Abstract
In zebrafish, the expression of long-wavelength cone (LC) opsin mRNA fluctuated rhythmically between the day and night. In a 24-h period, expression was high in the afternoon and low in the early morning. This pattern of fluctuation persisted in zebrafish that were kept in constant darkness, suggesting an involvement of circadian clocks. Functional expression of Clock, a circadian clock gene that contributes to the central circadian pacemaker, was found to play an important role in maintaining the circadian rhythms of LC opsin mRNA expression. In zebrafish embryos, in which the translation of Clock was inhibited by anti-Clock morpholinos, the circadian rhythms of LC opsin mRNA expression diminished. CLOCK may regulate the circadian rhythms of LC opsin mRNA expression via cyclic adenosine monophosphate (cAMP)-dependent signaling pathways. In control retinas, the concentration of cAMP was high in the early morning and low in the remainder of the day and night. Inhibition of Clock translation abolished the fluctuation in the concentration of cAMP, thereby diminishing the circadian rhythms of opsin mRNA expression. Transient increase of cAMP concentrations in the early morning (i.e. by treating the embryos with 8-bromo-cAMP) restored the circadian rhythms of LC opsin mRNA expression in morpholino-treated embryos. Together, the data suggest that Clock plays important roles in regulating the circadian rhythms in photoreceptor cells.
Animals display robust day-night rhythms in behavior and physiology. The machinery that regulates the daily rhythms is similar across species. In vertebrates, the daily rhythms are generated by interlocked transcription-translation of core circadian genes, which include Clock, Bmal1, Period, and Cryptochrome. The transcription-translation of some circadian clock genes fluctuates in ∼24-h cycles. In mammals, the central circadian pacemaker is located in the suprachiasmatic nuclei. In lower vertebrates, such as bird, frog, or fish, pacemakers are located in the pineal gland. The output of the central pacemaker transmits rhythmic signals to other tissues via cell-cell communication or circulation (1–3).
The neural retina is considered an important part of the circadian system. The retina is capable of generating self-sustained and robust circadian oscillations, including rhythms of photoreceptor outer segment disk shedding, retinomotor movements, melatonin synthesis, and opsin expression (4–6). In frogs, the expression of rod opsin mRNA fluctuates between the day and night (7). This pattern of fluctuation in opsin expression persists in animals kept in constant darkness (DD),2 suggesting an involvement of circadian clocks. In chick retinas, the expression of cone opsin shows a robust circadian rhythm that persists in cultured cells in DD (8). Thus, the circadian oscillators that control the rhythms of cone opsin expression appear to be located in the photoreceptor cells.
The circadian rhythms of photoreceptor cells may be entrained by a variety of environmental factors, such as light exposure (9, 10). However, intrinsic mechanisms that regulate the circadian rhythms of photoreceptor cell functions remain to be determined. Recent studies have suggested that the expression of Clock may play a role in photoreceptor cell circadian rhythms (11). However, in some tissues NPAS2 may substitute for CLOCK in the circadian oscillator (12, 13), suggesting that the expression of Clock may not be essential for generating rhythms. In this study, we examined the roles of Clock in the regulation of the circadian rhythms of opsin mRNA expression in zebrafish retinal photoreceptor cells. We demonstrate that functional expression of Clock is required for maintaining the circadian rhythms of the expression of long-wavelength cone (LC) opsin mRNA in photoreceptor cells. The effect is likely mediated by cyclic adenosine monophosphate (cAMP)-dependent signal transduction pathways. The data suggest that Clock plays important roles in the regulation of the circadian rhythms of oscillators, such as those in photoreceptor cells.
EXPERIMENTAL PROCEDURES
Animals and Maintenance—Zebrafish (Danio rerio) were maintained as described (14). Normally, the embryos were maintained under cyclic light and darkness (LD; fluorescent light, 07:00 to 21:00). For DD experiments, the embryos were removed from LD at 21:00 the day before the experiment and thereafter kept in DD. All of the experimental procedures adhered to the National Institutes of Health Guidelines for Animals in Research.
Total RNA Extraction and Real Time Reverse Transcription (RT)-PCR—Total RNA was extracted from whole embryos. Embryos (5–7 embryos per sample) were homogenized in 500 μl of RNAwiz (Ambion, Austin, TX), followed by chloroform extraction. The RNA was precipitated with isopropyl alcohol, washed with 75% ethyl alcohol, and resuspended in 20 μl of distilled H2O (RNase-free). LC opsin-specific primers and probes (GenBank™ sequence accession number AF109371, 5′-TGG AGC AGA TAC TGG CCT CAT-3′ and 5′-GGG TCC TCG CTT CCA CTG A-3′, TaqMan probe 5′-TCT GAA GAC CTC CTG TGG CCC TGA TG-3′) and Per3-specific primers and probes (GenBank™ sequence accession number NM131584, 5′-GAC GAC CGG CTG CTC ATG-3′ and 5′-GAG GCT GAC CCG CGT ACT T-3′, TaqMan probe 5′-TGG CCA TGC ACC GCA AAA TTG T-3′) were designed according to known sequences using the Primer Express System (Applied Biosystems, Foster City, CA).
Real time RT-PCR was performed using the TaqMan One-Step RT-PCR Master Mix reagents kit (Applied Biosystems). The reaction (25 μl) contained 2 ng of total RNA, 300 nm primers, and 250 nm probe. Each sample was run in duplicate along with control reactions, which did not include reverse transcriptase and template. TaqMan ribosomal RNA was used as an internal control. The thermal cycling conditions were 30 min at 48 °C, 10 min at 95 °C, and 45 cycles of 15 s at 95 °C and 1 min at 60 °C. Standard dilution curves of cDNA were generated for both opsin mRNA and rRNA as previously described (15). To normalize the data to the endogenous control rRNA, the amount of LC opsin mRNA, Per3 mRNA, and rRNA was determined from the standard curve for each sample. Relative LC opsin mRNA expressions were determined by dividing the concentration of mRNA obtained at each time in the day and night by the concentration of mRNA obtained at 07:00.
Radioimmunoassays—Retinas (10–15 eyes per sample) were homogenized in 6% trichloroacetic acid. To determine the recovery of cAMP during extraction, ∼4,000 dpm of [3H]cAMP marker (Perkin-Elmer Life Sciences) was added to each sample. Homogenates were centrifuged at 2,500 × g at 4 °C for 15 min, and the supernatant fluid was removed and extracted three times with 5 ml of ethyl ether saturated in water. The extracted aqueous phase was evaporated, and the residue was dissolved in sodium acetate buffer, pH 6.2. Proteins in the pellet were solubilized in 0.1 n sodium hydroxide and quantified using bovine serum albumin as a standard. cAMP levels were measured by radioimmunoassay according to the manufacturer's instructions, normalized to the protein concentration, and expressed as pmol of cAMP per mg of protein.
Morpholino Injections—Morpholinos (MOs) were designed according to the 5′ sequence of the untranslated region of the Clock gene (16). The sequence for MO was 5′-CAT CCC GGT CTA TGC TGG AGG TCA T-3′ and for mismatch MO 5′-CAT GCC GCT CTA AGC TGG ACG TGA T-3′. MOs were diluted in Danieau buffer at a final concentration of 0.5 μg/μl. MOs or mismatch MOs were injected (4.6 ng) into 1-cell stage wild-type embryos. Mismatch MOs did not inhibit Clock mRNA translation because they contained five nucleotides that did not match the sequence of 5′ Clock mRNA. When injected at concentrations of 1–5 ng, MOs did not appear to disrupt the course of eye development. In most cases, the embryos showed normal eye morphology. Individuals that showed obvious defects in eye morphology were excluded from this study. The embryos were allowed to grow to 6 days post-fertilization and sacrificed for PCR, cAMP, or Western blot analyses.
Western Blot—Embryos (5–7 embryos per sample) were homogenized in a buffer containing 150 mm NaCl, 20 mm HEPES (pH 8), 1 mm EDTA, 1 mm EGTA, 1 mm dithiothreitol, 1 mm NaVO4, 1 mm phenylmethylsulfony fluoride, 1 μg/ml leupeptin, 1 μg/ml pepstatin, and 1 mM PMSF. Homogenates were centrifuged at 10,000 × g, and the supernatants were collected. Total proteins (35 μg) were separated in a 6% SDS-polyacrylamide gel and transferred to a nitrocellulose membrane. The membrane was blocked with Tris/NaCl/Tween 20 buffer for 1 h at room temperature and incubated with goat anti-CLOCK polyclonal antibody (1:100; Santa Cruz Biotechnology, Santa Cruz, CA) and goat anti-actin polyclonal antibody (1:1,000; Santa Cruz Biotechnology) overnight at 4 °C. The membrane was then incubated in horseradish peroxidase-conjugated rabbit anti-goat IgG (1:7,500; Jackson ImmunoResearch Laboratories, West Grove, PA) for 1 h at room temperature. CLOCK immunoreactivity was detected with an ECL Western blotting detection analysis system (Amersham Biosciences) with subsequent exposures to autoradiographic films.
RESULTS
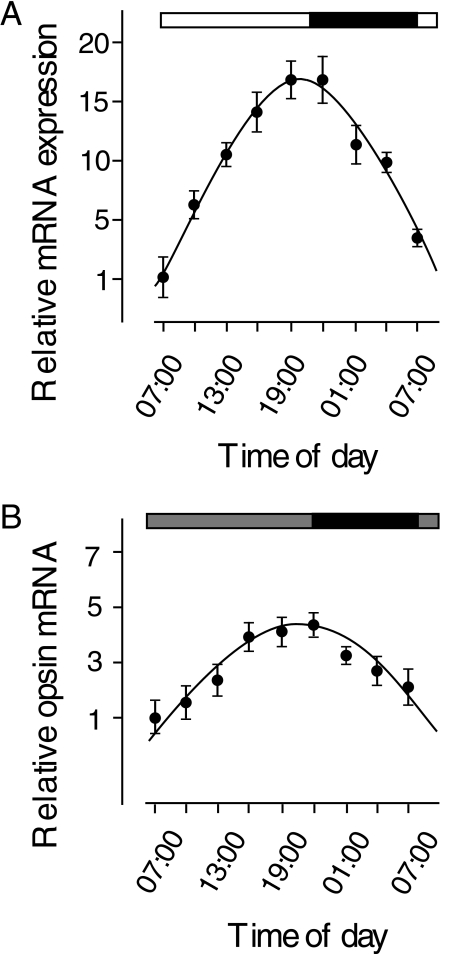
Circadian Rhythms of LC mRNA Expression—LC opsin mRNA is specifically expressed in red cone photoreceptor cells. We measured the expression of LC opsin mRNA at different times in the day and night while the zebrafish embryos (6 days post-fertilization) were kept in either LD or DD conditions. Under either condition, the expression of LC opsin mRNA fluctuated rhythmically between the day and night (Fig. 1). In LD, the expression was low in the early morning and high in the late afternoon and early evening. In the evening, the expression of LC mRNA increased ∼15-fold compared with the expression in the early morning. At night, the expression of LC opsin mRNA gradually decreased. The expression reached the lowest level in the early morning on the following day. The same pattern of fluctuation in LC mRNA expression persisted in zebrafish embryos kept in DD, except that the increase in LC opsin mRNA in the afternoon was not as great as the increase seen in embryos that were kept in LD conditions. In DD, the expression of LC opsin mRNA was ∼4-fold higher in the afternoon than the expression in the early morning.
FIGURE 1.
Circadian rhythms of LC opsin mRNA expression in developing zebrafish embryos (6 days post-fertilization) that were kept in LD (A) or DD (B). Relative expression at different times in the day and night was normalized to the lowest expression in a 24-h period (07:00). In both LD and DD conditions, the expression of LC opsin mRNA fluctuated rhythmically between the day and night. Horizontal bars indicate lighting conditions; black bar, night; white bar, daytime in light; gray bar, subjective day without light. Data represent the means ± S.E. (n = 6).
CLOCK Is Required for Maintaining the Circadian Rhythms of LC Opsin mRNA Expression—Previous studies have shown that most tissues express circadian clock genes (1–3). We examined the expression of some core circadian clock genes (Clock and Per3) in zebrafish red cone photoreceptor cells. In zebrafish, red cone photoreceptor cells are the principal member of double cone photoreceptors cells (17), which can be readily identified by their morphology (Fig. 2, A–C). RT-PCR analyses revealed that in ∼40% of the red cone cells examined both Clock and Per3 were expressed (Fig. 2D). LC opsin mRNA expression was detected in the eye but not in other tissues (Fig. 2E).
FIGURE 2.
Coexpression of Clock, Per3, and LC opsin mRNA in red cone photoreceptor cells. A, diagram showing the principal (red cone) and associate (green cone) members of double cone photoreceptor cells. B, C, images that show the morphology of double cone cells and immunoreactivity of red cone opsin antibodies in the outer segment of red cone cell. Scale bar, 30 μm. D, single-cell RT-PCR showing the expression of Clock, Per3, and LC opsin in a red cone photoreceptor cell. Molecular markers are shown in the left lane of the gel. E, RT-PCR showing that LC opsin mRNA is expressed in the eye but not in embryos in which the eyes were enucleated. Actin (control) was detected in both samples.
To determine whether Clock regulates the circadian rhythms of LC opsin mRNA expression, we measured LC opsin mRNA expression at different times in the day and night in which the translation of Clock was inhibited by anti-Clock MOs. MOs block gene translation by annealing to the 5′ untranslated region of mRNA (16). MOs or mismatch MOs were injected into one- to four-cell stage embryos. The embryos were sacrificed at different times in the subjective day and night at 6 days post-fertilization, and the translation of Clock and the expression of LC opsin mRNA were assessed.
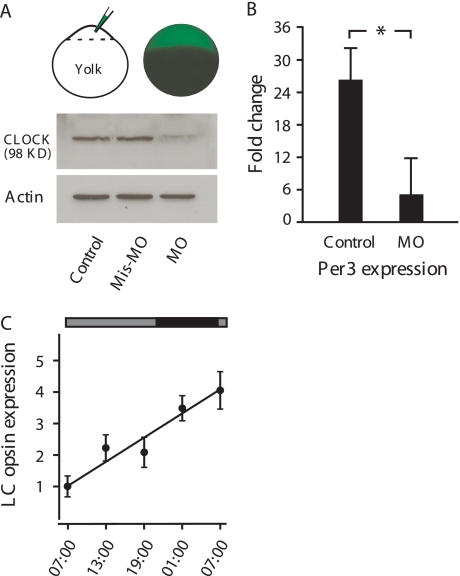
The injection of MOs efficiently blocked the translation of Clock mRNA into protein. In MO-injected embryos, Western blot analyses showed no CLOCK expression. In mismatch MO-injected embryos, the expression of CLOCK was evident and was similar to the expression seen in untreated embryos (Fig. 3A). Due to the inhibition of Clock mRNA translation, the expression of the central circadian machinery appeared to be disrupted. For example, the circadian rhythms of Per3 expression, which are normally antiphasic to the expression of Clock, were no longer evident. In control embryos, Per3 expression was ∼25-fold higher in the early morning than in the evening (Fig. 3B). In MO-injected embryos, the differences in Per3 expression decreased to 5-fold (Fig. 3B).
FIGURE 3.
Effects of MOs on Clock, Per3, and LC opsin mRNA expression. A, MOs were injected into 1- to 4-cell stage embryos. At 8 h, MOs (tagged with green fluorescent proteins) were seen in all of the dividing cells. Western blot analyses show that in control and mismatch MO-injected embryos CLOCK was detected by CLOCK antibodies. In MO-injected embryos, no obvious CLOCK expression was detected. Actin (control) was detected in all of the samples. B, fold changes in Per3 mRNA expression between early morning and late evening in control and MO-injected embryos are shown. In control embryos, Per3 expression was 25-fold higher in the morning than in the evening. In MO-treated embryos, no significant differences in Per3 expression were detected. *, p < 0.005. C, relative LC opsin mRNA expression in the subjective day and night in MO-injected embryos is shown. Horizontal bars on the top of the figure indicate lighting conditions; black bar, night; gray bars, subjective day without light. Data represent the means ± S.E. (n = 6).
Blocking the translation of Clock resulted in disruption of the circadian rhythms of LC opsin mRNA expression (Fig. 3C). Whereas the level of basal LC mRNA expression in the early morning remained unaffected, the expression showed no pattern of fluctuation. Instead, during the subjective day and night, the expression of LC opsin mRNA increased steadily. No decreases in the expression were observed during subjective night. By the early morning on the following day, the expression of LC mRNA increased ∼4-fold compared with the expression measured in the early morning on the previous day.
CLOCK May Regulate the Circadian Rhythms of LC Opsin mRNA Expression via cAMP—In the retina, the concentrations of cAMP fluctuated between the day and night, apparently generated by clock-controlled circadian expression of the Type 1 adenylyl cyclase (18–20). Because cAMP influences opsin mRNA expression (21, 22), we hypothesized that CLOCK may regulate the circadian rhythms of LC opsin mRNA expression via cAMP signaling pathways. For example, it is possible that the loss of the regulatory effect of CLOCK on cAMP synthesis leads to a disruption of the circadian rhythms of cAMP production. Consequently, the circadian rhythms of LC opsin mRNA expression are disrupted.
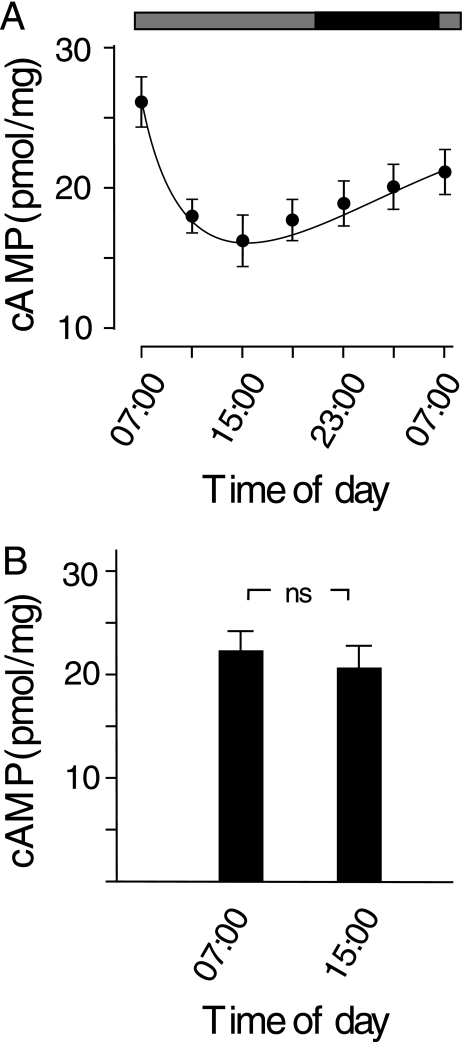
We measured the concentration of cAMP in the retina of control and MO-treated embryos at different times in the day and night in DD. In control embryos, the concentration of cAMP fluctuated rhythmically: cAMP concentrations were high in the subjective early morning hours and low in the remainder of the day. After the subjective midnight, the concentration of cAMP gradually increased (Fig. 4A). In embryos treated with MOs, however, the fluctuation of cAMP concentrations between the subjective day and night was no longer evident. When measured at 07:00 and 15:00 in DD, for example, the concentration of cAMP was 22.3 ± 2.2 and 20.2 ± 2.3 pmol/mg, respectively (Fig. 4B).
FIGURE 4.
Fluctuations in cAMP concentrations (pmol/mg) in developing eyes in DD. A, in control samples, cAMP concentrations were high in the subjective morning (07:00) and low in the subjective day and early night. Horizontal bars indicate lighting conditions; black bar, night; gray bars, subjective day without light. B, retinal cAMP concentrations are shown in MO-treated embryos measured at subjective 07:00 and 15:00, respectively. n.s., not significant. Data represent the means ± S.E. (n = 6).
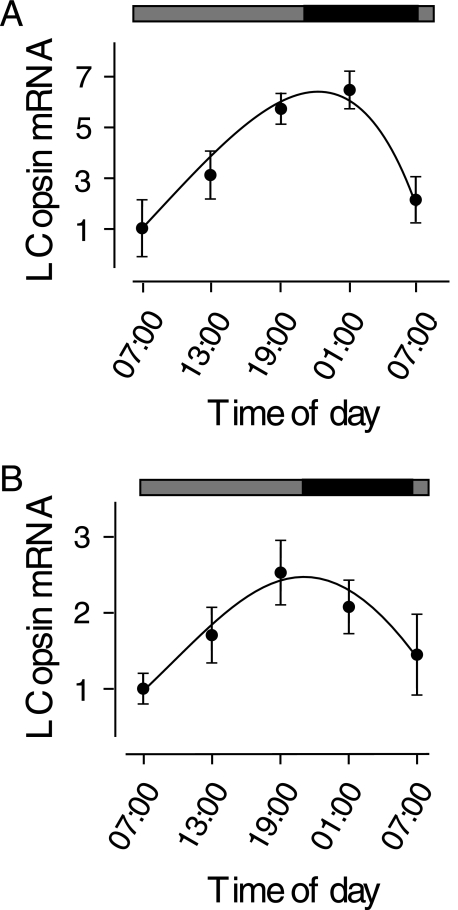
To determine whether the fluctuation of cAMP concentrations leads to circadian expression of LC opsin mRNA, we measured the expression of LC opsin mRNA in control and MO-injected embryos that were treated with 8-bromo-cAMP (8Br-cAMP, a membrane-permeable cAMP analog) in the early morning hours, when cAMP concentrations are high in control animals. In control embryos, treatment with 8-Br-cAMP (5 μm) did not alter the overall circadian pattern of LC opsin mRNA expression, except that the peak expression of LC opsin mRNA shifted a few hours in comparison to the peak expression in untreated control embryos (Fig. 5A). In MO-treated embryos, transient increases in cAMP concentrations (by administering 5 μm 8-Br-cAMP between 01:00 and 02:00 the night prior to sampling) resulted in restoration of the circadian rhythms of LC opsin mRNA expression (Fig. 5B). After 8-Br-cAMP treatment, the expression of LC opsin mRNA in MO-treated embryos gradually increased. The expression peaked in the late afternoon (19:00) and was ∼2.5-fold higher than the expression in the early morning. In the evening, the expression of LC opsin mRNA gradually decreased. The decrease in LC opsin expression continued at night. By 07:00 on the second day, the expression decreased to levels that were similar to those seen in the morning of the first day.
FIGURE 5.
Effects of 8-Br-cAMP (5 μm) treatment on circadian rhythms of LC opsin mRNA expression in control and MO-treated embryos. A, in control animals, the circadian rhythms of LC opsin mRNA expression remained relatively unchanged following 8-Br-cAMP treatment, except that the peak of expression was slightly delayed relative to the peak expression in untreated controls. B, in MO-injected animals, treatment with 8-Br-cAMP resulted in restoration of the circadian rhythms of LC opsin mRNA expression. The expression of LC opsin mRNA increased in the day, peaked in the evening, and decreased at night, similar to the expression pattern in control embryos. Black bar, night; gray bars, subjective day without light. Data represent the means ± S.E. (n = 6).
DISCUSSION
Retinal photoreceptor cells may function as independent circadian oscillators. In a recent study, Yu et al. (23) demonstrated that in isolated retinas the expression of the zebrafish rhodopsin promoter displayed robust day-night rhythms. However, the rhythms were not synchronized among individual photoreceptor cells. In some cells, the expression peaked in the early morning, whereas in other cells the expression peaked in midday or at night. The data suggest that in isolated retinal tissues individual oscillators, which no longer receive output from the central pacemaker, may act under their own circadian control. In the present study, we have demonstrated that the endogenous circadian control is from the expression of some core circadian genes, such as Clock and that the amplitude of relative opsin expression is reduced more in DD than in LD (Fig. 1). Decreases in the circadian expression of opsin genes in DD have been previously reported. The expression of opsin is regulated not only by endogenous mechanisms (i.e. circadian clocks) but also by environmental factors, particularly light. It has been reported that either light exposure or prolonged darkness may alter the expression of opsin genes (5).
Although the circadian clock genes are expressed in most tissues, LC opsin mRNA is expressed specifically in photoreceptor cells. Functional expression of Clock is required for maintaining the circadian rhythms of LC opsin mRNA expression in photoreceptor cells. When the translation of Clock mRNA was inhibited, the circadian rhythms of LC opsin mRNA expression diminished. The regulatory effect of Clock on LC opsin mRNA expression is mediated by cAMP-dependent cascades; i.e. Clock regulates rhythmic synthesis of cAMP, which in turn regulates circadian expression of LC opsin mRNA. It must be noted that the expression of cAMP and LC opsin mRNA was antiphasic. When cAMP concentrations in the retina were high in the early morning (07:00), the expression of LC opsin mRNA was low. Peak expression of LC opsin mRNA was detected ∼12 h after the peak expression of cAMP. The delay between the peak expressions of cAMP and opsin mRNA is consistent with previous evidence that protein kinase A inhibits opsin expression (21).
In control embryos, transient increases in cAMP concentrations resulted in increases in the expression of LC opsin mRNA. The effect was only intermediate, however (i.e. 1 to 2 h). Treatment with 8-Br-cAMP had little effect on the circadian rhythms of opsin mRNA expression. By contrast, in MO-treated embryos, in which the endogenous control of cAMP synthesis was disrupted, transient increases in cAMP concentration might have had a significant impact on the expression of opsin mRNA, i.e. by resetting the circadian rhythms of the expression of opsin mRNA.
Among the red cone photoreceptor cells examined (n = 17), we found coexpression of Clock, Per3, and opsin mRNA in seven cells. Lack of the coexpression of circadian clock genes in photoreceptor cells has been reported in mice (24). This may be due to several factors; for example, some of the circadian clock genes may not be expressed in photoreceptor cells, or they may be expressed but at levels too low to detect by single-cell RT-PCR. In such cases, the circadian rhythms of photoreceptor cells may be influenced by signals from other retinal cell types, such as dopaminergic amacrine cells (25, 26). Using a transgenic approach, Yu et al. (23) demonstrated that light exposures, via dopaminergic signaling pathways, synchronized the circadian rhythms of zebrafish rhodopsin promoter expression via cAMP-dependent protein kinase A pathways. Recently, Ruan et al. (24) suggested that, in mouse retinas, the expression of circadian clock genes may only play regulatory roles in gene expression, but exogenous signals control the circadian rhythmicity in photoreceptor cells.
In summary, this study provides evidence that peripheral clock cells, such as retinal photoreceptor cells, may function under the influence of the expression of core circadian clock genes. It is interesting that not every retinal cell type expresses the core circadian genes. The cell types that do express core circadian genes may set the pace and synchronize the circadian rhythms of photoreceptor cells via dopaminergic- or cAMP-dependent signal transduction pathways.
Acknowledgments
We thank Deb Bang of the Center for Zebrafish Research at the University of Notre Dame for maintaining zebrafish colonies and C. Yu, B. Belliveau, and L. Huang for technical assistance.
This work was supported, in whole or in part, by National Institutes of Health Grants R01 EY013147 (to L. L.), EY004864 and EY 014764 (to P. M. I.), and Core Grant for Vision Research P30 EY03630. The costs of publication of this article were defrayed in part by the payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 U.S.C. Section 1734 solely to indicate this fact.
Footnotes
The abbreviations used are: DD, constant darkness; LD, cyclic light and darkness; LC, long-wavelength cone; cAMP, cyclic adenosine monophosphate; 8-Br-cAMP, 8-bromo-cAMP; MO, morpholino; RT, reverse transcription.
References
- 1.Takahashi, J. S. (1995) Annu. Rev. Neurosci. 18 531-553 [DOI] [PubMed] [Google Scholar]
- 2.Young, M. W., and Kay, S. A. (2001) Nat. Rev. Genet. 2 702-715 [DOI] [PubMed] [Google Scholar]
- 3.Reppert, S. M., and Weaver, D. R. (2002) Nature 418 935-941 [DOI] [PubMed] [Google Scholar]
- 4.Green, C. B. (2003) J. Neuroendocrinol. 15 350-354 [DOI] [PubMed] [Google Scholar]
- 5.Green, C. B., and Besharse, J. C. (2004) J. Biol. Rhythms 19 91-102 [DOI] [PubMed] [Google Scholar]
- 6.Iuvone, P. M., Tosini, G., Pozdeyev, N., Haque, R., Klein, D. C., and Chaurasia, S. S. (2005) Prog. Retin. Eye Res. 24 433-456 [DOI] [PubMed] [Google Scholar]
- 7.Korenbrot, J. I., and Fernald, R. D. (1989) Nature 337 454-457 [DOI] [PubMed] [Google Scholar]
- 8.Pierce, M. E., Sheshberadaran, H., Zhang, Z., Fox, L. E., Applebury, M. L., and Takahashi, J. S. (1993) Neuron 10 579-584 [DOI] [PubMed] [Google Scholar]
- 9.Besharse, J. C., Hollyfield, J. G., and Rayborn, M. E. (1977) Science 196 536-538 [DOI] [PubMed] [Google Scholar]
- 10.Cahill, G. M., and Besharse, J. C. (1993) Neuron 10 573-577 [DOI] [PubMed] [Google Scholar]
- 11.Hayasaka, N., LaRue, S. I., and Green, C. B. (2002) J. Neurosci. 22 1600-1607 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Reick, M., Garcia, J. A., Dudley, C., and McKnight, S. L. (2001) Science 293 506-509 [DOI] [PubMed] [Google Scholar]
- 13.DeBruyne, J. P., Weaver, D. R., and Reppert, S. M. (2007) Nat. Neurosci. 10 543-545 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Westerfield, M. (1995) The Zebrafish Book, University of Oregon Press, Eugene, OR
- 15.Li, P., Temple, S., Gao, Y., Haimberger, T. J., Hawryshyn, C. W., and Li, L. (2005) J. Exp. Biol. 208 497-504 [DOI] [PubMed] [Google Scholar]
- 16.Corey, D. R., and Abrams, J. M. (2001) Genome Biol. 2 1015-1015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Robinson, J., Schmitt, E. A., Hárosi, F. I., Reece, R. J., and Dowling, J. E. (1993) Proc. Natl. Acad. Sci. U. S. A. 90 6009-6012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ivanova, T. N., and Iuvone, P. M. (2003) Brain Res. 991 96-103 [DOI] [PubMed] [Google Scholar]
- 19.Fukuhara, C., Liu, C., Ivanova, T. N., Chan, G. C., Storm, D. R., Iuvone, P. M., and Tosini, G. (2004) J. Neurosci. 24 1803-1811 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Chaurasia, S. S., Haque, R., Pozdeyev, N., Jackson, C. R., and Iuvone, P. M. (2006) J. Neurochem. 99 1142-1150 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Alfinito, P. D., and Townes-Anderson, E. (2001) J. Neurochem. 76 881-891 [DOI] [PubMed] [Google Scholar]
- 22.Yu, C. J., Gao, Y., Willis, C. L., Li, P., Tiano, J. P., Nakamura, P. A., Hyde, D. R., and Li, L. (2007) J. Neurosci. Res. 85 488-496 [DOI] [PubMed] [Google Scholar]
- 23.Yu, C. J., Gao, Y., Li, P., and Li, L. (2007) J. Exp. Biol. 210 676-684 [DOI] [PubMed] [Google Scholar]
- 24.Ruan, G. X., Zhang, D. Q., Zhou, T., Yamazaki, S., and McMahon, D. G. (2006) Proc. Natl. Acad. Sci. U. S. A. 103 9703-9708 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ribelayga, C., and Mangel, S. C. (2003) J. Comp. Neurol. 467 243-253 [DOI] [PubMed] [Google Scholar]
- 26.Zhang, D. Q., Zhou, T. R., and McMahon, D. G. (2007) J. Neurosci. 27 692-699 [DOI] [PMC free article] [PubMed] [Google Scholar]