Abstract
The leukocyte integrin, lymphocyte function-associated antigen 1 (LFA-1) (CD11a/CD18), mediates cell adhesion and signaling in inflammatory and immune responses. To support these functions, LFA-1 must convert from a resting to an activated state that avidly binds its ligands such as intercellular adhesion molecule 1 (ICAM-1). Biochemical and x-ray studies of the Mac-1 (CD11b/CD18) I domain suggest that integrin activation could involve a conformational change of the C-terminal α-helix. We report the use of NMR spectroscopy to identify CD11a I domain residues whose resonances are affected by binding to ICAM-1. We observed two distinct sites in the CD11a I domain that were affected. As expected from previous mutagenesis studies, a cluster of residues localized around the metal ion-dependent adhesion site (MIDAS) was severely perturbed on ICAM-1 binding. A second cluster of residues distal to the MIDAS that included the C-terminal α-helix of the CD11a I domain was also affected. Substitution of residues in the core of this second I domain site resulted in constitutively active LFA-1 binding to ICAM-1. Binding data indicates that none of the 20 substitution mutants we tested at this second site form an essential ICAM-1 binding interface. We also demonstrate that residues in the I domain linker sequences can regulate LFA-1 binding. These results indicate that LFA-1 binding to ICAM-1 is regulated by an I domain allosteric site (IDAS) and that this site is structurally linked to the MIDAS.
Lymphocyte function-associated antigen 1 (LFA-1) is a leukocyte integrin that supports inflammatory and immune responses by mediating cell adhesion, the trafficking of leukocytes, and the augmentation of signaling through the T cell receptor (1). This integrin consists of an α (CD11a) and β (CD18) chain and binds to the cell surface ligands intercellular adhesion molecule 1 (ICAM-1), ICAM-2, and ICAM-3. Mutational studies indicate that ICAM-1 interacts with LFA-1 through a module of approximately 200 residues designated the A or I domain that is located in the amino-terminal region of CD11a (2, 3). Integrin I domains are homologous to the A-domains present in von Willebrand factor, several collagen and complement proteins, and cartilage matrix protein (1).
There are nine integrin α chains that contain I domains including the leukocyte integrins LFA-1 and Mac-1 (CD11b/CD18). X-ray crystal structures of the I domains of CD11a (4, 5), CD11b (6–8), α2 (9), and α1 (10) have been solved, as have structures for the A1 and A3 domains of von Willebrand factor (11, 12). An A domain has also been predicted to occur in a conserved region of all integrin β chains (6). The N and C termini of the I domain are adjacent to each other in the x-ray structures and have been proposed in modeling studies (13) to be connected by short linker sequences to the rest of the α chain. A metal ion-dependent adhesion site (MIDAS) was identified in the structural studies that is located in the upper face of the I domain, most distal to the N and C termini (6).
The I domain plays an important role in integrin function. In vitro, the isolated CD11a I domain binds to ICAM-1 (14). Mutagenesis (15–19), epitope mapping (20), and structural (6) studies suggest that the ICAM-1 binding site is located on the upper face of the I domain. Mutational studies of ICAM-1 have found that E34 is an essential residue for binding to LFA-1 (21, 22). This supports the hypothesis that E34 coordinates the Mg+2 ion in the I domain. Models of the I domain/ICAM-1 complex have been proposed in which the relatively flat MIDAS of the I domain interacts with a complementary flat surface on ICAM-1 that surrounds E34 (17, 23).
The binding of LFA-1 to its ligands is stimulated by T-cell receptor engagement (24, 25), chemokines (26), certain CD18 monoclonal antibodies (mAbs) (27), or high concentrations of metal (28). The mechanism by which LFA-1 is converted to this active state may involve an increase in receptor clustering and avidity (29) or through a change in conformation resulting in increased affinity (4, 30). A conformational change in LFA-1 is supported by the ability of an antibody, mAb24, to bind specifically to an activation state (31).
There has been much controversy related to the potential changes in I domain conformation in forming an active binding state (32). Two different conformations (“open” and “closed” forms) of the C-terminal α-helix of the CD11b I domain have been observed in x-ray crystal structures (6–8, 32–34). In the open form, Mg2+ (6), or Mn2+ (34) in the MIDAS is ligated by a glutamate from a second I domain in the crystal. This indicates that conformation is not metal-dependent. A crystal contact is associated with a change in metal coordination and a shift in the position of the C-terminal α-helix that exposes several buried hydrophobic residues. Although the biological relevance of these changes has been questioned (8), it was hypothesized that this crystal contact mimics the integrin-ligand interaction and that this conformation is similar to the active state of the I domain (6, 33). Mutagenesis (34) and epitope mapping (35) studies of CD11b agree with the x-ray data and demonstrate that a conformational change in this I domain is physiologically relevant. Oxvig et al. (35) have shown that there is a link between the conformational change near the MIDAS and the lower face of the I domain. X-ray studies of CD11a (5) failed to reveal either structural changes in the MIDAS or conformational changes of buried hydrophobic residues. However, very different positions of α-helix 7 were observed in the metal-free (4) and Mn+2 bound (5) structures, suggesting that a conformational change could occur in this region.
A recent study has shown that a small molecule, lovastatin, can bind to the CD11a I domain and act as an inhibitor of ICAM-1 binding. Lovastatin was found to bind in a cleft adjacent to α-helix 7 of the I domain in a site distant from the MIDAS. No structural changes were observed for residues of the MIDAS, prompting the authors to conclude that lovastatin inhibits ICAM-1 binding by an indirect mechanism (36).
Here we report direct biophysical evidence that characterizes the ICAM-1 binding site on CD11a. Two-dimensional NMR studies demonstrate that ICAM-1 alters the chemical environments of two regions of the I domain, the MIDAS and a second site that we have termed the I domain allosteric site (IDAS). These results guided the selection of mutations to further demonstrate that residues in the IDAS play a functional role in ICAM-1 binding.
Materials and Methods
Molecular Biology.
The pET15b plasmids containing sequence encoding residues 127–309 or 127–307 of human CD11a were prepared by PCR amplification by using a cDNA template. Twenty-two CD11a I domain mutations were generated by using Stratagene's QuikChange Site-Directed Mutagenesis Kit, and following the manufacturer's protocols. Full-length CD11a template was used for COS cell studies and the CD11a I domain template for NMR studies.
COS Cell Transfections.
COS cells were seeded at 1.6 × 106 cells per 10-cm plate in DMEM + 10% FBS, grown for 18–24 h and transfected with CD18/pDC1 and CD11a/pDC1 (either wild-type or mutant). Transfectants were grown for 2 days in DMEM + 10% FBS, replated at a 1:2 dilution, and resuspended in adhesion buffer (RPMI + 5% heat-inactivated FBS) for the adhesion assay and FACS staining.
Adhesion Assay.
Adhesion assays were performed in 96-well Easy Wash plates (Corning) by using a modified procedure (26). Each well was coated with 50 μl of ICAM-1/Fc (5 μg/ml) in coating buffer (50 mM bicarbonate buffer, pH 9.6) overnight at 4°C. Some wells were coated with capture antibodies TS1/22 and TS1/18, to quantitate 100% input cell binding, or coating buffer alone to determine background binding. Plates were blocked with 1% BSA in PBS. COS cell transfectants (7.5 × 104 cells) were then added in triplicate with or without 20 μg/ml of blocking CD11a antibody (TS1/22) or isotype-matched control antibody (1B7). Plates (final volume 300 μl/well) were incubated at 37°C for 15–20 min. For mAb 240Q stimulation, transfectants were incubated with 30 μg/ml for 5 min before adding cells to the plates. Adherent cells were fixed by the addition of 50 μl of a 14% glutaraldehyde solution in PBS. Plates were washed with water, stained with 100 μl/well 0.5% crystal violet (Sigma) solution. Three hundred microliters per well of 70% ethanol was added, and adherent cells were quantitated by determining absorbance at 570 nm. Percentage of cell binding was determined by using the mean values for each triplicate in a given assay and the formula: [(A570 (binding to ICAM-1) − A570 (binding to BSA))/A570 (binding to CD11a + CD18 mAb)] × 100. To normalize the data from different assays, the percentage of wild-type binding was determined by using the formula: [(percentage of cell binding for mutant CD11a transfectant)/(percentage of cell binding for wild-type CD11a transfectant from the same assay)] × 100. Every mutation was tested in at least three independent transfections and assays, and the mean of means from all assays are reported.
MAb and FACs Staining.
Transfected cells (1–5 × 105 cells) were stained with antibodies to CD18 (TS1/18, ATCC), CD11a (TS1/22, ATCC), or an activating antibody to CD18 (mAb 240Q, ICOS, Bothell, WA) and were detected with sheep anti-mouse Ig -FITC (SIGMA F-2883). Controls included unstained cells, cells stained with secondary antibody only, and cells stained with an isotype-matched control antibody (1B7).
Production and Purification of Recombinant Human ICAM-1 Domains 1 and 2.
A 7-ml immunoaffinity column was created by coupling 2 mg of 18E3D antibody (D.E.S., unpublished work) per 1 ml of activated CNBr-Sepharose. Culture supernatant (2.5 liters) from CHO cells secreting recombinant human ICAM-1 domains 1 and 2 was loaded at 4°C, was washed with 20 mM Tris and 150 mM NaCl at pH 7.5, and was eluted with 2 M KSCN pH 8.0. Protein was dialyzed into 10 mM sodium phosphate (pH 7.2) and was concentrated to 0.26 mM by using an extinction coefficient of 1.0 AU280/1.4 mg protein.
NMR Samples.
Uniformly 15N,2H-labeled CD11a I domain (127–307) and 15N,13C-labeled CD11a I domain (127–309) were prepared by growing the Escherichia coli strain BL21(DE3) overexpressing the I domain on M9 medium containing 15NH4Cl in 100% D2O or containing 15NH4Cl and [U-13C]-glucose in H2O. The I domain was purified by using nickel affinity resin. 15N,1H-labeled I domain (127–309) mutants were prepared in a similar manner. For the assignment of apo I domain chemical shifts, the NMR samples contained 0.8 mM protein, 1.2 mM MgCl2, and 100 mM sodium phosphate at pH 7.2 in H2O/D2O (9:1) or 99.9% D2O. For the ICAM-1 binding studies, samples were prepared with 0.2 mM 15N,2H-I domain, 0.3 mM MgCl2, 10 mM sodium phosphate at pH 7.2 in H2O/D2O (9:1) with 0, 0.04, 0.1, and 0.17 mM ICAM-1 domains 1 and 2. 0.2 mM 13C,15N-I domain was also prepared with or without 0.1 mM ICAM-1.
NMR Spectroscopy.
NMR spectra for the assignment of apo-CD11a I domain were acquired at 30°C on Bruker DRX500 or DRX600 NMR spectrometers. Backbone resonances were assigned by using the HNCA, HN(CO)CA, HN(CA)CB, HN(COCA)CB, HNCO, and HN(CA)CO experiments, using ref. 37. Sidechain assignments were made by using the HACACO, HBHA(CO)NH, 15N Edited TOCSY, and the HCCH-TOCSY experiments (37). Nuclear Overhauser effect measurements were obtained from 13C-resolved three-dimensional nuclear Overhauser effect (spectroscopy) (37, 38) experiments. 15N HSQC and 13C constant-time HSQC spectra were recorded for the LFA-1 +/− ICAM-1 at 25°C. Amide protons with 50% or less signal remaining in the presence of 0.04 mM ICAM-1 and methyl groups with 7% or less signal remaining in the presence of 0.1 mM ICAM-1 were categorized as undergoing a larger than average decrease in peak height, which is attributable to chemical exchange broadening.
Results
NMR spectroscopy has proven to be a robust technique for the identification of residues that are important for protein-protein (39, 40) and protein-small molecule (41) binding interactions. The basis of the technique relies on the sensitivity of the chemical shifts and line widths of 13C, 15N, and 1H nuclei to changes in their chemical environment. Using this technique, we have evaluated the interaction between LFA-1 and ICAM-1. Previous studies have shown that the I domain of CD11a binds to the amino-terminal extracellular domain (domain 1) of ICAM-1 (14, 21). For the NMR studies, a minimized complex was prepared by using the I domain of human CD11a and domains 1 and 2 of human ICAM-1 (21). To identify the amino acids whose resonances were perturbed by the addition of soluble ICAM-1, the backbone and side chain signals of the uncomplexed I domain were assigned by using standard heteronuclear NMR experiments (Materials and Methods). The secondary structure of this construct of the LFA-1 I domain was found to be indistinguishable from that of the x-ray crystal structures of the CD11a I domain (4, 5), using both nuclear Overhauser effects and backbone chemical shifts (42). The conformation we have observed in solution is consistent with the x-ray crystal form of the protein in which α-helix 7 packs against the β sheets of the protein (5). Furthermore, no nuclear Overhauser effects were observed to suggest a dimer interface in which the C terminus of one monomer packs into a cleft between α-helix 7 and the β sheets (4).
When soluble ICAM-1 was titrated into a 0.2 mM solution of the CD11a I domain and 0.3 mM MgCl2, the 15N and 1H signals of CD11a amino acids are broadened because of the formation of a more slowly tumbling complex and because of chemical exchange. As more ICAM-1 was titrated, the resonances of residues in the interface completely disappeared. This result is consistent with the interaction being in intermediate to slow exchange on the NMR timescale and corresponds to a Kd of <100 μM. The reported Kd of activated LFA-1 for ICAM-1 has ranged from 133 nM (43) to 500 nM (44). For inactivated CD11a, a Kd of 100 μM has been reported (43). Thus, the NMR Kd for the interaction of the I domain and ICAM-1 domains 1 and 2 is consistent with previously reported affinities for the full length molecules. However, it was not possible to distinguish an activated versus inactivated state of the I domain given the inability to calculate a lower limit for the NMR Kd.
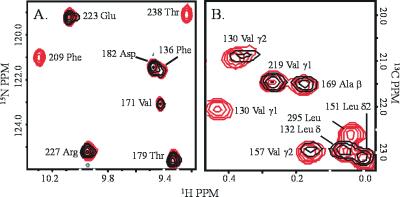
At submolar equivalents of ICAM-1, the chemical shifts of some residues broadened more than others. This is consistent with simulations of NMR spectra for a high molecular weight complex in intermediate to slow exchange (45). For residues whose environment changes because of contact with a ligand or, indirectly, because of a conformational change induced by the ligand, longer than average line broadening is observed due to chemical exchange. Fig. 1 depicts examples of the selective line broadening observed due to binding of the CD11a I domain to ICAM-1. Amide proton signals of residues at the MIDAS are broadened (e.g., T238 and F209) (Fig. 1A) whereas signals of many residues far from the MIDAS site are less affected by ICAM-1 (e.g., D182, T179, and E223) (Fig. 1A). Fig. 1B shows NMR signals of residues in a hydrophobic cleft that are also affected by ICAM-1. The methyl groups of V130, V157, and L295, which lie in the cleft, are affected. However, the methyl groups in the interior of the protein, such as V219 and A169, are less affected on the addition of ICAM-1.
Figure 1.
(A) Region of the 15N-HSQC spectrum showing an overlay of chemical shifts of 0.2 mM CD11a I domain in the presence (blue) and absence (red) of 0.04 mM ICAM-1 domains 1 and 2. (B) Region of the 13C-constant time HSQC spectrum showing an overlay of chemical shifts of 0.2 mM CD11a I domain in the presence (blue) and absence (red) of 0.1 mM ICAM-1 domains 1 and 2.
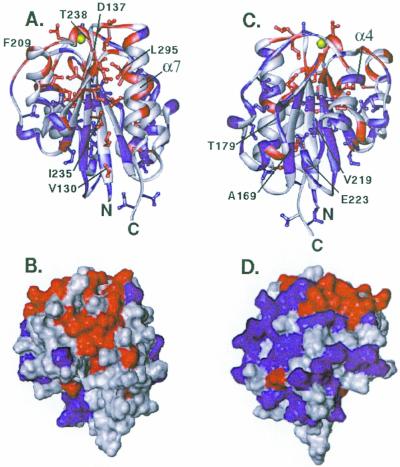
Residues that show a larger than average change on ICAM-1 binding are mapped onto ribbon and surface representations of the I domain (Fig. 2). A cluster of affected residues localizes around the MIDAS on the upper face of the I domain (Fig. 2A). An additional cluster of residues at a site distinct from the MIDAS also is markedly affected by ICAM-1 binding. This region of the I domain includes α-helix 7 as well as the opposing face of the core β sheet (Fig. 2 A and B). Residues on the face opposite to the cluster near α-helix 7 were less affected in the titration study (Fig. 2 C and D). These data suggest that two regions of the I domain are affected by ICAM-1 binding. One region corresponds to the MIDAS in the upper face of the I domain that forms a binding interface with ICAM-1. The second region could represent an unexpected binding site for ICAM-1. Alternatively, this second region of CD11a could be the site of a conformational change necessary for ICAM-1 binding and thus serve as a regulatory site.
Figure 2.
Ribbon and surface representations of the x-ray crystal structure of the CD11a I domain (4) showing residues affected by the addition of soluble ICAM-1 domains 1 and 2. Colors indicate the extent of line broadening: red, large decrease in peak height; blue, medium or no decrease in peak height; gray, unassigned in the complex due to spectral overlap. The magnesium ion of the MIDAS is shown in yellow. For the ribbon diagrams, the color coding of the ribbon indicates changes in peak intensity of amide proton signals, and colors of side chains indicate changes in peak intensity of methyl proton signals. For the surface representations, red indicates a large decrease in peak height of 15N-amide or 13C-methyl signals. (A and B) View of α-helix 7 and the opposing β-sheet. (C and D) Opposite face of the I domain showing α-helix 4.
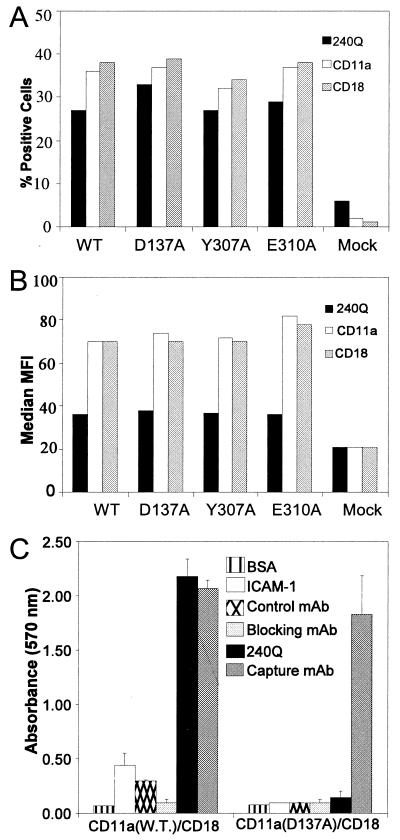
To determine whether residues in the second I domain site can regulate ICAM-1 binding, site-directed mutants were expressed in COS cells. Expression levels of LFA-1 on each transfectant were analyzed by FACS using both anti-CD11a (TS1/22) and anti-CD18 (TS1/18) mAb. The transfection efficiency (percent of positive cells) averaged ≈30% for all transfectants with a median MFI of ≈70 (Fig. 3 A and B; data not shown). The variability among all transfectants and between transfections was less than 20% of the mean. A COS cell-based static adhesion assay to immobilized ICAM-1 was established. COS cells transfected with CD18 and CD11a (wild type), but not those transfected with CD18 and a MIDAS-inactivating mutant, CD11a (D137A), readily adhered to ICAM-1. This binding was inhibited by a CD11a-specific blocking mAb (Blocking), but not an isotype-matched control mAb (Control) (Fig. 3C). Adhesion of LFA-1 (wild type) but not LFA-1 (D137A), was stimulated by addition of the activating anti-CD18 mAb, 240Q. Stimulation with 240Q was titratable (data not shown), and saturating concentrations were used in all experiments.
Figure 3.
Expression of LFA-1 mutants on COS cells and representative data for the ICAM-1 adhesion assay. Expression of the wild type and a subset of mutants that do not respond to mAb 240Q induction was determined by flow cytofluorometry. Data from a representative experiment are shown and were similar for all mutants and across all experiments. (A) Transfection efficacy, expressed as the percentage of cells staining positive for CD11a (TS1/22) and CD18 (TS1/18 and 240Q). (B) The relative expression levels of different mutants expressed as median MFI of CD11a and CD18 mAb. (C) Adhesion assay of COS cells expressing wild-type LFA-1 or LFA-1 with a D137A mutation in presence of Control mAb (1B7), Blocking mAb (TS1/22), or Activating mAb 240Q. The adhesion assay is representative of greater than seven independent experiments. Mean absorbency values and standard deviations from triplicate wells are shown.
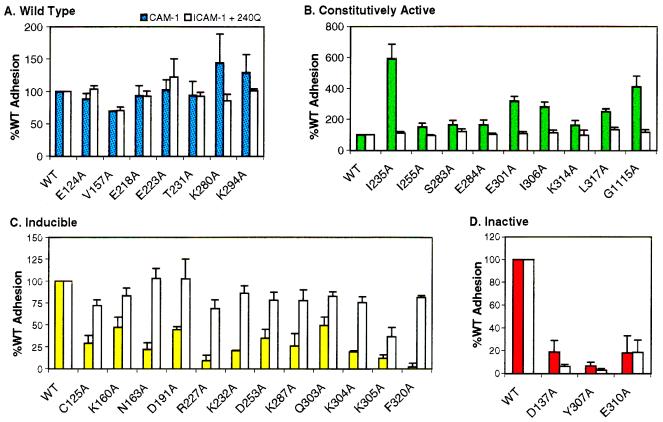
Twenty residues within and proximal to α-helix 7 and two control residues (E218 and K280), distal to α-helix 7 but within the CD11a I domain, were substituted with alanine. Many of these, including D137, K294, K287, V157, and I235, displayed broad NMR signals in the presence of ICAM-1. The effect of these specific mutations on cell adhesion and the ability of mAb 240Q to stimulate their binding was analyzed (Fig. 4). The mutants separated into three phenotypes: (i) mutants that demonstrated wild-type levels of binding with or without mAb 240Q induction (Fig. 4A), (ii) constitutively active mutants that supported 1.5- to 6-fold greater than wild-type levels of binding without mAb 240Q induction and wild-type levels with induction (Fig. 4B), and (iii) inducible mutants that possessed decreased levels of binding relative to the wild type in the absence of induction, but approximately 50–100% of wild-type levels with mAb 240Q induction (Fig. 4C). Two control mutations were also evaluated; D137A, which is in the MIDAS, and G1115A, which is located in the CD11a cytoplasmic GFFKR motif. As expected, substitution of a MIDAS residue resulted in an inactive phenotype with severely decreased binding that could not be rescued by mAb 240Q (Figs. 3C and 4D). This result is consistent with the hypothesis that the MIDAS is a point of contact with ICAM-1 rather than a regulatory site. G1115A is a mutation known to activate LFA-1 (46) and, in our binding studies, yielded a constitutively active phenotype with a 4-fold increase in binding (Fig. 4B).
Figure 4.
Average adhesion of COS cells expressing LFA-1 I domain mutants to ICAM-1. Filled bars indicate the percent of adhesion that occurs in the presence of ICAM-1. Open bars indicate the percentage of adhesion that occurs in the presence of ICAM-1 and the mAb 240Q. Data presented represent the mean values and standard error of at least three independent experiments. Data were first normalized to BSA and expressed relative to binding of wild-type LFA-1 under the same stimulation condition and within the same assay to compare data from multiple experiments (see Materials and Methods). (A) Alanine mutations that cause no significant change in adhesion both in the presence or absence of mAb 240Q. (B) Mutations that increased adhesion, but whose mAb 240Q response is equivalent to the wild-type response. (C) Mutations that decrease adhesion but whose adhesion is restored to wild-type levels (50–100% of wild type) by mAb 240Q. (D) Mutations that decrease adhesion relative to the wild type but whose effects cannot be reversed by mAb 240Q.
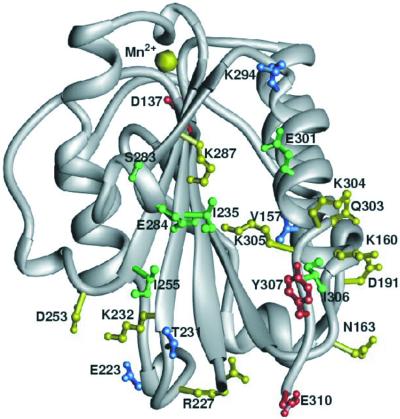
Changes in activity of I domain mutants were not caused by the lack of expression (Fig. 3 A and B; data not shown) or the misfolding of the I domain. Seven of the I domain mutants that significantly affected binding were 15N labeled, and their 1H-15N HSQC spectra were compared with that of wild-type I domain (data not shown). All of these mutant proteins had spectra that were very similar to that of the wild-type protein, indicating that the amino acid change had not caused the protein to misfold. Also, an I domain mAb (TS1/22) binds all mutant proteins at equivalent levels, indicating that substitutions did not disrupt the conformation of this epitope (Fig. 3 A and B; data not shown). The altered binding activities of these second site I domain mutants indicate that the spectral changes observed by NMR reflect a functionally relevant change with respect to ICAM-1 binding. The most constitutively active mutants involve residues that localize to α-helix 7 and the opposing β-sheet (Fig. 5). We designate this region the I domain allosteric site (IDAS).
Figure 5.
Location of I domain substitution mutations in the IDAS. The color coding indicates the affect on ICAM-1 binding: blue, wild-type levels of adhesion with or without mAb 240Q induction; green, greater than wild-type levels of adhesion without mAb 240Q and wild-type levels with the antibody; yellow, decreased adhesion without mAb 240Q and wild-type adhesion with the antibody; red, decreased adhesion with and without mAb 240Q.
The I domain is inserted into the rest of the α chain through amino and carboxy-terminal linker sequences not present in the recombinant I domain used in the NMR experiments described above. To determine whether these sequences might also function in LFA-1 activation, seven substitution mutants were generated and functionally characterized. One N-terminal mutation, C125A, yielded an inducible phenotype. Two C-terminal mutants, K314A and L317A, resulted in constitutive activation and two others, Y307A and E310A, resulted in inactivation (Fig. 4 B and D). The binding of mAb 240Q to these inactive mutants was not diminished (Fig. 3 A and B). These data suggest that amino acids in the linker are an important component of LFA-1 activation.
Discussion
We demonstrate the structural effects of an integrin I domain on binding to its natural ligand. Previous I domain antibody blocking (20) mutational studies (15–19) and models (17, 23) have indicated that the MIDAS face interacts with a corresponding flat surface on ICAM-1. The NMR data presented here is consistent with an interaction surface that includes the MIDAS. Seventy-five percent of the residues on the MIDAS face of CD11a that could be detected by NMR spectroscopy of the I domain/ICAM-1 complex were affected by binding.
The mechanism of integrin activation has been difficult to study, and conflicting theories have been proposed. One component of activation may involve a conformational change of the I domain. The disposition of α-helix 7 relative to the body of the I domain was found to differ in crystal structures of CD11a (4, 5) and CD11b (6–8, 34) suggesting that this helix undergoes a conformational change during activation. In CD11b, this change is associated with solvent exposure of F302. The NMR chemical shift data are direct evidence that residues in α-helix 7 are affected by ICAM-1 binding. Shift changes for residues in α-helix 7, the loop preceding it, and in the β sheet in which this helix packs were all observed. NMR data for F292, which is analogous to residue F302 of CD11b, and exact details of the changes that occur in the I domain due to ICAM-1 binding, such as alterations in metal coordination by side chains or water, or changes in the position of amino acid side chains, were not obtained in this study.
The functional importance of changes in I domain structure have been addressed by mutagenesis studies. Mutations on the bottom face of CD11a, CD11b, and α2 I domains, as well as vWF A domain near the region corresponding to the CD11a IDAS, can modulate binding (11, 17, 47, 48). Earlier reports have not ruled out the possibility that more than one part of an I domain, such as the IDAS, directly contacts ligand. Our results characterizing the effects of an extensive set of mutations in the IDAS indicate that structural changes in this region can indirectly promote ICAM-1 binding without directly contacting the ligand. It was found that certain mutations within the IDAS can constitutively activate LFA-1-dependent binding. The IDAS mutations that resulted in decreased binding could all be rescued with the activating CD18 mAb, 240Q. This is in contrast to the affect induced by the representative MIDAS mutant D137A (Figs. 3C and 4D). Of the 20 IDAS residues we mutated, none appear to contribute to a critical ligand binding interface, but rather, appear to regulate binding at the MIDAS.
Interestingly, the effects of IDAS mutations correlate with their chemical nature (Fig. 5). Mutations of hydrophobic residues at the center of the IDAS cleft (e.g., I235, I255, and I306) activate LFA-1 binding. Mutations of hydrophilic residues on the surface of or adjacent to the IDAS cleft (e.g., K232, K287, Q303, K304, and K305) inhibit the interaction with ICAM-1. One explanation is that replacement of larger hydrophobic amino acids by alanine creates cavities in the protein that lower the energy barrier for a conformational change in the I domain. Hydrophilic amino acids on the surface of the IDAS could stabilize an active state by interacting with other amino acids of LFA-1. Alanine substitutions would be expected to remove these polar interactions and thereby decrease the stability of the activated state.
Although the IDAS may regulate LFA-1 activation, residues that link the I domain to CD11a also seem to be involved. Alterations in ligand binding in response to linker mutations in von Willebrand factor have been reported (11). LFA-1 linker mutations can activate (K314A and L317A) (Fig. 4B) or severely decrease (Y307A and E310A) (Fig. 4D) adhesion to ICAM-1. These residues are not required for proper folding of the I domain based on the observation that an I domain that ends in K305 still yields a two-dimensional 15N-HSQC spectrum very similar to the longer construct (data not shown). One could infer, then, that a tertiary or quaternary change involved in activation depends on amino acid contacts between I domain linker and other CD11a or CD18 sequences. The later is supported by the failure of the CD18 mAb 240Q to rescue ICAM-1 binding to the linker mutants, Y307A and E310A. Activating CD18 mAb have been used as a surrogate for intracellular signaling leading to LFA-1 activation (27, 49). The inability of a CD18 mAb to activate ligand binding to the I domain mutants suggests that a CD18-dependent regulatory mechanism involving the linker residues has been disrupted.
The recent report of an x-ray structure of the LFA-1/lovastatin complex (36) demonstrates that a small molecule can inhibit LFA-1-mediated adhesion without binding to the MIDAS. This mechanism can be understood in light of our data. The drug binds to the IDAS that we have shown by NMR to change in response to ICAM-1 binding and that we have shown by mutagenesis to be functionally relevant. Structural differences in the IDAS of different I domains may allow the design of therapeutic agents with high integrin selectivity.
Abbreviations
- ICAM
intercellular adhesion molecule
- LFA
lymphocyte function-associated antigen
- MIDAS
metal ion-dependent adhesion site
- IDAS
I domain allosteric site
Footnotes
This paper was submitted directly (Track II) to the PNAS office.
References
- 1.Gahmberg C G. Curr Opin Cell Biol. 1997;9:643–650. doi: 10.1016/s0955-0674(97)80117-2. [DOI] [PubMed] [Google Scholar]
- 2.Diamond M S, Springer T A. J Cell Biol. 1993;120:545–556. doi: 10.1083/jcb.120.2.545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Landis R C, Bennett R I, Hogg N. J Cell Biol. 1993;120:1519–1527. doi: 10.1083/jcb.120.6.1519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Qu A, Leahy D J. Proc Natl Acad Sci USA. 1995;92:10277–10281. doi: 10.1073/pnas.92.22.10277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Qu A, Leahy D J. Structure (London) 1996;4:931–942. doi: 10.1016/s0969-2126(96)00100-1. [DOI] [PubMed] [Google Scholar]
- 6.Lee J O, Rieu P, Arnaout M A, Liddington R. Cell. 1995;80:631–638. doi: 10.1016/0092-8674(95)90517-0. [DOI] [PubMed] [Google Scholar]
- 7.Lee J O, Bankston L A, Arnaout M A, Liddington R C. Structure (London) 1995;3:1333–1340. doi: 10.1016/s0969-2126(01)00271-4. [DOI] [PubMed] [Google Scholar]
- 8.Baldwin E T, Sarver R W, Bryant G L, Jr, Curry K A, Fairbanks M B, Finzel B C, Garlick R L, Heinrikson R L, Horton N C, Kelley L L, et al. Structure (London) 1998;6:923–935. doi: 10.1016/s0969-2126(98)00093-8. [DOI] [PubMed] [Google Scholar]
- 9.Emsley J, King S L, Bergelson J M, Liddington R C. J Biol Chem. 1997;272:28512–28517. doi: 10.1074/jbc.272.45.28512. [DOI] [PubMed] [Google Scholar]
- 10.Rich R L, Deivanayagam C C, Owens R T, Carson M, Hook A, Moore D, Symersky J, Yang V W, Narayana S V, Hook M. J Biol Chem. 1999;274:24906–24913. doi: 10.1074/jbc.274.35.24906. [DOI] [PubMed] [Google Scholar]
- 11.Emsley J, Cruz M, Handin R, Liddington R. J Biol Chem. 1998;273:10396–10401. doi: 10.1074/jbc.273.17.10396. [DOI] [PubMed] [Google Scholar]
- 12.Bienkowska J, Cruz M, Atiemo A, Handin R, Liddington R. J Biol Chem. 1997;272:25162–25167. doi: 10.1074/jbc.272.40.25162. [DOI] [PubMed] [Google Scholar]
- 13.Springer T A. Proc Natl Acad Sci USA. 1997;94:65–72. doi: 10.1073/pnas.94.1.65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Randi A M, Hogg N. J Biol Chem. 1994;269:12395–12398. [PubMed] [Google Scholar]
- 15.Michishita M, Videm V, Arnaout M A. Cell. 1993;72:857–867. doi: 10.1016/0092-8674(93)90575-b. [DOI] [PubMed] [Google Scholar]
- 16.Edwards C P, Champe M, Gonzalez T, Wessinger M E, Spencer S A, Presta L G, Berman P W, Bodary S C. J Biol Chem. 1995;270:12635–12640. doi: 10.1074/jbc.270.21.12635. [DOI] [PubMed] [Google Scholar]
- 17.Edwards C P, Fisher K L, Presta L G, Bodary S C. J Biol Chem. 1998;273:28937–28944. doi: 10.1074/jbc.273.44.28937. [DOI] [PubMed] [Google Scholar]
- 18.Kamata T, Wright R, Takada Y. J Biol Chem. 1995;270:12531–12535. doi: 10.1074/jbc.270.21.12531. [DOI] [PubMed] [Google Scholar]
- 19.Huang C, Springer T A. J Biol Chem. 1995;270:19008–19016. doi: 10.1074/jbc.270.32.19008. [DOI] [PubMed] [Google Scholar]
- 20.Champe M, McIntyre B W, Berman P W. J Biol Chem. 1995;270:1388–1394. doi: 10.1074/jbc.270.3.1388. [DOI] [PubMed] [Google Scholar]
- 21.Staunton D E, Dustin M L, Erickson H P, Springer T A. Cell. 1990;61:243–254. doi: 10.1016/0092-8674(90)90805-o. [DOI] [PubMed] [Google Scholar]
- 22.Fisher K L, Lu J, Riddle L, Kim K J, Presta L G, Bodary S C. Mol Biol Cell. 1997;8:501–515. doi: 10.1091/mbc.8.3.501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Bella J, Kolatkar P R, Marlor C W, Greve J M, Rossmann M G. Proc Natl Acad Sci USA. 1998;95:4140–4145. doi: 10.1073/pnas.95.8.4140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Dustin M L, Springer T A. Nature (London) 1989;341:619–624. doi: 10.1038/341619a0. [DOI] [PubMed] [Google Scholar]
- 25.Stewart M P, McDowall A, Hogg N. J Cell Biol. 1998;140:699–707. doi: 10.1083/jcb.140.3.699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Sadhu C, Masinovsky B, Staunton D E. J Immunol. 1998;160:5622–5628. [PubMed] [Google Scholar]
- 27.Andrew D, Shock A, Ball E, Ortlepp S, Bell J, Robinson M. Eur J Immunol. 1993;23:2217–2222. doi: 10.1002/eji.1830230925. [DOI] [PubMed] [Google Scholar]
- 28.Dransfield I, Cabanas C, Craig A, Hogg N. J Cell Biol. 1992;116:219–226. doi: 10.1083/jcb.116.1.219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Stewart M, Hogg N. J Cell Biochem. 1996;61:554–561. doi: 10.1002/(sici)1097-4644(19960616)61:4<554::aid-jcb8>3.0.co;2-n. [DOI] [PubMed] [Google Scholar]
- 30.McDowall A, Leitinger B, Stanley P, Bates P A, Randi A M, Hogg N. J Biol Chem. 1998;273:27396–27403. doi: 10.1074/jbc.273.42.27396. [DOI] [PubMed] [Google Scholar]
- 31.Cabanas C, Hogg N. Proc Natl Acad Sci USA. 1993;90:5838–5842. doi: 10.1073/pnas.90.12.5838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Leahy D J. Annu Rev Cell Dev Biol. 1997;13:363–393. doi: 10.1146/annurev.cellbio.13.1.363. [DOI] [PubMed] [Google Scholar]
- 33.Liddington R, Bankston L. Structure (London) 1998;6:937–938. doi: 10.1016/s0969-2126(98)00094-x. [DOI] [PubMed] [Google Scholar]
- 34.Li R, Rieu P, Griffith D L, Scott D, Arnaout M A. J Cell Biol. 1998;143:1523–1534. doi: 10.1083/jcb.143.6.1523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Oxvig C, Lu C, Springer T A. Proc Natl Acad Sci USA. 1999;96:2215–2220. doi: 10.1073/pnas.96.5.2215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kallen J, Welzenbach K, Ramage P, Geyl D, Kriwacki R, Legge G, Cottens S, Weitz-Schmidt G, Hommel U. J Mol Biol. 1999;292:1–9. doi: 10.1006/jmbi.1999.3047. [DOI] [PubMed] [Google Scholar]
- 37.Clore G M, Gronenborn A M. Methods Enzymol. 1994;239:349–363. doi: 10.1016/s0076-6879(94)39013-4. [DOI] [PubMed] [Google Scholar]
- 38.Fesik S W, Zuiderweg E R P. J Magn Reson. 1988;78:588–593. [Google Scholar]
- 39.Chen Y, Reizer J, Saier M H, Jr, Fairbrother W J, Wright P E. Biochemistry. 1993;32:32–37. doi: 10.1021/bi00052a006. [DOI] [PubMed] [Google Scholar]
- 40.Garrett D S, Seok Y J, Peterkofsky A, Clore G M, Gronenborn A M. Biochemistry. 1997;36:4393–4398. doi: 10.1021/bi970221q. [DOI] [PubMed] [Google Scholar]
- 41.Shuker S B, Hajduk P J, Meadows R P, Fesik S W. Science. 1996;274:1531–1534. doi: 10.1126/science.274.5292.1531. [DOI] [PubMed] [Google Scholar]
- 42.Spera S, Bax A. J Am Chem Soc. 1991;113:5490–5492. [Google Scholar]
- 43.Lollo B A, Chan K W, Hanson E M, Moy V T, Brian A A. J Biol Chem. 1993;268:21693–21700. [PubMed] [Google Scholar]
- 44.Labadia M E, Jeanfavre D D, Caviness G O, Morelock M M. J Immunol. 1998;161:836–842. [PubMed] [Google Scholar]
- 45.Matsuo H, Walters K J, Teruya K, Tanaka T, Gassner G T, Lippard S J, Kyogoku Y, Wagner G. J Am Chem Soc. 1999;121:9903–9904. [Google Scholar]
- 46.Peter K, O'Toole T E. J Exp Med. 1995;181:315–326. doi: 10.1084/jem.181.1.315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Zhang L, Plow E F. J Biol Chem. 1996;271:29953–29957. doi: 10.1074/jbc.271.47.29953. [DOI] [PubMed] [Google Scholar]
- 48.Dickeson S K, Mathis N L, Rahman M, Bergelson J M, Santoro S A. J Biol Chem. 1999;274:32182–32191. doi: 10.1074/jbc.274.45.32182. [DOI] [PubMed] [Google Scholar]
- 49.Stephens P, Romer J T, Spitali M, Shock A, Ortlepp S, Figdor C G, Robinson M K. Cell Adhes Commun. 1995;3:375–384. doi: 10.3109/15419069509081292. [DOI] [PubMed] [Google Scholar]