Abstract
Relaxin peptides are important hormones for the regulation of reproductive tissue remodeling and the renal cardiovascular system during pregnancy. Recent studies demonstrated that two of the seven human relaxin family peptides, relaxin H2 (RLN2) and INSL3, signal exclusively through leucine-rich repeat-containing G protein-coupled receptors, LGR7 and LGR8. Although it was well characterized that an RXXXRXXI motif at the RLN2 B chain confers receptor activation activity, it is not clear what roles RLN2 A chain plays in receptor interaction. Analyses of relaxin family genes on syntenic regions of model tetrapods showed that the A chain of RLN2 orthologs exhibited a greater sequence divergence as compared with the receptor-binding domain-containing B chain, foreshadowing a potential role in receptor interactions; hence, defining receptor selectivity in this fast evolving peptide hormone. To test our hypothesis that select residues in the human RLN2 A chain play key roles in receptor interaction, we studied mutant peptides with residue substitution(s) in the A chain. Here, we showed that alanine substitution at the A16 and A17 positions enhances LGR8-activation activity of RLN2, whereas mutation at the A22-23 region (RLN2A22-23) ablates LGR8, but not LGR7, activation activity. In addition, we demonstrated that the functional characteristics of the RLN2A22-23 mutant are mainly attributed to modifications at the PheA23 position. Taken together, our studies indicated that ThrA16, LysA17, and PheA23 constitute part of the receptor-binding interface of human RLN2, and that modification of these residues has led to the generation of novel human RLN2 analogs that would allow selective activation of human LGR7, but not LGR8, in vivo.
In humans, the relaxin gene family contains a total of seven members: relaxin H1 (RLN1),3 relaxin H2 (RLN2), relaxin 3/INSL7 (RLN3), INSL3/RLF, INSL4/EPIL, INSL5/RIF2, and INSL6/RIF1 (1-5). Among these family members, RLN2 and INSL3 signal through two leucine-rich repeat-containing GPCRs, LGR7 (RFXR1) and/or LGR8 (RFXR2). Whereas RLN2 is capable of activating both LGR7 and LGR8, INSL3 is a selective ligand for LGR8. In addition to the better characterized RLN2 and INSL3, RLN3 was shown to activate LGR7, GPCR135, and GPCR142, but not LGR8 (6-10); therefore, the relaxin family peptides exhibit overlapping specificity on the activation of LGR7 and LGR8. Although the physiological roles of RLN3 and other family peptides remain to be studied, analysis of null mice models showed that the ablation of relaxin and INSL3 genes caused phenotypes similar to that of LGR7- and LGR8-deficient mice, suggesting that LGR7 and LGR8 are the main receptors for the single relaxin and INSL3 peptides in rodents, respectively (11-15).
In addition to the role in remodeling of reproductive tissues, relaxin has been shown to effect endometrial differentiation during embryo implantation, nipple and mammary gland development, angiogenesis, wound healing, and renal cardiovascular responses (12, 13, 16-19). Furthermore, recent studies have shown that relaxin prolongs the survival of tumor-bearing mice by enhancing the degradation of the extracellular matrix, thereby slowing down tumor growth (20, 21). In contrast, INSL3 is essential for testis descent in rodents and contributes to the regulation of, 1) male germ cell apoptosis; 2) initiation of meiotic progression of arrested oocytes in preovulatory follicles (22); and 3) the positioning of the female gonad during development (23, 24). Although the importance of LGR7 and LGR8 in human RLN1 and RLN2 signaling remains to be studied, in vitro evidence indicated that human RLN2 could effect LGR8 signaling in vivo.
Based on its pleiotropic effects on tissue remodeling, relaxin has been the subject for clinical trials aimed to treat scleroderma, pre-eclampsia, congestive heart failure, and to enhance cervical ripening during the third trimester of pregnancy (25-28). The finding that human RLN2 is capable of activating both LGR7 and LGR8 raises the possibility that clinical applications of human RLN2 could pose unwanted responses in the LGR8 signaling pathway in patients, which could include effects on spermatogenesis and ovarian follicle development (14, 18, 29-32). Therefore, a better understanding of the molecular mechanisms underlying the interaction of human RLN2 and its receptors, as well as the generation of an LGR7-specific human relaxin analog are of great interest. Earlier studies showed that the replacement of 2 arginines and 1 isoleucine residue in the B chain of human RLN2 reduced the affinity by a 1000-fold, suggesting these three residues are critical for receptor interaction (33). X-ray crystallographic analysis indicates that these three residues are concentrated and form a binding surface for interaction with acidic residues in relaxin receptors (34). In addition, we have recently showed that a histidine residue in the B chain of INSL3 determines the receptor specificity of this peptide (35). Unlike the better characterized B chain, studies of porcine relaxin and human INSL3 variants with substitutions at the N terminus of the chain A indicated that no single amino acid in that region is functionally important (36-38). Nonetheless, the view that the A chain in relaxin family peptides has a minimal role in receptor interaction was challenged by recent studies showing that truncation of the N terminus of human RLN2 A chain leads to significantly reduced bioactivities (39).
Earlier analyses showed that human and porcine relaxins are capable of activating both LGR7 and LGR8 at nanomolar concentrations, whereas the orthologus relaxin from rodents are LGR7-specific (8, 40). This data suggested that the fast evolving relaxin family peptides and their receptor-interaction characteristics diverge according to lineage-specific adaptation. Based on the observation that relaxin peptides from mammals exhibit distinct receptor selectivity and that relaxin homologs underwent positive selection in mammals, we hypothesized that structural motifs tolerant of divergence in RLN2 (e.g. the C terminus of A chain) could harbor microdomains important for the regulation of receptor selectivity; hence, the receptor activation. Consistent with our hypothesis, studies of recombinant RLN2 peptides with point mutation(s) show that ThrA16, LysA17, and PheA23 residues in the RLN2 A chain play critical roles in interacting with LGR7 and LGR8. In addition, these analyses have led to the generation of novel human RLN2 analogs that exhibit a greater preference for LGR7 as compared with the wild type peptide as well as an analog that exhibits an enhanced bioactivity upon LGR8 activation.
EXPERIMENTAL PROCEDURES
Syntenic Mapping and Identification of Orthologous Relaxin Genes in Tetrapods—We analyzed the genomic DNA of human (Homo sapiens), chimpanzee (Pan troglodytes), the Rhesus monkey (Macaca mulatta), rat (Rattus norvegicus), mouse (Mus musculus), rabbit (Oryctolagus cuniculus), dog (Canis familiaris), cow (Bos taurus), pig (Sus scrofa), nine-banded armadillo (Dasypus novemcinctus), elephant (Loxodonta africana), Madagascar hedgehog (Echinops telfairi), gray short-tailed opossum (Monodelphis domestica), platypus (Ornithorhynchus anatinus), chicken (Gallus gallus), and the clawed frog (Xenopus tropicalis), and identified homologs of human relaxin family genes including RLN1, RLN2, RLN3, INSL3, INSL4, INSL5, and INSL6 (41). The identity of relaxin family genes from different species was determined by syntenic mapping and a series of reciprocal pairwise sequence comparisons using the BLAST server.
Chromosome fragments containing relaxin family genes syntenic to the RLN1/RLN2/INSL4/INSL6 locus on the human chromosome 9 from different tetrapod species were obtained from NCBI and Ensembl BioMart data bases based on syntenic mapping. The human-other mammal syntenic maps were downloaded from the Ensembl BioMart data mining tool (www.ensembl.org/multi/martview). The exact locations for relaxin family genes in the syntenic chromosome regions were also verified by BLAT searches using the UCSC Genome Bioinformatics webserver (genome.ucsc.edu/cgi-bin/hgBlat).
Subcloning of Wild Type Human Relaxin Family Genes and Variants—Full-length cDNA for human RLN2, INSL3, and RLN3 were subcloned from a human testis and an ovary Marathon-ready cDNA library (Clontech, Mountain View, CA). To allow efficient secretion of recombinant peptides into the conditioned media by transfected cells, cDNAs encoding the pro-protein region were appended with a signal peptide for secretion sequence from the prolactin precursor at the N terminus, and subcloned into the mammalian expression vector, pcDNA3.1 Zeo. Expression constructs for mutants with point mutation(s) and/or a truncated C domain were generated using overlapping PCR with specific primers in three steps. In the first PCR, forward and reverse primers were used to amplify a cDNA stretch encoding the signal peptide and the downstream site of the introduced mutation. In the second PCR, a set of primers were used to amplify the cDNA encoding the mutation point as well as the remaining C-terminal region of propeptide. Products from the first and the second reactions were designed to share a 20-30-bp overlap in the region of the introduced mutation. After purification, the mixed templates from the first and second PCRs were amplified with primers of the 5′- and 3′-ends of the propeptide coding region. The full-length mutant cDNAs were gel purified and ligated into the pcDNA3.1 Zeo vector. The Escherichia coli strain, Top10, was electroporated for transformation, and the integrity of the construct was confirmed by automated dye-terminator cycle DNA sequencing.
Expression of Relaxin Family Peptides and Variants—For efficient detection and purification of recombinant peptides, all expression constructs were tagged with a Myc epitope at the N terminus and a 6-histidine (His6) tag at the C terminus of the B chain. To allow efficient cleavage of the prepropeptide during post-translational modifications and the secretion of mature peptides, HEK293T cells were routinely cotransfected with the select expression construct and a one-tenth aliquot of a convertase expression vector (42). Cells were cultured in Dulbecco's modified Eagle's medium/F-12 media supplemented with 10% fetal bovine serum, 100 IU/ml penicillin, and 100 μg/ml streptomycin in a water-saturated atmosphere containing 5% CO2 at 37 °C.
Transfection was performed using the calcium-phosphate precipitation method. Four days after transfection, conditioned media were collected, centrifuged, and filtered to remove cell debris. Recombinant peptides were then purified using nickel affinity chromatography.
Nickel Affinity Chromatography—After cleared of cell debris, the conditioned medium was incubated with nickel-conjugated chelating Sepharose Fast flow resin (Amersham Biosciences) overnight at 4 °C with stirring. The resins bound with tagged peptides were washed with 10 column volumes of washing buffer (Tris-buffered saline (TBS), pH 7.5) containing 20 mm imidazole, followed by 20 column volumes of the washing buffer containing 40 mm imidazole. The bound peptides were eluted and fractionated with TBS containing 200 mm imidazole. Fractions with immunoreactive peptides were combined and concentrated with Amicon Ultra columns (Millipore, Billerica, MA; Mr cutoff, 5,000) after washing with 20 column volumes of phosphate-buffered saline. The integrity and purity of the peptides were verified by Western blotting analysis using a monoclonal anti-Myc or an anti-relaxin B chain polyclonal antibody (Calbiochem, San Diego, CA) as well as Coomassie Blue staining. Purified peptides were quantified based on Western blotting analysis or a histidine peptide enzyme-linked immunosorbent assay. To ensure the fidelity of mutant peptides, for each construct at least two different batches of peptides from one-half liter of conditioned medium were generated and characterized independently.
SDS-PAGE and Western Blotting Analysis—Cell extracts, conditioned media, and affinity column-purified peptides were electrophoresed using 18% SDS-PAGE before being electro-transfered to polyvinylidine difluoride membranes (Hybond-P, Amersham Biosciences) in a Trans-Blot® SD semi-dry transfer cell (Bio-Rad Laboratories). Samples were mixed with a loading buffer under nonreducing or reducing conditions (100 mm dithiothreitol and 5% β2-mercaptoethanol) before SDS-PAGE. Blots were then washed and blocked with 5% skim milk before immunoblotting using a primary antibody and a horseradish peroxidase-conjugated secondary IgG (1:5,000 final dilution in 3% skim milk). Signals were detected following immunofluorescent imaging using the ECL system (Amersham Biosciences).
Histidine Peptide Enzyme-linked Immunosorbent Assay—To quantify purified RLN2 peptides with different mutations, serial dilutions of peptides were bound to a His tag antibody plate (Novagen, Madison, WI) in 1× TBS with 3% bovine serum albumin. After a 3-h incubation, the plates were washed four times with 1× TBS containing 0.1% Tween 20, followed by incubation with a rabbit anti-relaxin antibody (1:2500 in TBS with 3% bovine serum albumin). After washing and incubating with a secondary horseradish peroxidase-conjugated goat anti-rabbit antibody (Amersham Biosciences), the signals were detected using TMB luciferase substrate (Calbiochem) with a luminometer (Bio-Rad).
Receptor-activation Analysis—The bioactivity of relaxin family peptides was determined based on the stimulation of adenylate cyclase activity in HEK293T cells stably expressing recombinant LGR7 or LGR8. Stable LGR7- or LGR8-expressing cells were maintained in Dulbecco's modified Eagle's medium/F-12 media supplemented with 200 ng/ml Zeocin (Invitrogen). After seeding at a density of 2 × 105 cells per well in 500 μl of Dulbecco's modified Eagle's medium/F-12 medium containing 0.1% bovine serum albumin in 48-well plates, cells were preincubated at 37 °C for 30 min in the presence of 0.25 mm 3-isobutyl-1-methylxanthine before treatment with increasing concentrations of peptides (from 0 to 100 nm) in quadruplicate. Twelve hours after treatment, the culture was reacted with acetic anhydride/triethylamine, and total cAMP content in cell lysates was measured using a specific cAMP radioimmunoassay. A nontagged human RLN2 peptide was used as a standard to verify the bioactivity of tagged RLN2 peptides (a kind gift from Dr. Elaine N. Unemori, Connectics, Mountain View, CA).
Receptor-binding Assay—To determine the binding characteristics of wild type and mutant peptides, HEK293T cells expressing LGR7 or LGR8 were grown to 90% confluence, collected by centrifugation at 3000 × g for 2 min, and washed two times in 2 ml of ice-cold binding buffer. Cells were resuspended in ice-cold binding buffer, and aliquots of cell suspension were incubated with increasing doses of purified recombinant peptides in the presence of 0.025 pmol of 125I-labeled RLN2 tracer (Phoenix Pharmaceuticals, Burlingame, CA) at room temperature for 2 h. After washing three times in an ice-cold binding buffer, radioactivity bound to the cells was measured using a γ-counter (EG&G Wallace, Gaithersburg, MD). Total binding was determined in the absence, and nonspecific binding in the presence, of 25 nmol of unlabeled RLN2. In regular assays, total binding was ∼30,000 cpm, and nonspecific binding amounted to less than 20% of the total binding. Data points were collected in triplicate, and similar results were obtained in at least two independent assays.
Mass Spectrometry Analysis of Recombinant Peptides—To confirm that the recombinant relaxin family peptides were processed similar to that of the native peptides and assumed a native conformation, the mass of purified peptides was determined by matrix-assisted laser desorption/ionization (MALDI) mass spectrometry. MALDI mass spectrometry was performed by the Pan facility at the Stanford University School of Medicine. Mass spectra were acquired on a Voyager DE-RP Model (Applied Biosystems, Foster City, CA).
Comparative Structure Modeling—To detect potential alterations in the surface motifs of RLN2 as a result of mutagenesis, we generated three-dimensional models of mutant peptides based on the human RLN2 crystal structure (Protein Data Bank 6RLN) using the SWISS-MODEL server (43) and refined by energy minimization. Molecular views were drawn with the Swiss-PdbViewer and Protein Explorer 2.4 programs (44).
Statistical Analyses—Receptor-activation and receptor-binding activity curves generated in nonlinear regression analyses (Prism; GraphPad Software, San Diego, CA) were evaluated relative to the samples without hormone treatment. In all cases data are reported as the mean ± S.E. of assays in triplicate or quadruplicate. Statistically significant responses (p < 0.05) were determined for each stimulated response to the average nonspecific response from control samples using analysis of variance and Student's t test.
RESULTS
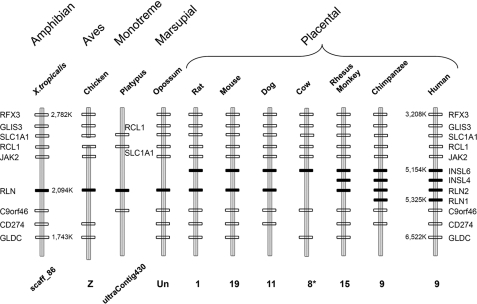
Identification of Relaxin Homologs Syntenic to Human RLN2 in Prototherian, Metatherian, and Eutherian Mammals—To better understand the functional motifs important for receptor interaction in human RLN2, we analyzed the sequence conservation of orthologous relaxin peptides from mammals including monotremes, marsupials, and placentals as well as nonmammalian tetrapods. Although earlier studies have identified putative relaxin orthologs from a number of vertebrates, whether some of those genes are orthologous to human RLN2 has not been fully clarified as a result of divergent evolution of this family of genes in mammals (35, 45). Our recent comparative genome analyses showed that relaxin family genes evolved from three independent loci including, 1) INSL5 or relaxin family locus A (RFLA) corresponding to human chromosome 1p31; 2) RLN1/RLN2/INSL4/INSL6 or relaxin family locus B (RFLB) corresponding to human chromosome 9p24; and 3) RLN3/INSL3 or relaxin family locus C (RFLC) corresponding to human chromosome 19p13 in the most recent common ancestor of vertebrates (35). The chromosomal regions syntenic to the human RFLB (RLN1/RLN2/INSL4/INSL6) locus could be identified in all studied species. In all therian mammals, this locus is marked by invariant flanking genes including RFX3, GLIS3, SLC1A1, RCL1, JAK2, C9orf46, CD274, and GLDC in neighboring loci (Fig. 1). Likewise, the single relaxin homolog at the syntenic regions of monotreme platypus and X. tropicalis is flanked by orthologous RCL1, SLC1A1, and C9orf46 genes.
FIGURE 1.
Syntenic mapping of the RLN1/RLN2/INSL4/INSL6 locus in tetrapods. Schematic representation of relaxin locus syntenic to the RLN1/RLN2/INSL4/INSL6 locus on human chromosome 9 in chimpanzee (P. troglodytes), Rhesus monkey (M. mulatta), cow (B. taurus), dog (C. familiaris), mouse (M. musculus), rat (R. norvegicus), the gray short-tailed opossum (M. domestica), platypus (O. anatinus), chicken (G. gallus), and the clawed frog (X. tropicalis). The relaxin family genes on the syntenic relaxin family locus B (RFLB) locus of different vertebrates are indicated by black rectangles, whereas, neighboring genes are indicated by blank rectangles (35). The chromosomal numbers or the genomic contig numbers are indicated at the bottom of the schematic representation of each genomic fragment. The positions of select genes in the genomes of human and X. tropicalis are also indicated (*), part of the syntenic region on chromosome 8 of cow is mapped to unknown chromosome in the GenBank™.
Similar to humans, the chimpanzee genome encodes one of each of the orthologus RLN1, RLN2, INSL4, and INSL6 genes that cluster within a 171-kb region in tandem. Unlike hominids that share a common ancestor 5-10 million years ago, the Rhesus monkey genome encodes only one of each of the RLN, INSL4, and INSL6. On the other hand, placental mammals including the dog, rat, and mouse each contain one RLN and one INSL6, whereas the cow contains only an INSL6 ortholog (Fig. 1). Unlike the placental mammals, opossum, platypus, chicken, and X. tropicalis each contain only a single relaxin family gene at the syntenic RFLB locus. These data indicate that the four human relaxin family paralogs in this locus were likely derived from nonallelic homologous recombinations at three separate geological times during the evolution of eutherians. The most likely scenario is that RLN1 and RLN2 evolved before the emergence of hominids. In contrast, INSL4 evolved before the divergence of old world monkeys and apes >15 million years ago, whereas the ancestral RLN and INSL6 genes evolved only after the emergence of eutherian mammals >90 million years ago.
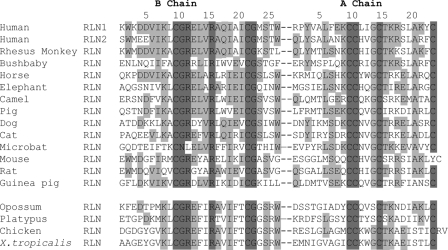
Sequence comparison of RLN homologs found at the syntenic RFLB locus of 15 mammals and that of chickens and X. tropicalis showed that the relaxin A chain exhibits a greater sequence divergence as compared with the RXXXRXXI motif-containing B chain (Fig. 2). Based on this observation and earlier studies showing that relaxin orthologs from rodents exhibit a more restricted preference upon the activation of LGR7 versus LGR8 as compared with human and pig relaxins (8, 40), we reasoned that the diverging C terminus of A chain could contain structural motifs important for interacting with LGR7 and/or LGR8, hence, receptor selectivity.
FIGURE 2.
Alignment of amino acid sequences of the B and A chains of relaxin peptides from the RFLB of 17 representative tetrapods. Residues that are completely conserved in all species analyzed are highlighted by a dark background. Resides that are conserved in at least five sequences are indicated by a gray background. Positions of residues are indicated by numbers on top of the alignment.
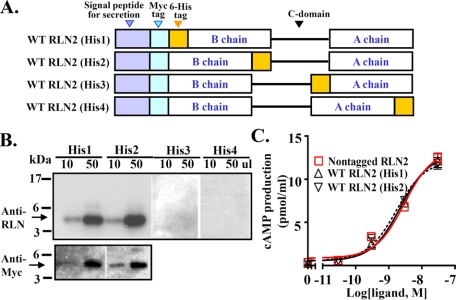
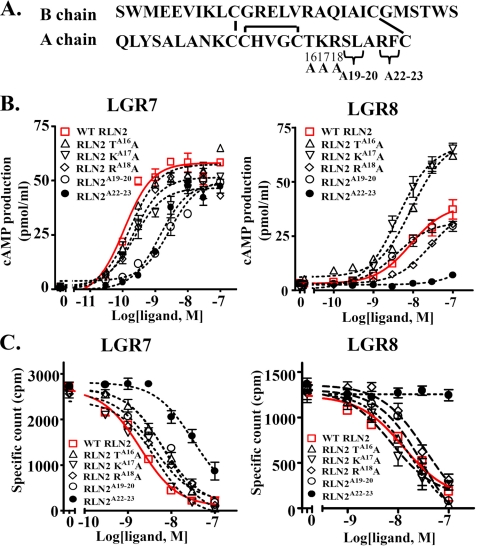
Generation of Relaxin Family Peptides Using the Recombinant Approach—To test the hypothesis that human RLN2 A chain is important for receptor interaction, we generated and analyzed mutant RLN2 peptides with point mutations at various positions of the C terminus of A chain. Although relaxin peptides were mainly generated by synthetic chemistry approaches in earlier studies (9, 46, 47), recent studies showed that recombinant techniques provide an alternative approach to generate functional relaxin family peptides and different analogs (6, 7, 42, 48). To allow routine generation of a variety of RLN2 mutants, we first analyzed the production of recombinant peptides with expression constructs in which the open reading frame of proRLN2 is tagged with a His6 epitope at four different positions of the mature peptide in addition to an N-terminal Myc epitope (WT RLN2 His1, His2, His3, and His4; Fig. 3A). Western blotting analysis shows that peptides tagged with a His6 epitope at the N or C terminus of the B chain were efficiently secreted into the conditioned media, and the majority of secreted peptides exhibited a predicted 5-kDa molecular mass under reducing conditions (Fig. 3B). Similar to the nontagged wild type RLN2 peptide, RLN2 His1 and RLN2 His2 peptides stimulated adenylate cyclase activity in LGR7-expressing HEK293T cells with similar potencies (Fig. 3C and Table 1). Based on these observations, we tagged all expression constructs with an N terminus Myc epitope and a C terminus His6 epitope flanking the mature B chain in subsequent studies.
FIGURE 3.
Generation and characterization of tagged recombinant human RLN2 peptide. A, schematic design of recombinant RLN2 peptides with an N-terminal Myc tag and a His6 tag at four different positions of the mature peptides (RLN2 His1-4). The mature B and A chains are indicated by a rectangle box, whereas the C domain is indicated by a line. Myc and His6 epitopes are indicated by a light blue rectangle and a yellow rectangle, respectively. B, Western blotting analysis of tagged RLN2 peptides in conditioned media of transfected HEK293T cells using an anti-relaxin antibody (Calbiochem; upper panel) or an anti-Myc antibody (lower panel) under reducing conditions. The molecular weight standard is shown on the left. C, stimulation of cAMP production by RLN2 His1 and RLN2 His2 peptides in LGR7-expressing HEK293T cells.
TABLE 1.
Potencies of recombinant wild type (WT) and mutant RLN2 peptides on the activation of LGR7 and LGR8
|
Ligand
|
pEC50
|
|
|---|---|---|
| LGR7 | LGR8 | |
| Nontagged RLN2 | 8.53 ± 0.10 | |
| WT RLN2 (His1) | 8.66 ± 0.09 | |
| WT RLN2 (His2) | 8.77 ± 0.10 | |
| RLN2 C-104 | 8.74 ± 0.05 | |
| RLN2 C-38 | 8.72 ± 0.10 | |
| RLN2 C-28 | 8.66 ± 0.07 | |
| RLN2 C-18 | 8.74 ± 0.09 | |
| RLN2 C-8 | 8.81 ± 0.10 | |
| WT RLN2 | 9.30 ± 0.04 | 7.41 ± 0.14 |
| WT RLN3 | 7.73 ± 0.07 | NDa |
| WT INSL3 | ND | 9.03 ± 0.05 |
| RLN2 RB12A | ND | ND |
| RLN2 RB16A | ND | ND |
| RLN2 TA16A | 9.75 ± 0.08 | 8.04 ± 0.06 |
| RLN2 KA17A | 9.18 ± 0.07 | 8.39 ± 0.06 |
| RLN2 RA18A | 9.67 ± 0.10 | 7.56 ± 0.04 |
| RLN2A19-20 | 8.56 ± 0.07 | 8.41 ± 0.07 |
| RLN2A22-23 | 8.87 ± 0.07 | ND |
| RLN2 RA22A | 10.03 ± 0.07 | 7.51 ± 0.09 |
| RLN2 FA23A | 9.45 ± 0.09 | ND |
| RLN2 KA17G | 9.04 ± 0.05 | 8.28 ± 0.05 |
| RLN2 KA17Q | 9.31 ± 0.11 | 7.77 ± 0.05 |
| RLN2 KA17D | 8.41 ± 0.06 | 8.36 ± 0.08 |
ND, the pEC50 values were not measurable.
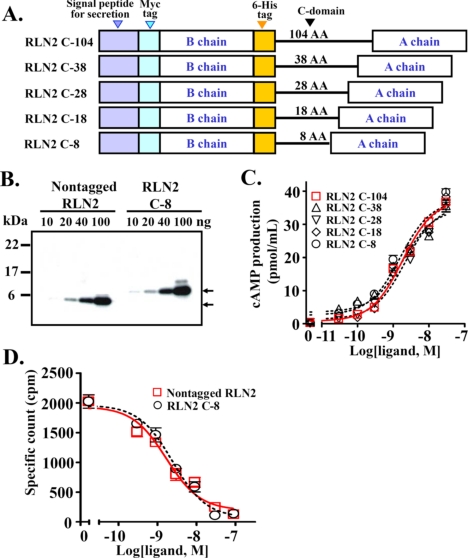
In addition, given that relaxin family peptides contain cryptic C domains of varying lengths and with multiple basic residues that could be subjected to alternative post-translational processing, we explored the possibility of generating relaxin peptides with a uniform C domain linker sequence to avoid differential processing of the secreted peptides. Studies of four expression constructs, in which the B and A chains of RLN2 were linked with a truncated C-domain of 8, 18, 28, or 38 amino acids (Fig. 4A, RLN2 C-8, RLN2 C-18, RLN2 C-28, and RLN2 C-38), revealed that the expression construct with as few as an 8-amino acid linker sequence allows efficient production of RLN2 in conditioned media (Figs. 4B and 5B). Functional analyses showed that RLN2 generated using the RLN2 C-8, RLN2 C-18, RLN2 C-28, or RLN2 C-38 constructs all have an EC50 upon LGR7 activation similar to that of RLN2 derived from the construct containing a 104-amino acid C domain sequence (Fig. 4C and Table 1). Similarly, receptor-binding analysis showed that the Myc epitope-tagged RLN2, derived from the RLN2 C-8 construct, competed for labeled nontagged RLN2 binding to LGR7 with a high affinity (Fig. 4D and Table 2). Subsequently, all our mutant expression constructs were engineered with an 8-amino acid miniature linker sequence between the B and A chains.
FIGURE 4.
Characterization of recombinant human RLN2 peptides generated using expression constructs with a truncated C domain linker sequence. A, schematic design of RLN2 expression constructs with a truncated C domain (RLN2 C-8 to C-38). The linker sequence was derived from the first 38-amino acids of the human RLN2 C domain and was flanked by a pair of dibasic cleavage sites for convertase cleavage. B, Western blotting analysis of a nontagged RLN2 peptide and the tagged RLN2 peptide generated using the expression construct with an 8-amino acid C domain sequence. Specific bands are detected using an anti-relaxin antibody, and are indicated by arrows. The molecular weight standard is shown on the left. C, stimulation of cAMP production by RLN2 peptides in LGR7-expressing HEK293T cells. Each data point represents the mean ± S.E. of quadruplicate samples. D, competitive LGR7-binding analysis of the Myc- and His6-tagged RLN2 C-8. Each data point represents the mean ± S.E. of triplicate samples.
FIGURE 5.
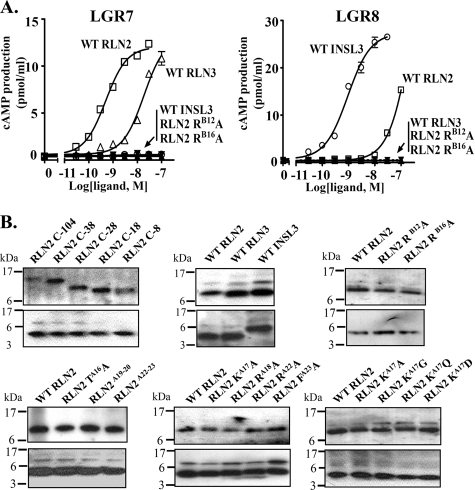
Stimulation of cAMP production in LGR7- and LGR8-expressing cells by recombinant RLN2, RLN3, and INSL3. A, recombinant RLN3 and INSL3 selectively activated LGR7 and LGR8, respectively. In contrast, RLN2 activates both receptors at the nanomolar range. Unlike the wild type RLN2, substitution of ArgB12 and ArgB16 residues with alanine abolished the LGR7- and LGR8-activation activities of RLN2. Each data point represents the mean ± S.E. of quadruplicate samples. B, Western blotting analysis of recombinant wild type and mutant relaxin peptides including, RLN2 C-104, C-38, C-28, C-18, C-8 (upper left), WT RLN2, WT RLN3, WT INSL3 (upper middle), WT RLN2, RLN2 RB12A, RLN2 RB16A (upper right), WT RLN2, RLN2 TA16A, RLN2A19-20, RLN2A22-23 (lower left), WT RLN2, RLN2 KA17A, RLN2 RA18A, RLN2 RA22A, RLN2 FA23A (lower middle), WT RLN2, RLN2 KA17A, RLN2 KA17G, RLN2 KA17Q, and RLN2 KA17D(lower right). Affinity column purified peptides (100 ng/lane) were resolved by 18% SDS-PAGE under nonreducing (upper panel) and reducing (lower panel) conditions. The positions of molecular mass markers are indicated on the left.
TABLE 2.
Potencies of wild type (WT) and mutant RLN2 peptides in competition for RLN2 binding to LGR7 and LGR8
|
Ligand
|
pIC50
|
|
|---|---|---|
| LGR7 | LGR8 | |
| Nontagged RLN2 | 8.78 ± 0.12 | |
| RLN2 C-8 | 8.61 ± 0.09 | |
| WT RLN2 | 8.72 ± 0.06 | 7.90 ± 0.16 |
| RLN2 TA16A | 8.40 ± 0.06 | 7.74 ± 0.07 |
| RLN2 KA17A | 8.36 ± 0.08 | 8.14 ± 0.18 |
| RLN2 RA18A | 8.42 ± 0.08 | 7.46 ± 0.17 |
| RLN2A19-20 | 8.21 ± 0.10 | 7.51 ± 0.12 |
| RLN2A22-23 | 7.50 ± 0.16 | NDa |
| RLN2 RA22A | 8.42 ± 0.08 | 6.80 ± 0.32 |
| RLN2 FA23A | 8.17 ± 0.06 | ND |
| RLN2 KA17G | 7.57 ± 0.22 | 7.40 ± 0.41 |
| RLN2 KA17Q | 7.93 ± 0.15 | 7.96 ± 0.11 |
| RLN2 KA17D | 7.75 ± 0.25 | 7.53 ± 0.24 |
ND, the pIC50 values were not measurable.
Using the same approach, we also generated recombinant INSL3 and RLN3. Mass spectrometry analysis of purified peptides confirmed that these recombinant peptides are processed as heterodimeric peptides after post-translational cleavage of the C domain sequences (Table 3). Importantly, functional analysis showed that recombinant peptides exhibit receptor-activation characteristics similar to that of purified or chemically synthesized peptides (49-52). Unlike RLN3 and INSL3, which selectively activated LGR7 and LGR8, respectively, RLN2 activates both receptors at the nanomolar range (Fig. 5A). Furthermore, we show that alanine substitution at either of the charged residues of the RXXXRXXI motif (ArgB12 and ArgB16) ablated both the LGR7 and the LGR8 activation activity of RLN2 (Fig. 5A).
TABLE 3.
Mass spectrometry analysis of purified recombinant peptides
| Peptide | Molecular weight measured | Expected molecular weight |
|---|---|---|
| WT RLN2 | 8224 | 8225 |
| WT RLN3 | 7888 | 7888 |
| WT INSL3 | 8270 | 8272 |
A16-17 and A22-23 Regions in the RLN2 A Chain Play Critical Roles in Receptor Interaction—To investigate the role of residues at the C terminus of A chain in receptor interaction, we generated five mutants with alanine substitution at five different positions (Fig. 6A, RLN2 TA16A, RLN2 KA17A, RLN2 RA18A, RLN2A19-20, and RLN2A22-23). Western blotting analysis showed that all five mutants were processed to the mature form (Fig. 5B). Whereas RLN2 TA16A, RLN2 KA17A, and RLN2 RA18A mutants exhibited an EC50 on LGR7 activation similar to that of the wild type peptides, RLN2A19-20 and RLN2A22-23 mutants exhibited decreased LGR7-activation activities (Fig. 6B, left panel, and Table 1). In addition, receptor-binding analyses shows that, except for RLN2A22-23, which exhibited a reduced affinity for LGR7, all other mutants competed for labeled RLN2 binding to LGR7 with affinities similar to that of wild type peptide (Fig. 6C, left panel, Table 2).
FIGURE 6.
Alanine substitution in the C terminus of A chain alters the receptor-activation activity of RLN2. A, schematic representation of RLN2 mutants with alanine substitution at five different positions of the A chain (RLN2 TA16A, RLN2 KA17A, RLN2 RA18A, RLN2A19-20, and RLN2A22-23). B, stimulation of cAMP production in LGR7- and LGR8-expressing cells by the wild type and mutant RLN2 peptides. Unlike the wild type peptide, RLN2 TA16A and RLN2 KA17A mutants exhibited a significantly enhanced LGR8-activation activity. C, competitive LGR7- and LGR8-binding assays of wild type and mutant RLN2 peptides. Each data point in the receptor-activation and receptor-binding assays represents the mean ± S.E. of quadruplicate and triplicate samples, respectively.
Although alanine substitution has minimal effects on maximum LGR7-activation activity of mutant peptides, mutation at the RLN2A16 and RLN2A17 positions leads to a 2-fold increase of the maximum LGR8-activation activity (Fig. 6B, right panel). In contrast, mutation of the RLN2A22-23 region almost abolished the LGR8-activation activity of RLN2. Consistent with analyses of receptor-activation activity, the RLN2 TA16A and RLN2 KA17A mutants exhibited an IC50 value similar to that of wild type RLN2 for both LGR7 and LGR8, whereas the RLN2A22-23 mutant lost its LGR8-binding activity (Fig. 6C and Table 2).
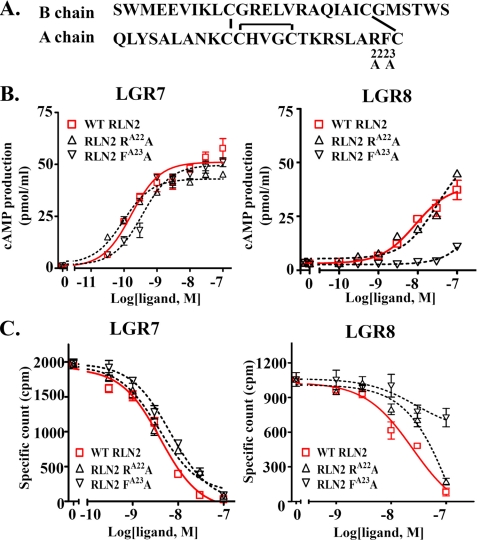
The Functional Characteristics of RLN2A22-23 Mutant Are Attributed to Mutation at the PheA23 Position—Given the findings that the A22-23 region plays important roles in receptor signaling, we then analyzed mutants with point mutation at ArgA22 and PheA23 positions (Figs. 5B and 7A). Functional analysis showed that alanine substitution at the ArgA22 position has minimal effect on the LGR7- or LGR8-activation activity (Fig. 7B). In contrast, the RLN2 FA23A mutant lacks an LGR8-activation activity, but retains an LGR7-activation activity similar to that of wild type peptides. Receptor-binding assays showed that the RLN2 RA22A and RLN2 FA23A mutants bind to LGR7 with affinities similar to that of wild type peptides (Fig. 7C). In contrast, alanine substitution at the PheA23 position significantly reduced the affinity for LGR8 (Fig. 7C).
FIGURE 7.
Alanine substitution at the PheA23 position alters the receptor-interaction activities of RLN2. A, schematic representation of RLN2 mutants with alanine substitution at the ArgA22 or PheA23 positions. B, stimulation of cAMP production in LGR7- and LGR8-expressing cells by RLN2 RA22A and RLN2 FA23A mutants. FA23A mutation ablated LGR8-activation activity. C, LGR7- and LGR8-binding activities of RA22A and FA23A mutant peptides. FA23A mutation significantly reduced LGR8-binding activity, but without an effect on LGR7-binding activity. Each data point in the receptor-activation and receptor-binding assays represents the mean ± S.E. of quadruplicate and triplicate samples, respectively.
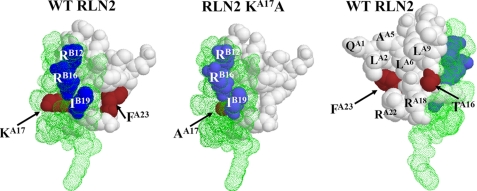
ThrA16, LysA17, and PheA23 Residues Constitute Part of the Receptor-interacting Interface in Human RLN2—To gain a better understanding of the molecular basis for the regulation of receptor activation by ThrA16, LysA17, and PheA23, we generated simulated structure models of RLN2 mutants using the SWISS-MODEL server (Fig. 8). Comparative structure analyses showed that the LysA17 residue (brown filled space) is embedded between the interface of the A chain (white filled space) and the B chain (green dot space), and is the only A chain residue with its side chain exposed on the same surface with ArgB12, ArgB16, and IleB19 of the RXXXRXXI motif (Fig. 8, left panel). Alanine substitution in the RLN2 KA17A mutant eliminated the extended side chain of lysine that protrudes toward the B chain surface (Fig. 8, middle panel).
FIGURE 8.
Comparative structure modeling of mutant peptides based on the crystal structure of human RLN2. Three-dimensional structure of the wild type human RLN2 and the KA17A mutant based on the RLN2 crystal structure (PDB 6RLN). The wild type RLN2 structure is shown on the left and right panels. The KA17A mutant is shown on the middle panel. The A and B chains are indicted by white filled space and green dot space, respectively. The amino acids at A16, A17, and A23 positions are indicated by the brown filled space. The ArgB12, ArgB16, and IleB19 residues in the RXXXRXXI motif of B chain are represented by the blue filled space.
Unlike the LysA17 residue, the ThrA16 and PheA23 residues are located opposite to the surface including LysA17 and the RXXX-RXXI motif (Fig. 8, right panel). Judging by the orientation and the distance of these residues to the binding motif on the B chain, it likely interacts with a distinct ligand-binding interface of LGR7 and LGR8.
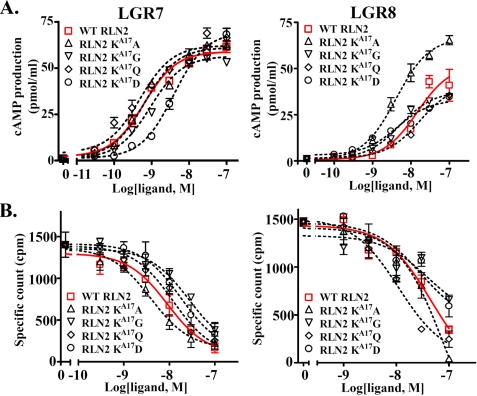
The finding that the elimination of the extended side chain of lysine at the LysA17 position leads to an increased LGR8-activation activity was of particular interest considering its close proximity to the RXXXRXXI motif. To further characterize the importance of the LysA17 position in receptor interaction, we analyzed the receptor-activation activity of mutants with an aspartic acid, glutamine, or glycine at this position (Fig. 5B). Unlike the pan-specific RLN2 KA17A mutant, peptides with a glycine or glutamine at the LysA17 position exhibited normal LGR7- and LGR8-activation activity (Fig. 9, A and B). Alternatively, substitution with an aspartic acid residue at the same position led to a reduced LGR7-activation activity (Fig. 9A). Receptor-binding assays showed that substitution with a glycine and aspartic acid at LysA17 reduced LGR7-binding activity by 3-fold (Fig. 9B). Conversely, glutamine substitution at LysA17 had a minimal effect on LGR7- or LGR8-binding activity.
FIGURE 9.
Stimulation of cAMP production in LGR7- and LGR8-expressing cells by mutant peptides with residue substitution at the LysA17 position. A, stimulation of cAMP production in LGR7- and LGR8-expressing cells by KA17A, KA17G, KA17D, and KA17Q mutants. Unlike the KA17A peptide, mutants with an aspartic acid, glycine, or glutamine at the LysA17 position did not exhibit an enhanced LGR8-activation activity. Each data point represents the mean ± S.E. of quadruplicate samples. B, LGR7- and LGR8-binding activities of KA17A, KA17G, KA17D, and KA17Q mutants. Each data point represents the mean ± S.E. of triplicate samples.
DISCUSSION
Based on analyses of the receptor-activation and receptor-binding activities, our study identified ThrA16, LysA17, and PheA23 of RLN2 as crucial residues in the interaction with LGR7 and LGR8, and suggested that these three residues act cooperatively with the well characterized RXXXRXXI motif in shaping the functional characteristics of human RLN2. These studies have also led to the generation of LGR7-specific human RLN2 analogs that could be useful for selective activation of LGR7 signaling pathways in patients as well as a pan-specific analog with an enhanced LGR8-activation activity.
Relaxin family peptides are structurally similar to insulin and are first synthesized as a single chain prorelaxin, in which a C domain connecting the B and A chains is excised during the post-translational modification. Based on studies of insulin, which shares a similar three disulfide-bridged two chain architecture, the C domain is thought to play a minimal role in the folding of a mature relaxin peptide. We show that, similar to insulin (53, 54), a minimal linker sequence with only 8 amino acids is sufficient for the generation of mature RLN2, INSL3, and RLN3 peptides in transfected cells. These results suggest that the conformational propensities for a mature two-chain relaxin family peptide does not require a native C domain, and are consistent with the observation that the C domain sequence of these genes diverged greatly even among closely related species.
Unlike the C domain sequences, earlier studies of insulin and relaxin biosynthesis have shown that the A chain may function mainly as a scaffold in the assembly of a bioactive disulfide-bridged mature peptide (55). On the other hand, a number of residues in the insulin B chain were shown to be essential for the folding of an active conformation. Mutation of these residues leads to impaired foldability and secretion of insulin (56). Consistent with this view, we show that RLN2 mutants with substitutions at various residues of the C terminus of the A chain were processed to mature forms, suggesting these residues play a minimal role in disulfide pairing and the folding of mature peptides. Although earlier studies have shown that relaxin interacts with its receptors via a tight receptor-binding interface in the B chain (57, 58), whether the B chain sequences are crucial for the folding of relaxin peptides remains to be investigated.
Earlier structural-functional relationships of relaxin family peptides have been inferred from the degree of residue conservation among mammalian species and studies of native receptors. An invariant RXXXRXXI motif in the B chain was found to be essential for receptor binding, and replacement of arginines or isoleucine in this motif greatly reduced the binding activity. Although the molecular basis for the interaction between RXXXRXXI motif and the receptor is not clear, x-ray crystallographic structure analysis shows that the conserved arginines and isoleucine form a tight receptor-binding surface (34). Based on these observations, it was proposed that interactions between these charged residues and select acidic residues within the concave face of the ectodomain of LGR7 are critical to the receptor activation (58). In contrast, it was hypothesized that the isoleucine residue of the RLN2 B chain could interact with a cluster of tryptophan, isoleucine, and leucine residues close to the ligand-interacting acidic residues through hydrophobic interactions (38, 58). Similar to relaxin, a pair of arginines and a histidine in the INSL3 B chain were shown to be critical for receptor interaction (35, 37, 38, 50, 59). Interestingly, relaxin orthologs from placental mammals display varied preference for the two relaxin receptors even though all these orthologs contain the invariant RXXXRXXI motif (8, 40). Based on this observation, we hypothesized that: 1) the expansion of family genes at the syntenic RFLB locus in placental mammals has allowed the divergence in functional characteristics of relaxin peptides unrelated to the core structure of these peptides; and 2) analysis of residues at sites tolerant of substitutions could reveal molecular determinants that are critical to the optimal signaling in a select lineage. Indeed, in contrast to the general notion that the A chain has a minimal role in receptor interaction, we show that mutations at ThrA16, LysA17, and PheA23 positions significantly alter the receptor-binding and receptor-activation activities of RLN2. Whereas PheA23 is important for high affinity binding to LGR8, ThrA16 and LysA17 play a more critical role in restricting the interaction with LGR8. These findings are consistent with the hypothesis that residues at sites tolerant of substitutions in a rapidly expanding protein family could represent critical motifs important for lineage-specific adaptations. However, whether the difference in receptor selectivity of relaxin peptides from different mammals was attributed to residue difference at the C terminus of A chain remains to be investigated.
Our study effectively expanded the known receptor-interacting interface of human RLN2, and indicated that ThrA16, LysA17, and PheA23 at the C terminus of the A chain, together with the RXXXRXXI motif, constitute a broad receptor-binding interface for optimal signal transmission. Of interest, structural modeling analyses show that LysA17 is the only residue with its side chain protruding onto the same surface with the RXXXRXXI motif and is in close proximity with ArgB12, ArgB16, and IleB19. Therefore, it is likely that LysA17 and the three receptor-binding residues in the RXXXRXXI motif form an extended binding interface to interact with the receptor. In contrast, ThrA16 and PheA23 are positioned on a surface opposite to that of the RXXXRXXI motif, suggesting that these residues could interact with a receptor interface distant from that for LysA17. It is important to note that depending on the amino acid introduced at the LysA17 position, the preference for LGR8 could be either enhanced or reduced. Whereas alanine substitution enhances LGR8-activation activity, corresponding substitution with a polar or a reverse charged residue reduces it. Therefore, the side chain at the LysA17 position could be conductive to conformational transformation and an alanine residue provides an induced fit for LGR8 activation. In contrast, a polar or a reversed charged residue at this position increases the hindrance for the adoption of an activation configuration for LGR8.
In the last few years, human RLN2 has been the subject of various clinical studies aimed to treat etiology in reproduction and other systems (25-28). Based on the understanding that: 1) RLN2 activates LGR8 at the nanomolar concentration; and 2) LGR8 signaling is involved in the regulation of oocyte maturation, ovarian follicle development, and the positioning of the ovary (10, 22, 23, 60), it is conceivable that clinical application of the indiscriminative wild type human RLN2 could impose unwanted side effects on LGR8-mediated physiological processes in patients. The finding that RLN2A22-23, and FA23A mutants preferentially activate LGR7 thus provides novel agents to allow selective activation of LGR7-mediated physiological processes in patients. Although the related RLN3 activates only LGR7, but not LGR8, it also functions as a potent agonist for GPCR135 and GPCR142. In contrast, the RLN2 TA16A and RLN2 KA17A mutants, which exhibit high potencies on the activation of both LGR7 and LGR8, function as pan-specific agonists for these two receptors. These reagents could be useful in therapeutic applications that aim to activate both LGR7 and LGR8 in patients.
Acknowledgments
We thank Caren Spencer and Cynthia Klein for editorial and technical assistance. We also thank Dr. Aaron Hsueh (Department of Obstetrics and Gynecology, Stanford University) for comments on the manuscript as well as Dr. Yang Jun (Phoenix Pharmaceuticals) for the kind gift of RLN2 tracers.
This work was supported, in whole or in part, by National Institutes of Health Grant R21 HD47606 (to S. Y. H.). This work was also supported by a March of Dimes research grant (to S. Y. H.). The costs of publication of this article were defrayed in part by the payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 U.S.C. Section 1734 solely to indicate this fact.
Footnotes
The abbreviations used are: RLN1, relaxin H1; RLN2, relaxin H2; RLN3, relaxin 3; INSL3, insulin-like 3; GPCR, G protein-coupled receptor; LGR7, leucine-rich repeat-containing GPCR 7; LGR8, leucine-rich repeat-containing GPCR 8; MALDI, matrix-assisted laser desorption/ionization; TBS, Tris-buffered saline.
References
- 1.Chassin, D., Laurent, A., Janneau, J. L., Berger, R., and Bellet, D. (1995) Genomics 29 465-470 [DOI] [PubMed] [Google Scholar]
- 2.Hsu, S. Y. (1999) Mol. Endocrinol. 13 2163-2174 [DOI] [PubMed] [Google Scholar]
- 3.Lok, S., Johnston, D. S., Conklin, D., Lofton-Day, C. E., Adams, R. L., Jelmberg, A. C., Whitmore, T. E., Schrader, S., Griswold, M. D., and Jaspers, S. R. (2000) Biol. Reprod. 62 1593-1599 [DOI] [PubMed] [Google Scholar]
- 4.Conklin, D., Lofton-Day, C. E., Haldeman, B. A., Ching, A., Whitmore, T. E., Lok, S., and Jaspers, S. (1999) Genomics 60 50-56 [DOI] [PubMed] [Google Scholar]
- 5.Bathgate, R. A., Samuel, C. S., Burazin, T. C., Layfield, S., Claasz, A. A., Reytomas, I. G., Dawson, N. F., Zhao, C., Bond, C., Summers, R. J., Parry, L. J., Wade, J. D., and Tregear, G. W. (2002) J. Biol. Chem. 277 1148-1157 [DOI] [PubMed] [Google Scholar]
- 6.Liu, C., Chen, J., Sutton, S., Roland, B., Kuei, C., Farmer, N., Sillard, R., and Lovenberg, T. W. (2003) J. Biol. Chem. 278 50765-50770 [DOI] [PubMed] [Google Scholar]
- 7.Liu, C., Eriste, E., Sutton, S., Chen, J., Roland, B., Kuei, C., Farmer, N., Jornvall, H., Sillard, R., and Lovenberg, T. W. (2003) J. Biol. Chem. 278 50754-50764 [DOI] [PubMed] [Google Scholar]
- 8.Hsu, S. Y., Nakabayashi, K., Nishi, S., Kumagai, J., Kudo, M., Sherwood, O. D., and Hsueh, A. J. (2002) Science 295 671-674 [DOI] [PubMed] [Google Scholar]
- 9.Sudo, S., Kumagai, J., Nishi, S., Layfield, S., Ferraro, T., Bathgate, R., and Hsueh, A. J. (2002) J. Biol. Chem. 278 7855-7862 [DOI] [PubMed] [Google Scholar]
- 10.Kumagai, J., Hsu, S. Y., Matsumi, H., Roh, J. S., Fu, P., Wade, J. D., Bathgate, R. A., and Hsueh, A. J. (2002) J. Biol. Chem. 277 31283-31286 [DOI] [PubMed] [Google Scholar]
- 11.Bogatcheva, N. V., Truong, A., Feng, S., Engel, W., Adham, I. M., and Agoulnik, A. I. (2003) Mol. Endocrinol. 17 2639-2646 [DOI] [PubMed] [Google Scholar]
- 12.Feng, S., Bogatcheva, N. V., Kamat, A. A., and Agoulnik, A. I. (2005) Ann. N. Y. Acad. Sci. 1041 82-90 [DOI] [PubMed] [Google Scholar]
- 13.Kamat, A. A., Feng, S., Bogatcheva, N. V., Truong, A., Bishop, C. E., and Agoulnik, A. I. (2004) Endocrinology 145 4712-4720 [DOI] [PubMed] [Google Scholar]
- 14.Zhao, L., Roche, P. J., Gunnersen, J. M., Hammond, V. E., Tregear, G. W., Wintour, E. M., and Beck, F. (1999) Endocrinology 140 445-453 [DOI] [PubMed] [Google Scholar]
- 15.Krajnc-Franken, M. A., van Disseldorp, A. J., Koenders, J. E., Mosselman, S., van Duin, M., and Gossen, J. A. (2004) Mol. Cell. Biol. 24 687-696 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Sherwood, O. D. (2004) Endocr. Rev. 25 205-234 [DOI] [PubMed] [Google Scholar]
- 17.Sherwood, O. D., Downing, S. J., Guico-Lamm, M. L., Hwang, J. J., O'Day-Bowman, M. B., and Fields, P. A. (1993) Oxf. Rev. Reprod. Biol. 15 143-189 [PubMed] [Google Scholar]
- 18.Weiss, G., and Goldsmith, L. T. (2001) Front. Horm. Res. 27 105-112 [DOI] [PubMed] [Google Scholar]
- 19.Dschietzig, T., Bartsch, C., Baumann, G., and Stangl, K. (2006) Pharmacol. Ther. 112 38-56 [DOI] [PubMed] [Google Scholar]
- 20.Ganesh, S., Gonzalez Edick, M., Idamakanti, N., Abramova, M., VanRoey, M., Robinson, M., Yun, C.-O., and Jooss, K. (2007) Cancer Res. 67 4399-4407 [DOI] [PubMed] [Google Scholar]
- 21.Kim, J.-H., Lee, Y.-S., Kim, H., Huang, J.-H., Yoon, A.-R., and Yun, C.-O. (2006) J. Natl. Cancer Inst. 98 1482-1493 [DOI] [PubMed] [Google Scholar]
- 22.Kawamura, K., Kumagai, J., Sudo, S., Chun, S. Y., Pisarska, M., Morita, H., Toppari, J., Fu, P., Wade, J. D., Bathgate, R. A., and Hsueh, A. J. (2004) Proc. Natl. Acad. Sci. U. S. A. 101 7323-7328 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Adham, I. M., Steding, G., Thamm, T., Bullesbach, E. E., Schwabe, C., Paprotta, I., and Engel, W. (2002) Mol. Endocrinol. 16 244-252 [DOI] [PubMed] [Google Scholar]
- 24.Koskimies, P., Suvanto, M., Nokkala, E., Huhtaniemi, I. T., McLuskey, A., Themmen, A. P., and Poutanen, M. (2003) Mol. Cell. Endocrinol. 206 159-166 [DOI] [PubMed] [Google Scholar]
- 25.Seibold, J. R., Korn, J. H., Simms, R., Clements, P. J., Moreland, L. W., Mayes, M. D., Furst, D. E., Rothfield, N., Steen, V., Weisman, M., Collier, D., Wigley, F. M., Merkel, P. A., Csuka, M. E., Hsu, V., Rocco, S., Erikson, M., Hannigan, J., Harkonen, W. S., and Sanders, M. E. (2000) Ann. Intern. Med. 132 871-879 [DOI] [PubMed] [Google Scholar]
- 26.Furst, D. E. (1998) Curr. Opin. Rheumatol. 10 123-128 [DOI] [PubMed] [Google Scholar]
- 27.Chen, S. A., Perlman, A. J., Spanski, N., Peterson, C. M., Sanders, S. W., Jaffe, R., Martin, M., Yalcinkaya, T., Cefalo, R. C., Chescheir, N. C., Mernard, M. K., and Mordenti, J. (1993) Pharm. Res. 10 834-838 [DOI] [PubMed] [Google Scholar]
- 28.Stewart, D. (2006) Society for the Study of Reproduction 39th Annual Meeting Omaha, NE, July 29 -August 1, 2006, University of Nebraska Medical Center, Omaha, NE
- 29.Nef, S., and Parada, L. F. (2000) Genes Dev. 14 3075-3086 [DOI] [PubMed] [Google Scholar]
- 30.Adham, I. M., Emmen, J. M., and Engel, W. (2000) Mol. Cell. Endocrinol. 160 11-16 [DOI] [PubMed] [Google Scholar]
- 31.Ivell, R., and Bathgate, R. A. (2002) Biol. Reprod. 67 699-705 [DOI] [PubMed] [Google Scholar]
- 32.Zhao, L., Samuel, C. S., Tregear, G. W., Beck, F., and Wintour, E. M. (2000) Biol. Reprod. 63 697-703 [DOI] [PubMed] [Google Scholar]
- 33.Bullesbach, E., Yang, S., and Schwabe, C. (1992) J. Biol. Chem. 267 22957-22960 [PubMed] [Google Scholar]
- 34.Eigenbrot, C., Randal, M., Quan, C., Burnier, J., O'Connell, L., Rinderknecht, E., and Kossiakoff, A. A. (1991) J. Mol. Biol. 221 15-21 [PubMed] [Google Scholar]
- 35.Park, J. I., Semyonov, J., Chang, C. L., Yi, W., Warren, W., and Hsu, S. Y. (2008) Genome Res. 18 974-985 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Bullesbach, E. E., and Schwabe, C. (1987) J. Biol. Chem. 262 12496-12501 [PubMed] [Google Scholar]
- 37.Bullesbach, E. E., and Schwabe, C. (2006) J. Biol. Chem. 281 26136-26143 [DOI] [PubMed] [Google Scholar]
- 38.Rosengren, K. J., Zhang, S., Lin, F., Daly, N. L., Scott, D. J., Hughes, R. A., Bathgate, R. A., Craik, D. J., and Wade, J. D. (2006) J. Biol. Chem. 281 28287-28295 [DOI] [PubMed] [Google Scholar]
- 39.Hossain, M. A., Rosengren, K. J., Haugaard-Jonsson, L. M., Zhang, S., Layfield, S., Ferraro, T., Daly, N. L., Tregear, G. W., Wade, J. D., and Bathgate, R. A. (2008) J. Biol. Chem. 283 17287-17297 [DOI] [PubMed] [Google Scholar]
- 40.Halls, M. L., Bond, C. P., Sudo, S., Kumagai, J., Ferraro, T., Layfield, S., Bathgate, R. A., and Summers, R. J. (2005) J. Pharmacol. Exp. Ther. 313 677-687 [DOI] [PubMed] [Google Scholar]
- 41.Park, J.-I., Chang, C., and Hsu, S. (2005) Rev. Endocr. Metab. Disorders 6 291-296 [DOI] [PubMed] [Google Scholar]
- 42.Marriott, D., Gillece-Castro, B., and Gorman, C. M. (1992) Mol. Endocrinol. 6 1441-1450 [DOI] [PubMed] [Google Scholar]
- 43.Schwede, T., Kopp, J., Guex, N., and Peitsch, M. C. (2003) Nucleic Acids Res. 31 3381-3385 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Martz, E. (2002) Trends Biochem. Sci. 27 107-109 [DOI] [PubMed] [Google Scholar]
- 45.Wilkinson, T. N., Speed, T. P., Tregear, G. W., and Bathgate, R. A. (2005) BMC Evol. Biol. 5 14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Bathgate, R. A., Lin, F., Hanson, N. F., Otvos, L., Jr., Guidolin, A., Giannakis, C., Bastiras, S., Layfield, S. L., Ferraro, T., Ma, S., Zhao, C., Gundlach, A. L., Samuel, C. S., Tregear, G. W., and Wade, J. D. (2006) Biochemistry 45 1043-1053 [DOI] [PubMed] [Google Scholar]
- 47.Fu, P., Layfield, S., Ferraro, T., Tomiyama, H., Hutson, J., Otvos, L., Jr., Tregear, G. W., Bathgate, R. A., and Wade, J. D. (2004) J. Pept. Res. 63 91-98 [DOI] [PubMed] [Google Scholar]
- 48.Zarreh-Hoshyari-Khah, R., Bartsch, O., Einspanier, A., Pohnke, Y., and Ivell, R. (2001) Regul. Pept. 97 139-146 [DOI] [PubMed] [Google Scholar]
- 49.Bullesbach, E. E., and Schwabe, C. (1986) Biochemistry 25 5998-6004 [DOI] [PubMed] [Google Scholar]
- 50.Bullesbach, E. E., and Schwabe, C. (1999) Biochemistry 38 3073-3078 [DOI] [PubMed] [Google Scholar]
- 51.Tan, Y. Y., Wade, J. D., Tregear, G. W., and Summers, R. J. (1999) Br. J. Pharmacol. 127 91-98 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Dawson, N. F., Tan, Y. Y., Macris, M., Otvos, L., Jr., Summers, R. J., Tregear, G. W., and Wade, J. D. (1999) J. Pept. Res. 53 542-547 [DOI] [PubMed] [Google Scholar]
- 53.Lee, H. C., Kim, S. J., Kim, K. S., Shin, H. C., and Yoon, J. W. (2000) Nature 408 483-488 [DOI] [PubMed] [Google Scholar]
- 54.Liu, M., Ramos-Castaneda, J., and Arvan, P. (2003) J. Biol. Chem. 278 14798-14805 [DOI] [PubMed] [Google Scholar]
- 55.Bullesbach, E. E., and Schwabe, C. (1994) J. Biol. Chem. 269 13124-13128 [PubMed] [Google Scholar]
- 56.Hua, Q. X., Liu, M., Hu, S. Q., Jia, W., Arvan, P., and Weiss, M. A. (2006) J. Biol. Chem. 281 24889-24899 [DOI] [PubMed] [Google Scholar]
- 57.Bullesbach, E. E., and Schwabe, C. (1991) J. Biol. Chem. 266 10754-10761 [PubMed] [Google Scholar]
- 58.Bullesbach, E. E., and Schwabe, C. (2005) J. Biol. Chem. 280 14051-14056 [DOI] [PubMed] [Google Scholar]
- 59.Bullesbach, E. E., and Schwabe, C. (1999) J. Biol. Chem. 274 22354-22358 [DOI] [PubMed] [Google Scholar]
- 60.Spanel-Borowski, K., Schafer, I., Zimmermann, S., Engel, W., and Adham, I. M. (2001) Mol. Reprod. Dev. 58 281-286 [DOI] [PubMed] [Google Scholar]