Abstract
Methylation of lysine 27 on histone H3 (H3K27) by the EZH2 complex is an epigenetic mark that mediates gene silencing. EZH2 is overexpressed in many cancers and correlates with poor prognosis in both breast and prostate cancers. However, the status of H3K27 methylation and its clinical implication in cancer patients have not been reported. We thus examined trimethylation of H3K27 (H3K27me3) by immunohistochemistry and its association with clinical variables and prognosis in breast, ovarian, and pancreatic cancers. We found that H3K27me3 expression was significantly lower in breast, ovarian and pancreatic cancers than in normal tissues (62% in breast cancer vs. 88% in normal breast tissue, P = 0.001; 38.4% in ovarian cancer vs. 83.3% in normal ovarian tissue, P < 0.05; and 26% in pancreatic cancer vs. 89% in normal pancreatic tissue, P < 0.001). H3K27me3 expression showed significant prognostic impact in breast, ovarian and pancreatic cancers in univariate survival analyses. In all three cancer types, patients with low expression of H3K27me3 had significantly shorter overall survival time when compared with those with high H3K27me3 expression. In a multivariate model, H3K27me3 expression was an independent prognostic value for overall survival in all three cancer types. These results suggest that H3K27me3 expression is a prognostic indicator for clinical outcome in patients with breast, ovarian, and pancreatic cancers.
Keywords: histone modification, cancer, prognosis
Introduction
Covalent histone modifications play an important role in regulating chromatin dynamics and function [1]. One such modification, methylation, occurs on both lysine and arginine residues. Methylation of lysine residues within the conserved N-terminal histone tail regulates transcriptional activity [2]. Histone methylation is mediated by methyltransferases that catalyze mono-, di-, or tri-methylation of specific lysine residues [3]. H3K27 methylation is a transcription-suppressive histone mark in chromatin with EZH2, a component of the Polycomb (PcG) complex, being the primary H3-K27 methyltransferase [4–7]. Recently, studies have shown that EZH2 is overexpressed in many cancers [8–11] and the expression level of EZH2 correlates with a worsened prognosis in both prostate [8] and breast cancers [10]. However, there is no report about changes of H3K27 methylation in cancers. Here, we show that H3K27me3 is lost in some breast, ovarian, and pancreatic cancers and loss of H3K27me3 is a predictor of poor outcome in cancer patients.
Materials and Methods
Patients
Paraffin-embedded primary tumors of 142 breast cancer patients were obtained from the Department of Pathology, Shanghai East Breast Disease Hospital, People's Republic of China. The median age was 51 yr (range, 26–87). Clinical follow-up data including overall survival were available for all cases. The median observation time for overall survival was 50 mo (range, 4–95 mo). In addition, 43 cases of normal tissue adjacent to breast cancer were obtained.
Tissue microarrays containing a total of 255 cases of ovarian cancer were provided by the Department of Pathology, The University of Texas M.D. Anderson Cancer Center with Institutional Review Board (IRB) approval. Clinical follow-up data were available for all cases. The median follow-up period was 35 mo (range, 1–182 mo). Six cases of normal ovarian tissue were also included.
Tissue microarrays containing 165 cases of invasive pancreatic adenocarcinoma were obtained from the Department of Pathology, The University of Texas M.D. Anderson Cancer Center and Department of Pathology, The Karmanos Cancer Institute and Harper University Hospital, Wayne State University, with IRB approval. Seventy-two cases of normal pancreatic epithelium adjacent to cancer were obtained. Clinical follow-up data were available for all cases. The median duration of follow-up time was 23.7 mo (range, 1–144 mo).
Immunohistochemical Staining
Samples were deparaffinized in xylene and rehydrated in a series of graded alcohols, and the antigen was retrieved in 0.01 M sodium-citrate buffer (pH 6.0) using a microwave oven. The sections were then treated with 1% hydrogen peroxide in methanol for 30 min to exhaust endogenous peroxidase activity. After 1 h preincubation in 10% normal horse serum to prevent unspecific staining, the samples were incubated with mouse anti-human H3K27me3 (#ab6002, Abcam, Inc., Cambridge, MA) (1:60) at 4°C overnight. The sections were thereafter treated with biotinylated horse anti-mouse immunoglobulin followed by incubations with avidin-biotin peroxidase complex solution for 1 h at room temperature. Color was developed with the 3-amino-9-ethylcarbazole solution. Counterstaining was carried out using Mayer's hematoxylin.
Evaluation of H3K27me3 Immunostaining
Two pathologists assessed the level of H3K27me3 staining independently and without knowledge of the clinicopathological variables. The tumor was considered positive for H3K27me3, if there was histological evidence of nuclear staining. Expression categories were based initially on percentage of positively stained cells, considering also the distribution plots, as well as number of cases and events when determining the final cutoff points between low and high expression (two categories). For each tumor type, the same cutoff point was used for analysis of associations and outcome; the median was used for all the tumor types (30% for breast and ovarian cancer, respectively; and 10% for pancreatic cancer). Thus, categories of high and low expression were consistently defined as groups with percentage of positive cells above or below/equal to the cutoff point used for each tumor.
Statistical Analysis
Statistical analysis was done using SPSS, version 10.0. Associations between the H3K27me3 expression and clinicopathological features were assessed using χ2 and Spearman tests. Univariate survival analysis was carried out according to Kaplan–Meier, whereas differences in survival curves were assessed with the log-rank test. Cox regression analysis was used for multivariate survival analyses. All P values were two sided and P < 0.05 was considered statistically significant.
Results
Immunohistochemical Analysis of H3K27me3 Expression in Human Cancer
H3K27me3 was detected in 88% (38/43) of normal breast tissue samples. In breast cancer expression of H3K27me3 was significantly lower (Figure 1). Of the breast cancer samples, 62% (88/142) showed positive staining for H3K27me3 (P = 0.001). The expression of H3K27me3 in normal pancreatic tissue and pancreatic cancer was 89% (64/72) and 26% (43/165), respectively (P < 0.001) (Figure 1). H3K27me3 was expressed in 83.3% (5/6) of normal ovarian tissue samples, whereas in ovarian cancer the expression of H3K27me3 was only 38.4% (98/255) (P < 0.05) (Figure 1).
Figure 1.
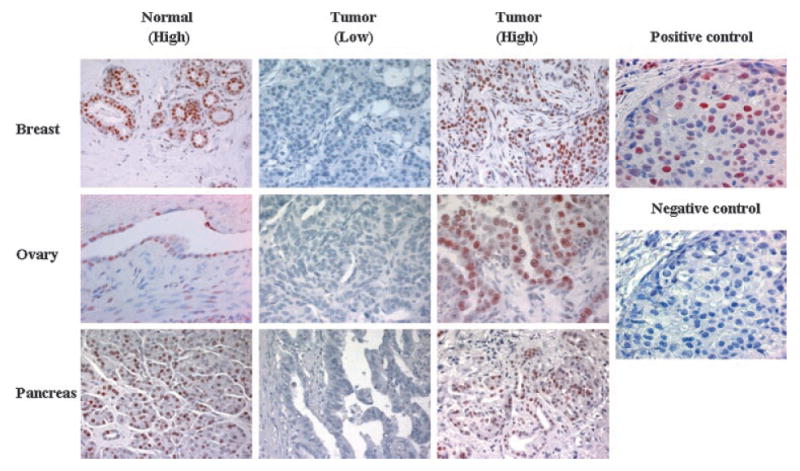
Immunohistochemical detection of H3K27me3 expression in normal and tumor tissues. Nuclear H3K27me3 immunostaining is detected in normal breast, ovarian, and pancreatic tissues. Low and high levels of nuclear H3K27me3 expression were shown for breast, ovarian, and pancreatic cancers. Positive (with H3K27me3 antibody) and negative control (without H3K27me3 antibody) staining from sequential slides of same case of breast cancer were shown in the right column. Original magnification, 400×.
Correlation Between H3K27me3 Expression and Prognostic Markers
Correlations between H3K27me3 expression and various prognostic markers are summarized in Table 1. In breast cancer, low H3K27me3 expression was associated with large tumor size (P < 0.001), ER negativity (P = 0.012), and lymph node positivity (P = 0.049), but there was no association with HER-2/Neu status (P = 0.13). In ovarian cancer, H3K27me3 expression was associated with high tumor grade (P = 0.031) and stage (P < 0.001), but not associated with histologic type (P = 0.124). In pancreatic cancer, H3K27me3 was associated with tumor grade (P = 0.016), but not associated with tumor size (P = 0.868) and lymph node status (P = 0.051). We did not find association between H3K27me3 and EZH2 expression in all three cancer types (P > 0.05).
Table 1.
Correlation of H3K27me3 Expression With Clinicopathological Variables
| H3K27me3 | ||||
|---|---|---|---|---|
| Variable | No. of cases | Low | High | P-value |
| Breast cancer | ||||
| Tumor size | ||||
| ≤2 cm | 61 | 14 | 47 | 0.001 |
| >2 cm | 81 | 40 | 41 | |
| Nodal status | ||||
| 0 | 80 | 25 | 55 | 0.049 |
| 1–4 | 28 | 12 | 16 | |
| >4 | 34 | 17 | 17 | |
| Her2/Neu status | ||||
| Negative | 85 | 27 | 58 | 0.13 |
| Positive | 49 | 22 | 27 | |
| ER status | ||||
| Negative | 39 | 20 | 19 | 0.012 |
| Positive | 43 | 10 | 33 | |
| EZH2 | ||||
| Low | 68 | 24 | 44 | 0.816 |
| High | 28 | 11 | 17 | |
| Ovarian cancer | ||||
| Histology | ||||
| Serous | 138 | 80 | 58 | 0.124 |
| Others | 117 | 77 | 40 | |
| Tumor grade | ||||
| Low | 28 | 12 | 16 | 0.031 |
| High | 227 | 145 | 82 | |
| Pathological stage | ||||
| Low | 37 | 13 | 24 | <0.001 |
| High | 218 | 144 | 74 | |
| EZH2 | ||||
| Low | 60 | 36 | 24 | 0.588 |
| High | 62 | 34 | 28 | |
| Pancreatic cancer | ||||
| Tumor size | ||||
| ≤3 cm | 61 | 46 | 15 | 0.868 |
| >3 cm | 97 | 72 | 25 | |
| Tumor grade | ||||
| Low | 111 | 76 | 35 | 0.016 |
| High | 51 | 44 | 7 | |
| Nodal status | ||||
| Negative | 50 | 32 | 18 | 0.051 |
| Positive | 112 | 88 | 24 | |
| EZH2 | ||||
| Low | 16 | 10 | 6 | 0.773 |
| High | 46 | 26 | 20 | |
H3K27me3 Expression and Patient Survival
The end point used in survival analyses was death as a result of disease, and other causes of death were censored at time of death. For breast cancer, 5-yr survival in cases with low H3K27me3 expression was 46%, compared with 72% among the rest (P = 0.005; Figure 2). In ovarian cancer, 5-yr survival was 27% and 47% (low and high expression, respectively) (P = 0.0028; Figure 2). Five-year survival regarding time to death of pancreatic cancer was 11% and 23%, for cases with low and high H3K27me3 expression, respectively (P < 0.001; Figure 2).
Figure 2.
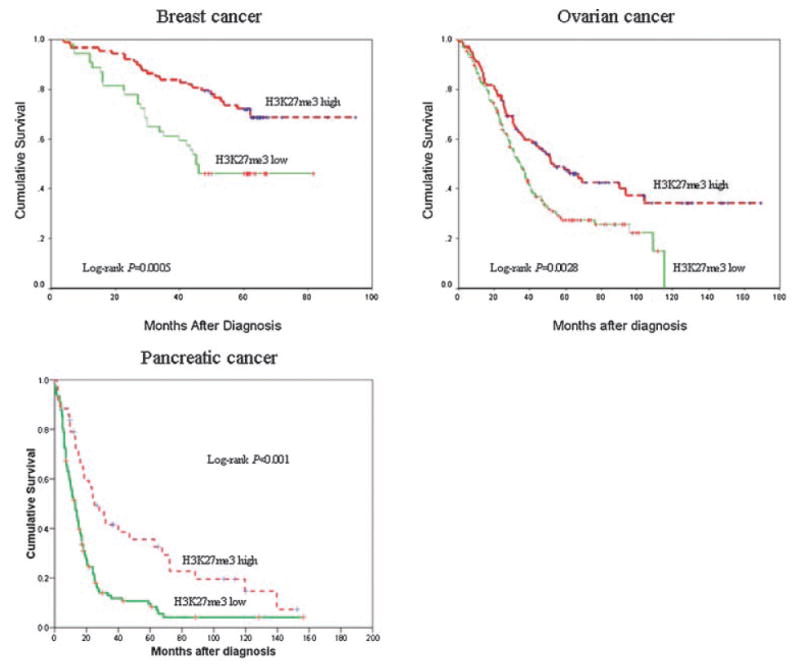
Kaplan–Meier analysis for overall survival according to H3K27me3 expression in patients with breast, ovarian, and pancreatic cancers.
Multivariate survival analysis was performed to evaluate the independence of H3K27me3 expression as a prognostic factor. In the breast cancer series, H3K27me3 expression had an independent prognostic influence (P = 0.045), when included together with multiple known prognostic variables such as tumor size, lymph node status, HER2/Neu status, and ER status (Table 2). In ovarian cancer, H3K27me3 expression independently predicted clinical outcome (P = 0.039), together with pathological stage (Table 2). H3K27me3 expression had a strong and independent prognostic influence (P = 0.001) in pancreatic cancer, together with tumor size and lymph node status (Table 2).
Table 2.
Multivariate Survival Analysis for Patients With Breast, Ovarian, and Pancreatic Cancer, Using Time to Death as End Point
| Variable | Hazard ratio | 95% CI | P-value |
|---|---|---|---|
| Breast cancer | |||
| Tumor size | 2.448 | 1.094–5.477 | 0.029 |
| Nodal status | 2.553 | 1.586–4.110 | <0.001 |
| HER-2/Neu status | 2.402 | 1.158–4.983 | 0.019 |
| ER status | 0.477 | 0.226–1.008 | 0.053 |
| H3K27me3 expression | 0.495 | 0.248–0.985 | 0.045 |
| Ovarian cancer | |||
| Histology | 0.839 | 0.605–1.165 | 0.294 |
| Tumor grade | 2.171 | 0.972–4.850 | 0.059 |
| Pathological stage | 2.843 | 1.411–5.728 | 0.003 |
| H3K27me3 expression | 0.699 | 0.497–0.981 | 0.039 |
| Pancreatic cancer | |||
| Tumor size | 1.652 | 1.140–2.394 | 0.008 |
| Tumor grade | 1.295 | 0.875–1.916 | 0.196 |
| Nodal status | 1.519 | 1.023–2.257 | 0.038 |
| H3K27me3 | 0.489 | 0.318–0.751 | 0.001 |
Discussion
Covalent modification of histones includes acetylation of lysines, methylation of lysines and arginines, phosphorylations of serines and threonines, ADP-ribosylation of glutamic acids, and ubiquitination and sumolyation of lysine residues [12]. The combination of these covalent modifications gives rise to what is known as the “histone code” [1]. Histone modifications appear to occur in specific patterns during neoplastic transformation. For example, it has been shown that the loss of monoacetylation at Lys16 and trimethylation at Lys20 of histone H4 is a common hallmark of human cancer [13]. Recently it has also been reported that histone modification patterns detected by immunohistochemistry, such as histone acetylation and dimethylation of H3 and H4 are predictive of clinical outcome in prostate cancer [14].
PcG proteins mediate heritable transcriptional silencing by generating and recognizing covalent histone modifications. One conserved PcG complex, PRC2, is composed of several proteins including the histone methyltransferase EZH2, the WD-repeat protein EED, and the Zinc-finger protein SUZ12. EZH2 methylates histone H3 on lysine 27[4–7]. Overexpression of EZH2 is associated with progression of prostate cancer [8] and aggressiveness of breast cancer [10].
We compared H3K27me3 expression in human breast, ovarian, and pancteatic cancers with that in corresponding normal tissue. We found the tumor samples had less trimethylated H3K27 than did normal tissue. We also found that there was no association between EZH2 and H3K27me3 expression in all three cancer types. The mechanisms underlying the loss of H3K27me3 in tumors are not known. Previous study from other group showed that increased EZH2 expression in normal prostatic epithelial cells by cDNA transfection leads to over-methylation of target gene chromatin and inhibition of target gene expression [15]. The result is from cell culture studies using normal cell, whether it reflects conditions of overexpression of EZH2 in cancer patients need to be further investigated. Another study indicated that overexpression of EZH2 promotes formation of different PRC complex, PRC4, and exhibits differential histone substrate specificities [16]. It is possible that loss of H3K27me3 in tumors is related to this new PRC complex formation. Given that EZH2 methyltransferase activity requires its association with other PRC subunits, it has been proposed that overexpression of EZH2 in tumors may result in the disruption of the integrity of the PRC complexes or may form new PRC complexes [17,18], therefore the methyltransferase activity or specificity toward H3K27 may be reduced. Another possibility is that loss of H3K27me3 in tumors may relate to the protein modification in components of PRC complexes [19].
The major finding of this study is the striking correlation between low H3K27me3 expression and poor prognosis in patients with breast, ovarian, and pancreatic cancers. Univariate survival analysis showed that, in all the three cancer types, patients whose cancers exhibited low expression of H3K27me3 had significantly shorter overall survival time when compared with those whose cancer had high H3K27me3 expression. When the Cox regression model was constructed for the entire series, low H3K27me3 expression remained an independent predictive factor of death in all the three cancer types. To our knowledge, our data presented here demonstrate for the first time a direct association of expression of H3K27me3 with clinical outcome in human cancers. As H3K27me3 serves as an epigenetic mark mediating silencing and represses target gene expression [4–7], loss of H3K27me3 expression may result in derepression of these silenced genes, such as some oncogenes; therefore, contributing to tumor progression. In a study using ovarian cancer cell lines, Abbosh et al. [20] found that dominant-negative histone H3 lysine 27 mutant can indeed derepress silenced genes in cancer cells.
In a recent study, Seligson et al. [14] reported that global histone modification patterns predict risk of tumor recurrence in low grade prostate cancer. However, variability in the levels of any single histone modification was not sufficient to predict outcome [14]. Interestingly, our study indicates that H3K27me3 expression, as a single histone modification, could serve as a prognostic factor for breast, ovarian, and pancreatic cancers.
In conclusion, we have shown in the present study that H3K27me3 is a novel prognostic marker in patients with breast, ovarian, and pancreatic cancers. The prognostic value of this marker is independent of conventional clinicopathological parameters in all the three cancer types. Additional studies should investigate the question as to whether H3K27me3 has a similar impact on prognosis in other forms of human cancer.
Acknowledgments
This work was supported by NIH grants RO1 CA109311, PO1 CA099031, NIH SPORE grant in Breast Cancer CA116199, NIH SPORE grant in Ovarian Cancer P50 CA83639, NIH SPORE grant in Pancreatic Cancer P20 CA101936, National Breast Cancer Foundation, Inc., Breast Cancer Research Foundation, and M.D. Anderson Cancer Center Support Grant CA16672. We would like to thank Drs. Jeng C. Cheng and Stephanie A. Miller for editing the manuscript.
References
- 1.Strahl BD, Allis CD. The language of covalent histone modifications. Nature. 2000;403:41–45. doi: 10.1038/47412. [DOI] [PubMed] [Google Scholar]
- 2.Kouzarides T. Histone methylation in transcriptional control. Curr Opin Genet Dev. 2002;12:198–209. doi: 10.1016/s0959-437x(02)00287-3. [DOI] [PubMed] [Google Scholar]
- 3.Bannister AJ, Schneider R, Kouzarides T. Histone methylation: Dynamic or static? Cell. 2002;109:801–806. doi: 10.1016/s0092-8674(02)00798-5. [DOI] [PubMed] [Google Scholar]
- 4.Cao R, Wang L, Wang H, et al. Role of histone H3 lysine 27 methylation in Polycomb-group silencing. Science. 2002;298:1039–1043. doi: 10.1126/science.1076997. [DOI] [PubMed] [Google Scholar]
- 5.Czermin B, Melfi R, McCabe D, Seitz V, Imhof A, Pirrotta V. Drosophila enhancer of Zeste/ESC complexes have a histone H3 methyltransferase activity that marks chromosomal Polycomb sites. Cell. 2002;111:185–196. doi: 10.1016/s0092-8674(02)00975-3. [DOI] [PubMed] [Google Scholar]
- 6.Muller J, Hart CM, Francis NJ, et al. Histone methyltransferase activity of a Drosophila Polycomb group repressor complex. Cell. 2002;111:197–208. doi: 10.1016/s0092-8674(02)00976-5. [DOI] [PubMed] [Google Scholar]
- 7.Kuzmichev A, Nishioka K, Erdjument-Bromage H, Tempst P, Reinberg D. Histone methyltransferase activity associated with a human multiprotein complex containing the Enhancer of Zeste protein. Genes Dev. 2002;16:2893–2905. doi: 10.1101/gad.1035902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Varambally S, Dhanasekaran SM, Zhou M, et al. The polycomb group protein EZH2 is involved in progression of prostate cancer. Nature. 2002;419:624–629. doi: 10.1038/nature01075. [DOI] [PubMed] [Google Scholar]
- 9.Bracken AP, Pasini D, Capra M, Prosperini E, Colli E, Helin K. EZH2 is downstream of the pRB-E2F pathway, essential for proliferation and amplified in cancer. EMBO J. 2003;22:5323–5335. doi: 10.1093/emboj/cdg542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kleer CG, Cao Q, Varambally S, et al. EZH2 is a marker of aggressive breast cancer and promotes neoplastic transformation of breast epithelial cells. Proc Natl Acad Sci USA. 2003;100:11606–11611. doi: 10.1073/pnas.1933744100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Raaphorst FM, Meijer CJ, Fieret E, et al. Poorly differentiated breast carcinoma is associated with increased expression of the human polycomb group EZH2 gene. Neoplasia. 2003;5:481–488. doi: 10.1016/s1476-5586(03)80032-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lund AH, van Lohuizen M. Epigenetics and cancer. Genes Dev. 2004;18:2315–2335. doi: 10.1101/gad.1232504. [DOI] [PubMed] [Google Scholar]
- 13.Fraga MF, Ballestar E, Villar-Garea A, et al. Loss of acetylation at Lys16 and trimethylation at Lys20 of histone H4 is a common hallmark of human cancer. Nat Genet. 2005;37:391–400. doi: 10.1038/ng1531. [DOI] [PubMed] [Google Scholar]
- 14.Seligson DB, Horvath S, Shi T, et al. Global histone modification patterns predict risk of prostate cancer recurrence. Nature. 2005;435:1262–1266. doi: 10.1038/nature03672. [DOI] [PubMed] [Google Scholar]
- 15.Chen H, Tu SW, Hsieh JT. Down-regulation of human DAB2IP gene expression mediated by Polycomb Ezh2 complex and histone deacetylase in prostate cancer. J Biol Chem. 2005;280:22437–22444. doi: 10.1074/jbc.M501379200. [DOI] [PubMed] [Google Scholar]
- 16.Kuzmichev A, Margueron R, Vaquero A, et al. Composition and histone substrates of polycomb repressive group complexes change during cellular differentiation. Proc Natl Acad Sci USA. 2005;102:1859–1864. doi: 10.1073/pnas.0409875102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Cao R, Zhang Y. The functions of E(Z)/EZH2-mediated methylation of lysine 27 in histone H3. Curr Opin Genet Dev. 2004;14:155–164. doi: 10.1016/j.gde.2004.02.001. [DOI] [PubMed] [Google Scholar]
- 18.Kuzmichev A, Jenuwein T, Tempst P, Reinberg D. Different EZH2-containing complexes target methylation of histone H1 or nucleosomal histone H3. Mol Cell. 2004;14:183–193. doi: 10.1016/s1097-2765(04)00185-6. [DOI] [PubMed] [Google Scholar]
- 19.Cha TL, Zhou BP, Xia W, et al. Akt-mediated phosphorylation of EZH2 suppresses methylation of lysine 27 in histone H3. Science. 2005;310:306–310. doi: 10.1126/science.1118947. [DOI] [PubMed] [Google Scholar]
- 20.Abbosh PH, Montgomery JS, Starkey JA, et al. Dominant-negative histone H3 lysine 27 mutant derepresses silenced tumor suppressor genes and reverses the drug-resistant phenotype in cancer cells. Cancer Res. 2006;66:5582–5591. doi: 10.1158/0008-5472.CAN-05-3575. [DOI] [PubMed] [Google Scholar]


