Abstract
Exposure of eukaryotic cells to extracellular stimuli results in activation of mitogen-activated protein kinase (MAPK) cascades composed of MAPKs, MAPK kinases (MAP2Ks), and MAPK kinase kinases (MAP3Ks). Mammals possess a large number of MAP3Ks, many of which can activate the c-Jun N-terminal kinase (JNK) MAPK cascade when overexpressed, but whose biological function is poorly understood. We examined the function of the MAP3K MEK kinase 1 (MEKK1) in proinflammatory signaling. Using MEKK1-deficient embryonic stem cells prepared by gene targeting, we find that, in addition to its function in JNK activation by growth factors, MEKK1 is required for JNK activation by diverse proinflammatory stimuli, including tumor necrosis factor α, IL-1, double-stranded RNA, and lipopolysaccharide. MEKK1 is also essential for induction of embryonic stem cell migration by serum factors, but is not required for activation of other MAPKs or the IκB kinase signaling cascade.
MEKK1 (MEK kinase 1) is one of the first identified members of the mitogen-activated protein kinase (MAPK) kinase kinase (MAP3K) group (1). Although MEKK1 was initially thought to be a specific activator of the extracellular signal-regulated kinase (ERK) MAPK cascade, it was found to be a more potent and preferential activator of the c-Jun N-terminal kinase (JNK) group of MAPKs (2), possibly through its high affinity toward the MAP2K, JNK kinase 1 (JNKK1)/SEK1/MKK4 (3). JNK activity is potently stimulated by a variety of physical and chemical stresses, most notably UV irradiation and osmotic stress, but also by the protein synthesis inhibitor anisomycin, arsenite, and heat shock (4–7). In addition, JNK is activated by a variety of proinflammatory stimuli, including tumor necrosis factor α (TNFα), IL-1, lipopolysaccharide (LPS), and double-stranded (ds)RNA (8–10). All of these stimuli are potent activators of innate immune responses (11), to which JNK activation makes an important contribution (10). JNK activity is also stimulated by certain growth factors and small G proteins, such as Ras and Rac (12, 13). Although only two MAPK kinases (MAP2Ks) function as JNK kinases, JNKK1/SEK1/MKK4 (14–16) and JNKK2/MKK7 (17–19), many MAP3Ks, in addition to MEKK1, can activate the JNK cascade (20–23). The exact physiological function of each of these MAP3Ks, which include MEKK2, MEKK3, MEKK4, transforming growth factor β (TFG-β)-activating kinase 1 (TAK1), and apoptosis signal-regulating kinase (ASK)1, is not known. Recently, however, gene-disruption experiments were used to generate embryonic stem (ES) cells deficient in MEKK1 (24). These studies revealed that MEKK1 plays a critical role in JNK activation by serum, lysophosphatidic acid (LPA), and nocodazole, a microtubule-disrupting agent (24, 25). MEKK1 is also partially involved in JNK activation by osmotic shock and plays an important role in JNK activation by oxidative stress, but is not required for responsiveness to heat shock, anisomycin, or UV radiation (24, 26). The role of MEKK1 in JNK activation by TNFα or other proinflammatory stimuli has not been investigated. The biological role of MEKK1 in cellular responses to serum growth factors has not been defined either.
Recently, we identified MEKK1 as a potential target for TNF receptor-associated factor 2 (TRAF2) and TRAF6 (27), two related signal transducers that are recruited to TNFα and IL-1 receptors, respectively (28, 29). The recruitment of TRAF2 and TRAF6 to proinflammatory receptors is essential for JNK activation (8, 27, 30). However, TNFα- and TRAF2-induced JNK activation was also suggested to be mediated by ASK1 (31), and another MAP3K, TAK1, was suggested to mediate JNK activation by IL-1 (32). MEKK1 was also suggested to be a critical mediator of NF-κB activation (33–35), acting by means of the IκB kinase (IKK) (36, 37). To investigate the function of MEKK1 in proinflammatory signaling, we generated MEKK1-deficient ES cells. Using these cells, we found that MEKK1 is required for JNK activation in response to diverse proinflammatory stimuli, including TNFα, IL-1, dsRNA, and LPS. MEKK1 is also required for induction of ES cell migration in response to serum factors. MEKK1, however, is not required for IKK activation.
Materials and Methods
Generation of Mekk1−/− ES Cells.
The targeting vector shown in Fig. 1 was constructed as previously described (38). The vector was linearized with NotI and electroporated into ES cells (Genomic System) (39). G418-resistant clones were analyzed by PCR and Southern blotting for the presence of a disrupted Mekk1 allele. One Mekk1+/− euploid ES cell clone was further cultured in elevated G418 concentration (2 mg/ml), and resistant clones were characterized by PCR and Southern blotting for disruption of both Mekk1 alleles.
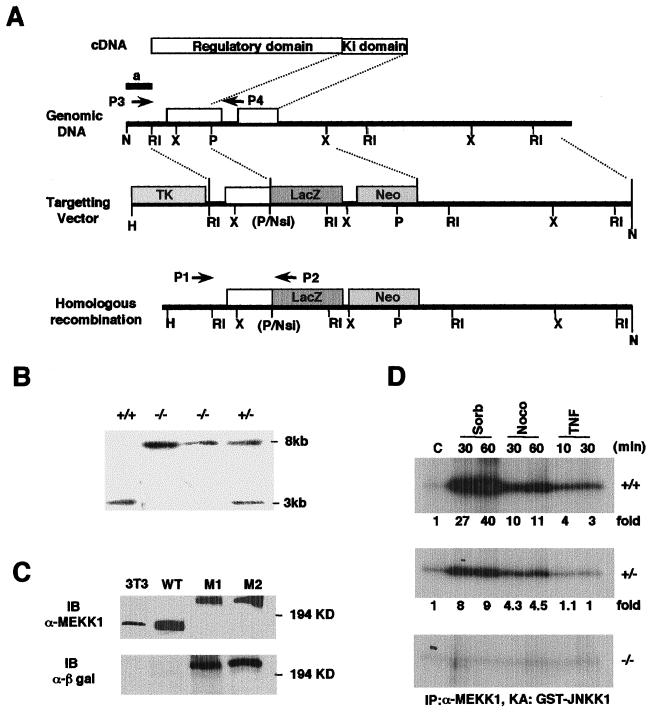
Figure 1.
Generation of Mekk1−/− ES cells. (A) Diagram of the targeting strategy. Schematic structures of Mekk1 cDNA, the relevant portion of the Mekk1 locus, the targeting vector, and the homologous recombinant. Indicated are locations of the N-terminal regulatory region and the C-terminal catalytic (Ki) domain of MEKK1, restriction sites (H, HindIII; RI, EcoRI; P, PstI; N, NotI; X, XbaI; B, BamHI), and the position of a 5′ external probe (a). Arrowheads labeled P1, P2, P3, and P4 indicate positions of primers used in genotyping. (B) Southern blot demonstrating homologous recombination within the Mekk1 locus. The 5′ external probe detects a 3-kb HindIII/PstI fragment for the wild-type locus and an 8-kb HindIII/PstI fragment for the targeted locus. (C) Immunoblot (IB) using MEKK1 and β-galactosidase antibodies revealing expression of 180-kDa mouse MEKK1 polypeptide in mouse fibroblasts (3T3) and ES cells (WT) and a 242-kDa MEKK1-β-galactosidase fusion protein in two homozygous recombinant clones. (D) MEKK1 kinase activity in Mekk1 wild-type (+/+), heterozygous (+/−), and homozygous (−/−) recombinant ES cells. Lysates were prepared from cells stimulated with sorbitol (300 mM), nocodazole (2 μM), or TNFα (10 ng/ml) for the indicated times. After immunoprecipitation (IP) with anti-MEKK1, immunocomplex kinase assays (KA) were performed by using GST-JNKK1 as a substrate. Fold-activation was calculated based on phosphorimager analysis.
Kinase and DNA Binding Assays.
To measure its kinase activity, MEKK1 was immunoprecipitated from cell lysates with rabbit antiserum to recombinant human MEKK1 (amino acids 1006–1170). The immunoprecipitates were subjected to kinase assays with bacterially expressed glutathione S-transferase (GST)-JNKK1 as a substrate, using standard protein kinase assay conditions (4). JNK was immunoprecipitated from cell lysates with an antibody against human full-length JNK1 (G151-333.8; PharMingen). The precipitates were subjected to in vitro kinase assay with GST-c-Jun (1–79) as a substrate (4). JNK, p38, and ERK activation were determined by immunoblotting total cell lysates separated by SDS/PAGE with anti-phospho-JNK (7932; Promega), anti-phospho-p38 (9211S; New England Biolabs), and anti-phospho-ERK (9101S; New England Biolabs), respectively, following suppliers' recommendations. These antibodies are specific to the activated forms of these MAPKs. Total loading of JNK, p38, and ERK was determined by reprobing stripped blots with anti-JNK (G151-666.13; PharMingen), anti-p38 (9212; New England Biolabs), or anti-ERK (9102; New England Biolabs) antibodies. IKK was immunoprecipitated from cell lysates with monoclonal antibody against recombinant IKKγ (c73-764; PharMingen), and its activity was measured by immunocomplex kinase assay with GST-IκBα (1–54) as a substrate (40). Electrophoretic mobility-shift assays (EMSAs) were performed as described (40). Briefly, 20 μg of whole cell extracts was incubated with 2 μg of poly(dI-dC) and 5,000 cpm of labeled oligonucleotide probes. After 30 min of incubation at room temperature, the samples were resolved on native 5% polyacrylamide gels.
Cell Migration Assays.
Cell migration was measured with modified Boyden chambers as described (41). There was no difference in the ability of wild-type and Mekk1−/− cells to adhere to fibronectin-coated plates.
MAPK Inhibitors.
The specific JNK inhibitor (SP 600125) was a gift from Signal Pharmaceuticals, San Diego. When used at 50 μM, this compound completely inhibits JNK activity but does not inhibit ERK1, ERK2, p38α, or p38β (unpublished results). The MEK inhibitor (PD 98059) was purchased from New England Biolabs, and the p38 inhibitor SB202190 was from Calbiochem. The inhibitors (20–50 μM) were added to cells in serum-free media for 0.5–1 h before the cell migration assay.
Results
Generation and Characterization of Mekk1−/− ES Cells.
To investigate the physiological functions of MEKK1, we used the targeting vector shown in Fig. 1A to disrupt the Mekk1 locus in mouse ES cells (39). Integration of the targeting vector into the Mekk1 locus should result in formation of an MEKK1-β-galactosidase fusion protein containing the first 1188 amino acids of MEKK1, but lacking its entire kinase domain. Indeed, heterozygote Mekk1+/− ES (Fig. 1B) cells express both full-length 180-kDa MEKK1 polypeptide and a 242-kDa MEKK1-β galactosidase fusion protein (Y.X., unpublished results). We obtained several cell lines in which both Mekk1 alleles were disrupted and no longer express wild-type MEKK1 by selecting Mekk1+/− ES cells for survival in high concentration of G418 (Fig. 1 B and C). These cells express only the 242-kDa MEKK1-β galactosidase fusion protein. Most of the work reported below was carried out with one such cell line (M1), although another cell line behaved in an identical manner (Y.X., unpublished results). MEKK1 kinase activity is stimulated by TNFα (27), nocodazole, and osmotic stress (Fig. 1D). No MEKK1 kinase activity could be detected in Mekk1−/− cells, whereas reduced kinase activity was present in lysates of Mekk1+/− cells (Fig. 1D). As the heterozygote cells express similar amounts of native MEKK1 and the MEKK1-β galactosidase fusion protein, it is unlikely that the latter acts as a dominant negative inhibitor of MEKK1. In addition, overexpression of the MEKK1 N-terminal fragment in HeLa or 293 cells did not reveal dominant negative activity (3).
MEKK1 Is Required for JNK but Not IKK Activation by Proinflammatory Stimuli.
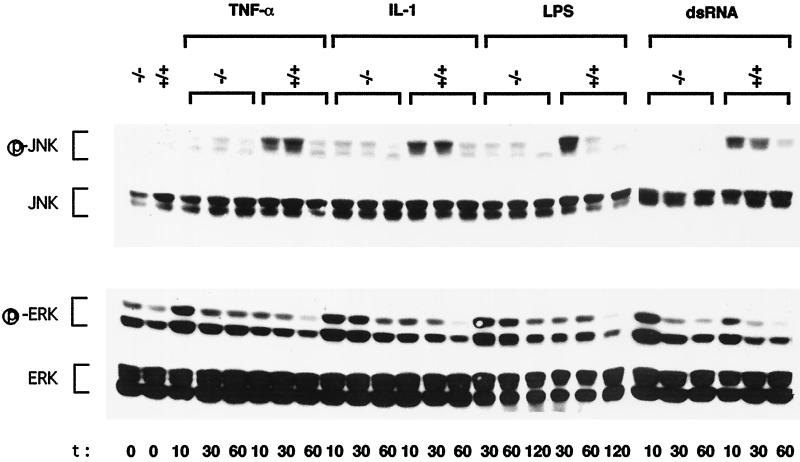
As previously described (24), we found that the absence of MEKK1 prevented JNK activation by nocodazole, but had only a small effect on the response to osmotic shock and anisomycin (42–45). Using a lower dose of UVC radiation than the one used previously (80 J/m2 vs. 100 J/m2), we found that JNK was activated with slower kinetics in Mekk1−/− cells in comparison to wild-type cells (data not shown). Thus, MEKK1 contributes to, but is not essential for, JNK activation in response to UVC exposure. We examined the response of JNK to four different proinflammatory stimuli, TNFα, IL-1, LPS, and dsRNA. Each stimulus causes substantial but transient JNK activation in wild-type cells (Fig. 2). However, none of these stimuli led to considerable JNK activation in Mekk1−/− cells. Exposure to proinflammatory stimuli also results in modest ERK activation (Fig. 2). In this case, however, the loss of MEKK1 resulted in more potent MAPK activation by all four stimuli: TNFα, IL-1, LPS, and dsRNA (Fig. 2). The increase in ERK activation was not because of higher levels of ERK1 or ERK2 in the mutant cells. It is possible, therefore, that JNK activation may exert a negative effect on activation of the ERK cascade by proinflammatory stimuli. Unfortunately, we were not able to determine the involvement of MEKK1 in p38 activation by these stimuli, as none of them led to considerable p38 activation, even in wild-type cells (data not shown).
Figure 2.
MEKK1 is required for JNK activation by proinflammatory stimuli. Wild-type (+/+) and MEKK1-deficient (−/−) ES cells were left untreated or incubated with TNFα (10 ng/ml), IL-1 (10 ng/ml), LPS (15 μg/ml), or dsRNA (5 μg/ml) for the indicated times (min) after which the cells were lysed. Equal amounts (100 μg) of lysates were resolved by SDS/PAGE, transferred to a nitrocellulose membrane, and sequentially probed with antibodies to activated (phosphorylated) JNK (p-JNK) and activated (phosphorylated) ERK (p-ERK). The membranes were stripped and reprobed with antibodies to all JNK and ERK isoforms. Shown are the results of one typical experiment of at least three similar and separate experiments.
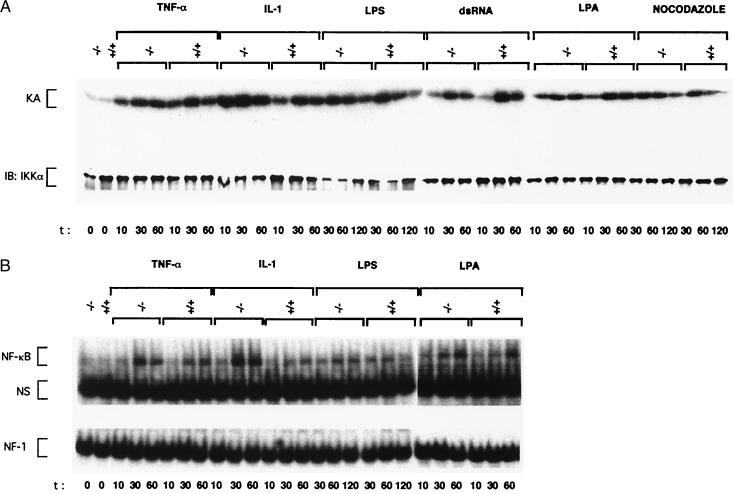
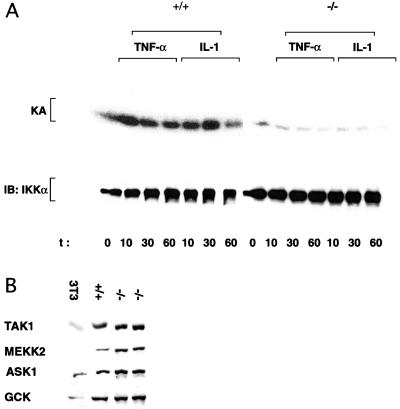
Proinflammatory stimuli are also potent activators of IKK and NF-κB (42–45). In contrast to the effect on JNK, the loss of MEKK1 did not reduce IKK or NF-κB activation in response to TNFα, IL-1, LPS, or dsRNA (Fig. 3 A and B). NF-κB activity is also stimulated by depolymerization of microtubules (46). Despite the large increase in MEKK1 catalytic activity in nocodazole-treated cells (Fig. 1D), MEKK1 is not required for nocodazole-induced IKK activation (Fig. 3A). We also found that LPA treatment resulted in IKK and NF-κB activation and this response was also not altered by the loss of MEKK1 (Fig. 3 A and B). To determine whether ES cells contain the same type of IKK activity characterized in differentiated cell types (40, 42, 43, 45), we generated ES cells that are deficient in the IKKγ regulatory subunit (C.M. and M.K., unpublished observations). As shown for other cell types (45), the loss of IKKγ expression prevented the activation of IKK (Fig. 4A) and NF-κB (data not shown) by TNFα or IL-1. Thus, there is nothing unusual about the IKK complex present in ES cells, and its activation requires the IKKγ regulatory subunit, but not the MAP3K MEKK1.
Figure 3.
MEKK1 is not involved in IKK and NF-κB activation in ES cells. (A) IKK activation. Wild-type (+/+) and MEKK1-deficient (−/−) ES cells were left untreated or stimulated with TNFα, IL-1, LPS, dsRNA, as described above, LPA (20 μM), or nocodazole (2 μM). IKK complexes were isolated from equal amounts of cell lysates by immunoprecipitation with anti-IKKγ monoclonal antibody, and immunocomplex kinase assays (KA) were performed by using GST-IκBα (1–54) as a substrate. Recovery of IKK was determined by immunoblotting (IB) with anti-IKKα. (B) NF-κB activation. Wild-type and MEKK1-deficient ES cells were treated as described above with TNFα, IL-1, LPS, and LPA. At the indicated times, cell extracts were prepared, and NF-κB and NF1 DNA binding activities were measured. NS, nonspecific protein–DNA complex.
Figure 4.
Dependence on IKKγ and expression of MAP3Ks and GCK in wild-type (+/+) and mutant (−/−) ES cells. (A) Wild-type and Ikkγ- (only a single Ikkγ allele was disrupted, but as Ikkγ is an x-linked gene and the ES cells we used were XO, this resulted in complete loss of IKKγ expression) ES cells were left untreated (0 time point) or treated with TNFα or IL-1 as described above. At the indicated times, cell lysates were prepared and IKK activity was determined by using an IKKα monoclonal antibody and immunocomplex kinase assay. (B) Exponentially growing ES cells or 3T3 fibroblasts were lysed, and equal amounts (50 μg) of total cell lysates were resolved by SDS/PAGE, transferred to a nitrocellulose membrane, and probed with antibodies to ASK1, TAK1, MEKK2, or GCK (Santa Cruz Biotechnology). All of these antibodies were found to specifically recognize their cognate antigens based on immunoblot analysis of cells transiently transfected with expression vectors for the different kinases (not shown).
At least two MAP3Ks, MEKK1 (3, 8, 27, 47) and ASK1 (31), were suggested to mediate JNK activation by TNFα. Another family member, TAK1, was suggested to mediate JNK activation in response to IL-1 (32). Our results indicate that, at least in ES cells, MEKK1 is required for JNK activation by TNFα, and this requirement is not compensated by other MAP3Ks, including MEKK2, ASK1, or TAK1, all of which are expressed at levels similar to MEKK1 in these cells (Fig. 4). Expression of all of these MAP3Ks, as well as the upstream kinase GCK, which was also proposed to be involved in TNFα signaling to JNK by MEKK1 (47), was unaltered in the Mekk1−/− cells.
MEKK1 Is Required for Serum-Induced JNK Activation and ES Cell Migration.
In addition to chemical and physical stressors and proinflammatory stimuli, JNK activity is stimulated by certain growth factors and small G proteins (2, 12, 13). As reported earlier (24), activation of JNK by serum or LPA was considerably reduced in Mekk1−/− cells (Fig. 5A). As these stimuli activated p38 in ES cells, we could examine the dependence of this response on MEKK1. In contrast to the defect in JNK activation, the activation of both p38 and ERK by serum or LPA was not considerably reduced by the loss of MEKK1 (Fig. 5A). However, we did find decreased activation of JNK by oncogenic Ha-Ras in Mekk1−/− cells, but expression of either activated Rac1 or full-length human MEKK1 resulted in the same JNK activation response as observed in wild-type cells (Fig. 5B). These results suggest that, in addition to TRAF proteins, which function as intermediates in proinflammatory signaling, MEKK1 is responsive to Ras proteins, which act as intermediates in growth factor signaling. However, despite previous expectations (12, 13), MEKK1 does not seem to act downstream to the Rac group of small G proteins in the pathway leading to JNK activation. Because stable expression of human MEKK1 in Mekk1−/− ES cells restored JNK activation (Y.X., unpublished results), the signaling defect in the Mekk1−/− ES cells is indeed caused by the MEKK1 deficiency and is fully reversible.
Figure 5.
MEKK1 is involved in serum- and Ras-induced JNK activation and induction of ES cell migration. (A) Serum- and LPA-induced MAPK activation. Exponentially growing wild-type (+/+) and MEKK1-deficient (−/−) ES cells were serum-starved for 2 h. The cells were either left untreated or incubated with serum (15%) or LPA (20 μM) for the indicated times (min). The cells were lysed and JNK, p38, and ERK activation and expression were determined as described in Fig. 2 by immunoblotting. (B) MEKK1 is involved in Ras-induced JNK activation. Wild-type or MEKK1-deficient ES cells were transfected with HA-JNK2 vector along with either empty vector or expression vectors for Ha-Ras(V12), Rac(L61) or full-length human MEKK1. JNK activation was determined by immunocomplex kinase assay (KA) by using GST-c-Jun (1–79) as a substrate. Fold-activation was determined after phosphorimager analysis. HA-JNK2 expression was examined by immunoblotting (IB) using anti-HA antibody. (C) MEKK1 is required for serum-stimulated ES cell migration on fibronectin. Wild-type and MEKK1-deficient ES cells were serum-starved for 2 h. Cells were allowed to migrate on fibronectin (10 μg/ml)-coated transwells (modified Boyden chambers) in the presence of BSA (10 μg/ml, control), serum (2%), TNFα (10 ng/ml), or LPA (20 μM). The numbers of migrated cells were determined after fixing and staining the cells with crystal violet. The results represent the average of three independent experiments.
Serum growth factors and Ha-Ras are important stimulators of cell proliferation. Although the loss of MEKK1 abolished JNK activation by these stimuli, it had little effect on serum-induced cell proliferation (Y.X., unpublished results), suggesting that JNK activation may not be critical for DNA synthesis and cell division, at least not in ES cells. However, serum growth factors and oncogenic Ha-Ras are also known to induce cell migration (48, 49). We considered the possibility that MEKK1 may be involved in this response. Indeed, we found that serum induced the migration of wild-type, but not Mekk1−/−, ES cells plated on fibronectin (Fig. 5C). The serum factor that induces ES cell migration was neither LPA nor TNFα. Using protein kinase inhibitors, we found that serum-induced ES cell migration was sensitive to a specific JNK inhibitor that does not inhibit other MAPKs, but not to a MEK inhibitor that prevents ERK activation (Y.X., unpublished results). Partial inhibition of cell migration was observed in response to high doses of a p38 inhibitor (Y.X. unpublished results). However, at such doses, p38 inhibitors partially interfere with JNK activity.
Discussion
Activation of JNK by TNFα requires recruitment of TRAF2 to TNF receptors (8, 30). The N-terminal effector domain of TRAF2 interacts with and activates MEKK1 on its oligomerization (27). IL-1 and LPS signaling to JNK, on the other hand, require a different TRAF protein, TRAF6 (50). Another proinflammatory stimulus that leads to JNK activation is dsRNA, which acts as a mimic of viral infection (10). dsRNA also activates NF-κB and IKK, and this activation depends on the RNA-dependent protein kinase PKR, but this enzyme is not involved in JNK activation (10). The dependence of JNK activation by dsRNA on MEKK1 suggests a similarity between dsRNA signaling and the pathways that mediate the responses to TNFα, IL-1, and LPS.
Using JNK2-deficient fibroblasts, we found that JNK activation by these proinflammatory stimuli is involved in induction of genes coding for cytokines and chemokines (10). JNK activity is also stimulated by growth factors, and this response also requires MEKK1. Although, in ES cells, JNK activation is not critically involved in stimulation of cell proliferation, it is required for induction of cell migration, another physiologically important cellular response to growth factors (48, 49). Interestingly, genetic evidence implicates the JNK pathway in induction of cell migration during Drosophila embryogenesis (51, 52).
Although MEKK1 appears to be a multifunctional MAP3K, mediating responses to proinflammatory stimuli, growth factors, and microtubule-disrupting agents, most of its downstream activity is channeled toward activation of the JNK MAPK pathway with little contribution, if any, to the ERK and p38 cascades. MEKK1 also does not contribute to activation of the IKK to NF-κB signaling cascade. Hence, the signaling function of MEKK1 is highly specific.
Acknowledgments
We thank Randall Johnson for providing reagents and allowing us to use his facilities to generate gene-targeted ES cells, Tony Manning for the JNK inhibitor, Jerold Chun for reagents, and Barbara Thompson for assisting in manuscript preparation. Y.X. and C.M were supported by postdoctoral fellowships from The Irvington Research Institute and Cancer Research Institute, respectively. This work was supported by grants from the National Institutes of Health (ES04151 and CA76188), the Department of Energy (DE-FG03-86ER60429), and the California Cancer Research Program. M.K. is the Frank and Else Schilling American Cancer Society Research Professor.
Abbreviations
- MEKK
MEK kinase
- JNK
c-Jun N-terminal kinase
- MAPK
mitogen-activated protein kinase
- MAP2K
MAPK kinase
- MAP3K
MAPK kinase kinase
- IKK
IκB kinase
- ERK
extracellular signal-regulated kinase
- TNF
tumor necrosis factor
- LPS
lipopolysaccharide
- ds
double-stranded
- ES
embryonic stem
- LPA
lysophosphatidic acid
- TRAF
TNF receptor-associated factor
- ASK
apoptosis signal-regulating kinase
- TAK
transforming growth factor β-activating kinase
- JNKK
JNK kinase
- GST
glutathione S-transferase
Footnotes
This paper was submitted directly (Track II) to the PNAS office.
References
- 1.Lange-Carter C A, Pleiman C M, Gardner A M, Blumer K J, Johnson G L. Science. 1993;260:315–319. doi: 10.1126/science.8385802. [DOI] [PubMed] [Google Scholar]
- 2.Minden A, Lin A, McMahon M, Lange-Carter C, Dérijard B, Davis R J, Johnson G L, Karin M. Science. 1994;266:1719–1723. doi: 10.1126/science.7992057. [DOI] [PubMed] [Google Scholar]
- 3.Xia Y, Wu Z, Su B, Murray B, Karin M. Genes Dev. 1998;12:3369–3381. doi: 10.1101/gad.12.21.3369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Hibi M, Lin A, Smeal T, Minden A, Karin M. Genes Dev. 1993;7:2135–2148. doi: 10.1101/gad.7.11.2135. [DOI] [PubMed] [Google Scholar]
- 5.Dérijard B, Hibi M, Wu I-H, Barrett T, Su B, Deng T, Karin M, Davis R J. Cell. 1994;76:1025–1037. doi: 10.1016/0092-8674(94)90380-8. [DOI] [PubMed] [Google Scholar]
- 6.Kyriakis J M, Banerjee P, Nikolakaki E, Dai T, Rubie E A, Ahmad M F, Avruch J, Woodgett J R. Nature (London) 1994;369:156–160. doi: 10.1038/369156a0. [DOI] [PubMed] [Google Scholar]
- 7.Cavigelli M, Li W W, Lin A, Su B, Yoshioka K, Karin M. EMBO J. 1996;15:6269–6279. [PMC free article] [PubMed] [Google Scholar]
- 8.Liu Z-G, Hu H, Goeddel D V, Karin M. Cell. 1996;87:565–576. doi: 10.1016/s0092-8674(00)81375-6. [DOI] [PubMed] [Google Scholar]
- 9.Ip Y T, Davis R J. Curr Opin Cell Biol. 1998;10:205–219. doi: 10.1016/s0955-0674(98)80143-9. [DOI] [PubMed] [Google Scholar]
- 10.Chu W-M, Ostertag D, Li Z-W, Chang L, Chen Y, Hu Y, Perrault J, Karin M. Immunity. 1999;11:1–20. doi: 10.1016/s1074-7613(00)80146-6. [DOI] [PubMed] [Google Scholar]
- 11.Hoffmann J A, Kafatos F C, Janeway C A J, Ezekowiz R A B. Science. 1999;284:1313–1318. doi: 10.1126/science.284.5418.1313. [DOI] [PubMed] [Google Scholar]
- 12.Minden A, Lin A, Claret F X, Abo A, Karin M. Cell. 1995;81:1147–1157. doi: 10.1016/s0092-8674(05)80019-4. [DOI] [PubMed] [Google Scholar]
- 13.Coso O A, Chiariello M, Yu J C, Teramoto H, Crespo P, Xu N, Miki T, Gutkind J S. Cell. 1995;81:1137–1146. doi: 10.1016/s0092-8674(05)80018-2. [DOI] [PubMed] [Google Scholar]
- 14.Sanchez I, Hughes R T, Mayer B J, Yee K, Woodgett J R, Avruch J, Kyriakis J M, Zon L I. Nature (London) 1994;372:794–798. doi: 10.1038/372794a0. [DOI] [PubMed] [Google Scholar]
- 15.Dérijard B, Raingeaud J, Barrett T, Wu I H, Han J, Ulevitch R J, Davis R J. Science. 1995;267:682–685. doi: 10.1126/science.7839144. [DOI] [PubMed] [Google Scholar]
- 16.Lin A, Minden A, Martinetto H, Claret F X, Lange-Carter C, Mercurio F, Johnson G L, Karin M. Science. 1995;268:286–290. doi: 10.1126/science.7716521. [DOI] [PubMed] [Google Scholar]
- 17.Wu Z, Jacinto E, Karin M. Mol Cell Biol. 1997;17:7407–7416. doi: 10.1128/mcb.17.12.7407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lu X H, Nemoto S, Lin A N. J Biol Chem. 1997;272:24751–24754. doi: 10.1074/jbc.272.40.24751. [DOI] [PubMed] [Google Scholar]
- 19.Holland P, Suzanne M, Campbell J, Noselli S, Cooper J. J Biol Chem. 1997;272:24994–24998. doi: 10.1074/jbc.272.40.24994. [DOI] [PubMed] [Google Scholar]
- 20.Ichijo H, Nishida E, Irie K, ten Dijke P, Saitoh M, Moriguchi T, Takagi M, Matsumoto K, Miyazono K, Gotoh Y. Science. 1997;275:90–94. doi: 10.1126/science.275.5296.90. [DOI] [PubMed] [Google Scholar]
- 21.Hirai S, Izawa M, Osada S, Spyrou G, Ohno S. Oncogene. 1996;12:641–650. [PubMed] [Google Scholar]
- 22.Shirakabe K, Yamaguchi K, Shibuya H, Irie K, Matsuda S, Moriguchi T, Gotoh Y, Matsumoto K, Nishida E. J Biol Chem. 1997;272:8141–8144. doi: 10.1074/jbc.272.13.8141. [DOI] [PubMed] [Google Scholar]
- 23.Gerwins P, Blank J L, Johnson G L. J Biol Chem. 1997;272:8288–8295. doi: 10.1074/jbc.272.13.8288. [DOI] [PubMed] [Google Scholar]
- 24.Yujiri T, Sather S, Fanger C R, Johnson G L. Science. 1998;282:1911–1914. doi: 10.1126/science.282.5395.1911. [DOI] [PubMed] [Google Scholar]
- 25.Yujiri T, Fanger G R, Garrington T P, Schlesinger T K, Gibson S, Johnson G L. J Biol Chem. 1999;274:12605–12610. doi: 10.1074/jbc.274.18.12605. [DOI] [PubMed] [Google Scholar]
- 26.Minamino T, Yujiri T, Papst P J, Chan E D, Johnson G L, Terada N. Proc Natl Acad Sci USA. 1999;96:15127–15132. doi: 10.1073/pnas.96.26.15127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Baud V, Liu Z-G, Bennett B, Suzuki N, Xia Y, Karin M. Genes Dev. 1999;13:1297–1308. doi: 10.1101/gad.13.10.1297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Hsu H, Shu H B, Pan M G, Goeddel D V. Cell. 1996;84:299–308. doi: 10.1016/s0092-8674(00)80984-8. [DOI] [PubMed] [Google Scholar]
- 29.Cao Z, Xiong J, Takeuchi M, Kurama T, Goeddel D V. Nature (London) 1996;383:443–446. doi: 10.1038/383443a0. [DOI] [PubMed] [Google Scholar]
- 30.Yeh W C, Shahinian A, Speiser D, Kraunus J, Billia F, Wakeham A, de la Pompa J L, Ferrick D, Hum B, Iscove N, et al. Immunity. 1997;7:715–725. doi: 10.1016/s1074-7613(00)80391-x. [DOI] [PubMed] [Google Scholar]
- 31.Nishitoh H, Saitoh M, Mochida Y, Takeda K, Nakano H, Rothe M, Miyazono K, Ichijo H. Mol Cell. 1998;2:389–395. doi: 10.1016/s1097-2765(00)80283-x. [DOI] [PubMed] [Google Scholar]
- 32.Ninomiya-Tsuji J, Kishimoto K, Hiyama A, Inoue J, Cao Z, Matsumoto K. Nature (London) 1999;398:252–256. doi: 10.1038/18465. [DOI] [PubMed] [Google Scholar]
- 33.Hirano M, Osada S, Aoki T, Hirai S, Hosaka M, Inoue J, Ohno S. J Biol Chem. 1996;271:13234–13248. doi: 10.1074/jbc.271.22.13234. [DOI] [PubMed] [Google Scholar]
- 34.Meyer C F, Wang X, Chang C, Templeton D, Tan T H. J Biol Chem. 1996;271:8971–8976. doi: 10.1074/jbc.271.15.8971. [DOI] [PubMed] [Google Scholar]
- 35.Lee F S, Hagler J, Chen Z J, Maniatis T. Cell. 1997;88:213–222. doi: 10.1016/s0092-8674(00)81842-5. [DOI] [PubMed] [Google Scholar]
- 36.Lee F S, Peters R T, Dang L C, Maniatis T. Proc Natl Acad Sci USA. 1998;95:9319–9324. doi: 10.1073/pnas.95.16.9319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Nemoto S, DiDonato J A, Lin A. Mol Cell Biol. 1998;18:7336–7343. doi: 10.1128/mcb.18.12.7336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Hu Y, Baud V, Delhase M, Zhang P, Deerinck T, Ellisman M, Johnson R, Karin M. Science. 1999;284:316–320. doi: 10.1126/science.284.5412.316. [DOI] [PubMed] [Google Scholar]
- 39.Nagy A, Rossant J, Nagy R, Abramow-Newerly W, Roder J C. Proc Natl Acad Sci USA. 1993;90:8424–8428. doi: 10.1073/pnas.90.18.8424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Rothwarf D M, Zandi E, Natoli G, Karin M. Nature (London) 1998;395:297–300. doi: 10.1038/26261. [DOI] [PubMed] [Google Scholar]
- 41.Klemke R L, Cai S, Giannini A L, Gallagher P J, deLanerolle P, Cheresh D A. J Cell Biol. 1997;137:481–492. doi: 10.1083/jcb.137.2.481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.DiDonato J A, Hayakawa M, Rothwarf D M, Zandi E, Karin M. Nature (London) 1997;388:548–554. doi: 10.1038/41493. [DOI] [PubMed] [Google Scholar]
- 43.Zandi E, Rothwarf D M, Delhase M, Hayakawa M, Karin M. Cell. 1997;91:243–252. doi: 10.1016/s0092-8674(00)80406-7. [DOI] [PubMed] [Google Scholar]
- 44.Mercurio F, Zhu H, Murray B W, Shevchenko A, Bennett B L, Li J, Young D B, Barbosa M, Mann M, Manning A, Rao A. Science. 1997;278:860–866. doi: 10.1126/science.278.5339.860. [DOI] [PubMed] [Google Scholar]
- 45.Yamaoka S, Courtois G, Bessia C, Whiteside S T, Weil R, Agou F, Kirk H E, Kay R J, Israël A. Cell. 1998;93:1231–1240. doi: 10.1016/s0092-8674(00)81466-x. [DOI] [PubMed] [Google Scholar]
- 46.Rosette C, Karin M. J Cell Biol. 1995;128:1111–1119. doi: 10.1083/jcb.128.6.1111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Yuasa T, Ohno S, Kehrl J H, Kyriakis J M. J Biol Chem. 1998;273:22681–22692. doi: 10.1074/jbc.273.35.22681. [DOI] [PubMed] [Google Scholar]
- 48.Bornfeldt K E, Raines E W, Graves L M, Skinner M P, Krebs E G, Ross R. Ann NY Acad Sci. 1995;766:416–430. doi: 10.1111/j.1749-6632.1995.tb26691.x. [DOI] [PubMed] [Google Scholar]
- 49.Anand-Apte B, Zetter B. Stem Cells. 1997;15:259–267. doi: 10.1002/stem.150259. [DOI] [PubMed] [Google Scholar]
- 50.Lomaga M A, Yeh W C, Sarosi I, Duncan G S, Furlonger C, Ho A, Morony S, Capparelli C, Van G, Kaufman S, et al. Genes Dev. 1999;13:1015–1024. doi: 10.1101/gad.13.8.1015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Glise B, Noselli S. Genes Dev. 1997;11:1738–1747. doi: 10.1101/gad.11.13.1738. [DOI] [PubMed] [Google Scholar]
- 52.Riesgo-Escovar J R, Jenni M, Fritz A, Hafen E. Genes Dev. 1996;10:2759–2768. doi: 10.1101/gad.10.21.2759. [DOI] [PubMed] [Google Scholar]