Abstract
The mechanism by which the Parkinson’s disease-related protein α-synuclein (α-syn) causes neurodegeneration has not been elucidated. To determine the genes that protect cells from α-syn, we used a genetic screen to identify suppressors of the super sensitivity of the yeast Saccharomyces cerevisiae expressing α-syn to killing by hydrogen peroxide. Forty genes in ubiquitin-dependent protein catabolism, protein biosynthesis, vesicle trafficking and the response to stress were identified. Five of the forty genes—ENT3, IDP3, JEM1, ARG2 and HSP82—ranked highest in their ability to block α-syn-induced reactive oxygen species accumulation, and these five genes were characterized in more detail. The deletion of any of these five genes enhanced the toxicity of α-syn as judged by growth defects compared with wild-type cells expressing α-syn, which indicates that these genes protect cells from α-syn. Strikingly, four of the five genes are specific for α-syn in that they fail to protect cells from the toxicity of the two inherited mutants A30P or A53T. This finding suggests that α-syn causes toxicity to cells through a different pathway than these two inherited mutants. Lastly, overexpression of Ent3p, which is a clathrin adapter protein involved in protein transport between the Golgi and the vacuole, causes α-syn to redistribute from the plasma membrane into cytoplasmic vesicular structures. Our interpretation is that Ent3p mediates the transport of α-syn to the vacuole for proteolytic degradation. A similar clathrin adaptor protein, epsinR, exists in humans.
INTRODUCTION
Parkinson’s disease (PD) is a common movement disorder that affects individuals who are >60 years old (1–3). The disease is characterized by slowness of movement, resting tremor, rigidity and disturbances of gait and posture. At a cellular level, dopamine-producing neurons in an area of the mid-brain called the substantia nigra pars compacta slowly degenerate, resulting in a deficit of dopamine. Intracytoplasmic inclusions termed Lewy bodies are found in the affected neurons, and this is a defining feature of the disease (4,5). The two main components of Lewy bodies are lipids and the protein α-synuclein (α-syn) (6). The identification of α-syn as the main component of Lewy bodies sparked research into this protein as a possible cause of PD, and this hypothesis was subsequently reinforced by discoveries that mutation of α-syn (A30P, A53T or E46K) causes rare, autosomal dominant forms of PD (7–9) and that triplication of the α-syn locus causes PD (10). Mutations in other genes such as PARKIN, DJ-1, PINK1 and LRRK2 also cause PD (3).
α-Syn is an intrinsically unfolded protein containing 140 amino acid (Mw = 14.5 kDa) that is thought to regulate cell differentiation, synaptic plasticity and dopaminergic neurotransmission. α-Syn binds to lipids (11), inhibits phospholipase D (12) and forms a myriad of differently sized protofibrils and fibers (13). By unknown mechanisms, it induces the accumulation of reactive oxygen species (ROS) (14,15), causes proteasome dysfunction (16,17) and blocks endoplasmic reticulum (ER)–Golgi traffic (18).
A variety of genetic screens have been conducted using Caenorhabditis elegans (19,20) and Saccharomyces cerevisiae (18,21,22) to identify genes that protect cells from α-syn toxicity. Having been done in different ways and with different organisms, not surprisingly, little overlap exists in the different sets of protective genes that have been reported. One theme, elucidated using yeast, is that genes involved in lipid metabolism and vesicle trafficking are over represented as suppressors of α-syn toxicity (21,23). In this study, a novel hydrogen peroxide-based genetic screen was used to identify suppressors of the super sensitivity of yeast cells expressing wild-type (WT) α-syn to killing by hydrogen peroxide. The finding that the cytoplasmic degradation of dopamine by monoamine oxidase generates hydrogen peroxide, and that the flux of hydrogen peroxide is especially high in the Parkinson’s brain (24,25), reinforces the usefulness of conducting a genome-wide screen with hydrogen peroxide. Here we report the identification of 40 genes that, in high copy, suppress the toxicity of human WT α-syn. The genes fall into the categories of ubiquitin-dependent protein catabolism, protein biosynthesis, vesicle trafficking and the response to stress.
RESULTS
A hydrogen peroxide-based genetic screen identifies suppressors of WT α-syn toxicity
We previously reported, using a high copy yeast genomic library to find suppressors of the super sensitivity of A30P, α-syn expressing cells to killing by hydrogen peroxide (22). This same screen was used herein to identify suppressors of the super sensitivity of WT α-syn expressing cells to killing by hydrogen peroxide. The basis of the screen is that α-syn (WT, A30P or A53T) causes oxidative stress in a variety of cells (14), including yeast (15), which in turn results in the accumulation of ROS in the cytoplasm. Because cells expressing α-syn (WT, A30P or A53T) are burdened with endogenous oxidants, they cannot tolerate exogenous oxidants. To exploit this phenomenon, cells harboring pTF201 (WT α-syn) and a high copy yeast library were pre-grown in non-inducing media until mid-log phase and then shifted into inducing media and plated on solid agar plates that contained a disk of concentrated hydrogen peroxide. Such cells exhibited a sizeable halo of inhibition (∼3 cm diameter) surrounding the peroxide disk where colonies failed to grow (Fig. 1A). In contrast, cells harboring pTF201 (WT α-syn) and the library often exhibited colonies within this halo of inhibition (Fig. 1B). Library plasmids from such colonies were isolated and sequenced as described in the Materials and Methods section. Strains and plasmids used in this study are given in Table 1.
Figure 1.
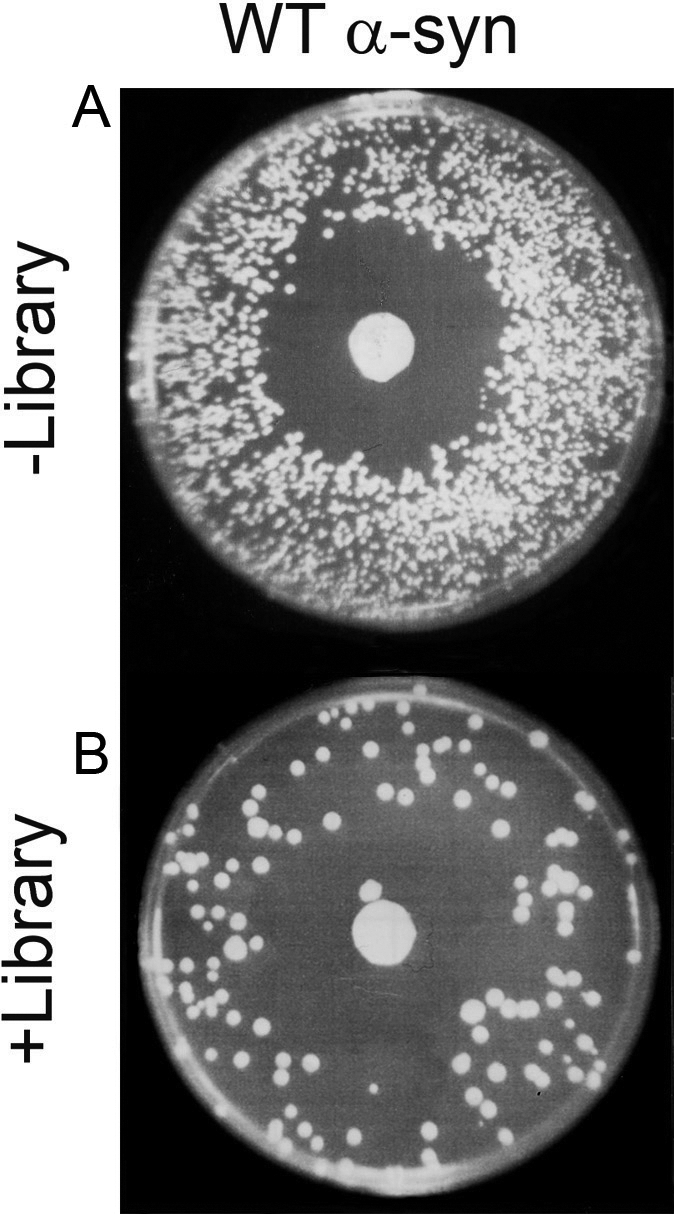
A genome-wide screen identifies suppressors of the super sensitivity of WT α-syn expressing cells to killing by hydrogen peroxide. (A) FY23 cells transformed with pTF201 (WT α-syn) were plated on SGal−Trp. The halo of inhibition is due to the concentrated hydrogen peroxide on the disk in the center of the plate. (B) Suppression of WT α-syn toxicity by a high-copy yeast genomic library. FY23 cells transformed with pTF201 (WT α-syn) and a 2µ yeast genomic library and plated on SGal−Trp−Leu. The disk in the center of each plate contained 10 µl of 8–10% hydrogen peroxide. Plates were incubated for 3 days at 30°C. In general, the relatively low transformation efficiency of the 2µ library resulted in fewer colonies per plate compared with the control plate.
Table 1.
Yeast strains and plasmids
| Strains | ||
| FY23 | MATa | ura3-52 trp1Δ63 leu2Δ1 |
| KT2253 | MATα | his3Δ1 leu2Δ0 met15Δ0 ura3Δ0 |
| arg2Δ | MATα | his3Δ1 leu2Δ0 met15Δ0 ura3Δ0 kanr::arg2 |
| ent3Δ | MATα | his3Δ1 leu2Δ0 met15Δ0 ura3Δ0 kanr::ent3 |
| hsp82Δ | MATα | his3Δ1 leu2Δ0 met15Δ0 ura3Δ0 kanr::hsp82 |
| idp3Δ | MATα | his3Δ1 leu2Δ0 met15Δ0 ura3Δ0 kanr::idp3 |
| jem1Δ | MATα | his3Δ1 leu2Δ0 met15Δ0 ura3Δ0 kanr::jem1 |
| Plasmids | ||
| pRS313 | Low copy CEN HIS3 ARS Ampr | ATCC |
| pRS314 | Low copy CEN TRP1 ARS Ampr | ATCC |
| pHY114-WT | GAL1 promoter GFP-WT α-syn in pRS314 | (15) |
| pTF200 | GAL1 promoter in pRS314 | (15) |
| pTF201 | GAL1 promoter WT α-syn in pRS314 | (15) |
| pTF202 | GAL1 promoter A30P α-syn in pRS314 | (15) |
| pTF203 | GAL1 promoter A53T α-syn in pRS314 | (15) |
| pJL100 | GAL1 promoter in pRS313 | This study |
| pJL101 | GAL1 promoter WT α-syn in pRS313 | This study |
| pJL102 | GAL1 promoter GFP in pRS313 | This study |
| pJL103 | GAL1 promoter GFP-WT α-syn in pRS313 | This study |
| pJL200 | 2µ URA3 AmprGAL1 promoter | This study |
| pARG2a | 2µ URA3 AmprGAL1 promoter ARG2 | Open Biosystems |
aSixty-six 2µ plasmids, which contained the 66 genes listed in Supplementary Material, Table S1, were purchased from Open Biosystems. Open Biosystems calls these plasmids BG1805. We refer to them by the gene name.
Using this approach, ∼100 protective library plasmids were recovered. Because library plasmids contained yeast chromosomal fragments, some of the library plasmids could contain more than one gene. Therefore, both strands from each protective plasmid were sequenced, and with this sequence information the identities of genes on each plasmid were determined using the Saccharomyces Genome Database (http://www.yeastgenome.org/). Some recovered plasmids contained more than one gene. Instead of subcloning all of the genes from the recovered library plasmids, many of the genes were available in expression vectors from Open Biosystems. In total, we purchased sixty-six 2µ expression vectors (Open Biosystems), which contained the genes of interest behind the GAL1 promoter (Supplementary Material, Table S1). To determine which genes conferred protection against WT α-syn toxicity, plasmids were introduced into the yeast strain FY23, and ROS accumulation was monitored using the dye dihydrorhodamine 123 (DHR123) (22). DHR123 is a non-fluorescent cell permanent dye that fluoresces when oxidized. The basis of this secondary screen was that any gene that protects against WT α-syn toxicity should decrease the amount of cytoplasmic ROS and therefore decrease the amount of DHR fluorescence. Conversely, any gene that does not protect should not affect the DHR fluorescence.
The ability of the 66 genes, identified in the hydrogen peroxide screen, to inhibit ROS accumulation in cells expressing WT α-syn was determined. Cells were transformed with pTF201 (WT α-syn) and with a 2µ plasmid (BG1805) containing one of the protective genes. Transformants were pre-grown in non-inducing media to mid-log phase and then shifted into inducing media. After 2 h, the DHR123 dye was added and cells were incubated for an additional hour. The controls consisted of cells transformed with pTF201 (WT α-syn) or various control plasmids with no inserts. Fluorescence, which correlates with cytoplasmic ROS accumulation, was detected using a standard plate reader.
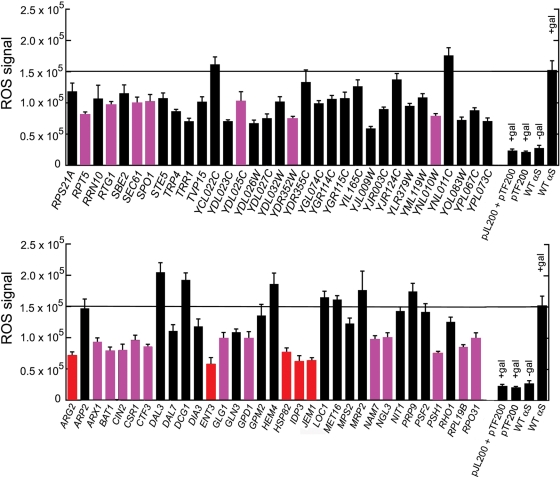
Figure 2 shows the results from the ROS fluorescence assay. First, yeast cells expressing WT α-syn (+gal) yielded an ROS fluorescence signal of 1.5 (±0.2) × 105 units, whereas yeast cells with no WT α-syn expression (−gal) yielded an ∼6-fold smaller ROS fluorescence signal of 2.7 (±0.5) × 104 units. Secondly, overexpression of 40 different genes decreased ROS accumulation by 30–60% (P << 0.05), compared with cells expressing WT α-syn only, and 24 of these have a human ortholog. These 24 protective yeast genes with human orthologs are given in Table 2. Thirdly, overexpression of several genes decreased ROS accumulation by 10–29%, compared with cells expressing WT α-syn only; however, we did not focus on these genes because of their relatively small effect. Fourthly, overexpression of nine genes had either no effect or slightly increased ROS accumulation, compared with cells expressing WT α-syn only. These nine genes were categorized as non-protective and were not evaluated any further. The complete list of genes and P-values is given in Supplementary Material, Table S1.
Figure 2.
ROS fluorescence assay of transformants with 66 yeast genes identified in the hydrogen peroxide screen as potential suppressors of WT α-syn toxicity. The y-axis is the fluorescence signal; the x-axis gives the gene name. The FY23 strain was transformed with pTF201 (WT α-syn) and pBG1805 (Open Biosystems 2µ plasmid with gene of interest) (see Supplementary Material, Table S1). Cells were pre-grown in sucrose drop out media to mid-log phase and then transferred into inducing media and incubated for 3 h at 30°C. The DHR123 dye (5 µg/ml) was added after 2 h in inducing media. The cells were washed several times in phosphate-buffered saline (PBS) and resuspended in PBS before measuring the ROS fluorescence signal. The excitation and emission wavelengths were 485 and 535 nm, respectively. As a control, FY23 was transformed with the pTF201 (WT α-syn), and these cells were treated identically to those described above. The ROS signal from these cells was 152 236 ± 15 177 and 27 382 ± 4710 units of fluorescence in inducing and non-inducing media, respectively. Note that FY23 cells transformed with pTF201 and pJL200 (empty vector) yielded 154 675 ± 2436 and 25 179 ± 895 units of fluorescence in inducing media and non-inducing media, respectively. Other empty vector controls (pTF200 or pTF200 and pJL200) are also shown. Values for the 24 genes with human orthologs are shown in color (magenta and red); values for the 5 genes that were characterized in more detail are shown in red.
Table 2.
Yeast genes with human orthologs that suppress WT α-syn toxicity
| Gene | |
|---|---|
| ENT3 | Vesicle trafficking (Golgi–vacuole) |
| IDP3 | Isocitrate dehydrogenase (peroxisomal variant) |
| JEM1 | DnaJ-like ER membrane protein |
| ARG2 | Mitochondrial enzyme in arginine synthesis |
| YDR352w | PQ-loop protein |
| PSH1 | Nuclear protein, putative RNA polymerase II elongation factor |
| HSP82 | Hsp90 chaperone |
| YNL010w | Possible phosphatase |
| BAT1 | Mitochondrial aminotransferase |
| CIN2 | Tubulin folding factor |
| RPT5 | One of six ATPases of the 19S regulatory particle of the 26S proteasome |
| RPL19b | Protein component of the large (60S) ribosomal subunit |
| CTF3 | Outer kinetochore protein; may bind the kinetochore to spindle microtubules |
| ARX1 | Shuttling pre-60S factor; involved in the biogenesis of ribosomal large subunit biogenesis |
| CSR1 | Phosphatidylinositol transfer protein with a potential role in lipid turnover |
| RTG1 | Transcription factor (bHLH) involved in interorganelle communication between organelles |
| NAM7 | ATP-dependent RNA helicase of the SFI superfamily, |
| SEC61 | Retrograde transport of misfolded proteins out of the ER |
| GPD1 | NAD-dependent glycerol-3-phosphate dehydrogenase, key enzyme of glycerol synthesis |
| RPO31 | RNA polymerase III subunit C160, part of core enzyme |
| GLG1 | Self-glucosylating initiator of glycogen synthesis |
| NGL3 | Putative endonuclease |
| SPO1 | Meiosis-specific protein with similarity to phospholipase B |
| YDL025c | Putative protein kinase, potentially phosphorylated by Cdc28p |
Deletion of protective genes enhances wt α-syn toxicity
ARG2, ENT3, IDP3, JEM1 and HSP82 were chosen for a more detailed analysis. These non-essential genes with human orthologs were chosen because four of them, ARG2, ENT3, IDP3 and JEM1, conferred the greatest protection against WT α-syn-induced ROS, and because HSP82 encodes for a ubiquitous chaperone (Hsp90p) that, in yeast, is required for pheromone signaling and negative regulation of heat shock factor, Hsf1p (26). Arg2p is a mitochondrial enzyme that catalyzes the first step in the biosynthesis of ornithine, which is a precursor of arginine (27). Ent3p is involved in clathrin recruitment and protein transport between the trans-Golgi network (TGN) and the vacuole (28). Idp3p is a peroxisomal variant of the enzyme isocitrate dehydrogenase (29). Jem1p is a DnaJ-like chaperone that localizes to the ER and is required for nuclear membrane fusion during mating (30).
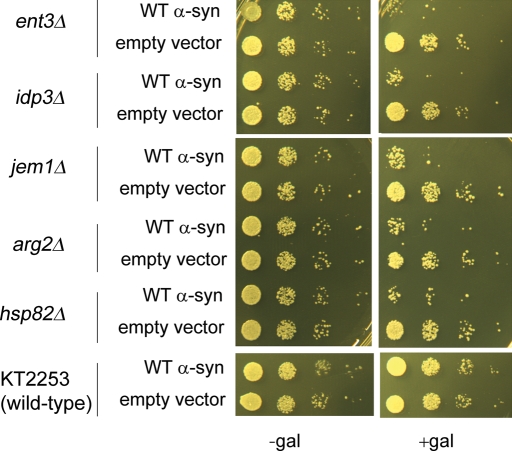
Given that these five genes in high copy suppressed the toxicity of WT α-syn, we reasoned that the deletion of any of them should enhance the toxicity of WT α-syn. Figure 3 shows a viability assay in which the five haploid deletion strains and the parental wild-type haploid strain KT2253 were transformed with WT α-syn (pJL101) or a control plasmid with no insert (pJL100), serially diluted and spotted on sucrose and galactose plates. In all cases, without WT α-syn expression (−gal), growth of cells that contained the WT α-syn plasmid resembled that of cells containing the control plasmid. In contrast, the five haploid deletion strains (ent3Δ, idp3Δ, jem1Δ, arg2Δ and hsp82Δ) expressing WT α-syn grew much more slowly than the deletion strains harboring the control plasmid (+gal). The five haploid deletion strains expressing WT α-syn also grew much more slowly compared with the wild-type strain (KT2253) expressing WT α-syn (+gal). No appreciable difference in growth was observed for the wild-type strain (KT2253) expressing WT α-syn (+gal) versus the control plasmid with no insert (+gal), indicating that cells can tolerate one copy of WT α-syn. The combined results demonstrate that the deletion of these various genes enhances the toxicity of WT α-syn and suggests a specific protective function of the various gene products.
Figure 3.
Growth properties of cells expressing WT α-synuclein. The effect of WT α-synuclein expression on the growth of five deletion strains (ent3Δ, idp3Δ, jem1Δ, arg2Δ and hsp82Δ) and the WT control strain KT2253 was evaluated. These strains were transformed with pJL101 (WT α-syn) or pJL100 (empty vector) and spotted in successive 10-fold serial dilutions on solid sucrose and solid galactose plates and incubated for 3 days at 30°C. Experiments were repeated three times with similar results.
The five deletion strains ent3Δ, idp3Δ, jem1Δ, arg2Δ and hsp82Δ expressing WT α-syn were also tested for ROS accumulation and for complementation of any ROS increase by expression of the corresponding gene. Similar to the experiment shown in Figure 2, cells were pre-grown in non-inducing media until mid-log phase and then shifted into inducing media. The ROS signal was measured after 3 h in inducing media. Each strain was transformed with pJL101 (WT α-syn) and a 2µ plasmid (Open Biosystems) with the gene of interest or pJL200 (empty 2µ control plasmid) (Table 1).
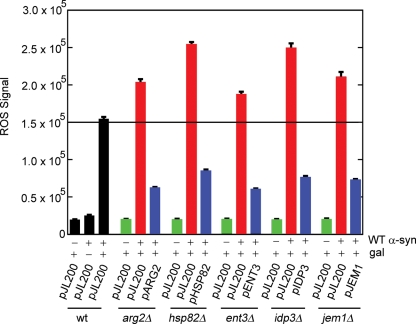
Figure 4 shows the results from the rescue experiments. First, the control strain KT2253 expressing WT α-syn (pJL101) and harboring a 2µ empty plasmid (pJL200) yielded an ROS signal of 154 675 ± 2436 units. In non-inducing media, this value decreased 84% to 25 179 ± ±895 units. Secondly, arg2Δ, ent3Δ, hsp82Δ, idp3Δ and jem1Δ deletion strains expressing WT α-syn each exhibited an ROS signal greater than the control strain. For example, arg2Δ cells expressing WT α-syn exhibited an ROS signal of 203 971 ± 3699 units, which is a 32% increase compared with the control strain (154 675→203 971). The percentage increases in ROS signal compared with the WT control strain were 32% (arg2Δ), 65% (ent3Δ), 21% (hsp82Δ), 62% (idp3Δ) and 37% (jem1Δ). This assay demonstrated that the deletion of genes that protect against WT α-syn toxicity indeed significantly increased the accumulation of ROS. Thirdly, expression of the gene of interest blocked accumulation of ROS, as expected. For example, arg2Δ cells expressing WT α-syn exhibited an ROS signal of 203 971 ± 3699 units, whereas the same cells but with ARG2 added back (pARG2) exhibited a 69% decrease in ROS signal to 62 690 ± ±680 units. On average, adding back the gene of interest in high copy caused a 67% decrease in ROS accumulation in the various deletion strains expressing WT α-syn. The combined results provided further support that these five genes protect cells from WT α-syn toxicity.
Figure 4.
ROS accumulation in ent3Δ, idp3Δ, jem1Δ, arg2Δ and hsp82Δ deletion strains expressing WT α-syn. The fluorescence assay using the dye DHR123 was used to measure the ROS accumulation in WT α-synuclein expressing cells after 3 h incubation in inducing media at 30°C. The WT haploid strain KT2253 transformed with pJL101 (WT α-syn) and pJL200 (empty vector) exhibited an ROS signal of 154 675 ± 2436 and 25 179 ± 895 in inducing media (+gal) and non-inducing media (−gal), respectively. KT2253 transformed with two empty vectors (pJL100 and PJL200) (+gal) exhibited an ROS signal of 19 763 ± 349. The arg2Δ strain transformed with pJL101 (WT α-syn) and pJL200 (red bar) or pARG2 (2µ Open Biosystems plasmid containing ARG2) (blue bar) were treated identically to the WT control strain, and ROS was measured at 3 h. The ROS signal from the arg2Δ strain transformed with two empty vectors (pJL100 and pJL200) is represented by the green bar. The other four deletion strains were prepared and analyzed in the same way as arg2Δ.
ARG2, ENT3, IDP3, HSP82 and JEM1 fail to inhibit ROS accumulation in cells expressing A30P or A53T
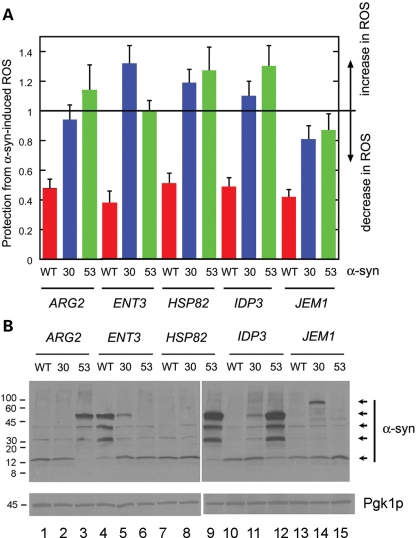
A recent study showed that WT α-syn and A53T, but not A30P, transit through the classical secretory pathway in yeast and target to the plasma membrane (31), whereas another study showed that the yeast gene YPT1 is a high copy suppressor of the toxicity of WT α-syn and A53T but not of A30P (18). In light of these findings, we hypothesized that ARG2, ENT3, IDP3, HSP82 and JEM1 would protect cells from WT α-syn and A53T, but not A30P. We thus tested whether these five genes in high copy would protect cells from ROS induced by these two inherited α-syn mutants. The other inherited mutant, E46K, was not evaluated. We have previously shown that A30P and A53T cause ROS accumulation in yeast (15). Cells were transformed with plasmids for WT α-syn (pTF201), A30P (pTF202) or A53T (pTF203) and with an Open Biosystems plasmid containing one of the five genes of interest, pre-grown in non-inducing media to mid-log phase, shifted into inducing media for 3 h, and assayed for ROS accumulation using DHR123. Matched controls were identically treated cells expressing WT α-syn but without the gene of interest in high copy. The bars in Figure 5 represent the ratio of the two signals: 1 indicates no decrease in ROS, <1 indicates a decrease in ROS and >1 indicates an increase in ROS.
Figure 5.
(A) ARG2, ENT3, HSP82, IDP3 and JEM1 in high copy fail to suppress the toxicity of A30P or A53T. The fluorescence assay using DHR123 was used to measure the ROS accumulation in cells expressing various α-synucleins (WT, A30P or A53T). For example, the FY23 strain was transformed with pARG2 (2µ plasmid with ARG2) and with pTF201 (WT α-syn), pTF202 (A30P) or pTF203 (A53T). Matched controls consisted of the FY23 strain transformed with pTF201 (WT α-syn), pTF202 (A30P) or pTF203 (A53T). Cells were pre-grown in non-inducing media, shifted into inducing media and then incubated for 3 h. The DHR123 dye (5 µg/ml) was added after 2 h in inducing media, and the ROS signal was measured 1 h later. Prior to measuring the fluorescence signal, the cells were washed and resuspended in PBS. Each bar in the graph represents the ratio: average ROS signal (multi-copy gene)/average ROS signal (single copy gene). A value of 1 indicates no protection; a value <1 indicates a decrease in ROS; and a value >1 indicates an increase in ROS. The average signals were obtained from triplicate samples recorded on three different days. Error bars were determined by to the method described in Bevington (64). (B) Western blot analysis of α-syn expression in the cultures as shown in (A). Lysates were prepared and then subjected to SDS–PAGE followed by western blot analysis. The cell-signaling monoclonal antibody against α-syn was used to visualize the three α-syns in yeast cells overexpressing the five genes of interest. The loading control was the yeast protein Pgk1p. Identical amounts of protein were loaded per well.
ARG2, ENT3, HSP82, IDP3 and JEM1 in high copy significantly blocked ROS in cells expressing WT α-syn (Fig. 5A). On average, high copy ARG2, ENT3, HSP82, IDP3 and JEM1 resulted in a 50–60% decrease in ROS accumulation. Strikingly, ARG2, ENT3, HSP82 and IDP3 in high copy had no protective effect on cells expressing A30P or A53T, i.e. for these samples the normalized ROS signal was ≥1. JEM1 in high copy produced a slight decrease in ROS accumulation in cells expressing A30P (S = 0.81 ± 0.09), whereas no appreciable decrease in ROS accumulation was observed for cells expressing A53T. We conclude that although the three α-syns (WT, A30P and A53T) each produce oxidative stress in cells, the pathobiology of these three α-syns must differ (see ‘ENT3 and the vacuole’ in the Discussion section).
To complement the above ROS experiments, western blot analysis was conducted to assess the expression levels of the various α-syns (WT, A30P or A53T) in the FY23 strain overexpressing Arg2p, Ent3p, Hsp82p, Idp3p or Jem1p. A monoclonal antibody that binds to each of the three forms of α-syn was used to visualize α-syn. Pgk1p was used as a loading control. The results are shown in Figure 5B.
First, consider the western blot of cells overexpressing Arg2p and expressing WT α-syn, A30P or A53T (Fig. 5B; lanes 1–3). The bands due to WT α-syn (lane 1) and A30P (lane 2) at 14.5 kDa have similar intensities; thus, it may be inferred that the expression levels of these two α-syns are very similar. In contrast, cells overexpressing Arg2p and expressing A53T display an intense band at 54 kDa and a less intense band at 39 kDa, which we assign to aggregated or modified A53T. Secondly, in three of the five experiments with genes in high copy (ARG2, HSP82 and IDP3), the A53T protein migrated as three species, i.e. 26, 39 and 54 kDa (lanes 3, 9 and 12). The intensities of these three A53T bands are greater than the intensity of the bands at 14.5 kDa due to monomeric α-syn. The results suggest that under some conditions A53T aggregates accumulate in cells compared with the other two α-syns (WT and A30P). A53T indeed rapidly forms large inclusions when expressed in yeast (15,32). Thirdly, in one case, specifically, for cells overexpressing Ent3p, the WT α-syn protein migrated as three high molecular mass species (26, 39 and 54 kDa) (lane 4), whereas A30P and A53T migrated as monomers (lanes 5 and 6). Note that oligomeric forms of WT α-syn have been previously detected using the yeast model (33). Overall, the western blot results demonstrate that α-syn (WT, A30P and A53T) is indeed expressed in the presence of the various genes in high copy, and that, in general, the A53T variant may accumulate to a greater extent than the other two α-syns. However, the five genes specifically protect only against ROS in cells expressing WT α-syn (Fig. 5A).
Ent3p alters the localization of GFP-WT α-syn
Given that ENT3 was the strongest suppressor and that it has a human ortholog (epsinR), we sought to determine the effect of Ent3p overexpression on the localization of GFP-WT α-syn by fluorescence microscopy. Yeast cells expressing GFP-WT α-syn displayed bright green fluorescence around the perimeter of the cells at 3 and 12 h of incubation in inducing media (Fig. 6; upper panels). This result is consistent with the association of the GFP-WT α-syn protein with the plasma membrane, as has been reported by numerous groups (15,31,32). Identically treated cells expressing GFP-WT α-syn, but also overexpressing Ent3p, displayed a similar membrane association of GFP-WT α-syn at 3 h (Fig. 6; lower left panel), whereas cells displayed numerous GFP-WT α-syn inclusions (puncta) at 12 h (Fig. 6; lower right panel). For these cells overexpressing Ent3p, it is noteworthy that only 0.5% (n = 714) of the cells displayed one or more GFP-WT α-syn inclusions at 3 h, whereas 84% (n = 1562 cells) of the cells displayed one or more GFP-WT α-syn inclusions at 12 h. A possible role of ENT3 in detoxifying yeast cells of WT α-syn is considered in the Discussion section.
Figure 6.
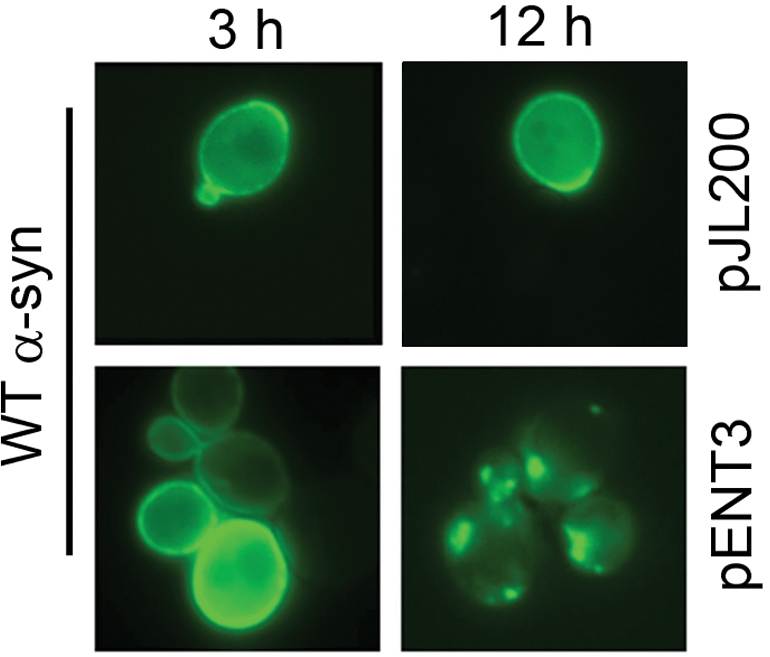
ENT3 alters the localization of WT α-syn. The KT2253 strain was transformed with pJL103 (GFP-WT α-syn) and pENT3 or the empty vector pJL200. Cells were pre-grown in non-inducing media until mid-log phase, washed and resuspended in inducing media, and incubated for 12 h at 30°C. Cells were visualized by fluorescence microscopy at 3 and 12 h as discussed in the Materials and Methods section. The top row shows cells expressing GFP-WT α-syn with pJL200 control plasmid; the bottom row shows cells expressing GFP-WT α-syn with ENT3 in high copy (pENT3).
DISCUSSION
We have identified 24 yeast genes with human orthologs that suppress the toxicity of WT α-syn. The genes fall into several categories, including ubiquitin-dependent protein catabolism, protein biosynthesis, vesicle trafficking and the response to stress.
Some of the genes we identified are similar to genes identified in an earlier genetic screen using S. cerevisiae. The pioneering study by Willingham et al. (21) used the yeast collection of 4852 haploid mutants containing deletions of non-essential genes to identify those mutants that were sick or synthetically lethal with WT α-syn. Although no overlap exists between the 86 genes identified by Willingham et al. and the 40 genes identified by us, intriguing similarities should be noted. They identified PEX2 and PEX8, which encode for peroxisome biogenesis proteins. We identified IDP3, which is an enzyme that functions to maintain high levels of the key co-factor NADPH in the interior of peroxisomes (see ‘IDP3 and the β-oxidation of lipids’ section). Willingham et al. identified four genes—SAC2, VPS24, VPS28 and VPS60—that are involved in protein sorting in the late Golgi, sorting to the prevacuolar endosomes and for protein sorting and trafficking to the vacuole, respectively. We identified ENT3, which is involved in protein transport between the trans-Golgi network (TGN) and the vacuole (see ‘ENT3 and the vacuole’ section). These combined results show the importance of vesicle-trafficking genes and peroxisomal genes as modifiers/regulators of α-syn toxicity.
Some of the genes we identified are similar to genes identified in a recent genetic screen using C. elegans. One study (19) identified a molecular chaperone of the DnaJ superfamily (T05C3.5, dnj-19) and genes associated with cargo transport and membrane trafficking (C54H2.5, sft-4 and F57B10.5, Emp24/gp25L/p24). We also indentified a molecular chaperone of the DnaJ superfamily (JEM1) and one gene associated with cargo transport and membrane traffic (ENT3). Another study using C. elegans (20) identified Hsp90 as a suppressor of WT α-syn toxicity; we identified the same gene.
Arg2
In yeast, Arg2p, which is located in the mitochondrial matrix, catalyzes the first step in the biosynthesis of the arginine precursor ornithine (27,34,35). Once synthesized, ornithine is transported into the cytosol, where it is converted by several enzymes into arginine. The transcription of the genes involved in arginine biosynthesis, including ARG2, is governed by the evolutionarily conserved transcription factor Gcn4p (36,37). It is noteworthy that Gcn4p also plays an essential role in the unfolded protein response (UPR) in yeast and higher eukaryotes. Specifically, Gcn4p and its activator Gcn2p are required for induction of a majority of UPR target genes during ER stress (38). WT α-syn transits through the classical secretory pathway in yeast and mammalian cells and induces ER stress and activates the UPR (31,39). We therefore reason that in response to α-syn-induced ER stress Gcn4p activates numerous UPR genes that serve to counteract such stress and propose that one of these genes is ARG2. We also point out that a recent screen of the S. cerevisiae yeast deletion collection for mutants that affect the telomere length identified 150 candidate genes not previously known to affect the telomere length, one of these mutants was arg2Δ (40). Telomeres are shorter in the arg2Δ strain compared with a WT strain. How arginine metabolism affects telomere length and α-syn toxicity is not known.
In mammals, NAGS synthesizes carbamyl phosphate synthetase and controls the formation of citrulline during the urea cycle (27). The urea cycle functions to remobilize nitrogen atoms from arginine (a nitrogen-storage amino acid), which may be important for cell rescue during oxidative stress (41), and perhaps even ER stress.
Chaperones
HSP82 and JEM1 were identified as high copy suppressors of the toxicity of WT α-syn. HSP82 encodes for a ubiquitous chaperone Hsp90 that, in yeast, is required for pheromone signaling and the negative regulation of heat shock factor (Hsf1p) (42,43). Hsp90 binds to transcription factors, kinases and other chaperones to regulate various signaling pathways (44). The overexpression of Hsp90 could protect yeast from the toxicity of WT α-syn in at least two different ways. First, Hsp90 might bind to WT α-syn, and this complexation could prevent the formation of toxic oligomers of WT α-syn (45). Note that the overexpression of HSP70 has been shown to protect against ROS induced by the expression of WT α-syn (15,25,46). Secondly, perhaps, Hsp90 overexpression protects cells from WT α-syn because of its ability to bind kinases (44). In some but not all models phosphorylation of WT α-syn at serine 129 enhances the toxicity of this protein (47–49). Perhaps, Hsp90, when overexpressed, binds to the kinase or kinases that phosphorylate WT α-syn, and this complexation prevents the phosphorylation of WT α-syn and thus protects cells from the toxic phosphorylated form of the protein.
The Jem1p protein is a member of the DnaJ superfamily, and it is located in the ER. DnaJ proteins bind unfolded proteins and deliver them to Hsp70 chaperones for holding or refolding (50). Yeast express a variety of different DnaJ proteins; these isoforms may reside in different cellular compartments and bind different substrates. Given that WT α-syn transits through the secretory pathway in yeast and human cells and is known to cause ER stress (31,39), as mentioned above, perhaps Jem1p functions to ameliorate the ER stress caused by WT α-syn.
IDP3 and the β-oxidation of lipids
Saccharomyces cerevisiae express mitochondrial (Idp1p), cytosolic (Idp2p) and peroxisomal (Idp3p) isoforms of isocitrate dehydrogenase (29,51). Two classes of this enzyme exist: one class uses NAD+ as an electron acceptor, whereas the other class uses NADP+ as an electron acceptor. Peroxisomal Idp3p is a NADP+-dependent isocitrate dehydrogenase that oxidizes isocitrate to yield α-ketoglutarate, NADPH and CO2. In yeast and many other organisms, the β-oxidation of lipids is confined to peroxisomes, whereas in mammals it occurs in peroxisomes and mitochondria. The oxidation of unsaturated fatty acids in yeast peroxisomes requires several enzymes, one of which is the NADPH-dependent 2,4-dienoyl-CoA reductase. As lipid molecules are oxidized within peroxisomes, the level of NADPH gradually decreases to a threshold concentration under which lipid oxidation ceases, unless NADPH is replenished. However, because the peroxisomal membrane is impermeable to pyrimidine nucleotides, there is no way to replenish NADPH. It was thus proposed that Idp3p replenishes peroxisomal NADPH through its oxidation of isocitrate (29); thus, IDP3 is linked to lipid metabolism.
Human cells also express mitochondrial, cytosolic and peroxisomal isoforms of isocitrate dehydrogenase. Human PICD was recently identified as an ortholog of yeast IDP3 (52). The PICD protein is 59% identical to Idp3p, it catalyzes the oxidative decarboxylation of isocitrate and it localizes to the cytosol and peroxisomes of human and rat liver cells. Similar to yeast, PICD probably replenishes peroxisomal pools of NADPH, which is a necessary co-factor for lipid oxidation (52). Given that cells expressing WT α-syn accumulate lipid droplets (32), perhaps IDP3 and PICD help cells metabolize accumulating lipids.
ENT3 and the vacuole
Ent3p mediates protein transport from the TGN to the vacuole in S. cerevisiae (28,53–56). Ent3p and its human counterpart epsinR (57) appear to be accessory proteins that interact with the clathrin adapter proteins AP-1/GGAs, which are proteins involved in clathrin-mediated anterograde and retrograde protein transport between the Golgi and endosomes. Ent3p and epsinR contain an amino-terminal phosphoinositide-binding domain, which enables binding to lipids, and each contains a clathrin-binding motif (58). One hypothesis to be tested in the future is that Ent3p, within the TGN, causes sorting of WT α-syn into endosomal compartments that then transit to the vacuole for degradation. Perhaps, the same role is played by epsinR in detoxifying human cells of WT α-syn, i.e. epsinR causes sorting of WT α-syn into endosomal compartments that then transit to the lysosome (mammalian equivalent of the vacuole), where WT α-syn is degraded.
We previously identified the essential yeast gene YPP1 as a high copy suppressor of A30P toxicity but not of WT α-syn or A53T (22). The Ypp1 protein, when over expressed, drives each of the three α-syns into vesicles that bud off the plasma membrane, but only A30P-containing vesicles traffic to and merge with the vacuole, where A30P is proteolytically degraded. We showed that Ypp1p binds to A30P but not the other two α-syns; that YPP1 interacts with genes involved in endocytosis/actin dynamics (SLA1, SLA2 and END3), protein sorting (class E vps), and vesicle–vacuole fusion (MON1 and CCZ1) to dispose of A30P and that YPP1 also participates in pheromone-triggered receptor-mediated endocytosis. We point out that Ypp1p and Ent3p are probably cargo proteins involved in protein transport to the vacuole but through different pathways. Ypp1p is specific for A30P and mediates its transport from the plasma membrane to the vacuole, whereas Ent3p is specific for WT α-syn and mediates its transport from the TGN to the vacuole.
WT α-syn and the inherited mutants
The five best suppressors against WT α-syn toxicity, as judged by ROS accumulation, did not appreciably decrease the ROS burden in cells expressing A30P or A53T (Fig. 5). This was unexpected in that WT α-syn and A53T (but not A03P) each appears to transit through the classical secretory pathway en route to the plasma membrane of yeast cells (31), and thus we had hypothesized that the genes protecting against WT α-syn would also protect against A53T. Instead, ARG2, ENT3, HSP82, IDP3 and JEM1 exhibit significant protection against ROS only in cells expressing WT α-syn. One interpretation of this finding is that WT α-syn, A53T and perhaps even A30P cause toxicity to cells by the same mechanism. In this model, overexpression of any one of these five genes inhibits ROS in cells expressing WT α-syn but not A30P or A53T because of subtle differences between the two mutants and the WT protein. For example, perhaps because of structural differences in WT α-syn compared with A53T, Ent3p sorts WT α-syn but not A53T from the TGN into endosomes that then transit to the vacuole. The overexpression of Ent3p would then protect cells from WT α-syn but not the two mutants—although the mechanism by which the three proteins cause toxicity to cells is the same. Another interpretation is that these three α-syns cause toxicity to cells by different mechanisms. Thus, three different diseases would be associated with α-syn (WT, A30P or A53T), with perhaps three different treatments. It would not be surprising in this model that genes that protect against WT α-syn, for example, would not protect cells from the toxicity due to A30P or A53T.
In summary, we have identified several novel genes that in high copy suppress the toxicity of WT α-syn in yeast cells. Many of these genes have human orthologs. In the future, we hope to use yeast to elucidate in more detail the pathways available to cells to degrade WT α-syn and its inherited mutants. Perhaps, novel small molecules can be discovered using yeast that will stimulate the various degradative pathways.
MATERIALS AND METHODS
Strains and media
The yeast strains used in this study were FY23 (59), KT2253 and various deletion strains from the haploid yeast deletion collection (Table 1; Invitrogen, Carlsbad, CA, USA). KT2253 is a meiotic spore clone from diploid strain Y23175 (60) in the BY4743 strain background; KT2253 is congenic to the mutant and parental strains of the yeast deletion collection. Cells transformed with galactose-inducible plasmids were pre-grown in synthetic sucrose (2% w/v) drop out media to maintain the selection for plasmids. The medium containing sucrose is referred to as non-inducing medium. α-Syn expression was induced in the same drop out media with galactose (2% w/v) replacing sucrose (61). The medium containing galactose is referred to as inducing medium. The various deletion strains used in this study were cultured in non-inducing medium with selection for plasmids supplemented with 0.4 g/l G418 (Geneticin) or inducing media with selection for plasmids supplemented with 0.4 g/l G418. The high copy yeast genomic library was YEp13 based (LEU2) (62). In every experiment in this study, cells were pre-grown in non-inducing media (sucrose) with selection for plasmids to mid-log phase and then shifted to inducing media (galactose) with selection for plasmids and induced for 3 h. In some cases, 12 h inductions were used. Cells were grown at 30°C.
Plasmids
Plasmids harboring untagged or GFP-tagged α-syn (WT, A30P or A53T) are described elsewhere (15). Plasmids were constructed for this study as follows. The control plasmid pJL100 (no insert) was made by amplifying the GAL1 promoter from the plasmid pHY114-WT α-syn using the forward and reverse primers 5′-TTGTCGACATGGATATCAAAAAAGAGGA-3′ and 5′-GGCGCTTAAGCTTCATGAAAGTGAAACA-3′, which contained SalI and EcoRI restriction sites, respectively. The PCR product was subcloned into pRS313 using these same restriction sites. The plasmid pJL101 (WT α-syn) was made by PCR amplifying the WT α-syn gene from pHY114-WT α-syn (Table 1) using the forward and reverse primers 5′-GTGACTAGTATGGATGTATTCATG-3′ and 5′-TCTCTCGAGTAAGGCTTCAGGTTCG-3′, which contained SpeI and SacI restriction sites, respectively. The PCR product was then subcloned into these sites in the pRS313 vector. The plasmid pJL102 (GFP) was made by PCR amplifying the GFP gene from pHY114-WT α-syn using the forward and reverse primers 5′-TTGTCGACATGGATATCAAAAAAGAGGA-3′ and 5′-TTACCTAGGATTAAAACAAATTGAAATTC-3′, which contained SalI and BamHI restriction sites, respectively. The PCR product was then subcloned into the pRS313 vector using the same restriction sites. The pJL103 (GFP-WT α-syn) plasmid was made by PCR amplifying the GFP-WT-α-syn insert from pHY114 (GFP-WT α-syn) using the forward and reverse primers 5′-GTGACTAGTATGGATGTATTCATG-3′ and 5′- TCTCTCGAGTAAGGCT-TCAGGTTCG-3′, which contained SpeI and SacI restriction sites, respectively. The PCR product was then subcloned into pRS313 using the same sites. The control plasmid pJL200 (no insert) was made by digesting the pARG2 Open Biosystems plasmid using the EagI restriction enzyme (New England Biolabs, Ipswich, MA, USA); this digestion released ARG2. The purified linear plasmid was ligated using T4 ligase and then propagated in DH5α according to the standard protocols (63). Sixty-six 2µ plasmids harboring the genes of interest (see Supplementary Material, Table S1) were purchased from Open Biosystems; each of these plasmids was subjected to DNA sequencing at the Iowa State University DNA facility (http://www.dna.iastate.edu/) to verify the authenticity of the insert. Likewise, the plasmids constructed for this study were each subjected to DNA sequencing to verify the authenticity of the inserts.
Hydrogen peroxide-based genetic screen
Cells transformed with pTF201 (WT α-syn) and the high copy yeast genomic library and induced with galactose were grown to 1.5 × 107 cells/ml and then plated on SGal–Leu–Trp. A 3/8 in. disk of sterile Whatman filter paper containing 10 µl of 8% H2O2 was placed in the center of each plate, and plates were incubated for 3 days. Colonies growing within ∼3 cm diameter from the peroxide disk were selected, cultured overnight and the plasmid DNA was isolated and then amplified in DH5α cells. Both strands of each protective plasmid were sequenced. Approximately 400 plates were screened.
Microscopy
Fluorescence microscopy was performed with an Olympus AX70 microscope, including an Olympus UPlanFl 100×/1.35 NA objective with Roper CoolSNAP HQ CCD camera. The acquisition software was IPLab v.3.6 from Scanalytics Inc. An Olympus U-MWG (510–550) filter set was used for detecting GFP. Data were collected at room temperature.
ROS plate reader assay
A Perkin Elmer Wallac Victor3 1420 multilabel counter was used to screen α-syn expressing yeast cells for ROS accumulation. Cells transformed with the plasmid for α-syn (WT, A30P or A53T) were pre-grown in non-inducing media until mid-log phase and then shifted into galactose-inducing media and induced for 3 h. After the second hour of induction, the DHR123 dye was added to a final concentration of 5 µg/ml. Prior to adding the cells to the 96-well plates, cells were washed twice in phosphate-buffered saline (PBS), and the assay was conducted in PBS. The excitation and emission wavelengths were 485 and 535 nm, respectively. The optical density of the culture in each well was also measured at 560 nm. The ‘ROS signal’ from each well is the ratio of the fluorescence signal divided by the optical density signal, i.e. S = F(535 nm)/OD(560 nm). Experiments were conducted in three independent experiments, each in triplicate.
Western blot analysis
Western blots were conducted as described previously (15), with minor modifications. In brief, yeast cells transformed with plasmids for the various α-syns (pTF201-WT, pTF202-A30P or pTF203-A53T) and the Open Biosystem plasmid harboring the gene of interest were cultured in non-inducing media to mid-log phase and then shifted into inducing media for 3 h at 30°C. Cells were lysed (and all subsequent manipulations were carried out at ice temperature), protease inhibitors were added and then the protein concentration was determined (Bio-Rad protein assay). Samples (80 µg/well) were subjected to SDS–PAGE and then transferred to a membrane for detection of the various α-syns. The monoclonal antibody against human α-syn was purchased from Cell Signaling Technologies (Beverly, MA, USA). The monoclonal antibody against Pgk1p was purchased from Invitrogen. Secondary antibodies were purchased from Jackson ImmunoResearch Laboratories, Inc (West Grove, PA, USA). Chemoluminescence detection (GE Health Care) was used to visualize α-syn and the loading control Pgk1p. After detecting α-syn, the membrane was stripped of antibody and probed for Pgk1p. Western blot analysis was repeated three times.
Statistical analysis
Figure 2 shows the effect of the identified genes on ROS accumulation in cells expressing WT α-syn. ROS signals from cells transformed with pTF201 (WT α-syn) and the 2µ plasmid containing the genes of interest were compared with cells expressing WT α-syn only (pTF201), using a paired Student’s t-test. To adjust for multiple comparisons, the Bonferroni correction method was used, with a family-wise error rate of 0.05 or 0.001. For example, since 66 genes were tested, the highest accepted P-value was 7.6 × 10−4 (=0.05/66) for a family error rate of 0.05 or 1.5 × 10−5 (=0.001/66) for a family error rate of 0.001 (Supplementary Material, Table S1).
SUPPLEMENTARY MATERIAL
FUNDING
This study was supported by the National Institutes of Health (1R21NS053678 to S.N.W.) and the Parkinson’s Disease Resource of Northwest Louisiana (to S.N.W.). Funding to pay the Open Access Charge was provided by National Institutes of Health R01NS057656.
Supplementary Material
Acknowledgments
Conflict of Interest statement. None declared.
REFERENCES
- 1.Abou-Sleiman P.M., Healy D.G., Wood N.W. Causes of Parkinson’s disease: genetics of DJ-1. Cell Tissue Res. 2004;318:185–188. doi: 10.1007/s00441-004-0922-6. [DOI] [PubMed] [Google Scholar]
- 2.Recchia A., Debetto P., Negro A., Guidolin D., Skaper S.D., Giusti P. Alpha-synuclein and Parkinson’s disease. FASEB J. 2004;18:617–626. doi: 10.1096/fj.03-0338rev. [DOI] [PubMed] [Google Scholar]
- 3.Moore D.J., West A.B., Dawson V.L., Dawson T.M. Molecular pathophysiology of Parkinson’s disease. Annu. Rev. Neurosci. 2005;28:57–87. doi: 10.1146/annurev.neuro.28.061604.135718. [DOI] [PubMed] [Google Scholar]
- 4.Lucking C.B., Brice A. Alpha-synuclein and Parkinson’s disease. Cell Mol. Life Sci. 2000;57:1894–1908. doi: 10.1007/PL00000671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Dawson T.M., Dawson V.L. Molecular pathways of neurodegeneration in Parkinson’s disease. Science. 2003;302:819–822. doi: 10.1126/science.1087753. [DOI] [PubMed] [Google Scholar]
- 6.Baba M., Nakajo S., Tu P.H., Tomita T., Nakaya K., Lee V.M., Trojanowski J.Q., Iwatsubo T. Aggregation of alpha-synuclein in Lewy bodies of sporadic Parkinson’s disease and dementia with Lewy bodies. Am. J. Pathol. 1998;152:879–884. [PMC free article] [PubMed] [Google Scholar]
- 7.Polymeropoulos M.H., Lavedan C., Leroy E., Ide S.E., Dehejia A., Dutra A., Pike B., Root H., Rubenstein J., Boyer R., et al. Mutation in the alpha-synuclein gene identified in families with Parkinson’s disease. Science. 1997;276:2045–2047. doi: 10.1126/science.276.5321.2045. [DOI] [PubMed] [Google Scholar]
- 8.Kruger R., Kuhn W., Muller T., Woitalla D., Graeber M., Kosel S., Przuntek H., Epplen J.T., Schols L., Riess O. Ala30Pro mutation in the gene encoding alpha-synuclein in Parkinson’s disease. Nat. Genet. 1998;18:106–108. doi: 10.1038/ng0298-106. [DOI] [PubMed] [Google Scholar]
- 9.Zarranz J.J., Alegre J., Gomez-Esteban J.C., Lezcano E., Ros R., Ampuero I., Vidal L., Hoenicka J., Rodriguez O., Atares B., et al. The new mutation, E46K, of alpha-synuclein causes Parkinson and Lewy body dementia. Ann. Neurol. 2004;55:164–173. doi: 10.1002/ana.10795. [DOI] [PubMed] [Google Scholar]
- 10.Singleton A.B., Farrer M., Johnson J., Singleton A., Hague S., Kachergus J., Hulihan M., Peuralinna T., Dutra A., Nussbaum R., et al. Alpha-synuclein locus triplication causes Parkinson’s disease. Science. 2003;302:841. doi: 10.1126/science.1090278. [DOI] [PubMed] [Google Scholar]
- 11.Narayanan V., Scarlata S. Membrane binding and self-association of alpha-synucleins. Biochemistry. 2001;40:9927–9934. doi: 10.1021/bi002952n. [DOI] [PubMed] [Google Scholar]
- 12.Jenco J.M., Rawlingson A., Daniels B., Morris A.J. Regulation of phospholipase D2: selective inhibition of mammalian phospholipase D isoenzymes by alpha- and beta-synucleins. Biochemistry. 1998;37:4901–4909. doi: 10.1021/bi972776r. [DOI] [PubMed] [Google Scholar]
- 13.Caughey B., Lansbury P.T. Protofibrils, pores, fibrils, and neurodegeneration: separating the responsible protein aggregates from the innocent bystanders. Annu. Rev. Neurosci. 2003;26:267–298. doi: 10.1146/annurev.neuro.26.010302.081142. [DOI] [PubMed] [Google Scholar]
- 14.Xu J., Kao S.Y., Lee F.J., Song W., Jin L.W., Yankner B.A. Dopamine-dependent neurotoxicity of alpha-synuclein: a mechanism for selective neurodegeneration in Parkinson’s disease. Nat. Med. 2002;8:600–606. doi: 10.1038/nm0602-600. [DOI] [PubMed] [Google Scholar]
- 15.Flower T.R., Chesnokova L.S., Froelich C.A., Dixon C., Witt S.N. Heat shock prevents alpha-synuclein-induced apoptosis in a yeast model of Parkinson’s disease. J. Mol. Biol. 2005;351:1081–1100. doi: 10.1016/j.jmb.2005.06.060. [DOI] [PubMed] [Google Scholar]
- 16.Lindersson E., Beedholm R., Hojrup P., Moos T., Gai W., Hendil K.B., Jensen P.H. Proteasomal inhibition by alpha-synuclein filaments and oligomers. J. Biol. Chem. 2004;279:12924–12934. doi: 10.1074/jbc.M306390200. [DOI] [PubMed] [Google Scholar]
- 17.Betarbet R., Sherer T.B., Greenamyre J.T. Ubiquitin-proteasome system and Parkinson’s diseases. Exp Neurol. 2005;191(Suppl 1):S17–S27. doi: 10.1016/j.expneurol.2004.08.021. [DOI] [PubMed] [Google Scholar]
- 18.Cooper A.A., Gitler A.D., Cashikar A., Haynes C.M., Hill K.J., Bhullar B., Liu K., Xu K., Strathearn K.E., Liu F., et al. Alpha-synuclein blocks ER–Golgi traffic and Rab1 rescues neuron loss in Parkinson’s models. Science. 2006;313:324–328. doi: 10.1126/science.1129462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hamamichi S., Rivas R.N., Knight A.L., Cao S., Caldwell K.A., Caldwell G.A. Hypothesis-based RNAi screening identifies neuroprotective genes in a Parkinson’s disease model. Proc. Natl. Acad. Sci. USA. 2008;105:728–733. doi: 10.1073/pnas.0711018105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.van Ham T.J., Thijssen K.L., Breitling R., Hofstra R.M., Plasterk R.H., Nollen E.A. C. elegans model identifies genetic modifiers of alpha-synuclein inclusion formation during aging. PLoS Genet. 2008;4:e1000027. doi: 10.1371/journal.pgen.1000027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Willingham S., Outeiro T.F., DeVit M.J., Lindquist S.L., Muchowski P.J. Yeast genes that enhance the toxicity of a mutant Huntingtin fragment or alpha-synuclein. Science. 2003;302:1769–1772. doi: 10.1126/science.1090389. [DOI] [PubMed] [Google Scholar]
- 22.Flower T.R., Clark-Dixon C., Metoyer C., Yang H., Shi R., Zhang Z., Witt S.N. YGR198w (YPP1) targets A30P alpha-synuclein to the vacuole for degradation. J. Cell Biol. 2007;177:1091–1104. doi: 10.1083/jcb.200610071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Scherzer C.R., Feany M.B. Yeast genetics targets lipids in Parkinson’s disease. Trends Genet. 2004;20:273–277. doi: 10.1016/j.tig.2004.05.005. [DOI] [PubMed] [Google Scholar]
- 24.Cohen G., Farooqui R., Kesler N. Parkinson disease: a new link between monoamine oxidase and mitochondrial electron flow. Proc. Natl. Acad. Sci. USA. 1997;94:4890–4894. doi: 10.1073/pnas.94.10.4890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Witt S.N., Flower T.R. Alpha-synuclein, oxidative stress and apoptosis from the perspective of a yeast model of Parkinson’s disease. FEMS Yeast Res. 2006;6:1107–1116. doi: 10.1111/j.1567-1364.2006.00135.x. [DOI] [PubMed] [Google Scholar]
- 26.Louvion J.F., Abbas-Terki T., Picard D. Hsp90 is required for pheromone signaling in yeast. Mol. Biol. Cell. 1998;9:3071–3083. doi: 10.1091/mbc.9.11.3071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Abadjieva A., Pauwels K., Hilven P., Crabeel M. A new yeast metabolon involving at least the two first enzymes of arginine biosynthesis: acetylglutamate synthase activity requires complex formation with acetylglutamate kinase. J. Biol. Chem. 2001;276:42869–42880. doi: 10.1074/jbc.M103732200. [DOI] [PubMed] [Google Scholar]
- 28.Wendland B., Steece K.E., Emr S.D. Yeast epsins contain an essential N-terminal ENTH domain, bind clathrin and are required for endocytosis. EMBO J. 1999;18:4383–4393. doi: 10.1093/emboj/18.16.4383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Henke B., Girzalsky W., Berteaux-Lecellier V., Erdmann R. IDP3 encodes a peroxisomal NADP-dependent isocitrate dehydrogenase required for the beta-oxidation of unsaturated fatty acids. J. Biol. Chem. 1998;273:3702–3711. doi: 10.1074/jbc.273.6.3702. [DOI] [PubMed] [Google Scholar]
- 30.Nishikawa S., Endo T. The yeast JEM1p is a DnaJ-like protein of the endoplasmic reticulum membrane required for nuclear fusion. J. Biol. Chem. 1997;272:12889–12892. doi: 10.1074/jbc.272.20.12889. [DOI] [PubMed] [Google Scholar]
- 31.Dixon C., Mathias N., Zweig R.M., Davis D.A., Gross D.S. Alpha-synuclein targets the plasma membrane via the secretory pathway and induces toxicity in yeast. Genetics. 2005;170:47–59. doi: 10.1534/genetics.104.035493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Outeiro T.F., Lindquist S. Yeast cells provide insight into alpha-synuclein biology and pathobiology. Science. 2003;302:1772–1775. doi: 10.1126/science.1090439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Zabrocki P., Bastiaens I., Delay C., Bammens T., Ghillebert R., Pellens K., De Virgilio C., Van Leuven F., Winderickx J. Phosphorylation, lipid raft interaction and traffic of alpha-synuclein in a yeast model for Parkinson. Biochim. Biophys. Acta. 2008;1783:1767–1780. doi: 10.1016/j.bbamcr.2008.06.010. [DOI] [PubMed] [Google Scholar]
- 34.Jauniaux J.C., Urrestarazu L.A., Wiame J.M. Arginine metabolism in Saccharomyces cerevisiae: subcellular localization of the enzymes. J. Bacteriol. 1978;133:1096–1107. doi: 10.1128/jb.133.3.1096-1107.1978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Wipe B., Leisinger T. Regulation of activity and synthesis of N-acetylglutamate synthase from Saccharomyces cerevisiae. J. Bacteriol. 1979;140:874–880. doi: 10.1128/jb.140.3.874-880.1979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Hinnebusch A.G. The general control of amino acid biosynthetic genes in the yeast Saccharomyces cerevisiae. CRC Crit. Rev. Biochem. 1986;21:277–317. doi: 10.3109/10409238609113614. [DOI] [PubMed] [Google Scholar]
- 37.Hinnebusch A.G. Translational regulation of GCN4 and the general amino acid control of yeast. Annu. Rev. Microbiol. 2005;59:407–450. doi: 10.1146/annurev.micro.59.031805.133833. [DOI] [PubMed] [Google Scholar]
- 38.Patil C.K., Li H., Walter P. Gcn4p and novel upstream activating sequences regulate targets of the unfolded protein response. PLoS Biol. 2004;2:E246. doi: 10.1371/journal.pbio.0020246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Smith W.W., Jiang H., Pei Z., Tanaka Y., Morita H., Sawa A., Dawson V.L., Dawson T.M., Ross C.A. Endoplasmic reticulum stress and mitochondrial cell death pathways mediate A53T mutant alpha-synuclein-induced toxicity. Hum. Mol. Genet. 2005;14:3801–3811. doi: 10.1093/hmg/ddi396. [DOI] [PubMed] [Google Scholar]
- 40.Askree S.H., Yehuda T., Smolikov S., Gurevich R., Hawk J., Coker C., Krauskopf A., Kupiec M., McEachern M.J. A genome-wide screen for Saccharomyces cerevisiae deletion mutants that affect telomere length. Proc. Natl. Acad. Sci. USA. 2004;101:8658–8663. doi: 10.1073/pnas.0401263101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Yokota A., Kawasaki S., Iwano M., Nakamura C., Miyake C., Akashi K. Citrulline and DRIP-1 protein (ArgE homologue) in drought tolerance of wild watermelon. Ann. Bot. (Lond) 2002;89(Spec):825–832. doi: 10.1093/aob/mcf074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Nathan D.F., Vos M.H., Lindquist S. In vivo functions of the Saccharomyces cerevisiae Hsp90 chaperone. Proc. Natl. Acad. Sci. USA. 1997;94:12949–12956. doi: 10.1073/pnas.94.24.12949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Picard D. Heat-shock protein 90, a chaperone for folding and regulation. Cell Mol. Life Sci. 2002;59:1640–1648. doi: 10.1007/PL00012491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Zhao R., Davey M., Hsu Y.C., Kaplanek P., Tong A., Parsons A.B., Krogan N., Cagney G., Mai D., Greenblatt J., et al. Navigating the chaperone network: an integrative map of physical and genetic interactions mediated by the hsp90 chaperone. Cell. 2005;120:715–727. doi: 10.1016/j.cell.2004.12.024. [DOI] [PubMed] [Google Scholar]
- 45.Volles M.J., Lansbury P.T., Jr Vesicle permeabilization by protofibrillar alpha-synuclein is sensitive to Parkinson’s disease-linked mutations and occurs by a pore-like mechanism. Biochemistry. 2002;41:4595–4602. doi: 10.1021/bi0121353. [DOI] [PubMed] [Google Scholar]
- 46.Auluck P.K., Chan H.Y., Trojanowski J.Q., Lee V.M., Bonini N.M. Chaperone suppression of alpha-synuclein toxicity in a Drosophila model for Parkinson’s disease. Science. 2002;295:865–868. doi: 10.1126/science.1067389. [DOI] [PubMed] [Google Scholar]
- 47.Fujiwara H., Hasegawa M., Dohmae N., Kawashima A., Masliah E., Goldberg M.S., Shen J., Takio K., Iwatsubo T. Alpha-synuclein is phosphorylated in synucleinopathy lesions. Nat. Cell Biol. 2002;4:160–164. doi: 10.1038/ncb748. [DOI] [PubMed] [Google Scholar]
- 48.Chen L., Feany M.B. Alpha-synuclein phosphorylation controls neurotoxicity and inclusion formation in a Drosophila model of Parkinson’s disease. Nat. Neurosci. 2005;8:657–663. doi: 10.1038/nn1443. [DOI] [PubMed] [Google Scholar]
- 49.Gorbatyuk O.S., Li S., Sullivan L.F., Chen W., Kondrikova G., Manfredsson F.P., Mandel R.J., Muzyczka N. The phosphorylation state of Ser-129 in human alpha-synuclein determines neurodegeneration in a rat model of Parkinson disease. Proc. Natl. Acad. Sci. USA. 2008;105:763–768. doi: 10.1073/pnas.0711053105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Walsh P., Bursac D., Law Y.C., Cyr D., Lithgow T. The J-protein family: modulating protein assembly, disassembly and translocation. EMBO Rep. 2004;5:567–571. doi: 10.1038/sj.embor.7400172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Haselbeck R.J., McAlister-Henn L. Function and expression of yeast mitochondrial NAD- and NADP-specific isocitrate dehydrogenases. J. Biol. Chem. 1993;268:12116–12122. [PubMed] [Google Scholar]
- 52.Geisbrecht B.V., Gould S.J. The human PICD gene encodes a cytoplasmic and peroxisomal NADP(+)-dependent isocitrate dehydrogenase. J. Biol. Chem. 1999;274:30527–30533. doi: 10.1074/jbc.274.43.30527. [DOI] [PubMed] [Google Scholar]
- 53.Duncan M.C., Costaguta G., Payne G.S. Yeast epsin-related proteins required for Golgi-endosome traffic define a gamma-adaptin ear-binding motif. Nat. Cell Biol. 2003;5:77–81. doi: 10.1038/ncb901. [DOI] [PubMed] [Google Scholar]
- 54.Eugster A., Pecheur E.I., Michel F., Winsor B., Letourneur F., Friant S. Ent5p is required with Ent3p and Vps27p for ubiquitin-dependent protein sorting into the multivesicular body. Mol. Biol. Cell. 2004;15:3031–3041. doi: 10.1091/mbc.E03-11-0793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Costaguta G., Duncan M.C., Fernandez G.E., Huang G.H., Payne G.S. Distinct roles for TGN/endosome epsin-like adaptors Ent3p and Ent5p. Mol. Biol. Cell. 2006;17:3907–3920. doi: 10.1091/mbc.E06-05-0410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Copic A., Starr T.L., Schekman R. Ent3p and Ent5p exhibit cargo-specific functions in trafficking proteins between the trans-Golgi network and the endosomes in yeast. Mol. Biol. Cell. 2007;18:1803–1815. doi: 10.1091/mbc.E06-11-1000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Hirst J., Motley A., Harasaki K., Peak Chew S.Y., Robinson M.S. EpsinR: an ENTH domain-containing protein that interacts with AP-1. Mol. Biol. Cell. 2003;14:625–641. doi: 10.1091/mbc.E02-09-0552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Legendre-Guillemin V., Wasiak S., Hussain N.K., Angers A., McPherson P.S. ENTH/ANTH proteins and clathrin-mediated membrane budding. J. Cell Sci. 2004;117:9–18. doi: 10.1242/jcs.00928. [DOI] [PubMed] [Google Scholar]
- 59.Winston F., Dollard C., Ricupero-Hovasse S.L. Construction of a set of convenient Saccharomyces cerevisiae strains that are isogenic to S288C. Yeast. 1995;11:53–55. doi: 10.1002/yea.320110107. [DOI] [PubMed] [Google Scholar]
- 60.Brachmann C.B., Davies A., Cost G.J., Caputo E., Li J., Hieter P., Boeke J.D. Designer deletion strains derived from Saccharomyces cerevisiae S288C: a useful set of strains and plasmids for PCR-mediated gene disruption and other applications. Yeast. 1998;14:115–132. doi: 10.1002/(SICI)1097-0061(19980130)14:2<115::AID-YEA204>3.0.CO;2-2. [DOI] [PubMed] [Google Scholar]
- 61.Burke D., Dawson D., Stearns T. Methods in Yeast Genetics. Cold Spring Harbor: Cold Spring Harbor Laboratory Press; 2000. [Google Scholar]
- 62.Nasmyth K.A., Tatchell K. The structure of transposable yeast mating type loci. Cell. 1980;19:753–764. doi: 10.1016/s0092-8674(80)80051-1. [DOI] [PubMed] [Google Scholar]
- 63.Sambrook J., Fritsch E.F., Maniatis T. Molecular Cloning: A Laboratory Manual. Cold Spring Harbor, NY: Cold Spring Harbor Press; 1989. [Google Scholar]
- 64.Bevington P.R. Data Reduction and Error Analysis for the Physical Sciences. McGraw-Hill; 1969. p. 336. New York. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.