Abstract
Loss-of-function mutations in progranulin (GRN) cause ubiquitin- and TAR DNA-binding protein 43 (TDP-43)-positive frontotemporal dementia (FTLD-U), a progressive neurodegenerative disease affecting ∼10% of early-onset dementia patients. Here we expand the role of GRN in FTLD-U and demonstrate that a common genetic variant (rs5848), located in the 3′-untranslated region (UTR) of GRN in a binding-site for miR-659, is a major susceptibility factor for FTLD-U. In a series of pathologically confirmed FTLD-U patients without GRN mutations, we show that carriers homozygous for the T-allele of rs5848 have a 3.2-fold increased risk to develop FTLD-U compared with homozygous C-allele carriers (95% CI: 1.50–6.73). We further demonstrate that miR-659 can regulate GRN expression in vitro, with miR-659 binding more efficiently to the high risk T-allele of rs5848 resulting in augmented translational inhibition of GRN. A significant reduction in GRN protein was observed in homozygous T-allele carriers in vivo, through biochemical and immunohistochemical methods, mimicking the effect of heterozygous loss-of-function GRN mutations. In support of these findings, the neuropathology of homozygous rs5848 T-allele carriers frequently resembled the pathological FTLD-U subtype of GRN mutation carriers. We suggest that the expression of GRN is regulated by miRNAs and that common genetic variability in a miRNA binding-site can significantly increase the risk for FTLD-U. Translational regulation by miRNAs may represent a common mechanism underlying complex neurodegenerative disorders.
INTRODUCTION
Frontotemporal lobar degeneration (FTLD) is a progressive neurodegenerative disorder representing ∼5% of all dementia patients (1). It is the second most common form of early-onset neurodegenerative dementia after Alzheimer’s disease (AD), affecting 10–20% of patients with an onset of dementia before 65 years. FTLD patients present with prominent behavioral and personality changes, often accompanied by language impairment, which evolve gradually into cognitive impairment and dementia (2,3). FTLD may occur alone or in combination with motor neuron disease (MND) (4). The most common neuropathology associated with clinical FTLD is frontal and anterior temporal lobe atrophy with neuronal inclusions immunoreactive for ubiquitin and TAR DNA-binding protein 43 (TDP-43), but negative for tau and α-synuclein (FTLD-U) (5–7). Neuronal cytoplasmic inclusions (NCIs) in the neocortex, striatum and the dentate fascia of the hippocampus are the pathological hallmarks of FTLD-U. Up to four subtypes of FTLD-U have been delineated that are based on the distribution of NCIs, dystrophic neurites and the presence of neuronal intranuclear inclusions (NIIs) (8–10). Interestingly, all cases with GRN mutations have a common FTLD-U subtype, characterized by NCIs, short, thin neurites in layer II of the cortex and lentiform NIIs (11–13). This subtype is referred to as Type 1 by Mackenzie et al. (8) and Type 3 by Sampathu et al. (10).
FTLD has a high familial incidence with up to 50% of patients reported to have a family history of dementia. Recent molecular genetic advances in the field of FTLD have revealed that the genetic basis of FTLD-U is heterogeneous, and the causative mechanisms are just starting to be unraveled (14). Loss-of-function mutations in the gene encoding the secreted growth factor progranulin (GRN) on chromosome 17 have been identified as a major cause of familial FTLD-U and are present in up to 25% of familial FTLD-U patients worldwide (15–17). In addition, mutations in the valosin containing protein gene (VCP) and the gene encoding the charged multivesicular body protein (CHMP2B) have been reported in a small number of FTLD-U families (18,19). Despite these recent developments, it is evident that additional FTLD-U genes and genetic risk factors remain to be identified to explain the disease in the majority of the familial patients and in apparently sporadic FTLD-U patients.
We previously reported the identification of 23 different pathogenic loss-of-function mutations in GRN in 10% of the patients in our Mayo Clinic FTLD series, including a small number of apparently sporadic patients (17). In the present study, we expand the spectrum of mutational mechanisms that can lead to the loss of functional GRN. Using cell-based systems and in vivo studies we demonstrate that a common genetic variant (rs5848), located in the 3′-untranslated region (UTR) of GRN in a binding-site for micro-RNA (miRNA) miR-659 significantly increases the risk of developing FTLD-U most likely through suppressed translation of GRN. Our findings suggest that translational regulation by miRNAs may present a common mechanism underlying complex neurodegenerative disorders.
RESULTS
Association study of rs5848 with FTLD-U
We previously performed sequencing analyses of GRN in an extended population of FTLD patients (n = 378) derived from the Mayo Clinic FTLD series to assess the genetic contribution of GRN mutations to FTLD (17). A close inspection of our sequencing results in the subgroup of non-GRN mutation carriers (n = 339) revealed a statistically significant deviation from the expected Hardy–Weinberg equilibrium (HWE) for the common polymorphism rs5848 (P = 0.002), which was attributable to an excess of homozygous patients (Table 1). Deviations from HWE were not observed for any of the other genetic variants in GRN. We re-genotyped rs5848 using a pre-designed Taqman genotyping assay and confirmed all rs5848 genotypes in the FTLD patient series. Subsequent analyses of rs5848 in a large cohort of control individuals ascertained at Mayo Clinic Jacksonville (MCJ) and Mayo Clinic Scottsdale (MCS) showed a selective increase in the TT genotype frequency in FTLD patients (16%) compared with control individuals (9%) (Pgenotypic = 0.002) (Table 2).
Table 1.
Hardy–Weinberg calculation of rs5848 in Mayo Clinic FTLD patients
| Non-PGRN FTLD patients (n = 339) |
|||
|---|---|---|---|
| rs5848 genotypes | Observed | Expected | Overall P-value |
| n | n | ||
| CC | 160 | 145.4 | 0.002 |
| CT | 124 | 153.2 | |
| TT | 55 | 40.4 | |
Table 2.
Genotype frequencies of rs5848 in Mayo Clinic FTLD case–control series
| rs5848 genotypes | Controls (n = 934) |
Patients (n = 339) |
Overall P-value | ||
|---|---|---|---|---|---|
| n | % | n | % | ||
| CC | 463 | 49.6 | 160 | 47.2 | 0.002 |
| CT | 384 | 41.1 | 124 | 36.6 | |
| TT | 87 | 9.3 | 55 | 16.2 | |
To further confirm the genetic contribution of rs5848 to the development of FTLD, we focused our analyses on a homogeneous series of patients with a primary neuropathological diagnosis of FTLD-U with confirmed TDP-43-positive neuronal inclusions, derived from the MCJ brain bank and an age- and gender-matched control group ascertained at MCJ and MCS. Of the 81 genealogically unrelated FTLD-U patients identified in our brain bank, 19 (23.5% of the FTLD-U population) carried a pathogenic loss-of-function GRN mutation and were excluded from the study. One VCP and one LRRK2 mutation carrier were also excluded, resulting in a total of 59 FTLD-U patients for the genetic studies. Using logistic regression analyses of rs5848, we showed a highly significant association of rs5848 with FTLD-U (Padjusted = 0.003), resulting from an increase in the TT genotype frequency of rs5848 in FTLD-U patients (25.4%) compared with control individuals (9.9%) (Table 3). We calculated that within our series, the odds ratio (OR) to develop FTLD-U for carriers homozygous for the minor T-allele of rs5848 compared with homozygous C-allele carriers was 3.18 [Padjusted = 0.003; 95% confidence interval (CI): 1.50–6.73]. In contrast, individuals heterozygous for rs5848 did not show an increased risk to develop FTLD-U (Padjusted = 0.74; OR = 1.12; 95% CI: 0.59–2.10) (Table 3). Since MND pathology is rare or absent in GRN loss-of-function mutation carriers, we also re-analyzed the association excluding patients with MND pathology (n = 11), which further increased the OR for homozygous T-allele carriers to 3.76 (95% CI: 1.69–8.39; Padjusted = 0.001). Comparison of gender, age at death and brain weight of FTLD-U patients by rs5848 genotype groups did not show significant differences (mean age at death was 71.6 ± 7.4 years in CC, 76.0 ± 10.5 years in CT and 75.5 ± 11.3 years in TT carriers).
Table 3.
Logistic regression analyses of rs5848 in FTLD-U patient–control series
| rs5848 genotypes | Controls (n = 433) |
Patients (n = 59) |
Padjusted-value | OR | 95% CI | ||
|---|---|---|---|---|---|---|---|
| n | % | n | % | ||||
| CC | 199 | 46.0 | 21 | 35.6 | — | — | — |
| CT | 191 | 44.1 | 23 | 39.0 | 0.74 | 1.12 | 0.59–2.10 |
| TT | 43 | 9.9 | 15 | 25.4 | 0.003 | 3.18 | 1.50–6.73 |
Detailed genetic analyses in GRN genomic region
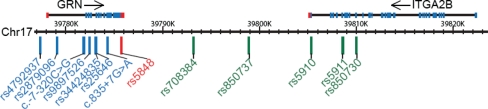
To study whether rs5848 is the likely functional variant underlying the association with FTLD-U or whether another genetic variant in linkage disequilibrium (LD) with rs5848 could be responsible for the observed association, we determined the LD structure underlying the GRN genomic region and performed single SNP and haplotype association analyses (Supplementary Material, Results). A panel of 12 additional SNPs was selected for single SNP and haplotype association analyses in the FTLD-U patient-control series: seven tagging SNPs identified in the genomic sequencing analyses that together with rs5848 capture 94% of the genetic diversity in the GRN region (Supplementary Material, Fig. S1; Fig. 1, SNPs in blue) and five SNPs in considerable LD with rs5848 selected from the downstream haplotype block (Supplementary Material, Fig. S2: Fig. 1, SNPs in green). Single SNP association analyses did not reveal SNPs that were more strongly associated with FTLD-U than rs5848 (Supplementary Material, Table S1). Moreover, haplotype analyses in the GRN genomic region and in a downstream haplotype block containing rs5848 only showed significant association when the risk T-allele of rs5848 was included (Supplementary Material, Table S2). Sequencing and genotyping analyses further revealed multiple GRN genetic backgrounds for the risk T-allele of rs5848, further favoring rs5848 as the potential functional variant underlying the observed association (Supplementary Material, Results and Table S3).
Figure 1.
Overview of genetic variants included in the GRN association studies. Each of the 13 genetic variants included in the association studies are shown relative to their position on chromosome 17q21.31. SNP rs5848 located in the 3′-UTR of GRN and strongly associated with FTLD-U is shown in red, while all other SNPs included in the GRN genomic region are in blue. SNPs from the downstream haplotype block are in green.
rs5848 is located in a predicted miRNA binding site of GRN
The rs5848 single base change (c.*78C>T) is located 78 nucleotides downstream of the translation termination codon in the 3′-UTR of the GRN transcript in a predicted binding site for the human specific miRNA miR-659 (Supplementary Material, Table S4). miRNAs are small non-coding RNAs that bind via imperfect base-pairing with target mRNAs to posttranscriptionally modulate their expression (20). We hypothesized that rs5848 may increase the risk for FTLD-U by altering the miRNA regulation of GRN, similar to previous studies in which a single nucleotide change in a miRNA target site had been shown sufficient to affect miRNA regulation (21,22).
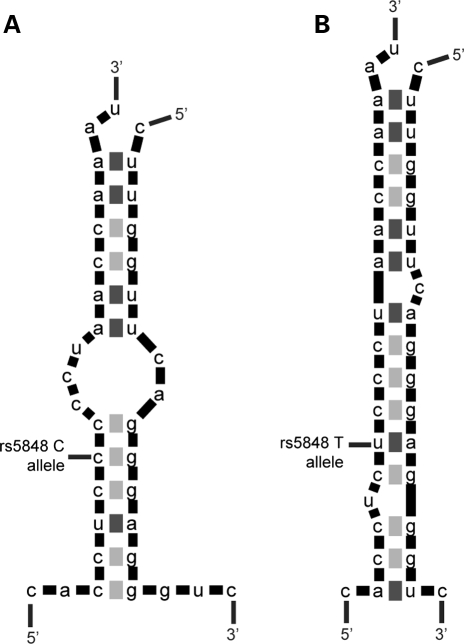
By means of in silico analyses using the RNA folding and two-state hybridization servers (http://frontend.bioinfo.rpi.edu/applications/mfold/) we predicted that miR-659 binds to the GRN 3′-UTR through a perfect complementarity of the ‘seed’ region at position 2–7 of the miRNA and an additional 3′match of an adenosine anchor at position 1 (23). However, depending on the presence of the C-allele or the T-allele at rs5848, the positioning of miR-659 with respect to the miRNA binding site in GRN was expected to shift, resulting in the formation of three additional base-pairs at the 5′end of the miRNA when the risk T-allele of rs5848 was present (Fig. 2). The stronger binding of miR-659 to the GRN mRNA containing the T-allele was expected to result in a more efficient inhibition of GRN translation leading to reduced GRN expression levels.
Figure 2.
In silico analyses of the pairing of miR-659 to the predicted binding site in the 3′-UTR of GRN. (A) Base-pairing in the presence of wild-type C-allele at rs5848 or (B) risk T-allele at rs5848. The presence of the risk T-allele is expected to lead to the formation of three additional base-pairs at the 5′end of miR-659 compared with the wild-type C-allele, resulting in a stronger binding and more efficient inhibition of GRN translation.
rs5848 affects GRN protein levels but not mRNA levels in FTLD-U patients
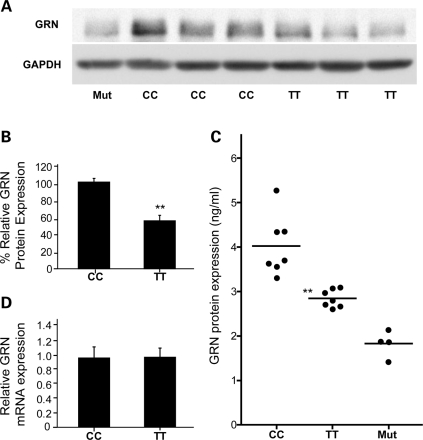
To determine the effect of rs5848 on GRN expression, we performed GRN immunoblot analyses using brain extracts derived from cerebellum of FTLD-U patients homozygous for the C- or T-allele. Using western blot analyses we observed a significant decrease in GRN protein levels in TT carriers compared with CC carriers (P < 0.001) (Fig. 3A and B). These data were further supported by immunohistochemical analysis, which showed a significant reduction in GRN burden in the granular cell layer of the cerebellum in TT compared with CC patients (P < 0.001) (Supplementary Material, Fig. S3). To further quantify the reduction in GRN, we re-extracted brain homogenates from the cerebellum of FTLD-U rs5848 CC, CT and TT carriers and determined GRN expression using an enzyme-linked immunoassay (ELISA). A significant ∼30% decrease in GRN levels was observed in rs5848 TT carriers compared with CC carriers in the TBS-X fraction (P < 0.001) (Fig. 3C). Intermediate levels of GRN protein were observed in FTLD-U patients heterozygous for rs5848 supporting a dose-dependent decrease of GRN with each T-allele (data not shown).
Figure 3.
Correlation of rs5848 genotypes with GRN expression levels in vivo. (A) Representative immunoblot analyses of six cerebellar tissue samples of FTLD-U patients with the indicated genotypes, showing reduced expression of GRN in rs5848 TT compared with CC carriers. A loss-of-function GRN mutation carrier (Mut) is included for comparison. (B) Quantification of immunoblotted GRN in cerebellar tissue samples of FTLD-U rs5848 TT or CC carriers (n = 7 in each group). GRN expression was normalized to GAPDH. Data represent mean ± SEM. (**indicates P < 0.001; two sample t-test). (C) Quantification of GRN protein levels by ELISA in the TBS-X fraction of cerebellar tissue samples of FTLD-U rs5848 TT and CC carriers (n = 7 in each group). GRN expression was ∼30% lower in FTLD-U rs5848 TT compared with CC carriers. Four loss-of-function GRN mutation carriers are included for comparison. (**indicates P < 0.001; two sample t-test). (D) Relative GRN mRNA expression determined by quantitative RT-PCR in cerebellar brain samples of FTLD-U rs5848 TT or CC carriers (n = 7 in each group). No significant difference in mRNA expression levels was observed. Data represent mean ± SEM.
As expected from a translational repression by miR-659, real-time mRNA expression analyses did not show a significant difference in GRN mRNA levels between rs5848 CC and TT carriers (Fig. 3D).
miR-659 inhibits GRN translation at rs5848
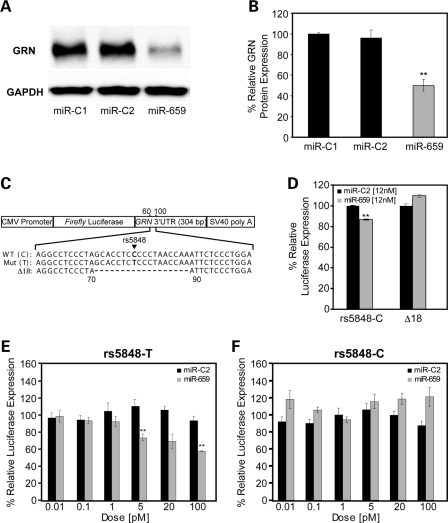
To provide evidence that miR-659 binds to GRN and can regulate the expression of GRN levels in vitro, we transiently transfected human M17 cells with 12 nm of miR-659 to mimic the expression of endogenous miR-659, or 12 nm negative control miRNAs, miR-C1 and miR-C2 (Ambion). Forty-eight hours post-transfection, cells were harvested and the endogenous human GRN levels produced by the M17 cells were measured by immunoblot analyses. A highly significant decrease in the expression of endogenous GRN was observed in M17 cells treated with miR-659 (P < 0.001) (Fig. 4A and B).
Figure 4.
Functional analyses of the GRN regulation by miR-659 in vitro. (A) Representative immunoblot analyses of human M17 cells transfected with miR-659 or control miRNAs (miR-C1 and miR-C2). (B) Quantification of immunoblotted GRN in human M17 cells transfected with miR-659 or control miRNAs. GRN expression was normalized to GAPDH. Data are from four independent experiments and represent mean ± SEM. (**indicates P < 0.001; two sample t-test). (C) Schematic overview of the pMIR-REPORT vector containing a CMV promoter, firefly luciferase gene, full-length 3′-UTR of GRN and SV40 poly A-tail. Constructs with wild-type C-allele and risk T-allele of rs5848 at position 78 in 3′-UTR were created as well as a Δ18 construct in which the complete predicted binding site of miR-659 was deleted (position 71–88 in the 3′-UTR). (D) Luciferase expression in N2A cells transfected with pMIR-REPORT-rs5848C or pMIR-REPORT- Δ18 and co-transfected with high dose (12 nm) miR-659 or miR-C2. Relative luciferase activity was determined as firefly luciferase activity normalized to Renilla luciferase activity. Each experiment was repeated six times. miR-659 significantly decreased the expression of firefly luciferase using the wild-type pMIR-REPORT-rs5848C vector, but not using the pMIR-REPORT-Δ18 in which the miR-659 binding site was deleted. Data represent mean ± SEM. (**indicates P < 0.001; two sample t-test). (E–F) pMIR-REPORT-rs5848C and pMIR-REPORT-rs5848T vectors were transfected in N2A cells and co-transfected with variable low doses (0.01–100 pM) of miR-659 or miR-C2. Each experiment was repeated three times. A significant reduction in the expression of firefly luciferase was observed in the presence of 5 and 100 pM of miR-659 from the rs5848T construct (D, **P < 0.02; two-tailed t-test), while no reduction in the expression of firefly luciferase from the rs5848C construct was observed at any of the doses. Data represent mean ± SEM.
To further study the regulation of GRN expression by miR-659, we inserted the full-length 304 bp GRN 3′-UTR sequence containing the wild-type C-allele at position 78 downstream of the luciferase reporter gene in the pMIR-REPORT miRNA expression reporter vector system and transiently transfected the construct into mouse N2A neuroblastoma cells (Fig. 4C). Co-transfection of a high dose (12 nm) of miR-659 or negative control miR-C2 in these cells resulted in significantly reduced expression of luciferase (representing GRN protein) in the presence of miR-659 further confirming the functional potential of the mRNA–miRNA duplex (Fig. 4D). We next constructed a luciferase construct with the GRN 3′-UTR sequence in which the complete 18 bp predicted binding site of miR-659 was deleted (position 71–88 downstream of the termination codon; vectorΔ18) (Fig. 4C). Addition of 12 nM miR-659 to N2A cells transfected with vectorΔ18 failed to repress the luciferase activity (Fig. 4D), supporting the hypothesis that miR-659 binds to the predicted binding-site in the 3′-UTR of GRN, overlapping with rs5848 at position 78.
To further study the differential regulation of GRN expression resulting from the presence of the wild-type ‘C’ or risk ‘T’ allele at rs5848, we created a luciferase construct containing the GRN 3′-UTR including the T-allele at position 78 (Fig. 4C). Co-transfection of the luciferase construct containing either the C-allele or the T-allele with variable low doses (0.01–100 pm) of miR-659 or negative control miR-C2 showed a dose-dependent reduction of luciferase activity derived from constructs containing the T-allele, reaching statistical significance at doses of 5 and 100 pM (P < 0.02) (Fig. 4E). A strong translational repression of the T-allele construct was also observed at 20 pM, however this result was non-significant (P = 0.06). Translational repression of luciferase activity from the wild-type rs5848-C construct was not observed for cells treated with miR-659 at any of these low doses (0.01–100 pM) (Fig. 4F).
miR-659 is expressed in human brain
To confirm the expression of miR-659 in human cells and in brain, we used a Taqman expression assay specific for miR-659 (Applied Biosystems). Positive expression of mature miR-659 was observed in both M17 cells and cerebellum (Supplementary Material, Fig. S4). Additional analyses using RNA extracted from seven different brain regions of a control brain (amygdala, occipital lobe, temporal lobe, frontal lobe, hippocampus, caudate and cerebellum) showed miR-659 expression in all analyzed regions, including frontal and temporal neocortex, which is most affected in FTLD-U (data not shown).
Correlation of rs5848 genotypes with FTLD-U pathological subtype
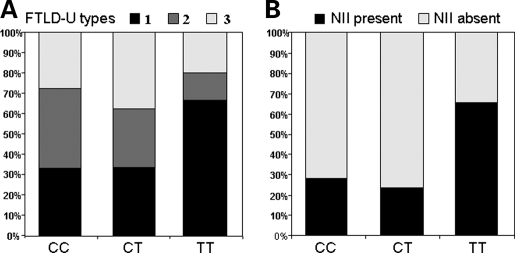
To determine whether the neuropathology of rs5848 TT carriers resembles the pathology of GRN loss-of-function mutation carriers, we determined the FTLD-U pathological subtype as proposed by the classification scheme of Mackenzie et al. (8) for all FTLD-U cases included in the genetic analyses of rs5848 with paraffin-embedded tissue blocks available for additional studies (n = 57). In addition, we determined the absence or presence of lentiform NIIs. Overall, FTLD-U subtypes could be determined for 54 FTLD-U patients resulting in 23 patients with FTLD-U type 1 (42.6%), 15 patients with FTLD-U type 2 (27.8%) and 16 patients with FTLD-U type 3 (29.6%). For the remaining three patients, the FTLD-U subtype could not be unambiguously assigned in part because inclusions were sparse (FTLD-U type 1 versus type 3). Stratification of FTLD-U patients by rs5848 genotype showed a non-significant increase in the frequency of FTLD-U type 1 pathology (resembling GRN mutation carriers) in TT carriers (66.7% in TT versus 33.3% in CT and CC carriers; P = 0.26; Fisher exact test) (Fig. 5A). Moreover, NIIs were significantly more common in the subgroup of TT carriers compared with CT or CC carriers (66.7% in TT versus 23.8% in CT and 27.8% in CC carriers; P = 0.02; Fisher exact test) (Fig. 5B). Finally, MND pathology, which is rare or absent in GRN mutation carriers, was only present in one of the 15 FTLD-U patients homozygous for the risk T-allele (6.7%) compared with five out of 23 CT carriers (21.7%) and five out of 21 CC carriers (23.8%).
Figure 5.
Pathological characterization of 54 FTLD-U patients stratified by rs5848 genotype. (A) Stratified by rs5848 genotype, the percentage of patients with each FTLD-U pathological subtype is shown. FTLD-U patients homozygous for the risk T-allele show the highest frequency of FTLD-U type 1. (B) Stratified by rs5848 genotype, the percentage of patients with characteristic lentiform NIIs is shown. FTLD-U patients homozygous for the risk T-allele present significantly more frequent with NIIs compared with heterozygous CT and homozygous CC carriers (P = 0.02, Fisher exact test).
DISCUSSION
We previously showed that heterozygous loss-of-function mutations in GRN are an important cause of FTLD, explaining up to 25% of familial patients with the FTLD-U pathological subtype. In this study we focused our attention on the role of the common genetic variant rs5848, located in the 3′-UTR of GRN. This was stimulated by our observation of a deviation from the expected HWE for rs5848 in an extensive Mayo Clinic FTLD patient series, due to a selective increase in the TT genotype frequency in patients compared with control individuals. We hypothesized that if rs5848 is a true genetic risk factor for FTLD, the genetic association would be strongest in a homogeneous population of patients with FTLD-U pathology. To test this hypothesis, we performed genetic association analyses in a pathology-confirmed FTLD-U series derived from the MCJ brain bank and confirmed the genetic risk associated with rs5848. In our series, the OR to develop FTLD-U for carriers homozygous for the minor T-allele of rs5848 compared with homozygous C-allele carriers was 3.18 (95% CI: 1.50–6.73; Padjusted = 0.003). Detailed genetic analyses of GRN and its flanking genomic regions further showed multiple haplotype backgrounds for the risk T-allele of rs5848 and did not reveal other genetic variants or haplotypes that were more strongly associated with FTLD-U, favoring rs5848 as the functional variant underlying the association.
We determined that rs5848 is located in the 3′-UTR of GRN within a predicted binding site for the human specific miRNA miR-659. miRNAs are a widely distributed class of non-coding RNAs that play an integral role in gene regulation by binding to partially complementary sites in the 3′-UTR of target mRNA transcripts, thereby inducing translational repression (20). Using in silico analyses we predicted a stronger binding (reducing GRN expression) of miR-659 to the GRN 3′-UTR containing the risk T-allele compared with the wild-type C-allele of rs5848.
To support the hypothesis that rs5848 is indeed a functional variant regulating GRN expression, we determined GRN expression levels in brain extracts derived from FTLD-U rs5848 CC and TT carriers by western blot analyses, ELISA and immunohistochemistry. These in vivo analyses confirmed the functional capacity of rs5848. Compared with CC carriers, a statistically significant ∼30% decrease in GRN protein levels was observed in FTLD-U rs5848 TT carriers. Furthermore, intermediate levels of GRN protein were observed in FTLD-U patients heterozygous for rs5848 supporting a dose-dependent decrease of GRN protein levels with each T-allele. As expected from a translational suppression of GRN by miRNAs, GRN mRNA expression levels were not significantly different between FTLD-U rs5848 CC and TT carriers.
Using cell-based systems we further demonstrated that miR-659 binds to the predicted binding site in the 3′-UTR of GRN and is able to suppress GRN expression in vitro. To determine the differential regulation of GRN expression resulting from the wild-type C-allele or risk T-allele of rs5848, we generated luciferase reporter constructs expressing firefly luciferase under the regulation of the GRN 3′-UTR carrying both rs5848 alleles. Our data suggested that miR-659 binds more efficiently to the risk T-allele than the wild-type C-allele, as demonstrated by the dose-dependent translational inhibition of the luciferase reporter (representing GRN protein) from the T-allele construct at low miRNA doses ranging from 5 to 100 pm (Fig. 4E). No translational repression was observed using the wild-type C-allele constructs, further supporting the differential effect of rs5848 (Fig. 4F). Together with the confirmation of positive expression of miR-659 in human brain, these results strongly support the functional potential of the miR-659/GRN mRNA complex. Of note, neither the binding site of miR-659 in the 3′-UTR of GRN, nor the gene encoding miR-659 on human chromosome 22q13, are conserved in other vertebrate or invertebrate species, prohibiting the modeling of the regulation of GRN by miR-659 in non-human systems.
If rs5848 increases FTLD-U risk by miRNA-mediated translational repression of GRN, one would expect a significant proportion of homozygous T-allele carriers to develop a pathological FTLD-U phenotype resembling GRN loss-of-function mutation carriers. Indeed, histological subtyping of FTLD-U patients from our cohort, blinded to their rs5848 genotype status, revealed that >65% of patients homozygous for the risk T-allele (10/15 patients) had FTLD-U consistent with Mackenzie type 1 (12). In addition, compared with the other rs5848 genotype groups, the frequency of lentiform NIIs was significantly higher in homozygous T-allele carriers (P = 0.02). Finally, MND pathology, which is rare or absent in GRN loss-of-function mutation carriers, was only present in one of the 15 FTLD-U patients homozygous for the risk T-allele (6.6%). Re-analyses of the genetic association study excluding patients with a pathological diagnosis of FTD- MND increased the OR for homozygous T-allele carriers to 3.76 (95% CI: 1.69–8.39; Padjusted = 0.001). The strong similarity in pathological presentation between GRN loss-of- function mutation carriers and patients homozygous for the rs5848 T-allele provides additional support for the hypothesis that rs5848 increases FTLD-U risk by reducing the expression of GRN.
In 2 years, 57 different pathogenic GRN loss-of-function mutations have been reported in 160 genealogically unrelated FTLD families (Alzheimer Disease and Frontotemporal Dementia Mutation Database, http://www.molgen.ua.ac.be/FTDmutations/). The majority of these mutations are nonsense, frameshift and splice-site mutations that introduce a premature stop codon leading to the degradation of mutant RNA by the process of nonsense-mediated decay and the subsequent loss of 50% functional GRN (15). The identification in this study of homozygosity of the T-allele of rs5848 as a major risk factor for FTLD-U, represents yet another way in which genetic variation in GRN may lead to FTLD-U via a decrease in the levels of functional GRN. In this case, haploinsufficiency is predicted to result from increased suppression of GRN translation through altered miRNA regulation. Our findings suggest a scenario whereby a decrease in GRN expression below a critical threshold results in the development of FTLD-U. While a single GRN null allele can sufficiently reduce GRN expression levels to cause FTLD-U, two copies of the risk T-allele of rs5848 are required (but may not be sufficient) to reduce the GRN expression below this critical threshold. We propose that a homozygous state of the risk T-allele at rs5848 combined with other environmental and/or genetic factors, will ultimately determine which individuals develop FTLD-U. In this respect, the association of rs5848 with FTLD-U is similar to many other neurodegenerative diseases where genetic variability at the same loci implicated in Mendelian pathogenic forms of the disease predispose to sporadic forms of the disease (24–29). Whether rs5848 may also contribute to the development of other TDP43-proteinopathies (such as amyotrophic lateral sclerosis) or may determine the presence of TDP-43 pathology in more common neurodegenerative conditions such as AD (30) needs further investigation.
The assumption of a role for miRNA pathways in neurodegeneration is intriguing and has been previously suggested based on experiments in mice, flies and cultured neurons in which the enzyme Dicer (required for miRNA maturation) was genetically inactivated (31–33). A role for specific miRNAs in the development of common neurodegenerative diseases has further been suggested by comparing changes in miRNA expression profiles in brain samples derived from patients and controls (32,34). Based on the possible role of miRNA dysregulation of GRN in FTLD presented in this study and the knowledge that overexpression of neurodegenerative disease proteins, such as the amyloid precursor protein in AD (35) and α-synuclein in Parkinson’s disease (36), is sufficient to cause disease, we speculate that regulatory mutations affecting the interaction between miRNAs and their targets may present a common mechanism underlying complex neurodegenerative disorders.
MATERIALS AND METHODS
FTLD patient and control populations
Mayo Clinic FTLD case–control series
The Mayo Clinic FTLD patient series in which the initial observation of a deviation from HWE for rs5848 was observed has been previously described in detail (17). A total of 934 control individuals (mean age at inclusion 64.8 ± 10.9 years) ascertained through MCJ and MCS were employed to determine initial rs5848 control genotype frequencies.
FTLD-U case–control series
The MCJ brain bank comprises >2500 neurodegenerative brain samples primarily ascertained through The State of Florida Alzheimer’s Disease Initiative funded through the Department of Elder Affairs, The Einstein Aging Study (P01-AG03949), The Udall Center for Excellence in Parkinson’s Disease Research (P50-NS40256), CurePSP/The Society for Progressive Supranuclear Palsy, Mayo Clinic ADRC (P50-AG16574), the Alzheimer’s Disease Patient Registry (ADPR) (P30-AG19610) and the Florida ADRC (P50-25711).
We selected all 81 patients from the MCJ brain bank with the neuropathological diagnosis of FTLD-U and positive TDP-43 immunostaining. Of these, 19 patients with a GRN loss-of- function mutation, one VCP mutation carrier and one LRRK2 mutation carrier were excluded from the study, resulting in a total of 59 FTLD-U patients for genetic and functional analyses. In this series, the mean age at death was 74.4 ± 9.8 years (range 56–97 years). A selection of 433 control individuals matched for age and gender to the FTLD-U patient population were drawn from two large cohorts of unrelated control individuals collected at MCJ and MCS. The average age at inclusion of the study was 74.5 ± 5.7 years (range 65–87 years).
Genotyping analyses
Genotyping of all additional SNPs selected for association and LD structure analyses, with the exception of rs34424835, were genotyped using pre-designed and custom TaqMan SNP genotyping assays (Applied Biosystems) and analyzed on an ABI 7900HT Fast Real Time PCR system using the SDSv2.2.2 software (for assay information see Supplementary Material, Table S5). A pre-designed TaqMan SNP genotyping assay was also used to confirm the rs5848 genotyping observed by sequencing the Mayo Clinic FTLD patient series and to determine the genotype frequencies of rs5848 in the FTLD-U and control individuals.
To genotype the rs34424835 deletion polymorphism, marker D17S1860 and the newly developed marker GRN_GT15, each marker was PCR amplified with one fluorescently labeled primer, and analyzed on an automated ABI3100 DNA-analyzer (for primers see Supplementary Material, Table S6). Alleles were scored using the GENOTYPER software (Applied Biosystems).
Single SNP and haplotype association analyses
In the FTLD-U patient–control series, single SNP age- and gender adjusted logistic regression analyses were performed using the StatsDirect statistical software program (http://www.statsdirect.com). A genotypic model was used to determine the risk associated with carrying one or two rare alleles, using homozygote carriers of the frequent allele as a reference. Haplotype association was analyzed using the score method of Schaid et al. (37) with adjustments made for age and gender. Only haplotypes with an estimated overall frequency of ≥5% were considered in the analyses. The level of significance was defined as P < 0.05.
Estimation of haplotype frequencies in GRN genomic region
The expectation maximization algorithm provided by the Arlequin package was applied in order to estimate GRN haplotype frequencies in 35 FTLD patients homozygous for the rs5848 T-allele.
Immunoblot analyses
Fourteen cerebellar brain samples of FTLD-U cases (seven CC and seven TT carriers) were selected for immunoblot analyses. The quality of each of these brain samples was previously assessed by determining the RNA Integrity Number (RIN) using an Agilent 2100 Bioanalyzer, and only samples with RIN values >6 were included in this study. Brain samples were sonicated in PARIS cell disruption buffer (Ambion/Applied Biosystems, Austin, TX) supplemented with protease and phosphatase inhibitors, centrifuged and the protein concentrations determined with a BCA protein assay (Pierce, Rockford, IL). Forty micrograms of protein were resolved by SDS–PAGE using pre-cast 8% Tris–Glycine gels (Invitrogen, Carlsbad, CA). Separated proteins were transferred to PVDF membranes and blocked for 1 h at RT with 5% skim milk/TBST. After incubation with anti-human PCDGF (1:1000; Zymed, South San Francisco, CA) primary antibody under blocking conditions, proteins were detected with anti-rabbit HRP conjugated secondary antibody (1:5000; Southern Biotech, Birmingham, AL) and ECL-Plus (Perkin Elmer, Waltham, MA). Quantification of immunoreactive bands was performed by densitometry (Image J, Research Services Branch, NIMH, Bethesda, MD). GRN protein levels were normalized to GAPDH by re-probing blots with anti-GAPDH (1:50 000; Biodesign International, Saco, ME) and anti-mouse HRP conjugated secondary (1:25 000: Southern Biotech) antibody. To compare GRN protein levels assessed on separate immunoblots, a further normalization was performed using a reference sample included on each blot. In addition, all immunoblots were run in parallel on the same day, to minimize variability in experimental conditions.
To determine GRN expression in human M17 cells transiently transfected with miR-659 or negative control miRNAs (Cye-3 dye labeled miRNA Negative Control #1 (miR-C1) and miRNA Negative Control #2 (miR-C2), Ambion), M17 cells were plated on TC-treated Costar® 6-well cell culture plates (Corning, Inc., Corning, NY) at 3.0×105 cells per well in antibiotic-free Opti-mem reduced serum medium supplemented with 10% fetal bovine serum (Invitrogen). Four hours after plating, cells were transfected with Lipofectamine 2000 (Invitrogen) using manufacturer’s instructions, with miR-659, miR-C1 or miR-C2, all at 12 nm. Three replicates were performed for each treatment. Forty-eight hours after transfection cells were harvested, lysed in a detergent-free buffer containing protease and phosphatase inhibitors, sonicated and centrifuged. SDS–PAGE and immunoblot analyses were performed as described above using 6 µg of protein. Immunoblot analysis was performed four times for each treatment group. Statistical analyses were performed using two-tailed t-tests.
GRN ELISA assay
To further quantify GRN expression in human brain, cerebellar brain samples of 25 FTLD-U cases (7 FTLD-U cases with each of the rs5848 genotypes and 4 loss-of-function GRN mutation carriers) were homogenized in 1× TBS (Boston Bioproducts Inc., Worcester, MA) supplemented with protease and phosphatase inhibitors and centrifuged for 5 min at 16 000g at 4°C. The supernatant was saved as the TBS fraction and the pellet was resuspended and sonicated in TBS-X (1× TBS with 0.1% Triton X-100). After centrifugation (5 min at 16 000g at 4°C), the supernatant was saved as the TBS-X fraction. The protein concentration of each fraction was determined using a BCA protein assay (Pierce). For each sample, 100 µg of protein of each fraction was analyzed in duplicate using a GRN ELISA assay (Human Progranulin ELISA Kit, AdipoGen Inc., Seoul, Korea). Recombinant human GRN provided with the ELISA kit was used as a standard.
Immunohistochemical analyses
FTLD-U subtyping
Glass-mounted sections (5 µm thick) of formalin-fixed, paraffin-embedded tissue from multiple brain regions (frontal cortex, temporal cortex, hippocampus, amygdala, basal ganglia and medulla) were immunostained for TDP-43 (rabbit polyclonal antibody; 1:3000; ProteinTech Group, Inc., Chicago, IL) with a DAKO-Autostainer (DAKO-Cytomaton, Carpinteria, CA) and 3,3′-diaminobenzidine as the chromogen. Sections were lightly counterstained with hematoxylin. The distribution of NCIs, dystrophic neurites and NIIs were noted in each section, and an FTLD-U subtype was assigned according to a scheme proposed by Mackenzie et al. (8). Presence or absence of NIIs and MND was also recorded. Type 1 cases had pleomorphic NCIs and short, thin neurites in upper cortical layers, as well as pleomorphic NCIs in the hippocampal dentate fascia, amygdala and striatum. There was variable fine neuritic pathology in the hippocampal pyramidal layer. Hypoglossal neurons were not affected in sections of the medulla, but NCIs were sometimes found in the inferior olive. Most type 1 cases had lentiform NIIs. Type 2 cases had long thick dystrophic neurites in the cortex that did not show a clear laminar distribution, with sparse cortical NCIs. In contrast to paucity of NCIs in the cortex, round NCIs were prominent in the dentate fascia and striatum, but not in lower levels of the neuraxis. Motor neuron pathology was absent. Type 3 cases had pleomorphic NCIs in the cortex, including neurons with granular cytoplasmic TDP-43 immunoreactive that did not form a discrete inclusion (‘pre-inclusions’). The amygdala was often affected, but basal ganglia pathology was minimal. Neurons in the hypoglossal nucleus often had TDP-43 immunoreactivity.
The frequency of pathological FTLD-U subtypes and number of FTLD-U patients with NIIs in each of the rs5848 genotype groups were compared using Fisher exact tests.
GRN immunohistochemistry and image analysis
Transverse sections of the cerebellum, including cortex, white matter and dentate nucleus were sampled in 10 FTLD-U cases (5 homozygous C-allele carriers and 5 homozygous T-allele carriers) previously included in the immunoblot analyses. To maintain staining consistency, paraffin-embedded sections (5 µm thick) were immunostained with a DAKO-Autostainer using a primary antibody to human GRN (anti-human progranulin, 1:600, R&D Systems, Inc., Minneapolis, MN) and 3, 3′-diaminobenzidine as the chromogen. Sections were lightly counterstained with hematoxylin. To obtain a quantitative measure of GRN immunoreactivity by image analysis, immunostained slides were converted into high-resolution digital images using an Aperio slide scanner (Aperio Technologies, Vista, CA). Blinded to genotypic information, GRN immunoreactivity was quantified in the granular cell layer of the cerebellum, using a positive pixel count algorithm (Imagescope version 8; Aperio Technologies, Vista, CA) and expressed as a percentage of the total area (GRN burden). Statistical comparisons were performed using a two-tailed t-test.
Real-time GRN mRNA expression analyses
Total RNA was isolated from cerebellum using the Trizol Plus RNA Purification System (Invitrogen), and its quality assessed on an Agilent 2100 Bioanalyzer. Only RNA samples with RNA integrity (RIN) values >6 were included in the analyses. RNA samples from 14 FTLD-U patients (7 CC and 7 TT carriers; same samples as included in the immunoblot analyses) were normalized to 500 ng/μl and using 3 µg as template, a reverse transcription reaction was performed using a 2:1 mix of random hexamers and oligo(dT) primers and the SuperScript III system (Invitrogen). For expression analyses Applied Biosystems assays were used for GRN (Hs00173570_m1), and for the endogenous controls GAPDH (Hs00266705_g1), YWHAZ (Hs00852925_sH) and HPRT1 (Hs99999909_m1). Real-time PCR was performed on an ABI 7900 using the TaqMan® method. Reactions contained 1 µl cDNA amplified with 0.25µl primer/probe mix and 2.5 µl TaqMan 2X Universal PCR Master Mix. The cycling parameters as recommended by the manufacturer were followed; 50°C for 2 min, 95°C for 10 min, followed by 40 cycles of 95°C for 15 s/ 60°C for 1 min. All samples were run in triplicate and normalized to the geometric mean of the three endogenous controls as described previously (38). The FAM-fluorescent signal was analyzed using SDSv2.2.2 software, and relative quantities of GRN mRNA were determined using the ΔΔct method.
Generation of human GRN 3′-UTR luciferase constructs
The 3′-UTR of GRN was amplified from genomic DNA from individuals homozygous for the C-allele or T-allele of rs5848 using the following primers: 3′-UTR-SpeI-F: AATTACTAGTGGGACAGTACTGAAGACTCTGC and 3′-UTR-HindIII-R: AATTAAGCTTAGTGTACAAACTTTATTGAAACGC. The 5′ end of each primer was designed to include restriction enzyme digest sites for subsequent digestion and ligation into the multiple cloning site of the pMIR-REPORT Luciferase vector (Ambion) to create pMIR-REPORT-rs5848C and pMIR-REPORT-rs5848T vectors. PCR reactions were performed in 100 µl using 50 ng genomic template and 20 pmol of each primer. Initial denaturation at 95°C was followed by 35 cycles of 94°C for 30 s, 60–50°C touchdown annealing for 30 s and 72°C for 30 s with a final extension at 72°C for 10 min. PCR products were purified using the Qiaquick purification system (Qiagen) both before and after digestion with SpeI and HindIII (New England Biolabs, Ipswich, MA).
To create the pMIR-REPORT-Δ18 luciferase vector in which the 18 bp predicted binding site of miR-659 was deleted, we performed site-directed mutagenesis using the QuikChange protocol (Stratagene, La Jolla, CA) on a full length GRN cDNA clone (Invitrogen) using the following primers Δ18-F: CAGGCCTCCCTAATTCTCCCTGGAC and Δ18-R: GTCCAGGGAGAATTAGGGAGGCCTG. The mutated construct was subsequently used as template for PCR amplification using the 3′-UTR-SpeI-F and 3′-UTR-HindIII-R primers, digestion and ligation into the pMIR-REPORT Luciferase vector as described earlier. Inserts of all constructs were verified by direct sequencing.
Luciferase assays
N2A neuroblastoma cells were plated on TC-treated Costar® 6-well cell culture plates (Corning) at 1.0 × 105 cells per well in antibiotic-free Opti-mem reduced serum medium supplemented with 10% fetal bovine serum and 2 mm l-Glutamine (Invitrogen). 4 h after plating, cells were transfected with Lipofectamine 2000 (Invitrogen) using the manufacturer’s protocol. For the first experiment, each well was co-transfected with 100 ng of either pMIR-REPORT-rs5848C or 100 ng pMIR-REPORT-Δ18 and 100 ng of pRL-CMV-renilla luciferase (Promega, Madison, WI), and treated with either miR-659 or negative control miR-C2 (Ambion) at 12 nM. Six replicates were performed for each treatment. Twenty-four hours after transfection, cells were lysed using 250 µl of Reporter Gene Assay lysis buffer (Roche, Indianapolis, IN) and Luciferase Firefly (LAF) and Luciferase Renilla (LAR) activities were measured in triplicate using the dual-luciferase reporter assay (Promega) on a Veritas microplate luminometer using manufacturer’s instructions. To determine the differential regulation of the rs5848 C- or T-allele constructs by miR-659, luciferase experiments were further performed by co-transfection of pMIR-REPORT-rs5848C or pMIR-REPORT-rs5848T with 100 ng of pRL-CMV-renilla luciferase and treatment with increasing but low doses of miR-659 and negative control miR-C2 (0.01–100 pM). Three replicates were performed for each treatment. For each well the relative luciferase activity (RLA) was calculated as RLA=LAF/LAR using the average from three independent measurements. Next, for each quantity of miRNA, the mean RLA was calculated based on all replicates. Statistical analyses using t-tests were performed for each quantity of miRNA by comparing the mean RLA in cells treated with miR-659 with the mean RLA in cells treated with negative control miR-C2.
Real-time miR-659 expression analyses
Tissue from the cerebellum of a control brain (pathologically normal) was dissected and processed using the mirVana PARIS system (Ambion) to extract RNA enriched for small RNA species. RNA from an M17 cell pellet was prepared similarly. A TaqMan microRNA reverse transcription reaction was performed following the manufacturer’s protocol and using the primers specific for miR-659 (RT 1514) and for human control RNU48 (RT1006) (Applied Biosystems). Real-time PCR was performed on the reverse transcription products to confirm the presence of miR-659 in the M17 cells and the control brain; each 5 µl reaction contained 0.33 µl reverse transcription product with 0.25 µl primer/probe mix and 2.5 µl TaqMan 2X Universal PCR Master Mix (Applied Biosystems) and was cycled as recommended by the protocol: 95°C for 10 min, followed by 40 cycles of 95°C for 15 s/60°C for 1 min. Samples were run in triplicate and analyzed using SDSv2.2.2 software. This procedure was repeated on enriched RNA extracted from amygdala, occipital lobe, temporal lobe, frontal lobe, hippocampus, caudate and cerebellum tissue from the same control brain to confirm the presence of miR-659 in these various brain regions.
SUPPLEMENTARY MATERIAL
FUNDING
The work was supported by NIH grants P50-NS40256 (D.W.D., R.J.U. and Z.K.W.), P01-AG17216 (D.W.D. and Z.K.W.), P50-25711 (D.W.D.), P01-AG03949 (D.W.D.), P30-AG19610 (R.J.C.), R01-MH57899 (R.J.C.) and P50 AG16574 (B.F.B., D.S.K., D.W.D., J.E.P., M.L.H., N.R.G.-R. and R.R.). Additional support was obtained from Arizona Alzheimer’s Research Consortium (R.J.C.), the Robert and Clarice Smith Fellowship in Neurodegenerative Disease (J.E.), the Pacific Alzheimer’s Disease Research Foundation grant #C06-01 (D.W.D., H.F., I.R.M., R.J.U., R.R. and Z.K.W.) and the Canadian Institutes of Health Research Operating grant #74580 (I.R.M. and H.F.). Funding to pay the Open Access publication charges for this article was provided by The Mayo Foundation.
Supplementary Material
ACKNOWLEDGEMENTS
The authors wish to acknowledge and thank the families who contributed samples that were critically important to the completion of this study. We further thank John Gonzalez, David Stroh, Linda Rousseau, Virginia Phillips and Monica Casey-Castanedes for brain banking and histologic support and Magdalena Cichon and Eric T. Blair (Applied Biosystems) for technical assistance.
Conflict of Interest statement. None declared.
REFERENCES
- 1.Graff-Radford N.R., Woodruff B.K. Frontotemporal dementia. Semin. Neurol. 2007;27:48–57. doi: 10.1055/s-2006-956755. [DOI] [PubMed] [Google Scholar]
- 2.McKhann G.M., Albert M.S., Grossman M., Miller B., Dickson D., Trojanowski J.Q. Clinical and pathological diagnosis of frontotemporal dementia: report of the Work Group on Frontotemporal Dementia and Pick’s Disease. Arch. Neurol. 2001;58:1803–1809. doi: 10.1001/archneur.58.11.1803. [DOI] [PubMed] [Google Scholar]
- 3.Neary D., Snowden J.S., Gustafson L., Passant U., Stuss D., Black S., Freedman M., Kertesz A., Robert P.H., Albert M., et al. Frontotemporal lobar degeneration: a consensus on clinical diagnostic criteria. Neurology. 1998;51:1546–1554. doi: 10.1212/wnl.51.6.1546. [DOI] [PubMed] [Google Scholar]
- 4.Lomen-Hoerth C., Anderson T., Miller B. The overlap of amyotrophic lateral sclerosis and frontotemporal dementia. Neurology. 2002;59:1077–1079. doi: 10.1212/wnl.59.7.1077. [DOI] [PubMed] [Google Scholar]
- 5.Josephs K.A., Holton J.L., Rossor M.N., Godbolt A.K., Ozawa T., Strand K., Khan N., Al-Sarraj S., Revesz T. Frontotemporal lobar degeneration and ubiquitin immunohistochemistry. Neuropathol. Appl. Neurobiol. 2004;30:369–373. doi: 10.1111/j.1365-2990.2003.00545.x. [DOI] [PubMed] [Google Scholar]
- 6.Lipton A.M., White C.L., III, Bigio E.H. Frontotemporal lobar degeneration with motor neuron disease-type inclusions predominates in 76 cases of frontotemporal degeneration. Acta Neuropathol. (Berl) 2004;108:379–385. doi: 10.1007/s00401-004-0900-9. [DOI] [PubMed] [Google Scholar]
- 7.Mackenzie I.R., Shi J., Shaw C.L., Duplessis D., Neary D., Snowden J.S., Mann D.M. Dementia lacking distinctive histology (DLDH) revisited. Acta Neuropathol. 2006;112:551–559. doi: 10.1007/s00401-006-0123-3. [DOI] [PubMed] [Google Scholar]
- 8.Mackenzie I.R., Baborie A., Pickering-Brown S., Du Plessis D., Jaros E., Perry R.H., Neary D., Snowden J.S., Mann D.M. Heterogeneity of ubiquitin pathology in frontotemporal lobar degeneration: classification and relation to clinical phenotype. Acta Neuropathol. 2006;112:539–549. doi: 10.1007/s00401-006-0138-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Neumann M., Mackenzie I.R., Cairns N.J., Boyer P.J., Markesbery W.R., Smith C.D., Taylor J.P., Kretzschmar H.A., Kimonis V.E., Forman M.S. TDP-43 in the ubiquitin pathology of frontotemporal dementia with VCP gene mutations. J. Neuropathol. Exp. Neurol. 2007;66:152–157. doi: 10.1097/nen.0b013e31803020b9. [DOI] [PubMed] [Google Scholar]
- 10.Sampathu D.M., Neumann M., Kwong L.K., Chou T.T., Micsenyi M., Truax A., Bruce J., Grossman M., Trojanowski J.Q., Lee V.M. Pathological heterogeneity of frontotemporal lobar degeneration with ubiquitin-positive inclusions delineated by ubiquitin immunohistochemistry and novel monoclonal antibodies. Am. J. Pathol. 2006;169:1343–1352. doi: 10.2353/ajpath.2006.060438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Cairns N.J., Neumann M., Bigio E.H., Holm I.E., Troost D., Hatanpaa K.J., Foong C., White C.L., III, Schneider J.A., Kretzschmar H.A., et al. TDP-43 in familial and sporadic frontotemporal lobar degeneration with ubiquitin inclusions. Am. J. Pathol. 2007;171:227–240. doi: 10.2353/ajpath.2007.070182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Mackenzie I.R., Baker M., Pickering-Brown S., Hsiung G.Y., Lindholm C., Dwosh E., Gass J., Cannon A., Rademakers R., Hutton M., et al. The neuropathology of frontotemporal lobar degeneration caused by mutations in the progranulin gene. Brain. 2006;129:3081–3090. doi: 10.1093/brain/awl271. [DOI] [PubMed] [Google Scholar]
- 13.Josephs K.A., Ahmed Z., Katsuse O., Parisi J.F., Boeve B.F., Knopman D.S., Petersen R.C., Davies P., Duara R., Graff-Radford N.R., et al. Neuropathologic features of frontotemporal lobar degeneration with ubiquitin-positive inclusions with progranulin gene (PGRN) mutations. J. Neuropathol. Exp. Neurol. 2007;66:142–151. doi: 10.1097/nen.0b013e31803020cf. [DOI] [PubMed] [Google Scholar]
- 14.Rademakers R., Hutton M. The genetics of frontotemporal lobar degeneration. Curr. Neurol. Neurosci. Rep. 2007;7:434–442. doi: 10.1007/s11910-007-0067-6. [DOI] [PubMed] [Google Scholar]
- 15.Baker M., Mackenzie I.R., Pickering-Brown S.M., Gass J., Rademakers R., Lindholm C., Snowden J., Adamson J., Sadovnick A.D., Rollinson S., et al. Mutations in progranulin cause tau-negative frontotemporal dementia linked to chromosome 17. Nature. 2006;442:916–919. doi: 10.1038/nature05016. [DOI] [PubMed] [Google Scholar]
- 16.Cruts M., Gijselinck I., van der Zee J., Engelborghs S., Wils H., Pirici D., Rademakers R., Vandenberghe R., Dermaut B., Martin J.J., et al. Null mutations in progranulin cause ubiquitin-positive frontotemporal dementia linked to chromosome 17q21. Nature. 2006;442:920–924. doi: 10.1038/nature05017. [DOI] [PubMed] [Google Scholar]
- 17.Gass J., Cannon A., Mackenzie I.R., Boeve B., Baker M., Adamson J., Crook R., Melquist S., Kuntz K., Petersen R., et al. Mutations in progranulin are a major cause of ubiquitin-positive frontotemporal lobar degeneration. Hum. Mol. Genet. 2006;15:2988–3001. doi: 10.1093/hmg/ddl241. [DOI] [PubMed] [Google Scholar]
- 18.Skibinski G., Parkinson N.J., Brown J.M., Chakrabarti L., Lloyd S.L., Hummerich H., Nielsen J.E., Hodges J.R., Spillantini M.G., Thusgaard T., et al. Mutations in the endosomal ESCRTIII- complex subunit CHMP2B in frontotemporal dementia. Nat. Genet. 2005;37:806–808. doi: 10.1038/ng1609. [DOI] [PubMed] [Google Scholar]
- 19.Watts G.D., Wymer J., Kovach M.J., Mehta S.G., Mumm S., Darvish D., Pestronk A., Whyte M.P., Kimonis V.E. Inclusion body myopathy associated with Paget disease of bone and frontotemporal dementia is caused by mutant valosin-containing protein. Nat. Genet. 2004;36:377–381. doi: 10.1038/ng1332. [DOI] [PubMed] [Google Scholar]
- 20.Bartel D.P. MicroRNAs: genomics, biogenesis, mechanism, and function. Cell. 2004;116:281–297. doi: 10.1016/s0092-8674(04)00045-5. [DOI] [PubMed] [Google Scholar]
- 21.Abelson J.F., Kwan K.Y., O’Roak B.J., Baek D.Y., Stillman A.A., Morgan T.M., Mathews C.A., Pauls D.L., Rasin M.R., Gunel M., et al. Sequence variants in SLITRK1 are associated with Tourette’s syndrome. Science. 2005;310:317–320. doi: 10.1126/science.1116502. [DOI] [PubMed] [Google Scholar]
- 22.Clop A., Marcq F., Takeda H., Pirottin D., Tordoir X., Bibe B., Bouix J., Caiment F., Elsen J.M., Eychenne F., et al. A mutation creating a potential illegitimate microRNA target site in the myostatin gene affects muscularity in sheep. Nat. Genet. 2006;38:813–818. doi: 10.1038/ng1810. [DOI] [PubMed] [Google Scholar]
- 23.Zuker M. Mfold web server for nucleic acid folding and hybridization prediction. Nucleic Acids Res. 2003;31:3406–3415. doi: 10.1093/nar/gkg595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Clarimon J., Asgeirsson H., Singleton A., Jakobsson F., Hjaltason H., Hardy J., Sveinbjornsdottir S. Torsin A haplotype predisposes to idiopathic dystonia. Ann. Neurol. 2005;57:765–767. doi: 10.1002/ana.20485. [DOI] [PubMed] [Google Scholar]
- 25.Farrer M., Maraganore D.M., Lockhart P., Singleton A., Lesnick T.G., de Andrade M., West A., de Silva R., Hardy J., Hernandez D. alpha-Synuclein gene haplotypes are associated with Parkinson’s disease. Hum. Mol. Genet. 2001;10:1847–1851. doi: 10.1093/hmg/10.17.1847. [DOI] [PubMed] [Google Scholar]
- 26.Rademakers R., Melquist S., Cruts M., Theuns J., Del-Favero J., Poorkaj P., Baker M., Sleegers K., Crook R., De Pooter T., et al. High-density SNP haplotyping suggests altered regulation of tau gene expression in progressive supranuclear palsy. Hum. Mol. Genet. 2005;14:3281–3292. doi: 10.1093/hmg/ddi361. [DOI] [PubMed] [Google Scholar]
- 27.Singleton A., Myers A., Hardy J. The law of mass action applied to neurodegenerative disease: a hypothesis concerning the etiology and pathogenesis of complex diseases. Hum. Mol. Genet. 2004;13(Spec. no. 1):R123–R126. doi: 10.1093/hmg/ddh093. [DOI] [PubMed] [Google Scholar]
- 28.Brouwers N., Sleegers K., Engelborghs S., Bogaerts V., Serneels S., Kamali K., Corsmit E., De Leenheir E., Martin J.J., De Deyn P.P., et al. Genetic risk and transcriptional variability of amyloid precursor protein in Alzheimer’s disease. Brain. 2006;129:2984–2991. doi: 10.1093/brain/awl212. [DOI] [PubMed] [Google Scholar]
- 29.Theuns J., Marjaux E., Vandenbulcke M., Van Laere K., Kumar-Singh S., Bormans G., Brouwers N., Van den Broeck M., Vennekens K., Corsmit E., et al. Alzheimer dementia caused by a novel mutation located in the APP C-terminal intracytosolic fragment. Hum. Mutat. 2006;27:888–896. doi: 10.1002/humu.20402. [DOI] [PubMed] [Google Scholar]
- 30.Amador-Ortiz C., Lin W.L., Ahmed Z., Personett D., Davies P., Duara R., Graff-Radford N.R., Hutton M.L., Dickson D.W. TDP-43 immunoreactivity in hippocampal sclerosis and Alzheimer’s disease. Ann. Neurol. 2007;61:435–445. doi: 10.1002/ana.21154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Bilen J., Liu N., Burnett B.G., Pittman R.N., Bonini N.M. MicroRNA pathways modulate polyglutamine-induced neurodegeneration. Mol. Cell. 2006;24:157–163. doi: 10.1016/j.molcel.2006.07.030. [DOI] [PubMed] [Google Scholar]
- 32.Kim J., Inoue K., Ishii J., Vanti W.B., Voronov S.V., Murchison E., Hannon G., Abeliovich A. A MicroRNA feedback circuit in midbrain dopamine neurons. Science. 2007;317:1220–1224. doi: 10.1126/science.1140481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Schaefer A., O’Carroll D., Tan C.L., Hillman D., Sugimori M., Llinas R., Greengard P. Cerebellar neurodegeneration in the absence of microRNAs. J. Exp. Med. 2007;204:1553–1558. doi: 10.1084/jem.20070823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Hebert S.S., Horre K., Nicolai L., Papadopoulou A.S., Mandemakers W., Silahtaroglu A.N., Kauppinen S., Delacourte A., De Strooper B. Loss of microRNA cluster miR-29a/b-1 in sporadic Alzheimer’s disease correlates with increased BACE1/beta-secretase expression. Proc. Natl Acad. Sci. USA. 2008;105:6415–6420. doi: 10.1073/pnas.0710263105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Rovelet-Lecrux A., Hannequin D., Raux G., Le Meur N., Laquerriere A., Vital A., Dumanchin C., Feuillette S., Brice A., Vercelletto M., et al. APP locus duplication causes autosomal dominant early-onset Alzheimer disease with cerebral amyloid angiopathy. Nat. Genet. 2006;38:24–26. doi: 10.1038/ng1718. [DOI] [PubMed] [Google Scholar]
- 36.Singleton A.B., Farrer M., Johnson J., Singleton A., Hague S., Kachergus J., Hulihan M., Peuralinna T., Dutra A., Nussbaum R., et al. alpha-Synuclein locus triplication causes Parkinson’s disease. Science. 2003;302:841. doi: 10.1126/science.1090278. [DOI] [PubMed] [Google Scholar]
- 37.Schaid D.J., Rowland C.M., Tines D.E., Jacobson R.M., Poland G.A. Score tests for association between traits and haplotypes when linkage phase is ambiguous. Am. J. Hum.Genet. 2002;70:425–434. doi: 10.1086/338688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Vandesompele J., De Preter K., Pattyn F., Poppe B., Van Roy N., De Paepe A., Speleman F. Accurate normalization of real-time quantitative RT-PCR data by geometric averaging of multiple internal control genes. Genome Biol. 2002;3:RESEARCH0034. doi: 10.1186/gb-2002-3-7-research0034. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.