Abstract
Mutations in the aristaless-related homeobox (ARX) gene are associated with multiple neurologic disorders in humans. Studies in mice indicate Arx plays a role in neuronal progenitor proliferation and development of the cerebral cortex, thalamus, hippocampus, striatum, and olfactory bulbs. Specific defects associated with Arx loss of function include abnormal interneuron migration and subtype differentiation. How disruptions in ARX result in human disease and how loss of Arx in mice results in these phenotypes remains poorly understood. To gain insight into the biological functions of Arx, we performed a genome-wide expression screen to identify transcriptional changes within the subpallium in the absence of Arx. We have identified 84 genes whose expression was dysregulated in the absence of Arx. This population was enriched in genes involved in cell migration, axonal guidance, neurogenesis, and regulation of transcription and includes genes implicated in autism, epilepsy, and mental retardation; all features recognized in patients with ARX mutations. Additionally, we found Arx directly repressed three of the identified transcription factors: Lmo1, Ebf3 and Shox2. To further understand how the identified genes are involved in neural development, we used gene set enrichment algorithms to compare the Arx gene regulatory network (GRN) to the Dlx1/2 GRN and interneuron transcriptome. These analyses identified a subset of genes in the Arx GRN that are shared with that of the Dlx1/2 GRN and that are enriched in the interneuron transcriptome. These data indicate Arx plays multiple roles in forebrain development, both dependent and independent of Dlx1/2, and thus provides further insights into the understanding of the mechanisms underlying the pathology of mental retardation and epilepsy phenotypes resulting from ARX mutations.
INTRODUCTION
Mutations in aristaless-related homeobox (ARX) result in at least nine distinct neurological disorders. They can be categorized as (i) those diseases that include structural malformations: hydrancephaly with abnormal/ambiguous genitalia (HYD-AG; OMIM #300215) (1), X-linked lissencephaly with ambiguous genitalia (XLAG; OMIM #300215) (1–5), and agenesis of the corpus callosum with abnormal genitalia (ACC-AG; OMIM #300004; also known as Proud’s syndrome) (1), and (ii) those diseases that include no obvious structural malformations: infantile epileptic-dyskinetic encephalopathy (IEDE; OMIM #308350) (6), early infantile epileptic encephalopathy with suppression-burst pattern (EIEE; also known as Ohtahara’s syndrome; OMIM #308350) (7), West syndrome (WS; also known as X-linked infantile spasm syndrome; OMIM #308350) (8–10), X-linked myoclonic epilepsy with mental retardation and spasticity (XMEMRS; OMIM #300432) (8,11), Partington syndrome (PRTS; OMIM #309510) (8,12) and X-linked mental retardation with or without seizures (NS-XLMR; OMIM #300419) (8,13–18).
The ARX protein contains a paired-related homeodomain, a highly conserved octapeptide domain, a C-terminal aristaless domain and four polyalanine (polyA) tracts (19). In general, the malformation phenotypes are associated with protein truncation mutations and missense mutations in the homeobox domain, whereas the non-malformation phenotypes are associated with missense mutations outside of the homeobox or an expansion in the first or second polyA tracts (1,20).
Mice deficient for Arx exhibit a complex, pleiotrophic phenotype, generally reflective of the expression domains of Arx. Within the forebrain Arx is expressed in the developing ganglionic eminences (GEs), the cerebral cortical ventricular zone (VZ), the striatonigral neurons of the striatum, the hippocampus, the ventral thalamus and the hypothalamus (2,13,21–24). In the cerebral cortex, hypothalamus and olfactory bulbs, Arx is primarily localized in GABAergic interneurons where its expression is maintained throughout adulthood (2,21,23,24). During early development (E9.0–E14.5) Arx is expressed in Emx2+ cortical VZ cells, apparently under the transcriptional control of Pax6 (25); likely in progenitor cells that will give rise to the excitatory neurons of the cortex (21,23). Curiously, colabeling of Arx has not been observed in mature excitatory neurons, potentially due to rapid downregulation of expression upon exit from the cell cycle (21,23). Outside the forebrain, it is expressed in the floor plate of the developing spinal cord, somites, testes, pancreas and skeletal muscle (2,8,13,23,26,27).
Arx-deficient mice exhibit decreased proliferation of cortical neuroepithelial cells, dysgenesis of the thalamus, hippocampus, striatum and olfactory bulbs, as well as, abnormal interneuron migration and subtype differentiation (2,28). How loss of Arx in mice results in these phenotypes is unknown but must reflect alterations in Arx-dependent transcriptional networks.
Given that Arx is a transcription factor, defining the transcriptional targets and gene regulatory networks (GRNs) in which Arx participates is essential to further elucidating its role in normal brain development and how disruptions in ARX lead to human disease. Using Arx deficient mice, combined with expression profiling, we have defined the GRN regulated by Arx within the subpallium, validated a subset of these target genes and used these data to gain new insights into the genetic regulation of interneuron development.
RESULTS
Generation of ARX mutant mice
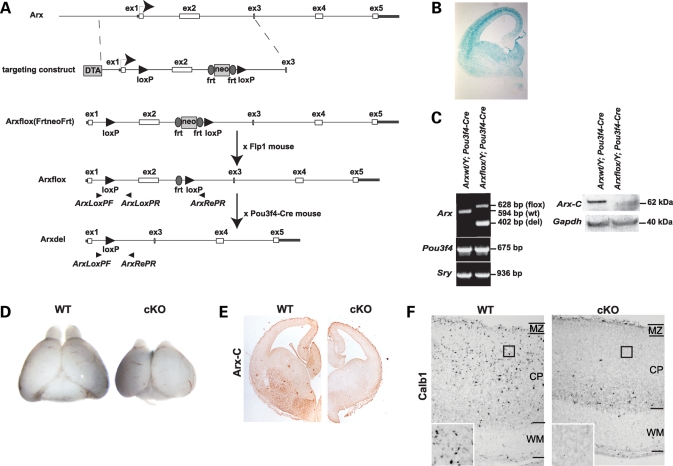
To generate mice carrying a floxed Arx allele we used the strategy summarized in Figure 1A (also see Materials and Methods). Arxflox/+ and Arxflox/Y (Arx is on the X-chromosome) mice were bred to homozygosity (Arxflox/flox female mice) and mated with Pou3f4-Cre male mice which express Cre throughout the pallium and subpallium at E14.5 (kindly provided by Dr E.B. Crenshaw, Children’s Hospital of Philadelphia) (Fig. 1B) (29,30). Western blots using an antibody specific to the C-terminus of Arx confirmed the absence of Arx protein in the GEs dissected from conditional mutant mice (Fig. 1C). At E18.5, the whole brain of conditional mutant mice was smaller than that of the wild-type and the olfactory bulbs were nearly absent, similar to that observed by others in Arx mutant mouse (Fig. 1D) (2,24). Immunohistochemistry using the C-terminal α-Arx antibody detected a near complete loss of Arx expression in the forebrain of Arx−/y mice (Fig. 1E). Furthermore, immunohistochemistry using an antibody directed to calbindin (Calb1) showed a loss of the normal distribution of Calb1+ interneurons throughout the cerebral cortex, being restricted to the subcortical and SVZ regions in Arx−/y;Pou3f4-Cre mice (Fig. 1F), again similar to that previously reported in Arx mutant mice (2,28).
Figure 1.
(A) Genomic organization of mouse Arx and details of the targeting construct. Arrowheads denote the location of primers used for genotyping. (B) Brain section from a E14.5 Pou3f4-Cre×B6.129S4-Gt(ROSA)26Sortm1Sor/J stained with X-gal reveals the ubiquitous expression of the Pou3f4-Cre transgene throughout the telencephalon. (C) PCR of genomic DNA from the hindbrains of E14.5 Arx+/Y; Pou3f4-Cre+ and Arx−/Y; Pou3f4-Cre+ mice with primer sets specific to Arx, Pou3f4 and Sry (left). Western blotting of whole cell lysate from E14.5 subpallia of Arx+/Y; Pou3f4-Cre+ and Arx−/Y; Pou3f4-Cre+ mice, using an antibody specific to the C-terminus of Arx (Arx-C), reveals the absence of Arx in conditionally mutant Arx mice (right). (D) Gross morphology of prosencephalon from E18.5 Arx+/Y; Pou3f4-Cre+ (left) and Arx−/Y; Pou3f4-Cre+ (right) mice. Total brain size appeared smaller and olfactory bulbs were nearly absent in the conditional mutant (KO). (E) Lack of Arx protein within the subpallium of conditional knockouts. Telencephalon sections from E14.5 were used for immunohistochemistry using the same anti-Arx antibody used for western blots. (F) Immunohistochemistry with an antibody specific for Calb1 on sagittal sections of E18.5 brain revealed the presence of Calb1+ interneurons throughout the cortical plate of Arx+/Y; Pou3f4-Cre+ mice (left), while Calb1+ interneurons were confined to the ventricular and subventricular zones of Arx−/Y; Pou3f4-Cre+ mice. MZ, marginal zone; CP, cortical plate; WM, white matter. Inset represents 4× magnification at the location represented by the black square.
Identification of putative Arx target genes
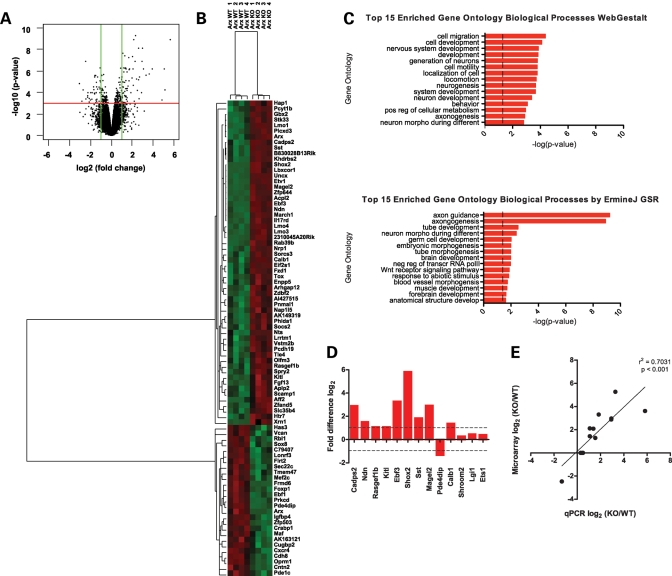
The strong subpallial expression of Arx, along with the well-defined phenotype in GE-derived cells in Arx mutant mice, prompted us to focus on the role of Arx in subpallial development. Thus, all subsequent studies were based on a genome-wide microarray analysis of the subpallia microdissected from E14.5 Arx−/y; Pou3f4-Cre+ (mutant embryos) and Arx+/y; Pou3f4-Cre+ (control embryos). Interrogating 39 000 transcripts and variants from 34 000 murine genes (Affymetrix MOE430-2) resulted in the identification of 84 genes with at least a 2-fold change in expression within the subpallium and with a false discovery rate (FDR) of <5% (Fig. 2A). Fifty-seven genes were increased and 27 genes decreased in conditional Arx-null subpallium when compared with control subpallium (Supplementary Material, Table S1). Hierarchical centroid-linkage and unsupervised clustering grouped differentially expressed genes according to relative variation in gene expression patterns (Fig. 2B).
Figure 2.
(A) Volcano plot of log2 fold-change versus −log10 FDR-corrected P-value for all probes deemed present in at least four of the eight microarrays analyzed. Green lines denote the selected 2-fold change cutoff, while the red line denotes the 0.05 selected FDR-corrected P-value cutoff. Points in the upper left and upper right quadrants represent probes that met the requirements to be called differentially expressed. (B) Hierarchical clustering of the 84 genes deemed as differentially expressed was performed using a correlation matrix and centroid linkage. Red represents increased expression, while green represents decreased expression. (C) Bar chart representing the −log(p-values) for the top 15 overrepresented Gene Ontology (GO) Biological Processes as determined by WebGestalt (top) and ErmineJ Gene Set Resampling (bottom) algorithms. (D) qPCR on E14.5 subpallia from control and conditionally mutant mice for genes deemed differentially expressed (Cadps2, Ndn, Rasgef1b, Kitl, Ebf3, Shox2, Sst, Magel2, Pde4dip and Calb1) and three control genes that were not differentially expressed (Shroom2, Lgi1 and Ets1). Data were compared with Actb expression using the ΔΔCT method and presented as the log2 fold ratio difference between mutant and control mice. (E) Microarray expression data (log2) and qPCR data (log2) were found to be positively correlated (P < 0.001, r2 = 0.7031).
The microarray data set was validated by performing quantitative polymerase chain reaction (qPCR) on three independent pairs of subpallia from E14.5 mutant (Arx−/Y; Pou3f4-Cre+) and control (Arx+/Y; Pou3f4Cre+) embryos. A subset of 10 genes identified as differentially expressed, along with three genes that did not change significantly, were selected for testing. All 10 genes designated as differentially expressed by microarray analysis changed at least 2-fold by qPCR, whereas the three control genes that failed to change on the microarray also showed no difference in expression by qPCR (Fig. 2D). Log2 expression observed by microarray analysis was found to positively and significantly correlate to the log2 expression observed by qPCR (Pearson r2 = 0.7031, P < 0.001) (Fig. 2E).
Functional classification of Arx target genes
To gain insight into the functions of identified Arx target genes, we used two methods to identify Gene Ontology (GO) biological processes (31) represented in the population of differentially expressed genes. WebGestalt (32), considering only the differentially expressed genes relative to all expressed genes, and ErmineJ Gene Set Resampling (GSR) (33), which is a non-threshold based method that looks for enrichment across the entire ranked expression set, were used to gain insight into the functions of identified Arx target genes. These analyses identified 38 enriched biological processes (FDR-corrected P-value <0.01) in the differentially expressed genes relative to all expressed genes (Fig. 2C). The identified genes included those implicated in cell/neuronal migration (P = 4.3 × 10−5, P = 4.3 × 10−2), axonogenesis (P = 1.3 × 10−3, 1.2 × 10−9), neuron morphogenesis during differentiation (1.6 × 10−3, 4.0 × 10−3) and regulation of transcription (P = 4.8 × 10−3; negative 1.1 × 10−2 and positive 3.6 × 10−2), with P-values reported for WebGestalt and GSR, respectively (Fig. 2C and Supplementary Material, Tables S2 and S3).
Genes differentially expressed in Arx mutants show spatial expression changes in the subpallium
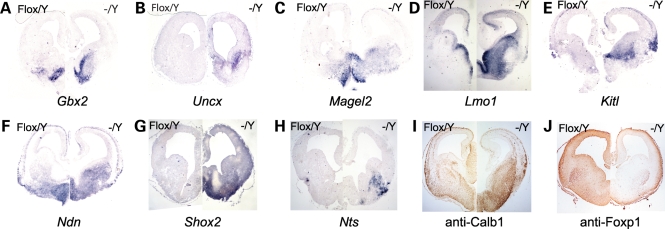
To further validate the microarray results, we performed in situ hybridization or immunohistochemistry for 10 of the differentially expressed genes (Gbx2, Uncx, Magel2, Lmo1, Kitl, Ndn, Shox2, Nts, Calb1 and Foxp1) on E14.5 brains from mutant and control embryos. We observed Gbx2 expression to be restricted to the basal forebrain in sections from the telencephalon of control mice. In sections from mutant mice, we found a dorsal expansion of Gbx2 from the basal forebrain into the medial GE (Fig. 3A). No expression of Uncx was observed in the telencephalon of control mice; however, in the absence of Arx, ectopic expression of Uncx was identified in the anterior portion of the lateral GE (Fig. 3B). In control sections, Magel2 expression was restricted to the septum and anterior hypothalamus, while expression expanded laterally to fill most of the medial ganglion eminence and striatum in mutant brains (Fig. 3C). Magel2 expression was additionally observed in the intermediate zone of the pallium in the mutant telencephalon. Lmo1 expression within the control telencephalon primarily localized to the VZ of the GEs. In contrast, Lmo1 expression was greatly expanded ventrolaterally to encompass nearly the entire volume of the GEs in mutant mouse brains (Fig. 3D). Lmo1 expression was additionally found in the medial cortex of both control and mutant mice. In control mice, Kitl was strongly expressed within the septum, the boundary between the lateral GE and striatum, in the lateral cortex at the entry point of interneurons into the pallium and within the pallial intermediate zone (Fig. 3E). The mutant telencephalon showed similar Kitl expression; however, within the subpallium, Kitl expression expanded medially through the medial GE. In addition, we noted a stronger expression of Kitl in the lateral cerebral cortex of mutant mice when compared with control mice. Ndn was expressed within the septum, basal forebrain, striatum and the ventricular and intermediate zones of both control and mutant telencephalon (Fig. 3F). However, within the mutant subpallium, the Ndn domain expanded into the medial and lateral GEs. Shox2 exhibited no expression within the forebrain of control mice (Fig. 3G), although expression was evident in the dorsal thalamus (data not shown). Within the mutant forebrain, a wide array of Shox2 expression was detected in the septum and basal forebrain. Nts was observed in a discrete region in the ventrolateral cortex in control mice; however, within mutant mice Nts expression expanded medially into the LGE (Fig. 3H). Calb1 was observed throughout the subpallium and in streams of presumptive interneurons within the cortex of control and mutant mice (Fig. 3I). We observed a significantly stronger expression of Calb1 in mutant subpallium when compared with control subpallium. Finally, Foxp1 was expressed in the striatum, basal forebrain, cortical plate and cortical VZ of both control and mutant telencephalon (Fig. 3J). However, within the mutant striatum and basal forebrain we found a decrease in Foxp1 expression.
Figure 3.
In situ hybridization on E14.5 mouse brain sections using probes directed to (A) Gbx2, (B) Uncx, (C) Magel2, (D) Lmo1, (E) Kitl, (F) Ndn, (G) Shox2 and (H) Nts. Immunohistochemistry on E14.5 mouse brain sections using (I) anti-Calb1 and (J) anti-Foxp1 antibodies. For each panel, Arxflox/Y; Pou3f4-Cre− is presented on the left, while Arx−/Y; Pou3f4-Cre+ is presented on the right.
Identification of Arx target genes involved in interneuronal function
Expression of the homeobox transcription factors Dlx1 and Dlx2 in GE cells is necessary for tangential migration of interneurons from the basal forebrain to the neocortex (34). Dlx1 and Dlx2 are thought to promote interneuron differentiation by repressing oligodendroglial cell fate (35) and promote interneuron migration by repressing neurite outgrowth (36). Furthermore, early expression of Dlx genes matches the expression of interneuron obligate genes, such as Gad1 and Gad2, and is sufficient (although not necessary) for their expression (37–40). These data suggest that activation of the Dlx transcriptional pathway is shared by most, if not all, telencephalic interneurons. Given that Dlx1/2 expression appears to overlap that of Arx and be sufficient, and within some brain regions necessary, for Arx expression, it has been suggested that Arx itself may be a transcriptional target of Dlx1/2 (19,36,41).
As the loss of Arx results in a pleiotrophic phenotype (2) and the genes identified in our screen could underlie any of the various observed phenotypes, we sought to determine whether there exists an identifiable gene signature shared between genes significantly altered in Arx conditional mutant and Dlx mutant mice. This signature would represent those genes downstream of Arx that likely play a role in interneuron differentiation and/or migration. We reasoned that genes defining such a signature would (i) be enriched within the GE-derived interneuron population and GABAergic projection neurons and, given Arx’s putative position downstream of Dlx1/2 (41), (ii) show expression changes in the absence of Dlx1/2 similar to those we observed in the absence of Arx.
To identify this gene signature, we took advantage of the availability of two previously published data sets. The first data set was derived from FAC-sorting GFP-positive and -negative cells from the subpallium and pallium of E14.5 Dlx5/6Cre-IRES-eGFP embryos, which were then used in microarray analyses to compare the expression profiles between the different populations of sorted cells (42). In the second data set, Rubenstein and coworkers (36) compared the expression profiles between E14.5 subpallium-derived cells from either heterozygous or homozygous Dlx1/2 double mutant mice. This data set was similar, but not identical, to that previously published. We used gene set enrichment analysis (GSEA) (43), a computational method for assessing whether a predefined gene set is statistically enriched in one biological state when compared with another (43), to determine (i) whether and which Arx targets genes have differential expression in the subpallium in the absence of Dlx1/2, (ii) whether and which Arx target genes exhibit enriched expression in interneurons, and (iii) of those Arx target genes expressed in interneurons, which gene sets overlap with those expressed in cells migrating into cortical regions and which overlap with those expressed in cells migrating into subcortical regions.
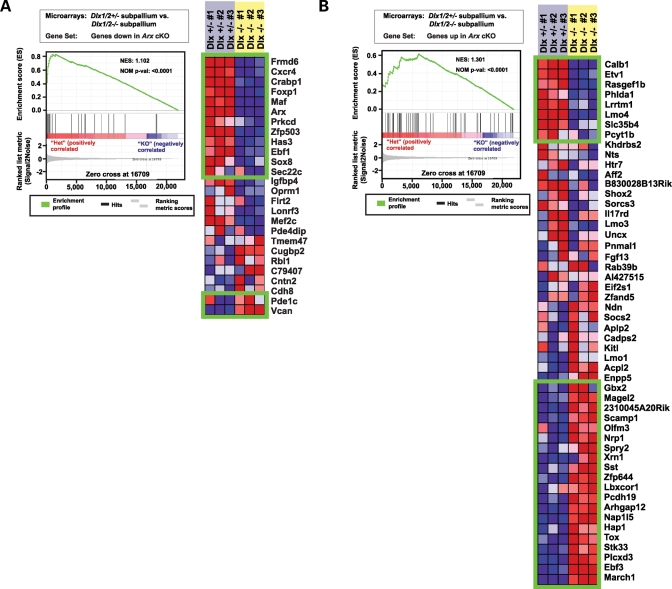
In the first analysis, we sought to determine whether the gene sets that corresponded to genes that increased and decreased in the absence of Arx were enriched in microarray experiments comparing the gene expression changes in subpallia from Dlx1/2 heterozygous and homozygous mice. We observed that the gene sets both downregulated (NES = 1.102; P < 0.0001; FDR = 0.105) (Fig. 4A) and upregulated (NES = 1.301; P < 0.0001; FDR = 0.299) (Fig. 4B) in the absence of Arx are enriched in the population of genes differentially expressed in the subpallium in the absence of Dlx1/2. Of the genes that decreased in the subpallium in the absence of Arx, 12/26 also decreased in the subpallium of Dlx1/2 mutant mice, while 2/26 increased in the subpallium of Dlx1/2 mutant mice (Pde1c and Vcan). The remaining 12/26 genes decreased in the subpallium of the Arx mutant but not the Dlx1/2 mutant. Of those genes that increased in the subpallium in the absence of Arx, 20/52 also increased in the subpallium of Dlx1/2 mutant mice. Interestingly, while Dlx1/2 appeared to enhance expression of Calb1, Etv1, Rasgef1b, Phlda1, Lrrtm1, Lmo4, Slc35b4 and Pcyt1b, Arx appeared to repress expression of these genes. Alternatively, it is possible that we instead observed an accumulation of a particular migratory cell subpopulation(s) that failed to exit the subpallium in the absence of Arx. The remaining 24/52 genes increased in the subpallium of the Arx mutant mouse but not the Dlx1/2 mutant mouse. Collectively, these data indicate Arx is not simply a downstream target for Dlx genes in subpallial cell development, but also regulates a GRN that is independent of its role downstream of Dlx, i.e. gene expression signatures were identified in the subpallium of Arx mutant mice that were shared with those of Dlx1/2 mutant mice; however, there also existed a gene expression signature specific to the Arx mutant mice.
Figure 4.
GSEA revealed a shared GRN within the subpallium of Arx and Dlx1/2 mutants. (A) Graph denotes the Absolute Enrichment (NES = 1.102; P < 0.0001) of the gene set containing those genes whose expression decreased in the subpallium in the absence of Arx within the Rubenstein et al. data set representing those genes whose expression changed in the absence of Dlx1/2 (left). Green boxes contain those genes deemed as statistically enriched (right). The genes contained within the top box are those genes whose expression decreased in the subpallium in both Arx and Dlx1/2 mutant mice. The lower box contains genes whose expression increased in the subpallium of the Arx mutant mouse, but increased in the subpallium of Dlx1/2 mutant mice. (B) Graph denotes the Absolute Enrichment (NES = 1.301, P < 0.0001) of the gene set containing those genes whose expression increased in the subpallium in the absence of Arx within the Rubenstein et al. data set (left). Green boxes contain those genes deemed as enriched (right). The genes contained within the top box are those genes whose expression increase in the subpallium of Arx mice but that decrease in the subpallium of the Dlx1/2 mutant mouse. The lower box contains genes whose expression increased in the subpallium of both Arx and Dlx1/2 mutant mice.
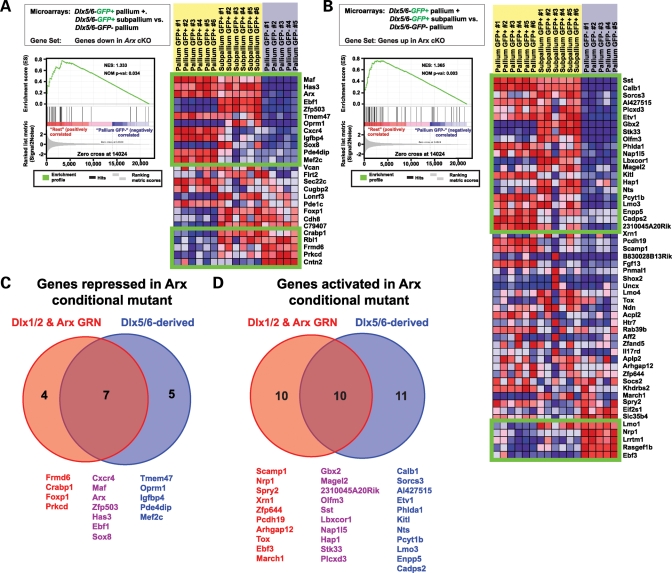
We next asked which Arx target genes were enriched in Dlx5/6-derived versus non-Dlx5/6-derived cell populations. Using GSEA we tested whether gene sets corresponding to genes that increased or decreased in the absence of Arx were enriched within microarray experiments comparing gene expression profiles from Dlx5/6-GFP-negative cells of the pallium (primarily principal projection neurons and neuronal progenitors of the VZ) to the combined populations of Dlx5/6-GFP-positive cells of the pallium (interneurons) and subpallium (interneurons and striatal GABAergic projection neurons). We observed that both genes whose expression decreased (NES = 1.333; P = 0.034; FDR = 0.002) and those whose expression increased (NES = 1.365; P = 0.003; FDR = 0.069) in the absence of Arx were enriched in that population of genes belonging to the Dlx5/6-GFP-positive cells of the pallium and subpallium. Among those genes whose expression increased in the absence of Arx, a set of 21/52 genes was enriched in Dlx5/6-derived cells (Fig. 5B), while 12/26 of the genes whose expression decreased in the absence of Arx were also enriched in Dlx5/6-derived cells (Fig. 5A). Five genes that increased in the subpallium in the absence of Arx were normally expressed in non-Dlx5/6-derived cells (Ebf3, Rasgef1b, Lrrtm1, Nrp1 and Lmo1) (Fig. 5B), while five genes that decreased in the subpallium in the absence of Arx were normally expressed in non-Dlx5/6-derived cells (Cntn2, Prkcd, Frmd6, Rbl1 and Crabp1) (Fig. 5A). Among those genes enriched in Dlx5/6-derived cells and that exhibited decreased expression in the subpallium of Arx mutant mice, 7/12 were also observed to decrease in the Dlx1/2 mutant subpallium (Cxcr4, Maf, Arx, Zfp503, Has3, Ebf1 and Sox8) (Fig. 5C).
Figure 5.
GSEA revealed that the gene sets corresponding to genes differentially expressed in the subpallium of Arx mutant mice are enriched in those genes differentially expressed between Dlx5/6-derived and non-Dlx5/6-derived cells. (A) Graph denotes the Absolute Enrichment (NES = 1.333, P = 0.034) of the gene set containing those genes whose expression decreased in the subpallium in the absence of Arx within a subset of the Marsh et al. data set (genes expressed in GFP+ telencephalic cells versus those expressed in GFP- cortical cells FAC-sorted from Dlx5/6Cre-IRES-eGFP mice) (left). Green boxes contain those genes deemed as enriched (right). The genes contained within the top box are those genes whose expression decreased in the subpallium Arx mutant mice and were enriched in GFP+ interneurons. The lower box contains genes whose expression decreased in the subpallium of the Arx mutant mouse and were enriched in GFP- non-interneuron cell populations. (B) Graph denotes the Absolute Enrichment (NES = 1.365, P = 0.003) of the gene set containing those genes whose expression increased in the subpallium in the absence of Arx within a subset of the Marsh et al. data set (genes expressed in GFP+ telencephalic cells versus those expressed in GFP− cortical cells FAC sorted from Dlx5/6Cre-IRES-eGFP mice) (left). Green boxes contain those genes deemed as enriched (right). The genes contained within the top box are those genes whose expression increased in the subpallium Arx mutant mice and were enriched in GFP+ interneurons. The lower box contains genes whose expression increased in the subpallium of the Arx mutant mouse and were enriched in GFP− non-interneuron cell populations. (C) Venn diagram illustrates the overlap between genes whose expression was observed to decrease in the subpallium of conditional Arx mutant and Dlx1/2 mutant mice (red) and those genes whose expression was observed to decrease in the subpallium in the absence of Arx and were enriched in Dlx5/6-derived cells (blue). (D) Venn diagram illustrates the overlap between genes whose expression was observed to increase in the subpallium of conditional Arx mutant and Dlx1/2 mutant mice (red) and those genes whose expression was observed to increase in the subpallium in the absence of Arx and were enriched in Dlx5/6-derived cells (blue).
Among those genes enriched in Dlx5/6-derived cells and that were observed to increase expression in the subpallium of Arx mutant mice, 10/21 were also observed to increase in the subpallium of Dlx1/2 mutant mice (2310045A20Rik, Gbx2, Hap1, Lbxcor1, Magel2, Nap1l5, Olfm3, Plcxd3, Sst and Stk33) (Fig. 5D). Two genes (Ebf3 and Nrp1) were enriched in non-Dlx5/6-derived cells and increased expression in the subpallium of both Dlx1/2 and Arx mutant mice. Interestingly, 5/21 genes enriched in Dlx5/6-derived cells that exhibited increased expression in the subpallium of Arx mutant mice were decreased in the subpallium of Dlx1/2 mutant mice (Calb1, Etv1, Pcyt1b and Phlda1).
We next asked whether Arx target genes were preferentially expressed in Dlx5/6-derived cells that migrated out of the subpallium or those that migrate within the subpallium. To do this, we looked for enrichment of gene sets corresponding to genes that increased or decreased in the absence of Arx within those genes differentially expressed in microarray experiments comparing Dlx5/6-GFP+ cells within the pallium relative to Dlx5/6-GFP+ cells within the subpallium. Neurons that migrate from the LGE give rise to interneurons of the olfactory bulb and the medium spiny neurons of the striatum, while neurons that migrate from the MGE become Calb1+, Sst+ and Pval+ interneurons of the neocortex, and to a lesser extent of the anterior olfactory nucleus, piriform cortex, medium spiny neurons of the striatum, multiple nuclei of the amygdala and the CA1 and caudal aspect of the CA3 of the hippocampus (44–50). Cells that arise in the interganglionic region contribute to interneurons of the preoptic and hypothalamic regions (51). CGE-derived interneurons populate the cortex (primarily those of layer 5 and those of the Calb2+ and Npy+ subclasses), multiple nuclei of the amygdala, CA1 of the hippocampus, and medium spiny neurons of the striatum (45,52).
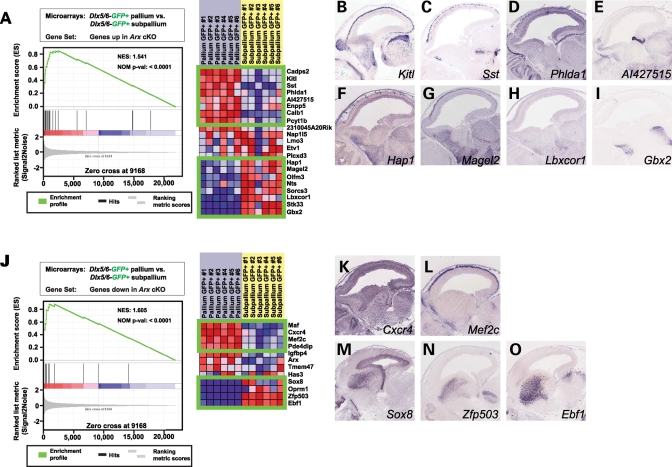
Of the genes that exhibited decreased expression in the subpallium of Arx mutant mice and enrichment in Dlx5/6-derived cells, Ebf1, Zfp503, Sox8 and Oprm1 were enriched in Dlx5/6-GFP+ cells that remain in the subpallium (interneurons and striatal GABAergic projection neurons), while Maf, Cxcr4, Mef2c and Pde4dip were enriched in Dlx5/6-GFP+ cells contributing to the pallium (interneurons) (Fig. 6J). In situ hybridization images of E14.5 wild-type brains retrieved from the GenePaint database (53) confirmed the predicted subpallial expression of Sox8 (Fig. 6M), Zfp503 (Fig. 6N) and Ebf1 (Fig. 6O) and the pallial expression of Cxcr4 (Fig. 6K) and Mef2c (Fig. 6L). Of the genes that exhibited increased expression in the subpallium of Arx mutant mice and enrichment in Dlx5/6-derived cells, Gbx2, Stk33, Lbxcor1, Sorcs3, Nts, Olfm3, Magel2 and Hap1 were enriched in Dlx5/6-GFP+ cells that remain in the subpallium, while Cadps2, Kitl, Sst, Phlda1, AI427515, Enpp5, Calb1 and Pcyt1b were enriched in Dlx5/6-GFP+ cells contributing to the pallium (Fig. 6A). In situ hybridization images confirm the predicted pallial expression of Kitl (Fig. 6B), Sst (Fig. 6C) and Phlda1 (Fig. 6D) and the subpallial expression of Hap1 (Fig. 6F), Magel2 (Fig. 6G), Lbxcor1 (Fig. 6H) and Gbx2 (Fig. 6I). A pattern emerged where genes whose expression is normally confined to the subpallium and that decreased in the absence of Arx localized to the striatum in wild-type mice (Fig. 6M–O), while those that increased in the absence of Arx tended to localize to the basal forebrain and septum (Fig. 6F–I).
Figure 6.
(A) Graph denotes the Absolute Enrichment (NES = 1.541, P < 0.001) of the gene set containing those genes whose expression increased in the subpallium in the absence of Arx and were enriched in interneurons within a subset of the Marsh et al. data set (genes expressed in GFP+ subpallial cells versus those expressed in GFP+ cortical cells FAC sorted from Dlx5/6Cre-IRES-eGFP mice) (left). Green boxes contain those genes deemed as enriched (right). The genes contained within the top box are those genes whose expression increased in the subpallium of Arx mutant mice and were enriched in GFP+ interneurons within the cortex. The lower box contains genes whose expression increased in the subpallium of the Arx mutant mouse and were enriched in GFP+ neurons that remain within the subpallium. To validate the predicted expression data, in situ hybridization images of E14.5 embryonic brain were downloaded from the GenePaint web site (53) for (B) Kitl, (C) Sst, (D) Phlda1, (E) AI427515, (F) Hap1, (G) Magel2, (H) Lbxcor1 and (I) Gbx2. (J) Graph denotes the Absolute Enrichment (NES = 1.605, P < 0.001) of the gene set containing those genes whose expression decreased in the subpallium in the absence of Arx and were enriched in Dlx5/6-derived cells within a subset of the Marsh et al. data set (genes expressed in GFP+ subpallial cells versus those expressed in GFP+ cortical cells FAC-sorted from Dlx5/6Cre-IRES-eGFP mice) (left). Green boxes contain those genes deemed as enriched (right). The genes contained within the top box are those genes whose expression decreased in the subpallium of Arx mutant mice and were enriched in GFP+ interneurons within the cortex. The lower box contains genes whose expression decreased in the subpallium of the Arx mutant mouse and were enriched in GFP+ neurons that remain within the subpallium. To validate the predicted expression data, in situ hybridization images of E14.5 embryonic brain were downloaded from the GenePaint web site for (K) Cxcr4, (L) Mef2c, (M) Sox8, (N) Zfp503 and (O) Ebf1.
Homeobox transcription factor binding sites are enriched in the promoter regions of genes differentially expressed in the absence of Arx
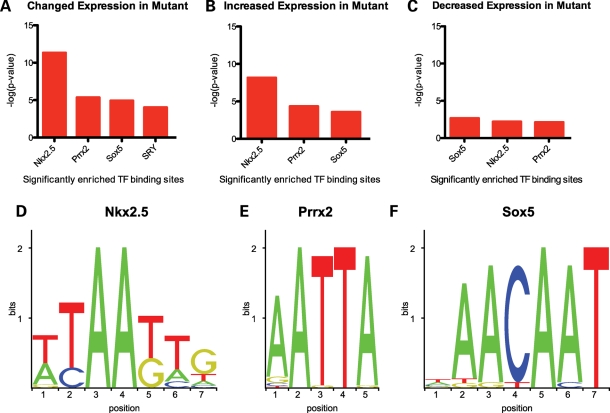
In an attempt to determine which of the modulated genes may be direct targets of Arx, we used oPOSSUM (54), a program that determines the overrepresentation of transcription factor binding sites (TFBSs), as determined by sliding position weight matrices obtained from the JASPER database along aligned and conserved sequences from given regions upstream of a defined transcriptional start site (TSS), within a set of coexpressed genes when compared with a precompiled background set, to search for enrichment of conserved (TFBSs) −5 kb upstream of the TSS of genes whose expression was modulated in the absence of Arx. We reasoned that, given that transcription factors within families often share similar binding sites (55–57), it remained a formal possibility that, although the binding site of Arx remains to be discovered, it might be highly similar to that of transcription factors that have already been annotated. We considered a site enriched if the Z-score was >10 and the Fisher P-value cutoff was <0.01, as these conditions result in an average false positive rate of <15% (54). Additionally, we required that there be at least five target gene hits. When all differentially expressed genes were considered, enrichment of the following TFBSs were observed: Nkx2-5 (36.25% target gene hits, Z-score=40.55, P = 4.70 × 10−12); Prrx2 (62.50% target gene hits, Z-score=35.68, P = 4.36 × 10−6); Sox5 (30% target gene hits, Z-score=13.41, P = 1.16 × 10−5) and Sry (15% target gene hits, Z-score=10.69, P = 9.42 × 10−5) (Fig. 7A). When only the genes whose expression increased in the subpallium in the absence of Arx are considered, enrichment of Nkx2.5 (40.74% target gene hits, Z-score=37.44, P = 7.02 × 10−9), Prrx2 (62.96% target gene hits, Z-score=40.78, P = 4.63 × 10−5) and Sox5 (27.78% target gene hits, Z-score=10.13, P = 2.69 × 10−4) motifs is observed (Fig. 7B). If we only consider those genes whose expression decreased in the subpallium in the absence of Arx, enrichment is observed of the Sox5 (34.62% target gene hits, Z-score=11.32, P = 2.15 × 10−3), Nkx2.5 (26.92% target gene hits, Z-score=12.13, P = 6.28 × 10−3) and Prrx2 (61.54% target gene hits, Z-score=17.15, P = 7.43 × 10−3) binding sites (Fig. 7C). Analysis of the weblogos (58) corresponding to the position weight matrices for the three enriched binding sites across the three sets of genes revealed a similarity between the enriched binding sites (Fig. 7D–F). Both Nkx2.5 and Prrx2 are homeodomain transcription factors, and the reverse complement of the Prrx2 site (TAATT) is contained within the Nkx2.5 site. Prrx2 is a paired-boxed, homeodomain transcription factor of the aristaless family as is Arx (59).
Figure 7.
(A) JASPAR TFBSs significantly enriched in the region −5000 bp upstream of the TSS of all differentially expressed genes relative to those regions upstream of all genes present in at least four of the eight arrays where P < 0.01 and Z-score >10. (B) TFBSs significantly enriched in the regions −5000 bp upstream of the TSS of genes whose expression increased within the subpallium in the absence of Arx. (C) TFBSs significantly enriched in the regions −5000 bp upstream of the TSS of genes whose expression decreased within the subpallium in the absence of Arx. Weblogos of the position weight matrices corresponding to the JASPAR (D) Nkx2.5, (E) Prrx2 and (F) Sox5 binding sites found to be enriched.
Genes containing E2F family TFBSs are enriched in subpallium of wild-type mice when compared with the subpallium of Arx mutants
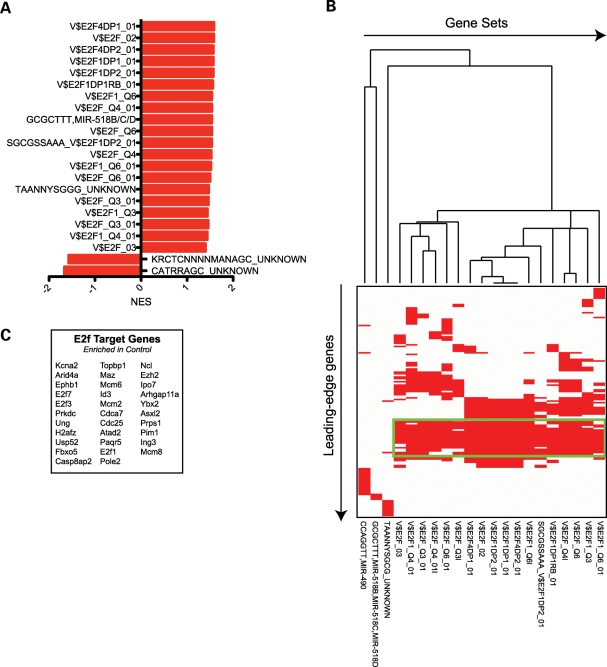
In addition to using oPOSSUM to identify enriched TFBS in those genes that were identified as differentially expressed, we also used GSEA to look for enrichment, within the ranked list of all gene expression values from the Arx array experiments, of precomputed gene sets that share a cis-regulatory motif contained within 4000 bp regions centered at the predicted TSS that is conserved across the human, mouse, rat and dog genomes (60). We identified 20 gene sets enriched (P < 0.05; FDR < 25%) in the wild-type phenotype and 2 gene sets enriched in the mutant phenotype (Fig. 8A). The two gene sets enriched in the mutant phenotype correspond to highly conserved motifs that correspond to an unknown transcription factor. To our surprise, 17/20 gene sets enriched in the wild-type phenotype corresponded to conserved motifs that represented binding sites of E2F family member proteins. We next clustered the leading-edge genes corresponding to the 20 gene sets enriched in the wild-type phenotype (Fig. 8B), which lead to the discovery of a cluster of leading-edge genes shared between the 17 enriched motifs corresponding to binding sites for E2F family proteins (Fig. 8C). These results are highly suggestive that loss of Arx function results in a loss of E2F-family proteins ability to bind their target genes.
Figure 8.
(A) GSEA of microarray data obtained from analysis of control versus Arx mutant subpallia, using a gene set including putative TF and microRNA binding sites conserved within human, mouse, rat and dog (60), revealed TF and microRNA binding site motifs significantly enriched (P < 0.05 and FDR < 0.25) in control (positive NES values) or conditional mutant (negative NES values) subpallia. (B) Clustering of significant gene sets and genes within those gene sets enriched in control subpallia reveals a cluster of genes shared among 18 of the enriched gene sets. All 18 of these gene sets correspond to genes with various known E2F family binding sites within 4000 bp centered on the TSS of the genes. (C) List of genes contained within the cluster.
Ebf3, Lmo1 and Shox2 are direct transcriptional targets of Arx
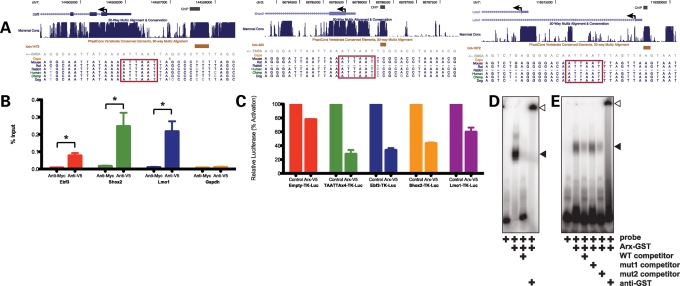
We first tested whether Arx could be recruited to the putative enhancers of Arx target genes by performing chromatin immunoprecipitation (ChIP) assays on Neuro2a neuroblastoma cells transfected with a V5-tagged Arx construct and using an anti-V5 antibody (or anti-myc as a control) to immunoprecipitate Arx-chromatin complexes. After the ChIP procedure, immunoprecipitated chromatin was tested for the presence of Ebf3 −2064 to −1854 (Refseqs NM_010096, NM_001113414, NM_001113415), Lmo1 −126 to −295 (Aceview splice variant Lmo1.b; −5326 to −5495 relative to Refseq NM_057173) and Shox2 −850 to −748 (Refseq NM_013665) putative enhancer regions by qPCR with specific primer pairs spanning the identified phastCon regions containing the predicted Arx consensus binding sites (Fig. 9A). Primer pairs designed to bind within the promoter region of murine Gapdh were used as a control. Significant enrichment was found for the Ebf3, Lmo1 and Shox2 putative enhancer fragments when an anti-V5 antibody was used but not when an anti-myc antibody was used (Fig. 9B). Furthermore, no enrichment was observed at the Gapdh promoter. These data indicate Arx can bind to the putative enhancers of Ebf3, Lmo1 and Shox2.
Figure 9.
(A) Schematics from the UCSC Genome Browser (135) demonstrate the presence of conserved S8/Prrx2 binding sites upstream of the Ebf3, Shox2 and Lmo1 genes. Genes are oriented from right to left. The region chosen for ChIP for each of the genes is annotated and overlaps a PhastCon Vertebrate Conserved Element (132). Below the PhastCon, in gray, the EMSA probe, which is located within the ChIP region, is annotated. Below the annotated EMSA probe, the nucleotide conservation is demonstrated for several mammals with the S8/Prrx2 binding site designated by a red box. (B) Quantitative ChIP assays demonstrate that anti-V5, but not anti-myc control, antibody immunoprecipitates the putative enhancer regions upstream of Ebf3, Shox2 and Lmo1, but not the negative control Gapdh promoter in Neuro2a cells transfected to express V5-tagged Arx. Data are presented as percentage of input chromatin (±SEM). (C) Luciferase reporter assays demonstrate that Arx, in Neuro2a cells, represses heterlogous expression of an engineered site containing four copies of the putative Arx binding site, as well as, the putative enhancers for Ebf3, Shox2 and Lmo1, cloned upstream of TK-luciferase, to a greater extent than that observed for the empty vector control. Luciferase data were normalized to Renilla expression. Data for the Arx-transfected data points are presented as percentage activation (±SEM) relative to empty vector-transfected cells. (D) EMSA assays demonstrate that GST-Arx can bind a 30 bp oligonucleotide containing the putative Arx binding site and corresponding to a region upstream of Shox2 (black arrowhead; lane 2), and that this binding is readily competed with cold competitor (lane 3). The complex was supershifted in the presence of an anti-GST antibody (open arrowhead; lane 4). (E) EMSA assays demonstrate that GST-Arx can bind a 30 bp oligonucleotide containing the putative Arx binding site and corresponding to a region upstream of Lmo1 (black arrowhead; lane 2), and that this binding is readily competed with cold competitor (lane 3). The binding is not readily competed, however, with cold competitor where the TAATTA site has been mutated to TCCTTA (lane 4), but is readily competed with cold competitor where a dinucleotide mutation is made outside of the putative Arx binding site (lane 5). The complex was supershifted in the presence of an anti-GST antibody (open arrowhead; lane 6).
Given that expression of Ebf3, Lmo1 and Shox2 were all increased in the absence of Arx, we next asked whether Arx binds and transcriptionally represses the Ebf3, Lmo1 and Shox2 enhancers. We generated luciferase reporter constructs by cloning the regions identified by ChIP assays upstream of TK-luciferase. As a positive control, we generated a luciferase reporter with four copies of the predicted Arx binding site, TAATTA, and as a negative control we used the empty TK-luciferase-containing vector. Reporter constructs were transiently transfected into Neuro2a cells along with Arx or an empty expression plasmid. Although Arx could minimally repress the empty vector (22.2%), the TAATTA×4, Ebf3, Shox2 and Lmo1 vectors were all significantly repressed by Arx (72.2, 66.8, 56.8 and 43.4% repression, respectively). These data suggest that TAATTA is likely the binding site for Arx, and the regions we identified by ChIP were sufficient to bind Arx and functionally repress transcription.
To determine whether Arx is capable of binding to the S8/Prrx2 motif, EMSA was performed. 32P-labeled oligonucleotide (double-stranded 30mers) spanning the TAATTA found in the Ebf3 (−1862 to −1892), Lmo1 (−217 to −247) and Shox2 (−780 to −810) putative enhancers were probed with purified GST-Arx. We observed that GST-Arx bound the 32P-labeled Shox2 30mer (Fig. 9D, lane 2), and this binding was attenuated by competition with a 50-fold molar excess of unlabeled template (Fig. 9D, lane 3). Furthermore, the addition of an anti-Arx antibody to the binding reaction supershifted the complex (Fig. 9D, lane 4). We observed that GST-Arx bound to the Lmo1 template (Fig. 9E, lane 2), and this binding was attenuated by competition with a 50-fold molar excess of unlabeled template (Fig. 9E, lane 3). GST-Arx binding of Lmo1, however, was not competed by 50-fold molar excess of an unlabeled template where TAATTA was mutated to TCCTTA (Fig. 9E, lane 4). Excess unlabeled template where a TG dinucleotide upstream of the TAATTA was mutated to CC readily competed for binding of GST-Arx (Fig. 9E, lane 5), suggesting that the TAATTA site within the oligonucleotide was necessary for binding of GST-Arx. The addition of an anti-Arx antibody to the binding reaction again supershifted the complex (Fig. 9E, lane 6). Similar results were obtained with the Ebf3 template (data not shown). These data, along with the other data presented, indicate Ebf3, Lmo1 and Shox2 are direct transcriptional targets of Arx whose transcription is repressed by Arx.
DISCUSSION
Mutations in ARX have been identified in multiple human neurodevelopmental disorders, yet its mechanistic role as a transcription factor remains obscure. To gain insight into ARX’s role in brain development we sought to define the downstream targets of ARX in the developing brain. Based on the clinical phenotypes observed in patients with ARX mutations, we were specifically interested in evaluating the role of ARX in cerebral cortical interneurons (61). Using microarray assays on tissue derived from the subpallium of mice in which Arx has been conditionally abrogated, we identified 57 genes significantly over expressed in the mutant mice and 27 genes whose expression was downregulated when compared with litter mate controls. These data were validated by quantitative PCR and in situ hybridization for a subset of genes. Based on our results we conclude Arx functions as both a transcriptional activator and as a transcriptional repressor.
Although we observed both the activation and repression of distinct sets of genes in the absence of Arx, our data indicate Arx functions primarily as a transcriptional repressor in the ventral forebrain. We base this on the fact that a greater number of genes were over-expressed in the absence of Arx when compared with those showing a decrease in expression. A greater magnitude in change of gene expression was also found for the over-expressed group of genes. Finally a greater enrichment of putative Arx binding sites was present in those genes that are overexpressed in the absence of Arx.
Arx was previously shown to regulate Pax4 during pancreas development through binding to a conserved sequence upstream of Pax4 and repress its expression (62). The identified sequence, however, lacks a canonical homeobox binding site, suggesting that either (1) Arx can bind directly to sequences other than canonical homeobox binding sites or (2) Arx forms heterodimeric transcriptional complexes with non-homeobox or homeobox transcription factors that recognize a variant of the canonical homeobox binding site. The Drosophila homolog of Arx, al, can bind clawless (mammalian homologs are Tlx1, Tlx2 and Tlx3) and Chip (mammalian homologs are Ldb1 and Ldb2) (63,64). Tlx1, Tlx2 and Tlx3 are homeobox transcription factors that bind TAAGTG (65). Additionally, Arx has been suggested to interact with Nr2f1 and/or Nr2f2 (66) which bind the nuclear receptor binding site, (A/G)G(G/T)TCA (67,68), Finally, in developing murine muscle, Arx appears to exists within a transcriptional complex that includes Myod1 and Mef2c during myocyte differentiation and mediates the activation of downstream targets (26). Thus, while Pax4 was previously the only known target for Arx, several lines of evidence support Arx functioning within the telencephalon as a transcriptional activator and/or repressor. Our data indicate Arx binds, at least within the subpallium, to ATTAAT in its role as a transcriptional repressor. The fact that mutations in this sequence competed out binding in our EMSA assays support this conclusion.
GSEA analyses were utilized to search for genes belonging to specific sets of subpallial-derived neurons. The Arx-dependent GRN identified in the current study was compared with the expression profiles of Dlx5/6-derived and non-Dlx5/6-derived cell populations within the telencephalon previously elucidated by our laboratory (42) and the Dlx1/2 GRN previously identified by the Rubenstein laboratory (NCBI GEO GSE2161). First, we found statistically significant overlap between the Arx and Dlx1/2 GRNs, which is in agreement with previous data, suggesting that Arx is downstream of Dlx1/2 in the subpallium (41), and we have identified the individual genes that are shared between these two networks. However, we have also identified genes belonging to the Arx GRN whose expression appears not to be affected by loss of Dlx1/2. In fact, we find a small subset of genes that appear to be activated in the absence of Arx but are repressed in the absence of Dlx1/2. These data suggest that the Arx and Dlx appear to be in both direct pathways as well as parallel or divergent pathways.
We were able to determine that subsets of genes altered in the absence of Arx were enriched in Dlx5/6-derived versus non-Dlx5/6-derived expression profiles. Those genes shared in the two data sets, define a set of genes localized to subpallial destined interneurons and GABAergic projection neurons versus interneurons that migrate into the pallium. These analyses allow for the prediction of which elements of the Arx GRN underlie specific aspects of the Arx phenotype. For example Maf, Cxcr4, Mef2c and Pde4dip normally localized to pallial interneurons, and the expression of these genes was lost in the absence of Arx, suggesting that the activation of these genes by Arx may be necessary for either the migration of interneurons into the pallium or differentiation of interneuron subtypes that are pallial-specific. Cxcr4, for example, is expressed on the surface of interneurons that migrate into the cortex and appears to be required for normal interneuron positioning from ventral–lateral to dorsal–medial (69–73).
In contrast, genes such as Hap1, Magel2, Olfm3, Nts, Sorcs3, Lbxcor1, Stk33 and Gbx2 normally localized to septal and basal forebrain neurons and were ectopically expressed in the GE in the absence of Arx. This suggests that Arx may be required to repress this set of genes to allow for maintenance of specific subpallial interneuron subtypes or, for example, the cholinergic neurons which normally arise from this region (47) and are lost in Arx knockout (28). Our GSEA analyses also indicate that a subpopulation of genes whose expression decreases in the absence of Arx normally localized to the striatum. Zfp503 (74,75), Sox8 (76), Ebf1 (77,78) and Oprm1 (79) have all been shown to be enriched in, or involved in development, of the striatum. Ebf1 expression in the mantle of the LGE is required for maintenance of Cdh8 and Crabp1 mantle expression (77). We observed Ebf1, Cdh8 and Crabp1 to all be decreased in the subpallium of Arx conditional mutants. Ebf1 expression was previously observed to be normal in the striatum of P0 Arx−/Y mice, but is lost in the globus pallidus at P0 (28). At earlier time points, Ebf1 was excluded from its normal expression pattern in the LGE corridor, and Ebf1- cells accumulated in the SVZ (28). Colombo et al. (28) also observed an ectopic expression of Etv1 throughout the globus pallidus and accumulation of Calb1 positive cells in the MGE. These findings are consistent with the increase in Etv1 and Calb1 expression and decrease in Ebf1, Cdh8 and Crabp1 expression observed in the subpallium of our Arx conditional mutant mice in the current study. Furthermore, Foxp1, whose expression decreased in the absence of Arx, is expressed in the post-mitotic neurons of the striatum and is specific to the Ppp1r1b- and Gria2/3-expressing medium spiny projection neurons, rather than the interneuron population (80–82). Similarly, Zfp503 and Ebf1 also are expressed in the Ppp1r1b-expressing medium spiny projection neuron population, but appears to be turned on earlier than Foxp1 (75,77,78,83). Collectively, these data suggest that Arx may be required to activate genes that play a role in the specification of striatal GABAergic projection neurons.
GSEA enrichment analysis of TFBSs suggests that the expression of a large cohort of E2F target genes is decreased in the subpallium of Arx conditional mutant mice. This implies that Arx may play a role in controlling cell cycle exit of neural progenitors within the subpallium. The change in expression of a majority of these E2F target genes fell below our 2-fold cutoff, suggesting that these genes themselves are not likely direct targets of Arx; however, the expression of the E2F family members was not altered greater than our cutoff in the conditional Arx mutant. However, two genes known to be involved in E2f1-signaling were altered—Ndn and Rbl1, also known as p107. Ndn binds to the transactivational domain of E2f1 and E2f4, and represses E2f1-induced transactivation (84,85). Ectopic expression of Ndn in neuroblastoma results in the induction of neuronal markers (84). Furthermore, Ndn binds Dlx2 and Dlx5 via Maged1 to promote the differentiation of Calb1+ interneurons, and Ndn-deficient mice exhibit impaired interneuron differentiation (86). Rbl1, normally expressed in the VZ of developing brain, is rapidly downregulated at the onset of neural differentiation (87–89). Further support for this possibility comes from data indicating Rbl1 also regulates the expansion of stem cell populations in brain and regulates expression of Notch1 (90). Alternatively, it remains a formal possibility that Arx could directly bind Rb, Rbl1 or Rbl2, as has been observed, at least in vitro, for other paired-type homeodomain proteins, including Prrx1, Pax3 and Vsx2 (91).
Finally, disruptions in interneuron development have been implicated or linked to a number of neurodevelopmental disorders (reviewed in 92–94). Mutations in Arx that cause neurodevelopmental disorders, such as epilepsy, mental retardation, dystonia and autism spectrum disorder, may have no morphological defects, favoring a primary interneuronopathy (61). Thus, knowing the genes regulated by Arx in interneurons provides an important resource for identifying additional human disease genes and potentially better targets for therapy. Evidence that our analyses has likely identified genes relevant to human disorders comes from the recognition that many of the genes whose expression was altered in the subpallium in the absence of Arx are known to be involved, or have been linked to, other CNS disorders associated with mental retardation or autistic-like phenotypes. These include MAGEL2 and NDN (95–97), CADPS2 (98), SOX8 (99,100), GBX2 (101), AFF2 (102,103), PCDH19 (104) and FGF13 (105). Two other genes, LRRTM1 and HTR7, have been predicted to be susceptibility factors for schizophrenia (106,107). It is likely that many of the other genes identified in this study are also important in human disease. Further studies designed to understand their function will unquestionably bring new insights into our understanding of these complex human disorders.
MATERIALS AND METHODS
Animal assurance
All experiments were approved by and carried out in accordance with the Animal Care and Use Committee of the Children’s Hospital of Philadelphia.
Generation of floxed Arx animals
The targeting vector was generated from a C57BL/6J BAC containing Arx (ATCC, Manassas, VA). A fragment containing exons 1–3 was subcloned into pBS-SK+. An frt-neoR-frt cassette was obtained from Dr Susan Dymecki (Harvard Medical School), and a loxP site was ligated into the 3′ end of this cassette. The resulting frt-neoR-frt-loxP cassette was ligated into a unique AgaI site of intron 2. A second loxP site was introduced, into intron 1, using a unique Nsi1 site. Finally, diphtheria toxin (DTA) was cloned onto the 5′ end of the targeting vector. The resulting construct recombines to generate an allele of Arx with loxP sites flanking exon 2. The floxed construct was electroporated into 129 Sv6(T4) ES cells, as previously described (108), and 576 NeoR colonies were isolated (data not shown). Southern blots on 450 of these clones identified 6 recombinant clones (data not shown). Two independent ES cell lines were selected and injected into C57BL/6 blastocysts. Chimeric mice were obtained, confirmed by Southern blot, and mated to C57BL/6 mice. To prevent unwanted ectopic gene expression of adjacent genes, triggered by the neoR promoter, we removed the frt-neoR-frt cassette by mating germline mice with a floxed Arx allele to those expressing Flp1 (B6;SJL-Tg(ACTFLPe)9205Dym/J; The Jackson Laboratory, Bar Harbor, ME). For the experiments described herein Arxflox/X or Arxfloxflox females were mated to Pou3f4Cre+; Arx+/Y (30) males to generate conditional Arx−/Y; Pou3f4-Cre+ mutant male mice. Hindbrain genomic DNA from animals was genotyped by PCR using the following primers: ArxLoxPF 5′-AAGAACCCCAAAAGCTAAGGAATG-3′; ArxLoxPR 5′-TGGAGCGGGGACAGGGGTGAGGTT-3′, and ArxRePR 5′-GGCCGGTCTCTTTCTTTCTACTCA-3′ to amplify the Arx locus; Brn4Fw 5′-CAATGCTGTTTCACTGGTTATG-3′ and 5′-CATTGCCCCTGTTTCACTATC-3′; SryFw 5′-CAGAAATGAACTACTGCATCCC-3′ and SryRev 5′-AACTTGTGCCTCTCACCACG-3′. The Arx LoxPF, ArxLoxPR and ArxRePR were run as a multiplex PCR. The combination of primers ArxLoxPF and ArxLoxPR yields a 594 bp product for the wild-type allele. The combination of primers ArxLoxPF and ArxLoxPR amplifies a 637 bp product from the floxed allele. If recombination occurs, then ArxLoxPF and ArxRePR amplify a 402 bp product. The Brn4 and Sry primer sets yield 675 and 936 bp products, respectively.
X-gal histochemistry
Pou3f4-Cre mice were mated with B6.129S4-Gt(ROSA)26 Sortm1Sor/J mice (The Jackson Laboratory). Brains from E14.5 embryos were harvested, fixed for 2 h at 4ºC, dropped in 30% sucrose overnight and then snap frozen and cut at 50 µm on a cryostat. Sections were stained as free-floating sections in 1 mg/ml 5-bromo-4-chloro-3-indolyl-β-D-galactopyranoside (X-gal) in 5 mm K3[Fe(CN)6], 5 mm K4[Fe(CN)6]·3H2O, 2 mm MgCl2 in PBS overnight at room temperature. Sections were then rinsed with several washes of PBS and mounted to SuperFrost slides (Fisher Scientific) and mounted with aqueous mounting medium.
Western blotting
GEs from E14.5 Arx−/Y; Pou3f4Cre+ or Arx+/Y; Pou3f4Cre+ brains were lysed in 10 mm Tris–HCl, pH 7.4; 150 mm NaCl; 0.1% Triton X-100, with complete EDTA-free protease inhibitor mixture (Roche Diagnostics), passed six times through a 27-G needle to shear nucleic acids and spun at 14 000g for 20 min. The supernatant was then diluted with 4× LDS sample buffer and 10× reducing agent (Invitrogen). For whole cell extracts, 20 µl of lysate was used. Lysates were separated using NuPAGE 10% Bis–Tris gels, transferred to PVDF membranes, and after blocking were incubated with a C-terminal anti-Arx antibody (1:1000; #sc-48843; Santa Cruz Biotechnology, Santa Cruz, CA). Following incubation with secondary antibody, blots were developed using ECL (GE Healthcare) chemiluminescent reagent and exposed to film. Blots were then stripped using Restore (Pierce) and reprobed with anti-Gapdh (1:5000; #MAB374; Millipore) antibody as a protein loading control.
Extraction and amplification of cRNA
Each biological replicate consisted of a pair of GEs from a single animal. A total of four animals from each genotype (Arx+/Y; Pou3f4Cre+ or Arx−/Y; Pou3f4Cre+) were taken from three separate litters. For microarray experiments, we chose to use Arx+/Y; Pou3f4Cre+, rather than Arxflox/Y; Pou3f4-, mice as controls to eliminate any gene expression changes that may result from any toxicity that may result from Cre activity alone (50,109–113). After sacrificing, all tissue was rapidly dissected from E14.5 GEs (MGE and LGE from both left and right hemispheres). Total RNA was extracted using Trizol (Invitrogen), followed by purification using RNeasy mini columns (Qiagen), and then labeled according to the Affymetrix GeneChip. Expression Analysis Technical Manual (114). Briefly, total RNA was converted to first-strand cDNA using Superscript II reverse transcriptase, primed by a poly(T) oligomer that incorporates the T7 promoter. Second-strand cDNA synthesis was followed by in vitro transcription for linear amplification of each transcript and incorporation of biotinylated CTP and UTP. The cRNA products were then fragmented and hybridized to eight GeneChip. Mouse Genome 430 2.0 microarrays (Affymetrix, Santa Clara, CA), each containing 45 101 probes. Arrays were washed and scanned according to the manufacturer’s recommended protocol.
Microarray data analysis
Statistical analyses were performed using open source R software packages available as part of the Bioconductor project (http://www.bioconductor.org) (115). After normalization and summarization of gene expression data by GC Robust Multi-array Averaging (116) using the affy package (117), MAS5.0 presence/absence calls were used to filter out all probe sets lacking a present call in at least four of the eight samples. Differential gene expression, defined by the microarrays, from Arx−/Y; Pou3f4Cre+ and Arx+/Y; Pou3f4Cre+ tissues was then determined by computing empirical Bayes moderated t-statistics with the limma package (118,119). The moderated t-statistic P-values were adjusted for multiple testing by Benjamini and Hochberg’s method to control FDR (120). Differentially expressed genes were defined as those having a FDR-corrected P-value <0.05 and a log2 fold change in expression ≥1 or ≤ − 1. For clustering, log2-transformed expression data for differentially expressed genes were gene-centered (mean) and normalized. Hierarchical clustering was performed using a correlation matrix and centroid linkage using the Cluster 3.0 algorithm (121) and visualized using the Java TreeView program (122). Microarray data generated for this study has been deposited to and is freely available from NCBI Gene Expression Omnibus (GEO) as record GSE12609.
Quantitative real-time polymerase chain reaction
Total RNA was extracted from the GEs of Arx+/Y; Pou3f4Cre+ (n = 5) and Arx−/Y; Pou3f4Cre+ mice (n = 5) as described earlier and DNase-treated using DNA-free (Ambion, Foster City, CA). Approximately 500 ng of total RNA was reverse transcribed with SuperScript II reverse transcriptase with random primers (Invitrogen, Carlsbad, CA), and 10 ng of cDNA was used for real-time PCR. Real-time PCR was carried out using Taqman Gene Expression Assays (Applied Biosystems, Foster City, CA). Assay IDs are available upon request. Each sample was run in triplicate, along with probes for Actb on the same plate, on a Stratagene MX3005P real-time PCR machine (La Jolla, CA) following the manufacturer’s recommended protocol. Ten probes sets corresponding to genes that changed on the arrays were chosen (Pde4dip, Magel2, Cadps2, Ndn, Rasgef1b, Kitl, Calb1, Ebf3, Shox2 and Sst) along with three negative control genes (Shroom2, Ets1 and Cntnap4) that did not change on the array. Specificity of the amplification was checked by melting-curve analysis. Relative levels of mRNA expression were calculated according to the ΔΔCT method (123), normalized by comparison to Actb mRNA expression.
GO term enrichment analysis
GO term enrichment analysis was performed using the online version of WebGestalt (32). Enrichment was assessed by using the hypergeometric test to compare the frequency of GO Biological Process categories represented in the non-redundant list of Gene IDs corresponding to putative Arx target genes (n = 76) versus the global frequency of GO categories in the reference set containing non-redundant Gene IDs corresponding to all genes present in at least four out of the eight arrays. We chose a P-value cutoff of P < 0.01 and required that all categories tested contain at least five genes. Additionally, we used ErmineJ (33) to assess the overrepresentation of GO Biological Process categories, which allows testing across the entire population of genes without requiring a threshold to divide genes into ‘selected’ and ‘nonselected’ genes. Raw P-values corresponding to all 45 038 probes, as determined by Bayes moderated t-statistics, were entered into the program. The GSR method was used with the following parameters: maximum gene set size = 100; minimum gene set size = 5; with the mean of replicates, 100 000 iterations and full resampling. Significant GO terms were identified using a Benjamini–Hochberg FDR of <0.05.
Gene set enrichment analysis
GSEA analysis was performed, using the GSEA version 2.0 program (124). Two sets of microarray data were downloaded (as .CEL files) from the NCBI Gene Expression Omnibus (GEO) (125) and GC-RMA normalized as described earlier. The first data set, generated by the Rubenstein laboratory (GSE2161), contains data in which cRNA, derived from E14.5 subpallium dissected from either Dlx1/2 heterozygous (n = 3) or Dlx1/2 double mutant (n = 3) mice, was hybridized to Affymetrix MOE430-2 microarrays. A similar, companion data set (GSE8311) was previously described (36). The second data set (GSE5817), generated by our own laboratory (42), contains a data set from microarrays hybridized to cRNA derived from GFP+ pallial cells (n = 6), GFP- pallial cells (n = 5) and GFP+ subpallial cells (n = 6) FAC sorted from the dissected brains of E14.5 Dlx5/6Cre-IRES-eGFP mice. The GSEA program first collapsed the 45 037 probe sets represented by the microarrays to 21 890 individual genes, and the GC-RMA normalized expression values for those genes were ranked with respect to the phenotype (Dlx1/2 heterozygote versus Dlx1/2 KO; GFP-negative pallial cells versus GFP-positive pallial and subpallial cells; or GFP-positive pallial cells versus GFP-positive subpallial cells) by using an absolute value (rather than positive or negative) SNR (126). With this approach, the final position in the ranked gene list depended only on the strength of the gene in discriminating between phenotypes rather than specific up- or down-regulation in a given phenotype. Represented members of the Arx target gene sets, divided into one gene set comprising those genes whose expression increased in the absence of Arx and a second gene set comprising those genes whose expression decreased in the absence of Arx, were then located within the ranked gene list, and the proximity of the Arx target gene sets to the most differentially expressed genes according to phenotype (i.e. those with the highest absolute SNR value) was measured with a weighted Kolmogorov–Smirnov statistic [ES, enrichment score (43)], with a higher score corresponding to a higher proximity. The observed ES score was then compared with the distribution of 1000 permuted ES scores after permutation of phenotype.
Identification of enriched TFBSs
The gene set including putative Arx target genes was analyzed for enrichment of TFBSs using the oPOSSUM 2.0 program (54). For each transcript, genes were mapped to EnsEMBL gen. identifiers and submitted to Human SSA. The top 10% of conserved regions (minimum conservation of 70%) within 5000 bp upstream and matrix match threshold 95% was scanned for TFBSs in the JASPAR database (127) using a position weight matrices algorithm. Enriched TFBSs were identified using Fisher’s exact test at P < 0.01 by comparing the putative Arx target genes with a random set of 1000 genes as background. Additionally, we performed GSEA, as described earlier, on the entire Arx microarray data set, testing for enrichment of the C3 motif gene set included with the GSEA package (60). Briefly, this gene set includes 174 candidate TFBS motifs and 106 putative miRNA binding motifs located within a 4 kb window centered at the annotated TSS or the 3′-UTR, respectively, and conserved in human, mouse, rat and dog genomes.
In situ hybridization
In situ hybridization was performed on 12 μm thick, paraformaldehyde fixed cryostat sections from mutant (Arx−/Y; Pou3f4-Cre+) and control (Arxflox/Y; Pou3f4-Cre-) as previously described (128). Briefly, cDNA for genes of interest was obtained by PCR from an E10.5 whole-embryo mouse cDNA library and cloned into the pBluescript-SK(−) plasmid (Stratagene). Primers used for PCR amplification are listed in Supplementary Material, Table S5. Digioxigenin (DIG)-labeled cRNA probes were generated by in vitro transcription and hybridized to tissue overnight at 70°C. Hybridization signals were detected with alkaline phosphatase-conjugated, anti-DIG Fab fragments (1:2000; Roche Applied Science), followed by color development with NBT and BCIP (Roche Applied Science). In situ hybridizations, corresponding to E14.5 embryos and as presented in Figure 6, were obtained from http://www.genepaint.org, a digital atlas of gene expression patterns in the mouse (53): Kitl (Set ID# HD11), Sst (Set ID# MH430), Phlda1 (Set ID# EH2968), AI427515 (Set ID# EB1326), Hap1 (Set ID# MH834), Magel2 (Set ID# EH1944), Lbxcor1 (Set ID# EH934), Gbx2 (Set ID# MH867), Cxcr4 (Set ID# EN1117), Mef2c (Set ID# MH765), Sox8 (Set ID# EN1112), Zfp503 (Set ID# MH819) and Ebf1 (Set ID# MH1369).
Immunohistochemistry
Immunohistochemistry was performed as previously described (129). Briefly, sections were washed in 0.1 M tris buffer saline (TBS), endogenous peroxidases were quenched and non-specific binding was blocked with 5% normal horse serum and 0.3% Triton-X (Sigma) in TBS. The sections were incubated overnight with the primary antibodies that were detected using a secondary biotin-conjugated antibody (1:2000; Jackson Immunoresearch, West Grove, PA) applied for 1 h at room temperature. The sections were then incubated with the ABC reagent (Vector Laboratories, Burlington, CA, USA) for 1 h at room temperature, washed and developed with 3,3′-diaminobenzidine (DAB; Vector Laboratories). The sections were mounted, dehydrated in graded ethanols, cleared in xylene, coverslipped and analyzed with a light microscope. Primary antibodies included anti-Calb1 (1:2000, CB38; Swant, Bellinzona, Switzerland), anti-Arx (1:100, #sc-48843; Santa Cruz Biotechnology, Santa Cruz CA), anti-Foxp1 (1:400; Dr Edward E. Morrisey of the Department of Cell and Developmental Biology, University of Pennsylvania) (130).
Quantitative ChIP assays
Chromatin immunoprecipitation assays were performed using the ChIP Assay Kit (Millipore) as described by the manufacturer with minor modifications. Neuro2a cells were transfected with pCDNA3.1-Arx-V5 (131) using FuGENE6 (Roche) according to the manufacturer’s suggested protocol. After 48 h, transfected Neuro2a cells (2 × 107) were dissociated and fixed in 1% formaldehyde at room temperature for 10 min. Cells were lysed in SDS lysis buffer (ChIP Assay Kit, Upstate USA, Charlottesville, VA), homogenized and sonicated six times for 30 s using a Fisher Scientific Model 100 Sonic Dismembrator (Fisher) at power settings 3, 3, 4, 5, 6 and then 6 so as to fragment the DNA (‘input DNA’). Subsequently, immunoprecipitation of chromatin equivalents of 2 × 106 cells was carried out at 4°C overnight using 5 µg of either anti-V5 (Invitrogen) or anti-myc (Sigma) antibodies, and immune complexes pulled down using protein A-agarose beads blocked with salmon sperm DNA (Upstate). Following washes according to manufacturer instructions (Upstate), complexes were eluted in 1% SDS/100 mm NaHCO3, and cross-links were reversed by adding NaCl to 200 mm and heating to 65°C for 14 h. DNA was extracted and subjected to quantitative PCR using primers flanking the phastCon (132) site containing the conserved Arx binding sites identified by our in silico analysis. Amplicons were measured by SYBR Green fluorescence (Qiagen) in 25 µl reactions. Reactions were performed in triplicate. The amount of product was determined relative to a standard curve of input chromatin. The primers used were as follows: Shox2-Fw: 5′-TCCAGTTCCCCAGTGTTTTACTAAGT-3′ and Shox2-Rev: 5′-GCTCTTGGCCATTAATCCAGGATT-3′; Lmo1-Fw: 5′- TAAGCTAATGGCGGGCACCT-3′ and Lmo1-Rev: 5′-CTCGCTCTCACCAGAGTGCA-3′; and Ebf3-Fw: 5′-CCGTAATGGATTTTGAGATGGGA-3′ and Ebf3-Rev: 5′-TGAATTGGTGGTGTGTGTGC-3′; Gapdh-Fw: 5′- TACTCGCGGCTTTACGGG-3′ and Gapdh-Rev: 5′- TGGAACAGGGAGGAGCAGAGAGCA-3′. For statistical analyses, student’s t-test was used.
Luciferase reporter assays
Neuro2a mouse neuroblastoma cells (0.8 × 10−5) were transfected 24 h after plating with 200 ng of luciferase reporter plasmid DNA, 50 ng of pCDNA3.1-Arx-V5-His (131) or pCDNA3.1-V5-His (Invitrogen) and 50 ng of pRL-TK-Renilla luciferase plasmid DNA (Promega) using FuGENE 6 (Roche Diagnostics, Alameda, CA). Forty-eight hours post-transfection, cell lysis and measurement of firefly and Renilla luciferase activity was performed using the Dual-Glo Luciferase Assay System (Promega) according to the manufacturer’s instructions using a Veritus Microplate Luminometer (Turner BioSystems, Sunnyvale, CA). Transfections were performed in quadruplicate, and three independent experiments were performed. The firefly luciferase activity was normalized according to the corresponding Renilla luciferase activity, and luciferase activity was reported as mean (±SEM) relative to pCDNA3.1-V5-His/luciferase transfection. Luciferase reporter constructs were generated using primer sequences identical to those used for ChIP, modified to include a HindIII on the 5′ end of the sense primer and a BamHI site on the 3′ end of the antisense primer. These primers were used to PCR amplify product from E14.5 mouse genomic DNA, and the amplified products were cut with BamHI and HindIII and cloned into the BamHI and HindIII sites of PPREX3-TK-Luc (Addgene plasmid 1015) (133), which contains the thymidine kinase promoter upstream of luciferase, replacing the PPAR response element.
Expression and purification of Arx (286–430) in Escherichia coli
Sequences corresponding to amino acids 286–430 of Arx were amplified by PCR using primers 286F-E (5′-cgGAATTCga GAGGGCGGGGAGCTGTCGCC-3′) and 430R-Xh (5′-ccgCTCGAGTTA CGAATCAAGCGCAGGGTGATGCG-3′), digested with EcoRI and XhoI and then cloned into the EcoRI and XhoI sites of pGEX-5X-3 (Amersham Biosciences). GST-Arx (286–430) protein was expressed in TOP10 competent cells (Invitrogen), and its expression was induced by addition of 0.3 mm IPTG to late logarithmic cultures (OD = 0.5) for 3 h at 30° C. Cells were then harvested, resuspended in PBS (phosphate buffer saline pH 7.4) containing 0.5% triton X-100 and disrupted by sonication in the presence of protease inhibitor-cocktail (Sigma). After centrifugation, the supernatants were applied to glutathione–Sepharose beads (Amersham Bioscience), and the beads were washed with PBS. The GST fusion protein was finally eluted with 30 mm glutathione in 100 mm Tris–Cl, pH 7.4.
Electrophoretic mobility-shift assay
Electrophoretic mobility-shift assays (EMSAs) were performed as previously described with slight modifications (134). Oligonucleotides (5 pmol) were labeled by T4 polynucleotide kinase in the presence of [γ-32P]ATP (6000 Ci/mmol; Amersham Bioscience). Unincorporated [γ-32P]ATP was removed by column filtration using MicroSpin G-25 columns (Amersham Biosciences). The purified probe was annealed in Buffer H (Roche) by heating for 1 min and cooling to room temperature. Five hundred nanograms of GST-Arx (286–430) was incubated with 0.1 pmol of labeled-probe in total 20 µl of binding buffer [20 mm HEPES pH 7.4, 50 mm KCl, 1 mm MgCl2, 1 mm DTT, 5% glycerol, 1 µg poly(dI-dC) (Roche)] for 30 min on ice. In competition experiments, 10 pmol cold probes were added. In antibody supershift experiment, 2 µl of anti-GST antibody (Amersham Biosciences) was added. The mixture was loaded on 5% polyacrylamide gel and electrophoresized (constant 33 mA) at 4°C in 1× TBE. Gels were dried and visualized on a phosphorimager (Molecular Dynamics). Oligonucleotides used were as follows: Ebf3: 5′-GCGATTTTCCCGATTAATTAAAATATTAACGCA-3′ and 5′-GTGCGTTAATATTTTAATTAATCGGGAAAATCG-3′; Ebf3mut1: 5′-GCGATTTTCCCGATTCCTTAAAATATTAACGCA-3′ and 5′-GTGCGTTAATATTTTAAGGAATCGGGAAAATCG-3′; Ebf3mut2: 5′-GCGATCCTCCCGATTAATTAAAATATTAACGCA-3′ and 5′-GTGCGTTAATATTTTAATTAATCGGGAGGATCG-3′. Lmo1: 5′-GTAATGAATTGATTTAATTAACAGGGGAGTCTGA-3′ and 5′-GTCAGACTCCCCTGTTAATTAAATCAATTCATTA-3′; Lmo1mut1: 5′- GTAATGAATTGATTTCCTTAACAGGGGAGTCTGA -3′ and 5′-GTCAGACTCCCCTGTTAAGGAAATCAATTCATTA-3′; Lmo1mut2: 5′-GTAATGAACCGATTTAATTAACAGGGGAGTCTGA-3′ and 5′-GTCAGACTCCCCTGTTAATTAAATCGGTTCATTA-3′; Shox2: 5′-GCAAAATCCACGCTTAATTAAATTAATTAGGGA-3′ and 5′-GTCCCTAATTAATTTAATTAAGCGTGGATTTTG-3′.
FUNDING
This work was supported by NIH NS46616 (J.A.G.) and the MRDDRC at the Children’s Hospital of Philadelphia (P30HD26979).
SUPPLEMENTARY MATERIAL
Supplementary Material
ACKNOWLEDGEMENTS
We would like to thank Dr E. Bryan Crenshaw III, The Children’s Hospital of Philadelphia, for providing the Pou3f4-Cre mice. We thank Dr Edward E. Morrisey, University of Pennsylvania School of Medicine for the kind gift of anti-Foxp1 antibody; Dr Susan Dymecki, Harvard Medical School for kindly providing the frt-neoR-frt-containing plasmid; Dr Bruce Spiegelman, Harvard Medical School, for kindly providing the PPREX3-TK-Luc plasmid; Dr Gail Martin, University of California, San Francisco for kindly providing the pGbx2-HA-FL plasmid; and Dr John Cobb, University of Calgary, for kindly providing the pGEM4-Shox2cds plasmid. We also thank Dr Steven S. Scherer, University of Pennsylvania, for kindly providing the Neuro2a neuroblastoma cell line. The microarray work was completed through the National Institute of Health Neuroscience Microarray Consortium (http://arrayconsortium.tgen.org) with the assistance of Sheila Westman at the W.M. Keck Foundation Biotechnology Resource Laboratory at Yale University. Finally, we would like to thank Mr Jeremy C. Minarcik for assisting us with animal husbandry.
Conflict of Interest statement. None declared.
REFERENCES
- 1.Kato M., Das S., Petras K., Kitamura K., Morohashi K., Abuelo D.N., Barr M., Bonneau D., Brady A.F., Carpenter N.J., et al. Mutations of ARX are associated with striking pleiotropy and consistent genotype-phenotype correlation. Hum. Mutat. 2004;23:147–159. doi: 10.1002/humu.10310. [DOI] [PubMed] [Google Scholar]
- 2.Kitamura K., Yanazawa M., Sugiyama N., Miura H., Iizuka-Kogo A., Kusaka M., Omichi K., Suzuki R., Kato-Fukui Y., Kamiirisa K., et al. Mutation of ARX causes abnormal development of forebrain and testes in mice and X-linked lissencephaly with abnormal genitalia in humans. Nat. Genet. 2002;32:359–369. doi: 10.1038/ng1009. [DOI] [PubMed] [Google Scholar]
- 3.Bhat S.S., Rogers R.C., Holden K.R., Srivastava A.K. A novel in-frame deletion in ARX is associated with lissencephaly with absent corpus callosum and hypoplastic genitalia. Am. J. Med. Genet. A. 2005;138:70–72. doi: 10.1002/ajmg.a.30892. [DOI] [PubMed] [Google Scholar]
- 4.Hahn A., Gross C., Uyanik G., Hehr U., Hügens-Penzel M., Alzen G., Neubauer B.A. X-linked lissencephaly with abnormal genitalia associated with renal phosphate wasting. Neuropediatrics. 2004;35:202–205. doi: 10.1055/s-2004-817955. [DOI] [PubMed] [Google Scholar]
- 5.Uyanik G., Aigner L., Martin P., Gross C., Neumann D., Marschner-Schäfer H., Hehr U., Winkler J. ARX mutations in X-linked lissencephaly with abnormal genitalia. Neurology. 2003;61:232–235. doi: 10.1212/01.wnl.0000079371.19562.ba. [DOI] [PubMed] [Google Scholar]
- 6.Guerrini R., Moro F., Kato M., Barkovich A.J., Shiihara T., McShane M.A., Hurst J., Loi M., Tohyama J., Norci V., et al. Expansion of the first PolyA tract of ARX causes infantile spasms and status dystonicus. Neurology. 2007;69:427–433. doi: 10.1212/01.wnl.0000266594.16202.c1. [DOI] [PubMed] [Google Scholar]
- 7.Kato M., Saitoh S., Kamei A., Shiraishi H., Ueda Y., Akasaka M., Tohyama J., Akasaka N., Hayasaka K. A longer polyalanine expansion mutation in the ARX gene causes early infantile epileptic encephalopathy with suppression-burst pattern (Ohtahara syndrome) Am. J. Hum. Genet. 2007;81:361–366. doi: 10.1086/518903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Strømme P., Mangelsdorf M.E., Shaw M.A., Lower K.M., Lewis S.M., Bruyere H., Lütcherath V., Gedeon A.K., Wallace R.H., Scheffer I.E., et al. Mutations in the human ortholog of Aristaless cause X-linked mental retardation and epilepsy. Nat. Genet. 2002;30:441–445. doi: 10.1038/ng862. [DOI] [PubMed] [Google Scholar]
- 9.Kato M., Das S., Petras K., Sawaishi Y., Dobyns W.B. Polyalanine expansion of ARX associated with cryptogenic West syndrome. Neurology. 2003;61:267–276. doi: 10.1212/01.wnl.0000068012.69928.92. [DOI] [PubMed] [Google Scholar]
- 10.Wohlrab G., Uyanik G., Gross C., Hehr U., Winkler J., Schmitt B., Boltshauser E. Familial West syndrome and dystonia caused by an Aristaless related homeobox gene mutation. Eur. J. Pediatr. 2005;164:326–328. doi: 10.1007/s00431-005-1622-2. [DOI] [PubMed] [Google Scholar]
- 11.Scheffer I.E., Wallace R.H., Phillips F.L., Hewson P., Reardon K., Parasivam G., Stromme P., Berkovic S.F., Gecz J., Mulley J.C. X-linked myoclonic epilepsy with spasticity and intellectual disability: mutation in the homeobox gene ARX. Neurology. 2002;59:348–356. doi: 10.1212/wnl.59.3.348. [DOI] [PubMed] [Google Scholar]
- 12.Frints S.G., Froyen G., Marynen P., Willekens D., Legius E., Fryns J.P. Re-evaluation of MRX36 family after discovery of an ARX gene mutation reveals mild neurological features of Partington syndrome. Am. J. Med. Genet. 2002;112:427–428. doi: 10.1002/ajmg.10628. [DOI] [PubMed] [Google Scholar]
- 13.Bienvenu T., Poirier K., Friocourt G., Bahi N., Beaumont D., Fauchereau F., Ben Jeema L., Zemni R., Vinet M.C., Francis F., et al. ARX, a novel Prd-class-homeobox gene highly expressed in the telencephalon, is mutated in X-linked mental retardation. Hum. Mol. Genet. 2002;11:981–991. doi: 10.1093/hmg/11.8.981. [DOI] [PubMed] [Google Scholar]
- 14.Poirier K., Lacombe D., Gilbert-Dussardier B., Raynaud M., Desportes V., de Brouwer A.P., Moraine C., Fryns J.P., Ropers H.H., Beldjord C., et al. Screening of ARX in mental retardation families: Consequences for the strategy of molecular diagnosis. Neurogenetics. 2006;7:39–46. doi: 10.1007/s10048-005-0014-0. [DOI] [PubMed] [Google Scholar]
- 15.Grønskov K., Hjalgrim H., Nielsen I.M., Brøndum-Nielsen K. Screening of the ARX gene in 682 retarded males. Eur. J. Hum. Genet. 2004;12:701–705. doi: 10.1038/sj.ejhg.5201222. [DOI] [PubMed] [Google Scholar]
- 16.Partington M.W., Turner G., Boyle J., Gécz J. Three new families with X-linked mental retardation caused by the 428-451dup(24 bp) mutation in ARX. Clin. Genet. 2004;66:39–45. doi: 10.1111/j.0009-9163.2004.00268.x. [DOI] [PubMed] [Google Scholar]
- 17.Stepp M.L., Cason A.L., Finnis M., Mangelsdorf M., Holinski-Feder E., Macgregor D., MacMillan A., Holden J.J., Gecz J., Stevenson R.E., et al. XLMR in MRX families 29, 32, 33 and 38 results from the dup24 mutation in the ARX (Aristaless related homeobox) gene. BMC Med. Genet. 2005;6:16. doi: 10.1186/1471-2350-6-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Van Esch H., Zanni G., Holvoet M., Borghgraef M., Chelly J., Fryns J.P., Devriendt K. X-linked mental retardation, short stature, microcephaly and hypogonadism maps to Xp22.1-p21.3 in a Belgian family. Eur. J. Med. Genet. 2005;48:145–152. doi: 10.1016/j.ejmg.2005.01.016. [DOI] [PubMed] [Google Scholar]
- 19.Miura H., Yanazawa M., Kato K., Kitamura K. Expression of a novel aristaless related homeobox gene ‘Arx’ in the vertebrate telencephalon, diencephalon and floor plate. Mech. Dev. 1997;65:99–109. doi: 10.1016/s0925-4773(97)00062-2. [DOI] [PubMed] [Google Scholar]
- 20.Sherr E.H. The ARX story (epilepsy, mental retardation, autism, and cerebral malformations): one gene leads to many phenotypes. Curr. Opin. Pediatr. 2003;15:567–571. doi: 10.1097/00008480-200312000-00004. [DOI] [PubMed] [Google Scholar]
- 21.Colombo E., Galli R., Cossu G., Gécz J., Broccoli V. Mouse orthologue of ARX, a gene mutated in several X-linked forms of mental retardation and epilepsy, is a marker of adult neural stem cells and forebrain GABAergic neurons. Dev. Dyn. 2004;231:631–639. doi: 10.1002/dvdy.20164. [DOI] [PubMed] [Google Scholar]
- 22.Kitamura K., Miura H., Yanazawa M., Miyashita T., Kato K. Expression patterns of Brx1 (Rieg gene), Sonic hedgehog, Nkx2.2, Dlx1 and Arx during zona limitans intrathalamica and embryonic ventral lateral geniculate nuclear formation. Mech. Dev. 1997;67:83–96. doi: 10.1016/s0925-4773(97)00110-x. [DOI] [PubMed] [Google Scholar]
- 23.Poirier K., Van Esch H., Friocourt G., Saillour Y., Bahi N., Backer S., Souil E., Castelnau-Ptakhine L., Beldjord C., Francis F., et al. Neuroanatomical distribution of ARX in brain and its localisation in GABAergic neurons. Brain Res. Mol. Brain Res. 2004;122:35–46. doi: 10.1016/j.molbrainres.2003.11.021. [DOI] [PubMed] [Google Scholar]
- 24.Yoshihara S., Omichi K., Yanazawa M., Kitamura K., Yoshihara Y. Arx homeobox gene is essential for development of mouse olfactory system. Development. 2005;132:751–762. doi: 10.1242/dev.01619. [DOI] [PubMed] [Google Scholar]
- 25.Visel A., Carson J., Oldekamp J., Warnecke M., Jakubcakova V., Zhou X., Shaw C.A., Alvarez-Bolado G., Eichele G. Regulatory pathway analysis by high-throughput in situ hybridization. PLoS Genet. 2007;3:1867–1883. doi: 10.1371/journal.pgen.0030178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Biressi S., Messina G., Collombat P., Tagliafico E., Monteverde S., Benedetti L., Cusella De Angelis M.G., Mansouri A., Ferrari S., Tajbakhsh S., et al. The homeobox gene Arx is a novel positive regulator of embryonic myogenesis. Cell Death Differ. 2008;15:94–104. doi: 10.1038/sj.cdd.4402230. [DOI] [PubMed] [Google Scholar]
- 27.Collombat P., Mansouri A., Hecksher-Sorensen J., Serup P., Krull J., Gradwohl G., Gruss P. Opposing actions of Arx and Pax4 in endocrine pancreas development. Genes Dev. 2003;17:2591–2603. doi: 10.1101/gad.269003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Colombo E., Collombat P., Colasante G., Bianchi M., Long J., Mansouri A., Rubenstein J.L., Broccoli V. Inactivation of Arx, the murine ortholog of the X-linked lissencephaly with ambiguous genitalia gene, leads to severe disorganization of the ventral telencephalon with impaired neuronal migration and differentiation. J. Neurosci. 2007;27:4786–4798. doi: 10.1523/JNEUROSCI.0417-07.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ahn K., Mishina Y., Hanks M.C., Behringer R.R., Crenshaw E.B., III BMPR-IA signaling is required for the formation of the apical ectodermal ridge and dorsal-ventral patterning of the limb. Development. 2001;128:4449–4461. doi: 10.1242/dev.128.22.4449. [DOI] [PubMed] [Google Scholar]
- 30.Heydemann A., Nguyen L.C., Crenshaw E.B., III Regulatory regions from the Brn4 promoter direct LACZ expression to the developing forebrain and neural tube. Brain Res. Dev. Brain Res. 2001;128:83–90. doi: 10.1016/s0165-3806(01)00137-7. [DOI] [PubMed] [Google Scholar]
- 31.Ashburner M., Ball C.A., Blake J.A., Botstein D., Butler H., Cherry J.M., Davis A.P., Dolinski K., Dwight S.S., Eppig J.T., et al. Gene ontology: tool for the unification of biology. The Gene Ontology Consortium. Nat. Genet. 2000;25:25–29. doi: 10.1038/75556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Zhang B., Kirov S., Snoddy J. WebGestalt: an integrated system for exploring gene sets in various biological contexts. Nucleic Acids Res. 2005;33:W741–W748. doi: 10.1093/nar/gki475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Lee H.K., Braynen W., Keshav K., Pavlidis P. ErmineJ: tool for functional analysis of gene expression data sets. BMC Bioinformatics. 2005;6:269. doi: 10.1186/1471-2105-6-269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Anderson S.A., Eisenstat D.D., Shi L., Rubenstein J.L. Interneuron migration from basal forebrain to neocortex: dependence on Dlx genes. Science. 1997;278:474–476. doi: 10.1126/science.278.5337.474. [DOI] [PubMed] [Google Scholar]
- 35.Petryniak M.A., Potter G.B., Rowitch D.H., Rubenstein J.L. Dlx1 and Dlx2 control neuronal versus oligodendroglial cell fate acquisition in the developing forebrain. Neuron. 2007;55:417–433. doi: 10.1016/j.neuron.2007.06.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Cobos I., Borello U., Rubenstein J.L. Dlx transcription factors promote migration through repression of axon and dendrite growth. Neuron. 2007;54:873–888. doi: 10.1016/j.neuron.2007.05.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Anderson S., Mione M., Yun K., Rubenstein J.L. Differential origins of neocortical projection and local circuit neurons: role of Dlx genes in neocortical interneuronogenesis. Cereb. Cortex. 1999;9:646–654. doi: 10.1093/cercor/9.6.646. [DOI] [PubMed] [Google Scholar]
- 38.Stühmer T., Anderson S.A., Ekker M., Rubenstein J.L. Ectopic expression of the Dlx genes induces glutamic acid decarboxylase and Dlx expression. Development. 2002;129:245–252. doi: 10.1242/dev.129.1.245. [DOI] [PubMed] [Google Scholar]
- 39.Stühmer T., Puelles L., Ekker M., Rubenstein J.L. Expression from a Dlx gene enhancer marks adult mouse cortical GABAergic neurons. Cereb. Cortex. 2002;12:75–85. doi: 10.1093/cercor/12.1.75. [DOI] [PubMed] [Google Scholar]
- 40.Anderson S.A., Qiu M., Bulfone A., Eisenstat D.D., Meneses J., Pedersen R., Rubenstein J.L. Mutations of the homeobox genes Dlx-1 and Dlx-2 disrupt the striatal subventricular zone and differentiation of late born striatal neurons. Neuron. 1997;19:27–37. doi: 10.1016/s0896-6273(00)80345-1. [DOI] [PubMed] [Google Scholar]
- 41.Cobos I., Broccoli V., Rubenstein J.L. The vertebrate ortholog of Aristaless is regulated by Dlx genes in the developing forebrain. J. Comp. Neurol. 2005;483:292–303. doi: 10.1002/cne.20405. [DOI] [PubMed] [Google Scholar]
- 42.Marsh E.D., Minarcik J., Campbell K., Brooks-Kayal A.R., Golden J.A. FACS-array gene expression analysis during early development of mouse telencephalic interneurons. Dev. Neurobiol. 2008;68:434–445. doi: 10.1002/dneu.20602. [DOI] [PubMed] [Google Scholar]
- 43.Subramanian A., Tamayo P., Mootha V.K., Mukherjee S., Ebert B.L., Gillette M.A., Paulovich A., Pomeroy S.L., Golub T.R., Lander E.S., et al. Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc. Natl Acad. Sci. USA. 2005;102:15545–15550. doi: 10.1073/pnas.0506580102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Anderson S.A., Marín O., Horn C., Jennings K., Rubenstein J.L. Distinct cortical migrations from the medial and lateral ganglionic eminences. Development. 2001;128:353–363. doi: 10.1242/dev.128.3.353. [DOI] [PubMed] [Google Scholar]
- 45.Fogarty M., Grist M., Gelman D., Marín O., Pachnis V., Kessaris N. Spatial genetic patterning of the embryonic neuroepithelium generates GABAergic interneuron diversity in the adult cortex. J. Neurosci. 2007;27:10935–10946. doi: 10.1523/JNEUROSCI.1629-07.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Jimenez D., Lopez-Mascaraque L.M., Valverde F., De Carlos J.A. Tangential migration in neocortical development. Dev. Biol. 2002;244:155–169. doi: 10.1006/dbio.2002.0586. [DOI] [PubMed] [Google Scholar]
- 47.Marin O., Anderson S.A., Rubenstein J.L. Origin and molecular specification of striatal interneurons. J. Neurosci. 2000;20:6063–6076. doi: 10.1523/JNEUROSCI.20-16-06063.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Pleasure S.J., Anderson S., Hevner R., Bagri A., Marin O., Lowenstein D.H., Rubenstein J.L. Cell migration from the ganglionic eminences is required for the development of hippocampal GABAergic interneurons. Neuron. 2000;28:727–740. doi: 10.1016/s0896-6273(00)00149-5. [DOI] [PubMed] [Google Scholar]
- 49.Wichterle H., Turnbull D.H., Nery S., Fishell G., Alvarez-Buylla A. In utero fate mapping reveals distinct migratory pathways and fates of neurons born in the mammalian basal forebrain. Development. 2001;128:3759–3771. doi: 10.1242/dev.128.19.3759. [DOI] [PubMed] [Google Scholar]
- 50.Schmidt E.E., Taylor D.S., Prigge J.R., Barnett S., Capecchi M.R. Illegitimate Cre-dependent chromosome rearrangements in transgenic mouse spermatids. Proc. Natl Acad. Sci. USA. 2000;97:13702–13707. doi: 10.1073/pnas.240471297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Tripodi M., Filosa A., Armentano M., Studer M. The COUP-TF nuclear receptors regulate cell migration in the mammalian basal forebrain. Development. 2004;131:6119–6129. doi: 10.1242/dev.01530. [DOI] [PubMed] [Google Scholar]
- 52.Nery S., Fishell G., Corbin J.G. The caudal ganglionic eminence is a source of distinct cortical and subcortical cell populations. Nat. Neurosci. 2002;5:1279–1287. doi: 10.1038/nn971. [DOI] [PubMed] [Google Scholar]
- 53.Visel A., Thaller C., Eichele G. GenePaint.org: an atlas of gene expression patterns in the mouse embryo. Nucleic Acids Res. 2004;32:D552–D556. doi: 10.1093/nar/gkh029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Ho Sui S.J., Mortimer J.R., Arenillas D.J., Brumm J., Walsh C.J., Kennedy B.P., Wasserman W.W. oPOSSUM: identification of over-represented transcription factor binding sites in co-expressed genes. Nucleic Acids Res. 2005;33:3154–3164. doi: 10.1093/nar/gki624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Schones D.E., Sumazin P., Zhang M.Q. Similarity of position frequency matrices for transcription factor binding sites. Bioinformatics. 2005;21:307–313. doi: 10.1093/bioinformatics/bth480. [DOI] [PubMed] [Google Scholar]
- 56.Luscombe N.M., Austin S.E., Berman H.M., Thornton J.M. An overview of the structures of protein-DNA complexes. Genome Biol. 2000;1 doi: 10.1186/gb-2000-1-1-reviews001. REVIEWS001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Sandelin A., Wasserman W.W. Constrained binding site diversity within families of transcription factors enhances pattern discovery bioinformatics. J. Mol. Biol. 2004;338:207–215. doi: 10.1016/j.jmb.2004.02.048. [DOI] [PubMed] [Google Scholar]
- 58.Crooks G.E., Hon G., Chandonia J.M., Brenner S.E. WebLogo: a sequence logo generator. Genome Res. 2004;14:1188–1190. doi: 10.1101/gr.849004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Meijlink F., Beverdam A., Brouwer A., Oosterveen T.C., Berge D.T. Vertebrate aristaless-related genes. Int. J. Dev. Biol. 1999;43:651–663. [PubMed] [Google Scholar]
- 60.Xie X., Lu J., Kulbokas E.J., Golub T.R., Mootha V., Lindblad-Toh K., Lander E.S., Kellis M. Systematic discovery of regulatory motifs in human promoters and 3′-UTRs by comparison of several mammals. Nature. 2005;434:338–345. doi: 10.1038/nature03441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Kato M., Dobyns W.B. X-linked lissencephaly with abnormal genitalia as a tangential migration disorder causing intractable epilepsy: proposal for a new term, ‘interneuronopathy’. J. Child Neurol. 2005;20:392–397. doi: 10.1177/08830738050200042001. [DOI] [PubMed] [Google Scholar]
- 62.Collombat P., Hecksher-Sørensen J., Broccoli V., Krull J., Ponte I., Mundiger T., Smith J., Gruss P., Serup P., Mansouri A. The simultaneous loss of Arx and Pax4 genes promotes a somatostatin-producing cell fate specification at the expense of the alpha- and beta-cell lineages in the mouse endocrine pancreas. Development. 2005;132:2969–2980. doi: 10.1242/dev.01870. [DOI] [PubMed] [Google Scholar]
- 63.Kojima T., Tsuji T., Saigo K. A concerted action of a paired-type homeobox gene, aristaless, and a homolog of Hox11/tlx homeobox gene, clawless, is essential for the distal tip development of the Drosophila leg. Dev. Biol. 2005;279:434–445. doi: 10.1016/j.ydbio.2004.12.005. [DOI] [PubMed] [Google Scholar]
- 64.Pueyo J.I., Couso J.P. Chip-mediated partnerships of the homeodomain proteins Bar and Aristaless with the LIM-HOM proteins Apterous and Lim1 regulate distal leg development. Development. 2004;131:3107–3120. doi: 10.1242/dev.01161. [DOI] [PubMed] [Google Scholar]
- 65.Tang S., Breitman M.L. The optimal binding sequence of the Hox11 protein contains a predicted recognition core motif. Nucleic Acids Res. 1995;23:1928–1935. doi: 10.1093/nar/23.11.1928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Melkman T., Sengupta P. Regulation of chemosensory and GABAergic motor neuron development by the C. elegans Aristaless/Arx homolog alr-1. Development. 2005;132:1935–1949. doi: 10.1242/dev.01788. [DOI] [PubMed] [Google Scholar]
- 67.Cooney A.J., Tsai S.Y., O’Malley B.W., Tsai M.J. Chicken ovalbumin upstream promoter transcription factor (COUP-TF) dimers bind to different GGTCA response elements, allowing COUP-TF to repress hormonal induction of the vitamin D3, thyroid hormone, and retinoic acid receptors. Mol. Cell Biol. 1992;12:4153–4163. doi: 10.1128/mcb.12.9.4153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Nakshatri H., Chambon P. The directly repeated RG(G/T)TCA motifs of the rat and mouse cellular retinol-binding protein II genes are promiscuous binding sites for RAR, RXR, HNF-4 and ARP-1 homo- and heterodimers. J. Biol. Chem. 1994;269:890–902. [PubMed] [Google Scholar]
- 69.Li G., Adesnik H., Li J., Long J., Nicoll R.A., Rubenstein J.L., Pleasure S.J. Regional distribution of cortical interneurons and development of inhibitory tone are regulated by Cxcl12/Cxcr4 signaling. J. Neurosci. 2008;28:1085–1098. doi: 10.1523/JNEUROSCI.4602-07.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.López-Bendito G., Sánchez-Alcañiz J.A., Pla R., Borrell V., Picó E., Valdeolmillos M., Marín O. Chemokine signaling controls intracortical migration and final distribution of GABAergic interneurons. J. Neurosci. 2008;28:1613–1624. doi: 10.1523/JNEUROSCI.4651-07.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Liapi A., Pritchett J., Jones O., Fujii N., Parnavelas J.G., Nadarajah B. Stromal-derived factor 1 signalling regulates radial and tangential migration in the developing cerebral cortex. Dev. Neurosci. 2008;30:117–131. doi: 10.1159/000109857. [DOI] [PubMed] [Google Scholar]
- 72.Tiveron M.C., Rossel M., Moepps B., Zhang Y.L., Seidenfaden R., Favor J., Konig N., Cremer H. Molecular interaction between projection neuron precursors and invading interneurons via stromal-derived factor 1 (CXCL12)/CXCR4 signaling in the cortical subventricular zone/intermediate zone. J. Neurosci. 2006;26:13273–13278. doi: 10.1523/JNEUROSCI.4162-06.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Stumm R.K., Zhou C., Ara T., Lazarini F., Dubois-Dalcq M., Nagasawa T., Hollt V., Schulz S. CXCR4 regulates interneuron migration in the developing neocortex. J. Neurosci. 2003;23:5123–5130. doi: 10.1523/JNEUROSCI.23-12-05123.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Arlotta P., Molyneaux B.J., Jabaudon D., Yoshida Y., Macklis J.D. Ctip2 controls the differentiation of medium spiny neurons and the establishment of the cellular architecture of the striatum. J. Neurosci. 2008;28:622–632. doi: 10.1523/JNEUROSCI.2986-07.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Chang C.W., Tsai C.W., Wang H.F., Tsai H.C., Chen H.Y., Tsai T.F., Takahashi H., Li H.Y., Fann M.J., Yang C.W., et al. Identification of a developmentally regulated striatum-enriched zinc-finger gene, Nolz-1, in the mammalian brain. Proc. Natl Acad. Sci. USA. 2004;101:2613–2618. doi: 10.1073/pnas.0308645100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Sock E., Schmidt K., Hermanns-Borgmeyer I., Bosl M.R., Wegner M. Idiopathic weight reduction in mice deficient in the high-mobility-group transcription factor Sox8. Mol. Cell Biol. 2001;21:6951–6959. doi: 10.1128/MCB.21.20.6951-6959.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Garel S., Marin F., Grosschedl R., Charnay P. Ebf1 controls early cell differentiation in the embryonic striatum. Development. 1999;126:5285–5294. doi: 10.1242/dev.126.23.5285. [DOI] [PubMed] [Google Scholar]
- 78.Lobo M.K., Karsten S.L., Gray M., Geschwind D.H., Yang X.W. FACS-array profiling of striatal projection neuron subtypes in juvenile and adult mouse brains. Nat. Neurosci. 2006;9:443–452. doi: 10.1038/nn1654. [DOI] [PubMed] [Google Scholar]
- 79.Moon Edley S., Herkenham M. Comparative development of striatal opiate receptors and dopamine revealed by autoradiography and histofluorescence. Brain Res. 1984;305:27–42. doi: 10.1016/0006-8993(84)91116-8. [DOI] [PubMed] [Google Scholar]
- 80.Ferland R.J., Cherry T.J., Preware P.O., Morrisey E.E., Walsh C.A. Characterization of Foxp2 and Foxp1 mRNA and protein in the developing and mature brain. J. Comp. Neurol. 2003;460:266–279. doi: 10.1002/cne.10654. [DOI] [PubMed] [Google Scholar]
- 81.Tamura S., Morikawa Y., Iwanishi H., Hisaoka T., Senba E. Expression pattern of the winged-helix/forkhead transcription factor Foxp1 in the developing central nervous system. Gene Expr. Patterns. 2003;3:193–197. doi: 10.1016/s1567-133x(03)00003-6. [DOI] [PubMed] [Google Scholar]
- 82.Tamura S., Morikawa Y., Iwanishi H., Hisaoka T., Senba E. Foxp1 gene expression in projection neurons of the mouse striatum. Neuroscience. 2004;124:261–267. doi: 10.1016/j.neuroscience.2003.11.036. [DOI] [PubMed] [Google Scholar]
- 83.Lobo M.K., Yeh C., Yang X.W. Pivotal role of early B-cell factor 1 in development of striatonigral medium spiny neurons in the matrix compartment. J. Neurosci. Res. 2008;86:2134–2146. doi: 10.1002/jnr.21666. [DOI] [PubMed] [Google Scholar]
- 84.Kobayashi M., Taniura H., Yoshikawa K. Ectopic expression of necdin induces differentiation of mouse neuroblastoma cells. J. Biol. Chem. 2002;277:42128–42135. doi: 10.1074/jbc.M205024200. [DOI] [PubMed] [Google Scholar]
- 85.Taniura H., Taniguchi N., Hara M., Yoshikawa K. Necdin, a postmitotic neuron-specific growth suppressor, interacts with viral transforming proteins and cellular transcription factor E2F1. J. Biol. Chem. 1998;273:720–728. doi: 10.1074/jbc.273.2.720. [DOI] [PubMed] [Google Scholar]
- 86.Kuwajima T., Nishimura I., Yoshikawa K. Necdin promotes GABAergic neuron differentiation in cooperation with Dlx homeodomain proteins. J. Neurosci. 2006;26:5383–5392. doi: 10.1523/JNEUROSCI.1262-06.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Callaghan D.A., Dong L., Callaghan S.M., Hou Y.X., Dagnino L., Slack R.S. Neural precursor cells differentiating in the absence of Rb exhibit delayed terminal mitosis and deregulated E2F 1 and 3 activity. Dev. Biol. 1999;207:257–270. doi: 10.1006/dbio.1998.9162. [DOI] [PubMed] [Google Scholar]
- 88.Ferguson K.L., Slack R.S. The Rb pathway in neurogenesis. Neuroreport. 2001;12:A55–A62. doi: 10.1097/00001756-200107030-00001. [DOI] [PubMed] [Google Scholar]
- 89.Gill R.M., Slack R., Kiess M., Hamel P.A. Regulation of expression and activity of distinct pRB, E2F, D-type cyclin, and CKI family members during terminal differentiation of P19 cells. Exp. Cell Res. 1998;244:157–170. doi: 10.1006/excr.1998.4197. [DOI] [PubMed] [Google Scholar]
- 90.Vanderluit J.L., Ferguson K.L., Nikoletopoulou V., Parker M., Ruzhynsky V., Alexson T., McNamara S.M., Park D.S., Rudnicki M., Slack R.S. p107 regulates neural precursor cells in the mammalian brain. J. Cell Biol. 2004;166:853–863. doi: 10.1083/jcb.200403156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Wiggan O., Taniguchi-Sidle A., Hamel P.A. Interaction of the pRB-family proteins with factors containing paired-like homeodomains. Oncogene. 1998;16:227–236. doi: 10.1038/sj.onc.1201534. [DOI] [PubMed] [Google Scholar]
- 92.Benes F.M., Berretta S. GABAergic interneurons: implications for understanding schizophrenia and bipolar disorder. Neuropsychopharmacology. 2001;25:1–27. doi: 10.1016/S0893-133X(01)00225-1. [DOI] [PubMed] [Google Scholar]
- 93.Di Cristo G. Development of cortical GABAergic circuits and its implications for neurodevelopmental disorders. Clin. Genet. 2007;72:1–8. doi: 10.1111/j.1399-0004.2007.00822.x. [DOI] [PubMed] [Google Scholar]
- 94.Uhlhaas P.J., Singer W. Neural synchrony in brain disorders: relevance for cognitive dysfunctions and pathophysiology. Neuron. 2006;52:155–168. doi: 10.1016/j.neuron.2006.09.020. [DOI] [PubMed] [Google Scholar]
- 95.Boccaccio I., Glatt-Deeley H., Watrin F., Roeckel N., Lalande M., Muscatelli F. The human MAGEL2 gene and its mouse homologue are paternally expressed and mapped to the Prader-Willi region. Hum. Mol. Genet. 1999;8:2497–2505. doi: 10.1093/hmg/8.13.2497. [DOI] [PubMed] [Google Scholar]
- 96.Jay P., Rougeulle C., Massacrier A., Moncla A., Mattei M.G., Malzac P., Roeckel N., Taviaux S., Lefranc J.L., Cau P., et al. The human necdin gene, NDN, is maternally imprinted and located in the Prader-Willi syndrome chromosomal region. Nat. Genet. 1997;17:357–361. doi: 10.1038/ng1197-357. [DOI] [PubMed] [Google Scholar]
- 97.MacDonald H.R., Wevrick R. The necdin gene is deleted in Prader-Willi syndrome and is imprinted in human and mouse. Hum. Mol. Genet. 1997;6:1873–1878. doi: 10.1093/hmg/6.11.1873. [DOI] [PubMed] [Google Scholar]
- 98.Sadakata T., Washida M., Iwayama Y., Shoji S., Sato Y., Ohkura T., Katoh-Semba R., Nakajima M., Sekine Y., Tanaka M., et al. Autistic-like phenotypes in Cadps2-knockout mice and aberrant CADPS2 splicing in autistic patients. J. Clin. Invest. 2007;117:931–943. doi: 10.1172/JCI29031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Gibson W.T., Harvard C., Qiao Y., Somerville M.J., Lewis M.E., Rajcan-Separovic E. Phenotype-genotype characterization of alpha-thalassemia mental retardation syndrome due to isolated monosomy of 16p13.3. Am. J. Med. Genet. A. 2008;146:225–232. doi: 10.1002/ajmg.a.32056. [DOI] [PubMed] [Google Scholar]
- 100.Pfeifer D., Poulat F., Holinski-Feder E., Kooy F., Scherer G. The SOX8 gene is located within 700 kb of the tip of chromosome 16p and is deleted in a patient with ATR-16 syndrome. Genomics. 2000;63:108–116. doi: 10.1006/geno.1999.6060. [DOI] [PubMed] [Google Scholar]
- 101.Stodgell C.J., Wanek N., O’Bara M., Hyman S.L., Bryson S.E., Rodier P.M. Association of HOXD1 and GBX2 allelic variants with autisms spectrum disorders. Am. J. Hum. Genet. 2003;71:2003. [Google Scholar]
- 102.Gecz J., Gedeon A.K., Sutherland G.R., Mulley J.C. Identification of the gene FMR2, associated with FRAXE mental retardation. Nat. Genet. 1996;13:105–108. doi: 10.1038/ng0596-105. [DOI] [PubMed] [Google Scholar]
- 103.Gu Y., Shen Y., Gibbs R.A., Nelson D.L. Identification of FMR2, a novel gene associated with the FRAXE CCG repeat and CpG island. Nat. Genet. 1996;13:109–113. doi: 10.1038/ng0596-109. [DOI] [PubMed] [Google Scholar]
- 104.Dibbens L.M., Tarpey P.S., Hynes K., Bayly M.A., Scheffer I.E., Smith R., Bomar J., Sutton E., Vandeleur L., Shoubridge C., et al. X-linked protocadherin 19 mutations cause female-limited epilepsy and cognitive impairment. Nat. Genet. 2008;40:776–781. doi: 10.1038/ng.149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Gecz J., Baker E., Donnelly A., Ming J.E., McDonald-McGinn D.M., Spinner N.B., Zackai E.H., Sutherland G.R., Mulley J.C. Fibroblast growth factor homologous factor 2 (FHF2): gene structure, expression and mapping to the Borjeson-Forssman-Lehmann syndrome region in Xq26 delineated by a duplication breakpoint in a BFLS-like patient. Hum. Genet. 1999;104:56–63. doi: 10.1007/s004390050910. [DOI] [PubMed] [Google Scholar]
- 106.Francks C., Maegawa S., Lauren J., Abrahams B.S., Velayos-Baeza A., Medland S.E., Colella S., Groszer M., McAuley E.Z., Caffrey T.M., et al. LRRTM1 on chromosome 2p12 is a maternally suppressed gene that is associated paternally with handedness and schizophrenia. Mol. Psychiatry. 2007;12:1129–1139, 1057. doi: 10.1038/sj.mp.4002053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Ikeda M., Iwata N., Kitajima T., Suzuki T., Yamanouchi Y., Kinoshita Y., Ozaki N. Positive association of the serotonin 5-HT7 receptor gene with schizophrenia in a Japanese population. Neuropsychopharmacology. 2006;31:866–871. doi: 10.1038/sj.npp.1300901. [DOI] [PubMed] [Google Scholar]
- 108.Tompers D.M., Labosky P.A. Electroporation of murine embryonic stem cells: a step-by-step guide. Stem Cells. 2004;22:243–249. doi: 10.1634/stemcells.22-3-243. [DOI] [PubMed] [Google Scholar]
- 109.Naiche L.A., Papaioannou V.E. Cre activity causes widespread apoptosis and lethal anemia during embryonic development. Genesis. 2007;45:768–775. doi: 10.1002/dvg.20353. [DOI] [PubMed] [Google Scholar]
- 110.Buerger A., Rozhitskaya O., Sherwood M.C., Dorfman A.L., Bisping E., Abel E.D., Pu W.T., Izumo S., Jay P.Y. Dilated cardiomyopathy resulting from high-level myocardial expression of Cre-recombinase. J. Card. Fail. 2006;12:392–398. doi: 10.1016/j.cardfail.2006.03.002. [DOI] [PubMed] [Google Scholar]
- 111.Lee J.Y., Ristow M., Lin X., White M.F., Magnuson M.A., Hennighausen L. RIP-Cre revisited, evidence for impairments of pancreatic beta-cell function. J. Biol. Chem. 2006;281:2649–2653. doi: 10.1074/jbc.M512373200. [DOI] [PubMed] [Google Scholar]
- 112.Schmidt-Supprian M., Rajewsky K. Vagaries of conditional gene targeting. Nat. Immunol. 2007;8:665–668. doi: 10.1038/ni0707-665. [DOI] [PubMed] [Google Scholar]
- 113.Forni P.E., Scuoppo C., Imayoshi I., Taulli R., Dastru W., Sala V., Betz U.A., Muzzi P., Martinuzzi D., Vercelli A.E., et al. High levels of Cre expression in neuronal progenitors cause defects in brain development leading to microencephaly and hydrocephaly. J Neurosci. 2006;26:9593–9602. doi: 10.1523/JNEUROSCI.2815-06.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Affymetrix. Santa Clara, CA: Affymetrix; 2004. [Google Scholar]
- 115.Gentleman R.C., Carey V.J., Bates D.M., Bolstad B., Dettling M., Dudoit S., Ellis B., Gautier L., Ge Y., Gentry J., et al. Bioconductor: open software development for computational biology and bioinformatics. Genome Biol. 2004;5:R80. doi: 10.1186/gb-2004-5-10-r80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Wu Z., Irizarry R.A., Gentleman R., Murillo F.M., Spencer F. A model based background adjustment for oligonucleotide expression arrays. J. Am. Stat. Assoc. 2004;99:909–917. [Google Scholar]
- 117.Gautier L., Cope L., Bolstad B.M., Irizarry R.A. affy—analysis of Affymetrix GeneChip data at the probe level. Bioinformatics. 2004;20:307–315. doi: 10.1093/bioinformatics/btg405. [DOI] [PubMed] [Google Scholar]
- 118.Smyth G.K. Linear models and empirical bayes methods for assessing differential expression in microarray experiments. Stat. Appl. Genet. Mol. Biol. 2004;3:Article3. doi: 10.2202/1544-6115.1027. [DOI] [PubMed] [Google Scholar]
- 119.Smyth G.K. In: Bioinformatics and Computational Biology Using R and Bioconductor. Gentleman R., Carey V.J., Dudoit S., Irizarry R., Huber W., editors. New York: Springer; 2005. pp. 397–420. [Google Scholar]
- 120.Benjamini Y., Hochberg Y. Controlling the false discovery rate: a practical and powerful approach to multiple testing. J R Statist Soc B. 1995;57:289–300. [Google Scholar]
- 121.de Hoon M.J., Imoto S., Nolan J., Miyano S. Open source clustering software. Bioinformatics. 2004;20:1453–1454. doi: 10.1093/bioinformatics/bth078. [DOI] [PubMed] [Google Scholar]
- 122.Saldanha A.J. Java Treeview—extensible visualization of microarray data. Bioinformatics. 2004;20:3246–3248. doi: 10.1093/bioinformatics/bth349. [DOI] [PubMed] [Google Scholar]
- 123.Livak K.J., Schmittgen T.D. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) Method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 124.Subramanian A., Kuehn H., Gould J., Tamayo P., Mesirov J.P. GSEA-P: a desktop application for Gene Set Enrichment Analysis. Bioinformatics. 2007;23:3251–3253. doi: 10.1093/bioinformatics/btm369. [DOI] [PubMed] [Google Scholar]
- 125.Barrett T., Troup D.B., Wilhite S.E., Ledoux P., Rudnev D., Evangelista C., Kim I.F., Soboleva A., Tomashevsky M., Edgar R. NCBI GEO: mining tens of millions of expression profiles—database and tools update. Nucleic Acids Res. 2007;35:D760–D765. doi: 10.1093/nar/gkl887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Saxena V., Orgill D., Kohane I. Absolute enrichment: gene set enrichment analysis for homeostatic systems. Nucleic Acids Res. 2006;34:e151. doi: 10.1093/nar/gkl766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Bryne J.C., Valen E., Tang M.H., Marstrand T., Winther O., da Piedade I., Krogh A., Lenhard B., Sandelin A. JASPAR, the open access database of transcription factor-binding profiles: new content and tools in the 2008 update. Nucleic Acids Res. 2008;36:D102–D106. doi: 10.1093/nar/gkm955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Cho G., Lim Y., Zand D., Golden J.A. Sizn1 is a novel protein that functions as a transcriptional coactivator of BMP signalling. Mol. Cell Biol. 2007;28:1565–1572. doi: 10.1128/MCB.01038-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Marklund N., Fulp C.T., Shimizu S., Puri R., McMillan A., Strittmatter S.M., McIntosh T.K. Selective temporal and regional alterations of Nogo-A and small proline-rich repeat protein 1A (SPRR1A) but not Nogo-66 receptor (NgR) occur following traumatic brain injury in the rat. Exp. Neurol. 2006;197:70–83. doi: 10.1016/j.expneurol.2005.08.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Lu M.M., Li S., Yang H., Morrisey E.E. Foxp4: a novel member of the Foxp subfamily of winged-helix genes co-expressed with Foxp1 and Foxp2 in pulmonary and gut tissues. Mech Dev. 2002;119(Suppl. 1):S197–S202. doi: 10.1016/s0925-4773(03)00116-3. [DOI] [PubMed] [Google Scholar]
- 131.Nasrallah I.M., Minarcik J.C., Golden J.A. A polyalanine tract expansion in Arx forms intranuclear inclusions and results in increased cell death. J. Cell Biol. 2004;167:411–416. doi: 10.1083/jcb.200408091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Siepel A., Bejerano G., Pedersen J.S., Hinrichs A.S., Hou M., Rosenbloom K., Clawson H., Spieth J., Hillier L.W., Richards S., et al. Evolutionarily conserved elements in vertebrate, insect, worm, and yeast genomes. Genome Res. 2005;15:1034–1050. doi: 10.1101/gr.3715005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Kim J.B., Wright H.M., Wright M., Spiegelman B.M. ADD1/SREBP1 activates PPARgamma through the production of endogenous ligand. Proc. Natl Acad. Sci. USA. 1998;95:4333–4337. doi: 10.1073/pnas.95.8.4333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Cho G., Kim J., Rho H.M., Jung G. Structure-function analysis of the DNA binding domain of Saccharomyces cerevisiae ABF1. Nucleic Acids Res. 1995;23:2980–2987. doi: 10.1093/nar/23.15.2980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Kent W.J., Sugnet C.W., Furey T.S., Roskin K.M., Pringle T.H., Zahler A.M., Haussler D. The human genome browser at UCSC. Genome Res. 2002;12:996–1006. doi: 10.1101/gr.229102. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.