Abstract
The PGC-1 coactivators are important regulators of oxidative metabolism. We previously demonstrated that LRP130 is a binding partner of PGC-1α, required for hepatic gluconeogenesis. LRP130 is the gene mutated in Leigh syndrome French Canadian variant, a rare neurodegenerative disease. The importance of LRP130 in other, non-hepatocyte biology remains obscure. To better understand PGC-1 coactivator function in brown fat development, we explored the metabolic role of LRP130 in brown adipocyte differentiation. We show that LRP130 is preferentially enriched in brown fat compared with white, and induced in a PGC-1-dependent manner during differentiation. Despite intact PGC-1 coactivator expression, brown fat cells deficient for LRP130 exhibit attenuated expression of several genes characteristic of brown fat, including uncoupling protein 1. Oxygen consumption studies support a specific defect in proton leak due to attenuated uncoupling protein 1 expression. Notably, brown fat cell development common to both PGC-1 coactivators is governed by LRP130. Conversely, the cAMP response controlled by PGC-1α is not regulated by LRP130. These data implicate LRP130 in brown fat cell development and differentiation.
Obesity is a global health concern, predisposing to insulin resistance, hypertension, dyslipidemia, type 2 diabetes, and certain cancers. Its pathogenesis is multifactorial, involving neurohormonal, behavioral, genetic, and adipocyte-specific elements. Integration of these factors establishes whole body energy balance.
Adipocytes constitute important regulators of systemic energy balance, and play a central role in obesity due to their inherent capacity to store or consume excess calories. The duality of the adipocyte lineage is illustrated in two different types of fat cells. White adipocytes store fat as triacylglycerol during caloric excess and release free fatty acids during fasting. In contrast, brown adipocytes oxidize triacylglycerol to generate heat (thermogenesis) (1, 2). Besides muscular shivering (2-5), thermogenesis in rodents and other mammals affords an additional level of thermoprotection. The importance of brown fat in adult humans is less clear. Some studies indicate that even adults have functional brown fat (6, 7).
Interest has focused on the development of brown fat due to clear anti-obesity effects in rodent models (8-12). Brown fat protects against obesity due to its capacity to dissipate calories as heat. Central to this process is uncoupling protein 1 (Ucp1),4 a proton transporter situated in the inner mitochondrial membrane (13-15). Flow of electrons through the electron transport chain establishes an electrochemical proton gradient. Protons utilized by Ucp1 traverse the mitochondrial membrane to generate heat instead of driving the ATP synthase (16).
The mechanism by which a brown fat cell acquires its unique features, including uncoupled respiration is incompletely understood. Several transcription factors and a few coregulators have been described. Many of these factors are extensively reviewed elsewhere (1, 2). Transcription factors involved in brown fat cell regulation include PPARγ2 (17, 18), PPARα (19), thyroid receptor, ATF-2, FoxC2 (20), and CREB. Coregulators include RIP140, PRDM16 (21), and the PGC-1 family of coactivators. RIP140 is a corepressor and represses Ucp1 expression in white adipocytes. White fat cells genetically ablated for RIP140 have increased Ucp1 gene expression (22, 23). Both sufficient and necessary for brown fat differentiation, PRDM16 potently drives expression of PGC-1α, Ucp1 and several other brown fat genes. The PGC-1 family of coactivators regulate Ucp1 gene expression and several brown fat selective genes during brown fat development, and following cold exposure or treatment with cAMP mimetic. PGC-1 (PPARγ coactivator-1α) is the founding member of the PGC-1 family and was isolated from a brown fat library to identify PPARγ-interacting proteins (24). PGC-1β is the closest homolog of PGC-1α (25) with PRC being more distally related (26). PGC-1α and PGC-1β exhibit complementary function during brown fat development (27). Complementation is shared at the level of several brown fat specific genes, including Ucp1, as well as, mitochondrial biogenesis. Either member of the family is singly sufficient to establish basal Ucp1 gene expression in cells and animals (27-31).
Complementation, however, does not extend to all aspects of brown fat function. In contrast to PGC-1β, PGC-1α regulates cold inducible Ucp1 gene expression in mature brown fat (27-31). How then do the PGC-1s share overlapping function during brown fat development, yet display differential function following cold exposure? The mechanism is unknown and many possibilities exist. One possibility is the existence of a specificity factor common to the PGC-1s, required for brown fat development, but not important for cold exposure. Identification of such a factor would help delineate developmental control from the environmental control, and shed insight on how the PGC-1s preferentially influence one program over another.
We previously demonstrated that LRP130 (leucine-rich protein 130), also known as LRPPRC (leucine-rich protein pentatricopeptide repeat-containing motif), is a component of the PGC-1α holo-complex (32). LRP130 is mutated in a rare neurological disorder called Leigh Syndrome French Canadian variant (33). Patients suffer a devastating neurological disease as well as liver dysfunction (34). LRP130 localizes to the nucleus and mitochondria, and has been shown to influence gene expression in both compartments (32, 35, 36). We recently demonstrated that LRP130 is an important coregulator of PGC-1α-dependent gluconeogenesis. Although LRP130 was shown to interact with PGC-1β (32), the functional importance of interaction with this PGC-1 member was not studied. We show here that LRP130 is a novel and important regulator of brown fat development. Notably it regulates the developmental program mediated by the PGC-1 coactivators, but does not influence the cAMP response. These results suggest that LRP130 is critical for the complementary actions of the PGC-1s during brown fat development.
EXPERIMENTAL PROCEDURES
Cell Culture and Adipocyte Differentiation—We digested brown fat from wild-type and PGC-1α KO newborn mice (28) with collagenase to isolate brown fat preadipocytes (37-39). The cells were immortalized by pBABE SV40 T retroviral infection, and selected in 2 μg/ml puromycin. Maintenance media contained 20% FBS, Dulbecco's modified Eagle's medium, 20 mm HEPES pH 7.55 (Invitrogen). Differentiation day 0: cells grown to 100% confluence were treated with 20 nm insulin, 1 nm T3, 500 nm dexamethasone, 125 μm indomethacin, 500 μm isobutylmethylxanthine in 10% FBS. Differentiation day 2: medium was changed to 20 nm insulin, 1 nm T3 in 10% FBS, and replenished every 48 h. Full differentiation happened between days 5 and 8. Treatment with 10 μm forskolin activated the thermogenic program. Stimulation for 4 h induced Ucp1 and PGC-1α, mitochondrial-encoded genes required 24 h.
We induced 3T3 L1 adipocyte differentiation as previously described (40, 41). Differentiation day 0: cells grown to 100% confluence were treated with 20 nm insulin, 1000 nm dexamethasone, 250 μm isobutylmethylxanthine in 10% FBS. Differentiation day 2: medium was changed to 20 nm insulin, 10% FBS, and replenished every 48 h. Full differentiation happened between days 5 and 7.
siLRP130 and LRP130 Stable Cell Lines—We stably expressed shRNA constructs in cell lines referred to as siCtrl and siLRP130. Briefly, we cloned LRP130 RNAi sequence, 5′-GAAGCTAGATGACCTGTTT-3′ (Dharmacon), into pSuperRetro GFP (42), and produced retrovirus in phoenix cells. Infected brown preadipocytes were selected by MoFlo cell sorting. We previously demonstrated the specificity of this LRP130 RNAi in several cell types and by comparing it to a separate LRP130 RNAi sequence, 5′-GCAGTTAGGTGTCGTATAT-3′ (32). A control shRNA cell line was generated for comparison and the sequence has been previously published (43). Uldry et al. (27) previously described generation of PGC-1 double deficient brown fat cells.
3T3-F442A cells were transduced with pMSCV human LRP130 retrovirus. After digestion with BamHI/PmeI, human LRP130-FLAG was cloned into the BglII/HpaI site of pMSCV puromycin. To generate stable human LRP130-FLAG cells, we infected cells with the retrovirus and selected in 2 μg/ml puromycin. Empty pMSCV retrovirus of similar titer was used to create a control cell line. Culture conditions and differentiation were identical to brown fat cells. Retroviral infection occasionally reduced adipogenesis in 3T3-F442A cells; therefore, 1 μm rosiglitazone (Sigma-Aldrich) was used in a few experiments to promote full differentiation. Results were comparable to treatments without rosiglitazone.
Animal Experiments—All animal experiments were performed according to procedures approved by the Institutional Animal Care and Use Committee at the Dana-Farber Cancer Institute. Mice were maintained on standard rodent chow with 12-h light-dark cycles. For acute cold exposure studies, we housed male C57BL/6 mice, ages 3-4 weeks, at 4 °C for 4 h. Brown adipose tissue (BAT) was harvested and mRNA isolated.
Immunodetection of Proteins—Whole cell protein lysates were prepared by 3 freeze/thaw cycles in FLAG lysis buffer: 50 mm Tris-Cl, pH 7.8, 137 mm NaCl, 10 mm NaF, 1 mm EDTA, 1% Triton X-100, 10% glycerol, and 0.2% sarkosyl (N-lauryl sarcosine). Samples were clarified at 14,000 × g for 10 min at 4 °C, and quantified using Bradford reagent (44). Rabbit polyclonal antibody raised against the C terminus of LRP130 was generated by PrimmBiotech Inc. We detected PGC-1β with affinity purified rabbit polyclonal antibody. Antibodies against, C-V-α and NDUFA9, were purchased from Mitosciences. FLAG M2 monoclonal antibody and Ucp1 antibody are available from Sigma-Aldrich, and tubulin antibody from Abcam. We probed with FLAG antibody per the manufacturer's protocol. All other antibodies were used at 1:1000 in TBS/0.1% Tween-20, 5% nonfat dry milk.
Electron Microscopy and Stereological Measurements—Samples were fixed, sectioned (∼60 nm), and visualized on a JEOL 1200EX (27). We laid a grid onto randomly selected micrographs (n > 30), and counted points falling onto mitochondria or cell area. The fraction of total points attributable to mitochondria was expressed as mitochondrial density (45).
Reporter Assays—After stable transduction with pMSCV control or hLRP130, 3T3-F442A cells were transiently transfected in 6-well dishes using Fugene (Roche Applied Sciences). We used 500 ng of the -4kbto1+ bp Ucp1 promoter fused to a luciferase reporter (46), and 1500 ng of RSV-β-galactosidase to normalize each transfection. To exclude nonspecific activation, we used empty reporter vector. Reporter activity was measured on days 2, 4, and 8 of differentiation.
Chromatin Immunoprecipitation—Mature brown fat cells grown in a 15-cm dish were used for 4 chromatin immunoprecipitation reactions. The reactions were processed using the EZ ChIP kit from Upstate. PPARγ (H-100 X), FLAG M2, mouse, and rabbit IgG were purchased from Sigma. For each reaction, we used either 6 μg of IgG, 3 μg of PPARγ, or 3 μg of FLAG antibody. ChIP experiments were performed three times; one representative experiment is shown. The primers for the promoters were: Ucp1 enhancer, forward 5′-TGAGGCTGATATCCCCAGAGA-3′, reverse 5′-TCTGTGTGTCCTCTGGGCATAA-3′; Ucp1-200 bp promoter, forward 5′-AATCAGGAACTGGTGCCAAATC-3′, reverse 5′-AGGTCTCCAAAGAGCTGCTAGTG-3′; aP2 (FABP4) DR-1 sites -4 kb promoter forward 5′-TTCCCAGCAGGAATCAGGTAG-3′, reverse 5′-CTGGGAACTCCATTTGCTCTC-3′; 18S gene, forward 5′-AGTCCCTGCCCTTTGTACACA-3′, reverse 5′-CGATCCGAGGGCCTCACTA-3′.
Oxygen Consumption—Cells in suspension were washed twice in 10% FBS, Dulbecco's modified Eagle's medium, and resuspended in respiration mediuim: DPBS, 2 mm glucose, 1 mm pyruvate, 2% bovine serum albumin. Total respiration and proton leak (“uncouple respiration”) were measured as previously described (45, 47). We normalized respiration rates to an aliquot of total cell protein lysate.
RNA Analysis—For Northern blots, total RNA was isolated using Trizol (Invitrogen). 10-20 μg was resolved on a formaldehyde gel, transferred to nylon membrane, and hybridized with gene-specific probes. We used a 1430-2523 bp probe for LRP130, 1-810 bp for PGC-1α, and 1500-2400 bp for PGC-1β.
For quantitative PCR, total RNA was isolated with Trizol, and further purified with RNAesay columns (Qiagen). We reverse-transcribed RNA with a kit (Applied Biosystems), and measured signal intensity using SyberGreen (Applied Biosystems). Samples were normalized to TBP using the ΔΔCt method, and expressed as relative gene expression. Primer sequences are shown in supplemental Fig. S4.
Statistical Analysis—For bar graph analyses, we used two-way analysis of variance with Bonferroni post-test. One asterisk denotes p < 0.05, two: p < 0.01, three: p < 0.001.
RESULTS
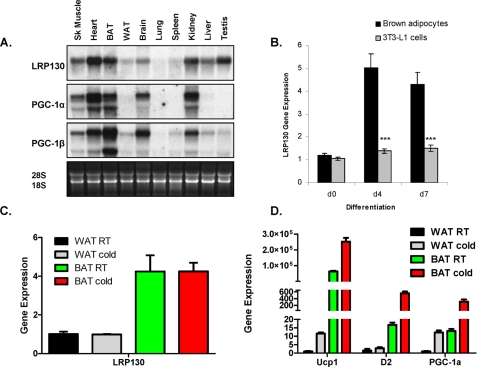
LRP130 Is Expressed in a Brown Fat-selective Manner—Certain regulators of brown fat development are selectively expressed in brown fat, and their expression may be further driven by differentiation or cold exposure. LRP130 mRNA is enriched in brown fat, compared with white. Its distribution across tissues is similar to PGC-1α and PGC-1β (Fig. 1A). LRP130 gene expression increases ∼5-fold during differentiation of immortalized brown fat cells, but does not change during differentiation of immortalized 3T3-L1 white fat cells (Fig. 1B). Upon cold exposure, however, LRP130 gene expression remains unchanged in brown fat (Fig. 1C). Notably Ucp1, PGC-1α and deiodinase 2 are all induced as expected (Fig. 1D). In parallel, stimulation of mature brown fat cells with forskolin does not induce LRP130 gene expression (data not shown).
FIGURE 1.
Spatiotemporal distribution of LRP130. A, tissue Northern of LRP130, PGC-1α, and PGC-1β. B, gene expression of LRP130 in differentiating brown fat cells versus 3T3 L1 cells. Differentiation mixture was added on day 0 (n = 3). C, gene expression of LRP130; D, select brown fat genes in white and brown fat depots derived from animals housed at room temperature or exposed to cold at 4 °C for 4 h (n = 5). Statistics: Panel D, no significant change in room temperature versus cold in WAT or BAT for LRP130. Panel C, p < 0.05; D2 changes in WAT not significant. Error bars represent mean (±S.E.). *, p < 0.05; **, p < 0.01; ***, p < 0.001.
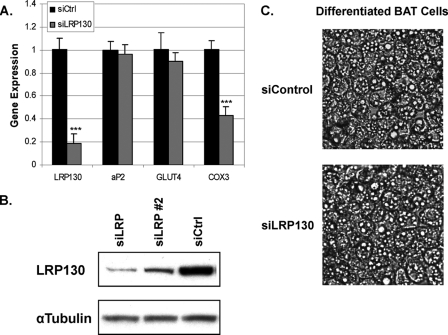
Effect of LRP130 Depletion on Adipogenesis and the Brown Fat Program—To assess the role of LRP130 in brown fat cell differentiation, we generated brown fat cells deficient for LRP130, using retroviral delivery of shRNA (Fig. 2A). To confirm that the shRNA against LRP130 is functional, we assessed protein level (Fig. 2B), and a target gene, Cox3 (Fig. 2A). Two genes, FABP4 (aP2) and Glut4, are induced during differentiation of brown or white fat cells. Expression of these genes were unaffected by depletion of LRP130, suggesting that adipogenesis is intact. Additionally, microscopic examination revealed no gross morphological defects (Fig. 2C), indicating LRP130 is not required for lipid accumulation.
FIGURE 2.
Adipogenic and morphological features of brown fat cells deficient for LRP130. A, gene expression of select genes at day 7 of differentiation (n = 3). B, protein expression of LRP130 in stably depleted brown fat cells at day 7 differentiation. A detailed gene expression profile for RNAi no. 2 is in supplemental Fig. S1. C, phase contrast of cells stably transduced with control shRNA (siControl) or shRNA against LRP130 (siLRP130). Error bars represent mean (±S.E.). *. p < 0.05; **, p < 0.01; ***, p < 0.001.
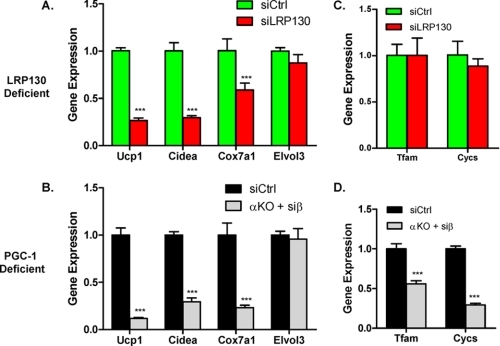
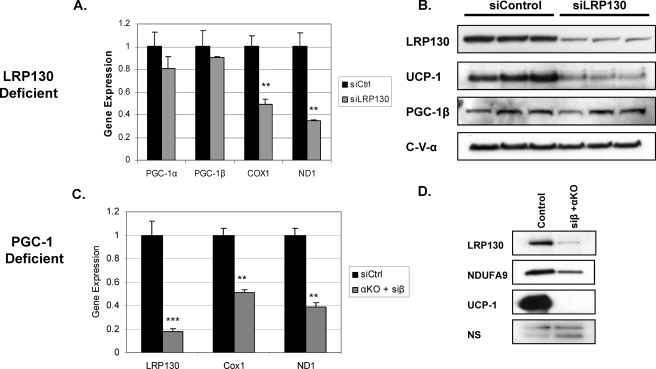
Next we evaluated the capacity of LRP130-deficient cells to regulate the brown adipocyte phenotype. As shown in Fig. 3A, several brown fat genes regulated by the PGC-1 coactivators are attenuated in LRP130-deficient cells, Ucp1, Cidea, Cox7a1, but not Elovl3. This is very similar to the pattern observed in PGC-1 double-deficient cells (Fig. 3B). Although moderately blunted in PGC-1 double-deficient cells, mRNA expression of mitochondrial transcription factor A (TFAM) is unaffected in LRP130-deficient cells (Fig. 3C). Similarly, the gene expression of cytochrome c (cycs), a component of the electron transport chain and marker of mitochondrial biogenesis, was unaffected by LRP130 depletion. Unlike cyctochrome c, Cox7a1 is a component of the electron transport chain that is also a brown fat-specific gene (21). Its gene expression is attenuated by LRP130 depletion, highlighting the importance of LRP130 in the brown fat program. Similar genetic findings using a second RNAi against LRP130 were observed (supplemental Fig. S1).
FIGURE 3.
LRP130 phenocopies certain features of PGC-1 double-deficient brown fat cells. Designated αKO + siβ, PGC-1-deficient cells are PGC-1α-null cells with stable shPGC-1β knock-down. Designated siCtrl, the control cells contain a control shRNA. Gene expression profile of brown fat genes and mitochondrial genes in LRP130-deficient (A and C) versus PGC-1 double-deficient mature brown fat cells (B and D) (n = 3). In comparison to siCtrl cell, PGC-1α-null cells stably expressing a control shRNA showed no difference (data not shown). Error bars represent mean (±S.E.). *, p < 0.05; **, p < 0.01; ***, p < 0.001.
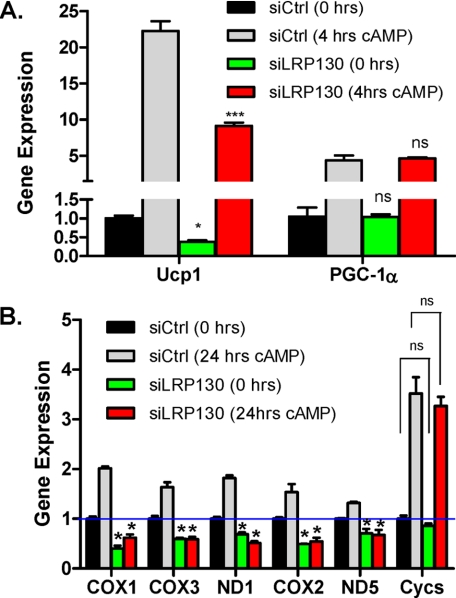
Prior studies have demonstrated that cAMP treatment or cold exposure induces PGC-1α, which then drives the expression of Ucp1 and deiodinase 2. To determine the effect of LRP130 on this program, we stimulated mature brown fat cells deficient for LRP130 with forskolin, an adenylate cyclase agonist. As shown in Fig. 4A, absolute expression of Ucp1 is reduced ∼70% in untreated and cAMP-treated LRP130-deficient cells; however, the fold change from baseline is identical to control cells, about 20-25-fold. Fold change and absolute level of PGC-1α was similar between both groups. Longer treatments with cyclic adenylate agonist induce several mitochondrial genes. As anticipated, cytochrome c (cycs), a nuclear encoded mitochondrial gene, is unaffected, but the mitochondrial-encoded genes are attenuated in LRP130-deficient cells (Fig. 4B). Although not required for Ucp1 induction following cAMP stimulation, LPR130 is required for the induction of the mitochondrial-encoded genes.
FIGURE 4.
Depletion of LRP130 does not alter cAMP mediated induction of Ucp1. A, gene expression of Ucp1 and PGC-1α in mature brown fat cells treated with forskolin for 4 h. Untreated siCtrl cells were statistically compared with untreated siLRP130 cells. cAMP-treated siCtrl cells were statistically compared with cAMP-treated siLRP130 cells (n = 4). B, gene expression of several mitochondrial-encoded genes, and nuclear-encoded cytochrome c (Cycs). 24 h of cAMP treatment proved optimal for inducing these genes (n = 4). Untreated siCtrl cells were statistically compared with untreated siLRP130 cells. cAMP-treated siCtrl cells were statistically compared with cAMP-treated siLRP130 cells. Note, except for ND5 all mitochondrial genes in the siCtrl cells were significantly induced (p < 0.05) following cAMP treatment. Statistical analysis: Two-way analysis of variance with Bonferroni post-test. Error bars represent mean (±S.E.). *, p < 0.05; **, p < 0.01; ***, p < 0.001.
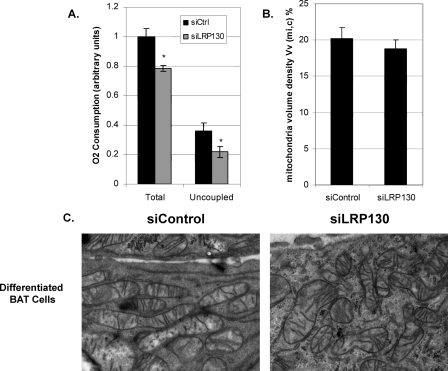
Depletion of LRP130 Attenuates Uncoupled Respiration without Impacting Mitochondrial Density—Increased oxygen consumption and mitochondrial biogenesis are important components of the brown fat program. Because LRP130 regulates Ucp1 and mitochondrial-encoded gene expression, we evaluated oxygen consumption and mitochondrial density. LRP130 depletion resulted in a modest 20% reduction in total respiration (Fig. 5A). This is comparable to the depletion of a single PGC-1 coactivator (27). Quantitative morphometry revealed that the decline was not attributable to decreased mitochondria (Fig. 5, B and C). These results indicate that LRP130 imposes qualitative, not quantitative, changes on mitochondria.
FIGURE 5.
LRP130-deficient brown fat cells have a specific block in proton leak. A, total respiration and proton leak (“uncoupled”) in intact mature brown fat cells (n = 6). B, mitochondrial volume density and C, electron micrographs of mitochondria from stable shControl (siControl) or shLRP130 (siLRP130) mature brown fat cells. Error bars represent mean (±S.E.). *, p < 0.05; **, p < 0.01; ***, p < 0.001.
Proton leak represents uncoupled respiration, largely attributable to Ucp1 action in brown adipose cells. In LRP130-deficient cells, uncoupled respiration is reduced by ∼44% compared with control (Fig. 5A). This effect is substantial and distinct from the modest defect in total respiration, supporting a specific role for LR130 in Ucp1 regulation. In summary, LRP130 is critical for respiratory uncoupling, but not mitochondrial biogenesis per se.
LRP130 Action on Brown Fat Genes Is Downstream of the PGC-1 Coactivators—As LRP130-deficient cells phenocopy several features of the PGC-1 double-deficient cells, perturbations in the expression of the PGC-1 coactivators might explain our findings. To address this possibility, we assessed the effect of LRP130 on the PGC-1 coactivators. No significant perturbation was observed in PGC-1 expression at the gene expression or protein level in LRP130-deficient cells (Fig. 6, A and B). The corresponding PGC-1α immunoblot is not included (Fig. 6B), since current commercial antibodies do not reliably detect endogenous PGC-1α in these cells. We also considered the possibility that the PGC-1 coactivators might regulate LRP130 expression. In cells null for PGC-1α or deficient in PGC-1β, LPR130 gene expression is not affected (supplemental Fig. S2). This indicates that single loss of PGC-α or PGC-1β has no impact on LRP130 gene expression. In contrast, LRP130 mRNA and protein were dramatically reduced in PGC-1 double-deficient cells (Fig. 6, C and D). These data indicate that LRP130 expression in brown fat cells is downstream of either PGC-1α or PGC-1β. Depletion of both coactivators severely attenuates LRP130 expression, a deficiency which likely attenuates certain brown fat genes.
FIGURE 6.
The PGC-1 coactivators regulate LRP130 gene expression. Expression of the PGC-1 coactivators, Ucp1 and some mitochondrial genes in LRP130-deficient cells. Left, mRNA (n = 3). Right, protein. (A, mRNA and B, protein) versus LRP130 and select mitochondrial gene expression in PGC-1 double-deficient cells (C, mRNA and D, protein). DesignatedαKO + siβ, PGC-1-deficient cells are PGC-1α-null cells with stable shPGC-1β knock-down. Designated siCtrl, control cells are wild-type cells stably expression a control shRNA. In comparison to siCtrl cell, PGC-1α-null cells stably expressing a control shRNA showed no difference (data not shown). In cells null for PGC-1α or deficient in PGC-1β, LPR130 gene expression was not affected (supplemental Fig. S2). Error bars represent mean (±S.E.). *, p < 0.05; **, p < 0.01; ***, p < 0.001.
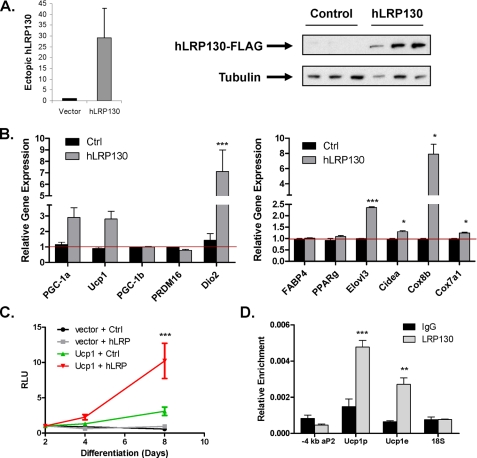
LRP130 Induces Ucp1 Gene Expression and Is Recruited to the Ucp1 Promoter—Our genetic studies indicate that LRP130 is required for certain brown fat pathways, including Ucp1 expression. The converse question is whether LRP130 is sufficient to drive these pathways. To investigate this question we used three strategies, one, gain-of-function, using forced expression of LRP130; two, reporter assays using the -4 kb Ucp1 promoter; three, chromatin immunoprecipitation assay assessing recruitment of LRP130 to the Ucp1 promoter. We chose 3T3-F442A cells (immortalized white adipocytes) for gain-of-function studies in lieu of brown fat cells (Fig. 7A), because the amount of LRP130 in brown fat cells is very high. As a result, LRP130 cannot be sufficiently increased to evaluate its effects. 3T3-F442A cells express comparably lower levels of LRP130, presumably because they represent white adipose cells (40, 48, 49). As shown in Fig. 7B, forced of expression of LRP130 induces several brown fat specific genes: deiodinase 2, PGC-1α, Cox8b, Elovl3, and Ucp1. Interestingly, PGC-1α gene expression is induced by LRP130 in 3T3-F442A cells, though unaffected in brown fat cells depleted of LRP130. Conversely, Cidea and Cox7a1 expression was not affected by forced expression of LRP130, though severely attenuated in brown fat cells depleted of LRP130. Thus, LRP130 induces part of the brown fat phenotype in white adipocytes These results support a model in which the PGC-1 coactivators drive LRP130 expression during differentiation; in turn, LRP130 regulates certain features of brown fat differentiation (Fig. 8).
FIGURE 7.
LRP130 induces Ucp1 and is enriched on the Ucp1 promoter. A, stable expression of exogenous human LRP130-FLAG in mature 3T3-F442A cells (n = 3). B, expression of brown fat genes in mature 3T3-F442A cells, stably expressing vector or human LRP130-FLAG (n = 3). C, luciferase reporter assay using the -4kbto 1+ bp Ucp1 promoter in mature 3T3-F442A cells. Measurements were performed on days 2, 4, and 8 of differentiation. RLU denotes relative luciferase units (n = 3). D, chromatin immunoprecipitation of LRP130 in mature brown fat cells. Q-PCR was used to quantify enrichment at the -4 kb PPRE of the aP2 promoter (-4kb aP2), + 1 to -200 bp region of the Ucp1 promoter (Ucp1 p), -2500 to -2300 bp region of the Ucp1 enhancer(Ucp1 e), and a random control site in the 18S gene. Error bars represent mean (±S.E.). *, p < 0.05; **, p < 0.01; ***, p < 0.001.
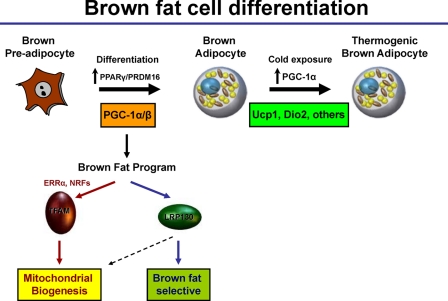
FIGURE 8.
Model depicting the role of LRP130 in brown fat cell differentiation and function. PPARγ and PRDM16 are important upstream regulators, induced after differentiation. Subsequently, the PGC-1 coactivators are induced to drive two programs, mitochondrial biogenesis and uncoupled respiration. The PGC-1s regulate ERRα and NRFs to induce TFAM and other factors critical for mitochondrial biogenesis. The PGC-1s induce LRP130, an induction important for remodeling of mitochondria. Inside the mitochondrion (dotted arrow), LRP130 regulates mitochondrial-encoded gene expression. Inside the nucleus it regulates Ucp1 expression, as well as, certain brown fat genes downstream of the PGC-1 coactivators. LPR130 may amplify the process by inducing PGC-1α in an autoregulatory loop. After basal Ucp1 expression is established, PGC-1α governs the cold response essential for thermogenesis.
The Ucp1 promoter is regulated by a number of factors, including PGC-1α. To assess the effect of LRP130, we used a region of the Ucp1 promoter fused to a luciferase reporter. 3T3-F442A cells were transfected with the murine -4kb to 1+ bp Ucp1 promoter or control vector, and differentiated for 8 days. Luciferase activity was measured on differentiation day 2, 4, and 8. As shown in Fig. 7C, luciferase activity is significantly induced at day 8 in cells expressing LRP130. These data indicate that LRP130 is sufficient to increase expression from the -4kb to 1+ bp Ucp1 promoter.
Presumably the regulatory effects of LRP130 on the Ucp1 promoter occur via a transcriptional mechanism. We therefore analyzed whether LRP130 is enriched on the Ucp1 promoter, using chromatin immunoprecipitates from brown fat cells. As shown in Fig. 7D, LRP130 is preferentially recruited to the promoter (-200 to 1+ bp) and enhancer (-2500 to -2300 bp) region of the Ucp1 -4kbto1+ bp promoter; not the aP2 promoter or 18S gene (negative controls). PPARγ served as positive control, since it is a strong regulator of aP2 expression and is also recruited to the Ucp1 promoter (supplemental Fig. S3). Overall, these findings indicate LRP130 is recruited to the Ucp1 promoter, likely to specify key transcriptional events.
DISCUSSION
In this report, we investigated the role of LRP130 in brown fat cell differentiation and gene expression. LRP130 functions downstream of the PGC-1 coactivators, and exhibits a spatiotemporal pattern coincident with brown fat and its cellular differentiation. Depletion of LRP130 impairs expression of several brown fat genes including Ucp1. Notably depletion does not impact adipogenesis, mitochondrial biogenesis or the relative induction of Ucp1, following cAMP activation. Exogenous expression of LRP130 in immortalized white adipocytes induces several brown fat specific genes. The effects are observed on the Ucp1 -4kbto1+ bp promoter, to which LRP130 is recruited. We propose a novel role for LRP130 in brown fat differentiation, and its requirement for the PGC-1-dependent brown fat program.
Brown fat differentiation is much more complex than previously understood. The PGC-1 coactivators play complementary roles in mitochondrial biogenesis and certain features of the brown fat program, including Ucp1 induction during differentiation. Thermogenesis in response to cold or cyclic AMP is an exception to complementary action of the PGC-1s: PGC-1α is the crucial family member that regulates thermogenesis (27). LRP130 is critical for the induction of Ucp1 during differentiation and essential for the complementary action of the PGC-1 coactivators. It is not required for mitochondrial biogenesis per se. Depletion of LRP130 in cAMP-treated cells impaired the absolute expression of Ucp1, not the fold change. This differs with our observations in primary hepatocytes, where LRP130 depletion severely impaired PGC-1α action on gluconeogenic genes following cAMP stimulation (32). Therefore, PGC-1α remains the only coactivator that regulates the cold response, despite striking similarity with PGC-1β. On the other hand, LRP130 is important in several complementary functions of the PGC-1 coactivators. The relationship of LRP130 to the PGC-1α is more complex than anticipated.
Although the exact mechanism whereby LRP130 regulates certain brown fat genes is not entirely clear yet, several possibilities exist. Transcriptional control is one potential candidate, as the PGC-1 proteins coactivate several transcription factors implicated in Ucp1 regulation. We analyzed immunoprecipitates of LRP130 for co-immunoprecipitation with CREB, PPARγ, ERRα, and other factors, without success (data not shown). Alternatively, other factors may be involved or LRP130 acts via another mechanism. One study proposed LRP130 might stabilize certain RNA transcripts (35). Because LRP130 interacts with the PGC-1 coactivators (32), drives brown fat gene expression in immortalized white fat cells, induces luciferase activity from the -4 kb Ucp1 promoter and is enriched on the promoter and enhancer of Ucp1, we favor a transcriptional mechanism. Nonetheless, we cannot rule out RNA trafficking and binding, or effects on protein translation. In fact, PGC-1α plays role in RNA processing, and LRP130 has been found to bind nuclear and mitochondrial RNA (50).
LRP130, the gene defective in Leigh Syndrome French Canadian variant (LSFC), has been reported as a mitochondrial disorder. Our study is the first to report the respiratory consequences of LRP130 deficiency in cells and the impact on mitochondrial density. Depletion of LRP130 modestly attenuated total mitochondrial respiration by 20%, with no impact on mitochondrial density. Surprisingly, this is similar to the ablation of one PGC-1 coactivator (27), despite global suppression of mitochondrial-encoded genes. Most of the mitochondrial-encoded proteins are likely stable, with a few being limited. Lack of severe respiratory defects highlights the importance of LRP130 nuclear function. Perhaps compounds that enhance interaction of LRP130 and the PGC-1 coactivators might favorably affect energy balance in obesity.
Supplementary Material
Acknowledgments
The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.
This work was supported, in whole or in part, by National Institutes of Health Grants R01DK61562 (to B. M. S.) and K08DK071017 (to M. P. C.). The costs of publication of this article were defrayed in part by the payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 U.S.C. Section 1734 solely to indicate this fact.
The on-line version of this article (available at http://www.jbc.org) contains supplemental Figs S1-S4.
Footnotes
The abbreviations used are: Ucp, uncoupling protein; BAT, brown adipose tissue; WAT, white adipose tissue; cyc, cytochrome c; FBS, fetal bovine serum.
References
- 1.Hansen, J. B., and Kristiansen, K. (2006) Biochem. J. 398 153-168 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Cannon, B., and Nedergaard, J. (2004) Physiol. Rev. 84 277-359 [DOI] [PubMed] [Google Scholar]
- 3.Hull, D., and Segall, M. M. (1965) J. Physiol 181 468-477 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Simon, H. E. (1950) Am. J. Surg. 80 127-130 [DOI] [PubMed] [Google Scholar]
- 5.Dawkins, M. J., and Hull, D. (1964) J. Physiol. 172 216-238 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Huttunen, P., Hirvonen, J., and Kinnula, V. (1981) Eur. J. Appl. Physiol. Occup. Physiol. 46 339-345 [DOI] [PubMed] [Google Scholar]
- 7.Ricquier, D., Nechad, M., and Mory, G. (1982) J. Clin. Endocrinol. Metab. 54 803-807 [DOI] [PubMed] [Google Scholar]
- 8.Lowell, B. B., Susulic, V., Hamann, A., Lawitts, J. A., Himms-Hagen, J., Boyer, B. B., Kozak, L. P., and Flier, J. S. (1993) Nature 366 740-742 [DOI] [PubMed] [Google Scholar]
- 9.Hamann, A., Flier, J. S., and Lowell, B. B. (1996) Endocrinology 137 21-29 [DOI] [PubMed] [Google Scholar]
- 10.Melnyk, A., Harper, M. E., and Himms-Hagen, J. (1997) Am. J. Physiol. 272 R1088-R1093 [DOI] [PubMed] [Google Scholar]
- 11.Kopecky, J., Clarke, G., Enerback, S., Spiegelman, B., and Kozak, L. P. (1995) J. Clin. Investig. 96 2914-2923 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kopecky, J., Hodny, Z., Rossmeisl, M., Syrovy, I., and Kozak, L. P. (1996) Am. J. Physiol. 270 E768-E775 [DOI] [PubMed] [Google Scholar]
- 13.Heaton, G. M., Wagenvoord, R. J., Kemp, A., Jr., and Nicholls, D. G. (1978) Eur. J. Biochem. 82 515-521 [DOI] [PubMed] [Google Scholar]
- 14.Ricquier, D., and Kader, J. C. (1976) Biochem. Biophys. Res. Commun. 73 577-583 [DOI] [PubMed] [Google Scholar]
- 15.Ricquier, D., Thibault, J., Bouillaud, F., and Kuster, Y. (1983) J. Biol. Chem. 258 6675-6677 [PubMed] [Google Scholar]
- 16.Murray, R., Granner, D., and Rodwell, V. (2006) Harper's Illustrated Biochemistry, 27 Ed., McGraw-Hill, New York
- 17.Rosen, E. D., Sarraf, P., Troy, A. E., Bradwin, G., Moore, K., Milstone, D. S., Spiegelman, B. M., and Mortensen, R. M. (1999) Mol. Cell 4 611-617 [DOI] [PubMed] [Google Scholar]
- 18.Tai, T. A., Jennermann, C., Brown, K. K., Oliver, B. B., MacGinnitie, M. A., Wilkison, W. O., Brown, H. R., Lehmann, J. M., Kliewer, S. A., Morris, D. C., and Graves, R. A. (1996) J. Biol. Chem. 271 29909-29914 [DOI] [PubMed] [Google Scholar]
- 19.Barbera, M. J., Schluter, A., Pedraza, N., Iglesias, R., Villarroya, F., and Giralt, M. (2001) J. Biol. Chem. 276 1486-1493 [DOI] [PubMed] [Google Scholar]
- 20.Cederberg, A., Gronning, L. M., Ahren, B., Tasken, K., Carlsson, P., and Enerback, S. (2001) Cell 106 563-573 [DOI] [PubMed] [Google Scholar]
- 21.Seale, P., Kajimura, S., Yang, W., Chin, S., Rohas, L. M., Uldry, M., Tavernier, G., Langin, D., and Spiegelman, B. M. (2007) Cell Metab. 6 38-54 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Debevec, D., Christian, M., Morganstein, D., Seth, A., Herzog, B., Parker, M., and White, R. (2007) Mol. Endocrinol. 21 1581-1592 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kiskinis, E., Hallberg, M., Christian, M., Olofsson, M., Dilworth, S. M., White, R., and Parker, M. G. (2007) EMBO J. 26 4831-4840 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Puigserver, P., Wu, Z., Park, C. W., Graves, R., Wright, M., and Spiegelman, B. M. (1998) Cell 92 829-839 [DOI] [PubMed] [Google Scholar]
- 25.Lin, J., Puigserver, P., Donovan, J., Tarr, P., and Spiegelman, B. M. (2002) J. Biol. Chem. 277 1645-1648 [DOI] [PubMed] [Google Scholar]
- 26.Andersson, U., and Scarpulla, R. C. (2001) Mol. Cell. Biol. 21 3738-3749 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Uldry, M., Yang, W., St Pierre, J., Lin, J., Seale, P., and Spiegelman, B. M. (2006) Cell Metab. 3 333-341 [DOI] [PubMed] [Google Scholar]
- 28.Lin, J., Wu, P. H., Tarr, P. T., Lindenberg, K. S., St Pierre, J., Zhang, C. Y., Mootha, V. K., Jager, S., Vianna, C. R., Reznick, R. M., Cui, L., Manieri, M., Donovan, M. X., Wu, Z., Cooper, M. P., Fan, M. C., Rohas, L. M., Zavacki, A. M., Cinti, S., Shulman, G. I., Lowell, B. B., Krainc, D., and Spiegelman, B. M. (2004) Cell 119 121-135 [DOI] [PubMed] [Google Scholar]
- 29.Leone, T. C., Lehman, J. J., Finck, B. N., Schaeffer, P. J., Wende, A. R., Boudina, S., Courtois, M., Wozniak, D. F., Sambandam, N., Bernal-Mizrachi, C., Chen, Z., Holloszy, J. O., Medeiros, D. M., Schmidt, R. E., Saffitz, J. E., Abel, E. D., Semenkovich, C. F., and Kelly, D. P. (2005) PLoS. Biol. 3 e101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lelliott, C. J., Medina-Gomez, G., Petrovic, N., Kis, A., Feldmann, H. M., Bjursell, M., Parker, N., Curtis, K., Campbell, M., Hu, P., Zhang, D., Litwin, S. E., Zaha, V. G., Fountain, K. T., Boudina, S., Jimenez-Linan, M., Blount, M., Lopez, M., Meirhaeghe, A., Bohlooly, Y., Storlien, L., Stromstedt, M., Snaith, M., Oresic, M., Abel, E. D., Cannon, B., and Vidal-Puig, A. (2006) PLoS. Biol. 4 e369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Sonoda, J., Mehl, I. R., Chong, L. W., Nofsinger, R. R., and Evans, R. M. (2007) Proc. Natl. Acad. Sci. U. S. A. 104 5223-5228 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Cooper, M. P., Qu, L., Rohas, L. M., Lin, J., Yang, W., Erdjument-Bromage, H., Tempst, P., and Spiegelman, B. M. (2006) Genes Dev. 20 2996-3009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Mootha, V. K., Lepage, P., Miller, K., Bunkenborg, J., Reich, M., Hjerrild, M., Delmonte, T., Villeneuve, A., Sladek, R., Xu, F., Mitchell, G. A., Morin, C., Mann, M., Hudson, T. J., Robinson, B., Rioux, J. D., and Lander, E. S. (2003) Proc. Natl. Acad. Sci. U. S. A. 100 605-610 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Morin, C., Mitchell, G., Larochelle, J., Lambert, M., Ogier, H., Robinson, B. H., and De Braekeleer, M. (1993) Am. J. Hum. Genet. 53 488-496 [PMC free article] [PubMed] [Google Scholar]
- 35.Xu, F., Morin, C., Mitchell, G., Ackerley, C., and Robinson, B. H. (2004) Biochem. J. 382 331-336 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Tsuchiya, N., Fukuda, H., Nakashima, K., Nagao, M., Sugimura, T., and Nakagama, H. (2004) Biochem. Biophys. Res. Commun. 317 736-743 [DOI] [PubMed] [Google Scholar]
- 37.Fasshauer, M., Klein, J., Kriauciunas, K. M., Ueki, K., Benito, M., and Kahn, C. R. (2001) Mol. Cell. Biol. 21 319-329 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Klein, J., Fasshauer, M., Klein, H. H., Benito, M., and Kahn, C. R. (2002) Bioessays 24 382-388 [DOI] [PubMed] [Google Scholar]
- 39.Tseng, Y. H., Kriauciunas, K. M., Kokkotou, E., and Kahn, C. R. (2004) Mol. Cell. Biol. 24 1918-1929 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Green, H., and Meuth, M. (1974) Cell 3 127-133 [DOI] [PubMed] [Google Scholar]
- 41.Spiegelman, B. M., Frank, M., and Green, H. (1983) J. Biol. Chem. 258 10083-10089 [PubMed] [Google Scholar]
- 42.Brummelkamp, T. R., Bernards, R., and Agami, R. (2002) Science 296 550-553 [DOI] [PubMed] [Google Scholar]
- 43.Fan, M., Rhee, J., St Pierre, J., Handschin, C., Puigserver, P., Lin, J., Jaeger, S., Erdjument-Bromage, H., Tempst, P., and Spiegelman, B. M. (2004) Genes Dev. 18 278-289 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Bearden, J. C., Jr. (1978) Biochim. Biophys. Acta 533 525-529 [DOI] [PubMed] [Google Scholar]
- 45.St Pierre, J., Lin, J., Krauss, S., Tarr, P. T., Yang, R., Newgard, C. B., and Spiegelman, B. M. (2003) J. Biol. Chem. 278 26597-26603 [DOI] [PubMed] [Google Scholar]
- 46.Cao, W., Daniel, K. W., Robidoux, J., Puigserver, P., Medvedev, A. V., Bai, X., Floering, L. M., Spiegelman, B. M., and Collins, S. (2004) Mol. Cell. Biol. 24 3057-3067 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Mootha, V. K., Handschin, C., Arlow, D., Xie, X., St Pierre, J., Sihag, S., Yang, W., Altshuler, D., Puigserver, P., Patterson, N., Willy, P. J., Schulman, I. G., Heyman, R. A., Lander, E. S., and Spiegelman, B. M. (2004) Proc. Natl. Acad. Sci. U. S. A. 101 6570-6575 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Green, H., and Kehinde, O. (1975) Cell 5 19-27 [DOI] [PubMed] [Google Scholar]
- 49.Green, H., and Kehinde, O. (1976) Cell 7 105-113 [DOI] [PubMed] [Google Scholar]
- 50.Mili, S., and Pinol-Roma, S. (2003) Mol. Cell. Biol. 23 4972-4982 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.