Abstract
15-Lipoxygenase-1 (15-LOX-1) contributes significantly to inflammation regulation and terminal cell differentiation. 15-LOX-1 is transcriptionally silenced in cancer cells, and its transcriptional reactivation (e.g. via histone deacetylase inhibitors (HDACIs)) is essential for restoring terminal cell differentiation to cancer cells. STAT-6 acetylation via the histone acetyltransferase KAT3B has been proposed to be necessary for 15-LOX-1 transcriptional activation. However, the exact mechanism underlying 15-LOX-1 transcriptional reactivation in cancer cells is still undefined, especially in regard to the contribution of 15-LOX-1 promoter histone modifications. We therefore examined the relative mechanistic contributions of 15-LOX-1 promoter histone modifications and STAT-6 to 15-LOX-1 transcriptional reactivation by HDACIs in colon cancer cells. We found that: 1) histone H3 and H4 acetylation in the 15-LOX-1 promoter through KAT3B was critical to 15-LOX-1 transcriptional activation; 2) 15-LOX-1 transcription was activated independently from STAT-6; and 3) dimethyl-histone H3 lysine 9 (H3K9me2) demethylation in the 15-LOX-1 promoter via the histone lysine demethylase KDM3A was an early and specific histone modification and was necessary for activation of transcription. These findings demonstrate that histone modification in the 15-LOX-1 promoter is important to 15-LOX-1 transcriptional silencing in colon cancer cells and that HDACIs can activate gene transcription via KDM3A demethylation of H3K9me2.
15-Lipoxygenase-1 (15-LOX-1)2 is a critical enzyme for the production of various inflammation-regulatory lipid signaling mediators, including 13-S-hydroxyoctadecadienoic acid from linoleic acid (1, 2), lipoxins from arachidonic acid (3), and resolvins and protectins from docosahexaenoic acid (4). 15-LOX-1 has an important regulatory function in terminal cell differentiation (5–11) and inflammation (3, 12), and its expression is inducible and highly regulated in normal human cells (13). 15-LOX-1 is down-regulated in various human cancers, including colon cancer (14–16), esophageal cancer (17), breast cancer (18), and pancreatic cancer (19). 15-LOX-1 re-expression in cancer cells via pharmaceutical agents or plasmid or adenoviral vectors induces growth inhibition and re-establishes terminal cell differentiation and apoptosis (10, 15–17, 20–26).
15-LOX-1 expression, regulated at the translational level in normal cells (27), is transcriptionally silenced in cancer cells (28–32). Reversal of 15-LOX-1 transcriptional silencing in cancer cells has been proposed to occur via various mechanisms: global histone H4 (H4) acetylation (28), STAT-6 acetylation and phosphorylation (33), inhibition of GATA-6 transcriptional repression of the 15-LOX-1 promoter (29, 34), and 15-LOX-1 promoter DNA demethylation (30, 35). Nonetheless, the exact mechanisms underlying 15-LOX-1 transcriptional reactivation in cancer cells remain unknown. For example, it is unclear whether global H4 acetylation activates 15-LOX-1 transcription through direct 15-LOX-1 promoter chromatin modification or through the expression of transcriptional factors that subsequently influence the 15-LOX-1 promoter. Suberoylanilide hydroxamic acid (SAHA) down-regulates STAT-6 expression (36) but still activates 15-LOX-1 expression (23), raising the question of whether STAT-6 is necessary for 15-LOX-1 transcriptional activation. GATA-6 knockdown inhibits GATA-6 binding to the 15-LOX-1 promoter but fails to activate 15-LOX-1 transcription (31). 15-LOX-1 promoter DNA demethylation is insufficient to reactivate 15-LOX-1 expression (32).
To identify the crucial molecular mechanisms underlying 15-LOX-1 transcriptional reactivation in cancer cells, we examined the mechanisms by which HDACIs activate 15-LOX-1 transcription, given that HDACIs are the most potent class of agents known to induce this activation (23, 28, 32). In particular, we examined whether HDACIs activated 15-LOX-1 transcription via promoter histone modification or STAT-6, which has been proposed to be crucial to 15-LOX-1 transcriptional activation (33). In a series of experiments, we identified novel chromatin modification events in the 15-LOX-1 promoter that activate 15-LOX-1 transcription independently of STAT-6 via KAT3B acetylation of histone H3 and H4 and KDM3A demethylation of histone H3 dimethyllysine 9 (H3K9me2).
EXPERIMENTAL PROCEDURES
Materials—SAHA was provided by Merck and the National Cancer Institute. Depsipeptide was provided by Gloucester Pharmaceuticals, Inc., and the National Cancer Institute. Antibodies against acetyl-histone H3, acetyl-histone H4, acetyl-histone H3 lysine 9 (H3K9ac), dimethyl-histone H3 lysine 4 (H3K4me2), monomethyl-histone H3 lysine 9 (H3K9me1), dimethyl-histone H3 lysine 9 (H3K9me2), trimethyl-histone H3 lysine 9 (H3K9me3), dimethyl-histone H3 lysine 27 (H3K27me2), trimethyl-histone H3 lysine 27 (H3K27me3), and trimethyl-histone H4 lysine 20 (H4K20me3) were purchased from Upstate Cell Signaling Solutions (Lake Placid, NY). Rabbit anti-human KDM3A polyclonal antibody was purchased from Abcam, Inc. (Cambridge, MA). Anti-STAT-6 antibodies were purchased from Santa Cruz Biotechnology (Santa Cruz, CA). Rabbit polyclonal antiserum to recombinant human 15-LOX-1 protein was obtained as described previously (21). Recombinant human interleukin 4 (IL-4) was purchased from Bio-source (Camarillo, CA). Caco-2, HT29, SW480, and HCT-116 colon cancer cells were purchased from the American Type Culture Collection (Manassas, VA), and RKO and DLD-1 colorectal cancer cell lines were obtained as described previously (21). Other reagents, molecular grade solvents, and chemicals were obtained from commercial manufacturers or as specified.
Cell Cultures and Treatments—HT29, SW480, HCT-116, RKO, and DLD-1 cells were grown in RPMI 1640 medium, and Caco-2 cells were grown in Eagle's minimal essential medium (Cambrex, Walkersville, MD) with l-glutamine in a humidified atmosphere containing 5% CO2 at 37 °C. Both types of medium contained 10% fetal bovine serum and were supplemented with 1% penicillin-streptomycin as described previously (21). Depsipeptide, a selective HDAC1 and HDAC2 inhibitor (37, 38), and SAHA, a nonselective HDAC inhibitor (38), were dissolved in dimethyl sulfoxide and added to the culture media at the indicated concentrations.
Small Interfering RNA (siRNA) Transfection—SW480 and Caco-2 cells were cultured to 40–50% confluence and then transfected with 100 nm of a pooled mixture of four siRNA duplexes (SMARTselected or On Target; Dharmacon, Inc., Lafayette, CO) for the targeted gene (e.g. STAT-6, KAT3B) or with nonspecific control siRNA (Dharmacon, Inc.) using Lipofectamine 2000 (Invitrogen).
RNA Extraction and Real-time PCR—Total RNA was extracted from cells using TRI reagent (Molecular Research Center Inc., Cincinnati, OH). RNA from each sample was reverse transcribed and then measured quantitatively by real-time PCR using a comparative Ct method, as described previously (31).
Chromatin Immunoprecipitation (ChIP) and ChIP/Real-time PCR Assays—Cross-linking was performed by adding formaldehyde to the cell culture medium to a final concentration of 1% and incubating for 10 min at 37 °C. ChIP assays were performed using a commercial kit according to the manufacturer's protocol (Upstate Cell Signaling Solutions). Chromatin was immunoprecipitated using the indicated antibodies. ChIP assays using an agarose gel electrophoresis method were performed similarly to what was described previously (31). For studying STAT-6 binding to the 15-LOX-1 promoter, we used primers that amplified a 182-bp fragment of the human 15-LOX-1 promoter that covers a specific STAT-6 binding site located 952 bp upstream of the 15-LOX-1 start codon (39): 5′-TAA TTC ACT CTG GTG GGG TGG-3′ (sense) and 5′-CAG TTT CTT TTT GGG CTG GA-3′ (antisense).
The relative enrichment of transcription factors or histone modifications (e.g. KDM3A, H3K9me2) was measured using ChIP/real-time PCR with the following primers and FAM dye-labeling probe: forward, GTGTTTTCGGTCCAAATCCTTTTCT; reverse, GAGAGCAGGGAGTGGAAACC; probe, CCTCCCGTCAAGATAGT to amplify the –280 bp to –212 bp region of the 15-LOX-1 promoter relative to the ATG site. Histone acetylation levels in the 15-LOX-1 promoter were measured by ChIP/real-time PCR with the following primers and FAM labeling probe: forward, GAGACCTGCCACCCACATT; reverse, GAGCTAAATAACCCAGCCTGAGA; probe, CCGACCCCAAGCGAC to amplify the –120 bp to –47 bp region of the 15-LOX-1 promoter relative to the ATG site. Real-time PCR was performed as described previously (32).
Statistical Analyses—We used the t test for two-group comparisons. For analyses involving single factors and more than two groups, we performed a one-way analysis of variance. If the overall analysis of variance test result was significant, we performed pairwise comparisons, adjusting for multiplicities using the Bonferroni correction. We analyzed data involving the simultaneous consideration of two factors using two-way analysis of variance. We first tested the interaction effect, and if it was statistically significant, we subsequently performed specific comparisons to investigate which differences were driving this effect, using the Bonferroni correction to adjust for the multiple testing problem. An individual comparison was considered significant only if the p value was less than 0.05/k, with k representing the number of comparisons performed. If the interaction effect was not significant, we tested the individual main effects. Then, if those were significant, we determined which pairwise comparisons were significant, again adjusting for multiplicities using the Bonferroni correction. We used the Pearson correlation method to correlate differences in the levels of the studied variables (e.g. 15-LOX-1 mRNA expression level, histone H3 acetylation in the 15-LOX-1 promoter) between depsipeptide- or SAHA-treated and control cells. All of the tests were two-sided and conducted at the p ≤ 0.05 level. Quantitative data log transformation was used to account for the normal distributional assumptions underlying the methods. The data were analyzed using SAS software (SAS Institute, Cary, NC).
RESULTS
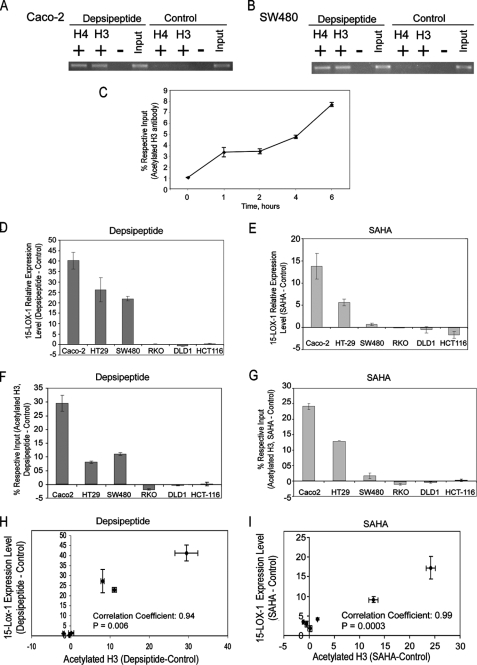
15-LOX-1 Promoter Histone Acetylation and 15-LOX-1 Transcriptional Reactivation—Depsipeptide induced acetylation of histones H3 and H4 in the 15-LOX-1 promoter (Fig. 1, A and B). The level of acetylated histone H3 in the 15-LOX-1 promoter increased significantly within the first 60 min and continued to increase during 6 h after depsipeptide treatment in Caco-2 cells (Fig. 1C; p < 0.0001). Depsipeptide and SAHA differentially induced 15-LOX-1 expression (Fig. 1, D and E, p < 0.0001) and histone H3 acetylation in the 15-LOX-1 promoter (Fig. 1, F and G, p < 0.0001) in the six tested colorectal cancer cell lines. The induction of 15-LOX-1 expression correlated highly with the increase in histone H3 acetylation in the 15-LOX-1 promoter by both depsipeptide (Fig. 1H; Pearson correlation coefficient, 0.94; p = 0.006) and SAHA (Fig. 1I; Pearson correlation coefficient, 0.99; p = 0.0003) in the six tested colorectal cancer cell lines.
FIGURE 1.
15-LOX-1 promoter histone acetylation and 15-LOX-1 transcriptional activation. A and B, effects of depsipeptide on acetylation of histones H3 and H4 in the 15-LOX-1 promoter. Caco-2 (A) and SW480 (B) cells were treated with depsipeptide (5 nm) or the vehicle solvent for depsipeptide (control). The cells were harvested 48 h after treatment and subjected to ChIP assays using specific antibodies against acetylated histone H3 (H3) or acetylated histone H4 (H4). + and – indicate immunoprecipitation with and without H3 or H4 antibodies, respectively, and Input indicates total DNA before immunoprecipitation. Depsipeptide induced H3 and H4 acetylation in the 15-LOX-1 promoter. C, kinetics of effects of depsipeptide on H3 acetylation. Caco-2 cells were treated with depsipeptide (5 nm), harvested at the indicated times, and subjected to ChIP/real-time PCR assays using specific acetylated H3 antibodies. The results are percentages of the respective input genomic DNA for the 15-LOX-1 promoter. The values are the means ± S.D. of triplicate measurements. D, effects of depsipeptide on 15-LOX-1 expression in colorectal cancer cells. Colorectal cancer cells were treated with either depsipeptide (5 nm) or solvent (control). The cells were harvested 24 h later and processed for 15-LOX-1 mRNA by real-time reverse transcription-PCR. The data are presented as the difference in 15-LOX-1 relative expression levels between depsipeptide- and control-treated cells. The values shown are the means ± S.D. of triplicate experiments. E, effects of SAHA on 15-LOX-1 expression in colorectal cancer cells. Colorectal cancer cells were treated with either SAHA (1 μm) or solvent (control) and processed for 15-LOX-1 mRNA measurements as in D. The values shown are the means ± S.D. of triplicate experiments. F and G, effects of depsipeptide and SAHA on H3 acetylation in the 15-LOX-1 promoter. Colorectal cancer cells were treated with depsipeptide (F), SAHA (G), or solvent only (control). The cells were harvested and subjected to ChIP/real-time PCR using specific antibodies against acetylated histone H3. The data are presented as the difference in the H3 acetylation levels in the 15-LOX-1 promoter between depsipeptide- or SAHA-treated and control-treated cells. The values shown are the means ± S.D. of triplicate experiments. H and I, correlation between 15-LOX-1 expression and H3 acetylation in the 15-LOX-1 promoter in colorectal cancer cell lines. Scatter plots of the differences in 15-LOX-1 mRNA expression (treated – control) in relation to those in 15-LOX-1 promoter H3 acetylation levels (treated – control) in colorectal cancer cell lines treated with depsipeptide (H) or SAHA (I). The values shown are the means ± S.D. of triplicate experiments.
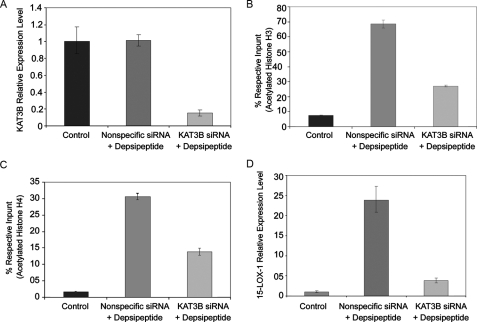
15-LOX-1 Promoter Histone Acetylation Is Required for 15-LOX-1 Transcriptional Activation—KAT3B is a very well known histone acetyltransferase (40, 41). We used KAT3B knockdown by siRNA to examine whether 15-LOX-1 promoter histone acetylation is necessary for 15-LOX-1 transcriptional activation. Transfection of KAT3B siRNA reduced the level of KAT3B expression by 85% in Caco-2 cells (Fig. 2A; p < 0.0001) and 78% in SW480 cells (supplemental Fig. S1; p < 0.0001). In Caco-2 cells, depsipeptide markedly increased 15-LOX-1 promoter enrichment with acetylated histone H3, and KAT3B siRNA reduced this increase by 60% (Fig. 2B; p < 0.0001). In SW480 cells, depsipeptide increased 15-LOX-1 promoter enrichment with acetylated histone H3, and KAT3B siRNA reduced this increase by 70% (supplemental Fig. S1; p < 0.0001). Similarly, in Caco-2 cells, depsipeptide markedly increased 15-LOX-1 promoter enrichment with acetylated H4, and KAT3B siRNA reduced this increase by 55% (Fig. 2C; p < 0.0001). In SW480 cells, depsipeptide increased the relative level of acetylated H4, and KAT3B siRNA reduced this increase by 82% (supplemental Fig. S1; p < 0.0001). In Caco-2 cells, depsipeptide increased 15-LOX-1 expression, and KAT3B siRNA reduced this increase by 84% (Fig. 2D; p < 0.0001). The same pattern was seen in SW480 cells (supplemental Fig. S1; p < 0.0001).
FIGURE 2.
Effects of KAT3B on 15-LOX-1 promoter histone acetylation and 15-LOX-1 transcriptional activation. A, effects of KAT3B siRNA transfection on KAT3B mRNA expression. Caco-2 cells were transfected with a pool of four siRNA duplexes for KAT3B (KAT3B siRNA), a nonspecific siRNA sequence (Nonspecific siRNA), or transfection medium only (Control) and then treated 24 h later with either depsipeptide (5 nm) or control solvent. The cells were harvested 48 h after transfection. KAT3B mRNA levels were measured by using real-time reverse transcription-PCR. The relative expression levels were calculated as the values relative to those of the calibrator samples (Nonspecific siRNA). B and C, effects of KAT3B knockdown on H3 (B) and H4 (C) acetylation in the 15-LOX-1 promoter. Caco-2 cells were transfected and treated as in A and then subjected to chromatin immunoprecipitation/real-time PCR. The results are presented as percentages of the respective input genomic DNA for the 15-LOX-1 promoter. The values are the means ± S.D. of triplicate measurements. D, effects of KAT3B knockdown on depsipeptide activation of 15-LOX-1 transcription. Caco-2 cells were transfected and treated as described in A, and 15-LOX-1 expression was measured by using real-time reverse transcription-PCR. The relative expression levels were calculated as the values relative to those of the calibrator samples (Control). The values shown are the means ± S.D. of triplicate experiments.
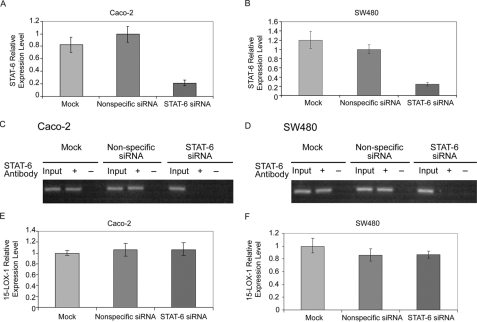
15-LOX-1 Transcriptional Activation in Colorectal Cancer Cells Is Independent of STAT-6—STAT-6 siRNA reduced STAT-6 expression relative to nonspecific siRNA by 79% in Caco-2 cells and 77% in SW480 cells (Fig. 3, A and B; p < 0.0001). STAT-6 binding to the 15-LOX-1 promoter was inhibited in STAT-6 siRNA-transfected cells but not in cells transfected with nonspecific siRNA (Fig. 3, C and D). 15-LOX-1 expression levels were similar for nonspecific siRNA and STAT-6 siRNA in both Caco-2 cells (p = 0.86) and SW480 cells (p = 0.8) (Fig. 3, E and F).
FIGURE 3.
Effects of STAT-6 on 15-LOX-1 transcriptional activation. A and B, effects of STAT-6 siRNA transfection on STAT-6 mRNA expression. Caco-2 (A) and SW480 (B) cells were transfected with a pool of four siRNA duplexes against STAT-6 (STAT-6 siRNA), a nonspecific siRNA sequence (Nonspecific siRNA), or transfection medium only (Mock) for 48 h and then treated with 5 nm depsipeptide for 24 h. STAT-6 mRNA levels were measured with real-time reverse transcription-PCR. Expression levels were calculated as the values relative to those of the calibrator samples (Nonspecific siRNA). The values are the means ± S.D. of triplicate experiments. C and D, effects of STAT-6 siRNA transfection on STAT-6 binding to the 15-LOX-1 promoter. Caco-2 (C) and SW480 (D) cells were transfected and treated as described in A and B. The cells were subjected to chromatin immunoprecipitation assays using a specific anti-STAT-6 antibody. STAT-6 antibody + and – indicate immunoprecipitation with and without STAT-6 antibody, respectively, and Input indicates total DNA before immunoprecipitation. E and F, effects of STAT-6 down-regulation by siRNAs on depsipeptide induction of 15-LOX-1 expression. Caco-2 (E) and SW480 (F) cells were transfected and treated as described in A and B. 15-LOX-1 mRNA relative expression levels, real-time reverse transcription-PCR, were calculated as the values relative to those of the calibrator samples (Mock). The values are the means ± S.D. of triplicate experiments.
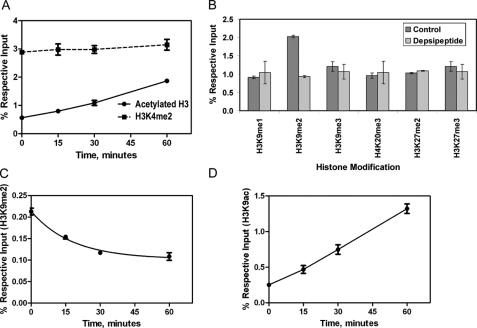
15-LOX-1 Promoter Chromatin Modification during 15-LOX-1 Transcriptional Activation—During the first hour after depsipeptide treatment, the level of acetylated histone H3 in Caco-2 cells increased rapidly (Fig. 4A; p = 0.0001), whereas the level of H3K4me2 remained unchanged (Fig. 4A; p = 0.188). Among the various other histone methylation markers we evaluated in the 15-LOX-1 promoter (H3K9me1, H3K9me2, H3K9me3, H3K27me2, H3K27me3, and H4K20me3), only the H3K9me2 level changed significantly, with a 54% reduction in recruitment level (p < 0.0001) measured 30 min after depsipeptide treatment (Fig. 4B). During the first 30 min after depsipeptide treatment, the level of H3K9ac increased, whereas the level of H3K9me2 decreased in a time-dependent manner (Fig. 4, C and D; p < 0.0001).
FIGURE 4.
Chromatin modification of 15-LOX-1 promoter during transcriptional activation. A, kinetics of depsipeptide effects on dimethyl-histone H3 lysine 4 (H3K4me2) in the 15-LOX-1 promoter. Caco-2 cells were treated with depsipeptide as indicated. ChIP/real-time PCR was performed with the use of specific H3K4me2 or acetylated histone H3 (acetylated H3) antibodies. The results are the means ± S.D. of four replicate experiments. B, effects of depsipeptide on H3 and H4 methylation patterns in the 15-LOX-1 promoter. Caco-2 cells were treated with depsipeptide (5 nm) or solvent control, harvested 30 min later, and processed for ChIP/real-time PCR using anti-H3K9me1, anti-H3K9me2, anti-H3K9me3, anti-H4K20me3, anti-H3K27me2, and anti-H3K27me3 antibodies. The results are the means ± S.D. of triplicate experiments. C and D, kinetics of depsipeptide effects on H3 acetylation and methylation. Caco-2 cells were treated with depsipeptide for the indicated times and processed for ChIP/real-time PCR using antibodies against H3K9me2 (C) and H3K9ac (D). The results are the means ± S.D. of four replicate experiments.
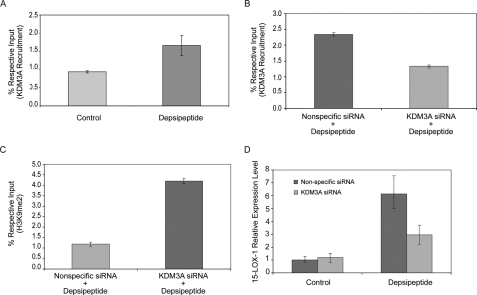
Recruitment of KDM3A to the 15-LOX-1 Promoter Is Required for 15-LOX-1 Transcriptional Activation—Depsipeptide significantly increased the recruitment of KDM3A, a demethylase of H3K9me2, to the 15-LOX-1 promoter in Caco-2 cells (Fig. 5A; p = 0.004). KDM3A siRNA transfection into Caco-2 cells significantly reduced KDM3A recruitment to the 15-LOX-1 promoter with depsipeptide compared with the recruitment seen with nonspecific siRNA (Fig. 5B; p < 0.0001). The H3K9me2 level was significantly increased in KDM3A siRNA-transfected cells treated with depsipeptide, compared with the level in cells transfected with nonspecific siRNA and treated identically with depsipeptide (Fig. 5C; p < 0.0001). KDM3A siRNA transfection reduced the 15-LOX-1 mRNA level in cells treated with depsipeptide compared with the level in cells transfected with nonspecific siRNA and treated with depsipeptide (Fig. 5D; p < 0.0001).
FIGURE 5.
Effects of KDM3A on 15-LOX-1 transcription. A, effects of depsipeptide on the recruitment of KDM3A to the 15-LOX-1 promoter. Caco-2 cells were treated with depsipeptide (5 nm) or control solvent, harvested 15 min later, and processed for ChIP/real-time PCR using specific anti-KDM3A antibody. The results are the means ± S.D. of triplicate experiments. B, effects of KDM3A siRNA transfection on recruitment of KDM3A to the 15-LOX-1 promoter. Caco-2 cells were transfected with a pool of four siRNA duplexes for KDM3A (KDM3A siRNA) or a nonspecific siRNA sequence (Nonspecific siRNA) and then treated with depsipeptide (5 nm). The cells were harvested 48 h later and then subjected to ChIP/real-time PCR using anti-KDM3A antibody. The results are the percentages of the respective input genomic DNA for the 15-LOX-1 promoter. The values are the means ± S.D. of triplicate measurements. C, effects of KDM3A siRNA on depsipeptide reduction of the dimethyl-histone H3 lysine 9 (H3K9me2) level. Caco-2 cells were treated as in B and processed for ChIP/real-time PCR using anti-H3K9me2 antibodies. The values are the means ± S.D. of triplicate measurements. D, effects of KDM3A siRNA on depsipeptide activation of 15-LOX-1 transcription. Caco-2 cells were transfected and treated as in B. The cells were processed for real-time reverse transcription-PCR of 15-LOX mRNA. The values are the means ± S.D. of triplicate measurements.
DISCUSSION
Our new findings demonstrate that: 1)15-LOX-1 transcriptional activation in colon cancer cells requires specific histone modification events in the 15-LOX-1 promoter: H3K9me2 demethylation by KDM3A and histone H3 and H4 acetylation by KAT3B; and 2) HDACIs can activate transcription via KDM3A demethylation of H3K9me2.
Histone acetylation in the 15-LOX-1 promoter but not STAT-6 was necessary for 15-LOX-1 transcriptional activation. 15-LOX-1 transcriptional activation highly correlated with histone H3 acetylation as demonstrated by our testing in six colorectal cancer cell lines using both specific (depsipeptide (38)) and nonspecific (SAHA) HDACIs. By using KAT3B knockdown via siRNA to inhibit histone H3 and H4 acetylation, we confirmed that acetylation of histone H3 and H4 in the 15-LOX-1 promoter was necessary for 15-LOX-1 activation. The authors of a prior report proposed that KAT3B activates 15-LOX-1 transcription via STAT-6 acetylation (33). However, we found that 15-LOX-1 transcriptional activation required KAT3B acetylation of 15-LOX-1 promoter histones but not STAT-6. We found that STAT-6 down-regulation via siRNA, although successful in inhibiting the binding of STAT-6 to the 15-LOX-1 promoter, failed to influence depsipeptide activation of 15-LOX-1 transcription. These findings agree with those in a recent report showing that SAHA down-regulated STAT-6 expression (36), which suggested that STAT-6 does not contribute to transcriptional activation by HDACIs. STAT-6 has been reported to be necessary for IL-4 transcriptional activation of 12/15-LOX (the mouse homologue of 15-LOX-1) in mice (42) and for 15-LOX-1 transcription in human BEAS-2B transformed airway epithelial cells and A534 lung cancer cells (33, 39). The fact that STAT-6 has been shown to be required for IL-4-induced but not HDACI-induced 15-LOX-1 transcriptional activation might reflect differences in the mechanisms of action between IL-4 and HDACIs. IL-4 binds to cell surface receptors and requires STAT-6 for signal transduction to influence nuclear events (43), whereas the HDACIs directly influence transcriptional events (38). In support of this notion, we found that depsipeptide activated 15-LOX-1 transcription much earlier than IL-4 did in A534 cells (supplemental Fig. S2).
Specific chromatin modification events in the 15-LOX-1 promoter activated 15-LOX-1 transcription. These events included very early removal of a transcriptional silencing histone modification, H3K9me2 (44, 45), and an increase in H3K9ac, a transcriptional activation marker (46). The specificity of these histone changes is supported by the lack of changes in the other histone markers of transcriptional activation (e.g. H3K4me2 (44)) or transcriptional silencing (e.g. H3K9me1, H3K9me3, H3K27me2, H3K27me3, or H4K20me3 (44, 47)) during 15-LOX-1 transcriptional activation. To our knowledge, the pattern we identified is the first specific chromatin modification pattern in the 15-LOX-1 promoter to have been identified, especially in regard to histone methylation. Previously published information on 15-LOX-1 promoter modifications is limited to nonspecific global H4 acetylation (28) and general histone H3 15-LOX-1 promoter acetylation (33).
We found that depsipeptide activated 15-LOX-1 transcription through demethylation of H3K9me2 by KDM3A, a recently identified specific H3K9me2 demethylase (48). Depsipeptide promoted KDM3A recruitment to the 15-LOX-1 promoter. Inhibition of this recruitment via KDM3A down-regulation by siRNA increased the H3K9me2 level in the 15-LOX-1 promoter and suppressed depsipeptide activation of 15-LOX-1 transcription. These findings indicate that depsipeptide recruitment of KDM3A to the 15-LOX-1 promoter and the subsequent H3K9me2 demethylation are necessary for activation of 15-LOX-1 transcription. Data are emerging to support the important role of histone demethylases in the transcriptional regulation of normal and pathologic molecular cellular events (49, 50). The only prior report of a pharmaceutical intervention that enhanced the recruitment of KDM3A for transcriptional activation was a report on the hormonal androgen analogue R1881 (48). Our current results show that KDM3A demethylation of H3K9me2 appears not to be limited to transcriptional activation by hormonal agents but also to contribute to transcriptional activation by the HDACIs.
The strong correlation between the degree of 15-LOX-1 transcription activation in cancer cell lines and histone acetylation, although it supports the notion that histone deacetylation is necessary for maintaining 15-LOX-1 transcriptional silencing, still leaves open the possibility that other mechanisms might contribute to 15-LOX-1 transcriptional silencing in cancer cells. For example, we have recently found that the dissociation of DNA methyltransferase-1 from the 15-LOX-1 promoter can significantly influence 15-LOX-1 transcriptional activation in colon cancer cells (32). DNA methyltransferase-1 contributes to H3K9me2 formation during replication (51). Further studies are needed to evaluate the contribution of DNA methyltransferase-1 to H3K9me2 formation in the 15-LOX-1 promoter.
In summary, our findings demonstrate the important role of specific chromatin modification in the regulation of 15-LOX-1 transcription. These results provide a new model for the orchestrated chromatin modification events in the 15-LOX-1 promoter that are involved in 15-LOX-1 transcriptional activation: KDM3A recruitment to demethylate H3K9me2 and histone H3 and H4 acetylation via KAT3B. Further studies of these chromatin modification events in the 15-LOX-1 promoter will not only help to improve our understanding of the mechanism of 15-LOX-1 silencing in cancer cells but also help to identify novel molecular targets for inhibiting colonic tumorigenesis.
Supplementary Material
Acknowledgments
We thank Mark T. Bedford for constructive discussions. We thank Merck and Company, Inc., Gloucester Pharmaceuticals, and the National Cancer Institute for providing SAHA and depsipeptide.
This work was supported, in whole or in part, by National Institutes of Health Grants CA106577 and CA104278. This work was also supported by the Caroline Wiess Law Endowment for Cancer Prevention and the National Colorectal Cancer Research Alliance. The costs of publication of this article were defrayed in part by the payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 U.S.C. Section 1734 solely to indicate this fact.
The on-line version of this article (available at http://www.jbc.org) contains supplemental Figs. S1 and S2.
Footnotes
The abbreviations used are: 15-LOX-1, 15-lipoxygenase-1; H3, histone H3; H4, histone H4; SAHA, suberoylanilide hydroxamic acid; HDACI, histone deacetylase inhibitor; H3K9me2, dimethyl-histone H3 lysine 9; H3K9ac, acetyl-histone H3 lysine 9; H3K4me2, dimethyl-histone H3 lysine 4; H3K9me1, monomethyl-histone H3 lysine 9; H3K9me3, trimethyl-histone H3 lysine 9; H3K27me2, dimethyl-histone H3 lysine 27; H3K27me3, trimethyl-histone H3 lysine 27; H4K20me3, trimethyl-histone H4 lysine 20; IL, interleukin; siRNA, small interfering RNA; ChIP, chromatin immunoprecipitation; STAT, signal transducers and activators of transcription; KAT3B, K-acetyltransferase 3B; KDM3A, K-demethylase 3A.
References
- 1.Baer, A. N., Costello, P. B., and Green, F. A. (1991) Biochem. Biophys. Res. Commun. 180 98–104 [DOI] [PubMed] [Google Scholar]
- 2.Brash, A. R., Boeglin, W. E., and Chang, M. S. (1997) Proc. Natl. Acad. Sci. U. S. A. 94 6148–6152 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Serhan, C. N., Jain, A., Marleau, S., Clish, C., Kantarci, A., Behbehani, B., Colgan, S. P., Stahl, G. L., Merched, A., Petasis, N. A., Chan, L., and Van Dyke, T. E. (2003) J. Immunol. 171 6856–6865 [DOI] [PubMed] [Google Scholar]
- 4.Ariel, A., and Serhan, C. N. (2007) Trends Immunol. 28 176–183 [DOI] [PubMed] [Google Scholar]
- 5.Kroschwald, P., Kroschwald, A., Kuhn, H., Ludwig, P., Thiele, B. J., Hohne, M., Schewe, T., and Rapoport, S. M. (1989) Biochem. Biophys. Res. Commun. 160 954–960 [DOI] [PubMed] [Google Scholar]
- 6.Hill, E. M., Eling, T., and Nettesheim, P. (1998) Am. J. Respir. Cell Mol. Biol. 18 662–669 [DOI] [PubMed] [Google Scholar]
- 7.van Leyen, K., Duvoisin, R. M., Engelhardt, H., and Wiedmann, M. (1998) Nature 395 392–395 [DOI] [PubMed] [Google Scholar]
- 8.Grullich, C., Duvoisin, R. M., Wiedmann, M., and van Leyen, K. (2001) FEBS Lett. 489 51–54 [DOI] [PubMed] [Google Scholar]
- 9.Spanbroek, R., Hildner, M., Kohler, A., Muller, A., Zintl, F., Kuhn, H., Radmark, O., Samuelsson, B., and Habenicht, A. J. R. (2001) Proc. Natl. Acad. Sci. U. S. A. 98 5152–5157 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Shureiqi, I., Wu, Y., Chen, D., Yang, X. L., Guan, B., Morris, J. S., Yang, P., Newman, R. A., Broaddus, R., Hamilton, S. R., Lynch, P., Levin, B., Fischer, S. M., and Lippman, S. M. (2005) Cancer Res. 65 11486–11492 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kim, K.-S., Chun, H.-S., Yoon, J.-H., Lee, J. G., Lee, J.-H., and Yoo, J.-B. (2005) Prostaglandins Leukot. Essent. Fatty Acids 73 77–83 [DOI] [PubMed] [Google Scholar]
- 12.Munger, K. A., Montero, A., Fukunaga, M., Uda, S., Yura, T., Imai, E., Kaneda, Y., Valdivielso, J. M., and Badr, K. F. (1999) Proc. Natl. Acad. Sci. U. S. A. 96 13375–13380 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kuhn, H., Walther, M., and Kuban, R. J. (2002) Prostaglandins Other Lipid Mediat. 68–69, 263–290 [DOI] [PubMed] [Google Scholar]
- 14.Shureiqi, I., Wojno, K. J., Poore, J. A., Reddy, R. G., Moussalli, M. J., Spindler, S. A., Greenson, J. K., Normolle, D., Hasan, A. A., Lawrence, T. S., and Brenner, D. E. (1999) Carcinogenesis 20 1985–1995 [DOI] [PubMed] [Google Scholar]
- 15.Nixon, J. B., Kim, K. S., Lamb, P. W., Bottone, F. G., and Eling, T. E. (2004) Prostaglandins Leukot. Essent. Fatty Acids 70 7–15 [DOI] [PubMed] [Google Scholar]
- 16.Heslin, M. J., Hawkins, A., Boedefeld, W., Arnoletti, J. P., Frolov, A., Soong, R., Urist, M. M., and Bland, K. I. (2005) Ann. Surg. 241 941–947 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Shureiqi, I., Xu, X., Chen, D., Lotan, R., Morris, J. S., Fischer, S. M., and Lippman, S. M. (2001) Cancer Res. 61 4879–4884 [PubMed] [Google Scholar]
- 18.Jiang, W. G., Watkins, G., Douglas-Jones, A., and Mansel, R. E. (2006) Prostaglandins Leukot. Essent. Fatty Acids 74 235–245 [DOI] [PubMed] [Google Scholar]
- 19.Hennig, R., Kehl, T., Noor, S., Ding, X. Z., Rao, S. M., Bergmann, F., Furstenberger, G., Buchler, M. W., Friess, H., Krieg, P., and Adrian, T. E. (2007) Neoplasia 9 917–926 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Shureiqi, I., Chen, D., Lee, J. J., Yang, P., Newman, R. A., Brenner, D. E., Lotan, R., Fischer, S. M., and Lippman, S. M. (2000) J. Natl. Cancer Inst. 92 1136–1142 [DOI] [PubMed] [Google Scholar]
- 21.Shureiqi, I., Jiang, W., Zuo, X., Wu, Y., Stimmel, J. B., Leesnitzer, L. M., Morris, J. S., Fan, H. Z., Fischer, S. M., and Lippman, S. M. (2003) Proc. Natl. Acad. Sci. U. S. A. 100 9968–9973 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Wu, J., Xia, H. H., Tu, S. P., Fan, D. M., Lin, M. C., Kung, H. F., Lam, S. K., and Wong, B. C. (2003) Carcinogenesis 24 243–247 [DOI] [PubMed] [Google Scholar]
- 23.Hsi, L. C., Xi, X., Lotan, R., Shureiqi, I., and Lippman, S. M. (2004) Cancer Res. 64 8778–8781 [DOI] [PubMed] [Google Scholar]
- 24.Deguchi, A., Xing, S. W., Shureiqi, I., Yang, P., Newman, R. A., Lippman, S. M., Feinmark, S. J., Oehlen, B., and Weinstein, I. B. (2005) Cancer Res. 65 8442–8447 [DOI] [PubMed] [Google Scholar]
- 25.Zuo, X., Wu, Y., Morris, J. S., Stimmel, J. B., Leesnitzer, L. M., Fischer, S. M., Lippman, S. M., and Shureiqi, I. (2006) Oncogene 25 1225–1241 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Wu, Y., Fang, B., Yang, X. Q., Wang, L., Chen, D., Krasnykh, V., Carter, B. Z., Morris, J. S., and Shureiqi, I. (2008) Mol. Ther. 16 886–892 [DOI] [PubMed] [Google Scholar]
- 27.Ostareck, D. H., Ostareck-Lederer, A., Wilm, M., Thiele, B. J., Mann, M., and Hentze, M. W. (1997) Cell 89 597–606 [DOI] [PubMed] [Google Scholar]
- 28.Kamitani, H., Taniura, S., Ikawa, H., Watanabe, T., Kelavkar, U. P., and Eling, T. E. (2001) Carcinogenesis 22 187–191 [DOI] [PubMed] [Google Scholar]
- 29.Shureiqi, I., Jiang, W., Fischer, S. M., Xu, X., Chen, D., Lee, J. J., Lotan, R., and Lippman, S. M. (2002) Cancer Res. 62 1178–1183 [PubMed] [Google Scholar]
- 30.Liu, C., Xu, D., Sjoberg, J., Forsell, P., Bjorkholm, M., and Claesson, H. E. (2004) Exp. Cell Res. 297 61–67 [DOI] [PubMed] [Google Scholar]
- 31.Shureiqi, I., Zuo, X., Broaddus, R., Wu, Y., Guan, B., Morris, J. S., and Lippman, S. M. (2007) FASEB J. 21 743–753 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Zuo, X., Shen, L., Issa, J.-P., Moy, O., Morris, J. S., Lippman, S. M., and Shureiqi, I. (2008) FASEB J. 22 1981–1992 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Shankaranarayanan, P., Chaitidis, P., Kuhn, H., and Nigam, S. (2001) J. Biol. Chem. 276 42753–42760 [DOI] [PubMed] [Google Scholar]
- 34.Kamitani, H., Kameda, H., Kelavkar, U. P., and Eling, T. E. (2000) FEBS Lett. 467 341–347 [DOI] [PubMed] [Google Scholar]
- 35.Hsi, L. C., Xi, X., Wu, Y., and Lippman, S. M. (2005) Mol. Cancer Ther. 4 1740–1746 [DOI] [PubMed] [Google Scholar]
- 36.Zhang, C., Richon, V., Ni, X., Talpur, R., and Duvic, M. (2005) J. Investig. Dermatol. 125 1045–1052 [DOI] [PubMed] [Google Scholar]
- 37.Furumai, R., Matsuyama, A., Kobashi, N., Lee, K. H., Nishiyama, M., Nakajima, H., Tanaka, A., Komatsu, Y., Nishino, N., Yoshida, M., and Horinouchi, S. (2002) Cancer Res. 62 4916–4921 [PubMed] [Google Scholar]
- 38.Bolden, J. E., Peart, M. J., and Johnstone, R. W. (2006) Nat. Rev. Drug Discov. 5 769–784 [DOI] [PubMed] [Google Scholar]
- 39.Conrad, D. J., and Lu, M. (2000) Am. J. Respir. Cell Mol. Biol. 22 226–234 [DOI] [PubMed] [Google Scholar]
- 40.Bannister, A. J., and Kouzarides, T. (1996) Nature 384 641–643 [DOI] [PubMed] [Google Scholar]
- 41.Ogryzko, V. V., Schiltz, R. L., Russanova, V., Howard, B. H., and Nakatani, Y. (1996) Cell 87 953–959 [DOI] [PubMed] [Google Scholar]
- 42.Heydeck, D., Thomas, L., Schnurr, K., Trebus, F., Thierfelder, W. E., Ihle, J. N., and Kuhn, H. (1998) Blood 92 2503–2510 [PubMed] [Google Scholar]
- 43.Mikita, T., Campbell, D., Wu, P., Williamson, K., and Schindler, U. (1996) Mol. Cell Biol. 16 5811–5820 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Martin, C., and Zhang, Y. (2005) Nat. Rev. Mol. Cell Biol. 6 838–849 [DOI] [PubMed] [Google Scholar]
- 45.Nightingale, K. P., O'Neill, L. P., and Turner, B. M. (2006) Curr. Opin. Genet. Dev. 16 125–136 [DOI] [PubMed] [Google Scholar]
- 46.Peterson, C. L., and Laniel, M.-A. (2004) Curr. Biol. 14 R546–R551 [DOI] [PubMed] [Google Scholar]
- 47.Bannister, A. J., and Kouzarides, T. (2005) Nature 436 1103–1106 [DOI] [PubMed] [Google Scholar]
- 48.Yamane, K., Toumazou, C., Tsukada, Y.-i., Erdjument-Bromage, H., Tempst, P., Wong, J., and Zhang, Y. (2006) Cell 125 483–495 [DOI] [PubMed] [Google Scholar]
- 49.Lee, D. Y., Teyssier, C., Strahl, B. D., and Stallcup, M. R. (2005) Endocr. Rev. 26 147–170 [DOI] [PubMed] [Google Scholar]
- 50.Shi, Y. (2007) Nat. Rev. Genet. 8 829–833 [DOI] [PubMed] [Google Scholar]
- 51.Esteve, P. O., Chin, H. G., Smallwood, A., Feehery, G. R., Gangisetty, O., Karpf, A. R., Carey, M. F., and Pradhan, S. (2006) Genes Dev. 20 3089–3103 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.