Abstract
Mutations in AIPL1 cause Leber congenital amaurosis (LCA), the most severe form of inherited blindness in children; however, the function of this protein in normal vision remains unknown. To determine amino acid subsequences likely to be important for function, we have compared the protein sequence of several species. Sequence conservation is highest across the three Aipl1 tetratricopeptide (TPR) motifs and extends across the protein, except for a proline-rich amino acid sequence present only at the C-terminus of primate Aipl1. The length of the proline-rich region varies within primates; however, the length differences between human and primate Aipl1 do not correlate with evolutionary distance. These observations reinforce the importance of the TPR domains for function, the similarity of Aipl1 to a family of proteins that act as molecular chaperones, and the importance of comparative sequencing data for determination of whether AIPL1 sequence variants in patients are likely to cause retinopathy.
Leber congenital amaurosis (LCA, Mendelian Inheritance in Man (MIM) No. 204000) is a severe, early-onset, inherited retinopathy that accounts for approximately 5% of all inherited retinal disease (Kaplan et al. 1990). To date, four genes associated with Leber congenital amaurosis have been identified, the most recent of which is the aryl-hydrocarbon interacting protein-like 1 gene, AIPL1 (Sohocki et al. 2000a). Mutations in AIPL1 are the cause of 9-11% of cases of autosomal recessive LCA and may cause dominant cone-rod dystrophy (Sohocki et al. 2000b and unpublished).
AIPL1 expression is limited to photoreceptors and the pineal gland, and the gene encodes a protein 384-amino acids in length. The precise functional role of AIPL1 within the photoreceptors remains unknown. However, the human protein sequence contains three tetratricopeptide (TPR) motifs, 34 amino acid motifs that are thought to serve as interfaces for protein-protein interactions. TPR motifs are found in proteins that mediate a variety of functions, including protein trafficking or protein folding, and these proteins are usually associated with multiprotein complexes (Blatch and Lässle 1999). In addition, a proline-rich region is present at the carboxyl-terminus of the protein in humans. Similar sequences are found in situations requiring rapid recruitment or interchange of several proteins, such as signaling cascades or initiation of transcription (Kay et al. 2000).
Comparative sequencing is a powerful method for determining important structural features in proteins with unknown function, such as AIPL1. In addition, this information is useful for determination of the likelihood that sequence variants identified in patients cause disease, for identification of appropriate animal models, and for information on appropriate systems for in vitro studies, such as yeast-two hybrid analysis.
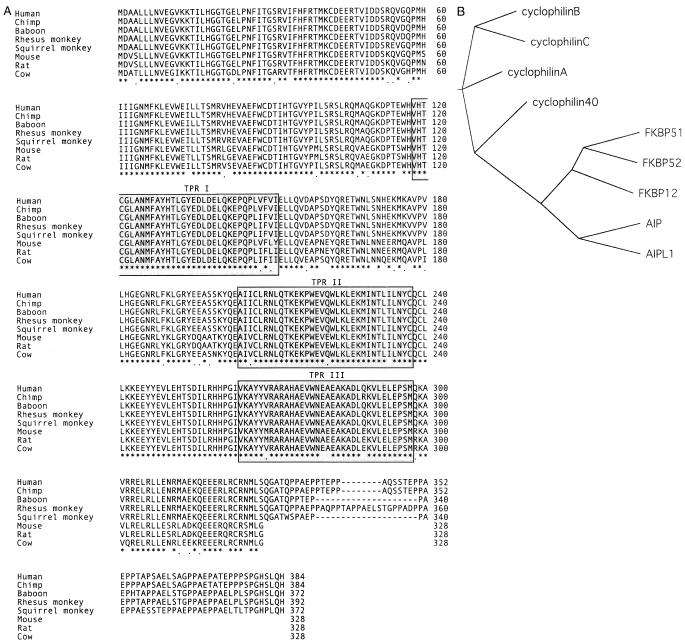
A zoo blot (EcoRI and BamHI digest) probed with a 200-bp fragment of AIPL1 indicated likely orthologs in dog, cow, and mouse. No signal was present in zebrafish or in chicken (data not shown). Sequencing of Aipl1 from cow and mouse retinal cDNA libraries revealed a high level of sequence conservation within mammals, outside of the 56-amino acid, proline-rich, carboxyl-terminal sequence that is present in human AIPL1 but lacking in non-primates (Fig. 1A). Genomic sequencing determined that the squirrel monkey Aipl1 proline-rich region is only 44 amino acids in length. In addition, a Drosophila protein sequence similar to AIPL1 (GenBank AE003487; 36% identity, 59% similarity) was identified by BLAST. This sequence, however, appears to be more highly related to AIP (39% identity, 59% similarity). As the complete Drosophila genomic sequence is known, with no other similar sequences, it is possible that this gene represents an ancestral Aip protein, predating the duplication event leading to separate Aip and Aipl1 in higher organisms.
Fig. 1.
(A) Protein sequence of Aipl1, indicating high degree of sequence conservation extending across the protein, outside of the proline-rich region present only at the C-terminus of primate Aipl1. Identical residues across all eight species are indicated with an asterisk. Residues with only conservative substitutions are indicated with a period. (B) Unrooted cladogram indicating the relationship of AIPL1 to the FK506-binding protein (FKBPs) and cyclophilin families of the human immunophilin proteins. Protein sequences used (GenBank number): cyclophilin A (P04374), cyclophilin B (P23284), cyclophilin C (NP_000934), cyclophilin 40 (BAA09923), FKBP12 (P20071), FKBP51 (Q13451), FKBP52 (Q02790), AIP (O00170), AIPL1 (AAF26708).
Owing to the difference in protein length between human and squirrel monkey, the Aipl1 sequences of several additional primates (chimpanzee, baboon, and rhesus monkey) were determined from genomic DNA (Fig. 1A). Although the chimpanzee and human proteins are identical in length, the difference or similarity in length of the proline-rich region in the other primates tested is inconsistent with evolutionary distance from humans. These data suggest that this region may be susceptible to gain or loss of repeats by replication slippage, perhaps owing to the repeat-type structure of the genomic sequence encoding the XXPP repeat within the proline-rich region. In addition, although several residues in this region are identical across the primates tested, the sequence identity/similarity to the human protein in this region is significantly lower than that within the TPR domains or across the length of the protein (Table 1).
Table 1.
AIPL1 protein homologies. Percents indicate identity/similarity for individual TPR domains, for the proline-rich region, and for the complete protein
| Species | TPRIa | TPRII | TPRIII | Proline-Rich Regionb | Complete Protein |
|---|---|---|---|---|---|
| Chimpanzee (Pan panisus) | 100/100 | 100/100 | 97/97 | 96/96 | 99/99 |
| Baboon (Papio cynocephalus) | 97/100 | 97/97 | 100/100 | 88/88 | 98/98 |
| Rhesus monkey (Macaca mulatta) | 97/100 | 97/97 | 100/100 | 78/78 | 96/96 |
| Squirrel monkey (Samiri bolvensis) | 97/100 | 100/100 | 100/100 | 73/75 | 94/96 |
| Mouse (Mus musculus) | 94/97 | 94/100 | 88/94 | — | 81/88 |
| Rat (Rattus norvegicus) | 94/100 | 94/100 | 88/94 | — | 87/96 |
| Cow (Bos Taurus) | 94/100 | 97/100 | 97/100 | — | 88/94 |
TPR = tetratricopeptide motif
Sequence identity/similarity within proline-rich region calculated without gap penalty.
These data require consideration in determining whether sequence variants identified in patients with inherited retinopathy are likely to cause disease. A 4-amino acid deletion, P351Δ12, has been identified within the proline-rich region of two unrelated patients with autosomal dominant cone-rod dystrophy (Sohocki et al. 2000b). Three of the four deleted amino acids (352-355) in these patients are identical across all primates tested, and the fourth deleted amino acid, although not conserved, is not involved in the loss or gain between primates. However, it will not be possible to determine whether the P351Δ12 variant causes the retinopathy in these patients without experimental assessment of the consequences of this variant in a model system.
The results of this study reveal a high degree of sequence conservation across mammalian Aipl1 proteins and reinforce the structural relationship of Aipl1 to the Fkbp family of proteins (Figure 1B), which function as molecular chaperones in steroid receptor signaling, heat shock responses, and immunosuppression. Therefore, Aipl1 is likely performing a similar function within the photoreceptors. The increased sequence conservation within the TPR motifs suggests an importance of these motifs for protein function. In addition, the conservation of certain residues within the proline-rich region suggests that they may be important for primate Aipl1 function.
Acknowledgements
We are particularly grateful to G. Aguirre, C. Craft, W. Baehr, and S. Semple-Rowland for providing retinal cDNA libraries, as well as to M.J. Aivaliotis and J.L. VandeBerg for providing primate genomic DNAs. We also thank K. Malone for expert technical assistance. Supported by grants from the Foundation Fighting Blindness and the George Gund Foundation, the William Stamps Farish Fund, the M.D. Anderson Foundation, the Hermann Eye Fund, and by grant EY07142 from the National Eye Institute-National Institutes of Health.
GenBank Accession numbers: Human AIPL1, AF148864; chimpanzee Aipl1, AF296413; baboon Aipl1, AF296414; rhesus monkey Aipl1, AF296411; squirrel monkey Aipl1, AF296415; Bovine Aipl1, AF296410; Rat Aipl1, AF180340; Mouse Aipl1, AF296412.
References
- Blatch GL, Lässle M. The tetratricopeptide repeat: a structural motif mediating protein-protein interactions. BioEssays. 1999;21:932–939. doi: 10.1002/(SICI)1521-1878(199911)21:11<932::AID-BIES5>3.0.CO;2-N. [DOI] [PubMed] [Google Scholar]
- Kaplan J, Bonneau D, Frezal J, Munnich A, Dufier JL. Clinical and genetic heterogeneity in retinitis pigmentosa. Hum Genet. 1990;85:635–642. doi: 10.1007/BF00193589. [DOI] [PubMed] [Google Scholar]
- Kay BK, Williamson MP, Sudol M. The importance of being proline: the interaction of proline-rich motifs in signaling proteins with their cognate domains. FASEB J. 2000;14:231–241. [PubMed] [Google Scholar]
- Sohocki MM, Bowne SJ, Sullivan LS, Blackshaw S, Cepko CL, et al. Mutations in the novel photoreceptor-pineal gene on 17p cause Leber congenital amaurosis. Nat Genet. 2000a;24:79–83. doi: 10.1038/71732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sohocki MM, Perrault I, Leroy BP, Payne AM, Dharmaraj S, et al. Prevalence of AIPL1 mutations in inherited retinal degenerative disease. Mol Genet Metab. 2000b;70:142–150. doi: 10.1006/mgme.2000.3001. doi: 10.1006/mgme.2000.3001. [DOI] [PubMed] [Google Scholar]