Abstract
Human plasma platelet-activating factor (PAF) acetylhydrolase functions by reducing PAF levels as a general anti-inflammatory scavenger and is linked to anaphylactic shock, asthma, and allergic reactions. The enzyme has also been implicated in hydrolytic activities of other pro-inflammatory agents, such as sn-2 oxidatively fragmented phospholipids. This plasma enzyme is tightly bound to low and high density lipoprotein particles and is also referred to as lipoprotein-associated phospholipase A2. The crystal structure of this enzyme has been solved from x-ray diffraction data collected to a resolution of 1.5Å. It has a classic lipase α/β-hydrolase fold, and it contains a catalytic triad of Ser273, His351, and Asp296. Two clusters of hydrophobic residues define the probable interface-binding region, and a prediction is given of how the enzyme is bound to lipoproteins. Additionally, an acidic patch of 10 carboxylate residues and a neighboring basic patch of three residues are suggested to play a role in high density lipoprotein/low density lipoprotein partitioning. A crystal structure is also presented of PAF acetylhydrolase reacted with the organophosphate compound paraoxon via its active site Ser273. The resulting diethyl phosphoryl complex was used to model the tetrahedral intermediate of the substrate PAF to the active site. The model of interface binding begins to explain the known specificity of lipoprotein-bound substrates and how the active site can be both close to the hydrophobic-hydrophilic interface and at the same time be accessible to the aqueous phase.
Platelet-activating factor (PAF, 1-O-alkyl-2-acetyl-sn-glycero-3-phosphocholine)2 is a phospholipid messenger synthesized by a variety of cells involved in host defense, such as endothelial cells, platelets, neutrophils, monocytes, and macrophages (1). High levels of PAF are responsible for a variety of human diseases such as inflammation, asthma, necrotizing enterocolitis, and sepsis (2). The enzyme PAF-AH (EC 3.1.1.47) was first identified from the plasma by its ability to hydrolyze and therefore inactivate PAF (3).
The enzyme plasma PAF-AH has been classified as group VIIA phospholipase A2 (PLA2) (4), and it hydrolyzes the ester bond at the sn-2 position of phospholipid substrates with a short sn-2 chain. In addition to its role in reducing PAF levels, PAF-AH functions by hydrolyzing other pro-inflammatory agents, such as oxidized lipids of LDL particles (5, 6). Many of these oxidized phospholipids have an oxidatively fragmented sn-2 chain that would orient away from the hydrophobic portion of an LDL particle. These oxidized phospholipid species are present at elevated levels at atherogenic lesions, and the PAF-AH hydrolysis of these species has attracted considerable attention recently as a potential therapeutic target (6–10).
Physiologically, plasma PAF-AH is associated to both LDL and HDL particles and therefore functions on the lipid-aqueous interface and can be considered a peripheral membrane protein. Another generally used name for plasma PAF-AH is lipoprotein-associated PLA2 (7). Kinetic studies have shown that although PAF-AH binds to interfaces, such as LDL particles and vesicles, this binding is not necessary for catalysis (11, 12). This is in contrast to the 14-kDa secreted PLA2 enzymes that are allosterically activated upon interface binding (13). The plasma PAF-AH and the homologous (42% identity) intracellular type II PAF-AH (also known as group VIIB PLA2) enzymes are calcium-independent and contain a GXSXG motif that is characteristic of neutral lipases and serine esterases (14). Structurally distinct from the 44-kDa plasma PAF-AH and type II PAF-AH enzymes is a 26-kDa intracellular brain catalytic domain that has been designated type I PAF-AH or more commonly PAF-AH-Ib (15). Although the structure of PAF-AH-Ib has been reported (16, 17), there is no sequence homology to the other PAF-AH enzymes. Additionally, the PAF-AH-Ib enzyme has been suggested to have a physiological function other than as a true PAF-AH enzyme (18, 19). Although a structure has been determined for a distantly related lipase from Streptomyces exfoliatus (20), the extent of sequence overlap between this enzyme and the mammalian plasma PAF-AH enzyme is limited to 19% identity over a subset of 228 aligned residues. Hence, a crystal structure of a mammalian PAF-AH was required to understand the relationship between structure and function.
Following identification of the enzyme in the plasma, it had been subjected to extensive biochemical characterization, including the determination of the limits of N- and C-terminal truncations that preserve native functions (14). Here, we report a 1.5-Å ligand-free crystal structure and a 2.1-Å diethyl phosphoryl (DEP) complex crystal structure of a 43.4-kDa construct (residues 47–429) of PAF-AH. This construct represents a form of the enzyme with native-like enzyme activity (14) toward the substrate PAF, as well as native-like LDL binding properties (12).
EXPERIMENTAL PROCEDURES
Protein Preparation and Crystallization—We obtained 160 mg of pure human plasma PAF-AH (PAFase, residues 47–429, NCBI accession Q13093) overexpressed from Escherichia coli from ICOS Corporation. Details of the protein preparation and initial crystallization of the ligand-free form of human PAF-AH have been reported (21) and will be summarized briefly. Prior to crystallization, PAF-AH samples were suspended in appropriate buffer/detergent solutions (see below). PAF-AH was assayed for kinetic activity using the ester substrate p-nitrophenyl acetate, and the reaction was followed by UV absorbance at 402 nm (ε402 nm = 17,700 m-1 cm-1).
Ligand-free PAF-AH Crystals—Our best crystals of ligand-free PAF-AH were obtained at 20 °C starting from a protein solution at 4 mg/ml that contained 10 mm Tris-HCl, 6 mm sodium citrate, 3% (w/v) sucrose, 1.0 mm dithiothreitol, 27 mm n-octyl-β-d-glucopyranoside, 0.04% (w/v) Pluronic F68, 0.002% (w/v) Tween 80, pH 6.7. The crystallization reservoir solution contained 98.5 mm MOPS buffer, pH 6.6, 44.3% (w/v) (NH4)2SO4, 0.394 m Li2SO4, 0.985 m sodium acetate, and 1.48% (v/v) 1,4-butanediol. Aliquots of 1.5 μl of protein and crystallization solutions were mixed to form each hanging drop, and protein crystals formed in 3–4 weeks.
To obtain phase information, the ligand-free crystals of PAF-AH were directly soaked with MeHgCl by adding a small grain of solid powder to the hanging drop and incubating it in the drop for 24 h. Derivative crystals were then flash cooled in liquid nitrogen, stored, and transported to the synchrotron for data collection.
Paraoxon Inhibited PAF-AH—PAF-AH protein received from ICOS Corp. was exchanged with the detergent Triton DF16 (0.01% w/v) using a Q-Sepharose column as described previously (21). The protein was then incubated overnight with the organophosphate compound paraoxon (ChemService) to covalently react with the enzyme active site Ser273. Protein was dialyzed to remove salt and excess paraoxon, and finally it was assayed to confirm a complete loss of enzyme activity. The protein sample was concentrated by centrifugal concentration using a Centricon 30 membrane (Millipore) at slow speed (2000 rpm) to avoid aggregation and precipitation. The protein component contained 3 mg/ml protein, 1.0 mm dithiothreitol, 27 mm n-octyl-β-d-glucopyranoside. The crystallization solution contained 41% (w/v) (NH4)2SO4, 400 mm Li2SO4, 800 mm sodium formate, pH 7.0, 1.24% (w/v) 1,4-butanediol. Aliquots of 1.5 μl of the protein and crystallization solutions were mixed and set as hanging drops. The crystals grew (150 × 120 × 90 μm) within 2 weeks.
X-ray Data Collection—Native and mercury-derivative data sets were collected at synchrotron beamlines using crystals frozen in liquid nitrogen; additional cryo-protectant was not required. The single- and multiple-wavelength anomalous dispersion data sets were collected at Beamline 19ID of the Advanced Photon Source (Argonne National Laboratory, Chicago, IL). The data sets for native ligand-free and paraoxon inhibited crystals were collected at Beamline X29 of the National Synchrotron Light Source (Brookhaven National Laboratory, Upton, NY). The diffraction data were indexed and processed using the program HKL2000 (22).
Crystal Structure Solution—A single-wavelength anomalous dispersion data set to a resolution of 2.71 Å of a MeHgCl derivative of PAF-AH was used to phase the structure. Eight mercury sites in the crystallographic asymmetric unit were identified and used to generate initial phases using the programs SOLVE and RESOLVE (23). Two subunits of PAF-AH are in the asymmetric unit. A noncrystallographic symmetry axis was identified from six of the mercury sites, with three similar sites per protein subunit. The noncrystallographic symmetry axis was used to improve initial electron density maps using the CCP4 suite program Dm (24). These medium resolution electron density maps, which contained regions of well defined α-helices and β-sheets, allowed the initial model building of roughly half the 766 amino acids/asymmetric unit with the program COOT (25). The partial model was then refined in the nonisomorphous native data set using data to a resolution of 2.6 Å. The partial model was next used as initial phases together with the native data set to a resolution of 1.5 Å for the automated program ARP/wARP (26). Within 60 cycles, the program had traced and auto-built side chains for 97% of the model. Refinement was initially carried out with the program REFMAC5 (24) and was finished using the program SHELXL (27).
Structure of PAF-AH-DEP Complex—A single subunit of the ligand-free PAF-AH structure was used as a search model. Molecular replacement was performed using the program MOLREP (24) to solve the structure of the PAF-AH-DEP complex, which had been crystallized using paraoxon inhibited PAF-AH. The structure of the PAF-AH-DEP complex was refined using the program REFMAC5 (24). A CIF parameter file for the DEP group, which is covalently attached to Ser273, was prepared using the monomer library sketcher module of the program CCP4 (24).
RESULTS
Overall Structure of PAF-AH—We have solved the crystal structures of human plasma PAF-AH in a ligand-free form and as a complex with the organophosphate compound paraoxon reacted with the active site Ser273 (Fig. 1). The protein displays a classic α/β-hydrolase fold, typical of other GXSXG lipases. The structure of plasma PAF-AH was submitted to the DALI website to obtain structural homologues (28); the S. exfoliatus lipase structure (20) was the top hit with a Z score of 23.8, Cα RMSD of 2.4 Å, and 19% identity over a subset of 228 amino acids aligned. The topology diagram of the PAF-AH structure is shown in supplemental Fig. S1. In both of the crystal structures reported here, there are two protein subunits in the asymmetric unit. The construct of PAF-AH crystallized contained 383 residues (47–429). A summary of the data collection, phasing and refinement of the crystal structures is presented in Table 1.
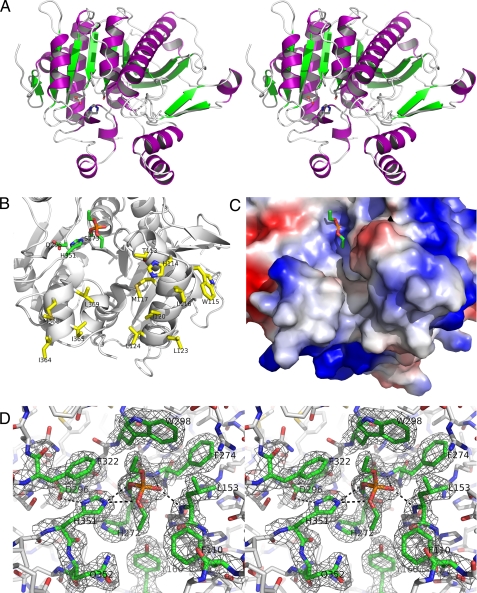
FIGURE 1.
A, stereo ribbon model of the PAF-AH structure (helix, purple, β-strands, green; loops, light gray). The catalytic triad of Ser273, His351, and Asp296 are rendered in ball and stick. The predicted LDL-binding surface is oriented to the bottom of the view. B, interface-binding region and active site of PAF-AH are shown from the crystal structure reacted with the organophosphate compound paraoxon. The view shown was rendered after a 60° rotation on the x axis from the view shown in A. The catalytic triad and bound DEP moiety are shown in green. Residues predicted to penetrate into the hydrophobic portion of the LDL interface (Thr113, His114, Trp115, Leu116, Met117, Ile120, Leu123, Leu124, Ile364, Ile365, Met368, and Leu369) are shown in yellow. C, electrostatic surface of the interfacial-binding region and active site pocket using the same view as B. The DEP ethoxy group pointing downward is sitting in a pocket where the sn-2 chain of PAF-AH substrates is predicted to bind. The DEP ethoxy group pointing to the top and outward is predicted to be where the larger lyso-PC would be oriented. D, stereo view of the active site S273 of PAF-AH covalently linked to DEP with an electron density map (coefficients 2Fo - Fc, 1.2 σ) around highlighted active site residues. The orientation shown is nearly identical to the views of B and C. The catalytic triad (Ser273, His351, and Asp296) and other active site residues (Trp298, Phe322, Phe274, Leu153, His272, Tyr160, Phe110, and Gln352) are shown in green ball and stick, with other neighboring protein atoms in gray. The figure was prepared using the program PyMOL (51).
TABLE 1.
Data collection, phasing, and refinement statistics of PAF-AH The values in parentheses are for the highest resolution shell. The RMSD bond angles are reported as angle distances using the program SHELXL and as degrees in the program REFMAC5.
| MeHgCl | Native | DEP complex | |
|---|---|---|---|
| Crystal parameters | |||
| Space group | C2 | C2 | C2 |
| Unit cell dimensions a, b, c (Å), β(°) | 114.7, 79.0, 96.5, 115.3 | 116.2, 83.1, 96.7, 115.1 | 117.2, 78.7, 97.3, 101.6 |
| Beamline | APS-19ID | NSLS-X29 | NSLS-X29 |
| Data collection | Peak | ||
| Wave length (Å) | 1.006a | 1.100 | 1.072 |
| Resolution (Å) | 50-2.7 (2.8-2.7)a | 50-1.5 (1.55-1.50) | 50-2.1 (2.18-2.10) |
| Completeness (%) | 98.6 (97.4)a | 96.4 (75.7) | 94.9 (73.8) |
| Redundancy | 2.1 (2.1)a | 3.6 (1.8) | 3.4 (2.1) |
| I/σI | 18.3 (2.1)a | 19.2 (1.9) | 19.6 (3.4) |
| Rmerge linear | 0.047 (0.387)a | 0.057 (0.283) | 0.053 (0.193) |
| Phasing statistics | |||
| Number of mercury sites | 8a | ||
| Phasing power anomalous | 0.603a | ||
| R factor (FC vs. FP)b | 0.298a | ||
| Figure of merit (acentric/centric/all) | 0.70/0.77/0.71a | ||
| Refinement statistics | |||
| Refinement program | SHELXL | REFMAC5 | |
| Resolution (Å) | 10.0-1.5 | 50.0-2.1 | |
| Rwork/Rfree | 0.131/0.179 | 0.208/0.264 | |
| Number of atoms (non-hydrogen) | 6692 | 6366 | |
| B-factor main chain | 24.0 | 28.3 | |
| B-factor side chain | 31.3 | 31.1 | |
| RMSD bond lengths (Å) | 0.01 | 0.02 | |
| RMSD bond angles | 0.03 Å | 1.87° | |
| Ramachandran plot (most favored) | 91.6% | 89.5% | |
| Ramachandran plot (most favored and additionally allowed region) | 99.5% | 99.7% |
These are the peak values
The R factor compares structure factors calculated from density modification (FC) to those measured (FP)
Quality of the Structures—As expected, the higher resolution 1.5 Å ligand-free structure has better statistics than the 2.1 Å DEP complex structure. The Ramachandran plots of the ligand-free complex and the DEP complex had 91.6 and 89.5% of residues in the most favored region, respectively. Only residue Ser273, which is the active site nucleophile, was observed to be outside of the generously allowed region (phi, 65; psi, -116). The electron density of this region, as shown for the DEP complex structure in Fig. 1D, is very distinct, supporting its proper modeling. The same holds true for the electron density difference map of the 1.5 Å ligand-free structure. The strained conformation of this serine is consistent with other α/β hydrolase enzymes that have nucleophilic serines in a strained ε-conformation (29). The overall B-factor and deviations from ideal geometry are likewise slightly smaller in the ligand-free structure. One other noteworthy outlier in each subunit of both structures reported is the presence of a cis-peptide bond between Phe72 and Asp73. The electron density of this region is very distinct, thereby supporting the presence of this unusual cis-peptide bond.
Disordered Residues and Side Chains—The N- and C-terminal limits of ordered residues observed in our crystal structures are consistent with functional observations of the protein. Plasma PAF-AH functions as a 43-, 44-, or 45-kDa protein with a heterogeneous N terminus starting at residue Ser35, Ile42 or Lys55 (14). In our structures, seven residues (positions 47–53) are missing from the N terminus in each subunit of both the ligand-free and DEP-bound structures. In each case, our structures begin with residue 54, which is close to the endogenous N-terminal start site at Lys55. In previous studies, the limits of N- and C-terminal truncations had been characterized that still ensure the presence of the full native functions of the enzyme (14). In our crystal structures, the C terminus has several disordered residues in the ligand-free structure (A, 426–429; B, 428–429) and DEP complex (A, 427–429; B, 424–429). The ordered C-terminal end of our structures is close to the functional limit tolerated by C-terminal truncation.
A subtle difference exists between the two subunits of the ligand-free crystal structure in the crystallographic dimer. In subunit B the electron density of residues 88–92 was weak; therefore their side chain atoms have been truncated to correspond to an alanine side chain. Similarly His114 and Trp115 are missing in subunit A and Trp115 and Leu116 are missing in subunit B; therefore each of these side chains have been truncated as well. In addition to these residues mentioned above, several other surface-accessible and polar side chains from the ligand-free crystal structure were disordered and therefore not modeled. In contrast, the DEP-bound crystal structure had well ordered side chain positions for all residues modeled, including residues His114–Leu116.
Interface Binding Surface of PAF-AH—The disorder of the residues His114–Leu116, described above, can be explained because of the flexibility of these residues on the surface of the protein. Additionally, these residues are on an α-helix (114–126) that is predicted to be a component of the interfacial binding surface of the protein that accesses the lipoprotein particle. Specific residues shown to be important for binding to LDL by site-directed mutagenesis were Tyr205, Trp115, Leu116, and to a lesser extent Met117 (30). A second short helix (residues 362–369) has hydrophobic residues positioned to insert into the hydrophobic portion of the aqueous-lipid interface. Recently, residues near this helix were predicted to be important for HDL binding (31). The predicted interfacial-binding residues of PAF-AH are displayed in Fig. 1B relative to the active site.
Active Site of PAF-AH—As originally predicted (14, 32) the active site of PAF-AH contains a catalytic triad of Ser273, His351, and Asp296. Ser273 is located at the N terminus of an α-helix and on the conserved GXSXG motif found in other lipases (14). The amide nitrogens of Phe274 and Leu153 are well poised to stabilize the negative charge of a tetrahedral intermediate of the reaction mechanism, thereby acting as the oxyanion hole of the enzyme. The other two catalytic triad residues are appropriately positioned to activate the nucleophilic Ser273 for catalysis; Asp296 is located on the C-terminal end of a β-sheet, and His351 is located at the N terminus of an α-helix. Like other lipases, the catalytic triad lies within a hydrophobic pocket that is oriented toward its substrate (Fig. 1, B and C). Notably, the orientation of the catalytic triad and hydrophobic residues Leu153 and Phe274 were previously predicted based on modeling from the distantly related S. exfoliatus lipase structure (32). From previous functional and kinetic characterization of PAF-AH, it was suggested that the active site would allow substrates to enter from lipoproteins and the aqueous phase (11, 12). The placement of the active site observed in the structure is consistent with these previous findings.
DEP Complex of PAF-AH—To gain insights into the active site properties of PAF-AH, we solved the crystal structure of a covalent complex with the organophosphate compound paraoxon. The enzyme was quickly inactivated by paraoxon, as determined by enzymatic assay. Crystal set-ups led to crystals in the C2 space group, just as with the ligand-free structure, but with slightly different cell dimensions (Table 1). An overall alignment of the ligand-free and DEP complex crystal structures shows only subtle differences of side chain positions. A comparison of the ligand-free and DEP complex structures shows an overall Cα RMSD of 0.3 Å. The active site residues are virtually unchanged, aside from the covalent attachment of DEP to Ser273 as depicted in Fig. 2; the complex with DEP-bound (panel A) serves as a mimic of the tetrahedral intermediate of the esterolysis reaction (panel B). The catalytic triad residues and neighboring active site residues are positioned around the PAF-AH-DEP complex in a manner consistent with a tetrahedral intermediate complex (Fig. 1D). Contact distances between active site residues and the bound DEP group are shown in Table 2. Notably, the amide nitrogens of Phe274 and Leu153 make H-bonds with the O-3 oxygen of the DEP moiety, which would correspond with the enolate oxygen of a tetrahedral intermediate. The positions of the two ethoxy groups of the bound DEP group are well ordered, as shown in the electron density maps shown in Fig. 1D. A view of these two groups are shown relative to the active site pocket, which is rendered with a space filling electrostatic surface in Fig. 1C. This complex will be further discussed below, as a means to begin to understand how substrates bind to PAF-AH.
FIGURE 2.
A, reaction of Ser273 of PAF-AH with the organophosphate compound paraoxon. The bound complex mimics the tetrahedral intermediate of the reaction. B, first step of the esterolysis reaction of Ser273 of PAF-AH with a substrate, such as PAF, where R1 represents lyso-PAF.
TABLE 2.
DEP and surrounding active site residue contacts within 4 Å Hydrogen bond contacts are shown in italics.
| DEP atom | Residue | Atom type | Distance |
|---|---|---|---|
| Å | |||
| P | His351 | NE2 | 3.74 |
| O-1 | Trp298 | CE3 | 3.41 |
| O-2 | His351 | NE2 | 3.08 |
| O-3 | Gly152 | C | 3.58 |
| Leu153 | N | 2.65 | |
| Phe274 | N | 2.90 | |
| C-1 | Phe322 | CZ | 3.62 |
| C-2 | Trp298 | CE2 | 3.69 |
| C-3 | Leu153 | O | 3.18 |
| His351 | NE2 | 3.93 | |
| C-4 | Leu153 | O | 3.72 |
| His272 | CE1 | 3.80 | |
| His351 | NE2 | 3.89 |
Two Subunits in Asymmetric Unit—Although PAF-AH does not show any oligomeric association in solution, the crystals contain two subunits in the asymmetric unit. A comparison of the A and B subunits shows a Cα RMSD of 0.3 Å for the ligand-free structure and 0.4 Å for the DEP complex structure. The largest subunit interface includes 13 contacts less than4Åand covers a surface area of 707 Å2/monomer (33). The PISA server evaluation of this interface predicts that it is due to crystal packing and does not play any role in protein function (33). Additionally, a sulfate ion is located at this interface, which is not surprising when you consider that the crystallization buffer contained high concentrations of ammonium sulfate and lithium sulfate. The active site opening faces the solvent channel and is not blocked by this or other crystal contacts. In the higher resolution ligand-free structure, two sulfate ions, 29 acetate ions, and 511 waters have been modeled. Additionally, many residues of this structure were modeled with alternate coordinates, which is typical of higher resolution structures. Despite the presence of several detergents in the crystallization conditions (n-octyl-β-d-glucopyranoside, Pluronic F68, and Tween 80), no ordered detergent molecules were observed in the reported crystal structures.
DISCUSSION
Recently, there has been a lot of attention focused on plasma PAF-AH because of its apparently contradictory anti-inflammatory, anti-atherogenic, pro-atherogenic, and pathological importance (8, 34, 35). Early work on the enzyme focused on its role to inactivate the G protein-coupled receptor ligand PAF, and therefore PAF-AH was believed to be a beneficial enzyme (3). The enzyme had been developed as a potential protein therapeutic by the company ICOS Corporation to treat patients deficient in the enzyme and who suffered severe inflammation conditions related to elevated PAF levels (3). In contrast, the lipoprotein-associated function of PAF-AH to hydrolyze oxidatively fragmented phospholipids led others to propose that the enzyme is a significant contributor to pathological conditions, such as atherosclerosis (7, 8, 36). Here, the development of specific and tight binding inhibitors of the enzyme would serve as effective treatments of these pathological conditions (10, 37, 38). In support of serving a pro-atherogenic role, PAF-AH has been shown to be an independent predictor of coronary heart disease (6, 39, 40). Additionally, epidemiological studies have demonstrated a correlation between the polymorphism at residue 379 and coronary artery disease (41) as well as the occurrence of heart attacks (42). Our crystal structure of PAF-AH lends a structural framework that enhances an understanding of previously published work and further opens the door for further studies to explore its functions.
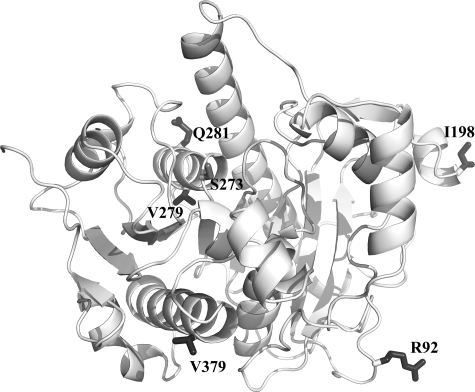
Polymorphism in PAF-AH—Several polymorphic alleles of PAF-AH have been discovered that have either shown a loss of plasma PAF-AH activity (V279F and Q281R) (43) or have been shown to be linked to human disease (I198T, A379V, and R92H) (41, 44). The position of each of these polymorphic sites is shown in Fig. 3. Our crystal structure of PAF-AH includes the most common variant at each polymorphic position, with the exception of the Val379 variant, which has a frequency of 0.24 in European populations (42). The two mutations (V279F and Q281R) that lead to a loss of function in 4% of the Japanese population (43) are core residues. The Val279 residue is a conserved hydrophobic core residue and is critical for proper folding of the enzyme and its function. A loss of function of plasma PAF-AH had also been observed in the Q281R variant. Residue 281 is in the middle of an α-helix that also contains the active site Ser273. Therefore, the Q281R variant is believed to disrupt the active site.
FIGURE 3.
Polymorphic sites of PAF-AH shown in gray ball and stick relative to the active site Ser273 and in a view looking directly at the interfacial binding surface of the enzyme. Three of the polymorphic sites (I198T, A379V, and R92H) are solvent-accessible. Two loss of function polymorphisms (V279F and Q281R) that lead to a loss of function in 4% of Japanese individuals are core residues. This figure was rendered using the program PyMOL (51).
Three polymorphic sites are solvent-exposed and distant from the active site (Fig. 3); therefore these polymorphisms most likely exhibit their phenotypic differences through interactions with lipoproteins or other binding partners. Notably, the Val379 variant is common in European populations (frequency of 0.24 overall) and has been implicated to be correlated with a decreased risk of heart disease (41, 42). In our PAF-AH structure, Val379 is located on a surface α-helix, is 39% solvent-accessible (45) and is 15 Å away from the catalytic His351. The Ile198 polymorphic site is 71% solvent-accessible and is 35 Å from the active site Ser273. Previous kinetic work suggested that the I198T and A379V variants show an increased apparent Km value (14 and 42 μm, respectively) when compared with the wild type (7 μm), although the Vmax parameter of these two variants essentially remained constant with a slight increase for the Val379 variant (44). Our PAF-AH structure raises doubts about whether the previously reported kinetics for the A379V and I198T variants reflect significant differences of these polymorphisms. We suggest that the phenotypic effect of these polymorphisms is a result of lipoprotein binding interactions and does not result directly from catalytic activity.
Lipoprotein-binding Regions—In humans, two-thirds of plasma PAF-AH associate with LDL, and one-third associates with HDL particles (46). There are two α-helices that help the enzyme associate with these lipoproteins. One α-helix (114–126) had been predicted to be a component of the interfacial binding surface of the protein that accesses the LDL particles, and specific mutations confirmed this (30). In addition to the interface positioning of several hydrophobic residues (Fig. 1B), this region has some other notable features. An apparently polar and charged residue His114 is predicted to be buried in the hydrophobic lipid phase. Previous studies showed that a pKa of LDL binding and mutagenesis of this residue supports the placement of the neutral form of His114 into the nonpolar phase (30). An oxidative inactivation role for Met117, and less so for the neighboring Tyr307 and Tyr335 residues, was suggested by site-directed mutagenesis and a species comparison of PAF-AH sequences (46). The second α-helix (residues 362–369), which presents hydrophobic residues to the interface-binding region, was recently shown to influence HDL binding (31). The predicted interfacial-binding residues of PAF-AH are displayed in Fig. 1B relative to the active site.
The cluster of hydrophobic residues discussed above lay the foundation for the development of models of interface binding. We have employed two methods to develop preliminary models of PAF-AH bound to an aqueous-lipid interface: the orientation of proteins in membranes (OPM) data base (47–49) and the membrane associated protein assessment (MAPAS) method (50). The result of the OPM method (prediction performed by A. Lomize) is presented in Fig. 4, as shown by a plane of gray spheres. The plane represents the interface between polar and nonpolar components. The polar head groups of the phospholipids would extend ∼10 Å above the plane shown (48). Following this observation we compared the predicted membrane binding orientation of PAF-AH by using the MAPAS method (50). In contrast to the OPM method, MAPAS proposed several potential orientations (data not shown), with one in agreement with the OPM method. In light of the functional data used to validate a model, we conclude that the OPM prediction shown in Fig. 4 serves as a suitable initial model to understand and rationalize previously published data, as well as to direct future studies. Each of the residues discussed above are buried below the plane predicted by OPM as shown in Fig. 4.
FIGURE 4.
A, model of the tetrahedral intermediate of C18-PAF (cyan) bound to PAF-AH with catalytic triad residues (Ser273, His351, and Asp296) in green and interfacial-binding residues (Thr113, His114, Trp115, Leu116, Met117, Ile120, Leu123, Leu124, Ile364, Ile365, Met368, and Leu369) in yellow. The coordinates of the tetrahedral intermediate were modeled based on the crystal structure of the DEP moiety complexed to PAF-AH (tetrahedral mimic). The C18-alkyl chain was oriented to penetrate into the hydrophobic portion of the LDL particle. The predicted plane of the hydrophilic-hydrophobic interface, which was predicted by the OPM method (47, 48), is displayed with small gray spheres. B, the view from A was rotated by 90° on the y axis to show a side view of the interface and substrate-bound model. A prominent cluster of 10 carboxylate residues (Asp374, Asp376, Asp382, Asp401, Asp403, Asp406, Glu410, Asp412, Asp413, and Glu414) are shown in red, and three basic residues (Lys55, Arg58, and Lys363) are shown in blue ball and stick. C, electrostatic surface view of A. D, electrostatic surface view of B. The figure was prepared using the program PyMOL (51).
Acidic/Basic Patch—The side view of the lipoprotein-bound PAF-AH shown in Fig. 4 reveals a potentially important region of PAF-AH for specific lipoprotein interactions. Fig. 4 (B and D) displays the neighboring acidic and basic patches. A cluster of 10 carboxylate residues (Asp374, Asp376, Asp382, Asp401, Asp403, Asp406, Glu410, Asp412, Asp413, and Glu414), which is shown in Fig. 4B, is adjacent to a group of basic residues (Lys55, Arg58, and Lys363). Although the functional and structural relevance of these residues is not clear, we believe the following are noteworthy. The cluster of 10 carboxylate residues and three basic residues are mainly conserved when compared across several mammalian sequences of plasma PAF-AH (supplemental Fig. S2). However, most of these residue are not present in the alignment with the intracellular homologue type II PAF-AH (42% identity, 62% similarity). This suggests a potential role of these residues toward LDL/HDL partitioning. Also, the A379V polymorphic site, which has been correlated with heart disease (41, 42), is located at the base of this acidic patch, adjacent to the predicted plane of lipoprotein binding. It is plausible that a switch between alanine and valine at this position may influence LDL/HDL partitioning. In addition to possible interactions with lipoproteins, these residues may maintain electrostatic interaction with charged or polar head groups of phospholipids on the surface of the lipoproteins, a process followed by hydrophobic interaction of the hydrophobic cluster discussed above. Studies are underway to explore the structural and functional relevance of the residues of these acidic and basic patches.
PAF-AH Substrate Specificity—The predicted mode of lipoprotein binding presented in Fig. 4 shows an open channel, and active site residues are positioned in such a way that they can access substrates from the aqueous phase. Gelb and co-workers (11, 13) hypothesized a mechanism in which plasma PAF-AH accesses its substrate from the aqueous phase, and only substrate in the aqueous phase can bind to the catalytic site of the enzyme at the membrane-water interface. Our model is consistent with this hypothesis if one merely adds that the long hydrophobic tail, of for example C18-PAF, remains partly embedded in the hydrophobic portion of the interface, as depicted in Fig. 4. In this model PAF-AH and lipophillic substrates are brought together by their common affinity for lipoproteins. However, the active site faces the aqueous phase and sits just above the interface.
The DEP complex crystal structure allows us to predict how a substrate would access and bind to the active site. The covalent complex of DEP with Ser273 from our crystal structure served as a tetrahedral intermediate mimic of the esterolysis reaction. We have substituted the DEP ligand from our crystal structure with a speculative model of the tetrahedral intermediate of C18-PAF, as depicted in Fig. 4. The lyso-PAF moiety, which would be the leaving group from the collapse of the tetrahedral intermediate, projects out of the active site toward the solvent and polar portion of the interface. A smaller pocket projects inward that has been modeled with the sn-2 chain of the substrate. Although this pocket may limit the size of the sn-2 chain of substrates and thereby explain the preference of the enzyme for a shortened and polar sn-2 chain (6, 11, 12), it is a fairly open pocket, which could accommodate larger chains. Another more likely explanation of why PAF-AH prefers substrates with shorter sn-2 chains could be merely due to its aqueous phase solubility. In our model, sn-2 chains that are flipped upward or away from the hydrophobic portion of the interface would be accessible to the PAF-AH active site.
In conclusion, the α/β-hydrolase fold of plasma PAF-AH is reminiscent of the fold found in lipases and esterases. Our crystal structures of the enzyme in both a ligand-free state and with a tetrahedral intermediate mimic have provided a working model of how plasma PAF-AH may bind to lipoproteins and catalyze the hydrolysis of both PAF and oxidized phospholipid substrates.
Supplementary Material
Acknowledgments
We thank ICOS Corp. for supplying us with the protein PAFase, which was used to screen crystallization conditions and solve the PAF-AH structure. We thank the staff at the Advanced Photon Source Beamline 19ID and National Synchrotron Light Source Beamline X29 for assistance with data collection. We thank Dr. Andrei Lomize for predicting the orientation of PAF-AH on a bilayer using methods of the OPM data base. Finally, we thank Prabhavathi Srinivasan for fruitful discussions.
The atomic coordinates and structure factors (codes 3D59 and 3D5E) have been deposited in the Protein Data Bank, Research Collaboratory for Structural Bioinformatics, Rutgers University, New Brunswick, NJ (http://www.rcsb.org/).
This work was supported, in whole or in part, by National Institutes of Health Grants 2P20RR015588 and 1R01HL084366-A1. The costs of publication of this article were defrayed in part by the payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 U.S.C. Section 1734 solely to indicate this fact.
The on-line version of this article (available at http://www.jbc.org) contains supplemental Figs. S1 and S2.
Footnotes
The abbreviations used are: PAF, platelet-activating factor; DEP, diethyl phosphoryl; AH, acetylhydrolase; PLA2, phospholipase A2; HDL, high density lipoprotein; LDL, low density lipoprotein; MOPS, 4-morpholinepropanesulfonic acid; RMSD, root mean square deviation; OPM, orientation of proteins in membranes; MAPAS, membrane associated protein assessment.
References
- 1.Prescott, S. M., Zimmerman, G. A., and McIntyre, T. M. (1990) J. Biol. Chem. 265 17381-17384 [PubMed] [Google Scholar]
- 2.Prescott, S. M., Zimmerman, G. A., Stafforini, D. M., and McIntyre, T. M. (2000) Annu. Rev. Biochem. 69 419-445 [DOI] [PubMed] [Google Scholar]
- 3.Tjoelker, L. W., Wilder, C., Eberhardt, C., Stafforini, D. M., Dietsch, G., Schimpf, B., Hooper, S., Le Trong, H., Cousens, L. S., Zimmerman, G. A., Yamada, Y., McIntyre, T. M., Prescott, S. M., and Gray, P. W. (1995) Nature 374 549-553 [DOI] [PubMed] [Google Scholar]
- 4.Schaloske, R. H., and Dennis, E. A. (2006) Biochim. Biophys. Acta 1761 1246-1259 [DOI] [PubMed] [Google Scholar]
- 5.Stremler, K. E., Stafforini, D. M., Prescott, S. M., and McIntyre, T. M. (1991) J. Biol. Chem. 266 11095-11103 [PubMed] [Google Scholar]
- 6.Davis, B., Koster, G., Douet, L. J., Scigelova, M., Woffendin, G., Ward, J. M., Smith, A., Humphries, J., Burnand, K. G., Macphee, C. H., and Postle, A. D. (2008) J. Biol. Chem. 283 6428-6437 [DOI] [PubMed] [Google Scholar]
- 7.Macphee, C. H., and Nelson, J. J. (2005) Eur. Heart J. 26 107-109 [DOI] [PubMed] [Google Scholar]
- 8.Macphee, C. H., Nelson, J., and Zalewski, A. (2006) Curr. Opin. Pharmacol. 6 154-161 [DOI] [PubMed] [Google Scholar]
- 9.Aiyar, N., Disa, J., Ao, Z. H., Ju, H. S., Nerurkar, S., Willette, R. N., Macphee, C. H., Johns, D. G., and Douglas, S. A. (2007) Mol. Cell Biochem. 295 113-120 [DOI] [PubMed] [Google Scholar]
- 10.Wilensky, R. L., Shi, Y., Zalewski, A., Mohler, E. R., Hamamdzic, D., Li, J. U., Pelcovitz, D., Webb, C., Burgett, M. E., Walker, M. C., and Macphee, C. H. (2007) Circulation 116 33-33 [Google Scholar]
- 11.Min, J. H., Jain, M. K., Wilder, C., Paul, L., Apitz-Castro, R., Aspleaf, D. C., and Gelb, M. H. (1999) Biochemistry 38 12935-12942 [DOI] [PubMed] [Google Scholar]
- 12.Min, J. H., Wilder, C., Aoki, J., Arai, H., Inoue, K., Paul, L., and Gelb, M. H. (2001) Biochemistry 40 4539-4549 [DOI] [PubMed] [Google Scholar]
- 13.Gelb, M. H., Min, J. H., and Jain, M. K. (2000) Biochim. Biophys. Acta 1488 20-27 [DOI] [PubMed] [Google Scholar]
- 14.Tjoelker, L. W., Eberhardt, C., Unger, J., Trong, H. L., Zimmerman, G. A., McIntyre, T. M., Stafforini, D. M., Prescott, S. M., and Gray, P. W. (1995) J. Biol. Chem. 270 25481-25487 [DOI] [PubMed] [Google Scholar]
- 15.Arai, H. (2002) Prostaglandins Other Lipid Mediat. 68–69, 83-94 [DOI] [PubMed] [Google Scholar]
- 16.Ho, Y. S., Swenson, L., Derewenda, U., Serre, L., Wei, Y., Dauter, Z., Hattori, M., Adachi, T., Aoki, J., Arai, H., Inoue, K., and Derewenda, Z. S. (1997) Nature 385 89-93 [DOI] [PubMed] [Google Scholar]
- 17.Tarricone, C., Perrina, F., Monzani, S., Massimiliano, L., Kim, M. H., Derewenda, Z. S., Knapp, S., Tsai, L. H., and Musacchio, A. (2004) Neuron 44 809-821 [DOI] [PubMed] [Google Scholar]
- 18.Derewenda, Z. S. (2002) Colloids Surfaces B: Biointerfaces 26 31-35 [Google Scholar]
- 19.Epstein, T. M. (2005) Structural and Kinetic Studies of Two Enzymes Catalyzing Phospholipase A2 Activity. Ph.D. thesis, University of Delaware, Newark, DE
- 20.Wei, Y., Swenson, L., Castro, C., Derewenda, U., Minor, W., Arai, H., Aoki, J., Inoue, K., Servin-Gonzalez, L., and Derewenda, Z. S. (1998) Structure 6 511-519 [DOI] [PubMed] [Google Scholar]
- 21.Samanta, U., Wilder, C., and Bahnson, B. J. (2008) Protein Peptide Lett., in press [DOI] [PMC free article] [PubMed]
- 22.Otwinowski, Z., and Minor, W. (1997) Methods Enzymol. 276 307-326 [DOI] [PubMed] [Google Scholar]
- 23.Terwilliger, T. C. (2003) Methods Enzymol. 374 22-37 [DOI] [PubMed] [Google Scholar]
- 24.CCP4. (1994) Acta Crystallogr. Sect. D Biol. Crystallogr. 50 760-763 [DOI] [PubMed] [Google Scholar]
- 25.Emsley, P., and Cowtan, K. (2004) Acta Crystallogr. Sect. D Biol. Crystallogr. 60 2126-2132 [DOI] [PubMed] [Google Scholar]
- 26.Perrakis, A., Morris, R., and Lamzin, V. S. (1999) Nat. Struct. Biol. 6 458-463 [DOI] [PubMed] [Google Scholar]
- 27.Sheldrick, G. M., and Schneider, T. R. (1997) Methods Enzymol. 277 319-343 [PubMed] [Google Scholar]
- 28.Holm, L., and Sander, C. (1996) Science 273 595-602 [DOI] [PubMed] [Google Scholar]
- 29.Ollis, D. L., Cheah, E., Cygler, M., Dijkstra, B., Frolow, F., Franken, S. M., Harel, M., Remington, S. J., Silman, I., Schrag, J., Sussman, J. L., Verschueren, K. H. G., and Goldman, A. (1992) Protein Eng. 5 197-211 [DOI] [PubMed] [Google Scholar]
- 30.Stafforini, D. M., Tjoelker, L. W., McCormick, S. P., Vaitkus, D., McIntyre, T. M., Gray, P. W., Young, S. G., and Prescott, S. M. (1999) J. Biol. Chem. 274 7018-7024 [DOI] [PubMed] [Google Scholar]
- 31.Gardner, A. A., Reichert, E. C., Topham, M. K., and Stafforini, D. M. (2008) J. Biol. Chem. 283 17099-17106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Derewenda, Z. S., and Derewenda, U. (1998) Cell. Mol. Life Sci. 54 446-455 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Krissinel, E., and Henrick, K. (2007) J. Mol. Biol. 372 774-797 [DOI] [PubMed] [Google Scholar]
- 34.Karasawa, K. (2006) Biochim. Biophys. Acta 1761 1359-1372 [DOI] [PubMed] [Google Scholar]
- 35.Tjoelker, L. W., and Stafforini, D. M. (2000) Biochim. Biophys. Acta 1488 102-123 [DOI] [PubMed] [Google Scholar]
- 36.Macphee, C. H., Nelson, J. J., and Zalewski, A. (2005) Curr. Opin. Lipidol. 16 442-446 [DOI] [PubMed] [Google Scholar]
- 37.Blackie, J. A., Bloomer, J. C., Brown, M. J., Cheng, H. Y., Hammond, B., Hickey, D. M., Ife, R. J., Leach, C. A., Lewis, V. A., Macphee, C. H., Milliner, K. J., Moores, K. E., Pinto, I. L., Smith, S. A., Stansfield, I. G., Stanway, S. J., Taylor, M. A., and Theobald, C. J. (2003) Bioorg. Med. Chem. Lett. 13 1067-1070 [DOI] [PubMed] [Google Scholar]
- 38.Mohler, E. R., 3rd, Ballantyne, C. M., Davidson, M. H., Hanefeld, M., Ruilope, L. M., Johnson, J. L., and Zalewski, A. (2008) J. Am. Coll. Cardiol. 51 1632-1641 [DOI] [PubMed] [Google Scholar]
- 39.Packard, C. J., O'Reilly, D. S., Caslake, M. J., McMahon, A. D., Ford, I., Cooney, J., Macphee, C. H., Suckling, K. E., Krishna, M., Wilkinson, F. E., Rumley, A., and Lowe, G. D. (2000) N. Engl. J. Med. 343 1148-1155 [DOI] [PubMed] [Google Scholar]
- 40.Mannheim, D., Herrmann, J., Versari, D., Gossl, M., Meyer, F. B., McConnell, J. P., Lerman, L. O., and Lerman, A. (2008) Stroke 39 1448-1455 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Ninio, E., Tregouet, D., Carrier, J. L., Stengel, D., Bickel, C., Perret, C., Rupprecht, H. J., Cambien, F., Blankenberg, S., and Tiret, L. (2004) Hum. Mol. Genet. 13 1341-1351 [DOI] [PubMed] [Google Scholar]
- 42.Abuzeid, A. M., Hawe, E., Humphries, S. E., Talmud, P. J., and Grp, H. S. (2003) Atherosclerosis 168 283-288 [DOI] [PubMed] [Google Scholar]
- 43.Stafforini, D. M., Numao, T., Tsodikov, A., Vaitkus, D., Fukuda, T., Watanabe, N., Fueki, N., McIntyre, T. M., Zimmerman, G. A., Makino, S., and Prescott, S. M. (1999) J. Clin. Investig. 103 989-997 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Kruse, S., Mao, X. Q., Heinzmann, A., Blattmann, S., Roberts, M. H., Braun, S., Gao, P. S., Forster, J., Kuehr, J., Hopkin, J. M., Shirakawa, T., and Deichmann, K. A. (2000) Am. J. Hum. Genet. 66 1522-1530 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Samanta, U., Bahadur, R. P., and Chakrabarti, P. (2002) Protein Engineering 15 659-667 [DOI] [PubMed] [Google Scholar]
- 46.MacRitchie, A. N., Gardner, A. A., Prescott, S. M., and Stafforini, D. M. (2007) FASEB J. 21 1164-1176 [DOI] [PubMed] [Google Scholar]
- 47.Lomize, A. L., Pogozheva, I. D., Lomize, M. A., and Mosberg, H. I. (2006) Protein Sci. 15 1318-1333 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Lomize, A. L., Pogozheva, I. D., Lomize, M. A., and Mosberg, H. I. (2007) BMC Struct. Biol. 7 1-30 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Lomize, M. A., Lomize, A. L., Pogozheva, I. D., and Mosberg, H. I. (2006) Bioinformatics 22 623-625 [DOI] [PubMed] [Google Scholar]
- 50.Sharikov, Y., Walker, R. C., Greenberg, J., Kouznetsova, V., Nigam, S. K., Miller, M. A., Masliah, E., and Tsigelny, I. F. (2008) Nat. Methods 5 119-119 [DOI] [PubMed] [Google Scholar]
- 51.DeLano, W. L. (2002) The PyMOL User's Manual, DeLano Scientific, Palo Alto, CA
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.