet. 1998;20:266–272. doi: 10.1038/3073. [DOI] [PubMed] [Google Scholar]
- 38.Ishov A M, Sotnikov A G, Negorev D, Vladimirova O V, Neff N, Kamitani T, Yeh E T H, Strauss J F, Maul G G. J Cell Biol. 1999;147:221–233. doi: 10.1083/jcb.147.2.221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Zhong S, Hu P, Ye T Z, Stan R, Ellis N A, Pandolfi P P. Oncogene. 1999;18:7941–7947. doi: 10.1038/sj.onc.1203367. [DOI] [PubMed] [Google Scholar]
- 40.Gray M D, Wang L, Youssoufian H, Martin G M, Oshima J. Exp Cell Res. 1998;242:487–494. doi: 10.1006/excr.1998.4124. [DOI] [PubMed] [Google Scholar]
- 41.Shiratori M, Sakamoto S, Suzuki N, Tokutake Y, Kawabe Y, Enomoto T, Sugimoto M, Goto M, Matsumoto T, Furuichi Y. J Cell Biol. 1999;144:1–9. doi: 10.1083/jcb.144.1.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Gharibyan V, Youssoufian H. Mol Carcinog. 1999;26:261–273. doi: 10.1002/(sici)1098-2744(199912)26:4<261::aid-mc5>3.0.co;2-a. [DOI] [PubMed] [Google Scholar]
- 43.Sun H, Karow J K, Hickson I D, Maizels N. J Biol Chem. 1998;273:27587–27592. doi: 10.1074/jbc.273.42.27587. [DOI] [PubMed] [Google Scholar]
- 44.Yeager T R, Neumann A A, Englezou A, Huschtscha L I, Noble J R, Reddel R R. Cancer Res. 1999;59:4175–4179. [PubMed] [Google Scholar]
- 45.Rosin M P, German J. Hum Genet. 1985;71:187–191. doi: 10.1007/BF00284570. [DOI] [PubMed] [Google Scholar]
- 46.Shimizu N, Itoh N, Utiyama H, Wahl G M. J Cell Biol. 1998;140:1307–1320. doi: 10.1083/jcb.140.6.1307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Matsumoto L, Kurek K, Larocque K, Gustafson G, Pires R, Zhang J, Tantravahi U, Suggs J W. Mutat Res. 1999;426:79–87. doi: 10.1016/s0027-5107(99)00042-1. [DOI] [PubMed] [Google Scholar]
- 48.Painter R B. In: Sister Chromatid Exchange. Sandberg A A, editor. New York: Liss; 1982. pp. 115–121. [Google Scholar]
- 49.Courcelle J, Hanawalt P C. Mol Gen Genet. 1999;262:543–551. doi: 10.1007/s004380051116. [DOI] [PubMed] [Google Scholar]