Abstract
Growth factor independent-1 (Gfi1) is a zinc finger protein with a SNAG-transcriptional repressor domain. Ajuba is a LIM domain protein that shuttles between the cytoplasm and the nucleus. Ajuba functions as a co-repressor for synthetic Gfi1 SNAG-repressor domain-containing constructs, but a role for Ajuba co-repression of the cognate DNA bound Gfi1 protein has not been defined. Co-immunoprecipitation of synthetic and endogenous proteins and co-elution with gel filtration suggest that an endogenous Ajuba·Gfi1·HDAC multiprotein complex is possible. Active histone deacetylase activity co-immunoprecipitates with Ajuba or Gfi1, and both proteins depend upon histone deacetylases for full transcriptional repression activity. Ajuba LIM domains directly bind to Gfi1, but the association is not SNAG domain-dependent. ChIP analysis and reciprocal knockdown experiments suggest that Ajuba selectively functions as a co-repressor for Gfi1 autoregulation. The data suggest that Ajuba is utilized as a corepressor selectively on Gfi1 target genes.
Group III LIM domain proteins, which have three to four tandem LIM domains at the C terminus, are involved in intracellular shuttling of interacting proteins. This suggests that these proteins may be involved in assembling multiple protein complexes with a wide range of functions such as cell development, differentiation, and signaling in different cellular compartments (for review see Refs. 1-3). The group III LIM protein family includes two subfamilies of proteins, which shuttle between the cytoplasm and the nucleus: Ajuba (Jub), LIMD1, WTIP, and Zyxin, LPP, and Trip6 (4-8). In the cytoplasm and at sites of cell adhesion, group-III LIM proteins function as adapter proteins in signal transduction (1-4, 9). For example, Ajuba directly interacts with Grb2 and stimulates Ras signaling (4). In the nucleus, group III LIM proteins are suggested to regulate transcription. Specifically, the Trip6 protein functions as a transcriptional co-activator for the REL oncoprotein (10). However, whereas nuclear Ajuba was shown to interact with the TTF1 transcription factor, the Ajuba and TTF1 interaction did not influence TTF1 transcriptional activity (11).
Growth Factor Independence-1 (Gfi1)4 is a 55-kDa nuclear transcriptional repressor protein that was identified as a target for proviral insertion of the Moloney murine leukemia virus (12). Gfi1 regulates genes involved in hemato-poietic stem cell maintenance, and myelopoiesis (reviewed in Ref. 13). Moreover, Gfi1 acts as a molecular switch in regulating granulopoiesis (14). Gfi1 target genes include GFI1, GFI1B (15-18), and CSF1 (14). Gfi1 contains six C2-H2 zinc fingers that bind to the core DNA sequence 5′-TAAATCAC(A/T)GCA-3′ (18). The N-terminal 20 amino acids of GFI1 encode a transferable repressor domain termed “SNAG,” because it is conserved between SNAIL and GFI1-related proteins (19). A yeast two-hybrid assay with the GFI1 SNAG domain identified Ajuba as an interacting protein (20). Synthetic constructs in which the Gfi1 SNAG domain was fused to a heterologous DNA binding domain suggested that Ajuba functions as a co-repressor for Gfi1 (20); however, analysis of the cognate DNA-bound Gfi1 protein was not performed, leaving open the question of Ajuba as a co-repressor for Gfi1. Here we show that Ajuba functions as an HDAC-dependent co-repressor for a subset of Gfi1 target genes. Specifically, Ajuba functionally mediates GFI1 autoregulation.
EXPERIMENTAL PROCEDURES
Cell Culture—EL4.IL-2 T cells (ATCC TIB-181) were grown in RPMI 1640 with 10% horse serum, 1% l-Gln, 1% Pen/Strep (Invitrogen). Jurkat T cells (clone E6-1, ATCC TIB-152) and human promyelocytic leukemia cells (HL60, ATCC CCL-240) were grown in RPMI 1640 with 10% fetal bovine serum, 1% l-Gln, 1% Pen/Strep (Invitrogen). Human embryonic kidney (HEK) 293T, and Phoenix cells were grown in Dulbecco's modified Eagle's medium with 10% fetal bovine serum, 1% l-Gln, 1% Pen/Strep (Invitrogen). All cell lines were kept at 37 °C with 5% CO2 in a humidified atmosphere.
Plasmids and Subcloning—Chloramphenicol acetyl transferase (CAT) reporter constructs and Gfi1 expression plasmids used in transient transcription assays were described previously (15, 19). Luciferase versions were generated by cloning the B30 × 2TK promoter and TK promoter fragments into the pGL3 vector (Promega, Madison, WI). MIEV-Gfi1-FLAG retroviral construct was generated by digesting CMV14-Gfi1-FLAG with XhoI to release the Gfi1-FLAG fragment. The Gfi1-FLAG insert was subcloned into SalI/XhoI-digested pMIEV. Plasmids were sequenced and screened by restriction endonucleases to corroborate the proper sequence and orientation of the insert. To generate the LexA-Ajuba expression vector and their mutants LexA-LIM and LexA-PreLIM we first PCR-amplified LexA with primers (5′-AGAATTCAACAGCCAGTCGCCGTTGCG-3′ and 5′-CCAAGCTTACCATGAAAGCGTTAACGGCC-3′) and subcloned it into the TOPO vector by blunt-end ligation generating the vector LexA-TOPO. Next, EcoRI/HindIII digestion released a LexA insert, which was agarose-purified and subcloned into EcoRI/HindIII-linearized pCS2 (4) to generate the plasmid pCS2-LexA. Finally, pCS2-Ajuba (4) was digested with EcoRI to release the cDNA for Ajuba, which was ligated into the EcoRI-linearized pCS2-LexA. The mutant pCS2-LexA-LIM was generated by the same strategy. To generate LexA-PreLIM, the vector pCS2-LexA-Ajuba was digested with StuI, and the agarose-purified vector band was self-ligated. Plasmids were sequenced and digested with restriction endonucleases to corroborate the correct orientation of the products.
Transient Transcription Assays—For transient transfections, 1.5 × 105 HEK 293T cells were plated on 24-well plates and transfected for 36 h with Lipofectamine 2000 reagent according to the manufacturer's directions (Invitrogen). Electroporation of EL4 cells was carried out with 20 μg of DNA, in 0.45-μm cuvettes (Bio-Rad), and 106 cells in 250 μl of media. Settings for pulses were 960 microfarads and 270 mV. Cells were cultured for 36 h before harvest. The chloramphenicol acetyltransferase assay was performed as previously described, utilizing a Gfi1-insensitive mutant of a cytomegalovirus-immediate-early promoter-driven β-galactosidase vector as a transfection efficiency control (19). Cellular extracts were adjusted for equivalent β-galactosidase activity, then analyzed by the scintillation method for CAT activity (19). Luciferase assay transfections were controlled by co-transfection of a Gfi1-insensitive Renilla luciferase vector (Promega). Firefly and Renilla luciferase activities were measured consecutively with Dual-Luciferase Reporter assay reagents (Promega) using Lumat LB 9507 (Berthold Technologies). A t statistic was calculated on the difference between the values of each measurement of CAT or Firefly luciferase activity to determine statistical significance for -fold repression. All assays shown were repeated at least three times with similar results unless other-wise stated.
Retroviral Transduction—Phoenix-Ampho cells were transiently transfected with retroviral constructs using the CaPO4 method, then co-cultured with Jurkat cells at 33 °C for 16 h. GFP+ cells were sorted 1 week after transduction, and single cell clones were obtained by limiting dilution. Clonal populations were analyzed for the presence of the FLAG-tagged proteins. Commercially available vectors were purchased for Gfi1 or Ajuba knockdown (Sigma). Lentiviral stocks were generated by three plasmid packaging in HEK 293 cells, concentrated by ultracentrifugation, and tittered on MEL cells. HL60 cells were transduced with non-targeting and Gfi1- or Ajuba-targeting shRNA viruses at an multiplicity of infection of 5. The transduced cells were cultured in RPMI media with 10% fetal bovine serum and puromycin (5 μg/ml).
Immunoblots and Immunoprecipitation—Nuclear extracts were prepared using a modified procedure of Dignam (21). Briefly, cells were rinsed twice in ice-cold phosphate-buffered saline (Invitrogen), trypsinized, and resuspended in buffer A (20 mm Tris-HCl, pH 7.4, 1.5 mm MgCl2, 10 mm KCl, 1 mm Complete inhibitor (Roche Applied Science), and 1 mm phenylmethylsulfonyl fluoride). After incubating 10 min on ice, cells were transferred into a glass Dounce homogenizer and processed for 30-40 strokes with pestle A. Nuclei were pelleted by centrifugation at 4 °C at 13,000 rpm for 10 min. Cytoplasmic extract was transferred to a clean tube and snap-frozen. The nuclei were resuspended in buffer C (20 mm Tris-HCl, pH 7.0, 25% glycerol, 0.42 m NaCl, 1.5 mm MgCl2, 0.2 mm EDTA, 1 mm Complete inhibitor (Roche Applied Science), and 1 mm phenylmethylsulfonyl fluoride), transferred into a glass Dounce homogenizer. Nuclear extract was prepared by processing with pestle B until the pellet was totally dispersed, then centrifuged for 10-min centrifugation at 13,000 rpm at 4 °C to pellet nuclear debris. For knockdown experiments, HL60 cell lysate was prepared using CompleteM lysis buffer (Roche Applied Science).
Protein concentrations were determined by BCA assay (Pierce), and 20 μg of protein extract was separated on 10% SDS-polyacrylamide gel and electroblotted onto an Immobilon-P membrane (Millipore). The membranes were blocked with 5% Carnation instant milk in TBS (50 mm Tris-HCl, 150 mm NaCl, pH 7.5) at 4 °C for 16 h. The blocked membranes were incubated with primary antibodies in 5% milk in TTBS (0.05% Tween 20 in TBS) between 1 and 4 h at room temperature. Gfi1 was detected using goat polyclonal N-20 (1:500, Santa Cruz Biotechnology) and mouse monoclonal (2.5D17) (22) antibody. Rabbit polyclonal antisera against the histone deacetylases HDAC1 (Upstate Biotechnologies), HDAC2 (H-54, Santa Cruz Biotechnology), HDAC3 (Upstate Biotechnologies), Ajuba (Cell Signaling), and a FLAG-horseradish peroxidase (HRP)-conjugated antibody (Sigma) were utilized according to the manufacturer's directions. The secondary antibodies were donkey anti-goat HRP-conjugated IgG (1:5000, Santa Cruz Biotechnology), Sheep anti-mouse Ig-HRP (1:2500) and donkey anti-rabbit Ig-HRP (1:5000, Amersham Biosciences). Secondary antibodies were prepared in 5% milk TTBS and incubated with blots for 1 h at room temperature. The detection was performed with ECL-plus detection reagent according to manufacturer's instructions (Amersham Biosciences).
For each immunoprecipitation, 200 μg of nuclear extracts was diluted to 150 mm NaCl and 5% glycerol. Extracts were precleared with a mix of protein A/G-agarose beads (Invitrogen) for 1 h under gentle rocking, then 15 μl of antisera was added, and the immunocomplexes were allowed to form for 1 h at 4 °C. To capture the immunocomplexes, 20 μl of protein A/G-agarose beads were added for 16 h at 4 °C. Beads were pelleted at 2000 rpm for 5 min and washed five times with 20 mm Tris-HCl, pH 7.0, 5% glycerol, 150 mm NaCl, 1.5 mm MgCl2, 0.2 mm EDTA, 0.5% Nonidet P-40. Samples were resuspended in 20 μl of 1× loading buffer, boiled for 10 min, and subjected to immunoblot analysis. Isotype-matched-control immunoglobulin was used as a control for the immunoprecipitations. FLAG and Myc-specific immunoprecipitations were carried out with M2 affinity resin (Sigma) and Myc-agarose beads (Santa Cruz Biotechnology), respectively, following the manufacturer's directions.
Size Exclusion Chromatography—Nuclear extracts from 3 to 5 billion Jurkat cells were applied to a Sephacryl S-300 column on an AKTA10 purifier (Amersham Biosciences). The column was run using Dignam buffer C with 150 mm NaCl at a flow rate of 0.5 ml/min. Fractions (1.0 ml) were collected, and 80 μl of every fifth fraction was used for immunoblot analysis.
HDAC Assays—Nuclear extracts from Jurkat cells transduced with Gfi1-FLAG or FLAG-Ajuba expressing or empty MSCV retroviral vectors were used for immunoprecipitation with the M2 affinity resin (Sigma) as described above. The beads were mixed with 200 μl of 50 μm HDAC assay substrate (Fluor-de-Lys HDAC assay, BioMol) and incubated for 1.5 h at room temperature. For each data point triplicate immunoprecipitations without and with TSA were analyzed. Aliquots were taken at 0.5 and 1.5 h, and the change in HDAC activity was determined according to the manufacturer's directions. TSA-sensitive relative-fluorescent units were generated by subtracting values for triplicate immunoprecipitations treated with TSA from values for triplicate immunoprecipitations treated with vehicle control.
Chromatin Immunoprecipitation—The ChIP assay was performed as previously described (14). Briefly, logarithmically growing HL60 cells (1 × 108 cells) were cross-linked using formaldehyde (final concentration, 1% v/v) in RPMI 1640 medium for 10 min on ice. Glycine was added to a final concentration of 0.125 m to stop cross-linking. Fixed cells were pelleted by centrifugation and sequentially washed three times with ice-cold phosphate-buffered saline with 1× Complete inhibitor (Roche Applied Science). The cells were then resuspended in lysis buffer (1% SDS, 50 mm Tris-HCl, pH 8.1, 10 mm EDTA, 1× Complete inhibitor) and were sonicated to make soluble chromatin using Sonicator 3000 cup horn (Misonix). An aliquot of total chromatin was taken at this point to use as a positive control in the PCRs (input chromatin). The cell lysates were precleared by incubation with protein A/G-Sepharose beads (Santa Cruz Biotechnology) and then incubated with Gfi1 monoclonal antibody (2.5D17) (22), Ajuba polyclonal antibody (4897) (Cell Signaling), and control mouse (GE Healthcare) or rabbit (sc-2027, Santa Cruz Biotechnology) IgG overnight at 4 °C. DNA·protein complexes were collected with protein A/G-Sepharose beads followed by several rounds of washing. Bound DNA·protein complexes were eluted from the antibodies with two incubations in elution buffer (100 mm NaHCO3, 1% SDS) at room temperature for 15 min. Cross-links were reversed by addition of sodium chloride followed by incubation at 65 °C overnight. RNase A and proteinase K were sequentially added and incubated for 1 h at 37 °C. DNA fragments were purified using a QIAquick PCR purification kit (Qiagen) and used for PCR amplifications. The PCR products were fractionated on 2% agarose gels, stained with ethidium bromide. PCR primer pairs for the ChIP were: GFI1 (5′-CACACCTTCATCCACACAGG-3′ and 5′-GATGAGCTTTGCACACTGGA-3′); GFI1B (5′-GGGCGATGCATTCATTTCC-3′ and 5′-CACCTCGATTTTGGATTTCTAG-3′); CSF1 (5′-GGGCCTCTGGGGTGTAGTAT-3′ and 5′-CCGAGGCAAACTTTCACTTT-3′). β-ACTIN (5′-AGCGCGGCTACAGCTTCA-3′ and 5′-CGTAGCACAGCTTCTCCTTAATGTC-3′) was used as the negative control. Each experiment was performed at least twice with similar results, and representative data are shown.
Quantitative PCR—TRIzol (Invitrogen) extracted RNA from shRNA vector-transduced cells was quantified, and equal amounts of RNA were utilized in first strand cDNA synthesis reaction and then applied to TaqMan probe sets for GFI1B (Hs00180261_m1), GFI1 (Hs00382207_m1), and CSF1 (Hs00174164_m1) according to the method of the manufacturer (ABI).
RESULTS
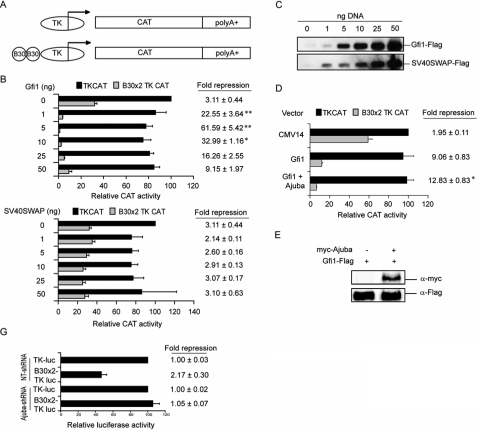
Gfi1 Mediates Transcriptional Repression through Titratable-associated Factors—Gfi1 transcriptional repression may require limiting and titratable-associated factors (14, 19). 293T cells are human epithelial kidney cells that express a low level of Gfi1 (15). 293T cells were co-transfected with reporter constructs with or without “B30” (18) high affinity Gfi1 binding sites (Fig. 1A), and increasing amounts of expression vectors encoding Gfi1 or a Gfi1·SNAG repressor domain mutant in which all 20 amino acids of the SNAG domain have been replaced with the SV40 nuclear localization motif (SV40SWAP) (19). Whereas low levels of the Gfi1 expression construct induced a significant increase in repression, expression construct levels in excess of 5 ng decreased repression in a dose-dependent manner (Fig. 1B, upper panel). Immunoblot analysis confirmed increased protein levels (Fig. 1C). Notably, a titration of the Gfi1 SV40SWAP expression vector resulted in increased protein levels (Fig. 1C) but did not lead to significant repression at any level (Fig. 1B, lower panel). Thus, excess Gfi1 may disrupt functional Gfi1 transcription complexes, perhaps by sequestering limiting proteins required for an active Gfi1·SNAG repressor complex.
FIGURE 1.
Ajuba increases Gfi1 transcriptional repression. A, map of the reporter constructs employed in transient transcription experiments. TK, herpes simplex virus thymidine kinase minimal promoter; CAT, chloramphenicol acetyltransferase; poly(A)+, polyadenylation sequences; B30, synthetic Gfi1 binding site (18). B, transient transcription assay in 293T cells with reporters in A, and titration of expression vectors encoding FLAG-epitope-tagged Gfi1 (top panel) or the SV40SWAP (Gfi1·SNAG domain) mutant (bottom panel). Data are expressed as -fold repression (values from TKCAT/values from B30TKCAT) ± S.E. Statistical analysis compared the -fold repression of samples to that of the preceding sample. C, immunoblot analysis of whole cell lysates corresponding to B with an anti-FLAG monoclonal antibody. D, transient transcription assay in 293T cells co-transfected with reporters and expression vectors encoding Gfi1 and Ajuba. The values for CAT activity were corrected for transfection efficiency, and the data are expressed as relative CAT activity with -fold repression. E, transient transcription assay in 293T cells co-transfected with the indicated constructs. F, immunoblot analysis of whole cell lysate corresponding to E with anti-FLAG and anti-Myc-monoclonal antibodies. G, transient transcription assay in 293T cells first transduced with non-targeting or Ajuba-targeting shRNA, then transfected with a control Renilla luciferase reporter, and TK- or B30 × 2-Firefly-luciferase reporter vectors. The values for Firefly luciferase activity were corrected for Renilla activity (for transfection efficiency). Data are expressed as relative Firefly luciferase activity and -fold repression. *, p < 0.05; **, p < 0.001.
The LIM Domain Protein Ajuba Increases Gfi1 Transcriptional Repression—Ajuba is a LIM domain protein that shuttles between the cytoplasm and the nucleus (6). In the nucleus, Ajuba interacted with a synthetic Gfi1·SNAG domain-containing construct to function as a corepressor (20). To determine if Ajuba affects cognate DNA-bound Gfi1 transcriptional repression functions, we performed transient transcription assays with Gfi1-responsive reporter constructs (Fig. 1A) and expression constructs encoding Gfi1 and Ajuba. Co-expression of Ajuba and Gfi1 significantly increased Gfi1-mediated repression when compared with the repression induced by GFI1 alone (Fig. 1D). Notably, the activity of the TK-CAT reporter, which lacks Gfi1 binding sites, was not altered by the presence of either Gfi1, or co-expression of Gfi1 and Ajuba (Fig. 1D). Thus, Ajuba effects on transcription repression in this assay are dependent upon the presence of Gfi1 DNA binding sites. Moreover, the level of Gfi1 protein was not affected by the overexpression of Ajuba (Fig. 1F).
In agreement with the low level of Gfi1 in 293T cells (15), transfected B30 × 2 TK reporters demonstrate modest endogenous Gfi1 repression activity (Fig. 1, B, C, and G). When Ajuba levels were decreased by shRNA knockdown, the activity of the Gfi1-responsive vector was equivalent to that of the non-responsive control (Fig. 1G). Thus, repression mediated by endogenous Gfi1 upon the B30 × 2 TK reporter is completely abrogated by shRNA against Ajuba, and endogenous Gfi1 requires endogenous Ajuba to mediate repression of the B30 × 2 TK reporter in 293T cells. These results strongly suggest that Ajuba acts as a co-repressor for cognate Gfi1 and validate the 293T system to determine the mechanism.
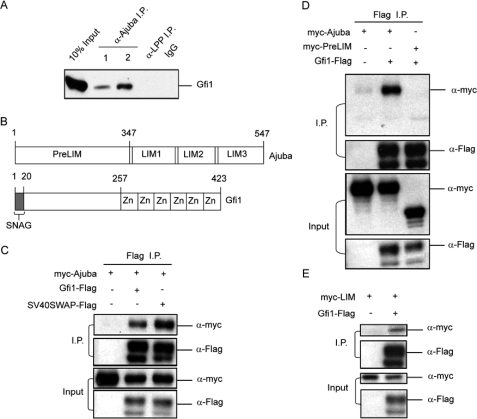
Gfi1 Associates with Ajuba—To further study the in vivo association of endogenous proteins, we performed immunoprecipitation from Jurkat cell line nuclear extracts. We employed two different antisera against Ajuba, an antisera against LPP (a related LIM domain protein), and isotype-matched IgG as controls. Immunoprecipitants were analyzed by immunoblot for the presence of Gfi1 (Fig. 2A). Notably, endogenous Gfi1 protein was co-immunoprecipitated with antiserum against Ajuba, but not LPP or the isotype IgG (Fig. 2A). The amount of Gfi1 that co-immunoprecipitated with Ajuba was <10% of the input. Thus, endogenous Ajuba and Gfi1 form a complex in Jurkat nuclear extracts.
FIGURE 2.
Ajuba associates with Gfi1. A, immunoprecipitation of Ajuba from Jurkat nuclear extracts with two different antisera against Ajuba, control antisera against LPP, or normal rabbit antisera, followed by immunoblot analysis to detect Gfi1. B, schematic of Ajuba and Gfi1 domains. C, immunoprecipitation of FLAG-epitope-tagged Gfi1 or the SV40SWAP mutant, followed by immunoblot analysis for myc-epitope-tagged Ajuba (I.P., upper panels). The blots were stripped and reprobed for FLAG to demonstrate the immunoprecipitated FLAG-tagged proteins (I.P., lower panel). Immunoblot analysis of epitope-tagged proteins from 10% of input whole cell extracts of 293T cells co-transfected with expression constructs encoding myc-Ajuba (Input, upper panel) and either Gfi1-FLAG or SV40SWAP-FLAG constructs (Input, lower panel). D, immunoprecipitation of FLAG-epitope-tagged Gfi1, followed by immunoblot analysis for myc-epitope-tagged Ajuba or the N-terminal pre-LIM region of Ajuba (upper panels). Immunoblot analysis of epitope-tagged proteins from 10% of input whole cell extracts of 293T cells co-transfected with expression constructs encoding Gfi1-FLAG and either myc-Ajuba or myc-Ajuba-preLIM (lower panels). E, immunoprecipitation of FLAG-epitope-tagged Gfi1, followed by immunoblot analysis for myc-epitope-tagged C-terminal LIM domains of Ajuba (upper panels). Immunoblot analysis of epitope-tagged proteins from 10% of input whole cell extracts of 293T cells co-transfected with expression constructs (lower panels).
Next, we broadly defined the protein domains (Fig. 2B) of interaction between Ajuba and Gfi1 by immunoprecipitating FLAG-epitope-tagged Gfi1 (or the SV40SWAP mutant), then analyzing by immunoblot for the presence of Myc-epitope-tagged Ajuba (or Ajuba mutants). Both Gfi1 and the SV40SWAP mutant co-immunoprecipitated with Ajuba (Fig. 2C). Unlike full-length Ajuba, the N-terminal Ajuba-PreLIM region did not co-immunoprecipitate with Gfi1 (Fig. 2D). However, the C-terminal Ajuba-LIM region co-immunoprecipitated with Gfi1 (Fig. 2E). Therefore, Ajuba associates with Gfi1 through the Ajuba LIM region, but this interaction is not dependent upon the Gfi1 SNAG domain.
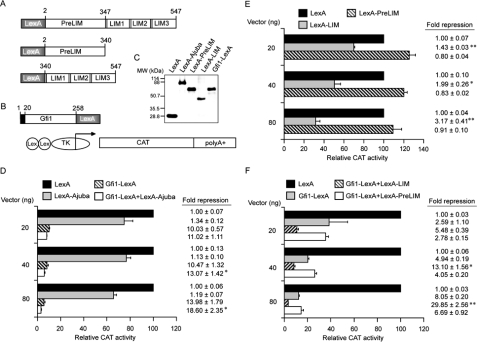
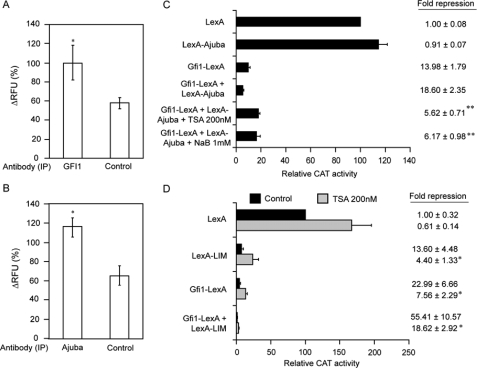
The LIM Domain of Ajuba Functions as a Corepressor—To confirm a co-repressor function for Ajuba, we performed transient transcription assays with chimeric proteins in which the N terminus of the bacterial DNA-binding protein LexA is fused to Ajuba, Ajuba mutants (Fig. 3A), or the N terminus of Gfi1 (Fig. 3B). Immunoblot analysis revealed that all the LexA fusion proteins were synthesized in 293T cells at the expected molecular weight (Fig. 3C). We previously demonstrated that the N terminus of Gfi1 transfers active transcriptional repression to LexA (19). Indeed, co-transfection of a TK-CAT reporter containing two Lex operons (Fig. 3B) and an expression vector encoding Gfi1-LexA resulted in transcriptional repression (Fig. 3D). In contrast, co-transfection of LexA-Ajuba with the reporter did not yield significant repression (Fig. 3D). However, co-transfection of LexA-Ajuba with Gfi1-LexA induced significant, additive, and dose-dependent repression of the reporter (Fig. 3D). These data could either indicate that Ajuba is not a co-repressor, or that the co-repressor function of Ajuba may only be revealed upon binding to a nuclear target.
FIGURE 3.
Ajuba LIM domains function as a co-repressor that synergizes with Gfi1. A, map of the Ajuba and Gfi1 expression and reporter constructs (B) employed in transient transcription experiments. “Lex” = LexA DNA binding site. C, immunoblot analysis with LexA-specific antisera on lysates from 293T cells transfected with expression constructs encoding LexA, or LexA fused to full-length Ajuba, the Ajuba-pre-LIM, Ajuba-LIM domains or the Gfi1 N terminus. D, transient transcription assay in 293T cells with the reporter and expression constructs in A and B encoding LexA, LexA-Ajuba, and Gfi1-N terminus-LexA fusions. Vector (ng) refers to the quantity of each expression vector plasmid transfected. Note that where only single expression vectors are indicated, the total amount of expression vector transfected has been controlled by the addition of equivalent amounts of empty vector control DNA. E, transient transcription assay in 293T cells with expression constructs encoding LexA, or LexA-Ajuba-LIM and LexA-Ajuba-pre-LIM region fusions. Vector (ng) as in D. F, transient transcription assay in 293T cells with expression constructs encoding LexA, Gfi1-LexA, LexA-Ajuba-LIM, and LexA-Ajuba-pre-LIM region fusions. The -fold repression of LexA-Gfi1 with LexA-Ajuba-LIM is compared with that of Gfi1-LexA with LexA control, or Gfi1-LexA with LexA-Ajuba-preLIM. Vector (ng) as in D. *, p < 0.05; **, p < 0.001.
To explore these possibilities we tested the function of LexA fusion to the Ajuba preLIM or LIM domains. Like LexA-Ajuba, LexA-PreLIM did not repress the activity of the reporter (Fig. 3E). In contrast, LexA-LIM induced significant and dose-dependent repression of the reporter (Fig. 3E). We reasoned that if the transcriptional activity of full-length Ajuba is hindered by intramolecular interactions, then LexA-LIM (which is only a portion of the protein) might be unhindered and potently synergize with Gfi1-LexA. In fact, LexA-LIM and Gfi1-LexA demonstrated significant dose-dependent and synergistic transcriptional repression (Fig. 3F). In contrast, LexA-PreLIM and Gfi1-LexA together resulted in reporter activity similar to Gfi1-LexA alone (Fig. 3F). Utilizing B30-TK-CAT reporter vectors and Gfi1 and Ajuba protein expression vectors (without LexA fusion), we find that the Pre-LIM domain does not increase Gfi1 repression. However, expression constructs encoding full-length Ajuba or isolated LIM domains increased Gfi1 repression (supplemental Fig. S1). Thus, the Ajuba LIM domains function as a Gfi1 co-repressor.
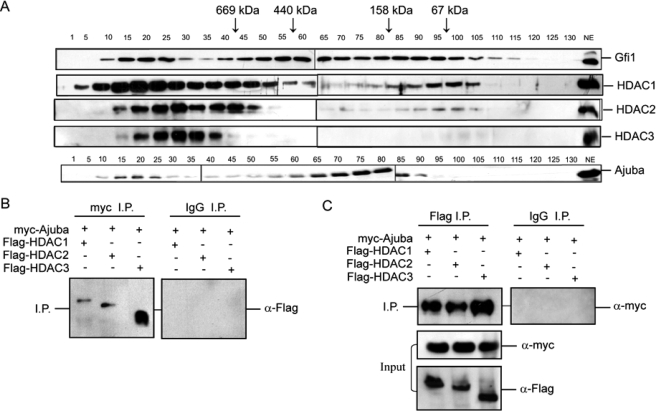
Gfi1 and Ajuba Interact Directly with HDACs—We next determined whether Gfi1 and Ajuba are in a multiprotein complex using gel-filtration chromatography. Gel filtration separates larger and smaller proteins in a complex mixture. Immunoblot analysis of nuclear extracts fractionated on a Sephacryl S-300 column revealed elution profiles. HDAC1, HDAC2, and HDAC3 are part of protein complexes that elute at large molecular size fractions (24-28). Indeed, HDAC1, HDAC2, and HDAC3 profiles corresponding to the expected high molecular weight complexes confirmed the validity of our gel-filtration technique (Fig. 4A). Gfi1 eluted in two major peaks; the first was larger than 800 kDa, and the other ran between the size of a Gfi1 monomer to the 669-kDa marker (Fig. 4A). The pattern of Gfi1 elution was similar in multiple runs and with different nuclear extract preps from both lymphoid and myeloid cell lines; however, the major (smaller) peak often ranged from approximately 200 to 600 kDa (data not shown). Thus, the monomers (Fig. 4A) may represent disrupted complexes. Notably, the larger Gfi1 complex was coincident with that of the HDAC proteins (Fig. 4A). Similar to Gfi1, a less abundant fraction of Ajuba eluted between 800 and 1.5 MDa, coincident with HDAC1, HDAC2, and HDAC3 (Fig. 4A). However, the bulk of Ajuba eluted between 150 and 440 kDa, coincident with part of the smaller Gfi1 complex (Fig. 4A). To confirm their interaction, Myc-tagged Ajuba and FLAG-tagged HDAC1, HDAC2, and HDAC3 proteins were co-transfected. Similar to our published data on Gfi1 (29), co-immunoprecipitates suggest that Ajuba is found in a complex with HDAC1, HDAC2, or HDAC3. Ajuba co-immunoprecipitates HDAC1, HDAC2, and HDAC3 (Fig. 4B) and immunoprecipitates of HDAC1, HDAC2, or HDAC3 contain Ajuba (Fig. 4C). These results illustrate the possibility that Gfi1, Ajuba, and HDACs form a complex; albeit a smaller proportion of the total Gfi1 and nuclear Ajuba protein.
FIGURE 4.
GFI1 and Ajuba associate with histone deacetylase enzymes. A, immunoblot analysis of Sephacryl S-300 gel-filtration fractions from Jurkat T cell nuclear extracts, with antisera against Gfi1, HDAC1, -2, or -3 and Ajuba. Elution peaks for calibration controls are indicated by arrows along with their molecular weight. B, co-immunoprecipitation of FLAG-epitope tagged HDAC1, -2, or -3 with antisera specific to a myc-epitope-tagged Ajuba, but not control IgG (top). C, co-immunoprecipitation of myc-epitope-tagged Ajuba with antisera specific to FLAG-epitope tagged HDAC1, -2, or -3 (top left panel), but not control IgG (top right panel). Immunoblot analysis reveals levels of 10% of input proteins (bottom panels).
Gfi1 and Ajuba Mediate Transcriptional Repression through HDACs—Histone deacetylase proteins may be part of the repression complex formed by overexpressed Gfi1 (29). Given the co-elution of a large Gfi1 complex with HDACs 1, 2, and 3 (Fig. 4A), and co-immunoprecipitation (Fig. 4, B and C), we next determined whether a histone deacetylase activity co-immunoprecipitates with Gfi1. Lymphoid cell line clones stably expressing low levels of Gfi1-FLAG were constructed. Gfi1 was immunoprecipitated from nuclear extracts with an anti-FLAG monoclonal antibody, then analyzed by a fluorescent histone-deacetylase assay. A murine isotype-matched immunoglobulin served as an immunoprecipitation control. Immunoprecipitants from Gfi1-FLAG-expressing (but not empty vector-transduced) cells had significantly higher TSA-sensitive histone deacetylase activity than that of the control (Fig. 5A). Thus, Gfi1 associates with endogenous histone deacetylase enzymes.
FIGURE 5.
HDAC activity is required for Gfi1 and Ajuba-mediated transcriptional repression. A, fluorescent histone-deacetylase enzymatic analysis of FLAG-monoclonal antibody or isotype-matched IgG-control immunoprecipitants from a Jurkat T cell clone stably expressing low levels of Gfi1-FLAG. TSA-sensitive relative-fluorescent units (values for triplicate immunoprecipitations treated with TSA subtracted from values for triplicate immunoprecipitations treated with vehicle control) ± absolute error are displayed with values from Gfi1-FLAG samples arbitrarily set to 100. B, fluorescent histone-deacetylase enzymatic analysis of FLAG monoclonal antibody or isotype-matched IgG-control immunoprecipitants from Jurkat T cells stably expressing Ajuba-FLAG. TSA-sensitive relative-fluorescent units (values calculated as in A) ± absolute error are displayed with values from Ajuba-FLAG samples arbitrarily set to 100. C, transient transcription assay in 293T cells transfected with the reporter and expression constructs in Fig. 3 (A and B) encoding LexA, LexA-Gfi1, or LexA-Ajuba, and treated with either TSA, sodium butyrate, or vehicle control. The relative CAT activity and -fold repression of TSA- or sodium butyrate-treated Gfi1-LexA with LexA-Ajuba are compared with those of vehicle-treated GFI1-LexA with LexA-Ajuba. D, transient transcription assay in 293T cells transfected with expression constructs encoding LexA, Gfi1-LexA, or LexA-Ajuba-LIM domains, and treated with TSA or vehicle control. The relative CAT activity and -fold repression of TSA-treated LexA-LIM, Gfi1-LexA, or Gfi1-LexA with LexA-LIM is compared with the corresponding samples treated with vehicle control. *, p < 0.05; **, p < 0.001.
Given that Ajuba co-elutes with HDAC1, HDAC2, or HDAC3 (Fig. 4A), and co-immunoprecipitates with the same proteins (Fig. 4B and 4C), we next sought to determine whether Ajuba mediates transcriptional repression through HDACs. First, a lymphoid cell line stably expressing low levels of FLAG-epitope-tagged Ajuba was constructed. Ajuba was immunoprecipitated and analyzed by a fluorescent histone deacetylase assay as in Fig. 5A. Immunoprecipitants from FLAG-Ajuba (but not empty vector-transduced) cells had significantly higher TSA-sensitive histone deacetylase activity than that of the control (Fig. 5B). Thus, Ajuba associates with endogenous histone deacetylase enzymes.
To determine if endogenous Gfi1 requires histone deacetylase activity to actively repress transcription, reporter constructs (Fig. 1A) were electroporated into a lymphoid cell line, then dosed with TSA for 16 h. The repression activity of Gfi1 significantly declined 1.3 units per nanomolar of TSA (supplemental Fig. S2A). TSA both inhibits histone deacetylase enzymatic activity and leads to protein accumulation. Specifically, HDAC1 levels increase upon TSA treatment (30). However, whereas 10 nm TSA increased the levels of Gfi1 and HDAC1, neither Gfi1 nor HDAC1 showed a further dose-dependent accumulation (supplemental Fig. S2B). Thus, the decrease in Gfi1 repression is likely to be due to inhibition of histone deacetylase enzymes as opposed to Gfi1 protein accumulation to levels unable to form a transcription complex.
To eliminate the possibility that TSA inhibited Gfi1 repression through a mechanism independent of histone deacetylase inhibition, we performed transient transcription assays with a different histone deacetylase inhibitor, sodium butyrate (NaB). 293T cells were co-transfected with Gfi1 reporter constructs (Fig. 1A), and expression vectors encoding FLAG-epitope-tagged Gfi1 (Gfi1-FLAG) or SV40SWAP. As expected, SV40SWAP mutant transcriptional repression is significantly impaired in comparison to Gfi1 (supplemental Fig. S2C). Similar to mutation of the SNAG domain, treatment with either 200 μm TSA or 1 mm NaB significantly inhibited Gfi1 repression. Immunoblot analysis of the extracts from the treated cells revealed TSA-induced accumulation of Gfi1; however, NaB-treated cells had Gfi1 expression levels similar to controls (supplemental Fig. S2D). Thus, the inhibition of Gfi1 repression is most likely due to drug inhibition of HDACs and not changes in protein levels. These data indicate that most of the repressor activity mediated by Gfi1 in 293T cells is dependent on histone deacetylases and suggest that Gfi1-mediated transcriptional repression requires the recruitment of HDACs in vivo.
Next, we performed transient transcription assays in 293T cells co-transfected with expression vectors encoding LexA-Ajuba and Gfi1-LexA chimeric proteins and dosed them with TSA and NaB. As expected, the presence of LexA-Ajuba increased Gfi1-LexA-mediated repression (Fig. 5C). Importantly, the addition of either TSA or NaB significantly impaired repression of the reporter (Fig. 5C). Thus, Ajuba-modulated Gfi1 repression is still dependent on histone deacetylases.
We next determined whether the corepressor function of the Ajuba LIM domains, and their effects on Gfi1, is sensitive to HDAC inhibitors. Similar to full-length Gfi1 (supplemental Fig. S2C), we note that the addition of TSA significantly impairs repression by the Gfi1-LexA fusion protein (Fig. 5C). Likewise, repression by LexA-LIM was significantly inhibited by TSA (Fig. 5D). When Gfi1-LexA and LexA-LIM are co-transfected, the proteins repressed transcription in a cooperative way, and this repression was also sensitive to TSA (Fig. 5D). However, we note that the addition of TSA failed to completely inhibit repression, and that this was exacerbated in cells expressing both Gfi1-LexA and LexA-LIM. Therefore, both HDAC-dependent and -independent mechanisms are integrated into Gfi1 and Ajuba transcriptional repression.
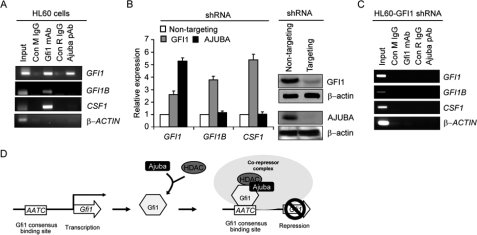
Ajuba Controls Gfi1 Autoregulation—Co-repressor proteins associate with DNA-bound transcription factors to mediate transcriptional repression of specific target genes. To determine if Ajuba and Gfi1 are bound to similar target genes in vivo, we first determined whether Ajuba is bound to Gfi1 target genes in a human myeloid cell line. Gfi1 target genes GFI1B (16, 17), GFI1 (15, 17), and CSF1 (14) were analyzed by chromatin immunoprecipitation (ChIP) analysis for the presence of both Gfi1 and Ajuba. The β-ACTIN gene was included as control, because it was not bound by Gfi1 in similar ChIP analyses (14). Notably, Gfi1 and Ajuba bound only to a subset of Gfi1 target genes (Fig. 6A). The specificity of this result is highlighted by the fact that β-ACTIN was not bound by either protein (Fig. 6A). Moreover, shRNA-mediated Gfi1 knockdown eliminated Gfi1 as well as Ajuba binding to GFI1 (Fig. 6C).
FIGURE 6.
Ajuba acts a Gfi1 co-repressor in living cells. A, ChIP analysis in HL60 cells with a Gfi1-specific monoclonal antibody (Gfi1 mAb), Ajuba-specific rabbit antisera (Ajuba pAb), or isotype IgG controls (Con M IgG and Con R IgG, respectively), and primers specific for Gfi1 target genes GFI1, GFI1B, and CSF1. β-ACTIN is used as the negative control. B, TaqMan analyses on HL60 cells transduced with three independent lentiviral shRNA specific for GFI1 or AJUBA or a non-targeting control. Representative immunoblot demonstrates lower levels of GFI1 and AJUBA with one of three independent shRNA constructs. C, schematic representation of GFI1 autoregulation by GFI1, AJUBA, and HDACs.
Next, we utilized shRNA-expressing lentiviral vectors to knock down endogenous GFI1 or AJUBA. The knockdown was confirmed by immunoblot analysis (Fig. 6B, inset). RNA from the same experimental samples served as template for TaqMan probes monitoring GFI1B, GFI1, and CSF1 steady-state mRNA levels. Notably, the GFI1 TaqMan probe targets the GFI1 mRNA exon 4-5 junction, which is upstream of the GFI1 shRNA 3′-untranslated region target. Thus, we reasoned that the TaqMan probe should report on initial GFI1 transcription, even if shRNA targeted the message for transcriptional or translational blockade, as has previously been demonstrated (31). In fact, down-regulation of GFI1 corresponded to deregulation of GFI1B, GFI1, and CSF1 message levels (Fig. 6B). Consistent with the presence of AJUBA only on GFI1, knockdown of AJUBA deregulated only GFI1 message levels (Fig. 6B). Similar results were seen in a different myeloid cell line (supplemental Fig. S3). We conclude that AJUBA functionally binds with GFI1 on the GFI1 promoter in living cells to mediate autoregulation (Fig. 6C).
DISCUSSION
LIM domain proteins play an important role in cellular differentiation, proliferation, and motility by transmitting signals from the cytoplasm to the nucleus. LIM proteins are involved in gene regulation through direct interaction with known transcription factors (32-34). Several lines of evidence support the existence of an Ajuba·Gfi1·HDAC complex. First, gel-filtration chromatography and co-immunoprecipitation indicate that both endogenous and overexpressed Ajuba bind to Gfi1 through Ajuba LIM domains. Second, similar to Gfi1 (29), Ajuba associated with HDACs. Gel-filtration chromatography revealed that Ajuba and Gfi1 co-elute with HDAC1, HDAC2, and HDAC3. Analysis of chromatin marks engendered by a synthetic Gfi1·SNAG domain fusion protein suggested that inducible DNA binding of this protein was coincident with deacetylation of histone H3 and H4 (20). Our co-immunoprecipitation analyses showed that Gfi1 (29) and Ajuba bind to HDAC1, -2, and -3 and that Gfi1 and Ajuba co-immunoprecipitate HDAC enzymatic activity. Moreover, the addition of drug inhibitors of HDAC activity impaired both Gfi1 repression and the synergy mediated by Ajuba LIM domains. We previously demonstrated that GFI1 and GFI1B transcriptionally regulate GFI1 (15) and now demonstrate that Ajuba functions as a corepressor for Gfi1 autoregulation. HDACs are found in the nucleus and cytoplasm. It is possible that Ajuba tethers HDACs in the cytoplasm, bringing them to the nucleus to stabilize the Gfi1-HDAC transcription complex and to facilitate Gfi1 autoregulation.
In sum, our data suggest the existence of a Gfi1-Ajuba-HDAC transcriptional complex; however, given our data on immunoprecipitation and gel filtration of endogenous proteins it is unlikely that this is the most prevalent Gfi1 protein complex. Clearly, our study shows that the majority of Gfi1 repression activity is dependent upon HDAC function. However, a residual repression activity is present in inhibitor-treated samples suggesting Gfi1 and Ajuba utilize as-yet unidentified non-HDAC-dependent repression mechanism(s).
We propose that the co-repressor function of Ajuba is not active until Ajuba binds a nuclear target. Specifically, pulldown assays reveal that Gfi1 co-immunoprecipitates with LIM domains but not with the Ajuba PreLIM region. Tethering full-length Ajuba alone to DNA did not provide significant transcriptional repression, whereas LexA-Ajuba increased the repression of Gfi1-LexA. Ajuba PreLIM region and LIM domains often interact with distinct proteins that contribute to a common cellular function (7, 35). Notably, the LIM domains, when trapped in the nucleus, affect cell proliferation (LIM1 or -2) and differentiation (LIM3), whereas the PreLIM region affects cell proliferation (6). The Ajuba PreLIM region interacts with the cytoplasmic signaling protein GRB2 (4), whereas the LIM domains interact with Aurora-A kinase (5), α-catenin (7), Snail (36), and now Gfi1. Because different individual LIM domains within a LIM region can interact with different targets, it is possible that distinct LIM domains of Ajuba interact with Gfi1 and HDACs. Transcriptional analysis also shows that the LexA fusion of Ajuba LIM (but not pre-LIM) domains were able to induce dramatic and synergistic transcriptional repression with Gfi1-LexA. In fact, LexA-PreLIM mildly interfered with Gfi1-LexA repressor activity. Langer et al. (36) suggested that the importance of the PreLIM region is in recruiting chromatin remodeling proteins; in contrast, our study shows that the LIM domains are sufficient for repression.
Ajuba has been shown to bind the Gfi1 SNAG (Snail + Gfi1) domain in a yeast two-hybrid assay (20). Ajuba also serves as a SNAG domain binding co-repressor for the Snail and Slug zinc finger proteins, which mediate repression of E-cadherin (36, 37). The SNAG domain mutant of Snail showed severe reduction in binding with Ajuba (36). In contrast to this, our study shows that Ajuba interaction with Gfi1 is not dependent upon the SNAG domain. Thus, unlike Snail and Slug, Ajuba interactions with Gfi1 are unlikely to be limited to the SNAG domain.
Severe congenital neutropenia (SCN) is characterized by a lack of neutrophils and subsequent recurrent bacterial and fungal infections (38). SCN is most commonly associated with mutations in elastase 2, neutrophil (ELA2), but mutations in GFI1, HAX1, and WAS have also been reported (39-41). Recently, we demonstrated that SCN-associated mutations in GFI1 generate dominant-negative-acting proteins (GFI1N382S), which selectively deprepress GFI1 target genes such as CSF1 (14). A SNAG domain mutant protein (Gfi1P2A (19)) also functioned in a dominant negative manner; however, a protein with both mutations (Gfi1P2A+N382S) lacked dominant negative activity. Thus, we hypothesized that Gfi1N382S sequesters limiting SNAG domain-associated factors. The requirement for SNAG-associated function in granulopoiesis is underscored by the Gfi1-/- phenotype of mice with homozygous targeted knock in of a Gfi1P2A mutation (42). The current study shows that Ajuba acts as a co-repressor for the cognate DNA-bound Gfi1 protein, but that this interaction is not dependent upon the SNAG domain. Moreover, although Ajuba is functionally bound to at least one Gfi1 target gene, it did not appear to regulate CSF1. Thus, it is unlikely that Ajuba is the critical limiting cofactor for GFI1N382S-associated SCN phenotypes. We note that CoREST and LSD1 have been reported to associate with Gfi1 via the SNAG repression domain (23) and thus represent alternative candidates for future analyses.
Supplementary Material
Acknowledgments
We thank F. Rauscher and K. Ayyanathan for scientific discussions.
This work was supported, in whole or in part, by National Institutes of Health Grants R01 HL079574 and CA105152 (to H. L. G.). This work was also supported by a Leukemia Lymphoma Society Scholar award (to H. L. G.). The costs of publication of this article were defrayed in part by the payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 U.S.C. Section 1734 solely to indicate this fact.
The on-line version of this article (available at http://www.jbc.org) contains supplemental Figs. S1-S3.
Footnotes
The abbreviations used are: GFI1, growth factor independent-1; HDAC, histone deacetylase; ELA2, elastase 2, neutrophil; CSF1, colony stimulating factor-1; SCN, severe congenital neutropenia; TSA, trichostatin A; NaB, sodium butyrate; TK, herpes simplex virus thymidine kinase minimal promoter; CAT, chloramphenicol acetyltransferase; SNAG, Snail + Gfi1 repressor domain; SV40SWAP, Gfi1 mutant with the SNAG domain replaced with the SV40 large T antigen nuclear localization motif; shRNA, short hairpin RNA; TBS, Tris-buffered saline; HRP, horseradish peroxidase; ChIP, chromatin immunoprecipitation.
References
- 1.Bach, I. (2000) Mech. Dev. 91 5-17 [DOI] [PubMed] [Google Scholar]
- 2.Dawid, I. B., Breen, J. J., and Toyama, R. (1998) Trends Genet. 14 156-162 [DOI] [PubMed] [Google Scholar]
- 3.Retaux, S., and Bachy, I. (2002) Mol. Neurobiol. 26 269-281 [DOI] [PubMed] [Google Scholar]
- 4.Goyal, R. K., Lin, P., Kanungo, J., Payne, A. S., Muslin, A. J., and Longmore, G. D. (1999) Mol. Cell. Biol. 19 4379-4389 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Hirota, T., Kunitoku, N., Sasayama, T., Marumoto, T., Zhang, D., Nitta, M., Hatakeyama, K., and Saya, H. (2003) Cell 114 585-598 [DOI] [PubMed] [Google Scholar]
- 6.Kanungo, J., Pratt, S. J., Marie, H., and Longmore, G. D. (2000) Mol. Biol. Cell 11 3299-3313 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Marie, H., Pratt, S. J., Betson, M., Epple, H., Kittler, J. T., Meek, L., Moss, S. J., Troyanovsky, S., Attwell, D., Longmore, G. D., and Braga, V. M. M. (2003) J. Biol. Chem. 278 1220-1228 [DOI] [PubMed] [Google Scholar]
- 8.Feng, Y., Zhao, H., Luderer, H. F., Epple, H., Faccio, R., Ross, F. P., Teitelbaum, S. L., and Longmore, G. D. (2007) J. Biol. Chem. 282 39-48 [DOI] [PubMed] [Google Scholar]
- 9.Ren, Y., Meng, S., Mei, L., Zhao, Z. J., Jove, R., and Wu, J. (2004) J. Biol. Chem. 279 8497-8505 [DOI] [PubMed] [Google Scholar]
- 10.Wang, Y., and Gilmore, T. D. (2001) Biochim. Biophys. Acta 1538 260-272 [DOI] [PubMed] [Google Scholar]
- 11.Missero, C., Pirro, M. T., Simeone, S., Pischetola, M., and Di Lauro, R. (2001) J. Biol. Chem. 276 33569-33575 [DOI] [PubMed] [Google Scholar]
- 12.Gilks, C., Bear, S., Grimes, H., and Tsichlis, P. (1993) Mol. Cell. Biol. 113 1759-1768 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kazanjian, A., Gross, E. A., and Grimes, H. L. (2006) Crit. Rev. Oncol./Hematol. 59 85-97 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Zarebski, A., Velu, C. S., Baktula, A. M., Bourdeau, T., Horman, S. R., Basu, S., Bertolone, S. J., Horwitz, M., Hildeman, D. A., Trent, J. O., and Grimes, H. L. (2008) Immunity 28 370-380 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Doan, L. L., Porter, S. D., Duan, Z., Flubacher, M. M., Montoya, D., Tsichlis, P. N., Horwitz, M., Gilks, C. B., and Grimes, H. L. (2004) Nucleic Acids Res. 32 2508-2519 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Duan, Z., and Horwitz, M. (2003) Proc. Natl. Acad. Sci. U. S. A. 100 5932-5937 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Yucel, R., Kosan, C., Heyd, F., and Moroy, T. (2004) J. Biol. Chem. 279 40906-40917 [DOI] [PubMed] [Google Scholar]
- 18.Zweidler-Mckay, P., Grimes, H., Flubacher, M., and Tsichlis, P. (1996) Mol. Cell. Biol. 16 4024-4034 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Grimes, H., Chan, T., Zweidler-McKay, P., Tong, B., and Tsichlis, P. (1996) Mol. Cell. Biol. 16 6263-6272 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ayyanathan, K., Peng, H., Hou, Z., Fredericks, W. J., Goyal, R. K., Langer, E. M., Longmore, G. D., and Rauscher, F. J., 3rd (2007) Cancer Res. 67 9097-9106 [DOI] [PubMed] [Google Scholar]
- 21.Dignam, J., Lebovitz, R., and Roeder, R. (1983) Nucleic Acids Res. 11 1475-1489 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kazanjian, A., Wallis, D., Au, N., Nigam, R., Venken, K. J., Cagle, P. T., Dickey, B. F., Bellen, H. J., Gilks, C. B., and Grimes, H. L. (2004) Cancer Res. 64 6874-6882 [DOI] [PubMed] [Google Scholar]
- 23.Saleque, S., Kim, J., Rooke, H. M., and Orkin, S. H. (2007) Mol. Cell 27 562-572 [DOI] [PubMed] [Google Scholar]
- 24.Hassig, C. A., Fleischer, T. C., Billin, A. N., Schreiber, S. L., and Ayer, D. E. (1997) Cell 89 341-347 [DOI] [PubMed] [Google Scholar]
- 25.Li, J., Wang, J., Wang, J., Nawaz, Z., Liu, J. M., Qin, J., and Wong, J. (2000) EMBO J. 19 4342-4350 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Wen, Y.-D., Perissi, V., Staszewski, L. M., Yang, W.-M., Krones, A., Glass, C. K., Rosenfeld, M. G., and Seto, E. (2000) Proc. Natl. Acad. Sci. U. S. A. 97 7202-7207 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Yoon, H. G., Chan, D. W., Huang, Z. Q., Li, J., Fondell, J. D., Qin, J., and Wong, J. (2003) EMBO J. 22 1336-1346 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Zhang, Y., Ng, H.-H., Erdjument-Bromage, H., Tempst, P., Bird, A., and Reinberg, D. (1999) Genes Dev. 13 1924-1935 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.McGhee, L., Bryan, J., Elliott, L., Grimes, H., Kazanjian, A., Davis, J., and Meyers, S. (2003) J. Cell. Biochem. 89 1005-1018 [DOI] [PubMed] [Google Scholar]
- 30.Gray, S. G., and Ekstrom, T. J. (2001) Exp. Cell Res. 262 75-83 [DOI] [PubMed] [Google Scholar]
- 31.Olsen, P. H., and Ambros, V. (1999) Dev. Biol. 216 671-680 [DOI] [PubMed] [Google Scholar]
- 32.Wadman, I. A., Osada, H., Grutz, G. G., Agulnick, A. D., Westphal, H., Forster, A., and Rabbitts, T. H. (1997) EMBO J. 16 3145-3157 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.McLoughlin, P., Ehler, E., Carlile, G., Licht, J. D., and Schafer, B. W. (2002) J. Biol. Chem. 277 37045-37053 [DOI] [PubMed] [Google Scholar]
- 34.Howard, P. W., and Maurer, R. A. (2001) J. Biol. Chem. 276 19020-19026 [DOI] [PubMed] [Google Scholar]
- 35.Pratt, S. J., Epple, H., Ward, M., Feng, Y., Braga, V. M., and Longmore, G. D. (2005) J. Cell Biol. 168 813-824 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Langer, E. M., Feng, Y., Zhaoyuan, H., Rauscher, F. J., 3rd, Kroll, K. L., and Longmore, G. D. (2008) Dev. Cell 14 424-436 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Peinado, H., Ballestar, E., Esteller, M., and Cano, A. (2004) Mol. Cell. Biol. 24 306-319 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Horwitz, M. S., Duan, Z., Korkmaz, B., Lee, H. H., Mealiffe, M. E., and Salipante, S. J. (2007) Blood 109 1817-1824 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Ancliff, P. J., Blundell, M. P., Cory, G. O., Calle, Y., Worth, A., Kempski, H., Burns, S., Jones, G. E., Sinclair, J., Kinnon, C., Hann, I. M., Gale, R. E., Linch, D. C., and Thrasher, A. J. (2006) Blood 108 2182-2189 [DOI] [PubMed] [Google Scholar]
- 40.Person, R. E., Li, F. Q., Duan, Z., Benson, K. F., Wechsler, J., Papadaki, H. A., Eliopoulos, G., Kaufman, C., Bertolone, S. J., Nakamoto, B., Papayannopoulou, T., Grimes, H. L., and Horwitz, M. (2003) Nat. Genet. 34 308-312 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Klein, C., Grudzien, M., Appaswamy, G., Germeshausen, M., Sandrock, I., Schaffer, A. A., Rathinam, C., Boztug, K., Schwinzer, B., Rezaei, N., Bohn, G., Melin, M., Carlsson, G., Fadeel, B., Dahl, N., Palmblad, J., Henter, J. I., Zeidler, C., Grimbacher, B., and Welte, K. (2007) Nat. Genet. 39 86-92 [DOI] [PubMed] [Google Scholar]
- 42.Fiolka, K., Hertzano, R., Vassen, L., Zeng, H., Hermesh, O., Avraham, K. B., Duhrsen, U., and Moroy, T. (2006) EMBO Rep. 7 326-333 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.