Abstract
Clearance of allergic inflammatory cells from the lung through matrix metalloproteinases (MMPs) is necessary to prevent lethal asphyxiation, but mechanistic insight into this essential homeostatic process is lacking. In this study, we have used a proteomics approach to determine how MMPs promote egression of lung inflammatory cells through the airway. MMP2- and MMP9-dependent cleavage of individual Th2 chemokines modulated their chemotactic activity; however, the net effect of complementing bronchoalveolar lavage fluid of allergen-challenged MMP2−/−/MMP9−/− mice with active MMP2 and MMP9 was to markedly enhance its overall chemotactic activity. In the bronchoalveolar fluid of MMP2−/−/MMP9−/− allergic mice, we identified several chemotactic molecules that possessed putative MMP2 and MMP9 cleavage sites and were present as higher molecular mass species. In vitro cleavage assays and mass spectroscopy confirmed that three of the identified proteins, Ym1, S100A8, and S100A9, were substrates of MMP2, MMP9, or both. Function-blocking Abs to S100 proteins significantly altered allergic inflammatory cell migration into the alveolar space. Thus, an important effect of MMPs is to differentially modify chemotactic bioactivity through proteolytic processing of proteins present in the airway. These findings provide a molecular mechanism to explain the enhanced clearance of lung inflammatory cells through the airway and reveal a novel approach to target new therapies for asthma.
The mechanisms that initiate allergic lung inflammation are relevant to understanding the pathophysiology of diseases such as asthma, but equally important are the factors underlying resolution of acute allergic inflammation. This poorly understood topic is important because failure to resolve allergic inflammation potentially results in irreversible airway remodeling and obstruction that are prominent features of chronic asthma (1, 2). Asthma occurs when exposure to respiratory allergens triggers a systemic immune response, characterized by activation of the adaptive immune cells that are biased toward Th2 cell-mediated airway inflammation (3, 4). Proinflammatory cytokines, in particular IL-4 and IL-13, induce up-regulation of chemokines and cytokines that allow homing of the activated Th2 cells to the site of inflammation (5, 6). Importantly, however, along with genes that are activated to promote inflammatory responses, programs of genes that act to suppress or limit inflammation are also activated (7, 8).
Integral to such suppressive gene programs, members of the matrix metalloproteinase (MMPs) 3 family of enzymes have been shown to play a significant role in the development and resolution of inflammatory lung diseases (9, 10). Up-regulation of MMPs is thought to be part of the innate immune response and host defense system, however, selected MMPs are also regulated by adaptive immunity. In particular, MMP2 and MMP9 have been shown to act downstream of Th2 cytokine signaling, but their presence is not required for the development of the allergic and obstructive lung phenotype (11–13).
Members of the serine and MMP family have been shown to cleave inflammatory mediators in vitro, and thus, proteolytic processing is hypothesized to alter the function of these proteins in vivo, resulting in a tightly regulated inflammatory response (14, 15). For instance, periodontal tissue destruction in Papillon-Lefevre syndrome may be in part due to failure of proper proteolysis of MIP-1αby neutrophil serine proteinases that can result in excess accumulation of this proinflammatory chemokine (16). Further supporting this hypothesis, truncation of human macrophage MCP-3 (CCL7), a potent CC chemokine, by MMP2 and MMP14 resulted in the formation of peptides that were able to bind the CCR and function as antagonists (17, 18). In addition, in vitro proteolytic processing of IL-8 can result in its loss of function, however, limited N-terminal processing of the same cytokine is shown to produce a more potent chemokine (19).
Proteolytic processing of inflammatory mediators in vitro has revealed important functional information regarding the possible biochemical behavior of molecules at sites of inflammation; however, despite these putative functions, little is known about the relevant in vivo substrates of proteinases, in particular MMPs (20). Because MMP2 and MMP9 are temporally expressed in the bron-choalveolar lavage (BAL) and lung in experimental asthma, and MMP2 and MMP9 double null (MMP2/9 −/−) mice show an exaggerated lung inflammatory response to inhaled allergens, predisposing them to lethal asphyxiation, we examined BAL fluid (BALF) of MMP2/9 −/− mice to gain insight into the role of MMPs in allergic inflammatory lung clearance. We have previously shown that several CC chemokines, in particular CCL7 (MARC), CCL17 (TARC), and CCL11 (eotaxin), are less abundant in the BAL of MMP2/9 −/− mice that were challenged with allergen and, consistent with this finding, that the BAL chemotactic activity of mice deficient in MMP2 and MMP9 is markedly reduced (11, 13). In this study, we tested the hypothesis that Th2-mediated up-regulation of these two gelatinases results in alteration of biological activity of several different classes of proteins in the BAL, thereby aiding in the clearance of lung inflammatory cells. Further, using a novel functional proteomics approach, we identified several proteins in the BALF that are cleaved by MMP2 and MMP9 and which are essential for regulating inflammatory pathways in experimental asthma.
Materials and Methods
Mice
MMP9 and MMP2 null mice (21, 22) (eight generations backcrossed to C57BL/6 background) were bred in the Association for Assessment and Accreditation of Laboratory Animal Care-accredited transgenic animal facility at Baylor College of Medicine. MMP2/MMP9 double null (MMP2/9−/−) mice were generated from F2 and F3 crosses of single null mice as we have described previously (11). Wild-type (WT) C57BL/6 mice were purchased from the Baylor College of Medicine vivarium. MMP2/9 +/− heterozygous littermates were used in limited experiments and showed no significant differences when compared with WT mice. All mouse studies were conducted in accordance with federal and institutional animal care and use guidelines.
In situ hybridization
To visualize mRNA expression, we used in situ hybridization on prepared paraffin sections of lung tissue. Antisense RNA probes for MMP9 and MMP2 were labeled as previously described (23). Briefly, plasmids pSP65, pSP65 92b, MMP2, and MMP9 probes were linearized and [35S]UTP-labeled probes (1000 Ci/nM; Amersham Biosciences) were transcribed from the SP6 promoter using a transcription kit (Promega). The probes were fractionated with Sephadex G-50 (Pharmacia), precipitated with ethanol, mixed with hybridization mixture, and placed on the pretreated lung sections. The sections were incubated overnight at 55°C, washed in high stringency conditions, and dipped in autographic emulsion (Kodak NTB2). After 4- to 8-day exposure, the emulsion was developed, and the sections were counterstained with hematoxylin and mounted.
Experimental model of asthma
Aspergillus fumigatus or Aspergillus oryzae allergens were prepared from a clinical isolate of A. fumigatus or purchased (Sigma-Aldrich) and mixed with OVA and will be referred to as complete Aspergillus allergen (CAA) (13, 24). A total of five CAA challenges was administered intranasally to MMP2/9 −/−, heterozygous WT littermates (MMP2/9 +/−) or WT C57BL/6 (n = 5 in each group) every 4 days as described (25). Because MMP2/9 −/+ heterozygotes and WT C57BL/6 showed identical responses to CAA (data not shown), we compared WT mice to the MMP2/9−/− mice.
Analysis of the asthma phenotype
All data were collected 24 h following the final allergen challenge. BAL cells were collected by serially instilling and withdrawing 0.8-ml aliquots of PBS from the tracheal cannula. Cells were washed, enumerated, and aliquots of 105 cells were centrifuged onto glass slides, stained using modified Giemsa, and used to determine the absolute numbers of BAL cells.
For histologic analysis, whole lungs were infused until distended with 4% formalin via the tracheal cannula and fixed for 24 h. Then, the tissues were embedded in paraffin, fin, and 4- to 5-µm sections were cut and either stained with periodic acid-Schiff (PAS) for viewing by light microscopy or prepared for immunohistochemistry.
Analysis of the BAL protein
BAL protein concentration was measured using the BCA assay (Pierce). BAL samples were then prepared for analysis using a two-dimensional (2D) cleanup kit (Bio-Rad) and pooled (n = 3 for WT and n = 3 for MMP2/9 −/−) just before separation and analysis using 2D differential in gel electrophoresis (DIGE) (26, 27). Resulting protein pellets were resuspended in sample buffer (7 mM urea, 2 mM thiourea, 4% CHAPS, 10 mM Tris (pH 8.8), and 5 mM magnesium acetate) to yield a final concentration of 5 mg/ml. To make the internal standard, equal amounts of protein from WT and knockout (KO) samples were mixed before CyDye labeling.
Protein samples were labeled with charge and molecular mass-matched fluorescent dyes (CyDye; Amersham Biosciences). A total of 400 pM of each CyDye was added to 50 µg of each unique protein sample and the resulting mixture was incubated on ice for 30 min. The reaction was quenched with the addition of 1 nM lysine and allowed to incubate an additional 10 min on ice. All CyDye-labeling reactions were prepared in the dark. BAL samples from WT, KO, and the internal standard (IS) were labeled and loaded for separation on triplicate gels as follows: gel 1, WT-Cy3, KO-Cy5, IS-Cy2; gel 2, WT-Cy5, KO-Cy3, IS-Cy2; gel 3, WT-Cy3, KO-Cy5, IS-Cy2.
The labeled protein samples for each gel were pooled and brought to a final volume of 250 µl with sample buffer that also contained 0.5% ampholytes (3–11 NL), 0.1% bromphenol blue, and 12 µl/ml DeStreak reagent (GE Healthcare).
First dimension separation (isoelectric focusing) was performed using the GE Healthcare IPGPhor II. The prepared sample was placed on the 13-cm strip holder and allowed to rehydrate dry pH 3–11 nonlinear, 13-cm immobilized pH gradient (IPG) strips (for 10 h at 20°C at 30 V; Bio-Rad). After rehydration, the IPG strips were focused in steps at 500 V for 1 h, 1,000 V for 1 h, and 8,000 V for a total of 20,000 V h. Temperature was kept constant at 20°C during focusing. After isoelectric focusing, the strips were equilibrated in SDS buffer (1.5 M Tris-Cl (pH 8.8) 8.8), 6 M Urea, 30% glycerol, 2% SDS, and 0.1% bromphenol blue) for 15 min with 1% DTT to reduce disulfide bonds in the sample. Reduction was followed by alkylation for 15 min in a 2.5% iodoacetamide solution in SDS buffer.
For second dimension separation, the IPG strips were secured in place with agarose on top of a 15% 18 × 16 cm SDS gel and were electrophoresed for 4 h at 20 mA. After electrophoresis, gels were scanned on a laser-based image (Typhoon 9400; GE Healthcare) and digitized using ImageQuant software (GE Healthcare). Gel images were imported into the DeCyder (version 5.5) Differential In-gel Analysis (GE Healthcare) program and resulting spots were matched among the three gels. Using DeCyder Biological Variation Analysis software (GE Healthcare), spot differences were quantified and protein spots that exhibited a significant normalized change in volume with respect to the internal standard sample (1.5-fold; p < 0.05) were picked for identification. For spot picking, spot map coordinates were transferred to the ETTAN Spot Handling Work Station (GE Healthcare) for automated picking, in-gel trypsin digestion, and MALDI plate spotting.
Gel plugs were digested with trypsin and washed sequentially with 200 µl of 50% methanol/water, 50% acetonitrile/0.5 mM NH4OH, and once with 75% acetonitrile. The gel cores were dried, mixed with 10 µl of trypsin in 20 mM NH4OH (200 ng), and incubated fo for 2 h a r at 37°C. After incubation, the tryptic peptides were eluted from the gel cores by the addition of 2 × 40 µl of 50% acetonitrile, 0.1% trifluoroacetic acid, and the peptides were transferred to a clean 96-well plate. The eluants were dried and resuspended in 3 µl of 50% acetonitrile, 0.5% trifluoroacetic acid. A total of 0.3 µl of this mixture was spotted onto a stainless steel target for tandem MALDI-TOF/TOF (Applied Biosystems MDS Sciex 4800). The AB/MDS Sciex 4800 MALDI-TOF/TOF uses the 4000 series Explorer software to control and collect data directly from the instrument. Spectra were analyzed with GPS Explorer (version 3.5) using an embedded MASCOT search engine. We identified proteins from peptide sequences using SwissProt and Celera data-bases. Protein identifications were considered significant if the database match yielded a protein score confidence interval >90%.
Immunohistochemistry
Slides were deparaffinized and washed in 3% H2O2 and PBST using standard protocols as previously described (28). Briefly, slides were blocked for 2 h at room temperature and then incubated in 4°C over-night with either rabbit anti-mouse Ym1 Ab (gift from Dr. P. Foster, Royal Newcastle Hospital, Newcastle, Australia; 1/100 dilution) or rabbit anti-mouse S100A8 Ab (29) (1/250 dilution), followed by biotinylated anti-rabbit secondary Ab (1/200 dilution) and Vectastain Elite ABC reagent (Vector Laboratories). Slides were counterstained with hematoxylin and mounted.
Protein analysis and N-terminal sequencing of S100 and Ym1 proteins
Western blotting of proteins identified in the BAL from each treatment group was performed using standard protocols. Briefly, BALF was concentrated using Microcon centrifugal filter units (Millipore). Thirty-five micrograms of protein was loaded per lane on 10, 12, or 15% SDS-PAGE gels, and, after transfer to nitrocellulose membrane, Western blotting was performed using 1/500 to 1/1000 dilution of Abs to S100A8 (29), S100A9 (R&D Systems), and Ym1 (R&D Systems).
In vitro cleavage
For in vitro proteolysis assays, we used recombinant, carrier-free mouse CC chemokines (CCL7, CCL11, CCL17), S100A9, Ym1, and MMPs (all from R&D Systems). S100A8 was made as previously described (29). Five micrograms of protein per 50, 100, or 500 ng of activated MMP2 and MMP9 was incubated at 37°C for 4 h. Equal volumes of samples (16.5 µl) were reduced and loaded onto a 16.5% Tricine gel and run for 3.5 h. Gels were developed using ProteoSilver kit (Sigma-Aldrich). Alternatively, cleaved proteins were separated as above, electroblotted in Tris-glycine buffer (25 mM Tris, 192 mM glycine, 10% MeOH (pH 8.3)) to a polyvinylidene difluoride membrane and rinsed with deionized water for 2–5 min, followed by staining with 0.05% Coomassie Blue in 1% acetic acid, 50% methanol. The polyvinylidene difluoride membranes were destained in 50% methanol until background was pale blue (5–15 min) and rinsed for 5–10 min in deionized water. The visible bands were cut out with a clean scalpel blade and air-dried, then N-terminal sequencing of each protein was performed using standard protein-sequencing methods (Applied Biosystems Procise 492cLC).
Chemotaxis assay
Inflammatory cells were extracted from lung or spleen of allergen (CAA)-challenged WT mice and used in chemotaxis assays as we have described previously (11, 13). Briefly, organs from CAA-immunized mice were harvested then minced and pressed through a fine nylon mesh. We purified the crude cell suspension by lysing RBC and washing in medium (5% heat-inactivated FCS, 1% l-glutamine, and 1% streptomycin in RPMI 1640). The final cell suspension was filtered through a 2-µm cell strainer and viable cells were resuspended to a final working concentration of 1 × 106 cell/ml for use in the chemotaxis assays. To assess the chemotactic activity of full-length and in vitro-cleaved proteins and BAL, we used two methods. First, we used transfilter assays in 48-well chemotaxis chambers (Neuroprobe). The chemotactic activity of the BALF from WT or MMP2/9−/− saline or CAA-challenged mice (n = 3) was assessed for each BAL sample in triplicate. A total of 27 µl of BAL or proteins (200 ng/ml) were loaded in the lower wells, and 53 µl of cells were loaded in upper wells. After a 30-min incubation in 37°C 5% CO2, the 5-µm filter was removed and stained. Cells were counted at ×40 magnification. Alternatively, we used 96-well disposable chemotaxis chambers (Neuroprobe). Disposable chemotaxis chambers were set up by loading 26 µl of either complete medium or chemokines (100 ng/ml) in the lower wells and 40 µl of cell suspension (1.8 × 106/ml) /above. After incubation in 37°C 5% CO2 for 4 h, filters were removed and fluid from the lower wells containing migrated cells was removed to a 3-ml round-bottom tube for preparation for flow cytometry. Each sample was fixed in 250 µl of 1% paraformaldehyde in PBS. After overnight fixation, we added 100,000 fluorospheres (Flow-check; Beckman Coulter) to each tube in 750 µl of PBS. Quantification of cell migration was accomplished by flow cytometry (Beckman Coulter Epic XL) as described (30). The following calculation was used to determine the number of cells migrated in each condition: cells/ml = ((events in cell gate)/(bead events)) ×(100,000 beads/ml).
In vivo analysis of function perturbing S100 Abs in asthma model
WT (C57BL/6) mice were immunized with i.p. injections of 50 µl of CAA on days 0, 4, 8, 12, and intranasal (IN) administration of CAA on day 16 as we have done previously (31). Two hours before the IN challenge, mice (n = 4) received i.p. injection of 2 mg of function-blocking Abs to S100A8 and S100A9 (29) or preimmune rabbit IgG (Ab control; n = 4). Airway hyperresponsiveness, lung, and BAL inflammatory cells were determined as described previously. Lung inflammation was determined using H&E stain of lung sections.
Statistics
All data are representative of at least three independent experiments with four to five mice in each in vivo experiment and are expressed as means ± SEM. We used Prism 4.0a for statistical analysis. We used two-way ANOVA and Bonferroni-corrected post hoc tests to identify significant differences (p < 0.05) between treatment groups.
Results
Spatiotemporal regulation of MMP2 and MMP9 in the allergic lung
We previously showed that MMP2 and MMP9 are secreted into the airways of mice that exhibit allergic lung disease (11). We next determined the mRNA expression of these two gelatinases in allergen-challenged lung by in situ hybridization. The in situ hybridization data confirmed that mRNA for MMP2 and MMP9 are up-regulated in mice that exhibit the asthma phenotype (Fig. 1) and are up-regulated in response to allergen challenge, as no signal was detected from lungs of naive animals (data not shown). In addition, lung mesenchymal cells, in particular endothelial cells, smooth muscle cells, and fibroblasts, expressed MMP2 while MMP9 was expressed primarily by eosinophils and macrophages. The airway epithelium expressed neither of these gelatinases (Fig. 1a; arrow heads point to the absence of mRNA in the epithelium).
FIGURE 1. MMP2 and MMP9 expression and modulation of the asthmatic phenotype.
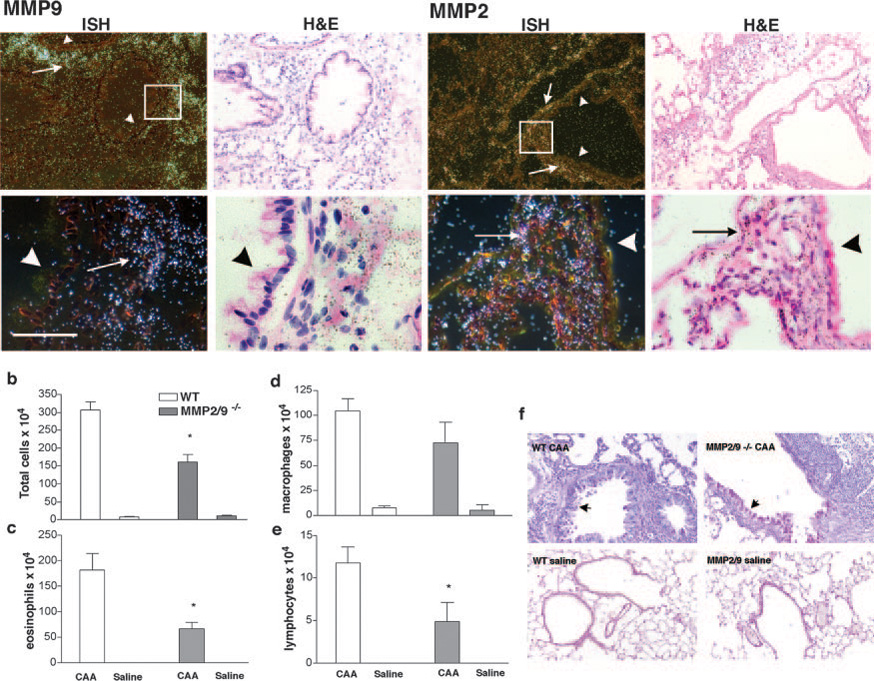
a, C57BL/6 mice were challenged with CAA or saline, and lung sections were hybridized with antisense MMP2 and MMP9 probes (left panels, dark field images) or sense probes (data not shown) in allergic mice. Short arrows point to negative expression of the gene in the airway epithelial cells and long arrows point to the presence of MMP2 and MMP9 in the inflammatory and mesenchymal cells. Boxes indicate the areas that are enlarged below each panel. Bar, 50 µm. b, Total cell count; c, eosinophil count; d, macrophage count; and e, lymphocyte count from bronchoalveolar lavage from WT or MMP2/9−/− mice (n = 5 in each group) that were intranasally challenged with either saline or CAA. There were no differences in neutrophil counts (data not shown).* ,Significant difference between WT and MMP2/9−/− asthma groups (p < 0.01). f, PAS stain of lung sections shows both WT and MMP2/9−/− mice have an intense allergic inflammatory reaction with extensive goblet cell metaplasia (arrows point to mucous producing goblet cells). Data are representative of three separate experiments.
Allergic lung inflammation in the absence of MMP2 and MMP9
To determine the role of MMP2 and MMP9 in the induction of acute allergic lung disease, we examined BALF of WT and MMP2/9 −/− mice that were challenged with CAA or saline (Fig. 1, b–f). We found that mice deficient in MMP2 and MMP9 developed all features of the allergic phenotype in response to IN CAA challenges, including increased total BALF cells and goblet cell metaplasia (Fig. 1, b–f; arrows point to PAS-positive goblet cells). For both mouse genotypes, the BALF cell population consisted of predominantly eosinophils followed by macrophages with few neutrophils and lymphocytes (Fig. 1, c– e). Nonetheless, MMP2/9−/− mice had significantly fewer eosinophils (p < 0.01) and lymphocytes (p < 0.05) in BALF relative to WT mice. Thus, consistent with our prior findings in MMP2 and MMP9 single null mice (11), MMP2 and MMP9 together are not required for allergic inflammation, but their absence results in decreased inflammatory cells in the BALF.
Dose-dependent cleavage of Th2 chemokines by MMP2 and MMP9
Th2 chemokines, especially the CCR3 and CCR4 ligands, MARC (CCL7), eotaxin (CCL11), and TARC (CCL17), are critical for the recruitment and clearance of allergic inflammatory cells (32, 33). We have previously shown that MMP2 and MMP9 play key roles in the formation of Th2-specific chemokine gradients in the BAL of mice that exhibit the asthma phenotype (11). Because many CC chemokines are known targets of cleavage by activated MMPs in vitro (14), we determined whether murine CCL7, CCL11, and CCL17 were cleaved by MMP2 and MMP9 in vitro. In a dose-dependent manner, both MMP2 and MMP9 cleaved recombinant CCL7, CCL11, and CCL17. At lower concentrations (5–50 nM), MMP2 was more efficient in cleaving the same chemokines than MMP9 (Fig. 2a, and data not shown). We verified our findings using N-terminal sequence analysis of the cleaved chemokines and found that MMP2 produced several different fragments of CCL7, CCL11, and CCL17 (Table I). These findings suggest that increased MMP2 and MMP9 activity and hence proteolytic processing in the airway during allergic inflammation alters the biochemical properties of chemokines.
FIGURE 2. Th2-specific chemokines are cleaved by activated MMP2 and MMP9 in vitro.
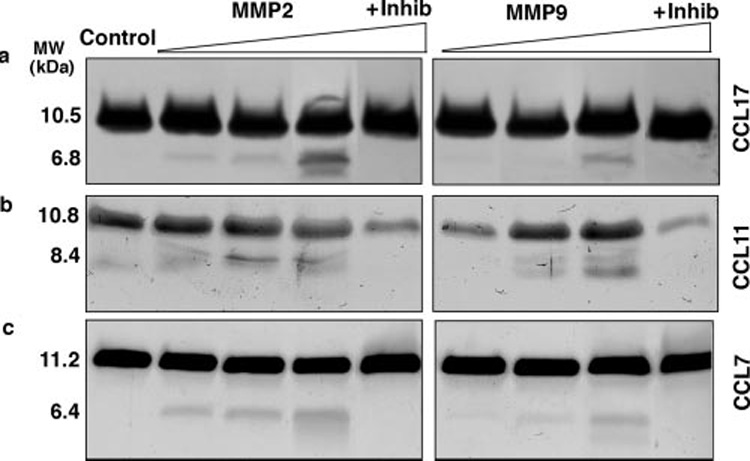
Recombinant (a) CCL17, (b) CCL11, and (c) CCL7 proteins (5 µg) were incubated for 4 h at 37°C with 5, 50, or 500 ng of p-aminophenylmercuric acetate-activated MMP2 and MMP9 in the presence or absence of 1′10-phenanthroline (100 nM), a potent inhibitor of MMPs. Proteins were resolved using 16.5% Tricine gels, and the native protein and cleaved fragments were detected with silver stain. Data are representative of three separate experiments.
Table I.
Amino acid sequences for select CC chemokinesa
| Protein | Sequence | |
|---|---|---|
| CCL7 | 1 | MRISATLLCL LLIAAAFSIQ VWAQPDGPNA STCCYVKKQK |
| 41 | IPKRNLKSYR RITSSRc→PWE AVIFKTKKGM EVCAEAHQKW | |
| 81 | VEEAIAYLDM KTPTPKP | |
| CCL11 | 1 | MQSSTALLFL LLTVTSFTSQ→VLAHPGSIPT SCCFIMTSKK |
| 41 | IPNTLLKSYK RITNNRCTLK AIVFKTRLGK EICADPKKKW | |
| 81 | VQDATKHLDQ KLQTPKP | |
| CCL17 | 1 | MRSLQMLLLA ALLLGTFLQH ARAARATNVG RECCLDYFKG |
| 41 | AIPIRKLVSW YKTSVECSRD→AIVFLTVQGK LICADPKDKH | |
| 81 | VKKAIRLVKN PRP | |
The indicated recombinant chemokines were incubated with either MMP2 or MMP9 in vitro and partial sequencing of cleavage products was performed using N-terminal sequencing to confirm their identity. Pairs of amino acids, underlined, are predicted cleavage sites for MMP2, whereas highlighted pairs shown in bold italics are also predicted cleavage sites for MMP9. We identified the actual cleavage site (→) using N-terminal sequencing.
Effect of MMP2 and MMP9 cleavage on chemotactic function
We next examined the biochemical function of chemokines that are also cleaved by MMP2 and MMP9 in vitro. We focused on their chemotactic activity because of the prior association of these proteins with chemotaxis and our previous finding that MMPs alter the chemotactic property of BALF (13). Using lung-derived Th2 inflammatory cells, in vitro processing of CCL11 by MMP2 and MMP9 resulted in a significant loss in chemotactic activity compared with the unmodified parent molecules (Fig. 3a).In contrast, processing of CCL7 resulted in significant enhancement of its chemoattractant properties (Fig. 3a).
FIGURE 3. Functional analysis of chemotactic activity of proteins cleaved by MMP2 and MMP9.
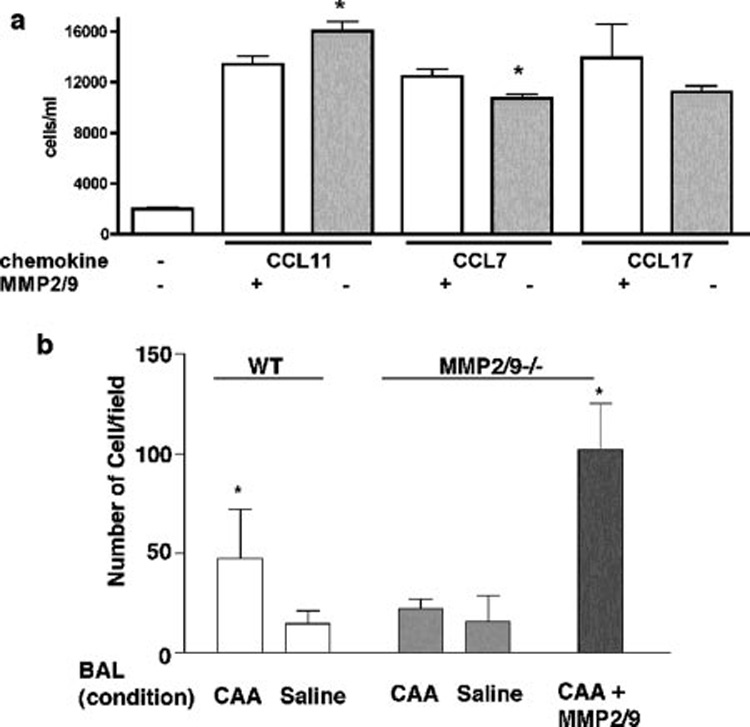
a, Recombinant (CCL11, CCL7, and CCL17; 5 µg/ml) proteins were used either intact or after cleavage with 100 ng of activated MMP2/MMP9 enzymes in a chemotaxis assay. Data represents average of six replicates in three different experiments. b, BAL (neat) of WT and MMP2/9 −/− (n = 3 in each group) CAA allergen-challenged or saline-treated mice were used in in vitro chemotaxis assays. Reconstitution of BAL from MMP2/9−/− allergen challenged mice with 100 ng of activated MMP2 and MMP9 restored chemotactic activity of the BAL. Data represents average of five independent fields from three replicates in two different experiments (* ,p < 0.05).
To understand the net effect of MMP2 and MMP9 on BALF proteins, we determined the overall chemotactic activity of BALF collected from allergen-challenged lungs before and after proteolysis with activated MMP2 and MMP9. As expected, BALF from WT mice challenged with allergen showed enhanced chemotactic activity compared with BALF from saline-challenged mice, whereas the chemotactic activity of BALF from MMP2/9−/− mice was not different from BALF of saline-challenged mice. However, addition of MMP2 and MMP9 to BALF of MMP2/9−/− mice challenged with allergen fully restored chemotactic activity (p < 0.05; Fig. 3b). Active MMP2 and MMP9 alone or in combination did not result in any increase in chemotactic activity (data not shown). These findings demonstrate that although the effect of MMP2 and MMP9 cleavage on individual prochemoattractants cannot be readily predicted, the net effect of their action on proteins secreted into the airway is to enhance chemotactic activity.
Identification of in vivo substrates for MMP2 and MMP9
The preceding studies indicate that MMP2 and MMP9 are capable of specifically cleaving chemokines in vitro, but the behavior of these airway MMPs in vivo has not been studied. To identify MMP2 and MMP9 substrates in vivo, we analyzed BALF from WT and MMP2/9−/− mice using 2D-DIGE (Fig. 4). We found significant differences in the 2D profile of proteins present in the two BALF samples (Fig. 4). In subsequent analysis with DeCyder software, we quantified protein isoform abundance in MMP2/9−/− BALF that differed significantly from WT BALF. Table II lists selected protein isoforms that were identified using mass spectrometry, and the 2D coordinates of the identified proteins are marked on Fig. 4d. The distinct differences in the proteome composition between samples are consistent with the different phenotypes of allergic inflammatory clearance seen in WT and MMP2/9−/− mice, thus, we further investigated the newly identified proteins as candidates for in vivo substrates of MMP2 and MMP9.
FIGURE 4. a–d, BAL proteins in CAA-challenged WT and MMP2/9−/− mice.
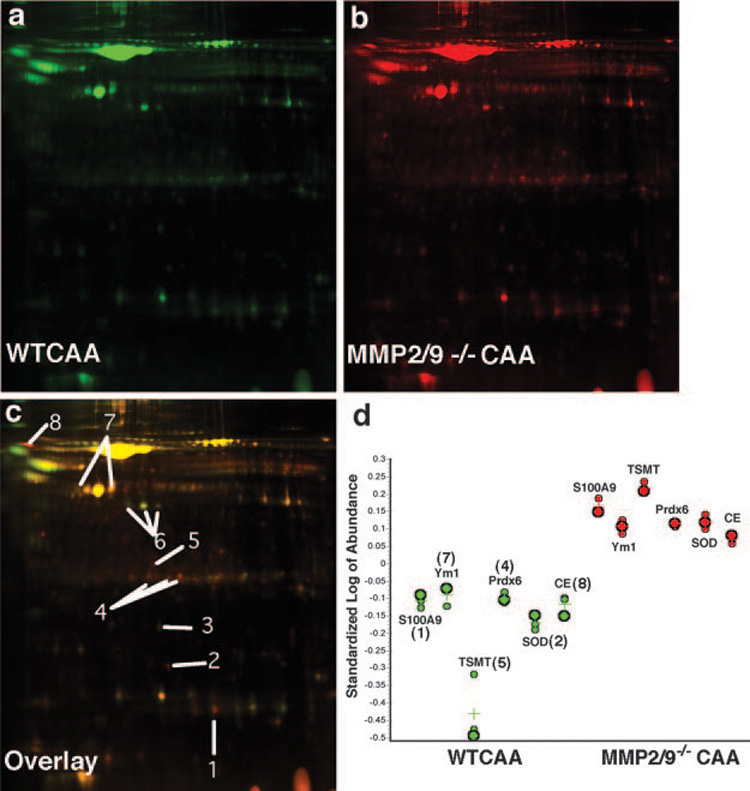
a, Two-dimensional gel electrophoresis of BALF from CAA-challenged WT (green) and (b) MMP 2/9−/− (red) reveals differential processing of several proteins (n = 3 per treatment). c, Overlaying green and red images highlights differences between WT and MMP2/9−/−. Yellow indicates no change, red spots were more abundant in KO, and green spots were more abundant in WT. Select identified proteins are labeled 1 through 8 and listed in Table II. These protein profiles are representative of those of three animals in each group. d, Quantification of proteins. Spot volumes were standardized and log transformed using DeCyder software, Biological Variation Analysis. WT (green) and MMP2/9−/− abundance values are shown for selected proteins; each point represents the spot volume from a pooled sample, and crosses represent the mean value (n = 3).
Table II.
Identification of selected proteins uniquely altered in BAL of MMP2/9null mice and their putative functionsa
| Molecular Mass | Abundance | |||||
|---|---|---|---|---|---|---|
| Spot No. | Protein | Alternative Name | Expression/Function | (kDa) | Ratio | t Test, p Value |
| 1 | Calgranulin B | S100A9, MRP14 | Inflammatory cell/chemotaxis | 13.5 | 1.87 | <0.001 |
| 2 | Superoxide dismutase | CuZn SOD | Alveolar macrophages/antioxidant | 32 | 1.96 | <0.0001 |
| 3 | CXCL15 | lungkine | Lung epithelial/neutrophil chemotaxis | 19.0 | −1.33 | <0.01 |
| 4.1 | Cys-Peroxiredoxin | prdx6, ADF | Macrophages/antioxidant enzyme | 24.8 | 1.46 | <0.001 |
| 4.2 | 1.18 | <0.05 | ||||
| 4.3 | 1.58 | <0.0001 | ||||
| 5 | Thio-e S Methylrans | S-adenosyl-L-met | Lung/selenium metabolism | 29.4 | 4.33 | <0.001 |
| 6.1 | Ym2 | Chitinase-3-like protein 4 | Epithelium/remodeling | 45.0 | −2.16 | <0.00001 |
| 6.2 | −2.16 | <0.00001 | ||||
| 6.3 | −1.66 | <0.0001 | ||||
| 7.1 | Ym1 | ECF-L | Macs and epithelium/chemotaxis | 44 | 1.54 | <0.001 |
| 7.2 | −1.15 | >0.05 | ||||
| 8 | Liver carboxylesterase | PES-N | Extracell metabo of lung surfactant | 61 | 1.53 | <0.001 |
Abundance ratio is the relative spot volume from WT to null mice, where positive values >1.5 were considered significantly higher in MMP2/9−/− BAL relative to WT, and negative values <1.5 were significantly lower in MMP2/9−/−BAL compared to WT. For proteins with multiple spots, abundance values listed from top to bottom (e.g., 4.1– 4.3) correspond to spot locations from left to right in Fig. 3.
Candidate MMP2 and MMP9 substrates are cleaved in vivo and in vitro
We next verified that the identified proteins from 2D gel electrophoresis contain peptide motifs that are known to favor cleavage by MMP2 or MMP9 based on published, predicted cleavage motifs identified by Turk et al. (34). Of the proteins that were identified in our proteomics studies, we focused on three: Ym1, S100A8, and S100A9, because of their putative chemotactic properties (35–37). For each protein, we found several potential cleavage sites for both MMP2 and MMP9 (Table III).
Table III.
Amino acid sequences for select identified proteinsa
| Protein | Sequence | |
|---|---|---|
| Ym1 | 1 | MAKLILVTGL AILLNVQLGS SYQLMCYYTS WAKDRPIEGS |
| 41 | FKPGNIDPCL CTHLIYAFAG MQNNEITYTH EQDLRDYEAL | |
| 81 | NGLKDKNTEL KTLLAIGGWK FGPASFSAMV STPQNRQIFI | |
| 121 | QSVIRFLRQY NFDGLNLDWQ YPGSRGSPPK DKHLFSVLVK | |
| 161 | EMRKAFEEES VEKDIPRLLL TSTGAGIIDV IKSGYKIPEL | |
| 201 | SQSIDYIQVM TYDLHDPKDG YTGENSPLYK SPYDIGKSAD | |
| 241 | LNVDSIISYW KDHGAASEKL IVGFPAYGHT FILSDPSKTG | |
| 281 | IGAPTISTGP PGKYTDESGLLAYYEVCTFL NEGATEVWDA | |
| 321 | PQEVPYA→YQG NEWVGYDNVR SFKLKAQWLK NNLGGAVVW | |
| 361 | PLDMDDFSGS FCHQRHFPLT STLKGDLNIH SASCKGPY | |
| S100A8 | 1 | MPSELEKALS NLIDVYHNYS N→IQGNHHALY KNDFKKMVTT |
| 41 | ECPQFVQNIN IENLFRELDI NSDNAINFEE FLAMVIKVGV | |
| 81 | ASHKDSHKE | |
| S100A9 | 1 | MANKAPSQME RSITTIIDTF HQYSRKEGHP DTLSKKEFRQ |
| 41 | MVEAQLATFM KKEKRNEALI NDIMEDLDTN QDNQLSFEEC | |
| 81 | MMLMAKLIFA CHEKLHENNP RGHGHSHGKG CGK | |
Pairs of amino acids, underlined, are predicted cleavage sites for MMP2, whereas underlined pairs that are bold and italicized are predicted cleavage sites for MMP9. We identified the actual cleavage site (→) using N-terminal sequencing.
Ym1 is an enzymatically inactive member of the chitinase family of proteins that has eosinophil chemoattractant properties (38, 39). Immunohistochemistry confirmed the presence of Ym1 in macrophages of saline-challenged mice and in macrophages and airway epithelium of allergen-challenged animals (Fig. 5a). Analysis of BALF of WT and MMP2/9−/− mice confirmed the presence of Ym1, but only in allergen-challenged animals (Fig. 5b). The BAL of WT CAA-challenged (asthmatic) mice showed multiple cleaved fragments of the Ym1 protein that are either completely absent or are present in small amounts in the BAL of asthmatic MMP2/9−/− mice (Fig. 5b). Intriguingly, using a different polyclonal Ab that only detects the full-length Ym1 protein, we found that much more Ym1 was in MMP2/9−/− BALF (data not shown), suggesting that these MMPs were required for the further processing and or clearance of Ym1. Silver staining of purified Ym1 after incubation with recombinant MMP2 and MMP9 confirmed that Ym1 is indeed an MMP substrate, with both enzymes yielding a 37-kDa peptide and an additional peptide of ~27 kDa (Fig. 5c). Furthermore, MMP9, but not MMP2, also cleaved Ym1 to produce an ~8-kDa peptide (Fig. 5c, asterisk). Interestingly, sequencing the 10 aa at the N terminus of the 8-kDa protein that was cleaved by MMP9 matched amino acids: YQGNEWVGYDN, a putative cleavage site of MMP2 (Table III). Sequencing the two larger proteins of 27 and 37 kDa, yielded YQLMCYYTSW at the predicted cleavage site for both MMP2 and MMP9 (Table III). These findings confirm that Ym1 is a substrate of MMP2 and MMP9 and probably additional proteinases.
FIGURE 5. Ym1 (eosinophil chemotactic factor; ECF-L) in allergic lung inflammation.
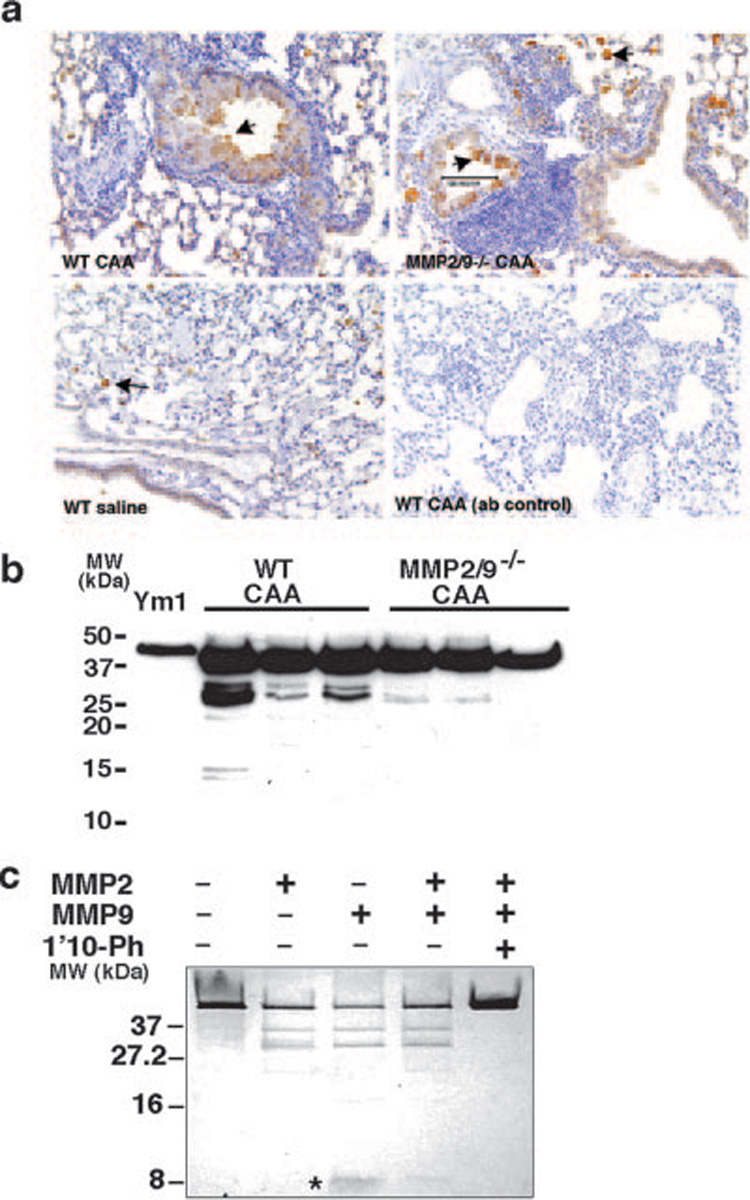
a, Expression of Ym1 in lung during allergic inflammation in vitro was detected by immunohistochemistry. Ym1 was up-regulated in the macrophages and epithelial cells in allergen-challenged mice. b, Western blot analysis of Ym1 in BAL of three representative WT and MMP2/9−/− mice showed increase in Ym1 in the BAL of MMP2/9−/− allergen-challenged mice. c, Purified Ym1 protein (5 µg) was cleaved by 100 ng of activated MMP2, MMP9, and their combination to yield peptides of ~37 and 27 kDa resolved on 16.5% Tricine gel and detected by sliver stain. MMP9, but not MMP2, resulted in formation of an ~8-kDa peptide (*). Data are representative of three independent experiments.
We next examined S100A8 and S100A9, two neutrophil and macrophage-specific, proinflammatory proteins (37, 40, 41) that were also processed differentially by MMP2 and MMP9 as suggested by 2D-DIGE analysis of BALF (Fig. 4). Immunohistochemical analysis of lung sections confirmed that S100A8 was present in the neutrophils of both WT and MMP2/9−/− CAA-challenged mice (Fig. 6a; arrows). Western blot analysis of the BALF protein from CAA-challenged mice identified several high molecular mass species representing mul-timers of the native 10-kDa protein; cleavage products of smaller molecular mass could not be distinguished with this technique most likely because S100A8 and S100A9 exist in hetero and homodimers in vivo (Fig. 6b). We therefore performed in vitro cleavage assays using MMP2 and MMP9 and purified S100 proteins separated on Tricine gels, which are capable of much finer resolution of variation in molecular mass. This analysis revealed that incubation of S100A8 and S100A9 with MMP2 slightly reduced the size of both molecules, resulting in additional cleavage bands for S100A9 (Fig. 6c,* );MMP9 appeared to not affect either S100 protein. Collectively, these findings demonstrate that Ym1, S100A8, and S100A9 are up-regulated in allergic inflammation and are substrates for MMP2, MMP9, or both enzymes.
FIGURE 6. S100A8 and S100A9 proteins in allergic lung inflammation.
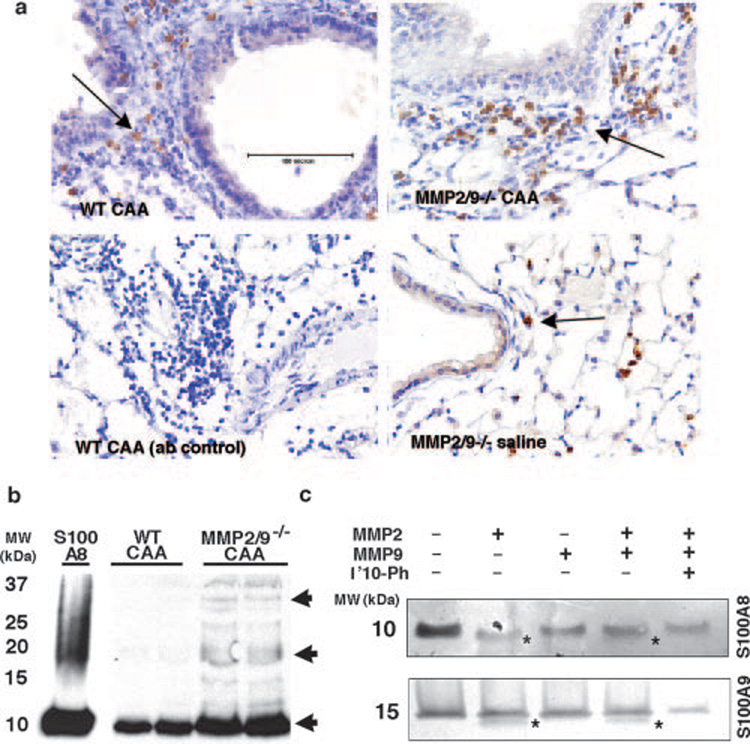
a, Expression of S100A8 in lung during allergic inflammation was determined by immunohistochemistry. S100A8 was expressed by neutrophils and macrophages under nonallergic conditions, and it was up-regulated during acute allergic inflammation. b, Western blot analysis of S100A8 in BAL of two representative WT and MMP2/9−/− mice showed several hetero-and homodimers of the S100 proteins. Arrow points to a dimerized form of the protein that was absent in the MMP2/9−/− CAA-challenged BAL. c, In vitro cleavage assay. Purified S100A8 and A100A9 (5 µg) were cleaved by 100 ng of activated MMP2 and MMP9 that resulted in cleaved S100A8 and S100A9 (*) in vitro.
Inhibition of S100 proteins alters migration of allergic inflammatory cells into the alveolar space
We next examined the functional role of S100A8 and S100A9 proteins, two of the newly identified substrates for MMP2 and MMP9, in allergic lung inflammation. Because deficiency in S100A8 results in embryonic lethality in mice (42), and as such we were not able to use a genetic approach to determine the function of this protein in asthma, we used function-blocking Abs to S100A8 and S100A9 in our asthma model (29). Mice that received S100A8 and S100A9 blocking Abs before intranasal challenge with allergen (CAA), showed a significant decrease in BAL cell numbers and differential analysis of these cells revealed that eosinophils were predominantly affected (Fig. 7, a and b). In contrast, there were no differences in airway hyperresponsiveness, BAL glycoprotein secretion, or IgE production (data not shown). S100A8 and S100A9 proteins were both weakly chemotactic in vitro, and S100A8, but not S100A9, cleaved by MMP2 and MMP9 altered its chemotactic activity (Fig. 7c).
FIGURE 7. In vivo function of S100 proteins in allergic lung inflammation.
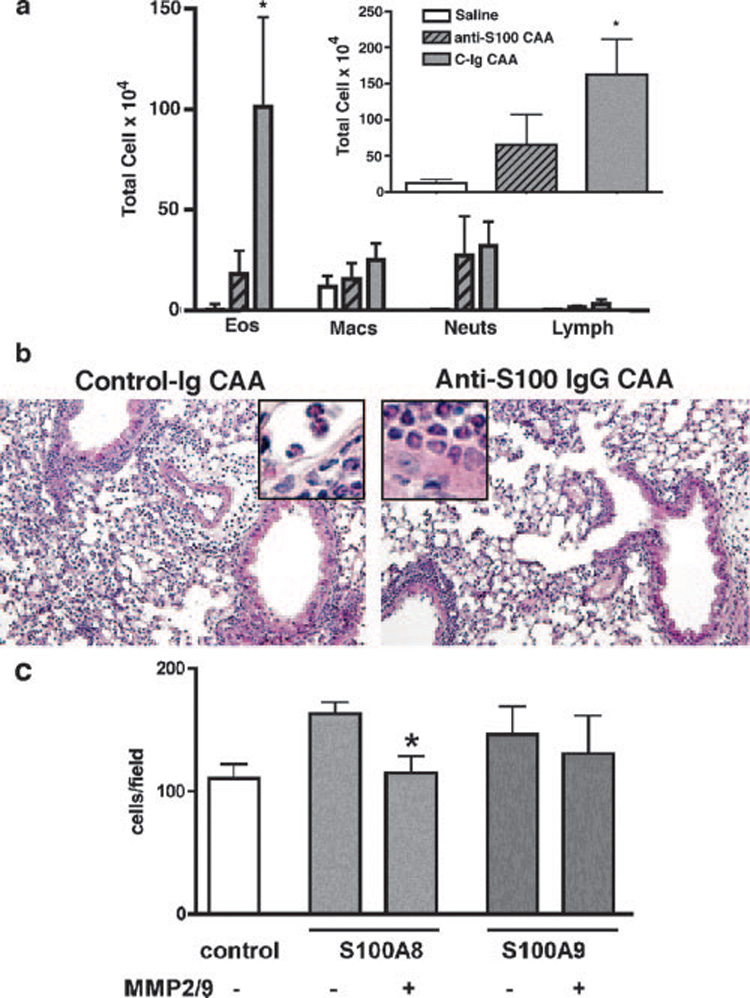
a, WT (C57BL/6) mice (n = 4) were challenged with CAA in the presence of function blocking Abs to S100A8, and S100A9 (anti-S100) or control Ab (C-Ig) and were compared with saline-treated mice. Differential cell counts in the BAL and total BAL cell count (inset) are shown.*, p < 0.005, n = 4 mice in each group. b, H&E stain of lung sections shows allergic inflammation in the lung parenchyma of mice that received anti-S100 blocking Abs. Insets, Eosinophils in the lung tissue. c, Th2 inflammatory cells from CAA-challenged mice were used to determine in vitro chemotactic activity of cleaved S100A8 and S100A9 proteins. Both S100 proteins (200 ng/ml) showed weak chemotactic activity in vitro, and S100A8, but not S100A9 showed reduced activity after in vitro cleavage with MMP2 and MMP9.*,p < 0.05. Data are representative of three separate experiments.
Discussion
In this study, we provide evidence that MMPs are present at multiple sites of inflammation in the lung parenchyma and identify several in vivo proteolytic substrates relevant to acute allergic airway inflammation. Before this study, knowledge of MMP substrate specificities, one of the most important aspects of MMP biology, was derived largely from in vitro biochemical analyses, with almost no studies assessing substrate specificities in vivo. Using mice deficient in MMPs and a well-characterized in vivo model of allergic inflammation, we determined several of the in vivo substrates of MMP2 and MMP9 as a means of explaining the biological role of these enzymes in allergic lung disease. We focused on MMP2 and MMP9 because their expression is controlled by Th2 cytokines, and they play a critical role in orchestrating the resolution of allergic lung inflammation in experimental asthma (9, 11, 13).
MMP2 and MMP9 have been implicated in many lung diseases, but the cells that express them have been unclear (43–46), information that may be important in determining enzyme function during inflammation. Previous reports on the expression of MMP9 protein using immunohistochemistry were conflicting, indicating its presence (44, 47) or absence (45) in airway epithelial cells. One possible reason for the disagreement is that MMPs can relocate from their site of production to distinct sites where Ab-based techniques detect them. In situ hybridization increases the spatiotemporal resolution of these gelatinases, allowing for more specific detection of MMP9 expression in the airway. Using this method, we show that in this model of allergic airway inflammation, there is no expression of MMP9 in airway epithelial cells.
Despite the lack of MMP2 and MMP9 mRNA expression in the airway epithelium, these MMPs are strongly present in the BALF of mice challenged with allergen and in humans with asthma indicating that their proteolytic action on proteins present in BAL may alter the behavior of allergic inflammatory cells, especially their egression through the airway. Using high-throughput proteomic analyses of BALF from allergen-challenged WT and MMP-deficient animals, we identified several substrates for MMP2 and MMP9 that play key roles in recruitment of inflammatory cells. Among identified molecules, three proteins of particular interest were S100A8, S100A9, and Ym1, all of which have known chemotactic activity in vitro (35–37, 40). N-terminal sequencing confirmed that these three proteins are substrates for either or both of the gelatinases. Using the proteomics approach described here, we have shown that even complex samples with thousands of proteins are amenable to identification and characterization of a small number of relevant molecules.
One of the possible mechanisms of action of MMPs in acute allergic inflammation is to modify the biochemical properties of proteins that are induced in response to inflammation. This is potentially a defense against the immunopathology that is the inevitable result of chronic inflammation. The first of the proteins we identified, Ym1 is a member of the chitinase family. Ym1 is secreted into the airway of mice that exhibit allergic lung inflammation and its expression is strongly up-regulated in murine primary macrophages in direct response to IL-4 and IL-13 in a STAT-6-dependent manner (35). Naturally occurring mutations in the active site of Ym1 render it devoid of chitinase activity, thus its role in allergic inflammation remained unclear (38). It has been speculated that Ym1 functions as an eosinophil chemotactic molecule and in support of this putative function, it contains a consensus CXC sequence near the NH2 terminus shared by many chemokines (38). We showed that Ym1 is cleaved in vitro by MMP2 and MMP9 and that the native molecule accumulates in the BAL of MMP2/9−/− mice. Current work is in progress to determine whether direct modification of this molecule could account for reduced eosinophils in BAL in MMP2/9−/− mice.
In addition to Ym1, we found two homologous myeloid-related proteins belonging to the S100 family, S100A8 and S100A9, small calcium-binding proteins that induce neutrophil chemotaxis and adhesion (37). Both were up-regulated in lung neutrophils and BAL of mice that exhibited the asthma phenotype and showed differential processing by MMP2 and MMP9 in vivo. S100A8 and S100A9 have been shown to form noncovalent homo- and heterodimers in biological fluids in a calcium-dependent manner, and their secretion from activated macrophages and neutrophils under inflammatory conditions in the lung has been reported. Most importantly, when we inhibited S100A8 and S100A9 proteins before intranasal challenge with allergen, mice retained all characteristics of the asthma phenotype, but had significant reduction in BALF eosinophils. These data are remarkable in that in vivo inhibition of S100A8 and S100A9 proteins produces a phenotype that is close to that of MMP2/9−/− mice in this mouse model of allergic inflammation, with the exception that the mechanism for reduced BALF eosinophilia is due to inhibition of novel chemotactic molecules distinct from the chemokines that are reduced in the BALF of MMP2/9−/− mice (11). In this study, we found that S100A8, but not S100A9 cleaved by MMP2, lost some of its weak chemotactic activity. In this case, the loss of chemotactic response following cleavage with MMP2 may be difficult to interpret since both intact and cleaved fragments compete for function. Also, since S100A8 and S100A9 proteins exist in dimer forms, single proteins may not represent true biological function.
MMPs may affect chemotaxis of inflammatory cells through multiple mechanisms. First, MMPs may inactivate a protein through cleavage or they may activate a protein by releasing an active fragment. These active/inactive proteins may form concentration gradients depending on the location of MMPS, but it is clear that the overall action of MMPs is to facilitate the egression of inflammatory cells into the airway lumen. In addition to the novel proteins we have identified as substrates, MMPs modify known chemokines. Using ELISA, we have previously shown that in the absence of MMP2 and MMP9, there is a decrease in the concentration of Th2 chemokines in the BALF. This finding may be perceived as inconsistent with the finding that chemokines are cleaved by MMPs (11). However, in this study, we show that biochemical detection of chemokines (i.e., via ELISA) and their functional activity in the intact or cleaved form (i.e., via chemotaxis assay) are quite distinct. Thus, while total concentration of chemokines by ELISA may be reduced, their cleaved forms may be more biologically active. Furthermore, chemokines in the BALF are bound to heparan sulfate proteoglycans, such as syndecans, and the action of MMP2 and MMP9 can modulate the function of chemokines through releasing the inhibited forms. In support of this idea, we have recently shown that syndecan-1 1null null mice have an exaggerated asthma phenotype, and shedding of syndecan-1regulates chemokine mobilization and transepithelial efflux of neutrophils in acute lung injury (48, 49). Although we can easily identify MMP-cleaved fragments of these chemokines in vitro, as yet, we have not been able to detect these cleaved fragments in the BAL of mice. Because these chemokines are present in such low concentrations in the BALF, neither the full-length chemokines nor their cleaved products are detectable using conventional methods, such as Western blot analysis. However, ongoing work in our laboratory using surface-enhanced laser desorption/ionization technology will allow us to detect and identify small (<3 kDa) fragments of protein, but these findings are beyond the scope of the current study.
More likely than the inevitable consequence of a gradient of chemotactic molecules, the process of chemotaxis appears to be dynamic and dependent on the activity of multiple proteolytic enzymes, not just the concentrations of chemokines. However, we have shown that optimal chemotaxis depends, in many cases, on appropriate cleavage of prochemotactic molecules (prochemokines) by regionally secreted MMPs. These findings raise fundamentally interesting questions regarding the fate of MMP-processed molecules and their precise role in inducing, or inhibiting chemotaxis. We have shown that MMP-mediated proteolysis of several molecules is required to induce their chemotactic activity. However, such processing may also affect the retention of such molecules in specific compartments such as the airway lumen or the ability of chemokines to ligate to their receptors.
In summary, we have identified several proteins that are novel, in vivo substrates for MMPs and these findings will aid in detection of new biochemical pathways regulating allergic lung disease. We expected to find significant substrate redundancy for these two structurally similar gelatinases, but our confirmatory in vitro cleavage assays showed that MMP2 and MMP9 also have significantly unique cleavage activity that most likely contributes to their complex biochemical behavior in vivo. Knowledge gained from our studies has not only identified mechanisms of allergic inflammatory cell recruitment, but has also provided a mechanism for the production of chemoattractants in the air spaces during allergic inflammation. Our findings potentially support the pharmacologic use of chemoattractant agents and other molecules to enhance inflammatory cell clearance as a means of relieving symptoms during allergic lung disease.
Acknowledgments
We acknowledge the help and support of Dr. Gabriela Garcia and the late Dr. Lili Feng. We would also like to thank Dr. Paul Foster for the gift of Ym1 Ab and protein.
Footnotes
This work was supported by grants from the National Institutes of Health (R01 HL072419 and K02 HL072062 (to F.K.), R01 HL069585 (to D.B.C.), T32 HL07747 (to K.J.G.), and P01 AI053194 (to Z.W.)), and by funds from the Sandler Family Sustaining Foundation (to Z.W. and D.B.C.). P.T. is a scholar from the Fonds de la Recherche en Santé du Québec.
Abbreviations used in this paper: MMP, matrix metalloproteinase; BAL, bronchoal-veolar lavage; WT, wild type; CAA, complete Aspergillus allergen; PAS, periodic acid-Schiff; 2D, two-dimensional; DIGE, differential in gel electrophoresis; KO, knockout; IS, internal standard; IN, intranasal; IPG, immobilized pH gradient.
Disclosures
The authors have no financial conflict of interest.
References
- 1.Barnes PJ. New drugs for asthma. Nat. Rev. Drug Discov. 2004;3:831–844. doi: 10.1038/nrd1524. [DOI] [PubMed] [Google Scholar]
- 2.Knight DA, Holgate ST. The airway epithelium: structural and functional properties in health and disease. Respirology. 2003;8:432–446. doi: 10.1046/j.1440-1843.2003.00493.x. [DOI] [PubMed] [Google Scholar]
- 3.Cohn L, Elias JA, Chupp GL. Asthma: mechanisms of disease persistence and progression. Annu. Rev. Immunol. 2004;22:789–815. doi: 10.1146/annurev.immunol.22.012703.104716. [DOI] [PubMed] [Google Scholar]
- 4.Prince JE, Kheradmand F, Corry DB. Immunologic lung disease. J. Allergy Clin. Immunol. 2003;111:S613–S623. doi: 10.1067/mai.2003.124. [DOI] [PubMed] [Google Scholar]
- 5.Corry DB, Kheradmand F. Biology and therapeutic potential of the interleukin-4/interleukin-13 signaling pathway in asthma. Am. J. Respir. Med. 2002;1:185–193. doi: 10.1007/BF03256608. [DOI] [PubMed] [Google Scholar]
- 6.Corry DB, Kheradmand F. The future of asthma therapy: integrating clinical and experimental studies. Immunol. Res. 2005;33:35–52. doi: 10.1385/IR:33:1:035. [DOI] [PubMed] [Google Scholar]
- 7.Gilroy DW, Lawrence T, Perretti M, Rossi AG. Inflammatory resolution: new opportunities for drug discovery. Nat. Rev. Drug Discov. 2004;3:401–416. doi: 10.1038/nrd1383. [DOI] [PubMed] [Google Scholar]
- 8.Gilroy DW, Newson J, Sawmynaden P, Willoughby DA, Croxtall JD. A novel role for phospholipase A2 isoforms in the checkpoint control of acute inflammation. FASEB J. 2004;18:489–498. doi: 10.1096/fj.03-0837com. [DOI] [PubMed] [Google Scholar]
- 9.Parks WC, Wilson CL, Lopez-Boado YS. Matrix metalloproteinases as modulators of inflammation and innate immunity. Nat. Rev. Immunol. 2004;4:617–629. doi: 10.1038/nri1418. [DOI] [PubMed] [Google Scholar]
- 10.Kheradmand F, Rishi K. The Role of Proteinases in Airway Remodeling. New York: Marcel Dekker; 2003. [Google Scholar]
- 11.Corry DB, Kiss A, Song LZ, Song L, Xu J, Lee SH, Werb Z, Kheradmand F. Overlapping and independent contributions of MMP2 and MMP9 to lung allergic inflammatory cell egression through decreased CC chemokines. FASEB J. 2004;18:995–997. doi: 10.1096/fj.03-1412fje. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.McMillan SJ, Kearley J, Campbell JD, Zhu XW, Larbi KY, Shipley JM, Senior RM, Nourshargh S, Lloyd CM. Matrix metalloproteinase-9 deficiency results in enhanced allergen-induced airway inflammation. J. Immunol. 2004;172:2586–2594. doi: 10.4049/jimmunol.172.4.2586. [DOI] [PubMed] [Google Scholar]
- 13.Corry DB, Rishi K, Kanellis J, Kiss A, Song Lz LZ, Xu J, Feng L, Werb Z, Kheradmand F. Decreased allergic lung inflammatory cell egression and increased susceptibility to asphyxiation in MMP2-deficiency. Nat. Immunol. 2002;3:347–353. doi: 10.1038/ni773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Overall CM, McQuibban GA, Clark-Lewis I. Discovery of chemokine substrates for matrix metalloproteinases by exosite scanning: a new tool for degradomics. Biol. Chem. 2002;383:1059–1066. doi: 10.1515/BC.2002.114. [DOI] [PubMed] [Google Scholar]
- 15.Lopez-Otin C, Overall CM. Protease degradomics: a new challenge for proteomics. Nat. Rev. Mol. Cell Biol. 2002;3:509–519. doi: 10.1038/nrm858. [DOI] [PubMed] [Google Scholar]
- 16.Ryu OH, Choi SJ, Firatli E, Choi SW, Hart PS, Shen RF, Wang G, Wu WW, Hart TC. Proteolysis of macrophage inflammatory protein-1α isoforms LD78β and LD78α by neutrophil-derived serine proteases. J. Biol. Chem. 2005;280:17415–17421. doi: 10.1074/jbc.M500340200. [DOI] [PubMed] [Google Scholar]
- 17.McQuibban GA, Gong JH, Tam EM, McCulloch CA, Clark-Lewis I, Overall CM. Inflammation dampened by gelatinase A cleavage of mono-cyte chemoattractant protein-3. Science. 2000;289:1202–1206. doi: 10.1126/science.289.5482.1202. [DOI] [PubMed] [Google Scholar]
- 18.McQuibban GA, Gong JH, Wong JP, Wallace JL, Clark-Lewis I, Overall CM. Matrix metalloproteinase processing of monocyte chemoattractant proteins generates CC chemokine receptor antagonists with anti-inflammatory properties in vivo. Blood. 2002;100:1160–1167. [PubMed] [Google Scholar]
- 19.Van Den Steen PE, Wuyts A, Husson SJ, Proost P, Van Damme J, Opdenakker G. Gelatinase B/MMP-9 and neutrophil collagenase/MMP-8 process the chemokines human GCP-2/CXCL6, ENA-78/CXCL5 and mouse GCP-2/LIX and modulate their physiological activities. Eur. J. Biochem. 2003;270:3739–3749. doi: 10.1046/j.1432-1033.2003.03760.x. [DOI] [PubMed] [Google Scholar]
- 20.Parks WC. Matrix metalloproteinases in repair. Wound Repair Regen. 1999;7:423–432. doi: 10.1046/j.1524-475x.1999.00423.x. [DOI] [PubMed] [Google Scholar]
- 21.Vu TH, Shipley JM, Bergers G, Berger JE, Helms JA, Hanahan D, Shapiro SD, Senior RM, Werb Z. MMP-9/gelatinase B is a key regulator of growth plate angiogenesis and apoptosis of hypertrophic chondrocytes. Cell. 1998;93:411–422. doi: 10.1016/s0092-8674(00)81169-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Itoh T, Ikeda T, Gomi H, Nakao S, Suzuki T, Itohara S. Unaltered secretion of β-amyloid precursor protein in gelatinase A (matrix metalloproteinase 2)-deficient mice. J. Biol. Chem. 1997;272:22389–22392. doi: 10.1074/jbc.272.36.22389. [DOI] [PubMed] [Google Scholar]
- 23.Kheradmand P, Rishi K, Werb Z. Signaling through the EGF receptor controls lung morphogenesis in part by regulating MT1-MMP-mediated activation of gelatinase A/MMP2. J. Cell Sci. 2002;115:839–848. doi: 10.1242/jcs.115.4.839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kheradmand F, Kiss A, Xu J, Lee SH, Kolattukudy PE, Corry DB. A protease-activated pathway underlying Th cell type 2 activation and allergic lung disease. J. Immunol. 2002;169:5904–5911. doi: 10.4049/jimmunol.169.10.5904. [DOI] [PubMed] [Google Scholar]
- 25.Corry DB, Grunig G, Hadeiba H, Kurup VP, Warnock ML, Sheppard D, Rennick DM, Locksley RM. Requirements for allergen- induced airway hyperreactivity in T and B cell-deficient mice. Mol. Med. 1998;4:344–355. [PMC free article] [PubMed] [Google Scholar]
- 26.Marouga R, David S, Hawkins E. The development of the DIGE system: 2D fluorescence difference gel analysis technology. Anal. Bioanal. Chem. 2005;382:669–678. doi: 10.1007/s00216-005-3126-3. [DOI] [PubMed] [Google Scholar]
- 27.Alban A, David SO, Bjorkesten L, Andersson C, Sloge E, Lewis S, Currie I. A novel experimental design for comparative two-dimensional gel analysis: two-dimensional difference gel electrophoresis incorporating a pooled internal standard. Proteomics. 2003;3:36–44. doi: 10.1002/pmic.200390006. [DOI] [PubMed] [Google Scholar]
- 28.Grumelli S, Corry D, Song L, Song L, Green L, Huh J, Hacken J, Espada R, Bag R, Lewis D, Kheradmand F. An immune basis for lung parenchymal destruction in chronic obstructive pulmonary disease and emphysema. PLoS. Med. 2004;1:74–83. doi: 10.1371/journal.pmed.0010008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Vandal K, Rouleau P, Boivin A, Ryckman C, Talbot M, Tessier PA. Blockade of S100A8 and S100A9 suppresses neutrophil migration in response to lipopolysaccharide. J. Immunol. 2003;171:2602–2609. doi: 10.4049/jimmunol.171.5.2602. [DOI] [PubMed] [Google Scholar]
- 30.Eustache F, Jouannet P, Auger J. Evaluation of flow cytometric methods to measure human sperm concentration. J. Androl. 2001;22:558–567. [PubMed] [Google Scholar]
- 31.Lee SH, Prince JE, Rais M, Kheradmand F, Shardonofsky F, Lu H, Beaudet AL, Smith CW, Soong L, Corry DB. Differential requirement for CD18 in T-helper effector homing. Nat. Med. 2003;9:1281–1286. doi: 10.1038/nm932. [DOI] [PubMed] [Google Scholar]
- 32.Baggiolini M. Chemokines in pathology and medicine. J. Intern. Med. 2001;250:91–104. doi: 10.1046/j.1365-2796.2001.00867.x. [DOI] [PubMed] [Google Scholar]
- 33.Moser B, Willimann K. Chemokines: role in inflammation and immune surveillance. Ann. Rheum. Dis. 2004;63:ii84–ii89. doi: 10.1136/ard.2004.028316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Turk BE, Cantley LC. Using peptide libraries to identify optimal cleavage motifs for proteolytic enzymes. Methods. 2004;32:398–405. doi: 10.1016/j.ymeth.2003.10.003. [DOI] [PubMed] [Google Scholar]
- 35.Welch JS, Escoubet-Lozach L, Sykes DB, Liddiard K, Greaves DR, Glass CK. TH2 cytokines and allergic challenge induce Ym1 expression in macrophages by a STAT6-dependent mechanism. J. Biol. Chem. 2002;277:42821–42829. doi: 10.1074/jbc.M205873200. [DOI] [PubMed] [Google Scholar]
- 36.Webb DC, McKenzie AN, Foster PS. Expression of the Ym2 lectin-binding protein is dependent on interleukin (IL)-4 and IL-13 signal transduction: identification of a novel allergy-associated protein. J. Biol. Chem. 2001;276:41969–41976. doi: 10.1074/jbc.M106223200. [DOI] [PubMed] [Google Scholar]
- 37.Ryckman C, Vandal K, Rouleau P, Talbot M, Tessier PA. Proin-flammatory activities of S100: proteins S100A8, S100A9, and S100A8/A9 induce neutrophil chemotaxis and adhesion. J. Immunol. 2003;170:3233–3242. doi: 10.4049/jimmunol.170.6.3233. [DOI] [PubMed] [Google Scholar]
- 38.Hung SI, Chang AC, Kato I, Chang NC. Transient expression of Ym1, a heparin-binding lectin, during developmental hematopoiesis and inflammation. J. Leukocyte Biol. 2002;72:72–82. [PubMed] [Google Scholar]
- 39.Tsai ML, Liaw SH, Chang NC. The crystal structure of Ym1 at 1.31 A resolution. J. Struct. Biol. 2004;148:290–296. doi: 10.1016/j.jsb.2004.07.002. [DOI] [PubMed] [Google Scholar]
- 40.Yang Z, Tao T, Raftery MJ, Youssef P, Di Girolamo N, Geczy CL. Proinflammatory properties of the human S100 protein S100A12. J. Leukocyte Biol. 2001;69:986–994. [PubMed] [Google Scholar]
- 41.Schmidt S, Linington C, Zipp F, Sotgiu S, de Waal Malefytqq R, Wekerle H, Hohlfeld R. Multiple sclerosis: comparison of the human T-cell response to S100 βand myelin basic protein reveals parallels to rat experimental autoimmune panencephalitis. Brain. 1997;120:1437–1445. doi: 10.1093/brain/120.8.1437. [DOI] [PubMed] [Google Scholar]
- 42.Passey RJ, Williams E, Lichanska AM, Wells C, Hu SP, Geczy CL, Little MH, Hume DA. A null mutation in the inflammation-associated S100 protein S100A8 causes early resorption of the mouse embryo. J. Immunol. 1999;163:2209–2216. [PubMed] [Google Scholar]
- 43.Hayashi T, Stetler-Stevenson WG, Fleming MV, Fishback N, Koss MN, Liotta LA, Ferrans VJ, Travis WD. Immunohistochemical study of metalloproteinases and their tissue inhibitors in the lungs of patients with diffuse alveolar damage and idiopathic pulmonary fibrosis. Am. J. Pathol. 1996;149:1241–1256. [PMC free article] [PubMed] [Google Scholar]
- 44.Betsuyaku T, Fukuda Y, Parks WC, Shipley JM, Senior RM. Gelatinase B is required for alveolar bronchiolization after intratracheal bleomycin. cin. Am. J. Pathol. 2000;157:525–535. doi: 10.1016/S0002-9440(10)64563-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Dunsmore SE, Saarialho-Kere UK, Roby JD, Wilson CL, Matrisian LM, Welgus HG, Parks WC. Matrilysin expression and function in airway epithelium. J. Clin. Invest. 1998;102:1321–1331. doi: 10.1172/JCI1516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Atkinson JJ, Senior RM. Matrix metalloproteinase-9 in lung remodeling. Am. J. Respir. Cell Mol. Biol. 2003;28:12–24. doi: 10.1165/rcmb.2002-0166TR. [DOI] [PubMed] [Google Scholar]
- 47.Yao PM, Delclaux C, d’Ortho MP, Maitre B, Harf A, Lafuma C. Cell-matrix interactions modulate 92-kD gelatinase expression by human bronchial epithelial cells. Am. J. Respir. Cell Mol. Biol. 1998;18:813–822. doi: 10.1165/ajrcmb.18.6.2984. [DOI] [PubMed] [Google Scholar]
- 48.Xu J, Park PW, Kheradmand F, Corry DB. Endogenous attenuation of allergic lung inflammation by syndecan-1. J. Immunol. 2005;174:5758–5765. doi: 10.4049/jimmunol.174.9.5758. [DOI] [PubMed] [Google Scholar]
- 49.Li Q, Park PW, Wilson CL, Parks WC. Matrilysin shedding of syndecan-1 regulates chemokine mobilization and transepithelial efflux of neutrophils in acute lung injury. Cell. 2002;111:635–646. doi: 10.1016/s0092-8674(02)01079-6. [DOI] [PubMed] [Google Scholar]


