Abstract
Trauma is a major cause of mortality in the United States. Death among those surviving the initial insult is caused by multiple organ failure (MOF) with the liver among the organs most frequently affected. We previously demonstrated in rodents that trauma complicated by hemorrhagic shock (trauma/HS) results in liver injury that can be prevented by IL-6 administration at the start of resuscitation; however, the contribution of the severity of HS to the extent of liver injury, whether or not resuscitation is required and the mechanism for the IL-6 protective effect have not been reported. In the experiments reported here, we demonstrated that the extent of liver apoptosis induced by trauma/HS depends on the duration of hypotension and requires resuscitation. We established that IL-6 administration at the start of resuscitation is capable of completely reversing liver apoptosis and is associated with increased Stat3 activation. Microarray analysis of the livers showed that the main effect of IL-6 was to normalize the trauma/HS-induced apoptosis transcriptome. Pharmacological inhibition of Stat3 activity within the liver blocked the ability of IL-6 to prevent liver apoptosis and to normalize the trauma/HS- induced liver apoptosis transcriptome. Genetic deletion of a Stat3β, a naturally occurring, dominant-negative isoform of the Stat3, attenuated trauma/HS-induced liver apoptosis, confirming a role for Stat3, especially Stat3α, in preventing trauma/HS-mediated liver apoptosis. Thus, trauma/HS-induced liver apoptosis depends on the duration of hypotension and requires resuscitation. IL-6 administration at the start of resuscitation reverses HS-induced liver apoptosis, through activation of Stat3α, which normalizes the trauma/HS-induced liver apoptosis transcriptome.
Keywords: Nucleosomes, TUNEL, expression Microarray, transcriptome
Introduction
Trauma is the leading cause of death for those under 45 years old in the United States [1]. While almost half of the deaths occur at the time of the injury, the leading cause of death among those surviving the initial insult is multiple organ failure (MOF) [2, 3]. The liver is one of the organs most frequently affected by trauma and hemorrhagic shock, and its central role in metabolism and homeostasis makes this organ a critical one for survival of the host after severe injury [4, 5].
We previously demonstrated in rats and mice that trauma complicated by hemorrhagic shock (trauma/HS) results in liver injury as evidence by hepatocyte apoptosis [6], liver necrosis [7] and elevated transaminases [8]; however, the contribution of the severity of hemorrhagic shock to the extent of liver injury and whether or not resuscitation is required for liver injury to occur have not been reported. We also previously demonstrated that administration of IL-6 at the start of resuscitation prevented liver apoptosis and necrosis [6, 7]. IL-6 activates two anti-apoptotic signaling pathways, one involving Akt and the other involving signal transducer and activator or transcription (STAT)3. Whether or not one or both pathways are involved in the anti-apoptotic effect of IL-6 has not been determined.
In the experiments reported here, we investigated the hypotheses: 1) that trauma/HS-induced liver apoptosis depends on the severity of hemorrhagic shock and requires resuscitation; and 2) that the protective effect of IL-6 administration is mediated by Stat3. We demonstrated that the extent of liver apoptosis induced by our model of trauma/HS depends on the duration of hypotension and requires resuscitation. We established that IL-6 administration at the start of resuscitation following the longest duration of hypotension is capable of completely reversing liver apoptosis and is associated with increased Stat3 activation. Microarray analysis of the livers showed that the main effect of IL-6 was to normalize the trauma/HS-induced apoptosis transcriptome. Pharmacological inhibition of Stat3 activity within the liver blocked the ability of IL-6 to prevent liver apoptosis and to normalize the trauma/HS-induced liver apoptosis transcriptome. Genetic deletion of a Stat3β, a naturally occurring, dominant-negative isoform of the Stat3, attenuated trauma/HS-induced liver apoptosis, confirming a role for Stat3, especially Stat3α, in preventing trauma/HS-mediated liver apoptosis. Thus, trauma/HS-induced liver apoptosis depends on the duration of hypotension and requires resuscitation. IL-6 administration at the start of resuscitation reverses HS-induced liver apoptosis, through activation of Stat3α, which normalizes the trauma/HS-induced liver apoptosis transcriptome.
Materials and Methods
Rat and mouse protocols for trauma plus hemorrhagic shock
These studies were approved by the Baylor College of Medicine Institutional Review Board for animal experimentation and conform to National Institutes of Health guidelines for the care and use of laboratory animals. Adult male Sprague-Dawley rats were obtained from Harlan (Indianapolis, IN). Stat3β homozygous-deficient (Stat3βΔ/Δ) mice were generated as described [9] and re-derived at Jackson labs. Pups from heterozygous matings were tailed and genotyped by PCR, as described, with minor modifications [9].
Eight-week old male Sprague-Dawley rats (200–250 gm) were used for all experiments in this study. Rats were subjected to the sham or hemorrhagic shock (HS) protocols, as described [10, 11] with modifications. Blood was withdrawn into a heparinized syringe episodically to maintain the target MAP at 35 mmHg until blood pressure compensation failed. Blood was then returned as needed to maintain the target MAP. The amount of shed blood returned (SBR) defined 5 different levels of shock severity reflected in the duration of hypotension: 0% SBR (SBR0) represented the lowest level of shock severity (duration of hypotension, 78 ± 2.5 minutes), 10% SBR (SBR10; duration of hypotension, 149 ± 41.4 minutes), 20% SBR (SBR20; duration of hypotension, 165 ± 32.7 minutes), 35% SBR (SBR35; duration of hypotension, 211 ± 7.6 minutes), and 50% SBR (SBR50; duration of hypotension, 273 ± 24.9 minutes). At the end of the hypotensive period, rats were resuscitated as described [10, 11] and humanely sacrificed 60 minutes after the start of resuscitation. Where indicated, rats received 10 μg/kg of recombinant human IL-6 in 0.1 ml PBS at the initiation of the resuscitation or PBS alone. Sham rats were anesthetized and cannulated for 250 minutes but were not subjected to hemorrhage or resuscitation. One group of rats (UHS) was subjected to the most severe hemorrhagic shock protocol (50% SBR), but not resuscitated and kept at the target MAP (35 mmHg) for an additional 60 minutes (duration of hypotension = 336 ± 10.3 minutes) before sacrifice.
Stat3βΔ/Δ mice and wild-type littermate mice were subjected to a trauma/HS protocol [8, 12], which was similar to the rat protocol except that the target MAP in the mouse was 30mm Hg and the duration of hypotension was 180 min in all mice. Sham mice were anesthetized and immobilized in a pair-wise fashion with HS mice and sacrificed at the same time as their HS companion.
Rat and mouse livers were harvested immediately after sacrifice. The right liver lobe was fixed with paraformaldehyde solution (2%) for histologic analysis and the left lobe was snap frozen in liquid nitrogen for protein and RNA extraction.
In vivo pharmacological inhibition of Stat3
To achieve pharmacological inhibition of Stat3 activity within the lungs of rats, rats were randomized to receive by tail vein injection the G-rich, quartet-forming oligodeoxynucleotide (GQ-ODN) T40214 or nonspecific (NS)-ODN (2.5 mg ODN/kg) complexed in polyethyleneimine, as described [13], 24 hours prior to subjecting them to the SBR50 protocol with IL-6 treatment. The half-life of T40214 in tissues is ≥ 48 hr [14].
Nucleosome ELISA
Levels of histone-associated DNA fragments (nucleosomes) were determined in liver homogenates with an ELISA method (Cell Death Detection ELISAplus; Roche Diagnostics, Manheim, Germany). Frozen livers were cut by cryotome into 5 micron sections and resuspended in cell lysis buffer using the reagents from the ELISA kit. The lysates were sonicated in ice 3 times, 10 seconds each, centrifuged and supernatants harvested. Total protein concentration of each supernatant was determined by Bradford assay (Bio-Rad Protein Assay, Bio-Rad Laboratories, Inc., Hercules, CA). Equal amounts of protein (200ug) were loaded into microtiter wells in duplicate. A positive control (lyophilized, stabilized nucleosome concentrate of known concentration, provided in the kit) and a negative control (water) were also loaded in duplicate. Serial dilutions of the positive control were loaded in duplicate and used to plot a standard curve. The nucleosome concentration for each sample was obtained by plotting each sample duplicate's OD against the standard curve. The final sample nucleosome concentration was the average of the duplicates [15]. The rest of the assay was performed according to manufacterer's instructions.
Terminal deoxynucleotidyl transferase (TdT) mediated nick end labeling (TUNEL) staining
TUNEL staining to enzymatically detect the free 3'-OH termini was performed using the ApopTag Plus Peroxidase in situ Apoptosis Detection Kit from Chemicon International. Slides were rehydrated from Xylene to PBS through a series of decreasing concentrations of ethanol and digested in proteinase K (20 ug/ml) for 3 minutes at 23°C. Endogenous peroxidases were quenched for 30 minutes in 3% hydrogen peroxide in PBS. TdT enzyme was diluted in TUNEL solution buffer then used as suggested by the manufacturer. Slides were counterstained with hematoxyllin. TUNEL positive cells were assessed microscopically by counting the total nuclei and the number of TUNEL-positive nuclei in twenty random 1000x fields by an experienced histologist, blinded to the treatment each rat received. Data is presented as the number of TUNEL positive cells per high power field (hpf).
Immunoblotting
Levels of STAT3 activation within the livers of rats were assessed by immunoblotting using whole-tissue extracts of liver sections with mouse monoclonal antibody to Tyr705 phosphorylated (p)STAT3 (Cell Signaling Technology, Inc., Danvers, MA; 1:1000 dilution). Briefly, frozen livers were cut by cryotome into 5 micron sections and resuspended in cell lysis buffer (Cell Death Detection ELISAplus Kit, Roche Diagnostics, Manheim, Germany). The supernatant was sonicated in ice 3 times, 10 seconds each. Samples were then centrifuged and the supernatant evaluated by Bradford assay for total protein quantification. Protein samples (60ug total protein) were separated by SDS- PAGE and transferred to a PVDF membrane. The membrane was incubated overnight with mouse monoclonal antibody and subsequently incubated with goat antimouse antibody with horseradish peroxidase (HRP) conjugate (Zymed, San Francisco, CA) for 1 hour. ECL agent (Amersham Biosciences, UK) was used for detection. The membrane was then stripped (using RestoreTM Western Blot Stripping Buffer, PIERCE, Rockford, IL) and immuoblotting performed to detect total STAT3 protein in the whole tissue extracts of livers using mouse IgG1 monoclonal antibody to STAT3 (BD Biosciences, Rockville, MD). Densitometry was performed using ImageQuant TL v2005 software (Amersham Biosciences, Buckinghamshire, England). Results are expressed as the ratio of pSTAT3 signal (after background signal subtraction) to total STAT3 signal (after background signal subtraction) for each sample.
RNA isolation and microarray hybridization and analysis procedures
Total RNA was isolated from 4–5 micron cryotome sections of liver using TRIzol® Reagent (Invitrogen, Carlsbad, California) single step RNA isolation protocol followed by purification with RNeasy® Mini Kit (QIAGEN, Hilden, Germany) as instructed by the RAE 230A following Affymetrix protocols used within the Baylor College of Medicine Microarray Core Facility. Gene expression profiling was performed with the Affymetrix Rat Array.
Microarray Analysis
We used Affymetrics GCOS, dChip and Array Analyzer (Insightful Corporation) software packages for quality assessment and statistical analysis and annotation. Expression estimation and group comparisons were done with Array Analyzer. Low-level analyses included background correction, quartile normalization and expression estimation using GCRMA [16]. One-way analysis of variance (ANOVA) with contrasts [17] was used for group comparisons on all genes and on the list of apoptosis related genes only. P-values were adjusted for multiple comparisons using the Benjamini-Hockberg method [18]. The adjusted p-values represent false discovery rates (FDR) and are estimates of the proportion of “significant” genes that are false or spurious “discoveries”. We used a FDR=10% as cut-off.
Statistical Analysis
Data are presented as mean ± standard error of the mean (SEM). Multiple group comparisons of means were done by one-way analysis of variance (ANOVA). Post hoc analysis was done by Student-Newman-Keuls test for 2-group comparisons of means. Correlation between duration of hypotension and nucleosome levels was done for each individual study animal by Pearson correlation coefficient. Goodness of fit was evaluated by R-square. All statistical analyses were done on SigmaStat 3.5 (SYSTAT Software Inc., Chicago, IL).
Results
HS-induced liver apoptosis depends on the severity of shock
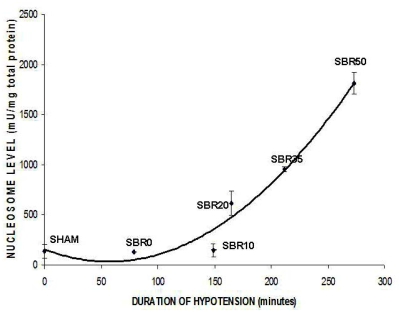
To confirm our previous findings that trauma/HS induces liver apoptosis, to determine if apoptosis is an early event following trauma/HS as well as to evaluate the contribution of the severity of shock in our rat model of trauma/HS, we measured histone-associated DNA fragments (nucleosomes) in the livers of rats subjected to increasing severity of shock 1 hr after the initiation of resuscitation. Nucleosome levels increased exponentially with increasing duration of shock (Pearson correlation coefficient 0.879, p<0.001) with the level of nucleosomes in the SBR50 group (1817.3 ± 105.9 units/ml) achieving a level 13.1 times higher than sham (139 ± 67 units/ml; p<0.001, ANOVA; Figure 1). Thus, trauma/HS-induced liver apoptosis occurs within 1 hr of resuscitation and depends on the severity of shock.
Figure 1.
Effect of shock severity on liver apoptosis. Rats were subjected to sham protocol (S) or to trauma/HS protocol with increasing duration of shock as indicated followed by resuscitation. The livers were harvested 60 minutes after the start of resuscitation. Nucleosome levels were measured in protein extracts of frozen sections of each liver and the results plotted after correction for total protein, as a function of the duration of the hypotensive period for each animal. Curve fitting was performed and the best-fitting curve shown; nucleosome levels increased exponentially with duration of hypotension (Pearson correlation coefficient=0.879, p<0.001).
Trauma/HS-induced liver apoptosis requires resuscitation
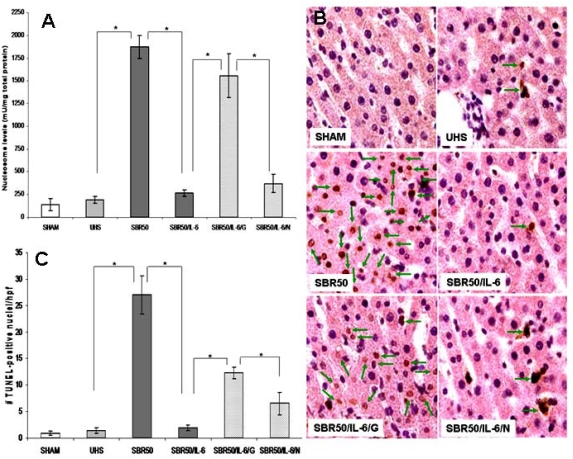
To determine the specific contribution of resuscitation to liver apoptosis, we assessed nucleosome levels as well as number of TUNEL-positive cells in the livers of rats subjected to HS without resuscitation (UHS group) and compared these results with those obtained in the sham and the resuscitated SBR50 groups. The level of nucleosomes in the UHS group (193 ± 36 units/mg total protein) was statistically indistinguishable from that of the sham group (139 ± 67 units/mg total protein; Figure 2A). Similar results were obtained when liver apoptosis was assessed by TUNEL staining. The number of TUNEL- positive nuclei/hpf in the UHS group (1.4 ± 0.5; Figure 2B, C) similar to that of the sham group (0.9 ± 0.4). In addition, histological evaluation of the cells containing TUNEL-positive nuclei revealed that more than 80% were hepatocytes, a key parenchyma cell. Thus, apoptosis within the liver following trauma/HS requires resuscitation. The fact that no liver apoptosis occurs without resuscitation suggests that complete prevention of liver apoptosis may occur with an appropriate intervention introduced at the start of resuscitation.
Figure 2.
Effect of resuscitation, IL-6 treatment and GQ-ODN pre-treatment on HSinduced liver apoptosis. Rats were subjected to sham protocol (Sham, n=3), unresuscitated hemorrhagic shock (UHS, n=3), HS treated with placebo at the beginning of resuscitation (SBR50, n=4), HS treated with IL-6 at the beginning of resuscitation (SBR50/IL-6, n=4), HS preceded by treatment with GQ oligodeoxynucleotide (GQ-ODN) 24 hours prior to resuscitation with IL-6 (SBR50/IL-6/G, n=3), or HS preceded by treatment with nonspecific-ODN (NS-ODN) 24 hours prior to resuscitation with IL-6 (SBR50/I-6/N, n=3). The livers were harvested 60 minutes after the start of resuscitation. Nucleosome levels were measured in protein extracts of frozen sections of each liver (Panel A). Data presented are mean + SEM of nucleosome level corrected for total protein for each group. Bars marked with an asterisk (*) differ significantly within the pair (p<0.05). In panel B, sections of paraformaldehyde-fixed liver were stained using the TUNEL assay. Representative photomicrographs of 1000x fields of liver specimens from each experimental group are shown. Apoptotic nuclei are indicated by arrows. In panel C, TUNEL-positive nuclei were counted; data shown are the mean ± SEM number of TUNELpositive nuclei per 1000x fields (20 fields counted). Bars marked with an asterisk (*) differ significantly within the pair (p<0.05).
IL-6 administration at the beginning of resuscitation prevents trauma/HS-induced liver apoptosis through activation of Stat3
In our mouse model of HS, we have previously demonstrated that IL-6 administration at the beginning of resuscitation prevented the development of HS-induced liver apoptosis detected 24 hrs after HS [6]. To confirm these findings and to gain an improved molecular and cellular understanding of the anti-apoptotic effects of IL-6, we measured apoptotic cell death in rats subjected to trauma/HS with the most severe HS protocol (50% SBR) and randomly assigned to receive either PBS (SBR50) or IL-6 (10 μg/kg, SBR50/IL-6) at the beginning of resuscitation. Nucleosome levels in the SBR50/IL-6 group (264 ± 36 units/ml) were decreased 3.3 times compared to those of the SBR50 group (874 ± 127 units/ml, p<0.001) and were similar to sham levels (139 ± 67 units/ml; Figure 2A). TUNEL staining confirmed these results. The number of TUNEL-positive nuclei/hpf in the SBR50/IL-6 group (1.9 ± 0.5) was decreased 14.2 times compared to the SBR50 group (27 ± 3.6, p<0.001), to levels statistically similar to those of the sham group (0.9 ± 0.4; Figures 2B and 2C). Thus, IL-6 administration at the beginning of resuscitation prevents trauma/HS-induced liver apoptosis occurring 1 hr after trauma/HS in rats as well as 24 hrs after trauma/HS in mice [6].
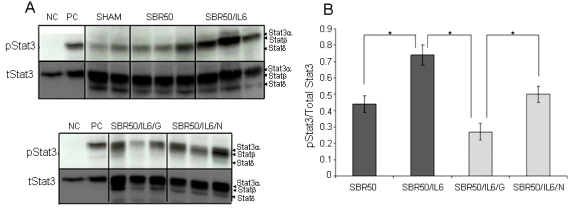
IL-6 binding to IL-Rα and gp130 results in gp130 dimerization and phosphorylation of gp130-associated protein-tyrosine kinases Jak1, Jak2, and Tyk2, which is followed by activation of two major signaling pathways within cells—Stat3 and SHP-2/Grb-2/ERK [19]. The SHP-2/Grb2/ERK pathway bifurcates resulting in activation of p38MAPK and PI-3K. Stat3 and P-I3K/Akt activation, but not p38MAPK, link to anti-apoptotic effects within cells. Stat3 mediates its anti-apoptotic effect in cancer cells through its ability to up-regulate anti-apoptotic genes such as Bcl-xL, Bcl-2 and Mcl-1 [20]. Akt is a highly promiscuous kinase with a large number of binding partners and targets [21] that posttranslationally modify transcription factor systems such as Forkhead [22, 23], IκB/NF-κB and cyclic AMP response element binding protein (CREB) [24], which together result in increased transcription of survival genes and decreased transcription of apoptotic genes [25]. To assess if the anti-apoptotic effects of IL-6 in the liver is mediated by Stat3 activation, we first determined if Stat3 is activated in the livers of rats resuscitated with IL-6. Extracts of cryotome sections of the liver harvested 1 hour after IL-6 treatment were examined by immunoblotting with mouse monoclonal antibody to Tyr705 phosphorylated (p)Stat3 (Figure 3A). Densitometric analysis of the signal intensity of the pStat3 bands normalized for total Stat3 indicated that Stat3 activity is increased 1.7 fold in the livers of IL- 6-treated rats compared to placebo-treated rats (p=0.002, ANOVA; Figure 3B).
Figure 3.
Effect of IL-6 treatment and GQ-ODN pre-treatment on Stat3 activity within the livers. Rats were subjected to the sham protocol or HS protocol and treated with placebo at the beginning of resuscitation (SBR50), HS treated with IL-6 at the beginning of resuscitation (SBR50/IL-6), HS preceded by treatment with GQ-oligodeoxynucleotide (GQ-ODN) 24 hours prior to resuscitation with IL-6 (SBR50/IL-6/G), or HS preceded by treatment with nonspecific-ODN (NS-ODN) 24 hours prior to resuscitation with IL-6 (SBR50/IL-6/N). The livers were harvested 60 minutes after the start of resuscitation. In panel A, protein extracts of whole liver were separated by SDS-PAGE and immunoblotted for phosphorylated (p)Stat3 and total Stat3 (NC = negative control, HepG2 cells incubated with PBS for 30 minutes prior to protein extraction; PC = positive control, HepG2 cells incubated with IL-6, 30 ng/ml for 30 minutes prior to protein extraction). In panel B, the pStat3 and total Sta3 bands were quantitated by densitometry and data presented as mean ± SEM of pStat3 signal corrected for total Stat3 signal for each group. Bands representing Stat3α, Stat3β and Stat3δ are indicated on the right [54, 55]. Bars marked with an asterisk (*) differ significantly within the pair (p<0.0001).
To further evaluate the role of Stat3 downstream of IL-6 in mediating its anti-apoptotic effects in the liver, we examined whether or not these effects of IL-6 could be reversed by pretreatment of rats with a G-rich oligodeoxynucleotide, G-quartet (GQ)-ODN, T40214, a novel Stat3 inhibitor, that forms a rigid G-quartet structure within cells and inhibits the growth of tumors in which Stat3 is constitutively activated [13, 14, 26, 27]. Rats were treated in a blinded fashion with GQ-ODN (SBR50/IL-6/G group) or nonspecific (NS) ODN (SBR50/IL-6/N group) 24 hours prior to being subjected to HS and resuscitation with IL-6. Pre-treatment with GQ-ODN reduced Stat3 activity within the livers of HS/I/G rats 1.9-fold compared to HS/I/N rats (Figures 3A, B). Importantly, the inhibition of Stat3 activation within the livers of the SBR50/IL-6/G rats was accompanied by a return of nucleosomes (1556 ± 241 units/ml) to levels similar to those of the placebo treated (SBR50) group (1874 ± 127 units/ml, p>0.05) and 11.2 fold higher that those of the IL-6 treated (SBR50/IL-6) group (264 ± 36 units/ml, p<0.001; Figure 2A). Similarly, the number of TUNEL-positive nuclei/hpf in livers of rats from the SBR50/IL-6/G group (12.3 ± 1.1) was 6 fold higher than that of the SBR50/IL-6 group (1.9 ± 0.5, p<0.0001); Figures 2B and C). Nucleosome levels and number of TUNEL- positive nuclei/hpf in livers of rats pre-treated with NS-ODN were indistinguishable from those of the SBR50/IL-6 group (Figures 2A and B). Thus, pharmacological inhibition of Stat3 using GQ-ODN in rats subjected to severe HS resuscitated with IL-6 markedly attenuated IL-6- mediated Stat3 activation and prevention of liver apoptosis.
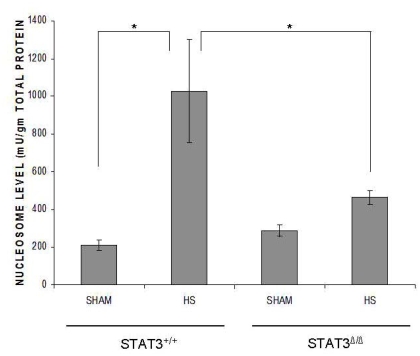
Two isoforms of Stat3 are expressed in all cells—α (p92) and β (p83)—both derived from a single gene by alternative mRNA splicing with Stat3α predominating [28]. Stat3α functions as an oncogene [29] in part through inhibiting apoptosis, while Statβ antagonizes the oncogenic function of Stat3α [30]. While mice deficient in both isoforms of Stat3 are embryonic lethal at day 6.5 to 7 [31] and mice deficient in Stat3α die within 24 hr of birth, mice deficient in Stat3β have normal survival and fertility [9]. To further support the hypothesis that Stat3, in particular Stat3α, contributes to resistance to apoptosis within the liver in the setting of HS, we subjected Stat3β homozygous-deficient (Stat3βΔ/Δ) mice and their littermate control wild type mice to a severe HS protocol (target MAP 30 mm Hg for 5 hr) and examined their livers for nucleosome levels 1 hr after the start of resuscitation. As expected, nucleosome levels in wild type HS mice (1027.3 ± 273.3 mU/mg total protein) were increased compared to wild type sham mice (210.3 ± 29.8; p < 0.01; Figure 4B). In contrast, however, nucleosome levels in the livers of Stat3βΔ/Δ HS (463.9 ± 3.9) mice were reduced 2.2 times compared to wild type HS mice and were similar to wild type sham mice (Figure 4B). These findings indicate that Stat3, in particular Stat3α, protects the liver from apoptosis in the setting of trauma/HS.
Figure 4.
Effect of Stat3β ablation on trauma/HS- induced liver apoptosis. Stat3β homozygous-deficient (Stat3βΔ/Δ) mice and their littermate control wild type mice were subjected to the murine trauma/HS protocol or sham protocol and their livers harvested 1 hr after the start of resuscitation. Nucleosome levels were measured in protein extracts of frozen sections of the liver and the results corrected for total protein. Data presented are the means ± SEM of each group (n ≥ 3). Significant differences are indicated (Student's t-test).
Microarray analysis of the liver transcriptome focusing on differential expression of apoptosis-related genes
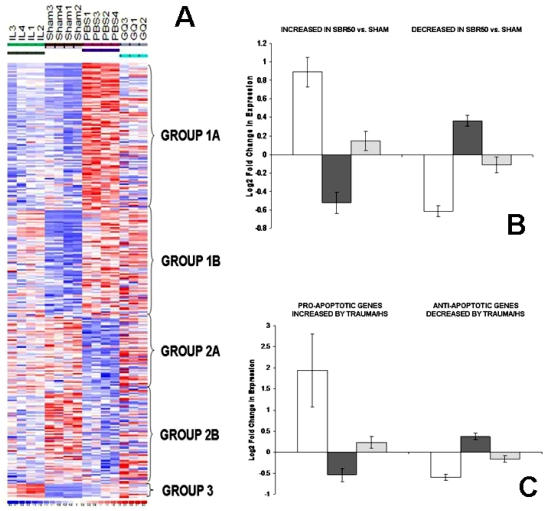
In addition to increasing the transcription of anti-apoptotic genes (Bcl-xL, Bcl-2, and Mcl-1) [29, 32–39], Stat3 has been shown to decrease transcription of pro-apoptotic genes (Bad, Bnip3l, Casp3). To evaluate the role of Stat3 downstream of IL-6 at the transcriptome level, and to identify genes altered within the livers of animals subjected to trauma/HS especially those involved in apoptosis in a global and unbiased manner, we performed Affymetrix oligonucleotide microarray analysis with RAE 230A chips. Fifteen chips were hybridized using mRNA isolated from 4 livers each from sham, SBR50, and SBR50/IL-6 groups, and 3 livers from SBR50/IL-6/G groups. All fifteen chips were included in the normalization and expression estimation steps of the analysis and were included in the statistical analysis and differential expression comparison. The 15,866 probesets on the RAE 230A chip represent 9,818 annotated genes or expressed sequence tags, including 860 apoptosis-related genes. The list of 860 apoptosis-related genes present on the RAE 230A (Table 1) was created by combining gene lists obtained by querying annotation databases provided in GeneSpring and dChip, which were derived from the Gene Ontology (GO) Consortium.
Table 1.
Apoptosis-related genes examined in the microarray experiments
| # | Accession | Gene Name | Gene Symbol |
|---|---|---|---|
| 1 | BF417479 | 24-dehydrocholestero reductase | Dhcr24 |
| 2 | NM_022225 | 5-hydroxtryptamine (serotomin) receptor IB | Etsl |
| 3 | NM_030870 | 8-oxoguanine DNA-glycosylase 1 | Oggl |
| 4 | NM_020306 | a distegnin and metalloproteinase domain 17 (tumor necrosis factor, alpha, converting enzyme) | Adaml7 |
| 5 | NM_131911 | acidic nuclear phosphoprotein 32 family, member B | Anp32b |
| 6 | NM_012912 | activating transcription factor 3 | Atf3 |
| 7 | BM391471 | activating transcription factor 5 | Atf5 |
| 8 | NM_019361 | activity regulated cytoskeletal-associated protein | Atf3 |
| 9 | AI600029 | activity-dependent neuroprotective protein | Adnp |
| 10 | NM_017155 | adenosine A1 receptor | Adoral |
| 11 | AF228684 | adenoaine A2a receptor | Adora2a |
| 12 | NM_012896 | adenosine A3 receptor | Adora3 |
| 13 | NM_031006 | adenosine deaminase, RNA-specific | Adar |
| 14 | AW523747 | adenosine molecule with Ig like domain 2 | Amigo2 |
| 15 | U07126 | adrenergic receptor, alpha 1a | Adralc |
| 16 | AY057895 | adreoergic receptor, beta 2 | Adrb2 |
| 17 | NM_012715 | adrenomedullin | Adm |
| 18 | NM_134326 | albumin | Alb |
| 19 | NM_022407 | aldehyde dehydrogenase family 1, member A1 | Aldhlal |
| 20 | NM_012498 | aldo-keto reductase family 1, member B4 (aldose reductase) | Akrlb4 |
| 21 | NM_017196 | allograft inflammatory factor 1 | Aifl |
| 22 | NM_012493 | alpha-feroprotein | Atp |
| 23 | NM_012892 | amiloride-sensitive cation channel 1, neuronal (degenerin) | Accnl |
| 24 | BM986220 | amyloid beta (A4) precursor protein | Afp |
| 25 | NM_053957 | amyloid beta (A4) precursor protein-binding, family B, member 3 | Apbb3 |
| 26 | U90829 | amyloid beta precursor protein binding protein 1 | Appbpl |
| 27 | NM_012502 | androgen receptor | Ar |
| 28 | AF275151 | androgen receptor-related apoptosis-associated protein CBL27 | Cbl27 |
| 29 | BI275292 | angiopoietin 2 | Angpt2 |
| 30 | AA818262 | angiopoietin-like 4 | Angptl4 |
| 31 | AF201331 | angiotensin I converting enzyme (peptictyl-dipeptidase A) 1 | Ace |
| 32 | BF552873 | angiotensin II receptor, type 2 | Agtr2 |
| 33 | NM_031009 | angiotensin receptor 1b | Agtr2 |
| 34 | NM_134432 | angiotensinogen (serpin pepidase inhibitor, cladle A. member 8) | Agt |
| 35 | AJ42B573 | ankyrin 3, epithelial | Ank3 |
| 36 | L81174 | ankyrin repeat domain 1 (cardiac muscle) | Ankrdl |
| 37 | NM_012904 | annexin A1 | Anxal |
| 38 | NM_024155 | annexin A4 | Anxa4 |
| 39 | NM_013132 | annexin A5 | Anxa5 |
| 40 | BI275921 | anterior pharynx defective la homolog (C. elegans) | Aphla |
| 41 | NM_133400 | apobec-1 complementation factor | Acf |
| 42 | J02582 | apolipoprotein E | Apoe |
| 43 | NM_053720 | apoptosis antagonizing transcription factor | Aatf |
| 44 | AI233249 | apoptosis inhibitor 5 (predicted) | Api5_predicted |
| 45 | AW144082 | Apoptosis, caspase activadon inhibitor (predicted) | Amid_predicted |
| 46 | AA894233 | apoptosis-inducing factor (AIF)-like mitochondrion-associated inducer of death (predicted) | Amid_predicted |
| 47 | BE116857 | apoptotic chromatin condensation inducer 1 | Acin1 |
| 48 | AF218388 | apoptotic peptidase activating factor 1 | Apaf1 |
| 49 | L07268 | aquaporin 1 | Aqpl |
| 50 | NM_019158 | anuaporin 8 | Aqp8 |
| 51 | NM_031010 | arachidonate 15-lipoxygenase | Aloxl5 |
| 52 | BF285345 | arrestin, beta 2 | Arrb2 |
| 53 | NM_013149 | aryl hydrocarbon receptor | Ahr |
| 54 | NM_012780 | aryl hydrocarbon receptor nuclear translocator | Amt |
| 55 | NM_021590 | aryl hydrocarbon receptor-interacting protein-like 1 | Aipll |
| 56 | BI274345 | ataxin 10 | Atxn10 |
| 57 | NM_058213 | ATPase, Ca++ transporting, cardiac muscle, fast twitch 1 | Atp2al |
| 58 | J04024 | ATPase, Ca++ transporting, cardiac muscle, slow twitch 2 | Atxn10 |
| 59 | AY082609 | ATP-binding cassette, sub-family B (MDR/TAP), member 1/ATP-binding cassette, sub-family B (MDR/TAP), member 1A | Abcb1 /// Abcb1a |
| 60 | NM_017228 | atrophin 1 | Atul |
| 62 | AI406520 | AXL receptor tyrosine kinase | Axl |
| 63 | NM_021752 | baculoviral IAP repeat-containing 2 | Bire2 |
| 64 | NM_023987 | baculoviral IAP repeat-containing 3 | Bire3 |
| 65 | AF304333 | baculoviral IAP repeat-containing 4 | Bire4 |
| 66 | NM_022274 | baculoviral IAP repeat-containing 5 | Bire5 |
| 67 | NM_031328 | B-cell CLL/lymphoma 10 | Bel10 |
| 68 | NM_016993 | B-cell leukemia/lynphoma 2 | Bc12 |
| 69 | NM_133416 | B-cell leukemia-lynphoma 2 related protein A1 | Bcl2a1 |
| 70 | AI172204 | B-cell receptor-associated protein 29 | Bcap29 |
| 71 | A1409930 | B-cell receptor-associated protein 31 | Bcap31 |
| 72 | NM_017258 | B-cell translocation gene 1, anti-proliferative | Btg1 |
| 73 | BI288701 | B-cell translocation gene 2, anti-proliferative | Btg2 |
| 74 | NM_139258 | Bcl2 modifying factor | Bmf |
| 75 | NM_053420 | BCL2/adeovirus E1B 19 kDa-interacting protein 3 | Bnip3 |
| 76 | NM_080888 | BCL2/adenovirus E1B 19 kDa-interacting protein 3-like | Bnip3l |
| 77 | NM_080897 | BCL2/adenovirus E1B 19 kDa-interacting protein 1 | Bnip1 |
| 78 | AI178277 | BCL2/adenovirus E1B 19kDa-interacting protein 1, NIP2 (predicted) | Buip2_predicted |
| 79 | NM_053812 | BCL2-antagonist/killer 1 | Bakl |
| 80 | BI280304 | Bcl2-associated athanogene 1 (predicted) | Bag1_predicted |
| 81 | AI231792 | Bcl2-associasd athanogene 3 | Bag3 |
| 82 | BI282898 | BC12-associated athanogene 5 | Bag5 |
| 83 | AF279911 | bcl2-associated death promoter | Bad |
| 84 | AI717547 | BCL2-associated transcription factor 1 | Bclafl |
| 85 | AF235993 | Bcl2-associated X protein | Bax |
| 86 | U72350 | Bcl2-like 1 | Bcl211 |
| 87 | NM_053733 | Bcl2-like 10 | Bcl2l10 |
| 88 | NM_022612 | BCL2-like 11 (apoptosis facilitator) | Bcl2111 |
| 89 | AI227978 | BCL2-like 12 (proline rich) (predicted) | Bcl2112_predicted |
| 90 | AA892271 | BCL2-like 13 (apoptosis facilitator) (predicted) | Bcl2113_predicted |
| 91 | NM_021850 | Bcl2-like 2 | Bcl212 |
| 92 | AF051093 | Bcl-2-related ovarian killer protein | Bok |
| 93 | NM_053739 | beclin 1 (coiled-coil, myosin-like BCL2-interacting protein) | Becul |
| 94 | AI008680 | benzodiazepine receptor, peripheral | Bzrp |
| 95 | NM_057130 | BH3 interacting (with BCL2 family) domain, apoptosis agonist | Bid3 |
| 96 | AF136282 | BH3 interacing domain death agonist | Bid |
| 97 | AI177631 | bifunctional apoptosis regulator | Bfar |
| 98 | NM_012827 | bone morphogenetic protein 4 | Bmp4 |
| 99 | BE118651 | bone morphogenic protein receptor, type II (serine/threonine kinase) | Bmpr2 |
| 100 | AA851481 | brain and reproductive organ-expressed protein | Bre |
| 101 | X67108 | brain derived neurotrophic factor | Bdnf |
| 102 | AI169085 | brain zinc finger protein | Zfpl79 |
| 103 | NM_017253 | branched chain aminotransferase 1, cytosolic | Bcat1 |
| 104 | NM_022622 | BRCA1 asociaced RING domain 1 | Bardl |
| 105 | BF404972 | Breast cancer 1 | Brcal |
| 106 | NM_012931 | breast cancer anti-estrogen resistance 1 | Bcarl |
| 107 | NM_134413 | BTB (POZ) domain containing 14B | Btbd14b |
| 108 | NM_031334 | cadherin 1 | Cdhl |
| 109 | NM_019161 | cadherin 22 | Cdh22 |
| 110 | AF061947 | calcineurin binding protein 1 | Cabin1 |
| 111 | BM958511 | calcium binding protein p22 | Chp |
| 112 | AB070350 | calcium binding protein p22 /// similar to calcium binding protein P22 (predicted) /// similar to calcium binding protein P22 (predicted) | Chp /// RGD1565588_predicted /// RGD1564956_predicted |
| 113 | BF404381 | Calcium/calmodulin-dependent protein kinase II, alpha | Chp /// RGD1565588_predicted /// RGD1564956_predicted |
| 114 | NM_016996 | calcium-sensing receptor | Casr |
| 115 | NM_019152 | calpain 1 | Capn1 |
| 116 | NM_053295 | calpastatin | Cast |
| 117 | NM_022399 | calreticulin | Calr |
| 118 | NM_032462 | Calsenilin, presenilin binding protein, EF hand transcription factor | Csen |
| 119 | NM_031017 | cAMP responsive element binding protein 1 | Crebl |
| 120 | NM_017334 | cAMP responsive element modulator | Crem |
| 121 | NM_012784 | cannabinoid receptor 1 (brain) | Cnrl |
| 122 | AW252112 | carbonic anhydrase 11 | Carll |
| 123 | BF281311 | casein kinase 2, beta subunit | Csnk2b |
| 124 | NM_057138 | CASP8 and FADD-like apoptosis regulator | Cflar |
| 125 | D85899 | caspase 1 | Casp 1 |
| 126 | NM_130422 | caspase 12 | Casp12 |
| 127 | AF136231 | caspase 2 | Casp2 |
| 128 | BM387008 | caspase 3, apoptosis related cysteine protease | Casp 3 |
| 129 | NM_053736 | caspase 4, apoptosis-related cysteine peptidase | Casp4 |
| 130 | NM_031775 | caspase 6 | Casp6 |
| 131 | BF283754 | caspase 7 | Casp7 |
| 132 | 1369262_at | caspase 8 | Casp8 |
| 133 | BF282281 | caspase 8 associated protein 2 (predicted) | Casp8ap2_predicted |
| 134 | AF262319 | caspase 9 | Casp9 |
| 135 | NM_022303 | caspase recruitment domain family, member 9 | Card9 |
| 136 | AI136555 | castration induced prostatic apoptosis-related protein 1 | Cipar1 |
| 137 | NM_022597 | cathepsin B | Ctsb |
| 138 | NM_134334 | cathepsin D | Ctsd |
| 139 | AI548979 | cationic trypsinogen | LOC286911 |
| 140 | NM_024125 | CCAAT/enhancer binding protein (C/EBP), beta | Cebpb |
| 141 | 1368813_at | CCAAT/enhancer binding protein (C/EBP), delta | Cebpd |
| 142 | NM_021744 | CD14 antigen | Cd14 |
| 143 | NM_017079 | CD1d1 antigen | Cd1d1 |
| 144 | NM_012830 | CD2 antigen | Cd2 |
| 145 | NM_013121 | CD28 antigen | Cd28 |
| 146 | 1389997_at | CD3 antigen, epsilon polypeptide (predicted) | Cd3e_predicted |
| 147 | AI044631 | CD3 antigen, gamma polypeptide | Cd3g_predicted |
| 148 | D30795 | CD38 antigen | Cd38 |
| 149 | AF065147 | CD44 antigen | Cd44 |
| 150 | NM_019295 | CD5 antigen | Cd5 |
| 151 | NM_012523 | CD53 antigen | Cd53 |
| 152 | NM_013069 | CD74 antigen (invariant polypeptide of major histocompatibility complex, class II antigen-associated) | Cd74 |
| 153 | NM_031755 | CEA-related cell adhesion molecule 1 | Ceacam 1 |
| 154 | U23056 | CEA-related cell adhesion molecule 1 /// CEA-related cell adhesion molecule 10 | Ceacaml /// Ceacam10 |
| 155 | BF284899 | cell death-inducing DNA fragmentation factor, alpha submit-like effector A (predicted) | Cidea_predicted |
| 156 | L24388 | cell division cycle 2 homolog (S.pombe)-like 1 | Cdc2 |
| 157 | AI059933 | Cell division cycle 25 homolog A (S. cerevisiae) | Cdc2 |
| 158 | NM_023026 | centaurin, gamma 1 | Cengla |
| 159 | NM_031530 | chemokine (C-C motif) ligand 2 | Cd2 |
| 160 | NM_031116 | chemokine (C-C motif) ligand 5 | Ccl5 |
| 161 | BE095824 | chemokine (C-C motif) ligand 6 | Cc16 |
| 162 | AA945737 | chemokine (C-X-C motif) receptor 4 | Cxcr4 |
| 163 | AI012221 | chloride intracellular channel 1 | clicl |
| 164 | NM_012829 | cholecystokinin | Cdk |
| 165 | NM_012832 | cholinergic receptor, nicotinic, alpha polypeptide 7 | Chma7 |
| 166 | AI171615 | chromosome segregation 1-like (S. cerevisiae) (predicted) | Csel1_predicted |
| 167 | NM_013092 | chymase 1, mast cell | Cmal |
| 168 | AA957183 | Citron | Cit |
| 169 | BG673439 | claudin 11 | Cldn1l |
| 170 | AF314657 | clusterin | Clu |
| 171 | NM_012950 | coagulation factor II (thrombin) receptor | F2r |
| 172 | NM_013057 | coagulation factor III | F3 |
| 173 | BM389673 | cofilin 1, non-muscle | Cfll |
| 174 | AF092207 | coiled-coil domain containing 5 | Ccdc5 |
| 175 | U00620 | colony stimulating factor 2 (granulocyte-macrophage) | Csf2 |
| 176 | NM_130825 | comparative gene identification transcript 94 | Cgi94 |
| 177 | NM_032060 | complement component 3a receptor 1 | C3ar1 |
| 178 | NM_053619 | complement component 5, receptor 1 | C3arl |
| 179 | AA819870 | complement component 8, beta polypeptide (mapped) | C8b |
| 180 | NM_057146 | complement component 9 | C9 |
| 181 | AW916366 | COP9 (constitutive photomorphogenic) homolog, subunit 3 (Arabidopsis thaliana) | Cops3 |
| 182 | NM_031019 | corticotropin releasing hormone | Crh |
| 183 | AW433973 | craniofacial development protein 1 | Cfdp1 |
| 184 | U47922 | crystallin, alpha A | Cryan |
| 185 | NM_012935 | crystallin, alpha B | Cryab |
| 186 | AF090695 | CUG triplet repeat, RNA binding protein 2 | Cugbp2 |
| 187 | BI284428 | cullin 1 (predicred) | Cnl1_predicted |
| 188 | BI295890 | cullin 2 (predicted) | Cul2_predicted |
| 189 | BI285751 | cullin 3 (predicted) | Cul3_predicted |
| 190 | NM_022683 | cullin 5 | Cul5 |
| 191 | X64589 | cyclin B1 | Ccnb1 |
| 192 | AW913890 | cyclin E | Ccne |
| 193 | NM_080885 | cyclin-dependent kinase 5 | Cdk5 |
| 194 | NM_053891 | cyclin-dependent kinase 5, regulatory submit 1 (p35) | Cdk5rl |
| 195 | H31766 | cyclin-dependent kinase 9 (CDC2-related kinase) | Cdk9 |
| 196 | AI010427 | cyclin-dependent kinase inhibitor 1A | Cdkn1a |
| 197 | AI013919 | cyclin-dependent kinase inhibitor 1C (P57) | Cdkn1c |
| 198 | AF474976 | cyclin-dependent kinase inhibitor 2A | Cdkn2a |
| 199 | AI409867 | cystatin B | Cstb |
| 200 | BG666933 | cystatin C | Cst3 |
| 201 | NM_031327 | cysteine rich protein 61 | Cyr61 |
| 202 | NM_023965 | cytochrome b-245, beta polypeptide | Cybb |
| 203 | NM_012839 | cytochrome c, somatic | Cycs |
| 204 | NM_012840 | cytochrome c, testis | Cyet |
| 205 | NM_012840 | cytochrome c, testis /// phosphodiesterase 11A | Cyctpd11 |
| 206 | X00469 | cytochrome P450, family 1, subfamily a, polypeptide 1 | Cpla1 |
| 207 | NM_031543 | cytochrome P450, family 2, subfamily e, polypeptide 1 | Cyp2el |
| 208 | BF285068 | cytokine induced apoptosis inhibitor 1 | Ciapin1 |
| 209 | BI298817 | Cytotoxic granule-associated RNA binding protein 1 | Tial |
| 210 | AI169146 | D4, zinc and double PHD fingers family 2 (predicted) | Dpf2_predicted |
| 211 | AI408110 | DEAD (Asp-Glu-Ala-Asp) box polypeptide 19 | Ddx19 |
| 212 | BM389310 | DEAD (Asp-Glu-Ak-Asp) box polypeptide 41 (predicted) | Ddx41_predicted |
| 213 | BI285645 | death associated protein 3 | Dap3 |
| 214 | AA818353 | death associated protein kinase 1 (predicted) | Dapk1_predicted |
| 215 | NM_031800 | death effector domain-containing | Dedd |
| 216 | NM_022526 | death-associated protein | Dap |
| 217 | NM_022546 | death-associated protein kinase 3 | Dapk3 |
| 218 | AI013627 | defender against cell death 1 | Dadl |
| 219 | NM_080482 | deleted in bladder cancer chromosome region candidate 1 (human) | Dbccr |
| 220 | NM_012841 | deleted in colorectal carcinoma | Dcc |
| 221 | NM_013097 | deoxyribonuclease I | Dnase1 |
| 222 | AF178975 | deoxyribonuclease II | Dnase 2 |
| 223 | NM_053907 | deoxyribonuclease I-like 3 | LOC681124 |
| 224 | NM_022531 | desmin | Des |
| 225 | BE110572 | diablo homolog (Drosophila) | Diablo |
| 226 | AI236726 | DNA fragmentation factor, alpha subunit | Dffa |
| 227 | NM_053362 | DNA fragmentation factor, beta subunit | Dffb |
| 228 | NM_024134 | DNA-damage inducible transcript 3 | Ddit3 |
| 229 | NM_080906 | DNA-damage-inducible transcript 4 | Ddit4 |
| 230 | BI282224 | DnaJ (Hsp40) homolog, subfamily A, member 3 | LOC294513 |
| 231 | BM384926 | DnaJ (Hsp40) homolog, subfamily B, member 1 (predicted) | Dnajbl_predicted |
| 232 | NM_012699 | DnaJ (Hsp40) homolog, subfamily B, member 9 | Dnajb9 |
| 233 | BI285682 | DnaJ (Hsp40) homolog, subfamily C, member 7 | Dnajc7 |
| 234 | BF406540 | DnaJ (Hsp40) related, subfamily B, member 13 | Dnajb13 |
| 235 | L12407 | dopamine beta hydroxylase | Dbh |
| 236 | NM_012547 | dopamine receptor 2 | Drd2 |
| 237 | M35077 | dopamine receptor D1A | Drdla |
| 238 | BE110108 | dual specificicy phosphatase 1 | Dusp1 |
| 239 | AI172067 | dual specificity phosphatase 22(predicted) | Dusp22_predicted |
| 240 | U23438 | dual specificity phosphatase 4 | Dusp4 |
| 241 | NM_133578 | dual specificity phosphatase 5 | Dusp5 |
| 242 | NM_053883 | dual specificity phosphatase 6 | Dusp6 |
| 243 | L24562 | dynamin 2 | Dnm2 |
| 244 | NM_053319 | dynein light chain LC8-type 1 | Dynll1 |
| 245 | NM_012551 | early growth response 1 | Egr1 |
| 246 | AF115249 | endothelial differentiation, sphingolipid G-protein-coupled receptor, 8 | Edg8 |
| 247 | NM_023090 | endothelial PAS domain protein 1 | Epasl |
| 248 | NM_053596 | endothelin converting enzyme 1 | Ecel |
| 249 | AB023896 | endothelin converting enzyme-like 1 | Ecell |
| 250 | X57764 | endothelin receptor type B | Ednrb |
| 251 | BI291645 | engulfment and cell motility 3, ced-12 homolog (C. elegans) | Elmo3 |
| 252 | NM_012842 | epidermal growth factor | Egfr |
| 253 | M37394 | epidermal growth factor receptor | Egfr |
| 254 | AF187818 | epidermal growth factor receptor /// peptidase D (mapped) | Egfr /// Pepd_mapped |
| 255 | BF564277 | epilepsy, progressive myoclonic epilepsy, type 2 gene alpha | Epme |
| 256 | NM_017001 | erythropoietin | Epo |
| 257 | AA866269 | Estrogen receptor 1 | Esr1 |
| 258 | AF042058 | estrogen receptor 2 beta | Esr2b |
| 259 | BF398331 | estrogen receptor-binding fragment-associated gene 9 | Ebag9 |
| 260 | AI412114 | etoposide induced 2.4 mRNA | Ei24 |
| 261 | NM_012660 | eukaryotic translation elongation factor 1 alpha 2 | Eefla2 |
| 262 | AI600237 | eukaryotic translation elongation factor 1 epsilon 1 (predicted) | Eeflel_predicted |
| 263 | NM_053950 | enkaryotic translation initiation factor 2B, subunit 4 delta | Eif2b4 |
| 264 | NM_053974 | eukaryotic translation initiation factor 4E | Eif4e |
| 265 | BI283681 | eukaryotic translation initiation factor 5A | Eif5a |
| 266 | BM388758 | excision repair cross-complementing rodent repair deficiency, complementation group 3 | Ercc3 |
| 267 | D13374 | expressed in non-metastatic cells 1 | Nme1 |
| 268 | AI385371 | extra spindle poles like 1 (S. cerevisiae) (predicted) | Espll_predicited |
| 269 | NM_080895 | Fas apoptotic inhibitory molecule | Faim |
| 270 | AF044201 | Fas apoptotic inhibitory molecule 2 | Faim2 |
| 271 | NM_080891 | Fas deith domain-associated protein | Daxx |
| 272 | NM_012908 | Fas ligand (TNF superfamily, member 6) | Faslg |
| 273 | AI227743 | Fas-activated serine/threonine kinase | Fastk |
| 274 | NM_130406 | Fas-associated factor 1 | Fafl |
| 275 | NM_053843 | Fe receptor, IgG, low affinity III /// Fe gamma receptor II beta | Fcgr3///LOC498276 |
| 276 | AA999104 | Feminization 1 homolog b (C. elegans) (predicred) | Fem1_predicted |
| 277 | NM_019305 | fibroblast growth factor 2 | Fgf2 |
| 278 | NM_130817 | fibroblast growth factor 3 | Fgf3 |
| 279 | AB079673 | fibroblast growth factor 4 | Fgf4 |
| 280 | NM_133286 | fibroblast growth factor 8 | Fgf8 |
| 281 | S54008 | Fibroblast growth receptor 1 | Fgfrl |
| 282 | NM_053429 | fibroblast growth factor receptor 3 | Fgfr3 |
| 283 | AA893484 | fibronectin 1 | Fnl |
| 284 | AI103600 | Filamin C, gamma (actin binding protein 280) (predicted) | Flnc_predicted |
| 285 | AF040256 | folate hydrolase | Folhl |
| 286 | M36804 | follicle stimulating hormone beta | Fshb |
| 287 | NM_012561 | follistatin | Fst |
| 288 | BI295511 | forkhead box O1A | Foxola |
| 289 | AI231684 | forkhead bos O3a(predicted) | Foxo3a_predicted |
| 290 | NM_012953 | fos-like antigen 1 | Fosl1 |
| 291 | NM_012954 | fos-like antigen 2 /// FBJ osteosarcoma oncogene B | Fos12 /// Fosb |
| 292 | NM_017181 | fumarylacetoacetate hydrolase | Fah |
| 293 | BE108192 | Gl to S phase transition 1 | Gsptl |
| 294 | NM_033237 | galanin | Gal |
| 295 | NM_019172 | galanin receptor 2 | Galr2 |
| 296 | NM_0533840 | gamma-glutamyltransferase 1 | Ggt1 |
| 297 | NM_019281 | gap junction membrane channel protein alpha 9 | Gja9 |
| 298 | NM_053388 | gap junction membrane channel protein beta 6 | Gja6 |
| 299 | NM_012849 | gastrin | Gast |
| 300 | AA945758 | gb:AA945758/DB_XREF=gi:3105674/DB_XREF=EST201257/CLONE=RLUAS87/FEA=EST/CNT-13/TID=Rn.7908.1/TIER=Stack/STK=8/UG=Rn.7908/UG_TITLE=ESTs | NS |
| 301 | AI230220 | gb:AI230220/DB_XREF=gi:3814107/DB_XREF=EST226915/CLONE=REMCT79/FEA=EST/CNT=9/TID=Rn.24381.1/TIER=Stack/STK=7/UG=Rn.24381/UG_TITLE=ESTs, Moderately similar to MLE3 RAT MYOSIN LIGHT CHAIN 3, SKELETAL MUSCLE ISOFORM (R.norvegicus) | NS |
| 302 | BF555051 | gb:BF555051/DB_XREF=gi:11664781/DB_XREF=UI-R-E0-cg-f-04-0-Ur1/CLONE=UI-R-E0-cg-f-04-0-UI/FEA=EST/CNT=3/TID=Rn.65517.1/TIER=ConsEnd/STK=1/UG=Rn.65517/UG_TITLE=ESTs, Weakly similar to VITAMIN K-DEPENDENT PROTEIN S PRECURSOR (R.norvegicus) | NS |
| 303 | BM384229 | gb:BM384229/DB_XREF=gi:18184282/DB_XREF=UI-R-DZO-cks-c-03-0-UI.s1/CLONE=UI-R-DZO-cks-c-03-0-UI/FEA=EST/CNT=12/TID=rn.14615.1/TIER=Stack/STK=11/UG=Rn.14615/UG_TITLE=ESTs, Highly similar to TRA2 MOUSE TNF RECEPTOR ASSOCIATED FACTOR 2 (M.musculus) | NS |
| 304 | J02582 | gb:J02582/DB_XREF=gi:202957/FEA=DNA_2/CNT=1/TID=Rn.64667.1/TIER=ConsEnd/STK=0/UG=Rn.64667/UG_TITLE=Rat apolipoprotein E gene, complete cds/DEF=Rat apolipoprotein E gene, complete cds | NS |
| 305 | BI285576 | gelsolin | Gsn |
| 306 | NM_021669 | ghrelin precursor | Ghrl |
| 307 | NM_019139 | glial cell line derived neurotrophic factor | Gdnf |
| 308 | BF281741 | glioma tumor suppressor candidate region gene 2 | Gltscr2 |
| 309 | NM_012728 | glucagon-like peptide 1 receptor | Glp1r |
| 310 | BI283882 | glucose phosphate isomerase | Gpi |
| 311 | NM_017006 | glucose-6-phosphate dehydrogenase X-linked | G6pdx |
| 312 | U08259 | glutamate receptor, ionotropic, NMDA2C | Grin2c |
| 313 | NM_017010 | glutamate receptor, ionotropic, N-methyl D-aspartate 1 | Grin1 |
| 314 | AF001423 | glutamate receptor, ionotropic, N-methyl D-aspartate 2A | Grin 2a |
| 315 | M91562 | glutamate receptor, ionotropic, N-methyl D-aspartate 2B | Grin2b |
| 316 | NM_017011 | glutamate receptor, metabotropic 1 | Gria2bml |
| 317 | M92075 | glutamate receptor, metabotropic 2 | Grm2 |
| 318 | AW522430 | glutamate receptor, metabotropic 3 | Grm3 |
| 319 | NM_022202 | glutamate receptor, metabotropic 8 | Gria2bm8 |
| 320 | J05181 | glutamate-cysteine ligase, catalytic subunit | Gclc |
| 321 | BG380882 | glutaminyl-tRNA synthetase /// similar to glutaminyl-tRNA synthetase (predicted) | Qars /// RGD1562301_predicted |
| 322 | NM_022278 | glutaredoxin 1 (thioltransferase) | Glrxl |
| 323 | S41066 | glutathione peroxidase 1 | Gpx1 |
| 324 | NM_017165 | glutathione peroxidase 4 | Gpx4 |
| 325 | NM_017013 | glutathione-S-transferase, alpha type 2 | Gsta2 |
| 326 | X02904 | glutathione-S-transferase, pi 1 /// glutathione S-transferase, pi 2 | Gstp1 /// Gstp 2 |
| 327 | NM_017008 | glyceraldehyde-3-phosphate dehydrogenase /// similar to glyceraldehyde-3-phosphate dehydrogenase (predicted) /// similar to glyceraldehyde-3-phosphate dehydrogenase (predicted) /// similar to glyceraldehyde-3-phosphate dehydrogenase (predicted) /// similar to glyceraldehyde-3-phosphate dehydrogenase (predicted) | Gapdh /// RGD1564688_predictated /// RGD1564351_predicted /// RGD1561683_predicted /// RGD1565368_predicted |
| 328 | BF287444 | glycogen synthase kinase 3 beta | Gsk3b |
| 329 | AI103970 | glyoxylase 1 | Glo1 |
| 330 | BM391371 | goliath | LOC652955 |
| 331 | NM_031038 | gonadotropin releasing hormone receptor | Gnrhr |
| 332 | NM_012767 | gonadotropin-releasing hormone 1 | Gnrh1 |
| 333 | M34097 | granzyme B | Gzmb |
| 334 | U57063 | granzyme G | Gzmg |
| 335 | NM_019282 | gremlin 1 homolog, cysteine knot superfamily (Xenopus laevis) | Grem1 |
| 336 | NM_024127 | growth arrest and DNA-damage-inducible 45 alpha | Gadd45a |
| 337 | B1287978 | growth arrest and DNA-damage-inducible 45 beta | Gadd45b |
| 338 | AI599423 | growth arrest and DNA-damage-inducible 45 gamma | Gadd45g |
| 339 | NM_057100 | growth arrest specific 6 | Gas6 |
| 340 | X62853 | Growth factor receptor bound protein 2 | Grb2 |
| 341 | V01238 | growth hormone 1 | Ghl |
| 342 | AI170771 | growth hormone receptor | Ghr |
| 343 | U94321 | growth hormone secretagogue receptor | Gshr |
| 344 | NM_024356 | GTP cyclohydrolase 1 | Gch |
| 345 | BM389208 | GTPase, IMAP family member 4 | Gimap4 |
| 346 | M12672 | guanine nucleotide binding protein, alpha inhibiting 2 | Gnai2 |
| 347 | BE117491 | gnanine nucleotide binding protein, alpha q polypeptide | Gnaq |
| 348 | BM390519 | GULP, engulfment adaptor PTB domain containing 1 | Gulp1 |
| 349 | BG379941 | Harvey rat sarcome viral (v-Ha-ras) oncogene homolog | Hras |
| 350 | NM_012966 | heat shock 10 kDa protein 1 (chaperonin 10) | Hspel |
| 351 | AI236601 | heat shock 105kDa/110kDa protein 1 | H2phl |
| 352 | NM_031970 | heat shock 27kDa protein 1 | Hspb1 |
| 353 | NM_031971 | heat shock 70kD protein 1A /// heat shock 70kD protein IB (mapped) | Hspala /// Hspalb_mapped |
| 354 | BI278231 | heat shock 70kD protein IB (mapped) | Hspalb_mapped |
| 355 | M14050 | heat shock 70kDa protein 5 (glucose-regulated protein) | Hspa5 |
| 356 | BI282281 | heat shock 70kDa protein 9A (predicted) | Hspa9a_predicted |
| 357 | AI237389 | heat shock 90kDa protein 1, beta | Hspcb |
| 358 | NM_022229 | heat shock protein 1 (chaperonin) | Hspd1 |
| 359 | BG671521 | heat shock protein 1, alpha | Hspca |
| 360 | AF077354 | heat shock protein 4 | Hspa4 |
| 361 | NM_012580 | heme oxygenase (decyclinj) 1 | Hmox1 |
| 362 | NM_013185 | hemopoietic cell kinase | Hck |
| 363 | 1387701_at | hepatocyte growth factor | Hgf |
| 364 | NM_012734 | hexokinase 1 | Hk1 |
| 365 | NM_012735 | hexokinase 2 | Hk2 |
| 366 | BG378885 | high mobility group AT-hook 1 | Hmga1 |
| 367 | BE107162 | high mobility group box 1 | Hmgb1 |
| 368 | AF275734 | high mobility group box 1 /// similar to High mobility group protein 1 (HMG-1) (predicted) /// similar to Hmgb1 protein (predicted) /// similar to High mobility group protein 1 (HMG-1) (predicted) | Hmgbl /// RGD1562312_predicted /// RGD1563786_predicted /// RGD1563012_predicted |
| 369 | AI180339 | histone deacetylase 1 (predicted) | Hdacl_predicted |
| 370 | NM_053448 | histone deacetylase 3 | Hdae3 |
| 371 | BF403027 | histone deacetylase 5 | Hdac5 |
| 372 | NM_053609 | HLA-B-associated transcript 3 | Bat3 |
| 373 | M37568 | homeo box C8 (mapped) | Hoxc8_mapped |
| 374 | BM392321 | homeodomain interacting protein kinase 2 (predicted) | Hipk2_predicted |
| 375 | NM_031787 | homeodomain interacting protein kinase 3 | Hipk3 |
| 376 | AB003726 | homer homolog 1 (Drosophila) | Homer1 |
| 377 | NM_053523 | homocysteine-inducible, endoplasmic reticulum stress-inducible, ubiquitin-like domain member 1 | Herpud1 |
| 378 | BE111733 | hormone-regulated proliferation associated protein 20 | Hrpap20 |
| 379 | AW253339 | huntingtin interacting protein 1 | Hipl |
| 380 | U18650 | Huntington disease gene homolog | Hdh |
| 381 | NM_019371 | hypothetical gene supported by NM_019371 | LOC497816 |
| 382 | BM390522 | hypothetical gene supported by NM_130426 | LOC497808 |
| 383 | H31665 | hypoxia induced gene 1 | Hig1 |
| 384 | NM_024359 | hypoxia inducible factor 1, alpha subunit | Hifla |
| 385 | BI282904 | hypoxia up-regulated 1 | Hyou1 |
| 386 | AI176519 | immediate early response 3 | Ier3 |
| 387 | AI411947 | immunoglobulin heavy chain 1a (serum IgG2a) | Igh-1a |
| 388 | NM_023973 | indoleamine-pyrrole 2,3 dioxygenase | Indo |
| 389 | NM_012590 | inhibin alpha | Inha |
| 390 | NM_017128 | inhibin beta-A | Inhba |
| 391 | NM_013060 | inhibitor of DNA binding 2 | Id2 |
| 392 | AF000942 | inhibitor of DNA binding 3 | Id3 |
| 393 | NM_053355 | inhibitor of kappaB kionase beta | Ikbkb |
| 394 | J05510 | inositol 1,4,5-triphosphate receptor 1 | Itpr1 |
| 395 | NM_019311 | inositol polyphosphate-5-phosphatase D | Inppd5 |
| 396 | NM_019129 | insulin 1 | Igf2bp1 |
| 397 | NM_032074 | insulin receptor substrate 3 | Irs3 |
| 398 | M15481 | insulin-like growth factor 1 | Igfl |
| 399 | NM_052807 | insulin-like growth factor 1 receptor | Igflr |
| 400 | NM_031511 | insulin-like growth factor 2 | Igf2 |
| 401 | NM_012588 | insulin-like growth factor binding protein 3 | Igfbp3 |
| 402 | BF282337 | integral membrane protein 2B | Itm2b |
| 403 | NM_017022 | integrin beta 1 (fibronectin receptor beta) | Itgbl |
| 404 | NM_133409 | integrin linked kinase | Ilk |
| 405 | NM_019127 | interferon beta 1, fibroblast | Ifnblf |
| 406 | AF010466 | interfereon gamma | Ifng |
| 407 | NM_012591 | interferon regulatory factor 1 | Irf1 |
| 408 | NM_017019 | interleukin 1 alpha | Il1a |
| 409 | NM_031512 | interleukin 1 beta | Il1b |
| 410 | L02926 | interleukin 10 | Il10 |
| 411 | AF347836 | interleukin 11 receptor, alpha chain 1 | Il11ra1 |
| 412 | NM_053828 | interleukin 13 | I113 |
| 413 | AF015718 | interleukin 15 | I115 |
| 414 | AJ222813 | interleukin 18 | I118 |
| 415 | NM_013163 | interleukin 2 receptor, alpha chain | I12ra |
| 416 | NM_013195 | interleukin 2 receptor, beta chain | I12rb |
| 417 | NM_031513 | interleukin 3 | I16 |
| 418 | X16058 | interleukin 4 | I14 |
| 419 | NM_012589 | interleukin 6 | I16 |
| 420 | AF367210 | interleukin 7 | I17 |
| 421 | BF405951 | Interleukin-1 receptor-associated kinase 4 (predicted) | Irak4_predicted |
| 422 | 1388184_at | isoprenylcysteine carboxyl methyltransferase | Icmt |
| 423 | NM_031514 | Janus kinase 2 | Jak2 |
| 424 | BE096021 | Jun D proto-oncogene | Jundl |
| 425 | BI288619 | Jun oncogene | Jun |
| 426 | NM_021836 | Jun-B oncogene | Junb |
| 427 | NM_012696 | kininogen 1 /// K-kininogen /// similar to alpha-1 major acute phase protein prepeptide | Kng1 /// LOC215087 /// MGC108747 |
| 428 | NM_031135 | Kruppel-like factor 10 | Klf10 |
| 429 | BM385790 | Kruppel-like factor 2 (lung) (predicted) | Klf2_predicted |
| 430 | NM_053394 | Kruppel-like factor 5 | Klf5 |
| 431 | NM_053902 | kynureninase (L-kynurenine hydrolase) | Kynu |
| 432 | NM_012594 | Lactalbiuain, alpha | Lalba |
| 433 | NM_019904 | lectin, galactose binding, soluble 1 | Lgals1 |
| 434 | NM_031832 | lectin, galactose binding, soluble 3 | Lgals3 |
| 435 | NM_022582 | lectin, galactose binding, soluble 7 | Lgals7 |
| 436 | NM_031048 | leukemia inhibitory factor receptor | Lifr |
| 437 | NM_031727 | LIM motif-containing protein kinase 1 | Limk1 |
| 438 | NM_130741 | lipocalin 2 | Lcn2 |
| 439 | BF289368 | lipopolysaccharide binding protein | Lbp |
| 440 | BI284739 | LPS-induced TN factor | Litaf |
| 441 | AA874924 | lymphocyte antigen 86 (predicted) | Lv86_predicted |
| 442 | AI137137 | lymphocyte protein tyrosine kinase (mapped) | Lck_mapped |
| 443 | AI012109 | lymphocyte specific 1 | Lsp1 |
| 444 | NM_080769 | lymphotoxin A | Lta |
| 445 | NM_053538 | Iysosomal-associated protein transmembrane 5 | Laptm5 |
| 446 | NM_031051 | macrophage migration inhibitory factor | Mif |
| 447 | NM_024352 | Macrophage stimulating 1 (hepatocyte growth factor-like) | Mst1 |
| 448 | NM_019191 | MAD homolog 2 (Drosophila) | Smad2 |
| 449 | AA997679 | MAD homolog 3 (Drosophila) | Smad3 |
| 450 | NM_019275 | MAD homolog 4 (Drosophila) | Smad4 |
| 451 | AW521447 | MAD homolog 7 (Drosophila) | Madh7 |
| 452 | NM_053585 | MAP-kinase acivating death domain | Madd |
| 453 | U65656 | matrix metallopeptidase 2 | Mmp2 |
| 454 | NM_031055 | matrix metallopeptidase 9 | Mmp9 |
| 455 | BI289109 | max binding protein (predicted) | Mnt_predicted |
| 456 | AW143154 | megakaryoblastic leukemia (translocation) 1 (predicted) | MKl1_predicted |
| 457 | NM_053409 | melanoma antigen, family D, 1 | Magedl |
| 458 | AF411318 | metallothionein 1a | Mtla |
| 459 | NM_053307 | methionine sulfoxide reductase A | Msrb2 |
| 460 | BI281702 | microtubule-associated protein 1b | Map1b |
| 461 | BE107978 | microtubule-associated protein tau /// hypothetical gene supported by NM_017212 | Mapt /// LOC497674 |
| 462 | BG685132 | mitochondrial carrier homolog 1 (C. elegans) | Mtchl |
| 463 | AA943734 | mitochondrial protein. 18 kDa | MGC94604 |
| 464 | BG378230 | mitochondrial ribosomal protein S30 (predicted) | Mrps30_predicted |
| 465 | NM_053842 | mitogen activated protein kinase 1 | Mapk1 |
| 466 | NM_012806 | mitogen activated protein kinase 10 | Mapk10 |
| 467 | AW254190 | mitogen activated protein kinase 14 | Mapk14 |
| 468 | AF155236 | mitogen activated protein kinase 3 | Mapk3 |
| 469 | NM_053777 | mitogen activated protein kinase 8 interacting protein | Mappk8ip |
| 470 | D13341 | mitogen activated protein kinase 1 | Map2k1 |
| 471 | D14592 | mitogen activated protein kinase kinase 2 | Map2k2 |
| 472 | NM_053887 | mitogen activated protein kinase kinase kinase 1 | Map3k1 |
| 473 | NM_013055 | mitogen activated protein kinase kinase kinase 12 | Map3k12 |
| 474 | AI146037 | Mitogen activated protein kinase kinase kinase 7 (predicted) | Map3k7_predicted |
| 475 | AI575972 | Mitogen-activaced protein kinase 8 interacting protein 2 | Mapk8ip2 |
| 476 | NM_017322 | mitogen-activated protein kinase 9 | Mapk9 |
| 477 | BI281589 | mitogen-activated protein kinase kinase kinase 11 | Map3k11 |
| 478 | NM_053847 | mitogen-activated protein kinase kinase kinase 8 | Map3k8 |
| 479 | D00688 | monoamine oxidase A | Maoa |
| 480 | NM_012982 | msh homeo box homolog 2 (Drosophila) | Msx2 |
| 481 | NM_053337 | Msx-interacting-zinc finger | Mizl |
| 482 | BI274326 | mucin 1, transmembrane | Muc1 |
| 483 | BM391100 | mucin 4 | Muc4 |
| 484 | NM_031053 | mutL homolog 1 (E. coli) /// hypothetical gene supported by NM_031053 | Mlh1 /// LOC407834 |
| 485 | NM_021837 | myc-like oncogene, s-myc protein | Myes |
| 486 | NM_012798 | myelin and lymphocyte protein. T-cell differentiation protein | Ma1 |
| 487 | BF281184 | myeloblastosis oncogene-like 2 (predicted) | Mybl2_predicted |
| 488 | NM_012603 | myelocytomatosis viral oncogene homolog (avian) | Myc |
| 489 | AI172056 | myeloid cell leukemia sequence 1 | Mcl1 |
| 490 | BI284349 | myeloid differentiation primary response gene 116 | Mydl16 |
| 491 | AI238590 | myeloid differentiation primary response gene 88 | Myd88 |
| 492 | NM_030860 | myocyte enhancer factor 2D | Mef2d |
| 493 | J02679 | NAD(P)H dehydrogenase, quinone 1 | Nqo1 |
| 494 | NM_053683 | NADPH oxidase 1 | Noxal |
| 495 | AI178285 | NCK-associated protein 1 | Nckap1 |
| 496 | NM_031069 | NEL-like 1 (chicken) | Nell1 |
| 497 | NM_012610 | nerve growth factor receptor (TNFR superfamily, member 16) | Ngfr |
| 498 | NM_053401 | nerve growth factor receptor (TNFRSF16) associated protein 1 | Ngfrap1 |
| 499 | BM388972 | nerve growth factor, beta (mapped) | Norb |
| 500 | U02323 | neuregulin 1 | Nrg1 |
| 501 | NM_023868 | neurotrophic Y receptor Y2 | Npy2r |
| 502 | NM_021589 | neurotrophic tryosine kinase, receptor, type 1 | Ntrk1 |
| 503 | NM_031073 | neutrophin 3 | Ntf3 |
| 504 | AI598730 | neurotrophin receptor a associated death domain | Nradd |
| 505 | NM_053734 | neutrophil cytosolic factor 1 | Ncf1 |
| 506 | BI285459 | nicastrin | Ncstn |
| 507 | L12562 | nitric oxide synthase 2, inducible | Nos2 |
| 508 | AJ011116 | nitric oxide synthase 3, endothelial cell | Nos3 |
| 509 | NM_053507 | non-metastatic cell expressed protein 3 | Nme3 |
| 510 | BF389398 | Notch gene homolog 1 (Drosophila) | Notchl |
| 511 | AI011448 | Notch gene homolog 2 (Drosophila) | Notch2 |
| 512 | NM_020087 | Notch gene homolog 3 (Drosophila) | Notch3 |
| 513 | BG377358 | nuclear factor of activated T-cells, cytoplasmic, calcineurin-dependent 4 | Nfatc4 |
| 514 | AA858801 | nuclear factor of kappa light chain gene enhancer in B-cells 1, p105 | Nfkb1 |
| 515 | AW672589 | nuclear factor of kappa light chain gene enhancer in B-cells inhibitor, alpha | Nfkbia |
| 516 | NM_030867 | nuclear factor of kappa light chain gene enhancer in B-cells inhibitor, beta | Nfkbib |
| 517 | NM_012991 | nuclear pore associated protein | Npap60 |
| 518 | NM_021745 | nuclear receptor subfamily 1, group H, member 4 | Nrlh4 |
| 519 | NM_05298O | nuclear receptor subfamily 1, group I, member 2 | Nrcli2 |
| 520 | NM_017323 | nuclear receptor subfamily 2, group C, member 2 | Nrc2c2 |
| 521 | AY066016 | nuclear receptor subfamily 3, group C, member 1 | Nr3c 1 |
| 522 | NM_013131 | nuclear receptor subfamily 3, group C, member 2 | Nr3c2 |
| 523 | NM_024388 | nuclear receptor subfamily 4, group A, member 1 | Nr4a1 |
| 524 | NM_031628 | nuclear receptor subfamily 4, group A, member 3 | Nr4a3 |
| 525 | NM_022799 | nuclear ubiquitous casein kinase and cyclin-dependent kinase substrate | Nucks |
| 526 | NM_053516 | nucleolar protein 3 (apoptosis repressor with CARD domain) | Nol3 |
| 527 | NM_012992 | nucleophosmin 1 | Npml |
| 528 | J04943 | nucleophosmin 1 /// similar to Nucleophosmin (NPM) (Nucleolar phosphoprotein B23) (Numatrin) (Nucleolar protein NO38) | Npml /// LOC300303 |
| 529 | BI286040 | nucleoporin 62 | Nup62 |
| 530 | NM_133525 | Nucleoside 2-deoxynbosyltransferase domain containing protein RGD620382 | RGD620382 |
| 531 | NM_012861 | O-6-methylgnanine-DNA methyltransferase | Mgmt |
| 532 | L20684 | opioid receptor, mu 1 | Oprl1 |
| 533 | NM_133585 | optic atrophy 1 homolog (human) | Opal |
| 534 | NM_053288 | orosomucoid 1 | Orml |
| 535 | NM_130402 | osteoclast inhibitory lectin | Ocil |
| 536 | NM_133306 | oxidized low density lipoprotein (lectin-like) receptor 1 | Oldlr1 |
| 537 | NM_019210 | p21 (CDKN1A)-activated kinase 3 | Pak3 |
| 538 | NM_053289 | pancreatitis-associated protein | Pap |
| 539 | NM_017044 | parathyroid hormone | Pth |
| 540 | BI281756 | Parkinson disease (autosomal recessive, early onset) 7 | Park7 |
| 541 | BG673589 | paxillin | Pxn |
| 542 | AI009656 | PEF protein with a long N-terminal hydrophobic domain | Peflin |
| 543 | BI291292 | peptidylprolyl isomerase C | Ppic |
| 544 | AA957342 | peptidylprolyl isomerase D (cyclophilin D) | Ppid |
| 545 | U68544 | peptidylprolyl isoinerase F (cyclophilin F) | Ppif |
| 546 | NM_017330 | perforin 1 (pore forming protein) | Prfl |
| 547 | NM_017169 | peroxiredoxin 2 | Prdx2 |
| 548 | NM_013196 | peroxisome proliferator activated receptor alpha | Ppara |
| 549 | U75918 | Peroxisome proliferator activated receptor delta | Ppard |
| 550 | NM_013124 | peroxisome proliferator activated receptor gamma | Pparg |
| 551 | AI598971 | PERP, TP53 apoptosis effector (predicted) | Perp_predicted |
| 552 | NM_021657 | PH domain and leucine rich repeat protein phosphatase | Phlpp |
| 553 | NM_031606 | phosphatase and tensin homolog | Pten |
| 554 | NM_053823 | phosphatidylinositol 3-kinase, C2 domain containing, gamma polypeptide | Pik3ca |
| 555 | BI290699 | phosphatidylinositol 3-kinase, catalytic, alpha polypeptide | Pik3ca |
| 556 | D64048 | phosphatidylinositol 3-kinase, regulatory subunit, polypeptide 1 | Pik3rl |
| 557 | NM_022185 | phosphatidylinositol 3-kinase, regulatory subunit, polypeptide 2 | Pik3r2 |
| 558 | AI232697 | phosphatidyhenrie receptor | Ptdsr |
| 559 | AI454840 | Photphodiesterase 1A, calmodulin-dependent | Pdela |
| 560 | AF327906 | phosphodiesterase 1B, Ca2+calmodulin dependant | Pdelb |
| 561 | NM_022958 | phosphoinositide-3-kinase class 3 | Pik3c3 |
| 562 | NM_133551 | phospholipase A2, group IVA (cytosolic, calcium-dependent) | Pla2g4a |
| 563 | U51898 | phopholipase A2, group VI | Pla2g6 |
| 564 | U69550 | phospholipase D1 | Pldl |
| 565 | BE112895 | phosphoprotein enriched in astrocytes 15 | Peal5 |
| 566 | NM_053491 | plasminogen | Plg |
| 567 | NM_013151 | plasminogen activator, tissue | Plat |
| 568 | NM_013085 | plasminogen activator, urokinase | Plau |
| 569 | AF007789 | plasminogen activator, urokinase receptor | Plaur |
| 570 | BE100812 | platelet derived growth factor, alpha | Pdgfa |
| 571 | BM392366 | platelet-activating factor acetylhydrolase, isoform 1b, alpha2 subunit | Pafahlb2 |
| 572 | AI009219 | Pleckstrin homology domain containing, family A member 5 | PlekhaS |
| 573 | NM_017180 | pleckstrin homology-like domain, family A, member 1 | Phlda1 |
| 574 | NM_012760 | pleiomorphic adenoma gene-like 1 | Pkgl1 |
| 575 | AB019366 | poly (ADP-ribose) glycohydrolase | Parp1 |
| 576 | NM_013063 | poly (ADP-ribose) polymerase family, member 1 | Parp1 |
| 577 | NM_017141 | polymerase (DNA directed), beta | Polb |
| 578 | AW531224 | polymerase (RNA) II (DNA directed) polypeptide A (mapped) | Polr2a_mapped |
| 579 | AW435212 | potassium channel, subfamily K, member 3 | Kcnk3 |
| 580 | NM_053405 | potassium channel, subfamily K, member 9 | Kcnk9 |
| 581 | NM_013186 | potassium voltage gated channel, Shab-related subfamily, member 1 | Kcnb1 |
| 582 | BM385544 | presenilin 1 | Psenl |
| 583 | AB004454 | presenilin 2 | Psen2 |
| 584 | AI232272 | presenilin enhancer 2 homolog (C. elegans) | Psenen |
| 585 | BI278802 | prion protein | Prnp |
| 586 | U05989 | PRKC, apoptosis WT1, regulator | Pawr |
| 587 | BI285575 | procollagen, type 1, alpha 1 | Colla1 |
| 588 | BE108058 | procollagen, type XVIII alpha 1 | Coll8a1 |
| 589 | AI599419 | Progesterone receptor | Pgr |
| 590 | AI704628 | programmed cell death 2 | Pdcd2 |
| 591 | NM_022265 | programmed cell death 4 | Pdcd4 |
| 592 | BF408447 | programmed cell death 5 (predicted) | Pdcd5_predicted |
| 593 | BI296393 | programmed cell death 6 (predicted) | Pdcd6_predicted |
| 594 | BE328942 | programmed cell death 6 interacting protein | Pdcd6ip |
| 595 | AF262320 | programmed cell death 8 | Pdcd8 |
| 596 | AI013847 | programmed cell death protein 7 (predicted) | Pdcd7_predicted |
| 597 | BI282863 | prohibition | Phb |
| 598 | NM_012629 | prolactin | Prl |
| 599 | L48060 | prolactin receptor | Prlr |
| 600 | BI290159 | proline-serine-threonine phosphatase-interacting protein 1 (predicted) | Pstpip1_predicted |
| 601 | NM_138857 | prominin 2 | Prom2 |
| 602 | NM_031644 | prostaglandin D2 synthase 2 | Ptgds2 |
| 603 | U03389 | prostaglandin-endoperoxide synthase 2 | Ptgs2 |
| 604 | A1600136 | protease, serine, 25 | Prss25 |
| 605 | NM_031978 | proteasome (prosome, macropain) 26S subunit, non-ATPase, 1 | Psmdl |
| 606 | NM_130430 | proteasome (prosome, macropain) 26S subunit nou-ATPase, 9 | Psmd9 |
| 607 | NM_012803 | protein C | Prc |
| 608 | AI639478 | protein disulfide isomerase associated 2 (predicted) | Pdia2_predicted |
| 609 | NM_017319 | protein disulfide isomerase associated 3 | Pdia3 |
| 610 | BF415343 | protein kinase C, alpha | Prkca |
| 611 | X04440 | protein kinase C, beta 1 | Prtcb1 |
| 612 | NM_133307 | protein kinase C, delta | Prkcd |
| 613 | AA799421 | protein kinase C, epsilon | Prkce |
| 614 | 1370197_a_at | protein kinase C, zeta | Prkcz |
| 615 | NM_019142 | protein kinase, AMP-activated, alpha 1 catalytic subunit | Prkaa1 |
| 616 | NM_023991 | protein kinase, AMP-activated, alpha 2 catalylic subunit | Prkaa2 |
| 617 | NM_013012 | protein kinase, cGMP-dependent type II | Prkg2 |
| 618 | BF400782 | protein kinase, DNA activated, catalytic polypeptide (predicted) | Prkdc_predicted |
| 619 | NM_019335 | Protein kinase, interferon-inducible double stranded RNA dependent | Prkr |
| 620 | NM_031527 | protein phosphatase 1, catalytic subunit, alpha isoform | Ppp1ca |
| 621 | NM_013065 | protein phosphatase 1, catalytic subunit, beta isoform | Ppp1cb |
| 622 | NM_022676 | protein phosphatase 1, regulatory (inhibitor) subunit 1 A | Ppp1rA |
| 623 | AI172276 | protein phosphatase 1, regulatory (inhibitor) subunit 2 | Ppp1r2 |
| 624 | AB023634 | protein phosphatase 1F (PP2C domain containing) | Ppmlf |
| 625 | BF408792 | protein phosphatase 2 (formerly 2A), catalytic subunit, alpha isoform | Ppp2ca |
| 626 | NM_017040 | protein phosphatase 2 (formerly 2A), catalytic subunit beta isoform | Ppp2cb |
| 627 | AA800669 | protein phosphatase 2 (formerly 2A), regulatory subunit A (PR 65), alpha isoform | Ppp2r1a |
| 628 | 1373959_at | protein phosphatase 2 (formerly 2A), regulatory subunit A (PR 65), beta isoform | Ppp2r1b |
| 629 | AI717081 | protein phosphatase 2 (formerly 2A), regulatory subunit B (PR 52), alpha inform | Ppp2r2a |
| 630 | BE113127 | Protein phosphatase 3, catalytic subunit, alpha isoform | Ppp3ca |
| 631 | NM_031729 | protein phosphatase 5, catalytic subunit | Ppp5c |
| 632 | U06230 | protein S (alpha) | Pros1 |
| 633 | U69109 | protein tyrosine kinase 2 beta | Ptk2b |
| 634 | NM_012637 | protein tyrosine phosphatase, non-receptor type 1 | Ptpn1 |
| 635 | AI172465 | Protein tyrosine phosphatase, non-receptor type 11 | Ptpn11 |
| 636 | NM_053908 | protein tyrosine phosphatase, non-receptor type 6 | Ptpn6 |
| 637 | M10072 | protein tyrosine phosphatase, receptor type, C | Ptprc |
| 638 | NM_022925 | protein tyrosine phosphatase, receptor type, Q /// hypothetical gene supported by NM_022925; NM_198323 | Ptprq |
| 639 | AI178772 | prothymosin alpha | Ptma |
| 640 | NM_017034 | proviral integration site 1 | Piml |
| 641 | BI294798 | PTK2 protein tyrosine kinase 2 | Ptk2 |
| 642 | AF231010 | purinergic receptor P2X, ligand-gated ion channel, 1 | P2rx1 |
| 643 | AF020757 | purinergic receptor P2X, ligand-gated ion channel, 2 | P2rx2 |
| 644 | NM_019256 | purinergic receptor P2X, ligand-gated ion channel, 7 | P2ry2 |
| 645 | NM_017255 | purinergic receptor P2Y, G-protein coupled 2 | P2ry2 |
| 646 | BI282953 | PYD and CARD domain containing | Pycard |
| 647 | NM_013018 | RAB3A, member RAS oncogene family | Rab3a |
| 648 | U70777 | rabaptin, RAB GTPase binding effector protein 1 | Rabep1 |
| 649 | AJ249986 | Rap guanine nucleotide exchange factor (GEF) 1 | Rapgef1 |
| 650 | AF002251 | Ras association (RalGDS/AF-6) domain family 5 | Rassfs |
| 651 | AF081196 | RAS guauyl releasing protein 1 | Rasgrp2 |
| 652 | AI408053 | ras homolog gene family, member A | Rhoa |
| 653 | NM_022542 | ras homolog gene family, member B | Rhob |
| 654 | NM_013135 | RAS p21 protein activator 1 | Rasa1 |
| 655 | BF414025 | Ras-induced senescence 1 | Ris1 |
| 656 | AA799542 | Ras-related C3 botulinum toxin substrate 1 | Rac1 |
| 657 | AF036537 | receptor-interacting serine-threonine kinase 3 | Ripk3 |
| 658 | NM_012641 | regenerating islet-derived 1 | Reg1 |
| 659 | L20869 | regenerating islet-derived 3 gamma | Reg3g |
| 660 | NM_031546 | regucalcin | Rgn |
| 661 | AJ299017 | ret proto-oncogene | Ret |
| 662 | AF051335 | reticulon 4 | Rtn4 |
| 663 | AI178012 | retinoblastoma 1 | Rb1 |
| 664 | NM_031094 | retinoblastoma-like 2 | Rb12 |
| 665 | NM_031528 | retinoic acid receptor, alpha | Rara |
| 666 | BF419646 | retinoic acid receptor, beta | Rarb |
| 667 | BI285959 | retinoid X receptor alpha | Rxra |
| 668 | AI408677 | Rho GDP dissociation inhibitor (GDI) alpha | Arhgdia |
| 669 | NM_031098 | Rho-associated coiled-coil forming kinase 1 | Rock1 |
| 670 | NM_013022 | Rho-associated coiled-coil forming kinase 2 | Rock2 |
| 671 | NM_022510 | ribosomal protein L4 | Rp14 |
| 672 | BI282255 | ribosomal protein S5 | Rps5 |
| 673 | M57428 | ribosomal protein S6 kinase, polypeptide 1 | Rps6kb1 |
| 674 | AI179991 | ring finger protein 34 | Rnf34 |
| 675 | AA858518 | ring finger protein 7 (predicted) | Rnf7_predicted |
| 676 | AI175966 | Rons sarcoma oncogene | Src |
| 677 | BF403180 | Runt related transcription factor 2 | Runx2 |
| 678 | NM_012618 | S100 calcium-binding protein A4 | S100a4 |
| 679 | NM_013191 | S100 protein, beta polypeptide | S100b |
| 680 | AA850867 | sarcoglycan, gamma (dystrophin-associated glycoprotein) | Sgcg |
| 681 | NM_031541 | scavenger receptor class B, member 1 | Scarb1 |
| 682 | BI294932 | SCF apoptosis response protein 1 | LOC499941 |
| 683 | NM_053587 | schlafen 3 | Slfn3 |
| 684 | BF394953 | SDA1 domain containing 1 | Sdad1 |
| 685 | NM_019364 | sec1 family domain containing 1 | Scfdl |
| 686 | AF220608 | secreted frizzled-related protein 4 | Sfrp4 |
| 687 | AB001382 | secreted phosphoprotein 1 | Sppl |
| 688 | NM_017310 | sema domain, immnuoglobulin domain (Ig), short basic domain, secreted, (semaphorin) 3A | Sema3a |
| 689 | BI299759 | sema domain, transmembrane domain (TM), and cytoplasmic domain, (semaphorin) 6A (predicted) | Sema6a_predicted |
| 690 | NM_133291 | seminal vehicle antigen-like 2 | Sva12 |
| 691 | M25590 | seminal vesicle protein 4 | Svp4 |
| 692 | BG663993 | sequestosome 1 | Sqstm1 |
| 693 | NM_012620 | serine (or cysteine) peptidase inhibitor, clade E, member 1 | Serpine1 |
| 694 | NM_021696 | serine (or cystsine) proteinase inhibitor, clade B, member 2 | Se2pinb2 |
| 695 | AA944455 | serine incorporator 3 | Serine3 |
| 696 | NM_133392 | serine/threonine kinase 17b (apoptosis-inducing) | Stk17b |
| 697 | NM_019349 | serine/threonine kinase 2 | Stk2 |
| 698 | NM_031735 | serine/threonine kinase 3 (STE20 homolog, yeast) | Stk3 |
| 699 | AF388527 | Seipine1 mRNA binding protein 1 | Serbp1 |
| 700 | NM_017170 | serum amyloid P-component | Apcs |
| 701 | NM_019232 | serum/glucocorticoid regulated kinase | Sgk |
| 702 | NM_080905 | seven in absentia 1A | Siah1a |
| 703 | NM_134457 | seven in absentia 2 | Siah2 |
| 704 | NM_012650 | sex hormone binding globulin | Shbg |
| 705 | BF284481 | SH3-domain GRB2-like B1 (endophilin) | Sh3glbl |
| 706 | AF255888 | SH3-domain kinase binding protein 1 | Sh3kbp1 |
| 707 | BF550890 | sialophorin | Spn |
| 708 | NM_032612 | signal transducer and activator of transcription 1 | Stat1 |
| 709 | B1285863 | signal transducer and activator of transcription 3 | Stat3 |
| 710 | NM_017064 | signal transducer and activator of transcription 5A | Stat5a |
| 711 | AI177626 | signal transducer and activator of transcription 5B | Stat5b |
| 712 | BE110607 | similar to apoptosis related protein APR-32; p18 protein (predicted) | RGD1311805_predicted |
| 713 | BF396386 | similar to cell division cycle and apoptosis regulator 1 (predicted) | RGD1560358_predicted |
| 714 | BI296385 | similar to chemokine (C-X-C motif) ligand 16 | Cxcl16 |
| 715 | BI294745 | similar to livin inhibitor of apoptosis isoform beta (predicted) | RGD1562883_predicted |
| 716 | NM_021846 | similar to MAP/microtubule affinity-regulating kinase 4 (MAP/microtubule affinity-regulating kinase like 1) (predicted) | RGD1561096_predicted |
| 717 | AA848545 | similar co programmed cell death 10 | MGC72992 |
| 718 | BF282636 | similar to RIKEN cDNA 1700023M03 | RGD1305457 |
| 719 | AW254416 | Similar to TGF-beta induced apoptosis protein 12 (predicted) | RGD130816_predicted |
| 720 | AA946199 | snail homolog 1 (Drosophila) | Snail |
| 721 | X89383 | SNF related kinase | Snrk |
| 722 | NM_012647 | sodium channel, voltage-gated, type 2, alpha 1 polypeptide | Scn2a |
| 723 | NM_013178 | sodium channel, voltage-gated, type IV, alpha polypeptide | Snc4a |
| 724 | BI284218 | solute carrier family 2(facilitated glucose transporter), member 1 | Slc2a1 |
| 725 | NM_017102 | solute carrier family 2(facilitated glucose transporter), member 3 | Slc2a3 |
| 726 | NM_017223 | solute carrier family 20, member 2 | Slc20a2 |
| 727 | BG666999 | solute carrier family 25 (mitochondrial carrier; adenine nucleotide translocator), member 4 | Slc25a4 |
| 728 | NM_017206 | solute carrier family 6 (neurotransmitter transporter, taurine), member 6 | Slc6a6 |
| 729 | NM_053442 | solute carrier family 7 (cationic amino acid transporter, y+ system), member 8 /// synaptic Ras GTPase activating protein 1 homolog (rat) | slc7a8 /// Syngap1 |
| 730 | U04933 | solute carrier family 8 (sodium/calcium exchanger), member 1 | Slc7a8 /// Syngap1 |
| 731 | NM_133522 | somatostatin receptcor 3 /// hypothetical gene supported by NM_133522 | SSTR |
| 732 | BI275248 | Son cell proliferation protein | Son |
| 733 | NM_017221 | sonic hedgehog homolog (Drosophila) | Shh |
| 734 | NM_012655 | Sp1 transcription factor | Sp1 |
| 735 | AB049572 | sphingosine kinase 1 | Sphk1 |
| 736 | BM386306 | sphingosine kinase 2 | Sphk2 |
| 737 | AI159638 | sphingosiue-1-phosphate phosphatase 1 | Sgpp1 |
| 738 | U53883 | ST8 alpha-N-acetyl-neuraminide alpha-2,8-sialyltransferase 1 | St8sia2 |
| 739 | AY083159 | Stam binding protein | Stambp |
| 740 | BM386683 | stanniocalcin 1 | Stc1 |
| 741 | NM_022230 | stanniocalcin 2 | Stc2 |
| 742 | NM_017166 | stathmin 1 | Stmn1 |
| 743 | AF335281 | STEAP family member 3 | Steap3 |
| 744 | AI235465 | steroid sensitive gene 1 | Ssg1 |
| 745 | AI009817 | succinate dehydrogenase complex, subunit C, integral membrane protein | Sdhc |
| 746 | NM_134378 | sulfatase 1 | Sdhc |
| 747 | NM_017050 | superoxide dismutase 1 | Sod1 |
| 748 | BG671549 | superoxide dismutase 2, mitochondrial | Sod2 |
| 749 | NM_058208 | suppressor of cytokine signaling 2 | Socs2 |
| 750 | NM_053555 | suppressor of cytokine signaling 3 | Socs3 |
| 751 | BM390864 | survival motor neuron domain containing 1 | Smndc1 |
| 752 | AI170385 | SWI/SNF related, matrix associated, actin dependent regulator of chromatin, subfamily a, member 2 | Smarca2 |
| 753 | BE329013 | SWI/SNF related, matrix associated, actin dependent regulator of chromatin, subfamily a, member 4 | Smarca4 |
| 754 | NM_053442 | synaptic Ras GTPase activating protein 1 homolog (rat) | Syngap1 |
| 755 | NM_013026 | syndecan 1 | Sdc1 |
| 756 | AA946430 | synovial apoptosis inhibitor 1, synoviolin | Syvn1 |
| 757 | NM_019169 | synuclein, alpha | Snca |
| 758 | BG671061 | tachykinin 1 | Tac1 |
| 759 | BM392226 | TAF10 RNA polymerase II, TATA box binding protein (TBP)-associated factor (predicted) | Taf10_predicted |
| 760 | NM_133615 | TAF9-like RNA polymerase II, TATA box binding protein (TBP)-associated factor, 31kDa | Taf91 |
| 761 | AI228250 | Tax 1 (human T-cell leukemia virus I) binding protein 1 | Taxlbp1 |
| 762 | AA957545 | T-box 3 | Tbx3 |
| 763 | AB029495 | TCF3 (E2A) fusion partner | Tfpt |
| 764 | AF247818 | telomerase reverse transcriptase | Tert |
| 765 | NM_019381 | testis enhanced gene transcript | Tegt |
| 766 | NM_133396 | testis-specific kinase 2 | Tesk2 |
| 767 | AA943723 | THAP domain containing, apoptosis associated protein 3 (predicted) | Thap3_piedicoed |
| 768 | NM_053800 | thioredoxin 1 | Txn1 |
| 769 | AA800180 | thioredoxin 2 | Txn2 |
| 770 | BM390196 | thioredoxin domain containing 5 (predicted) | Txndc5_predicted |
| 771 | BF555110 | thioredoxin-like 1 | Txnl1 |
| 772 | BM384228 | THO complex 1 | Thoc1 |
| 773 | AA998057 | thymoma viral proto-oncogene 1 | Akt1 |
| 774 | AI105076 | Thymoma viral proto-oncogene 2 | Akt2 |
| 775 | NM_031575 | thymoma viral proco-oncogene 3 | Akt3 |
| 776 | AI145313 | thymus cell antigen 1, theta | Thy1 |
| 777 | NM_012888 | thyroid stimulating hormone receptor | Tshr |
| 778 | AI101391 | Tial1 cytotoxic granule-associated RNA binding protein-like 1 (mapped) | Tial1 |
| 779 | NM_053819 | tissue inhibitor of metalloproteinase 1 | Timp1 |
| 780 | NM_012989 | tissue inhibitor of metalloproteinase 2 | Timp2 |
| 781 | NM_012886 | Tissue inhibitor of metalloproteinase 3 (Sorsby fundus dystrophy, pseudoinflammatory) | Timp3 |
| 782 | AI104533 | titin | Ttn |
| 783 | BI287742 | TM2 domain containing 1 (predicted) | Tm2dl_predicted |
| 784 | BM389034 | TNF receptor-associated protein 1 | Trap1 |
| 785 | BM386846 | TNFRSF1A-associated via death domain | Tradd |
| 786 | AF057025 | toll-like receptor 4 | Tlr4 |
| 787 | AI012419 | Transcribed locus | NS |
| 788 | AW533194 | Transcribed locus | NS |
| 789 | AI104523 | Transcribed locus | NS |
| 790 | AW251860 | transcription factor 7, T-cell specific (predicted) | Tcf7_predicted |
| 791 | NM_031326 | transcription factor A, mitochondrial | Tfam |
| 792 | BI284455 | transcription factor Pur-beta | pur-beta |
| 793 | NM_133317 | transducer of ErbB-2.1 | Tob1 |
| 794 | M58040 | transferrin receptor | Tfrc |
| 795 | BI297236 | Transformation related protein 53 inducible nuclear protein 1 | Trp53inp2 |
| 796 | AJ277449 | transformation related protein 63 | Trp63 |
| 797 | NM_012671 | transforming growth factor alpha | Tgfa |
| 798 | AW254561 | transforming growth factor beta regulated gene 4 | Tbrg4 |
| 799 | 1370082_at | transforming growth factor, beta 1 | TgSb1 |
| 800 | BF420705 | transforming growth factor, beta 2 | Tgfb2 |
| 801 | NM_012775 | transforming growth factor, beta receptor 1 | Tgfbr1 |
| 802 | BI275994 | transglutaminase 2, C polypeptide | Tgm2 |
| 803 | AB015231 | transient receptor potential cation channel, subfamily V, member 1 | Trpv1 |
| 804 | NM_023970 | transient receptor potenial cation channel, subfamily V, member 4 /// transient receptor potential cation channel, subfamily V, member 1 | Trpv4 |
| 805 | AB020967 | tribbles homolog 3 (Drosophila) | Trib3 |
| 806 | NM_080903 | tripartite motif protein 63 | Trim63 |
| 807 | AI104913 | tropomodulin 1 | Tmod1 |
| 808 | NM_031345 | TSC22 domain family 3 | Tsc22d3 |
| 809 | BI285434 | tubulin, alpha 1 /// tubulin, alpha 6 /// similar to Tubulin alpha-2-chain (Alpha-tubulin 2) (predicted) | Tuba1 /// Tuba6 /// RGD1565476_predicted |
| 810 | BI285434 | tubulin, gamma 1 | Tubg1 |
| 811 | AA819227 | tumor necrosis factor (TNF superfamily, member 2) | Tnf |
| 812 | BF283688 | tumor necrosis factor ligand superfamily member 12 | Tnfsf12 |
| 813 | NM_012870 | tumor necrosis factor receptor superfamily, member 11b (osteoprotegerin) | Tfrsf11b |
| 814 | BI303379 | tumor necrosis factor receptor superfamily, member 12a | Tnfrsfl2a |
| 815 | AI169601 | tumor necrosis factor receptor superfamily, member 14 (herpesvirus entry mediator) | Tnfrsf14 |
| 816 | NM_013091 | tumor necrosis factor receptor superfamily, member 1a | Tnfrsf1a |
| 817 | NM_013049 | tumor necrosis factor receptor superfamily, member 4 | Tnfrsf4 |
| 818 | AW433947 | tumor necrosis factor receptor superfamily, member 5 | Tofrsf5 |
| 819 | NM_139194 | Tumor necrosis factor receptor superfamily, member 6 | Tofrsf6 |
| 820 | BM387084 | tumor necrosis factor superfamily, member 5-induced protein 1 (predicted) | Tnfsf5ip1_predicted |
| 821 | AY009504 | tumor protein p53 | Tp53 |
| 822 | NM_053867 | tumor protein, translationally-controlled 1 | Tpt1 |
| 823 | BG057543 | tumor rejection antigen gp96 (predicted) | Tral_predicted |
| 824 | AI234654 | tumor susceptibility gene 101 | Tsg101 |
| 825 | NM_013052 | tyrosine 3-monooxygenase/tryptophan 5-monooxygenase activation protein, eta polypeptide | Ywhah |
| 826 | NM_019378 | tyrosine 3-monooxygenase/tryptophan 5-monooxygenase activation protein, gamma polypeptide | Ywhag |
| 827 | BF281342 | tyrosine 3-monooxygenase/trytohpan 5-monooxygenase activation protein, theta polypeptide | Ywhaq |
| 828 | AI228292 | tyrosine hydroxylase | Th |
| 829 | AI407490 | tyrosyl-tRNA synthetase | Yars |
| 830 | NM_053747 | Ubiquilin 1 | Ubqln1 |
| 831 | B1276086 | Ubiquitin specific protease 7 (herpes virus-associated) | Usp7 |
| 832 | BI296848 | ubiquitiuation factor E4B, UFD2 homolog (S. cerevisiae) (predicted) | Ube4b_predicted |
| 833 | AI102437 | ubiquitin-like 1 (sentrin) activating enzyme E1B | Uble1b |
| 834 | AF159706 | unc-13 homolog B (C. elegans) | Unc 13b |
| 835 | NM_022206 | unc-5 homolog A (C. elegans) | Uuc5a |
| 836 | NM_022207 | unc-5 homolog B (C. elegans) | Unc5b |
| 837 | BM388453 | uncharacterized protein family UPF0227 member RGD1359682 | RGD1359882 |
| 838 | U30789 | upregulated by 1,25-dihydroxyvitamin D-3 | Txnip |
| 839 | NM_053864 | valosin-containing protein | Vcp |
| 840 | AF080594 | vascular endothelial growth factor A | Vegfa |
| 841 | NM_012759 | vav 1 oncogene | Vav1 |
| 842 | NM_021687 | v-erb-a erythroblastic leukemia viral oncogene homolog 4 (avian) | Erbb4 |
| 843 | NM_017003 | v-erb-b2 erythro leukemia viral oncogene homolog 2, neuro/glioblastoma derived, oncogene homolog (avian) | Erbb2 |
| 844 | NM_012555 | v-ets ervthroblastosis virus E26 oncogene homolog 1 (avian) | Ets1 |
| 845 | NM_031140 | vimentin | Vim |
| 846 | AF268467 | voltage-dependent anion channel 1 | Vdac1 |
| 847 | NM_012639 | v-raf-1 murine leukemia viral oncogene homolog 1 | Raf1 |
| 848 | BF283772 | v-rel reticuloendotheliosis viral oucogene homolog A (avian) | Rela |
| 849 | NM_022548 | wild-type p53-induced gene 1 | Wig1 |
| 850 | NM_031534 | Wilms tumor 1 | Wt1 |
| 851 | NM_031590 | WNT1 inducible signaling pathway protein 2 | Wisp2 |
| 852 | BI300732 | WW domain-containing oxidoreductase (predicted) | RGD1565791_predicted |
| 853 | BI182111 | Y box protein 1 related, pseudogene 3 /// similar to nuclease sensitive element binding protein 1 (predicted) /// Y box protein 1 | YbKl-ps3 /// RGD1560265_predicted /// Ybx1 |
| 854 | NM_031615 | zinc finger protein 148 | Zfp148 |
| 855 | BE109605 | zinc finger protein 162 | Zfpl62 |
| 856 | BE111799 | Zinc finger protein 346 (predicted) | Zfp346_predicted |
| 857 | BM392399 | zinc finger protein 622 | Zfp622 |
| 858 | AA819804 | Zinc finger protein 91 | Zfp91 |
| 859 | BI289543 | zinc finger, MYND domain containing 11 | Znrynd11 |
| 860 | AI317880 | zinc responsive protein ZD7 | LOC474154 |
Signal detected above background for gene probeset in 20% or more of the chips.
“Yes” indicates significant differential gene expression within the Sham, SBR50, SBR50/IL-6, and SBR50/IL-6/G groups using False Discovery Rate (FDR) = 10%. “No” indicates not significant differential gene expression. “N/A” indicates genes not included in the analysis because of not being detected in at least 20% of the chips. “NS” indicates no gene symbol.
To identify genes differentially expressed among the experimental groups, the data were filtered to remove genes with nearly uniformly low expression (absent on ≥ 80% of chips). Of the 860 apoptosis-related genes represented on the chips, 731 genes met the requirement of this filtering process and were included in the analysis. One-way ANOVA (see Materials and Methods) was then performed which identified 350 apoptosis genes with differential expression among four experimental groups– sham, SBR50, SBR50/IL-6, and SBR50/IL-6/G– at a False Discovery Rate (FDR) = 10% (Table 2). Of the 350 apoptosis pathway genes whose expression was altered among the four groups, 311 were altered in the SBR50 vs. sham comparison (Figure 5A and Table 2). Among the genes whose differential expression was altered in the SBR50 vs. sham comparison, the transcripts of the majority of these genes (193 genes) were increased in SBR50 vs. sham by 4.2 ± 1.2 fold (range = 1.02 to 92.3 fold) while transcripts of 118 genes were decreased in SBR50 vs. sham by 1.5 ± 1.2 fold (range = 1.1 to 3.3 fold; Figure 5B). Importantly, 106 of the 193 genes (55%) that were increased in the SBR50 vs. sham group, were decreased significantly in the SBR50/IL- 6 vs. SBR50 group by 1.8 ± 0.9 fold (range = 1.3 to 14.3 fold) and 108 of the 118 genes (92%) that were decreased in SBR50 group, were increased significantly in the SBR50/IL-6 group by 1.4 ± 0.7 fold (range = 1.2 to 2.9 fold; Figure 5B). Thus, of the genes whose transcript levels were altered in SBR50 vs. sham group, 214 of 311 (69%) returned to sham level or were “normalized” in the SBR50/IL-6 group.
Table 2.
Apoptosis-related genes differentially expressed in the SBR50 vs. SHAM comparison
| SBR50 vs. Sham | SBR50/IL-6 vs. SBR50 | SBR50/IL/6/Gvs.SBR50/IL-6 | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| mRNA Probeset Number | Gene Name* | Gene Symbol | Fold P/Sham | FDR† | Fold IL-6/p | FDR† | Fold G/IL-6 | FDR† | ||||||
| GROUP IA | GENES INCREASED IN SBR50 vs. SHAM AND DECREASED IN SBR50/IL-6 vs. SBR50 | |||||||||||||
| NM_012620 | serine (or cysteine) peptidase inbitior, clade E, member 1 | Serpine1 | 29.09 | 0.000 | 0.25 | 0.004 | 1.73 | 0.147 | ||||||
| B1284218 | solute carrier family 2 (faciliated glucose transporter), member 1 | Slc2a1 | 20.29 | 0.000 | 0.26 | 0.002 | 1.72 | 0.079 | ||||||
| NM_012580 | heme oxygenase (decycling) 1 | Hmoxl | 19.97 | 0.002 | 0.13 | 0.020 | 1.20 | 0.846 | ||||||
| M58040 | transferrin receptor | Tfrc | 10.82 | 0.000 | 0.39 | 0.044 | 1.07 | 0.899 | ||||||
| D64048 | phosphatidylinositol 3-kinase, regulatory subunit, polypeptide 1 | Pik3rl | 6.17 | 0.003 | 0.21 | 0.014 | 0.64 | 0.433 | ||||||
| U02315 | neuregulin 1 | Nrg1 | 5.08 | 0.002 | 0.43 | 0.062 | 0.70 | 0.428 | ||||||
| NM_017334 | cAMP responsive element modulator | 3081 | 0.001 | 0.59 | 0.079 | 0.57 | 0.078 | |||||||
| AI172056 | myeloid cell leukemia sequence 1 | Mcl1 | 3.43 | 0.001 | 0.46 | 0.019 | 0.87 | 0.669 | ||||||
| NM_080902 | hypoxia induced gene 1 | Hig1 | 3032 | 0.001 | 0.36 | 0.005 | 1.09 | 0.773 | ||||||
| AF279286 | Bc12-like 1 | Bc1211 | 3023 | 0.001 | 0.40 | 0.007 | 1.36 | 0.300 | ||||||
| BI290699 | phosphatidylinositol 3-kinase, catalytic, alpha polypeptide | Pik3ca | 3.18 | 0.000 | 0.31 | 0.002 | 0.90 | 0.692 | ||||||
| NM_019142 | protein kinase, AMP-activated, alpha 1 catalytic subunit | Prkaa1 | 3.08 | 0.000 | 0.42 | 0.004 | 1.12 | 0.657 | ||||||
| L062638 | platelet derived growth factor, alpha | Pdgfa | 3.00 | 0.002 | 0.43 | 0.015 | 1.05 | 0.885 | ||||||
| U69550 | phospholipase D1 | PlaD1 | 2.72 | 0.000 | 0.54 | 0.009 | 0.98 | 0.945 | ||||||
| AB001382 | secreted phosphoprotein 1 | Sppl | 2.67 | 0.013 | 0.45 | 0.048 | 2.08 | 0.077 | ||||||
| NM_013052 | tyrosine 3-monooxygenase/tyrptophan 5-monooxygenase activation protein, eta polypeptide | Ywhah | 2.42 | 0.002 | 0.66 | 0.095 | 2.95 | 0.001 | ||||||
| NM_053843 | Fc receptor, IgG, low affinity III /// Fc gamma receptor II beta | Fcgr3 | 2.37 | 0.002 | 0.52 | 0.019 | 1.67 | 0.058 | ||||||
| NM_017039 | protein phosphate 2 (formerly 2A), catalytic subunit, alpha isoform | Ppp2ca | 2.33 | 0.001 | 0.59 | 0.018 | 1.76 | 0.015 | ||||||
| BE113920 | signal transducer and activator of transcription 3 | Stat3 | 2.33 | 0.086 | 0.38 | 0.068 | 1.57 | 0.395 | ||||||
| NM_019376 | tyrosine 3-monooxygenase/tryptophan 5-monooxygenase activation protein, gamma polypeptide | Ywhag | 2.30 | 0.000 | 0.57 | 0.004 | 1.35 | 0.059 | ||||||
| NM_053887 | mitogen activated protein kinase kinase kinase 1 | Map3k1 | 2.24 | 0.036 | 0.38 | 0.022 | 1.19 | 0.686 | ||||||
| NM_031140 | Vimentin | Vim | 2.19 | 0.001 | 0.51 | 0.004 | 1.66 | 0.018 | ||||||
| NM_031514 | Janus kinase 2 | Jak2 | 2.18 | 0.017 | 0.049 | 0.037 | 0.96 | 0.917 | ||||||
| NM_031832 | lectin, galactose binding, soluble 3 | Lgals3 | 2.03 | 0.066 | 0.46 | 0.061 | 2.58 | 0.033 | ||||||
| NM_019232 | serum/glucocorticoid regulated kinase | Sgk | 2.00 | 0.080 | 0.47 | 0.074 | 0.94 | 0.902 | ||||||
| AI409867 | cystatin B | CstB | 1.89 | 0.002 | 0.55 | 0.007 | 1.41 | 0.080 | ||||||
| NM_017255 | purinergic receptor P2Y, G-protein coupled 2 | P2ryl | 1.82 | 0.006 | 0.49 | 0.005 | 1.46 | 0.082 | ||||||
| NM_017022 | integrin beta 1 (fibronectin receptor beta) | Itgbl | 1.82 | 0.001 | 0.61 | 0.006 | 1.15 | 0.351 | ||||||
| NM_012886 | tissue inhibitor of metalloproteinase 3 (Sorsby fundus dystrophy, pseudoinflammatory) | Timp3 | 1.80 | 0.022 | 0.64 | 0.093 | 0.95 | 0.859 | ||||||
| AI600237 | eukaryotic translation elongation factor 1 epsilon 1 (predicted) | Eefle1 | 1.79 | 0.008 | 0.67 | 0.061 | 0.72 | 0.135 | ||||||
| AF228684 | adenosine A2a receptor | Adora2a | 1.76 | 0.006 | 0.60 | 0.016 | 1.93 | 0.005 | ||||||
| BE108192 | G1 to S phase transition 1 | Gspt1 | 1.74 | 0.000 | 0.70 | 0.004 | 1.03 | 0.772 | ||||||
| NM_053619 | complement component 5, receptor 1 | C5rl | 1.73 | 0.003 | 0.51 | 0.004 | 1.84 | 0.004 | ||||||
| B1285434 | tubulin, alpha 1 /// tubulin, alpha 6 /// similar to Tubulin alpha-2 chain (Alpha-tubulin 2) (predicted) | Tubal | 1.73 | 0.002 | 0.62 | 0.010 | 1.65 | 0.008 | ||||||
| BM385544 | presenilin 1 | Psen1 | 1.67 | 0.008 | 0.65 | 0.029 | 1.80 | 0.008 | ||||||
| BF281342 | tyrosine 3-monooxygenase/tryptophan 5-monooxygenase activation protein, theta polypeptide | Ywhaq | 1.63 | 0.001 | 0.69 | 0.009 | 1.09 | 0.463 | ||||||
| NM_021989 | tissue inhibitor of metalloproteinase 2 | Timp2 | 1.56 | 0.002 | 0.68 | 0.009 | 0.82 | 0.135 | ||||||
| AI236590 | myeloid differentiation primary response gene 88 | Myd88 | 1.56 | 0.044 | 0.56 | 0.019 | 1.30 | 0.272 | ||||||
| BM392321 | Homeodomain interacting protein kinase 2 (predicted) | Hipk2_predicted | 1.53 | 0.026 | 0.69 | 0.068 | 0.88 | 0.534 | ||||||
| BG379941 | Harvey rat sarcoma viral (v-Ha-Ias) oncogene homolog | HraS | 1.52 | 0.003 | 0.73 | 0.025 | 1.28 | 0.079 | ||||||
| BF417479 | 24-dehydrocholestrol reductase | Dhcr24 | 1.51 | 0.015 | 0.69 | 0.038 | 0.63 | 0.018 | ||||||
| NM_017141 | polymerase (DNA directed), beta | Polb | 1.51 | 0.075 | 0.61 | 0.048 | 1.46 | 0.134 | ||||||
| NM_031787 | Homeodomain interacting protein kinase 3 | Hipk3 | 1.50 | 0.093 | 0.52 | 0.019 | 0.59 | 0.060 | ||||||
| AA893484 | fibronectin 1 | Fank1 | 1.49 | 0.028 | 0.64 | 0.022 | 0.74 | 0.116 | ||||||
| AI172276 | protein phosphatase 1, regulatory (inhibitor) subunit 2 | Ppplr2 | 1.47 | 0.008 | 0.79 | 0.092 | 1.06 | 0.727 | ||||||
| NM_053720 | apoptosis antagonizing transcription factor | Aatf | 1.46 | 0.070 | 0.58 | 0.021 | 1.17 | 0.490 | ||||||
| M37394 | epidermal growth factor receptor | Hgfr | 1.44 | 0.092 | 0.52 | 0.012 | 0.52 | 0.014 | ||||||
| BG666933 | cystatin C | CstC | 1.40 | 0.002 | 0.77 | 0.014 | 0.71 | 0.004 | ||||||
| BM389673 | cofilin 1, non-muscle | Cfl1 | 1.39 | 0.086 | 0.61 | 0.021 | 1.33 | 0.171 | ||||||
| EE111972 | transforming growth factor, beta receptor 1 | Trfbr1 | 1.37 | 0.090 | 0.63 | 0.027 | 0.92 | 0.680 | ||||||
| AF388527 | Serpinel mRNA binding protein 1 | Serbp1 | 1.31 | 0.016 | 0.83 | 0.096 | 1.11 | 0.365 | ||||||
| NM_0131315 | RAS p21 protein activator 1 | Rasal | 1.28 | 0.033 | 0.66 | 0.005 | 1.04 | 0.771 | ||||||
| B1282863 | Prohibition | Phb | 1.26 | 0.092 | 0.76 | 0.069 | 0.70 | 0.026 | ||||||
| BI282281 | heat shock 70kDa protein 9A (predicted) | Hspa9a_predicted | 1.22 | 0.012 | 0.73 | 0.004 | 0.75 | 0.004 | ||||||
| AI103616 | ras-related C3 botulimum toxin substrate 1 | Rax1 | 1.21 | 0.029 | 0.74 | 0.007 | 1.31 | 0.012 | ||||||
| NM_019371 | EGL nine homolog 3 | Egin3 | 17.30 | 0.000 | 0.07 | 0.000 | 0.81 | 0.493 | ||||||
| AI602811 | dual specificity phosphatase 6 | Dusp6 | 15.48 | 0.000 | 0.44 | 0.012 | 1.27 | 0.418 | ||||||
| BI303379 | tumor necrosis factor receptor superfamily, member 12a | Tnfrsf12a | 9.82 | 0.002 | 0.29 | 0.064 | 2.08 | 0.277 | ||||||
| NM_012591 | interferon regulatory factor 1 | Irf1 | 9.32 | 0.000 | 0.39 | 0.005 | 1.33 | 0.311 | ||||||
| NM_133306 | oxidized low density lipoprotein (lectin-like) receptor 1 | Oldlr1 | 9.02 | 0.000 | 0.30 | 0.004 | 2.01 | 0.041 | ||||||
| NM_024127 | growth arrest and DNA-damage-inducible 45 alpha | Gadd45a | 6.25 | 0.001 | 0.50 | 0.098 | 1.95 | 0.130 | ||||||
| NM_053420 | BCL2/adenovirus E1B 19 kDa-interacting protein 3 | Bnip3 | 6.07 | 0.000 | 0.19 | 0.002 | 0.88 | 0.734 | ||||||
| AI232697 | phosphatidylserine receptor | Jmjd6 | 3.51 | 0.007 | 0.43 | 0.058 | 0.89 | 0.802 | ||||||
| NM_021846 | similar to MAP/microtubule affinity-regulating kinase 4 (MAP/microtubule affinity-regulating kinase like 1)(predicted) | RGD1561096 | 3.46 | 0.001 | 0.43 | 0.018 | 0.86 | 0.662 | ||||||
| B1287978 | growth arrest and DNA-damage-inducible 45 beta | Gadd45b | 3.20 | 0.002 | 0.54 | 0.061 | 1.33 | 0.383 | ||||||
| NM_021744 | CD14 antigen | Cd14 | 3.07 | 0.003 | 0.49 | 0.039 | 0.73 | 0.362 | ||||||
| NM_012637 | protein tyrosine phosphatase, non-receptor type 1 | Ptpn1 | 3.02 | 0.007 | 0.35 | 0.015 | 0.47 | 0.069 | ||||||
| B1275994 | transglutaminase 2, C polypeptide | Tgm2 | 2.98 | 0.000 | 0.45 | 0.004 | 2.99 | 0.000 | ||||||
| NM_013026 | syndecan 1 | Sdc1 | 2.91 | 0.000 | 0.48 | 0.005 | 2.92 | 0.001 | ||||||
| B1284739 | LPS-induced TN factor | Litaf | 2.70 | 0.000 | 0.50 | 0.005 | 1.23 | 0.289 | ||||||
| J04943 | nucleophosmin 1 /// similar to Nucleophosmin (NPM) (Nucleolar phosphoprotein B23) (Numatrin)(Nucleolar protein NO348) | Npm1 /// LOC300303 | 2.67 | 0.001 | 0.50 | 0.010 | 1.42 | 0.152 | ||||||
| AB049572 | sphingosine kinase 1 | Sphk1 | 2.63 | 0.002 | 0.61 | 0.067 | 0.60 | 0.074 | ||||||
| AI411788 | protein phosphatase 2 (formerly 2A), regulatory subunit A (PR 65), beta isoform | Ppp2f1b | 2.63 | 0.000 | 0.39 | 0.002 | 1.55 | 0.049 | ||||||
| B1289109 | max binding protein (predicted) | Mnt | 2.44 | 0.004 | 0.51 | 0.027 | 1.40 | 0.258 | ||||||
| X57764 | endothelin receptor type B | Ednrb | 2.43 | 0.002 | 0.58 | 0.028 | 1.11 | 0.673 | ||||||
| NM_134432 | angiotensinogen (serpin peptidase inhibitor, clade A, member 8) | Agt | 2.11 | 0.002 | 0.43 | 0.004 | 1.31 | 0.233 | ||||||
| AF136231 | caspase 2 | Casp2 | 2.04 | 0.009 | 0.38 | 0.004 | 0.86 | 0.592 | ||||||
| BM387008 | caspase 3, apoptosis related cysteine protease | Casp3 | 2.00 | 0.001 | 0.54 | 0.004 | 1.46 | 0.037 | ||||||
| DI3341 | mitogen activated protein kinase kinase 1 | Map2k1 | 1.96 | 0.007 | 0.62 | 0.055 | 1.81 | 0.026 | ||||||
| NM_053974 | eukaryotic translation initation factor 4E | Eif4eb 1 | 1.92 | 0.003 | 0.64 | 0.028 | 1.12 | 0.584 | ||||||
| AF066016 | nuclear receptor subfamily 3, group C, member 1 | Nr-3c1 | 1.85 | 0.006 | 0.59 | 0.022 | 1.10 | 0.680 | ||||||
| BF283772 | v-rel reticuloendotheliosis viral oncogene homolog A (avian) | Rela | 1.67 | 0.012 | 0.71 | 0.091 | 1.27 | 0.256 | ||||||
| NM_013091 | tumor necrosis factor receptor superfamily, member 1a | Trfrsf1a | 1.62 | 0.036 | 0.54 | 0.017 | 2.08 | 0.009 | ||||||
| NM_017040 | protein phosphatase 2 (formerly 2A), catalytic subunit, beta isoform | Ppp2cb | 1.60 | 0.002 | 0.64 | 0.005 | 0.98 | 0.899 | ||||||
| BM386683 | Stanniocalcin 1 | Stc1 | 1.59 | 0.016 | 0.65 | 0.033 | 0.80 | 0.264 | ||||||
| AI012221 | chloride intracellular channel 1 | Clic1 | 1.59 | 0.002 | 0.77 | 0.149 | 1.82 | 0.001 | ||||||
| B1282898 | BCL2-associated athanogene 5 | Bag5 | 1.57 | 0.002 | 0.61 | 0.004 | 0.78 | 0.077 | ||||||
| B1296393 | programmed cell death 6 (predicted) | Pdcd6 | 1.49 | 0.002 | 0.70 | 0.007 | 1.23 | 0.083 | ||||||
| AI169001 | antophagy-related 12 (yeast) | Atg12 | 1.47 | 0.004 | 0.65 | 0.006 | 0.90 | 0.387 | ||||||
| B1285751 | cullin 3 (predicted) | Cu13_predicted | 1.46 | 0.012 | 0.62 | 0.006 | 0.87 | 0.373 | ||||||
| NM_133307 | protein kinase C, delta | Prkcd | 1.42 | 0.025 | 0.73 | 0.058 | 1.23 | 0.207 | ||||||
| B1289418 | CD38 antigen | Cd38 | 1.42 | 0.091 | 0.68 | 0.088 | 1.19 | 0.433 | ||||||
| NM_012696 | kininogen 1 /// K-kininogen /// similar to alpha-1 major acute phase protein prepeptide | Kng1 | 1.42 | 0.019 | 0.68 | 0.019 | 1.84 | 0.002 | ||||||
| AA859665 | Nuclear pore associated protein | Npap60 | 1.40 | 0.012 | 0.69 | 0.012 | 0.89 | 0.401 | ||||||
| NM_080887 | thioredoxin-like 1 | Txnll | 1.39 | 0.002 | 0.74 | 0.007 | 1.24 | 0.034 | ||||||
| AA848545 | similar to programmed cell death 10 | Pdcd10 | 1.38 | 0.063 | 0.61 | 0.016 | 1.52 | 0.033 | ||||||
| NM_012839 | cytochrome c, somatic | Cycs | 1.38 | 0.001 | 0.77 | 0.007 | 1.03 | 0.772 | ||||||
| B1282255 | ribosomal protein S5 | Rps5 | 1.34 | 0.001 | 0.85 | 0.025 | 0.78 | 0.005 | ||||||
| NM_022510 | ribosomal protein L4 | Rp14 | 1.33 | 0.031 | 0.73 | 0.025 | 0.75 | 0.044 | ||||||
| AF411318 | metallothionein 1a | MtIa | 1.29 | 0.058 | 0.62 | 0.006 | 0.90 | 0.456 | ||||||
| NM_013065 | protein phosphatase 1, catalytic subunit, beta isoform | Ppp2cb | 1.26 | 0.051 | 0.75 | 0.028 | 0.98 | 0.865 | ||||||
| B1283681 | eukaryotic translation initation factor 5A | Eif5a | 1.25 | 0.031 | 0.77 | 0.023 | 1.29 | 0.031 | ||||||
| NM_080888 | BCL2/adenovirus E1B 19 kDa-interacting protein 3-like | Bnip31 | 1.19 | 0.077 | 0.73 | 0.010 | 0.60 | 0.001 | ||||||
| BF281741 | glioma tumor suppressor candidate region gene 2 | GItscr2 | 1.18 | 0.067 | 0.84 | 0.068 | 0.93 | 0.461 | ||||||
| BM389310 | DEAD (Asp-Glu-Ala-Asp) box polypeptide 41 (predicted) | Ddx41_predicted | 1.18 | 0.096 | 0.77 | 0.021 | 1.33 | 0.019 | ||||||
| GROUP 1B | GENES INCREASED IN SBR50 vs. SHAM AND UNCHANGED IN SBR50/IL-6 vs.SBR50 | |||||||||||||
| NM_012912 | activating transcription factor 3 | Atf3 | 92.25 | 0.000 | 1.36 | 0.435 | 0.45 | 0.054 | ||||||
| NM_031970 | heat shock 27kDa protein 1 | Hspbl | 11.79 | 0.006 | 4.12 | 0.100 | 0.44 | 0.360 | ||||||
| NM_053565 | suppressor of cytokine signaling 3 | Socs3 | 10.14 | 0.000 | 0.51 | 0.118 | 1.80 | 0.177 | ||||||
| NM_012603 | myeloctomatosis viral oncogene homolog (avain) | Myc | 9.53 | 0.000 | 0.65 | 0.357 | 1.08 | 0.894 | ||||||
| BM384926 | DnaJ (Hsp40) homolog, subfamily B, member 1 (predicted) | Dnajb1 | 9.09 | 0.005 | 2.79 | 0.155 | 0.26 | 0.081 | ||||||
| NM_013154 | CCAAT CCAAT/enhancer binding protein (C/EBP), delta | Cebpd | 6.32 | 0.000 | 0.60 | 0.151 | 1.38 | 0.387 | ||||||
| AF080594 | vascular endothelial growth factor A | Vegfa | 6.14 | 0.000 | 0.60 | 0.188 | 1.04 | 0.923 | ||||||
| BG671521 | heat shock protein 1, alpha | Hspca | 4.72 | 0.001 | 1.34 | 0.465 | 0.82 | 0.658 | ||||||
| BI284349 | myeloid differentiation primary response gene 116 | Myd116 | 4.65 | 0.004 | 1.71 | 0.270 | 0.98 | 0.969 | ||||||
| AF007789 | plasminogen activator, urokinase receptor | Plaur | 4.28 | 0.001 | 0.62 | 0.151 | 1.17 | 0.662 | ||||||
| NM_021836 | Jun-B oncogene | Junb | 4.12 | 0.000 | 1.31 | 0.303 | 0.94 | 0.845 | ||||||
| NM_022542 | ras homolog gene family, member B | Rhob | 4.06 | 0.001 | 1.19 | 0.629 | 1.05 | 0.900 | ||||||
| NM_012551 | early growth response 1 | Egrl | 3.89 | 0.001 | 1.15 | 0.693 | 0.92 | 0.839 | ||||||
| NM_058208 | suppressor of cytokine signaling 2 | Socs2 | 3.13 | 0.045 | 0.45 | 0.178 | 1.51 | 0.499 | ||||||
| NM_012699 | DnaJ (Hsp40) homolog, subfamily B, member 9 | Dnajb9 | 2.53 | 0.019 | 0.55 | 0.132 | 0.55 | 0.147 | ||||||
| NM_013085 | plasminogen activator, urokinase | Plau | 2.23 | 0.085 | 0.81 | 0.690 | 0.87 | 0.791 | ||||||
| NM_133416 | B-cell leukemia/lymphoma 2 related protein A1 | Bcl2a1 | 2.22 | 0.048 | 0.81 | 0.630 | 2.78 | 0.028 | ||||||
| BM392366 | platelet-activating factor acetylhtdrolase, isoform 1b, alpha2 subunit | Pafah1b2 | 2.13 | 0.007 | 0.84 | 0.511 | 1.26 | 0.398 | ||||||
| AI178012 | retinoblastoma 1 | Rb1 | 1.96 | 0.000 | 1.14 | 0.331 | 1.43 | 0.018 | ||||||
| BI294137 | Hexokinase 2 | Hk2 | 1.89 | 0.006 | 0.71 | 0.119 | 1.00 | 0.985 | ||||||
| AI008680 | benzodiazepine receptor, peripheral | Bzrp | 1.86 | 0.003 | 0.85 | 0.383 | 2.82 | 0.000 | ||||||
| NM_017034 | proviral integration site 1 | Pim1 | 1.85 | 0.012 | 0.88 | 0.586 | 1.09 | 0.736 | ||||||
| NM_012904 | annexi A1 | Anxa 5 | 1.82 | 0.036 | 0.97 | 0.939 | 0.78 | |||||||
| BE109605 | zinc finger protein 162 | Zfp162 | 1.74 | 0.016 | 0.75 | 0.199 | 0.95 | 0.837 | ||||||
| NM_013151 | plasminogen activator, tissue | Plat | 1.68 | 0.008 | 0.91 | 0.630 | 1.14 | 0.499 | ||||||
| AI178772 | prothymosin alpha | Ptma | 1.67 | 0.010 | 0.86 | 0.424 | 1.42 | 0.084 | ||||||
| NM_012734 | hexokinase 1 | Hk1 | 1.67 | 0.012 | 0.73 | 0.116 | 0.86 | 0.459 | ||||||
| NM_012523 | CD53 antigen | Cd53 | 1.66 | 0.013 | 0.73 | 0.117 | 1.14 | 0.517 | ||||||
| AI237389 | heat shock 90kDa protein 1, beta | Hspcb | 1.64 | 0.032 | 1.30 | 0.258 | 1.49 | 0.106 | ||||||
| NM_022531 | Desmin | Des | 1.60 | 0.032 | 0.82 | 0.380 | 1.53 | 0.076 | ||||||
| BI296385 | similar to chemokine (C-X-C motif) ligand 16 | Cxcl16 | 1.59 | 0.017 | 0.82 | 0.312 | 1.65 | 0.024 | ||||||
| AA945737 | chemokine (C-X-C motif) receptor 4 | Cxcr4 | 1.49 | 0.051 | 0.88 | 0.546 | 1.62 | 0.037 | ||||||
| NM_012715 | Adrenomedullin | Adm | 1.48 | 0.057 | 1.12 | 0.605 | 1.76 | 0.020 | ||||||
| J04024 | ATPase, Ca++ transporting, cardiac muscle, slow twitch 2 | Atp2a2 | 1.47 | 0.035 | 0.99 | 0.988 | 1.03 | 0.900 | ||||||
| NM_024359 | hypoxia inducible factor 1, alpha subunit | Hifla | 1.47 | 0.008 | 0.86 | 0.299 | 1.19 | 0.235 | ||||||
| M83297 | protein phosphatase 2 (formerly 2A), regulatory subunit B (PR 52), alpha isoform | Ppp2ca | 1.37 | 0.080 | 0.81 | 0.275 | 1.06 | 0.778 | ||||||
| BE116857 | apoptotic chormatin condensation inducer 1 | Acin1 | 1.30 | 0.089 | 0.92 | 0.605 | 1.14 | 0.421 | ||||||
| AI411586 | Sequestosome 1 | Sqstm1 | 1.29 | 0.040 | 0.94 | 0.673 | 1.21 | 0.153 | ||||||
| M25590 | seminal vesicle protein 4 | Svp4 | 1.28 | 0.072 | 0.92 | 0.601 | 0.93 | 0.625 | ||||||
| U69550 | phospholipase D1 | Pldl | 1.26 | 0.75 | 0.093 | 0.585 | 0.77 | 0.074 | ||||||
| NM_019161 | cadherin 22 | Cdh22 | 1.06 | 0.048 | 0.97 | 0.215 | 0.96 | 0.207 | ||||||
| NM_012641 | regenerating islet-derived 1 | Regl | 1.02 | 0.06 | 1.00 | 0.825 | 0.96 | 0.018 | ||||||
| BI288701 | B-cell translocation gene 2, anti-proliferative | Btg2 | 25.68 | 0.000 | 0.41 | 0.135 | 1.31 | 0.672 | ||||||
| NM_024388 | nuclear receptor subfamily 4, group A, member 1 | Nr4a1 | 9042 | 0.002 | 2.22 | 0.220 | 0.98 | 0.977 | ||||||
| NM_024134 | DNA_damage inducible transcript 3 | Ddit3 | 7.53 | 0.002 | 1.21 | 0.757 | 3.78 | 0.033 | ||||||
| NM_1335578 | dual specificity phosphatase 5 | Dusp5 | 6.41 | 0.000 | 1.08 | 0.857 | 0.96 | 0.909 | ||||||
| NM_013060 | inhibitor of DNA binding 2 | Id2 | 5.69 | 0.000 | 0.69 | 0.201 | 1.77 | 0.063 | ||||||
| NM_031628 | nuclear receptor subfamily 4, group A, member 3 | Nr4a3 | 4.96 | 0.036 | 1.34 | 0.726 | 0.68 | 0.653 | ||||||
| AI010427 | cyclin-dependent kinase inhibitor 1A | Cdkn1a | 2.90 | 0.013 | 0.55 | 0.151 | 1.83 | 0.171 | ||||||
| NM_053847 | mitogen-activated protein kinase kinase kinase 8 | Map3k8 | 2.89 | 0.008 | 0.59 | 0.170 | 1.52 | 0.289 | ||||||
| NM_057138 | CASP8 and FADD-like apoptosis regualtor | Cfr | 2.82 | 0.051 | 0.44 | 0.136 | 1.77 | 0.317 | ||||||
| NM_053319 | dynein light chain LC8-type 1 | Dynll1 | 2.80 | 0.001 | 1.26 | 0.375 | 1.35 | 0.255 | ||||||
| NM_053819 | tissue inhibitor of metalloproteinase 1 | Timp1 | 2.66 | 0.002 | 0.82 | 0.488 | 1.96 | 0.030 | ||||||
| NM_194457 | seven in absentia 2 | Siah2 | 2.47 | 0.005 | 0.83 | 0.527 | 0.87 | 0.662 | ||||||
| NM_012954 | fos-like antigen 2 /// FBJ asteosarcoma oncogene B | Fos12 | 2.42 | 0.008 | 1.02 | 0.974 | 1.49 | 0.223 | ||||||
| NM_012966 | heat shock 10 kDa protein 1 (chaperonin 10) | Hspel | 2.31 | 0.000 | 1.23 | 0.255 | 0.55 | 0.008 | ||||||
| BG671549 | superoxide dismutase 2, mitochondrial | Sod2 | 2.27 | 0.077 | 0.57 | 0.238 | 3.76 | 0.018 | ||||||
| U05989 | PRKC, apoptosis. WTI, regulator | Paur | 2.06 | 0.003 | 1.35 | 0.165 | 0.99 | 0.976 | ||||||
| NM_053957 | amyloid beta (A4) precursor protein-binding, family B, member 3 | Apbb3 | 2.03 | 0.014 | 0.69 | 0.184 | 1.24 | 0.463 | ||||||
| NM_017180 | pleckstrin homology-like domain, family A, member 1 | Phidal | 1.96 | 0.051 | 1.09 | 0.844 | 0.98 | 0.968 | ||||||
| AB020367 | tribbles homolog 3 (Drosophila) | Trib3 | 1.91 | 0.036 | 0.60 | 0.118 | 1.87 | 0.069 | ||||||
| NM_053394 | Kruppel-like factor 5 | Klf5 | 1.85 | 0.062 | 0.62 | 0.165 | 0.86 | 0.680 | ||||||
| NM_012953 | fos-like antigen 1 | Fosl1 | 1.84 | 0.061 | 0.65 | 0.204 | 0.82 | 0.595 | ||||||
| NM_017258 | B-cell translocation gene 1, anti-proliferative | Btg1 | 1.70 | 0.010 | 0.73 | 0.117 | 1.17 | 0.450 | ||||||
| NM_022612 | BCL2-like 11 (apoptosis faciliator) | Bcl2111 | 1.55 | 0.020 | 0.75 | 0.129 | 0.83 | 0.353 | ||||||
| AA892271 | BCL2-like 13 (apoptosis faciliator) (predicted) | Bcl2113 | 1.49 | 0.036 | 0.87 | 0.499 | 1.19 | 0.387 | ||||||
| AA957342 | peptidylprolyl isomerase D (cyclophilin D) | Ppid | 1.46 | 0.051 | 1.24 | 0.286 | 1.20 | 0.384 | ||||||
| NM_053289 | pancreatitis-associated protein | Pap | 1.43 | 0.100 | 0.70 | 0.120 | 1.11 | 0.672 | ||||||
| NM_017006 | glucose-6-phosphate dehydrogenase x-linked | G6pdx | 1.41 | 0.005 | 0.89 | 0.293 | 1.83 | 0.000 | ||||||
| NM_012829 | Cholecystokinin | Cck | 1.39 | 0.037 | 0.89 | 0.463 | 0.62 | 0.013 | ||||||
| AF235993 | Bc12-associated X protein | Box | 1.37 | 0.072 | 1017 | 0.399 | 2.76 | 0.000 | ||||||
| NM_019282 | gremlin 1 homolog cysteine knot superfamily (Xenopus laevis) | Grem1 | 1.26 | 0.087 | 0.84 | 0.215 | 0.92 | 0.568 | ||||||
| NM_131911 | acidic nuclear phosphoprotein 32 family, member B | Anp32b | 1.23 | 0.025 | 0.86 | 0.117 | 1.18 | 0.095 | ||||||
| B1280304 | Bc12-associated athanogene 1 (predicted) | Bag1 | 1.20 | 0.086 | 1.01 | 0.955 | 1.34 | 0.019 | ||||||
| BF287444 | glycogen synthase kinase 3 beta | Gsk3b | 1.18 | 0.067 | 0.92 | 0.349 | 1.04 | 0.669 | ||||||
| NM_031038 | Gonadotrophin releasing hormone receptor | Gnrhr | 1.15 | 0.050 | 0.89 | 0.113 | 0.97 | 0.736 | ||||||
| AF081196 | RAS guanyl releasing protein 1 | Rasgrp1 | 1.10 | 0.018 | 0.94 | 0.140 | 0.94 | 0.154 | ||||||
| NM_012650 | sex hormone binding globulin | Shbg | 1.02 | 0.065 | 0.99 | 0.566 | 0.97 | 0.028 | ||||||
| GROUP 2A | GENES DECREASED IN SBR50 vs. SHAM AND INCREASED IN SBR50/IL-6 vs. SBR50 | |||||||||||||
| NM_052980 | nuclear receptor subfamily 1, group I, member 2 | Nrli2 | 0.09 | 0.000 | 1.38 | 0.027 | 1.39 | 0.031 | ||||||
| NM_021745 | nuclear receptor subfamily 1, group H, member 4 | Nrlh4 | 0.40 | 0.002 | 2.99 | 0.003 | 1.35 | 0.251 | ||||||
| NM_022407 | aldehyde dehydrogenase family 1, member A1 | Aldhlal | 0.43 | 0.015 | 2.13 | 0.038 | 0.42 | 0.024 | ||||||
| NM_012805 | retinoid X receptor alpha | Rxra | 0.49 | 0.000 | 1.27 | 0.084 | 0.81 | 0.130 | ||||||
| NM_012493 | alpha-fetoprotein | Afp | 0.50 | 0.007 | 2.02 | 0.012 | 0.86 | 0.547 | ||||||
| AI235465 | steroid sensitive gene 1 | Ssgl | 0.50 | 0.012 | 2.07 | 0.015 | 0.53 | 0.030 | ||||||
| NM_133400 | apobec-1 complementation factor | Acf | 0.55 | 0.004 | 1.63 | 0.019 | 0.85 | 0.387 | ||||||
| BM392373 | CEA-related cell adhestion molecule 1 | Ceacaml | 0.58 | 0.003 | 1.58 | 0.013 | 0.69 | 0.038 | ||||||
| BG377107 | DnaJ (Hsp40) homolog, subfamily A, member 3 | Dnaja3 | 0.60 | 0.000 | 1.44 | 0.005 | 0.93 | 0.482 | ||||||
| NM_012931 | breast cancer anti-estrogen resistance 1 | Bcarl | 0.60 | 0.018 | 1.65 | 0.027 | 1.25 | 0.310 | ||||||
| NM_022614 | inhibin beta C | Inhbc | 0.61 | 0.020 | 1.64 | 0.028 | 0.94 | 0.809 | ||||||
| BF282337 | integral membrane protein 2B | Itm2b | 0.63 | 0.000 | 1.27 | 0.007 | 1.13 | 0.117 | ||||||
| BI276999 | testis enhanced gene transcript | Tegt | 0.63 | 0.000 | 1.39 | 0.002 | 0.79 | 0.005 | ||||||
| AY083159 | Stam binding protein | Stambp | 0.63 | 0.009 | 1.56 | 0.017 | 1.43 | 0.049 | ||||||
| BY275921 | anterior pharynx defective la homolog (C. elegans) | Aphla | 0.64 | 0.002 | 1.38 | 0.021 | 2.05 | 0.000 | ||||||
| AF347936 | interlekin 11 receptor alpha chain 1 | Ill1rals | 0.65 | 0.009 | 1.60 | 0.011 | 1.27 | 0.154 | ||||||
| AW672589 | nuclear factor of kappa light chain gene enhancer in B-cells inhibitor, alpha | Nfkbia | 0.65 | 0.003 | 1.82 | 0.002 | 2.10 | 0.000 | ||||||
| BI279735 | Ubiquilin 1 | Frapl | 0.68 | 0.001 | 1.41 | 0.004 | 1.34 | 0.008 | ||||||
| NM_017181 | fumarylac etoacetate hydrolase | Fah | 0.69 | 0.002 | 1.29 | 0.002 | 0.75 | 0.019 | ||||||
| NM_031546 | Regucalcin | Rgn | 0.69 | 0.004 | 1.57 | 0.004 | 0.31 | 0.000 | ||||||
| NM_022277 | caspase 8 | Casp8 | 0.70 | 0.013 | 1.32 | 0.058 | 1.92 | 0.001 | ||||||
| AW523747 | adhesion molecule with Ig like domain 2 | Amigo2 | 0.72 | 0.045 | 1.54 | 0.019 | 1.20 | 0.131 | ||||||
| BI274345 | ataxin 10 | Atxn10 | 0.73 | 0.008 | 1.22 | 0.079 | 0.90 | 0.387 | ||||||
| BI295511 | forkhead box O1A | Foxola | 0.73 | 0.020 | 1.39 | 0.022 | 0.88 | 0.353 | ||||||
| AI009817 | succinate dehydrogenase complex, subunit C, integral membrane protein | Sdhc | 0.73 | 0.034 | 1.37 | 0.043 | 1.02 | 0.925 | ||||||
| AI011448 | Notch gene homolog 2 (Drosophila) | Notch2 | 0.74 | 0.088 | 1.59 | 0.019 | 1.18 | 0.384 | ||||||
| BI285459 | Nicastrin | Ncstn | 0.76 | 0.003 | 1.20 | 0.037 | 1.34 | 0.005 | ||||||
| AB015946 | tubulin, gamma 1 | Tubgl | 0.78 | 0.090 | 1.79 | 0.004 | 1.31 | 0.104 | ||||||
| NM_022399 | Calreticulin | Calr | 0.80 | 0.016 | 1.31 | 0.012 | 1.05 | 0.595 | ||||||
| M15481 | insulin-like growth factor 1 | Igfl | 0.83 | 0.069 | 1.36 | 0.014 | 0.51 | 0.000 | ||||||
| NM_024352 | Macrophage stimulating 1 (hepatocyte growth factor-like) | Mstl | 0.83 | 0.038 | 1.28 | 0.017 | 0.84 | 0.077 | ||||||
| NM_053491 | Plasminogen | Parl | 0.83 | 0.009 | 1.13 | 0.073 | 0.82 | 0.014 | ||||||
| BI281756 | Parkinson disease (auosomal recessive, early onset) 7 | Park7 | 0.85 | 0.036 | 1.35 | 0.004 | 1.13 | 0.166 | ||||||
| NM_133317 | transducer of ErbB-2 1 | Tobl | 0.34 | 0.002 | 2.81 | 0.006 | 1.41 | 0.284 | ||||||
| AF335281 | STEAP family member 3 | Steap3 | 0.41 | 0.002 | 1.62 | 0.066 | 0.69 | 0.170 | ||||||
| AI412114 | etoposide induced 2–4 Mma | Ei24 | 0.46 | 0.000 | 2.14 | 0.002 | 0.98 | 0.917 | ||||||
| NM_012870 | retinoblastoma-like 2 | Rb12 | 0.49 | 0.006 | 1.54 | 0.079 | 1.03 | 0.909 | ||||||
| NM_012870 | tumor necrosis factor receptor superfamily, member 11b (osteoprotegerin) | Tnfrsf11b | 0.50 | 0.004 | 1.99 | 0.007 | 1.23 | 0.360 | ||||||
| NM_0530902 | kynurenimase (L-kynurenime hydrolase) | Kynu | 0.52 | 0.003 | 1.73 | 0.014 | 0.47 | 0.003 | ||||||
| AI169638 | sphingosine-l-phosphate phosphatase 1 | Sgppl | 0.53 | 0.002 | 1.51 | 0.028 | 1.47 | 0.048 | ||||||
| BM388758 | excision repair cross-complementing rodent repair deficiency, complementation group 3 | Ercc3 | 0.57 | 0.002 | 1.54 | 0.016 | 1.66 | 0.008 | ||||||
| BF398331 | estrogen receptor-binding fragment-associated gene 9 | Ebag9 | 0.61 | 0.013 | 1.41 | 0.082 | 0.81 | 0.300 | ||||||
| AB004454 | presenilin 2 | Psen 2 | 0.62 | 0.002 | 1.32 | 0.051 | 1.60 | 0.006 | ||||||
| NM_019158 | aquaporin 8 | Aqp8 | 0.64 | 0.018 | 1.66 | 0.016 | 0.92 | 0.692 | ||||||
| NM_057146 | complement component 9 | C9 | 0.64 | 0.002 | 1.29 | 0.042 | 1.36 | 0.022 | ||||||
| AF275151 | androgen receptor-related apoptosis-associated protein CBL27 | Cb127 | 0.65 | 0.001 | 1.36 | 0.011 | 1.03 | 0.772 | ||||||
| NM_017319 | protein disulfide isomerase associated 3 | Pdia3 | 0.66 | 0.001 | 1.67 | 0.002 | 1.22 | 0.062 | ||||||
| NM_053907 | deoxyribonuclease I-like 3 | Dnase112 | 0.66 | 0.005 | 1.36 | 0.034 | 0.66 | 0.012 | ||||||
| AF262320 | programmed cell death 8 | Pdcd8 | 0.69 | 0.000 | 1.19 | 0.009 | 0.92 | 0.164 | ||||||
| BM391371 | Goliath | LOC652955 | 0.71 | 0.001 | 1.26 | 0.016 | 0.82 | 0.034 | ||||||
| AA819870 | complement component 8, beta polypeptide (mapped) | C8b | 0.79 | 0.018 | 1.28 | 0.024 | 0.80 | 0.038 | ||||||
| AI409930 | B-cell receptor-associated protein 31 | Bcap31 | 0.81 | 0.007 | 1.28 | 0.006 | 1.08 | 0.333 | ||||||
| GROUP 2B | GENES DECREASED IN SBR50 vs. SHAM AND UNCHANGED IN SBR50/IL-6 vs. SBR50 | |||||||||||||
| NM_012842 | epidemal growth factor | Egfr | 0.32 | 0.000 | 1.29 | 0.204 | 0.59 | 0.021 | ||||||
| BM385790 | Kruppel-like factor 2 (lung) (predicted) | Klf2 | 0.40 | 0.006 | 1.10 | 0.789 | 1.33 | 0.384 | ||||||
| NM_032612 | signal transducer and activator of transcription 1 | Stat1 | 0.45 | 0.075 | 2.20 | 0.101 | 6.09 | 0.003 | ||||||
| NM_053923 | phosphatidylinositol 3-kinase, C2 domain containing, gamma polypeptide | Pic3c2g | 0.50 | 0.014 | 1.15 | 0.630 | 0.30 | 0.002 | ||||||
| NM_031541 | scavenger receptor class B, member 1 | Scarb1 | 0.52 | 0.002 | 1.29 | 0.151 | 0.77 | 0.152 | ||||||
| NM_053523 | homocysteine-inducible, endoplasmic reticulum stress-inducible, ubiquitin-like domain member 1 | Herpud 1 | 0.53 | 0.017 | 1.42 | 0.186 | 0.47 | 0.015 | ||||||
| NM_013063 | poly (ADP-ribose) polymerase family, member 1 | Parp12_predicted | 0.56 | 0.011 | 0.87 | 0.539 | 1.02 | 0.951 | ||||||
| NM_017206 | solute carrier family 6 (neurotransmitter transporter, taurine), member 6 | Slc6a6 | 0.56 | 0.078 | 0.81 | 0.539 | 1.60 | 0.185 | ||||||
| AA799421 | protein kinase C, epsilon | Prkce | 0.58 | 0.002 | 0.79 | 0.119 | 1.50 | 0.019 | ||||||
| NM_022185 | phosphatidylinositol 3-kinase, regulatory subunit, polypeptide 2 | Pik3r2 | 0.59 | 0.056 | 1.18 | 0.575 | 1.33 | 0.348 | ||||||
| AI177631 | bifunctional apoptosis regualtor | bflar | 0.59 | 0.086 | 0.93 | 0.857 | 1.52 | 0.230 | ||||||
| NM_017322 | mitogen-activated protein kinase 9 | Mapk9 | 0.60 | 0.011 | 1.15 | 0.468 | 1.24 | 0.289 | ||||||
| NM_017094 | growth hormone receptor | Ghr | 0.61 | 0.072 | 1.15 | 0.650 | 0.60 | 0.096 | ||||||
| AI716502 | spkingosine kinase 2 | Sphk2 | 0.64 | 0.046 | 1.44 | 0.119 | 1.06 | 0.837 | ||||||
| BG378230 | mitochondrial ribosomal protein S30 (predicted) | Mrps30 | 0.64 | 0.016 | 1.10 | 0.611 | 0.46 | 0.002 | ||||||
| NM_019364 | sec 1 family domain containing 1 | Scfd1 | 0.65 | 0.025 | 1.28 | 0.189 | 1.05 | 0.798 | ||||||
| BM385237 | annexin A4 | Anxa5 | 0.66 | 0.087 | 1.49 | 0.120 | 2.32 | 0.007 | ||||||
| BI289543 | zicn finger, MYND domain containing 11 | Zmynd11 | 0.67 | 0.006 | 1.05 | 0.744 | 1.24 | 0.137 | ||||||
| U68544 | peptidylprolyl isomerase F (cyclophilin F) | Ppif | 0.67 | 0.003 | 1.07 | 0.587 | 0.88 | 0.348 | ||||||
| NM_031053 | mutL homolog 1 (E. coli) /// hypothetical gene supported by NM_031053 | Mlhl | 0.68 | 0.45 | 1038 | 0.103 | 1.30 | 0.200 | ||||||
| X02904 | glutathione-S-transferase, pi 1///glutathione S-transferase, pi 2 | Gstpl | 0.71 | 0.012 | 1.22 | 0.136 | 0.79 | 0.089 | ||||||
| NM_053777 | mitogen activated protein kinase 8 interacting protein | Mapk8ip | 0.71 | 0.033 | 1.29 | 0.117 | 0.86 | 0.387 | ||||||
| BE110607 | similar to apoptosis related protein APR-3; p18 (predicted) | RGD1311605_predicted | 0.72 | 0.072 | 1.29 | 0.170 | 0.69 | 0.070 | ||||||
| NM_012803 | protein C | Sftpc | 0.72 | 0.005 | 1.18 | 0.123 | 0.70 | 0.007 | ||||||
| U69109 | protein tyrosine kinase 2 beta | Ptk2b | 0.74 | 0.081 | 1.11 | 0.561 | 1.21 | 0.321 | ||||||
| AA819804 | Zinc finger protein 91 | Zfp91 | 0.75 | 0.090 | 1.16 | 0.420 | 1.26 | 0.216 | ||||||
| BG673589 | paxillin | Pxn | 0.75 | 0.028 | 1.16 | 0.267 | 1.17 | 0.258 | ||||||
| BF419646 | retimoic acid receptor, beta | Ram | 0.75 | 0.086 | 1.14 | D.468 | 2.37 | 0.001 | ||||||
| NM_053739 | beclin 1 (coiled-coil, myosin-like BCL2-interacting protein) | Becn1 | 0.75 | 0.037 | 1.17 | 0.258 | 1.11 | 0.470 | ||||||
| AI171615 | chromosome segregation 1-like (S. cerevisiae) (predicted) | Csell_predicted | 0.78 | 0.062 | 1.26 | 0.104 | 1.45 | 0.019 | ||||||
| NM_053864 | valosin-containing protein | Vcpip1 | 0.78 | 0.010 | 1.06 | 0.501 | 1.14 | 0.176 | ||||||
| AIl70385 | SWI/SNF related, matrix associated, actin dependent regulator of chromatin, subfamily a, member 2 | Smarca2 | 0.80 | 0.089 | 1.21 | 0.170 | 0.99 | 0.923 | ||||||
| NM_012660 | eukaryotic translation elongation factor 1 alpha 2 | Eefl2a | 0.81 | 0.090 | 1.02 | 0.931 | 0.86 | 0.276 | ||||||
| NM_133409 | integrin linked kinase | Ilk | 0.83 | 0.087 | 1.07 | 0.582 | 1.27 | 0.054 | ||||||
| AA800669 | protein phosphatase 2 (formerly 2A), regulatory subunit A (PR 65), alpha isoform | Ppp2f1a | 0.88 | 0.091 | 1.00 | 0.979 | 1.23 | 0.024 | ||||||
| NM_013149 | aryl hydrocarbon receptor | Ahr | 0.32 | 0.001 | 1.21 | 0.502 | 0.92 | 0.776 | ||||||
| NM_022180 | hepatocyte nuclear factor 4, alpha | Hrf4a | 0.33 | 0.000 | 1.35 | 0.119 | 1.45 | 0.066 | ||||||
| BI297236 | Transformation related protein 53 inducible nuclear protein 1 | Trp53inp1 | 0.49 | 0.053 | 1.47 | 0.317 | 1.06 | 0.894 | ||||||
| BE1080508 | Procollagen, type XVIII, alpha 1 | Coll8a1 | 0.49 | 0.045 | 1.56 | 0.217 | 0.67 | 0.283 | ||||||
| AB019366 | poly (ADP-ribose) glycohydrolase | Parg | 0.52 | 0.004 | 1.12 | 0.611 | 1.49 | 0.072 | ||||||
| NM_012498 | aldo-keto reductase family 1,member B4 (aldose reductase) | Akr1b4 | 053 | 0.003 | 1.33 | 0.138 | 1.10 | 0.660 | ||||||
| NM_053307 | methionine sulfoxide reductase A | Msra | 0.60 | 0.002 | 1.26 | 0.125 | 0.47 | 0.001 | ||||||
| BF400782 | protein kinase, DNA activated, catalytic polypeptide (predicted) | Prkdc_predicted | 0.60 | 0.005 | 1.06 | 0.733 | 0.89 | 0.506 | ||||||
| NM_017079 | CDldl antigen | Cdldl | 0.60 | 0.031 | 1.46 | 0.118 | 1.28 | 0.317 | ||||||
| NM_053448 | histone deacetylase 3 | Hdac3 | 0.61 | 0.028 | 1.39 | 0.147 | 1.24 | 0.367 | ||||||
| A1408078 | BCL2-associated transcription factor 1 | Bclaf1 | 0.64 | 0.028 | 0.93 | 0.748 | 0.72 | 0.130 | ||||||
| BF283688 | tumor necrosis factor ligand superfamily member 12 | Tnflfl2 | 0.65 | 0.069 | 1.46 | 0.123 | 1.19 | 0.497 | ||||||
| B1286396 | PERP, TP53 apoptosis affector (predicted) | Perp | 0.69 | 0.042 | 1.13 | 0.540 | 0.51 | 0.005 | ||||||
| BE107162 | high mobility group box 1 | Hmgb1 | 0.69 | 0.005 | 1.17 | 0.199 | 0.89 | 0.374 | ||||||
| BF408447 | programmed cell death 5 (predicted) | Pdcd8 | 0.70 | 0.013 | 1.25 | 0.123 | 1.19 | 0.236 | ||||||
| U06230 | protein S (alpha) | Prosl | 0.70 | 0.019 | 1.14 | 0.405 | 0.70 | 0.034 | ||||||
| A1227743 | Fas-activated serine/threonine kinase | Fastk | 0.72 | 0.017 | 1.00 | 0.989 | 1.11 | 0.459 | ||||||
| AIl76519 | immediate early response 3 | Ier3 | 0.73 | 0.051 | 1.32 | 0.110 | 1.16 | 0.396 | ||||||
| NM_130406 | Fas-associated factor 1 | Fafl | 0.74 | 0.061 | 1.16 | 0.389 | 1.51 | 0.030 | ||||||
| BM389034 | TNF receptor-associated protein 1 | Trap 1 | 0.74 | 0.036 | 1.08 | 0.636 | 0.91 | 0.554 | ||||||
| AA800180 | thioredoxin 2 | Txn2 | 0.75 | 0.029 | 1.10 | 0.465 | 1.18 | 0.230 | ||||||
| D14592 | mitogen activated protein kinase kinase 2 | Map2k2 | 0.75 | 0.033 | 1.23 | 0.132 | 1.37 | 0.037 | ||||||
| BG665132 | mitochondrial carrier homolog 1 (C. elegans) | Mtch1 | 0.77 | 0.022 | 1.08 | 0.537 | 1.17 | 0.207 | ||||||
| BM390196 | thioredoxin domain containig 5 (predicted) | Txndc5_predicted | 0.77 | 0.043 | 1.22 | 0.126 | 0.65 | 0.008 | ||||||
| BF284481 | SH3-domain GRB2-like B1 (endophilin) | Sh3glbl | 0.77 | 0.087 | 0.93 | 0.673 | 1.23 | 0.213 | ||||||
| BM383722 | NCK-assocaited protein 1 | Nell1 | 0.80 | 0.070 | 1.06 | 0.646 | 0.95 | 0.719 | ||||||
| NM_031978 | proteasome (prosome, macropain) 26S subunit, non-ATPase, 1 | Psmd1 | 0.81 | 0.093 | 1.14 | 0.336 | 1.33 | 0.052 | ||||||
| AA946474 | Calpastatin | Cast | 0.82 | 0.089 | 0.96 | 0.781 | 1.15 | 0.288 | ||||||
| GROUP 3 | GENES INCREASED IN SBR50 vs. SHAM AND INCREASED IN SBR50/IL-6 vs. SBR50 | |||||||||||||
| B1278231 | heat shock 70kD protein 1B (mapped) | Hspalb | 51.80 | 0.002 | 7.75 | 0.072 | 0.06 | 0.026 | ||||||
| NM_031971 | heat shock 70kD protein 1A /// heat shock 70kD protein 1B (mapped) | Hspala | 45.41 | 0.002 | 7.32 | 0.061 | 0.08 | 0.026 | ||||||
| AI236601 | heat shock 105kDa/110kDa protein 1 | Hsph1 | 8.49 | 0.006 | 3.36 | 0.097 | 0.43 | 0.264 | ||||||
| B1288619 | Jun oncogene | Jun | 5.64 | 0.001 | 2.68 | 0.021 | 0.51 | 0.102 | ||||||
| NM_031327 | cysteine rich protein 61 | Cyr61 | 3.48 | 0.003 | 4.43 | 0.004 | 0.52 | 0.105 | ||||||
| AA818262 | angiopoietin-like 4 | Angpt14 | 2.98 | 0.000 | 1.50 | 0.028 | 1.37 | 0.089 | ||||||
| NM_012935 | crystallin, alpha B | Cryab | 1.82 | 0.016 | 1.84 | 0.021 | 0.88 | 0.619 | ||||||
| AF077354 | heat shock protein 4 | Hspa4 | 1.50 | 0.037 | l.46 | 0.063 | 1.20 | 0.381 | ||||||
| A1599423 | growth arrest and DNA-damage-inducible 45 gamma | Gadd45g | 8.66 | 0.000 | 2.50 | 0.005 | 0.80 | 0.411 | ||||||
| A1231792 | Bc12-associated athanogene 3 | Bag3 | 3.76 | 0.018 | 3.16 | 0.049 | 0.16 | 0.007 | ||||||
Genes listed in regular type are anti-apoptotic, while genes listed in italics are pro-apoptotic.
FDR: False discovery rate.
Figure 5.
Effect of trauma/HS without or with IL-6 treatment on liver apoptosisrelated gene expression; impact of Stat3 inhibition on the IL-6 effect. In panel A, a heat map of apoptosis pathway genes is shown containing those genes whose expression is altered within the 4 groups. Columns represent samples from the 4 groups examined as indicated (S, Sham; P, placebo-treated SBR50; I, IL-6-treated SBR50/IL-6; and G, animals pre-treated with G-quartet ODN prior to HS and IL-6 treatment, SBR50/IL-6/G). Rows represent genes as listed in Table 1. Red indicates a level of expression above the mean expression of a gene within the experimental group. White indicates a level of expression at the mean within the experimental group while blue indicates a level of expression below the mean within the experimental groups. Log2-fold changes in expression levels of subsets of apoptosis-related genes are shown in panels B and C comparing SBR50 vs. sham (open bars), SBR50/IL-6 vs. SBR50 (gray bars) and SBR50/IL-6/G vs. SBR50/IL-6/N (stippled bars). In panel B, the 308 apoptosis-related genes whose expression levels were changed in SBR50 vs. sham were separated into those genes whose transcript levels were increased in SBR50 vs. sham (134 genes; left side of panel) and those whose transcript levels were decreased in SBR50 vs. sham (90 genes; right side of the panel). Bars shown represent mean ± SD of the Log2-fold change in gene expression levels for each comparison. In panel C, the overall effect of trauma/HS in transcript levels of pro- and antiapoptotic genes is shown. In the left side of the panel, the mean ± SD of the Log2-fold change in gene expression levels of 87 proapoptotic genes whose expression was increased in the SBR50 vs. sham comparison is shown (open bar). The expression of 74 of 87 of these genes was decreased in the SBR50/IL-6 vs. SBR50 comparison (gray bar). In the right side of the panel, the mean ±SD of the Log2-fold change in gene expression levels of 68 anti-apoptotic genes whose expression was decreased in the SBR50 vs. sham comparison is shown (open bar). The expression of 63 of these genes was increased in the SBR50/IL-6 vs. SBR50 comparison (gray bar).
One hundred and twenty-five of the 214 genes altered in the SBR50 vs. sham comparison, and normalized in the SBR50/IL-6 vs. SBR50 comparison, were also altered in the SBR50/IL-6 vs. SBR50/IL-6/G comparison. Fifty seven of these 125 genes (46%) were altered in the opposite direction as the SBR50/IL-6 vs. SBR50 comparison, consistent with the hypothesis that IL-6 normalizes the trauma/HS-induced lung apoptosis transcriptome in part through activation of Stat3.
Apoptosis-related genes consist of those encoding proteins that prevent apoptosis (anti-apoptotic genes) and those encoding proteins that induce apoptosis (pro-apoptotic genes). To identify candidate apoptosis-related genes whose altered expression caused trauma/HS- induced AEC apoptosis, we focused on anti-apoptotic genes whose transcript levels were decreased by trauma/HS and on pro-apoptotic genes whose transcript levels were increased by trauma/HS. Among the genes differentially expressed in the SBR50 vs. sham comparison, 69 anti-apoptotic genes were decreased and 90 pro-apoptotic genes were increased (Figure 5C). Expression levels of 65 out of 69 (94%) anti-apoptotic genes decreased by trauma/HS were increased by IL-6 treatment; 76 of 90 (84%) of the pro-apoptotic genes that were increased by trauma/HS, were decreased by IL-6 treatment. Finally, the expression of 46% of anti-apoptotic genes increased by IL-6 treatment were decreased by pre-treatment with T40214; conversely 63% of pro-apoptotic genes decreased by IL-6 treatment were increased by T40214 pre-treatment (Figure 5C).
Pro-apoptotic genes whose expression was increased ≥ 6-fold by trauma/HS (≥2.5-fold upregulated) were EGL nine homolog 3 (Egln3; 17.3-fold), dual specificity phosphatase 6 (Dusp6; 15.5-fold), tumor necrosis factor receptor superfamily 12a (Tnfrsf12a; 9.8- fold), interferon regulatory factor 1 (Irf1; 9.3- fold), oxidized low density lipoprotein receptor 1 (Oldlr1; 9.0-fold), growth arrest and DNA- damage-inducible 45 alpha (Gadd45a; 6.3- fold), and BCL2 adenovirus E1B interacting protein 3 (Bnip3; 6.1-fold, Table 2). The expression of each was decreased in the IL-6- treated group by 1.7–14.3 fold (Table 2).
Discussion
In these studies, we demonstrated that trauma/HS-induced liver apoptosis occurs within 1 hr of resuscitation from trauma/HS and depends on the severity of shock with the degree of apoptosis increasing exponentially with increased duration of shock. There was an absolute requirement for resuscitation in order for apoptosis to occur; the finding that no apoptosis occurred in the absence of resuscitation suggested that complete prevention could be achieved by an appropriate intervention introduced at the start of resuscitation. IL-6 administration at the start of resuscitation completely prevented trauma/HS-induced liver apoptosis and was accompanied by increased levels of Stat3 activity within the liver. Pharmacological inhibition of Stat3 using the G-quartet oligodeoxynucleotide (GQ-ODN) T40214 markedly attenuated IL-6-mediated Stat3 activation and prevention of liver apoptosis. Mice deficient in Stat3β, an endogenous naturally occurring, dominant negative isoform of Stat3, were protected from trauma/HS- induced liver apoptosis confirming a role for Stat3, especially Stat3α, in protection against trauma/HS-induced liver apoptosis. Liver microarray analysis showed that 48% of known apoptosis–related genes were altered in trauma/HS. IL-6 “normalized” the expression of 69% of these genes; Stat3 was responsible for this normalization in 46% of the cases. Further examination of the microarray results indicated that the effect of IL-6 in the apoptosis transcriptome was two- fold; IL-6 increased levels of 96% of the anti-apoptotic gene transcripts whose levels were decreased by trauma/HS and also decreased transcript levels of 84% of the pro-apoptotic genes whose levels were increased by trauma/HS.
The liver is susceptible to injury following insults such as hemorrhagic shock. Since it is responsible for maintaining energy homeostasis, hepatic injury and dysfunction associated with hemorrhagic shock can affect other organs and lead to multiple organ failure and death [40–42]. We have previously demonstrated in mice that trauma/HS induces liver apoptosis detected 24 hrs after the resuscitation [6]. In the current study, using our rat model of trauma/HS we found that liver apoptosis occur as early as 1 hour after reperfusion, and for the first time, that its severity depends on the duration of hypotension and requires resuscitation. Liver apoptosis due to ischemia/reperfusion injury has been shown to occur in humans and in experimental models of liver transplantation [43], as well as during hemorrhagic shock [41, 42]. In these studies, release of ROS during reperfusion was implicated in the pathogenesis of liver apoptosis during reperfusion in models of liver transplantation [42, 43], and may be contributing to liver apoptosis in our model.
Interventions aiming at preventing liver apoptosis have been shown to prevent parenchymal injury and improve animal survival in ischemic injury during liver transplant [43]. We previously demonstrated that exogenous administration of IL-6 decreased liver apoptosis in mice following trauma/HS [6], however, the mechanism for the anti-apoptotic effects of IL-6 was not determined. Kovalovich et al. provided evidence that IL-6 protects hepatocytes from carbon tetrachloride-induced apoptosis [44] as well as from Fas-mediated apoptosis [45] through prevention of rapid degradation of the anti-apoptotic proteins Bcl-2, Bcl-xL and FLIP in the livers. While this may contribute to the anti-apoptotic effect of IL-6 administration in our rat trauma/HS model, our finding that the IL-6 preventive effect was blocked by a specific Stat3 inhibitor, T40214, and could be replicated in mice by genetic deletion of Stat3β, a naturally occurring, dominant-negative isoform of Stat3, indicates that a large portion of the effect of IL-6 in our trauma/HS model is being mediated by Stat3 which acts at the transcriptional and not the post-translational level.
IL-6-mediated activation of Stat3 has been shown to protect against toxin-mediated hepatocyte apoptosis through mechanisms that involve increased levels of anti-apoptotic proteins such as Bcl-xL and Bcl-2 in the liver [46, 47]. Stat3 activation mediates increased transcription of anti-apoptotic genes like Bcl- xL, Bcl-2, and Mcl-1, as well as decreased transcription of pro-apoptotic genes, including Bad, Bnip3 and Casp3 [29, 32–39]. Microarray analysis of the liver apoptosis transcriptome revealed that the expression levels of the pro- apoptotic genes Bnip3 and Casp3 were increased by 6- and 2-fold, respectively, following trauma/HS and were decreased by 5.4- and 4.1-fold respectively after IL-6 treatment. However, only the expression of Casp3 was increased by Stat3 pharmacological inhibition with T40214. None of the other apoptosis-related genes known to be modulated by Stat3 to promote apoptosis protection in other settings of liver injury were affected by IL-6 treatment or by pre-treatment with T40214, suggesting that other apoptosis-related genes are involved in the anti-apoptotic effects of IL-6-mediated Stat3 activation in our model of trauma/HS.
A complete assessment of the liver apoptosis transcriptome using oligonucleotide microarray analysis determined that trauma/HS altered the expression of 48% of apoptosis-related genes, of which 56% were anti-apoptotic and 44% were pro-apoptotic genes (Figure 5B). The overall effect of trauma/HS in the liver apoptosis transcriptome was to increase the expression of apoptosis-related genes. IL-6 administration induced the opposite effect by decreasing the expression of apoptosis-related genes whose expression was increased by trauma/HS, and by increasing the expression of apoptosis-related genes whose expression was decreased by trauma/HS (Figure 5C), suggesting a “normalizing” effect of IL-6 in the trauma/HS-induced liver apoptosis transcriptome. Stat3 inhibition, by GQ-ODN administration, reversed the IL-6 “normalizing” effect on gene expression in 46% of the apoptosis-related genes (Figure 5C). These results suggest that IL-6 administration promotes protection from trauma/HS-induced liver apoptosis by opposing the effects of trauma/HS in the liver apoptosis transcriptome in part by Stat3 activation. Anti- and pro-apoptotic subsets of genes were analyzed separately to determine the effect of trauma/HS in each subset of transcripts. Gene transcript levels in both, anti- and pro- apoptotic subsets were increased by trauma/HS. Interestingly, IL-6 normalized the expression of genes in both subsets of genes, an effect that was reversed by pre-treatment with Stat3 inhibitor (Figure 5C).
In our model of trauma/HS, pro-apoptotic genes upregulated by trauma/HS are likely to be mediators of trauma/HS-induced liver apoptosis, which is likely prevented by the IL-6 normalizing effect on their expression through Stat3 activation. Among the transcripts most up-regulated following trauma/HS and downregulated in the IL-6 treatment group were Egln3, Dusp6, Tnfrsf12, Irf1, Oldlr1, Gadd45a, and Bnip3. Egln3 is a prolyl hydroxylase which is induced in sympathetic neurons after nerve growth factor withdrawal, and induces apoptosis when overexpressed in pheochromocytoma cells [48]. Dusp6 is a cytosolic phosphatase with pancreatic tumor suppressive properties and mediates apoptosis and cell growth arrest by specifically inactivating extracellular signalregulated kinase (ERK) [49]. Tnfrs12 is known to sensitize melanoma cells to chemotherapy-induced apoptosis [50]. Irf1 is activated by serum from human patients with sepsis, and mediates apoptosis in fetal myocytes [51]. Oldlr1 is expressed in highly vascularized organs such as the lung and placenta, and in endothelial cells, smooth muscle cells, cardiomyocytes and activated macrophages. Oldlr1 is a type II glycoprotein and acts as a receptor for oxidized low-density lipoprotein (ox-LDL). Interaction with ox- LDL induces ROS, reduces NO and activates NFκB. It also increases expression of Bax and decreases expression of Bcl-2. Oldlr1 is known to induce apoptosis of vascular endothelial cells and vascular smooth muscle cells in a model of cardiac ischemia reperfusion [52]. Gadd45g belongs to a family of proteins involved in DNA damage response and cell growth arrest. It is ubiquitously expressed in all normal adult and fetal tissues. Gadd45g activates MTK1 kinase activity in response to environmental stresses leading to apoptosis through the p38/c-Jun kinase pathway. Downregulation of Gadd45g prevents apoptosis of cancer cells [53].
The protective effects of IL-6 in trauma/HS-induced liver injury were mediated in large measure by Stat3 through its ability to normalize the apoptosis transcriptome. Our findings provide evidence that support the use of IL-6 as a potential therapeutic agent to protect against liver injury and dysfunction by blocking apoptosis early after reperfusion. Such an intervention has the potential to prevent multiple organ failure and improve survival in the setting of severe hemorrhagic shock.
Acknowledgments
This work was supported, in part, by grant HL07619 (DJT) and T32-HL66991 (AM) from the National Heart, Lung and Blood Institute of the National Institutes of Health and H48839 (AM) from the American Lung Association.
References
- 1.Minino AM AR, Fingerhut LA, Boudreault MA, Warner M. Deaths: Injuries 2002. National Vital Statistics Reports. 2006;Vol. 54:1–125. [PubMed] [Google Scholar]
- 2.Moore FA, Sauaia A, Moore EE, Haenel JB, Burch JM, Lezotte DC. Postinjury multiple organ failure: a bimodal phenomenon. J Trauma. 1996;40:501–10. doi: 10.1097/00005373-199604000-00001. discussion 510–512. [DOI] [PubMed] [Google Scholar]
- 3.Ciesla DJ ME, Johnson JL, Burch JM, Cothren CC, Sauaia A. The role of the lung in postinjury multiple organ failure. Surgery. 2005;138:749–758. doi: 10.1016/j.surg.2005.07.020. [DOI] [PubMed] [Google Scholar]
- 4.Jarrar D, Wang P, Cioffi WG, Bland KI, Chaudry IH. Critical role of oxygen radicals in the initiation of hepatic depression after trauma hemorrhage. J Trauma. 2000;49:879–885. doi: 10.1097/00005373-200011000-00015. [DOI] [PubMed] [Google Scholar]
- 5.Heckbert SR, Vedder NB, Hoffman W, et al. Outcome after hemorrhagic shock in trauma patients. J Trauma. 1998;45:545–549. doi: 10.1097/00005373-199809000-00022. [DOI] [PubMed] [Google Scholar]
- 6.Arikan AA YB, Mastrangelo MA, Tweardy DJ. Interleukin-6 treatment reverses apoptosis and blunts susceptibility to intraperitoneal bacterial challenge following hemorrhagic shock. Crit Care Med. 2006;34:771–777. doi: 10.1097/01.ccm.0000201901.30292.c2. [DOI] [PubMed] [Google Scholar]
- 7.Meng ZH, Dyer K, Billiar TR, Tweardy DJ. Essential role for IL-6 in postresuscitation inflammation in hemorrhagic shock. Am J Physiol Cell Physiol. 2001;280:C343–351. doi: 10.1152/ajpcell.2001.280.2.C343. [DOI] [PubMed] [Google Scholar]
- 8.Hierholzer C, Harbrecht B, Menezes JM, et al. Essential role of induced nitric oxide in the initiation of the inflammatory response after hemorrhagic shock. J Exp Med. 1998;187:917–928. doi: 10.1084/jem.187.6.917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Maritano D, Sugrue ML, Tininini S, et al. The STAT3 isoforms alpha and beta have unique and specific functions. Nat Immunol. 2004;5:401–409. doi: 10.1038/ni1052. [DOI] [PubMed] [Google Scholar]
- 10.Hierholzer C HB, Menezes JM, Kane J, Macmicking J, Nathan CF, Peitzman AB, Billiar TR, Tweardy DJ. Essential role of induced nitric oxide in the initiation of the inflammatory response after hemorrhagic shock. J Exp Med. 1998;187:917–928. doi: 10.1084/jem.187.6.917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ono M YB, Hardison EG, Mastrangelo MA, Tweardy DJ. Increased susceptibility to liver injury after hemorrhagic shock in rats chronically fed ethanol: role of nuclear factor kappa-B, interlekin-6, and granulocyte colony-stimulating factor. Shock. 2004;21:519–525. doi: 10.1097/01.shk.0000126905.75237.07. [DOI] [PubMed] [Google Scholar]
- 12.Ono M, Yu B, Hardison EG, Mastrangelo MA, Tweardy DJ. Increased susceptibility to liver injury after hemorrhagic shock in rats chronically fed ethanol: role of nuclear factor-kappa B, interleukin-6, and granulocyte colony-stimulating factor. Shock. 2004;21:519–525. doi: 10.1097/01.shk.0000126905.75237.07. [DOI] [PubMed] [Google Scholar]
- 13.Jing N, Li Y, Xiong W, Sha W, Jing L, Tweardy DJ. G-quartet oligonucleotides: a new class of signal transducer and activator of transcription 3 inhibitors that suppresses growth of prostate and breast tumors through induction of apoptosis. Cancer Res. 2004;64:6603–6609. doi: 10.1158/0008-5472.CAN-03-4041. [DOI] [PubMed] [Google Scholar]
- 14.Jing N, Tweardy DJ. Targeting Stat3 in cancer therapy. Anticancer Drugs. 2005;16:601–607. doi: 10.1097/00001813-200507000-00002. [DOI] [PubMed] [Google Scholar]
- 15.SE SPaA. Mitochondria-associated apoptotic signalling in denervated rat skeletal muscle. J Physiol. 2005;565:309–323. doi: 10.1113/jphysiol.2004.081083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Wu JY, Feng L, Park HT, et al. The neuronal repellent Slit inhibits leukocyte chemotaxis induced by chemotactic factors. Nature. 2001;410:948–952. doi: 10.1038/35073616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Wang W, Goswami S, Lapidus K, et al. Identification and testing of a gene expression signature of invasive carcinoma cells within primary mammary tumors. Cancer Res. 2004;64:8585–8594. doi: 10.1158/0008-5472.CAN-04-1136. [DOI] [PubMed] [Google Scholar]
- 18.Benjamini Y, Drai D, Elmer G, Kafkafi N, Golani I. Controlling the false discovery rate in behavior genetics research. Behav Brain Res. 2001;125:279–284. doi: 10.1016/s0166-4328(01)00297-2. [DOI] [PubMed] [Google Scholar]
- 19.Ishihara K, Hirano T. Molecular basis of the cell specificity of cytokine action. Biochim Biophys Acta. 2002;1592:281–296. doi: 10.1016/s0167-4889(02)00321-x. [DOI] [PubMed] [Google Scholar]
- 20.Shirogane T, Fukada T, Muller JM, Shima DT, Hibi M, Hirano T. Synergistic roles for Pim-1 and c-Myc in STAT3-mediated cell cycle progression and antiapoptosis. Immunity. 1999;11:709–719. doi: 10.1016/s1074-7613(00)80145-4. [DOI] [PubMed] [Google Scholar]
- 21.Brazil DP, Park J, Hemmings BA. PKB binding proteins. Getting in on the Akt. Cell. 2002;111:293–303. doi: 10.1016/s0092-8674(02)01083-8. [DOI] [PubMed] [Google Scholar]
- 22.Brunet A, Bonni A, Zigmond MJ, et al. Akt promotes cell survival by phosphorylating and inhibiting a Forkhead transcription factor. Cell. 1999;96:857–868. doi: 10.1016/s0092-8674(00)80595-4. [DOI] [PubMed] [Google Scholar]
- 23.Kops GJ, de Ruiter ND, De Vries-Smits AM, Powell DR, Bos JL, Burgering BM. Direct control of the Forkhead transcription factor AFX by protein kinase B. Nature. 1999;398:630–634. doi: 10.1038/19328. [DOI] [PubMed] [Google Scholar]
- 24.Wang XJ, Liefer KM, Tsai S, O'Malley BW, Roop DR. Development of geneswitch transgenic mice that inducibly express transforming growth factor beta1 in the epidermis. Proc Natl Acad Sci USA. 1999;96:8483–8488. doi: 10.1073/pnas.96.15.8483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Nicholson KM, Anderson NG. The protein kinase B/Akt signalling pathway in human malignancy. Cell Signal. 2002;14:381–395. doi: 10.1016/s0898-6568(01)00271-6. [DOI] [PubMed] [Google Scholar]
- 26.Jing N, Li Y, Xu X, et al. Targeting Stat3 with G-quartet oligodeoxynucleotides in human cancer cells. DNA Cell Biol. 2003;22:685–696. doi: 10.1089/104454903770946665. [DOI] [PubMed] [Google Scholar]
- 27.Jing N, Sha W, Li Y, Xiong W, Tweardy DJ. Rational drug design of G-quartet DNA as anti-cancer agents. Curr Pharm Des. 2005;11:2841–2854. doi: 10.2174/1381612054546761. [DOI] [PubMed] [Google Scholar]
- 28.Huang Y, Qiu J, Dong S, et al. Stat3 isoforms, alpha and beta, demonstrate distinct intracellular dynamics with prolonged nuclear retention of Stat3beta mapping to its unique C-terminal end. J Biol Chem. 2007;282:34958–67. doi: 10.1074/jbc.M704548200. [DOI] [PubMed] [Google Scholar]
- 29.Bromberg JF WM, Devgan G, Zhao Y, Pestell RG, Albanese C, Darnell JE. Stat3 as an oncogene. Cell. 1999;98:295–303. doi: 10.1016/s0092-8674(00)81959-5. [DOI] [PubMed] [Google Scholar]
- 30.Turkson J, Bowman T, Garcia R, Caldenhoven R, Degroot RP, Jove R. Stat3 activation by Src induces specific gene regulation and is required for cell transformation. Molecular and Cellular Biology. 1998 May;18:2545–2552. doi: 10.1128/mcb.18.5.2545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Takeda K, Kishimoto T, Akira S. Signal transducer and activator of transcription protein (STAT): its relation to Th1/Th2-mediated diseases [editorial] Nutrition. 1997;13:987–8. doi: 10.1016/s0899-9007(97)00343-2. [DOI] [PubMed] [Google Scholar]
- 32.Chen CL HF, Lin J. Systemic evaluation of total Stat3 and Stat3 tyrosine phosphorylation in normal human tissues. Exp Mol Pathol. 2006;80:295–305. doi: 10.1016/j.yexmp.2005.11.003. [DOI] [PubMed] [Google Scholar]
- 33.Levy DaLC. What does Stat3 do? J Clin Invest. 2002;109:1143–1148. doi: 10.1172/JCI15650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Real P SA, Dejuan A, Segovia J, Lopez-Vega J, Fernandez-Luna J. Resistance to chemotherapy via Stat3-dependent overexpression of Bcl-2 in metastatic breast cancer cells. Oncogene. 2002;21:7611–7618. doi: 10.1038/sj.onc.1206004. [DOI] [PubMed] [Google Scholar]
- 35.Leslie K LC, Devgan G, Azare J, Berishaj M, Gerald W, Kim YB, Paz K, et al. Cyclin D1 is transcriptionally regulated by and required for transformation by activated signal transducer and activator of transcription 3. Cancer Res. 2006;66:2544–2552. doi: 10.1158/0008-5472.CAN-05-2203. [DOI] [PubMed] [Google Scholar]
- 36.Catlett-Falcone R, Landowski TH, Oshiro MM, et al. Constitutive activation of Stat3 signaling confers resistance to apoptosis in human U266 myeloma cells. Immunity. 1999;10:105–115. doi: 10.1016/s1074-7613(00)80011-4. [DOI] [PubMed] [Google Scholar]
- 37.Gritsko T WA, Tucson J, Kaneko S, et al. Persistent activation of Stat3 induces survivin gene expression and confers resistance to apoptosis in human breast cancer cells. Clin Cancer Res. 2006;12:11–19. doi: 10.1158/1078-0432.CCR-04-1752. [DOI] [PubMed] [Google Scholar]
- 38.Fukada T OT, Yoshida Y, Shirogane T, Nishida K, Nakajima K, Hibi M, Hirano T. Stat3 orchestrates contradictory signals in cytokine induced G1 to S cell cycle transition. The EMBO J. 1996;17:6670–6677. doi: 10.1093/emboj/17.22.6670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Taub R. Hepatoprotection via the IL-6/Stat3 pathway. J Clin Invest. 2003;112:978–980. doi: 10.1172/JCI19974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Kobelt F, Schreck U, Henrich HA. Involvement of liver in the decompensation of hemorrhagic shock. Shock. 1994;2:281–288. doi: 10.1097/00024382-199410000-00009. [DOI] [PubMed] [Google Scholar]
- 41.Sundar SV, Li YY, Rollwagen FM, Maheshwari RK. Hemorrhagic shock induces differential gene expression and apoptosis in mouse liver. Biochem Biophys Res Commun. 2005;332:688–96. doi: 10.1016/j.bbrc.2005.05.008. [DOI] [PubMed] [Google Scholar]
- 42.Ayuste EC, Chen H, Koustova E, et al. Hepatic and pulmonary apoptosis after hemorrhagic shock in swine can be reduced through modifications of conventional Ringer's solution. J Trauma. 2006;60:52–63. doi: 10.1097/01.ta.0000200156.05397.0b. [DOI] [PubMed] [Google Scholar]
- 43.Rudiger HA GR, Claiven PA. Liver ischemia: apoptosis as a central mechanism of injury. J Invest Surg. 2003;16:149–159. [PubMed] [Google Scholar]
- 44.Kovalovich K, DeAngelis RA, Li W, Furth EE, Ciliberto G, Taub R. Increased toxin-induced liver injury and fibrosis in interleukin-6-deficient mice. Hepatology. 2000;31:149–159. doi: 10.1002/hep.510310123. [DOI] [PubMed] [Google Scholar]
- 45.Kovalovich K, Li W, DeAngelis R, Greenbaum LE, Ciliberto G, Taub R. Interleukin-6 protects against Fas-mediated death by establishing a critical level of antiapoptotic hepatic proteins FLIP, Bcl-2, and Bcl-xL. J Biol Chem. 2001;276:26605–26613. doi: 10.1074/jbc.M100740200. [DOI] [PubMed] [Google Scholar]
- 46.Hong F, Jaruga B, Kim WH, et al. Opposing roles of STAT1 and STAT3 in T cellmediated hepatitis: regulation by SOCS. J Clin Invest. 2002;110:1503–1513. doi: 10.1172/JCI15841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Hong F, Kim WH, Tian Z, et al. Elevated interleukin-6 during ethanol consumption acts as a potential endogenous protective cytokine against ethanol-induced apoptosis in the liver: involvement of induction of Bcl-2 and Bcl-x(L) proteins. Oncogene. 2002;21:32–43. doi: 10.1038/sj.onc.1205016. [DOI] [PubMed] [Google Scholar]
- 48.Lee CK, Raz R, Gimeno R, et al. STAT3 is a negative regulator of granulopoiesis but is not required for G-CSF-dependent differentiation. Immunity. 2002;17:63–72. doi: 10.1016/s1074-7613(02)00336-9. [DOI] [PubMed] [Google Scholar]
- 49.Furukawa Y, Kawasoe T, Daigo Y, et al. Isolation of a novel human gene, ARHGAP9, encoding a rho-GTPase activating protein. Biochem Biophys Res Commun. 2001;284:643–649. doi: 10.1006/bbrc.2001.5022. [DOI] [PubMed] [Google Scholar]
- 50.Kokkinakis DM. Methionine-stress: a pleiotropic approach in enhancing the efficacy of chemotherapy. Cancer Lett. 2006;233:195–207. doi: 10.1016/j.canlet.2005.02.034. [DOI] [PubMed] [Google Scholar]
- 51.Kumar A, Kumar A, Michael P, et al. Human serum from patients with septic shock activates transcription factors STAT1, IRF1, and NF-kappaB and induces apoptosis in human cardiac myocytes. J Biol Chem. 2005;280:42619–42626. doi: 10.1074/jbc.M508416200. [DOI] [PubMed] [Google Scholar]
- 52.Kataoka K, Hasegawa K, Sawamura T, et al. LOX-1 pathway affects the extent of myocardial ischemia-reperfusion injury. Biochem Biophys Res Commun. 2003;300:656–660. doi: 10.1016/s0006-291x(02)02905-4. [DOI] [PubMed] [Google Scholar]
- 53.Zerbini LF, Libermann TA. Life and death in cancer. GADD45 alpha and gamma are critical regulators of NF-kappaB mediated escape from programmed cell death. CellCycle. 2005;4:18–20. doi: 10.4161/cc.4.1.1363. [DOI] [PubMed] [Google Scholar]
- 54.Chakraborty A, Tweardy DJ. Stat3 and G-CS induced myeloid differentiation. Leukemia and Lymphoma. 1998;30:433–442. doi: 10.3109/10428199809057555. [DOI] [PubMed] [Google Scholar]
- 55.Chakraborty A, White SM, Schaefer TS, Ball ED, Dyer KF, Tweardy DJ. Granulocyte colony-stimulating factor activation of Stat3 alpha and Stat3 beta in immature normal and leukemic human myeloid cells. Blood. 1996;88:2442–2449. [PubMed] [Google Scholar]