Abstract
The antiviral drug acyclovir is a guanosine nucleoside analog that potently inhibits herpes simplex virus (HSV) replication. Acyclovir treatment in patients coinfected with HSV and human immunodeficiency virus (HIV) has been observed to alter disease course and decrease HIV viral load, a finding that has been attributed to indirect effects of HSV suppression on HIV replication. Based on this hypothesis, several clinical studies have recently investigated the use of acyclovir for treatment of patients coinfected with HSV and HIV or for prophylaxis against HIV transmission. In this report, we use a single round HIV infectivity assay to show that acyclovir directly inhibits HIV infection with an IC50 of ∼5 μm. The target of acyclovir in HIV-infected cells is validated as HIV reverse transcriptase (RT) by the emergence of the RT variant V75I under the selective pressure of acyclovir. The V75I mutation is part of the multidrug resistance pathway that enhances viral resistance to many of the best RT inhibitors approved for the treatment of HIV. Biochemical analyses demonstrate that acyclovir triphosphate is a chain terminator substrate for HIV RT and can compete with dGTP for incorporation into DNA. Although acyclovir may prove a useful lead for development of new HIV treatments, the selection of resistant mutants raises a cautionary note to the use of acyclovir monotherapy in patients coinfected with HSV and HIV.
Herpes simplex virus-2 (HSV-2)2 and HIV infection are two of the most common sexually transmitted infections worldwide, sharing the pathogenic attribute of lifelong infection. Although the seroprevalence of HSV-2 ranges from more than 20% in the United States to as high as 60% in sub-Saharan Africa, the likelihood of HSV-2 infection in HIV-infected individuals can reach as high as 80% (1).
In addressing these high coinfection rates, both epidemiologic and biological studies have suggested an interaction between the two viruses. In HIV-infected patients, HSV can present with more severe mucocutaneous lesions and more frequent and persistent outbreaks (2, 3). HIV infection also increases the frequency of acquisition of HSV-2 (1). In turn, HSV has been shown to accelerate the natural course of HIV infection, with higher plasma and genital levels of HIV and more rapid progression to AIDS (4). Most disconcerting, HSV infection has been shown to increase both the frequency of HIV acquisition and the rate of transmission, demonstrating that these epidemics fuel one another, resulting in significant morbidity and mortality (5).
The association between these epidemics has been strengthened by evidence demonstrating a plausible biological connection between these viral infections. HSV-2 proteins are capable of activating the HIV long terminal repeat, resulting in HIV gene expression (6–8). Also, higher rates of HIV infection in HSV-2-positive patients have been explained by HSV-induced mucosal disruption and specific recruitment of CD4+ cells to these ulcers, providing a portal for HIV infection (9).
Currently, the guanosine analogue acyclovir provides one of the most effective therapies for treatment of HSV-2 infection during acute outbreaks or for long term prophylaxis against recurrent outbreaks. Within herpes virus-infected cells, the HSV thymidine kinase can phosphorylate the prodrug acyclovir at rates above those of cellular nucleoside kinases (10). After monophosphorylation, cellular kinases rapidly convert the drug to acyclovir triphosphate (ACVTP), which is incorporated into viral DNA by the HSV DNA polymerase but is not a substrate for human DNA polymerases (11). Because monophosphorylation and ACVTP incorporation are sequential steps at which selectivity is achieved and amplified, acyclovir is an efficacious and well tolerated drug. Mechanistic studies with the HSV DNA polymerase indicate that ACVTP competes with dGTP and is a chain terminator that results in the formation of a dead-end complex between the enzyme and DNA (11, 12).
The evidence for the interaction between the viruses has stimulated interest in modulation of HIV disease through therapy directed toward HSV. The guiding hypothesis behind these studies has been that decreasing HSV activation or the frequency of HSV lesions could indirectly modulate HIV progression or acquisition in coinfected patients. Most recently, two approaches to investigate the benefits of acyclovir or its prodrug, valacyclovir, have been reported (13, 14). In the first approach, coinfected patients not on highly active antiretroviral therapy (HAART) for HIV infection were given valacyclovir to suppress HSV-2 (13, 14). These studies found a decrease in the average plasma HIV viral load of 2–3-fold versus untreated controls (13, 14). Since viral load is correlated with rate of disease progression (15), valacyclovir therapy was suggested as a means to delay the initiation of HAART in coinfected patients. In principle, this approach has the benefit of avoiding cumulative toxicities associated with HAART. Acyclovir could also be beneficial for prophylaxis against acquisition of HIV in HSV-2-infected patients. However, in trials designed to examine this possibility, no significant protection against HIV acquisition was noted (13, 16), although concerns have been raised that the results may be the product of poor compliance (17).
Here we present findings that strongly indicate that the effects of acyclovir on HIV viral load arise directly from the inhibition of the HIV reverse transcriptase enzyme. Our findings raise significant concerns about the potential of generating cross-resistance to known HIV therapies by treatment of coinfected patients with acyclovir prior to a HAART regimen.
EXPERIMENTAL PROCEDURES
Infections and Phenotypic Analysis—Dose-response curves for the anti-HIV activity of acyclovir (Sigma) were generated from infection of phytohemagglutinin (PHA)-activated primary CD4+ lymphoblasts with a recombinant CXCR4 tropic HIV pseudovirus as described previously (18, 19). Pseudovirus was generated from cotransfection of HEK293T cells with a plasmid containing the HIV genome with part of the envelope gene replaced with GFP (pNL4-3-ΔE-eGFP) and a plasmid containing a CXCR4 tropic envelope gene. CD4+ T cells were isolated from PHA-activated peripheral blood mononuclear cells of healthy donors with selection using Miltenyi biotech magnetic beads. Experiments were repeated in three or more replicates each, with cells derived separately from more than five different healthy blood donors. Cells were preincubated with acyclovir for 16–20 h prior to infection to allow for cellular uptake. The V75I, T69N, and M184V mutant plasmids were generated by site-directed mutagenesis (Stratagene) of pNL4-3-ΔE-eGFP. Viruses were standardized using p24 enzyme-linked immunoabsorbancy assay (PerkinElmer Life Sciences).
Real-time PCR—CXCR4 tropic HIV pseudoviruses were DNase (Roche Applied Science)-treated and used to infect CD4+ lymphoblasts in the absence or presence of 400 μm acyclovir. Total DNA was isolated from cells (Qiagen) at 8 and 24 h after infection. Early, intermediate, and late reverse transcript products were quantified by real-time PCR using separate primer/probe sets as described previously (18, 20). A standard curve of pNL4-3-ΔE-eGFP was used to determine the copies of transcripts per cell.
In Vitro Selection—PHA-activated primary CD4+ lymphoblasts were infected with a replication-competent laboratory reference strain of HIV (Ba-L) in the absence or presence of acyclovir. Every 3–5 days, 4% of the culture supernatant was used to infect freshly isolated CD4+ lymphoblasts in the presence of three times the acyclovir concentration as the previous infection. After 11 passages, cultures were maintained at 400 μm acyclovir. Total DNA was isolated from cells (Qiagen) after every passage, and the proviral pol gene was amplified at multiple time points using limiting dilution digital PCR for sequencing of single viral genomes adapted from a previously described method (21, 22). A minimum of 25 isolated viral genomes were sequenced under each condition at every evaluated time point.
Reverse Transcriptase Assays—HIV RT produced in Escherichia coli and purified by affinity chromatography was kindly provided by Dr. Stuart Le Grice at the NCI, National Institutes of Health, Drug Resistance Program. For all assays, 5′-fluorescein-labeled DNA primer (Integrated DNA Technologies) and DNA templates (Invitrogen) were mixed at a 1:2 molar ratio in 10 mm Tris-Cl, pH 7.9, 50 mm NaCl, 10 mm MgCl2, 1 mm dithiothreitol and annealed by heating to 95 °C, stepwise cooling until 5 °C below the calculated Tm followed by cooling to reaction temperature of 37 °C. Reactions were initiated by the simultaneous addition of RT and dNTPs or ACVTP (Moravek Biochemicals) and quenched by the addition of an equal volume of gel loading buffer (95% formamide, 20 mm EDTA). Samples were denatured at 95 °C, and products were resolved on a 20% polyacrylamide gel containing 7 m urea run at 50 °C. Labeled DNA was visualized by the capture of fluorescein emission on a Typhoon scanner (Amersham Biosciences). Product was quantified using the ImageJ software.
RESULTS
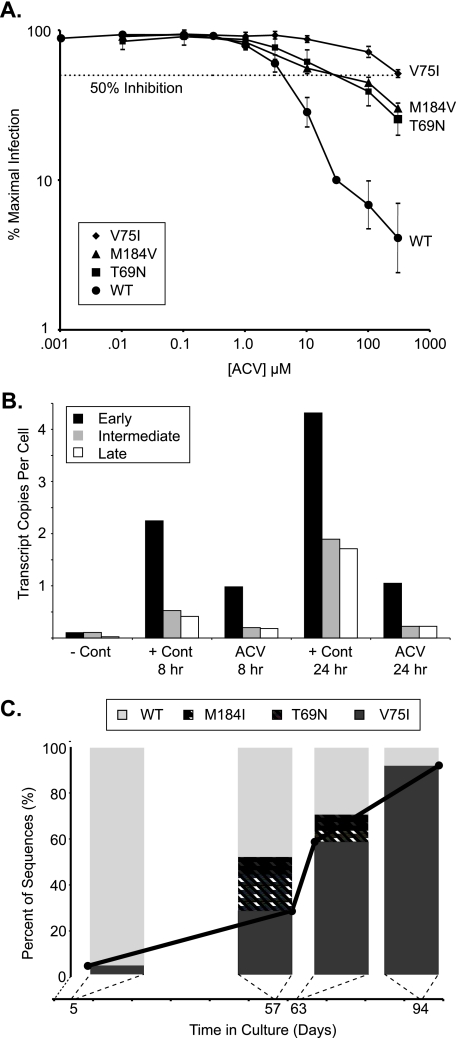
Acyclovir Inhibits HIV Replication—To determine whether acyclovir can inhibit HIV replication in vitro, we used a sensitive single cell HIV infectivity assay that has previously been used to confirm the anti-HIV activity of known antiviral drugs (19, 23) and revealed the unanticipated anti-HIV activity of the hepatitis B drug entecavir (18). The assay utilizes primary CD4+ lymphoblasts infected with a CXCR4 pseudotyped HIV virus capable of only a single round of infection due to replacement of part of the envelope gene with GFP. The infection was carried out in the presence of increasing concentrations of acyclovir, and productive infection was monitored by GFP expression (18, 19, 23). With the wild-type HIV reference strain, acyclovir inhibited HIV infection with a 50% inhibitory concentration (IC50) of ∼5 μm (Fig. 1A). These results were reproduced with primary CD4+ T cells derived from more than five donors, indicating that these observations are generalizable.
FIGURE 1.
Effects of acyclovir on HIV replication and identification of resistant variants. A, inhibition of HIV infectivity by acyclovir. HIV pseudoviruses derived from the wild-type (WT) clone, NL4-3, and the isogenic clones varying only at positions T69N, V75I, or M184V were used to infect primary CD4+ cells in the presence of varying amounts of acyclovir. GFP-expressing cells were analyzed 72 h after infection. The results are expressed as a percentage of maximal infection, which is determined from (the number of GFP-expressing cells in the presence of acyclovir divided by the GFP-expressing cells in the absence of a drug effect) × 100. The horizontal dotted line represents 50% inhibition. The IC50 values were ∼5 μm for wild-type virus, ∼50 μm for the T69N or M184V point mutants, and >300 μm for the V75I point mutant. When shown, error bars represent the standard deviation from three or more replicates from two or more normal blood donors. B, acyclovir prevents the appearance of reverse transcription products. Primary CD4+ T cells were infected with wild-type NL4-3 pseudovirus in the absence or presence of acyclovir. The amounts of early, intermediate, and late reverse transcripts were quantified by real-time PCR from DNA isolated at defined times and a standard curve generated from known concentrations of pNL4-3-ΔE-eGFP with appropriate primer pairs. The negative control (–Cont) infection was performed at 4 °C, and signal represents residual plasmid from the viral production step. Shown are representative data from one of three experiments with similar results. C, acyclovir selects for mutations in HIV RT. Replication-competent HIV was cultured in the presence of increasing concentrations of acyclovir. The graph presents quantification of proviral variants generated under the selective pressure of acyclovir. The percentage of variants was determined from a minimum of 25 independent sequences at the indicated days in culture. The solid black line denotes the percentage of V75I variant appearing as a function of time.
We hypothesized that the observed inhibition of HIV replication in vitro was due to direct anti-HIV activity. To narrow the range of potential molecular targets of acyclovir, we used a real-time PCR assay for detection of early, intermediate, and late reverse transcriptase products (18, 20). In this assay, drugs that inhibit at or before the reverse transcription step lead to a reduction in all transcription products. To perform this analysis, we infected CD4+ lymphoblasts in the absence or presence of acyclovir. When total DNA extracts were analyzed, the early, intermediate, and late reverse transcript products were all reduced (Fig. 1B), suggesting that inhibition by acyclovir takes place at or before reverse transcription.
Acyclovir Selects for Mutations in HIV Reverse Transcriptase—To further validate the molecular target of acyclovir, we cultured primary CD4+ lymphoblasts with a replication-competent CCR5 tropic HIV isolate in the presence of increasing concentrations of acyclovir. At various times during the selection process, we sequenced single proviral genomes amplified using a limiting dilution PCR assay. As early as 5 days after the initial infection, at an acyclovir concentration of only 0.01 μm, we identified 1 out of 25 sequences (4%) containing the V75I RT multidrug resistance mutation (Fig. 1C). This mutation expanded rapidly between day 57 and 63 and came to dominate sequences after 94 days in culture, constituting 92% of the total viral population. Interestingly, two other known HIV drug resistance mutations, T69N and M184I (which typically precedes emergence of the M184V variant of RT), also appeared in the presence of acyclovir (Fig. 1C). In our unbiased resistance selection protocol, no other known HIV drug resistance mutations were observed, and the predominant mutations were localized to the RT gene. Virus in control cultures, carried out in parallel in the absence of acyclovir, remained wild type at these positions at all time points, with the exception of 1 of 52 sequences at day 63 and 1 of 35 sequences at day 94 that contained the T69N variant. These findings provide strong support for the hypothesis that HIV RT is the molecular target of acyclovir.
Having identified RT mutations under acyclovir selective pressure, we sought to confirm their resistance phenotypes using our single cell HIV infectivity assay. Site-directed mutagenesis was used to construct isogenic strains differing only at each of the observed point mutant locations discovered through our selection protocol. The IC50 values for virus containing either the T69N or the M184V point mutations shifted ∼10-fold relative to wild-type virus (Fig. 1A). The V75I mutant virus, the dominant mutation noted in our selection protocol, had an IC50 shifted to >300 μm, approaching a nearly 2-log increase in resistance to acyclovir.
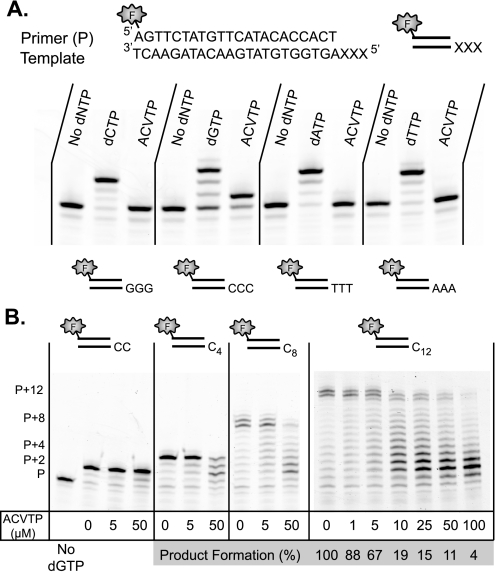
Acyclovir Triphosphate Is a Substrate of RT and Competes with dGTP—Since acyclovir inhibited HIV infection at the single cell level and resistance was associated with mutations in RT, we next explored the mechanism by which acyclovir inhibited RT. Because acyclovir lacks a 3′-OH to allow for DNA extension, it might be expected to act as an obligate chain terminator of HIV RT. We assayed for the ability of purified HIV RT to incorporate ACVTP using an oligonucleotide DNA template-primer. As suggested by the mutagenesis findings, ACVTP could indeed be incorporated by RT (Fig. 2A). Although each tested template could be extended to full-length product in the presence of the cognate dNTP, ACVTP was only incorporated opposite to cytosine. As expected for a chain terminator, only a single ACVTP was incorporated.
FIGURE 2.
ACVTP is a substrate and chain terminator of RT and competes with dGTP for incorporation. A, HIV RT can incorporate ACVTP. The primer (1 μm) was annealed to indicated template (2 μm) and incubated with 100 nm RT alone, RT and 100 μm cognate dNTP, or RT and 100 μm ACVTP for 5 min. For each primer/template pair, RT achieves extension with the cognate dNTP, yielding a +3 band representing consecutive nucleotide additions. Acyclovir is only incorporated opposite a cytosine-containing template and generates a +1 band suggesting single addition. B, ACVTP competes with dGTP for incorporation onto poly-C templates. The template names, Cn, denote the number of cytosine nucleotides repeated at the 5′-end of the template. The control lane lacks dGTP. For all other lanes, 100 nm indicated primer/template was incubated with 100 nm RT, 5 μm dGTP, and the indicated concentrations of ACVTP for 5 min at 37 °C. For reactions with C12-containing template, full-length product was taken as primer extended from 11 to 13 bases with dGTP, as alternate length results from the untemplated addition or sliding of the template relative to primer. Signal intensity was quantified for the full-length product relative to the total signal. The percentage of product formation was determined relative to the full-length product in the absence of ACVTP. With increasing ACVTP concentrations, product formation decreases and truncated products accumulate. XXX represents G-, C-, T-, or A- containing templates represented in panel A, respectively. P is primer.
An important question is whether ACVTP is an effective competitor in the presence of physiological concentrations of dGTP. We sought to mimic potential physiological conditions in a competition-based assay that depended on multiple dGTP incorporations, as would be required of RT in copying the HIV genome. Templates were constructed containing poly-C overhangs beyond the primer terminus. Competition was evaluated at varying concentrations of ACVTP and using a fixed concentration of 5 μm dGTP, similar to the physiological concentration found in activated T cells (24). It was observed that shorter templates provided limited opportunity for competitive chain termination, whereas full-length product formation was significantly diminished for the longer templates (Fig. 2B). Thus, ACVTP is an increasingly effective inhibitor for formation of full-length chains as the number of potential incorporation events increases. Since the HIV genome is ∼10 kb in size, ∼2500 opportunities exist for a chain terminator such as ACVTP to incorporate in lieu of dGTP.
DISCUSSION
This report demonstrates that the antiviral drug acyclovir inhibits HIV replication in primary CD4+ lymphoblasts and suggests that acyclovir is directly responsible for the decline in HIV plasma levels in HSV/HIV-coinfected patients on acyclovir monotherapy. Acyclic nucleoside or nucleotide analogs are used for the treatment of a variety of DNA and RNA viruses including the Herpes family of viruses (cidofovir and adefovir), hepatitis B virus (adefovir and tenofovir), and HIV (tenofovir). Based on these precedents, we evaluated the anti-HIV activity of acyclovir using an infectivity assay that has previously been used to evaluate currently approved anti-HIV drugs (19, 23). Our assay relies on a single round of infection, thereby avoiding complications of virus outgrowth in multiround infection assays (25). In addition, we can precisely measure infection events in individual cells by flow cytometry. Further, rather than using tissue samples or a cultured cell line, the assay uses primary CD4+ T cells, the in vivo target cells of HIV. The cells were isolated from multiple donors, demonstrating the reproducible and general nature of the inhibition by acyclovir. Our conclusion that acyclovir is a direct inhibitor of HIV infection is supported by a recent report that used an alternate approach to demonstrate the anti-HIV activity of acyclovir in a tissue based assay (26). Our results in a primary cell system are therefore almost certainly relevant to clinical treatments of HIV and HSV infections.
Our finding that acyclovir selects for resistance mutations in HIV-RT provides strong evidence that RT is the in vivo target of this agent and also raises concerns for the selection of cross-resistance to approved RT inhibitors. Our assay identified and confirmed the resistance of the V75I variant, a known HIV drug resistance mutation, and one of several mutations that constitute the Q151M multidrug resistance pathway that imparts resistance to most currently approved anti-HIV nucleoside analogs (27). Even in the absence of the Q151M complex, isolated Val-75 mutations have been shown to have reduced susceptibility to several nucleoside analogs (28). In addition to V75I, our selection protocol also identified the M184I (the M184V precursor) and T69N variants, both with ∼10-fold increased resistance to acyclovir. Although they appeared less fit than V75I in our single round infectivity assay, these mutations are of clinical significance. M184V confers near complete resistance to 3TC (lamivudine) or FTC (emtricitabine), drugs that form the foundation of most HAART regimens (29–31). The T69N mutation broadly reduces sensitivity to currently approved nucleoside analogs either by itself or, more significantly, in the context of other thymidine analog mutations (32, 33).
Although the full mechanism of RT inhibition by acyclovir remains to be explored, the dominant V75I mutation offers opportunity for speculation. Acyclovir inhibition of HSV DNA polymerase is due to the formation of a dead-end ternary complex involving the polymerase, the DNA substrate with acyclovir incorporated, and an incoming dNTP that is complementary to the template +1 position (11, 12). V75I is known to interact with the template +1 position (34). This correlation suggests the possibility that dead-end ternary complex formation may be compromised in the V75I RT variant. A more detailed study of this inhibition mechanism will likely provide a deeper understanding of the resistance mutations.
The generally accepted mechanism for activation of acyclovir requires action of the HSV thymidine kinase and thus raises the question of whether acyclovir works only in cells that are coinfected with herpes viruses. Supporting this notion, the report by Lisco et al. (26) suggested that the anti-HIV activity of acyclovir was dependent on herpes virus infection of the target tissues, and in particular, coinfection with the ubiquitous virus HHV-6. Our study used primary CD4+ T cells derived from normal donors, for which the herpes virus status is not known. It is notable, however, that low measurable levels of ACVTP are known to be generated even in non-HSV-infected cells (35). Although coinfection may increase the levels of ACVTP and result in efficient RT inhibition in such cells, it is feasible that low levels of ACVTP generated by host enzymes in non-HSV-infected cells could effectively compete with dGTP given that a chain terminator of HIV replication has numerous opportunities for incorporation during HIV replication.
Acyclovir is not as potent of an inhibitor of HIV infection as many nucleoside analogs. By comparison in our phenotypic assay, it is 50 times less potent than AZT (zidovudine) (IC50 ∼100 nm) and 500 times less potent than 3TC (lamivudine) (IC50 ∼10 nm) (18, 23). Pharmacokinetic studies suggest that the maximal clinically achieved plasma concentration (Cmax) of acyclovir can be greater than 20 μm (36). Although the weak inhibition we see in vitro would not likely make acyclovir an effective suppressive HIV drug, the IC50 of 5 μm is well within concentrations achieved clinically and may be responsible for the observed ∼0.5 log10 decline in plasma HIV RNA levels seen in coinfected patients on suppressive acyclovir therapy (13, 14). In this light, acyclovir could prove to be a useful scaffold for future drug development. At the same time, the use of acyclovir at subinhibitory concentrations against HIV, as well as the often episodic nature of treatment for HSV exacerbations, create prime conditions for development of resistance mutations. Since we have established that the RT variants selected by acyclovir in vitro are also known HIV drug resistance mutations, it will be important to obtain baseline genotypes for coinfected patients that have been maintained on suppressive acyclovir or patients who acquire HIV while taking acyclovir. Episodic antiviral use also raises the concern that a wild-type clinical genotype may not reflect drug resistant variants that could be archived in the latent reservoir generated during prior acyclovir therapy (37). Our studies suggest the need for caution in the use of acyclovir in HSV/HIV-coinfected patients not on a suppressive HAART regimen and should prompt reinterpretation of the observed effects of antiviral compounds on the interplay of these two epidemic viruses.
This work was supported, in whole or in part, by National Institutes of Health Grants AI43222 (to R. F. S.) and GM56834-12 (to J. T. S.). The costs of publication of this article were defrayed in part by the payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 U.S.C. Section 1734 solely to indicate this fact.
Footnotes
The abbreviations used are: HSV, herpes simplex virus; HIV, human immunodeficiency virus; RT, reverse transcriptase; ACVTP, acyclovir triphosphate; HAART, highly active antiretroviral therapy; PHA, phytohemagglutinin; GFP, green fluorescent protein; eGFP, enhanced GFP.
References
- 1.Corey, L., Wald, A., Celum, C. L., and Quinn, T. C. (2004) J. Acquir. Immune Defic. Syndr. 35 435–445 [DOI] [PubMed] [Google Scholar]
- 2.Schacker, T., Hu, H. L., Koelle, D. M., Zeh, J., Saltzman, R., Boon, R., Shaughnessy, M., Barnum, G., and Corey, L. (1998) Ann. Intern. Med. 128 21–28 [DOI] [PubMed] [Google Scholar]
- 3.Krone, M. R., Wald, A., Tabet, S. R., Paradise, M., Corey, L., and Celum, C. L. (2000) Clin. Infect. Dis. 30 261–267 [DOI] [PubMed] [Google Scholar]
- 4.Schacker, T., Zeh, J., Hu, H., Shaughnessy, M., and Corey, L. (2002) J. Infect. Dis. 186 1718–1725 [DOI] [PubMed] [Google Scholar]
- 5.Freeman, E. E., Weiss, H. A., Glynn, J. R., Cross, P. L., Whitworth, J. A., and Hayes, R. J. (2006) AIDS 20 73–83 [DOI] [PubMed] [Google Scholar]
- 6.Mosca, J. D., Bednarik, D. P., Raj, N. B., Rosen, C. A., Sodroski, J. G., Haseltine, W. A., Hayward, G. S., and Pitha, P. M. (1987) Proc. Natl. Acad. Sci. U. S. A. 84 7408–7412 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Margolis, D. M., Rabson, A. B., Straus, S. E., and Ostrove, J. M. (1992) Virology 186 788–791 [DOI] [PubMed] [Google Scholar]
- 8.Golden, M. P., Kim, S., Hammer, S. M., Ladd, E. A., Schaffer, P. A., DeLuca, N., and Albrecht, M. A. (1992) J. Infect. Dis. 166 494–499 [DOI] [PubMed] [Google Scholar]
- 9.Koelle, D. M., Posavad, C. M., Barnum, G. R., Johnson, M. L., Frank, J. M., and Corey, L. (1998) J. Clin. Investig. 101 1500–1508 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Keller, P. M., Fyfe, J. A., Beauchamp, L., Lubbers, C. M., Furman, P. A., Schaeffer, H. J., and Elion, G. B. (1981) Biochem. Pharmacol. 30 3071–3077 [DOI] [PubMed] [Google Scholar]
- 11.Reardon, J. E. (1989) J. Biol. Chem. 264 19039–19044 [PubMed] [Google Scholar]
- 12.Reardon, J. E., and Spector, T. (1989) J. Biol. Chem. 264 7405–7411 [PubMed] [Google Scholar]
- 13.Zuckerman, R. A., Lucchetti, A., Whittington, W. L., Sanchez, J., Coombs, R. W., Zuniga, R., Magaret, A. S., Wald, A., Corey, L., and Celum, C. (2007) J. Infect. Dis. 196 1500–1508 [DOI] [PubMed] [Google Scholar]
- 14.Nagot, N., Ouedraogo, A., Foulongne, V., Konaté, I., Weiss, H. A., Vergne, L., Defer, M. C., Djagbaré, D., Sanon, A., Andonaba, J. B., Becquart, P., Segondy, M., Vallo, R., Sawadogo, A., Van de Perre, P., Mayaud, P., and the ANRS 1285 Study Group (2007) N. Engl. J. Med. 356 790–799 [DOI] [PubMed] [Google Scholar]
- 15.Mellors, J. W., Rinaldo, C. R., Jr., Gupta, P., White, R. M., Todd, J. A., and Kingsley, L. A. (1996) Science 272 1167–1170 [DOI] [PubMed] [Google Scholar]
- 16.Watson-Jones, D., Weiss, H. A., Rusizoka, M., Changalucha, J., Baisley, K., Mugeye, K., Tanton, C., Ross, D., Everett, D., Clayton, T., Balira, R., Knight, L., Hambleton, I., Le Goff, J., Belec, L., Hayes, R., the HSV Trial Team, and the Steering and Data Monitoring Committees (2008) N. Engl. J. Med. 358 1560–1571 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lisco, A., and Vanpouille, C. (2008) N. Engl. J. Med. 359 535. [DOI] [PubMed] [Google Scholar]
- 18.McMahon, M. A., Jilek, B. L., Brennan, T. P., Shen, L., Zhou, Y., Wind-Rotolo, M., Xing, S., Bhat, S., Hale, B., Hegarty, R., Chong, C. R., Liu, J. O., Siliciano, R. F., and Thio, C. L. (2007) N. Engl. J. Med. 356 2614–2621 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Zhang, H., Zhou, Y., Alcock, C., Kiefer, T., Monie, D., Siliciano, J., Li, Q., Pham, P., Cofrancesco, J., Persaud, D., and Siliciano, R. F. (2004) J. Virol. 78 1718–1729 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Zack, J. A., Arrigo, S. J., Weitsman, S. R., Go, A. S., Haislip, A., and Chen, I. S. (1990) Cell 61 213–222 [DOI] [PubMed] [Google Scholar]
- 21.Bailey, J. R., Sedaghat, A. R., Kieffer, T., Brennan, T., Lee, P. K., Wind-Rotolo, M., Haggerty, C. M., Kamireddi, A. R., Liu, Y., Lee, J., Persaud, D., Gallant, J. E., Cofrancesco, J., Jr., Quinn, T. C., Wilke, C. O., Ray, S. C., Siliciano, J. D., Nettles, R. E., and Siliciano, R. F. (2006) J. Virol. 80 6441–6457 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Vogelstein, B., and Kinzler, K. W. (1999) Proc. Natl. Acad. Sci. U. S. A. 96 9236–9241 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Shen, L., Peterson, S., Sedaghat, A. R., McMahon, M. A., Callender, M., Zhang, H., Zhou, Y., Pitt, E., Anderson, K. S., Acosta, E. P., and Siliciano, R. F. (2008) Nat. Med. 14 762–766 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Traut, T. W. (1994) Mol. Cell. Biochem. 140 1–22 [DOI] [PubMed] [Google Scholar]
- 25.Ferguson, N. M., Fraser, C., and Anderson, R. M. (2001) Trends Pharmacol. Sci. 22 97–100 [DOI] [PubMed] [Google Scholar]
- 26.Lisco, A., Vanpouille, C., Tchesnokov, E. P., Grivel, J. C., Biancotto, A., Brichacek, B., Elliott, J., Fromentin, E., Shattock, R., Anton, P., Gorelick, R., Balzarini, J., McGuigan, C., Derudas, M., Götte, M., Schinazi, R. F., and Margolis, L. (2008) Cell Host Microbe 4 260–270 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Shirasaka, T., Kavlick, M. F., Ueno, T., Gao, W. Y., Kojima, E., Alcaide, M. L., Chokekijchai, S., Roy, B. M., Arnold, E., and Yarchoan, R. (1995) Proc. Natl. Acad. Sci. U. S. A. 92 2398–2402 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Matamoros, T., Kim, B., and Menendez-Arias, L. (2008) J. Mol. Biol. 375 1234–1248 [DOI] [PubMed] [Google Scholar]
- 29.Schinazi, R. F., Lloyd, R. M., Jr., Nguyen, M. H., Cannon, D. L., McMillan, A., Ilksoy, N., Chu, C. K., Liotta, D. C., Bazmi, H. Z., and Mellors, J. W. (1993) Antimicrob. Agents Chemother. 37 875–881 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Tisdale, M., Kemp, S. D., Parry, N. R., and Larder, B. A. (1993) Proc. Natl. Acad. Sci. U. S. A. 90 5653–5656 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Gao, Q., Gu, Z., Parniak, M. A., Cameron, J., Cammack, N., Boucher, C., and Wainberg, M. A. (1993) Antimicrob. Agents Chemother. 37 1390–1392 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Winters, M. A., and Merigan, T. C. (2001) Antimicrob. Agents Chemother. 45 2276–2279 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Johnson, V. A., Brun-Vezinet, F., Clotet, B., Gunthard, H. F., Kuritzkes, D. R., Pillay, D., Schapiro, J. M., and Richman, D. D. (2008) Top. HIV Med. 16 62–68 [DOI] [PubMed] [Google Scholar]
- 34.Huang, H., Chopra, R., Verdine, G. L., and Harrison, S. C. (1998) Science 282 1669–1675 [DOI] [PubMed] [Google Scholar]
- 35.Elion, G. B., Furman, P. A., Fyfe, J. A., de Miranda, P., Beauchamp, L., and Schaeffer, H. J. (1977) Proc. Natl. Acad. Sci. U. S. A. 74 5716–5720202961 [Google Scholar]
- 36.Jacobson, M. A. (1993) J. Med. Virol. Suppl. 1 150–153 [DOI] [PubMed] [Google Scholar]
- 37.Ruff, C. T., Ray, S. C., Kwon, P., Zinn, R., Pendleton, A., Hutton, N., Ashworth, R., Gange, S., Quinn, T. C., Siliciano, R. F., and Persaud, D. (2002) J. Virol. 76 9481–9492 [DOI] [PMC free article] [PubMed] [Google Scholar]