Abstract
Ankyrin-B targets ion channels and transporters in excitable cells. Dysfunction in ankyrin-B-based pathways results in defects in cardiac physiology. Despite a wealth of knowledge regarding the role of ankyrin-B for cardiac function, little is known regarding the mechanisms underlying ankyrin-B regulation. Moreover, the pathways underlying ankyrin-B targeting in heart are unclear. We report that alternative splicing regulates ankyrin-B localization and function in cardiomyocytes. Specifically, we identify a novel exon (exon 43′) in the ankyrin-B regulatory domain that mediates interaction with the Rho-GEF obscurin. Ankyrin-B transcripts harboring exon 43′ represent the primary cardiac isoform in human and mouse. We demonstrate that ankyrin-B and obscurin are co-localized at the M-line of myocytes and co-immunoprecipitate from heart. We define the structural requirements for ankyrin-B/obscurin interaction to two motifs in the ankyrin-B regulatory domain and demonstrate that both are critical for obscurin/ankyrin-B interaction. In addition, we demonstrate that interaction with obscurin is required for ankyrin-B M-line targeting. Specifically, both obscurin-binding motifs are required for the M-line targeting of a GFP-ankyrin-B regulatory domain. Moreover, this construct acts as a dominant-negative by competing with endogenous ankyrin-B for obscurin-binding at the M-line, thus providing a powerful new tool to evaluate the function of obscurin/ankyrin-B interactions. With this new tool, we demonstrate that the obscurin/ankyrin-B interaction is critical for recruitment of PP2A to the cardiac M-line. Together, these data provide the first evidence for the molecular basis of ankyrin-B and PP2A targeting and function at the cardiac M-line. Finally, we report that ankyrin-B R1788W is localized adjacent to the ankyrin-B obscurin-binding motif and increases binding activity for obscurin. In summary, our new findings demonstrate that ANK2 is subject to alternative splicing that gives rise to unique polypeptides with diverse roles in cardiac function.
Ankyrins are adapter proteins that facilitate the local organization of integral membrane proteins with cytoskeletal elements. Three human ankyrin genes ANK1, ANK2, and ANK3 encode polypeptides termed ankyrin-R, ankyrin-B, and ankyrin-G, respectively. Ankyrins-B and -G are ubiquitously expressed, whereas ankyrin-R has restricted distribution. Ankyrin-B is required for normal cardiac physiology, and ankyrin-B dysfunction in humans and mice results in disease. Specifically, mice lacking ankyrin-B die at postnatal day 1 (1). Mice heterozygous for a null mutation in ankyrin-B (ankyrin-B+/- mice) display bradycardia, heart rate variability, conduction defects, prolonged rate corrected QT intervals, catecholaminergic polymorphic ventricular tachycardia, syncope, and sudden cardiac death (2). ANK2 (encodes ankyrin-B) loss-of-function variants in humans result in dominantly inherited human arrhythmia termed “ankyrin-B syndrome” or “type 4 long QT syndrome.” Human disease phenotypes include bradycardia, atrial fibrillation, conduction block, ventricular arrhythmia, and risk of sudden cardiac death (2-4).
Ankyrin-B is a large polypeptide with three major structural domains (Fig. 1A). The ankyrin-B membrane-binding domain is comprised of 24 consecutive ANK repeats that associate with ion channels, transporters, and cell adhesion molecules, including the Na+/Ca2+ exchanger, Na/K-ATPase, InsP32 receptor, and L1 cell adhesion molecule (2, 5-8). The ankyrin-B spectrin-binding domain is a 62-kDa domain that displays binding activity for the β-spectrin family of actin-associated proteins (see Ref. 9). This domain is also a binding partner for protein phosphatase 2A regulatory subunit B56α (10). Finally, ankyrin-B contains a C-terminal regulatory domain (RD) comprised of a death domain and an ∼300-amino acid extended random coil (11).
FIGURE 1.
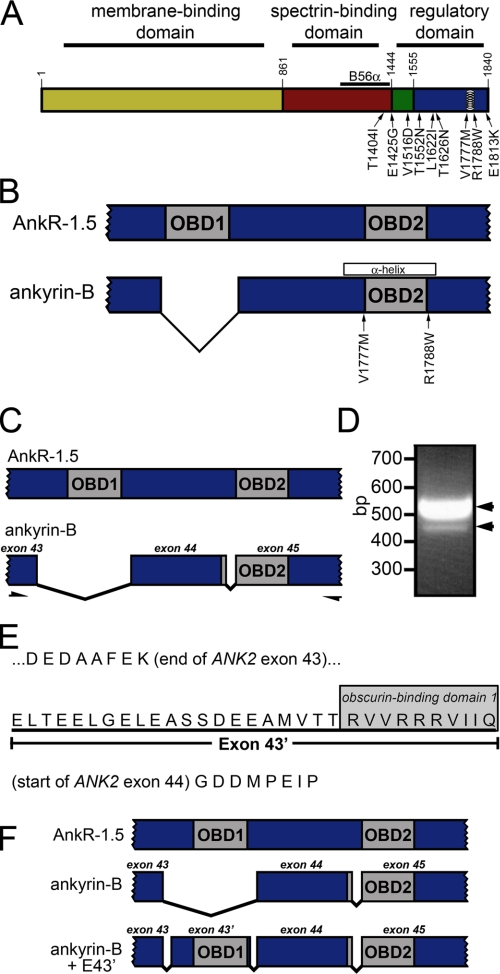
Novel ANK2 exon encodes putative obscurin-binding domain. A, ankyrin domain organization. Ankyrin-B contains three major domains, including the membrane-binding domain (yellow), spectrin-binding domain (red), and regulatory domain. The RD is comprised of the death domain (green) and C-terminal domain (blue). Identified ankyrin-B loss-of-function variants associated with human arrhythmia are noted on the diagram. Previously identified amphipathic α-helix in RD (28) is indicated with circles. B56α-binding site in the spectrin-binding domain is represented by a black line. B, both obscurin-binding domains (OBD1 and OBD2) in AnkR1.5 are required for AnkR/obscurin interaction. Ankyrin-B lacks OBD1. Ankyrin-B variants in this region are noted on the diagram. C, PCR-based strategy to identify novel ANK2 exon(s) encoding OBD1 in ankyrin-B. PCR primers were designed against ANK2 sequence in exons 43 and 45 to amplify cardiac ankyrin-B transcripts. D, ethidium bromide agarose gel demonstrates amplification of two PCR products (518 and 425 bp) from human heart cDNA. In the 518-bp PCR product, the additional 93 bp represent a new, previously unreported exon (exon 43′). E, exon 43′ encodes 31 amino acids that contain a homologous site to OBD1 in the terminal 10 amino acids. F, with the addition of exon 43′, human ankyrin-B + E43′ contains two homologous sites corresponding to OBD1 and OBD2 in small AnkR-1.5.
The ankyrin-B RD is essential for its function in cardiomyocytes (11, 12). Eight of the nine ANK2 human arrhythmia disease variants identified to date are located in this domain (Fig. 1A) (2-4). However, despite overwhelming evidence supporting the importance of this domain for ankyrin-B function, little is known regarding the cellular role for this domain in cardiomyocytes. Potential roles for this domain include regulation of ankyrin stability, folding, and targeting. Additionally, this domain may play critical roles for ankyrin molecular interactions (e.g. protein/protein, protein/lipid, etc.).
Obscurin is a large Rho-GEF that organizes sarcomeric proteins in striated muscle. Specifically, obscurin plays multiple roles in muscle myosin incorporation, A-band formation, and lateral alignment of M-lines in adjacent myofibrils (13-18). Recently, independent groups identified an interaction between obscurin and ankyrin-R (ANK1) isoforms 1.5 and 1.9 (19-23). This interaction was shown to be dependent on two unique structural motifs present in the RD of ankyrin-R (21). Interestingly, although their RDs are structurally quite similar, ankyrin-B lacks one of these two motifs (Fig. 1B) and was therefore dismissed as a potential obscurin-binding partner.
Here we report the identification of a new exon (exon 43′) in ANK2 that is expressed in human heart and modulates the novel interaction between ankyrin-B RD and the large cardiac Rho-GEF obscurin. Ankyrin-B containing exon 43′ is the predominant ankyrin-B isoform in heart. Moreover, ankyrin-B and obscurin are co-localized and associate in cardiac tissue. Ankyrin-B and obscurin directly interact, and this interaction is modulated by the insertion of key residues encoded by ANK2 exon 43′. We demonstrate that interaction with obscurin is critical for recruitment of the dominant population of ankyrin-B to the cardiac M-line. Moreover, we demonstrate that interaction of ankyrin-B with obscurin is essential for the subcellular targeting of a key cardiac signaling molecule, PP2A. These data define a novel ankyrin-B protein partner in cardiac muscle, identify the mechanism underlying ankyrin-B M-line targeting in heart, and describe the cellular pathway for PP2A targeting in myocytes. Finally, these new data illustrate the importance of ANK2 transcriptional regulation for ankyrin-B polypeptide function in vivo.
EXPERIMENTAL PROCEDURES
Human Tissue, RNA Isolation, and Reverse Transcription—RNA was isolated from pure left ventricular muscle tissue from healthy donor hearts that were not suitable for transplantation (subclinical atherosclerosis, advanced age, no matching recipients, etc.) through the Iowa Donors Network and through the National Disease Research Interchange, Inc. (Philadelphia). Age and sex were the only identifying data acquired from the tissue providers, and the University of Iowa Human Subjects Committee deemed that informed consent from each patient was not required. None of the patients died from cardiac related causes. RNA was isolated from 400 μg of the apex of flash-frozen left ventricular muscle samples (fat, coronary arteries excluded) using an RNeasy midi kit (Qiagen). 500 ng of RNA (with a 260/280 ratio of 1.9 to 2.1) was reverse-transcribed into cDNA using SuperScript III reverse transcriptase (Invitrogen) in 20-μl reactions. 1 μl of this reaction was used in subsequent PCR or qt-PCRs.
PCR Amplification and Isolation of Alternative ANK2 Transcripts—PCR products were amplified from reverse-transcribed ventricular mRNA and human heart cDNA library using Advantage 2 polymerase (Clontech). The RD primer set was designed to amplify a cDNA fragment ∼520 bp in length. The primer set was CCTGCCTGAAGAGTCATCTCTGG, GCCCTCTTCTGTGTGATGGCTTTACT. PCR products were separated and extracted from 2.5% agarose gel and then ligated into pCR2.1-TOPO vector (Invitrogen) for sequence analysis. Negative controls included PCR amplification of reactions minus reverse transcriptase and reactions using the 5′ or 3′ primer by itself.
Quantitative Real Time (qt)-PCR Analysis of Alternative ANK2 Transcripts—Based on the alternative ANK2 transcripts amplified from reverse-transcribed ventricular mRNA and human heart cDNA library, PCR primers that spanned exon boundaries were designed to selectively amplify particular ANK2 transcripts. For example, to amplify the ANK2 transcript that lacks exon 43′, we used a 5′-primer internal to exon 43 and a 3′-primer (24 bp) that spanned the junction of exons 43 (10 bp) and 44 (14 bp). The primer set (GCTCTCCATCATACAAGAAC and CTGTTAGCTCCTTTTCAAAAGCTG) selectively amplifies human ANK2 transcripts containing exon 43′ with an annealing temperature of 62.5 °C and a primer efficiency of 96%. The primer set (GCTCTCCATCATACAAGAAC and TATCGTCTCCCTTTTCAAAAGCTG) selectively amplifies human ANK2 transcripts lacking exon 43′ with an annealing temperature of 62.5 °C and a primer efficiency of 95.6%. Additionally, to test the specificity of exon-spanning primers, we repeated the PCR amplification using 3′-primers to each half of the exon-spanning primer (e.g. first 10 bp corresponding to exon 43 or last 14 bp corresponding to exon 44). No PCR products were amplified when half of the exon-spanning primers were used. Experiments were replicated twice, and each condition was performed in triplicate. Error bars represent standard deviation with a sample set of 3.
Generation of Obscurin C-terminal Domain cDNA Constructs—Obscurin C-terminal domain (CTD), corresponding to amino acid residues 6148-6460 (GenBank™ accession number NM_052843), was PCR-amplified from human heart cDNA library (Clontech) using the primer set (GCGGTACCATGCTGACCACTGGCAAC and GCGAATTCCTAGTTGTGCGTGAGGAT) and ligated into pcDNA3.1+ (Invitrogen) using the restriction sites KpnI and EcoRI. A GST fusion protein of obscurin CTD was PCR-amplified using the primer set (GCGGATCCATGCTGACCACTGGCAAC and GCGAATTCCTAGTTGTGCGTGAGGAT) and ligated into pGEX6p1 (GE Healthcare).
Generation of Ankyrin-B RD Constructs—Alanine-mutagenesis was performed using the QuikChange site-directed mutagenesis kit (Stratagene) to alter residues within ankyrin-B RD consisting of residues 1451-1840. Residues 1778V, 1780K, 1783R, and 1784K in ankyrin-B RD were changed to alanines using the primer set (TACCGCCTAATGATTGCCGCAGTAACCGCCTTTGCCACGGTGTGTCCAT and ATGGACACACCGTGGCAAAGGCGGTTACTGCGGCAATCATTAGGCGGTA). Residues 1745R, 1746R, and 1747R in ankyrin-B RD were changed to alanines using the primer set (ACTACCAGGGTTGTCGCCGCGGCAGTGATTATTCAGGGAandTCCCTGAATAATCACTGCCGCGGCGACAACCCTGGTAGT). Ankyrin-B RD residues 1743-1749 were removed by PCR amplifying the N terminus (upstream of residue 1743) with the primer set (GCGAATTCCCACAGGATGAGCAGGAA and GCACTAGTGGTAGTTACCATCGCCTC) and C terminus (downstream of residue 1749) with the primer set (GCACTAGTATTCAGGGAGACGATATG and GCCTCGAGCTACTCATTGTTGTCCTCTGA). Then both PCR products were ligated into pGEX6p1 using the restriction sites EcoRI, SpeI, and XhoI. Site-directed mutagenesis (Quikchange, Stratagene) was used to generate ankyrin-B R1788W and V1777M. GFP-tagged constructs of AnkB-RD±E43′ (residues 1451-1840) were subcloned into lentiviral vector pCDH1-MCS1 (System Biosciences). Pseudoviral particles were isolated from HEK293TN cells transfected with lentiviral constructs and packaging plasmids (System Biosciences). Viral particles were concentrated using centrifugal filtration devices (Centriplus YM-30, Amicon).
In Vitro Binding Assays—In vitro translated products of obscurin CTD were prepared using the TnT T7-Coupled Reticulate Lysate System (Promega) and labeled with [35S]methionine (GE Healthcare). In vitro translated products were incubated with GST fusion proteins at 4 °C in 500 μl of binding buffer (50 mm Tris, pH 7.4, 1 mm EDTA, 1 mm EGTA, 150 mm NaCl, 0.1% Triton X-100) overnight (for ankyrin-B RD). Binding reactions were washed five times in wash buffer (binding buffer supplemented with 1% Triton X-100 and 500 mm NaCl). Binding reactions were pelleted, resuspended in SDS-sample buffer, separated by SDS-PAGE, and visualized by PhosphorImaging (courtesy of Dr. Michael Welsh, University of Iowa).
Competition Assays—HEK293 cells were transduced with lentiviral GFP-AnkB RD±E43′. Cells were lysed in lysis buffer (150 mm NaCl, 5 mm EDTA, 5 mm EGTA, 1 mm phenylmethylsulfonyl fluoride, and 1× protease inhibitor mixture (Sigma)). Protein lysate from human left ventricular tissue was isolated as described previously (7). 200 μg of human heart lysate was incubated with GST-obscurin CTD conjugated to glutathione-Sepharose (GE Healthcare) overnight at 4 °C in the presence of increasing concentrations of competitor (GFP-AnkB RD or GFP-E43′ RD). Immunocomplexes were pelleted and washed five times in wash buffer, separated by SDS-PAGE, and visualized by chemiluminescence (Supersignal West Pico Chemiluminescent Substrate, Pierce) using antibodies to ankyrin-B (pAb 1.4 μg/ml) and GFP (pAb 3.4 μg/ml).
Primary Cardiomyocyte Cultures and Immunofluorescence—Neonatal hearts from ankyrin-B+/+, ankyrin-B+/-, and ankyrin-B-/- mice were extracted and enzymatically dispersed with trypsin and collagenase, plated on fibronectin-coated glass plates, and maintained in defined media (8). Adult cardiomyocytes were isolated as described (2, 24, 25). Cardiomyocytes were fixed in 2% paraformaldehyde and incubated with antibodies to obscurin (mAb 5H10 at 3 μg/ml, a generous gift of Dr. R. J. Bloch, University of Maryland) (16), B56α (mAb, BD Transduction Laboratories), α-actinin (Sigma), GFP (pAb at 3.4 μg/ml), and ankyrin-B (pAb at 2 μg/ml, mAb at 0.5 μg/ml) (12) in blocking buffer (1× phosphate-buffered saline, 2 mg/ml bovine serum albumin, 0.075% Triton X-100) overnight at 4 °C. Cells were washed and incubated with anti-mouse and anti-rabbit secondary antibodies (Alexa 488 and 568, Molecular Probes) that were visualized on a Zeiss LSM 510 Meta confocal microscope (63 power oil (NA 1.4) for neonatal cardiomyocytes and 40 power water (NA 1.2) for adult cardiomyocytes, pinhole equals 1.0 Airy Disc, room temperature) using Carl Zeiss Imaging software (Carl Zeiss, Inc.). Gain and offset values were held constant when comparing between samples.
Co-immunoprecipitation Assays—Rat hearts were lysed in 4 volumes of lysis buffer plus 1 mm phenylmethylsulfonyl fluoride, and 1× protease inhibitor mixture (Sigma) (7). Lysates (500 μl) were incubated with pAb to obscurin (123A, a generous gift of Dr. R. J. Bloch, University of Maryland) (16) coupled to protein A Affi-gel (Bio-Rad) (1 μg/10 μl Sepharose) overnight at 4 °C. Immunocomplexes were pelleted, washed three times with lysis buffer, separated by SDS-PAGE, and visualized using an alkaline phosphatase enzymatic reaction (Bio-Rad).
RESULTS
Identification of Novel ANK2 Regulatory Domain Exon—The ankyrin-B RD is essential for ankyrin-B function in cardiomyocytes (12). Moreover, nearly 90% of human ANK2 disease variants are located in this domain (2-4). Despite the importance of this domain for ankyrin-B function in vivo, there is no clear evidence for its cellular role. We were therefore interested in identifying novel ankyrin-B RD interacting proteins. Obscurin is a large Rho-GEF protein that organizes the cytoskeletal proteins underlying the sarcomeres of skeletal and cardiac muscle (14-16). Previous studies have shown that obscurin associates with small ankyrin-R isoforms 1.5 and 1.9 via two conserved motifs in the RD (see Fig. 1B) (19-22). Although ankyrin-R and ankyrin-B RDs are structurally similar, canonical 220-kDa ankyrin-B lacks the first of these two binding motifs (Fig. 1B), casting doubt on the possibility of an ankyrin-B/obscurin interaction in vivo.
Together, ANK2 exons 44 and 45 encode a homologous site corresponding to the second of two motifs required for obscurin binding in ankyrin-R (see Fig. 1, B-C). Based on the conservation of this second motif between ankyrin-R and ankyrin-B, we hypothesized that ANK2 genomic sequence might contain previously unidentified exon(s) that encode the first obscurin-binding motif in ankyrin-B. To identify the potential missing ANK2 exon(s), we performed reverse transcriptase PCR on mRNA isolated from left ventricular tissue from human hearts (see “Experimental Procedures”). PCR products were also amplified from a human heart cDNA library. Primer sets were designed against ANK2 exons 43 and 45 (see Fig. 1C). PCR products were gel-extracted, ligated into the vector pCR2.1-TOPO, and sequenced (Fig. 1D). Using this technique, we unexpectedly identified novel ANK2 sequence, now termed exon 43′, following exon 43. Sequencing revealed that exon 43′, consisting of 93 bp, maintains the open reading frame and encode 31 residues (see Fig. 1, E-F). We additionally identified numerous ankyrin-B cDNA clones lacking this 93-bp insertion (see lower band, Fig. 1D).
Analysis of human genomic sequence confirmed the presence of this sequence between ANK2 exons 43 and 44 (BC125235, BC125236, and BX538132). Moreover, the genomic sequence surrounding this cDNA was flanked by canonical acceptor (accag/GAGCTA...) and donor (...ATTCAG/gtaccc) splicing sites (26). As we hypothesized, translation of this novel exon revealed a homologous site similar to the obscurin-binding site 1 on ankyrin-R 1.5 (see Fig. 1E).
Primary Cardiac Ankyrin-B Isoform Contains Exon 43′—Reverse-transcriptase PCR data revealed the presence of two forms of ankyrin-B RD. One form contained exon 43′, whereas the other form lacked this exon. To measure relative mRNA expression levels of these two transcripts, we employed qt-PCR using exon boundary spanning primers to specifically recognize individual transcripts (see “Experimental Procedures” for list of qt-PCR primers) (27). Specifically, to amplify transcripts lacking exon 43′, we used a 3′-primer that spanned the splice junction of exons 43 and 44 (see arrows, Fig. 2A). Negative controls included PCR amplification of reactions minus reverse transcriptase and reactions using individual 5′- or 3′-primers. Moreover, to ensure the selective amplification of specific transcripts, we repeated PCR using each half of the exon-spanning primer individually (data not shown). PCR products were detected only when the full-length exon-spanning primer was used (see Fig. 2B). Annealing temperatures were optimized such that primer set efficiencies fell within the range of 90-110% as determined by standard curves of cycle threshold (Ct) values for a 10-fold dilution series of heart cDNA (1, 1:10, 1:100, and 1:1000) (see “Experimental Procedures” for annealing temperatures, primer set efficiencies, and a complete list of negative controls).
FIGURE 2.
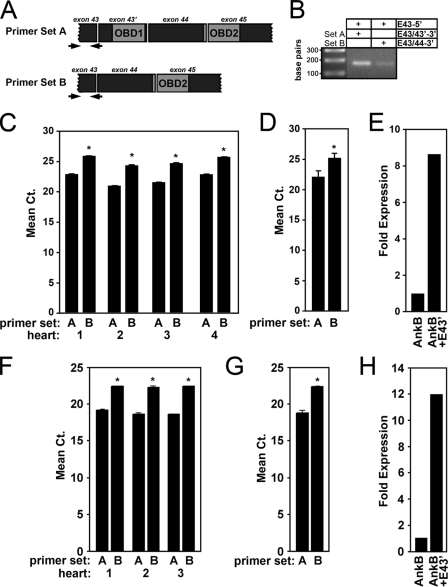
Ankyrin-B + E43′ is the primary ANK2 transcript in vertebrate heart. A, location of exon-exon spanning primers to PCR amplify ANK2 transcripts + exon 43′ (primer set A) and ANK2 transcriptsexon 43′ (primer set B). B, ethidium bromide-agarose gel demonstrating PCR amplification of ANK2 transcripts from mRNA isolated from human left ventricular tissue in the presence of the full-length exon-exon spanning primers (primer set A, 1st column; primer set B, 2nd column). In contrast, no PCR products are detected using half the boundary-spanning primer (data not shown). C, relative mRNA expression in four separate human hearts of ANK2 transcripts + exon 43′ (primer set A) and ANK2 transcriptsexon 43′ (primer set B) expressed as mean cycle threshold (Ct) values. D, average of the Ct values from four hearts (p < 0.05). E, fold expression for mRNA transcripts of human ANK2 ± exon 43′. Note that transcripts containing exon 43′ are expressed ∼8.5-fold greater than transcripts lacking exon 43′. F-H, relative mRNA expression in three mouse hearts of Ank2 ± exon 43′. Note that transcripts containing exon 43′ are expressed 12-fold greater than transcripts lacking exon 43′. Error bars represent standard deviations (n = 3, p < 0.05).
Relative mRNA expression levels of alternative splice variants were evaluated in ventricular tissue from four human and three mouse hearts. Experiments were replicated twice with each condition performed in triplicate. For each heart, results are displayed individually to observe the general trends between transcripts and to discern variability between hearts (Fig. 2, C and F). Additionally, mean data are shown for the human and mouse hearts (Fig. 2, D and G). We performed qt-PCR analysis of the reference gene hypoxanthine-guanine phosphoribosyltransferase to confirm relative cDNA amounts between hearts (data not shown).
Quantitative real time PCR revealed that human cardiac ankyrin-B transcripts that include exon 43′ are significantly more common (∼8.5-fold) than transcripts that exclude this exon (Fig. 2E). Consistent with human data, Ank2 transcripts containing exon 43′ in mouse hearts are expressed at levels ∼12-fold greater than transcripts lacking exon 43′ (Fig. 2, F-H). Together, our findings reveal a novel ANK2 exon (exon 43′) that is the primary ankyrin-B transcript in human and mouse hearts. Moreover, inclusion of this novel sequence in the ankyrin-B polypeptide potentially confers binding activity for the ankyrin-B RD with the cardiac cytoskeletal protein obscurin.
Alternative ANK2 Splicing Regulates Ankyrin-B/Obscurin Interaction—To assess the potential interaction between ankyrin-B and obscurin, we performed in vitro binding assays using a GST fusion protein of ankyrin-B RD ± exon 43′ with obscurin CTD (see Fig. 3A). Minimal obscurin binding activity was observed for ankyrin-B RD lacking exon 43′ (Fig. 3, B-C). In contrast, ankyrin-B RD containing exon 43′ displayed robust binding for obscurin at levels 10-fold greater than ankyrin-B RD lacking this exon (Fig. 3, B-C). These findings strongly suggest that ankyrin-B and obscurin are protein partners in heart and that alternative ANK2 splicing confers unique functional properties to distinct cardiac ankyrin-B isoforms. In fact, these findings establish a potential molecular mechanism by which enhanced binding activity may target specific ankyrin-B isoforms to domains enriched with obscurin, such as the M-line of sarcomeres in striated muscle.
FIGURE 3.
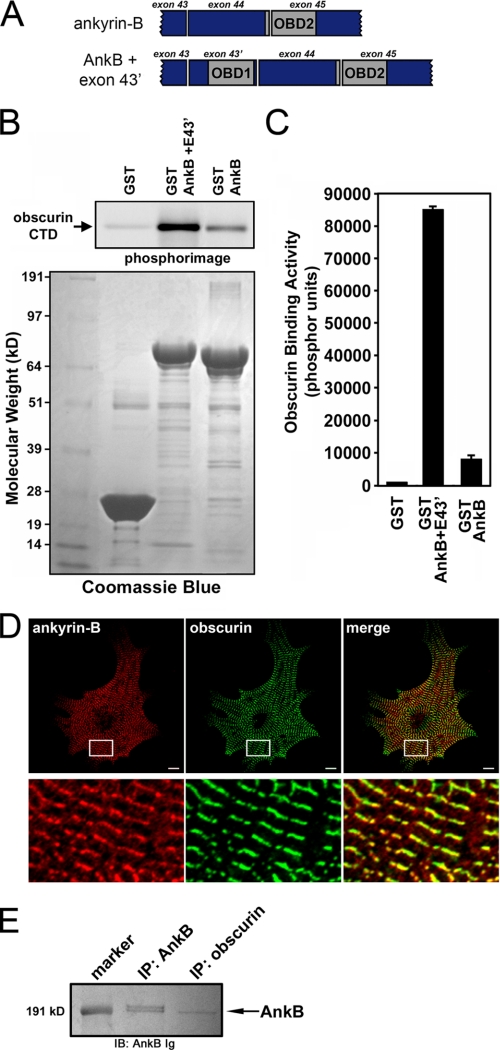
Binding and co-localization of ankyrin-B with obscurin; an association that requires novel obscurin-binding domain 1. A, organization for ankyrin-B RD constructs ± exon 43′. B, enhanced obscurin binding activity for ankyrin-B construct containing both OBD1 and -2. Upper panel, phosphorimage of obscurin CTD labeled with [35S]methionine bound to GST fusion proteins of AnkB RD ± exon 43′. Lower panel, Coomassie Blue stain of the same protein gel that demonstrates equal loading of GST and GST fusion proteins. C, quantification of obscurin binding activity. Error bars represent standard deviations (n = 3). D, upper panels, immunofluorescent localization of ankyrin-B (red), obscurin (green), and their co-localization in neonatal mouse cardiomyocytes. Scale bar is 10 μm. Regions boxed in white are magnified in the lower panels. E, ankyrin-B immunoblot (IB) demonstrating co-immunoprecipitation (IP) of ankyrin-B from rat heart lysate with antibodies to ankyrin-B or obscurin.
Ankyrin-B and Obscurin Are Binding Partners in Cardiomyocytes—To assess the potential in vivo relevance of an ankyrin-B/obscurin interaction, we determined the localization of ankyrin-B and obscurin in primary neonatal mouse cardiomyocytes using immunofluorescence and confocal microscopy (Fig. 3D). In agreement with prior reports, ankyrin-B is primarily localized in a striated pattern over neonatal cardiomyocyte M-lines (Fig. 3D) (2). Consistent with an interaction in cardiomyocytes, co-labeling of myocytes with obscurin Ig revealed similar pattern of M-line striations for obscurin (Fig. 3D).
Ankyrin-B RD containing exon 43′ directly associates with obscurin CTD (Fig. 3B). We next assessed whether ankyrin-B and obscurin were protein partners in vivo. As shown in Fig. 3E, ankyrin-B co-immunoprecipitated with obscurin from detergent-soluble fractions of rat heart lysate using an antibody to obscurin. Therefore, these findings strongly support an in vivo interaction between ankyrin-B and obscurin.
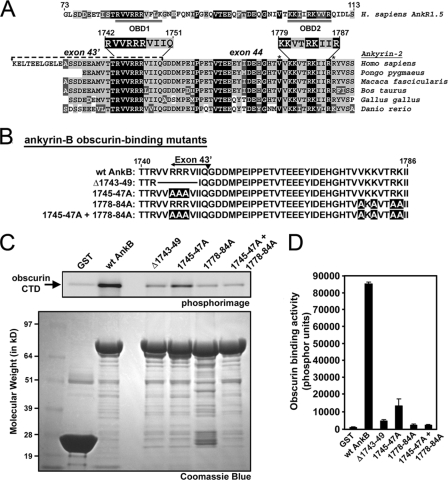
Identification of Structural Requirements for Obscurin-binding Sites on Ankyrin-B—Obscurin binds small ankyrin-R-1.5 (sAnk1.5) (19-21, 23). Bagnato et al. (20) proposed that the binding site consisted of sAnk1.5 residues Thr-99, Lys-101, Arg-104, and Lys-105 (Fig. 4A). A homologous site is present in ankyrin-B residing within residues 1779-1787 (Fig. 4A, OBD2). This region forms an amphipathic α-helix that is important for normal ankyrin-B function and Hdj-1 binding (28). Furthermore, missense variants located in this region (V1777M and R1788W) are human loss-of-function mutations associated with ankyrin-B syndrome (3, 4). This α-helical region also corresponds to an evolutionarily conserved module of 18 amino acids that is found in triplicate in ankyrin-B RD and is hypothesized to mediate obscurin binding activity (29). A recent study described the amino acid sequences comprising the two obscurin-binding sites in sAnk1.5 (21). Analysis of translated genomic sequences and expressed sequence tags for a number of organisms demonstrates that the putative first obscurin-binding site in ankyrin-B is highly conserved between human, monkey, cow, chicken, and fish (Fig. 4A).
FIGURE 4.
Mutations to OBD1 and -2 disrupt ankyrin-B/obscurin interactions. A, alignment of ankyrin-B RD from various species and human small AnkR1.5. Accession numbers for species-specific ankyrin-B transcripts are Q01484 (Homo sapiens), CR773816 (Pongo pygmaeus), CJ448751 (Macaca fascicularis), DV930431 (Bos taurus), AL588116 (Gallus gallus), and EB895593 (Danio rerio). All of the non-human sequences are derived from translated genomic sequences and have been confirmed in expressed sequence tags. Background color corresponds to the level of amino acid homology: black, identical; light gray, conserved; and dark gray, similar. For AnkR1.5, the double-underlined regions represent obscurin-binding domains 1 and 2 (21). The location of ANK2 exon 43′ is indicated in H. sapiens ankyrin-B. Asterisks denote location of ankyrin-B variants associated with human arrhythmia. B, diagram of ankyrin-B RD obscurin-binding mutants. C, mutations to specific residues in ankyrin-B OBD1 and -2 disrupt ankyrin-B/obscurin interactions. Upper panel, phosphorimage of obscurin CTD labeled with [35S]methionine bound to GST fusion proteins of ankyrin-B RD mutants. Lower panel, Coomassie Blue stain of the corresponding protein gel that demonstrates equal loading of GST fusion proteins. D, quantification of obscurin binding activity (phosphor units). Error bars represent standard deviations (n = 3).
Based on the similarity between the obscurin-binding motifs on sAnk1.5 and the newly identified site on ankyrin-B (encoded by exon 43′), we hypothesized that two regions in ankyrin-B (amino acids 1745-1749 and 1778-1784) were important for mediating interactions with obscurin. We generated a series of GST fusion ankyrin-B RD proteins with mutations in the two putative obscurin-binding sites (Fig. 4B). To disrupt the first binding site in exon 43′, we generated two constructs: one removed residues 1743-1749 (Δ1743-49) and the other converted the arginine residues at 1745-1747 to alanine residues (1745-47A). To disrupt the second binding site, we performed site-directed alanine mutagenesis to change the residues V1778A, K1780A, R1783A, and K1784A (1778-84A). Finally, we generated a construct that disrupted both putative obscurin-binding domains (1745-47A + 1778-84A).
Using mutant GST fusion ankyrin-B RD proteins, we performed in vitro binding assays with human obscurin CTD. As illustrated by Coomassie Blue stain, equal amounts of GST fusion proteins were incubated with [35S]methionine-labeled obscurin CTD (Fig. 4C, lower panel). As determined by PhosphorImager analysis, all mutations to the obscurin-binding sites reduced the interaction between obscurin and ankyrin-B (Fig. 4C, upper panel). The 1745-47A mutant retained residual binding activity for obscurin, similar to the level observed with ankyrin-B RD lacking exon 43′ (Fig. 3, B-C). The ankyrin-B Δ1743-49 mutant that removed residues flanking the three arginines further attenuated obscurin binding (Fig. 4, C and D). Interestingly, the 1778-84A mutant virtually abolished all obscurin binding. In fact, mutation of both obscurin-binding sites had no greater effect on loss of obscurin binding than mutating the second site alone. Together, our new findings demonstrate that ankyrin-B and obscurin are binding partners in vivo, and this binding is mediated by alternative splicing of exon 43′. Moreover, our findings demonstrate that vertebrate heart expresses two unique isoforms of ankyrin-B with unique functional properties (± obscurin-binding) based on alternative splicing. Finally, our data define the obscurin-binding sites within ankyrin-B as residues 1743-1749 and 1778-1784.
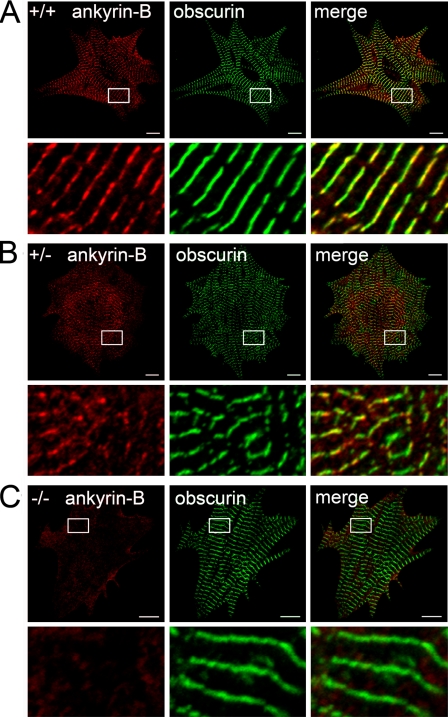
Obscurin Targeting Is Ankyrin-B-independent—Ankyrin-B is critical for the subcellular targeting of Na+/Ca2+ exchanger, Na/K-ATPase, InsP3R, and PP2A in heart (6-8, 10, 30). To determine whether ankyrin-B is also required for the proper subcellular targeting of obscurin, we evaluated obscurin localization by immunofluorescence in cardiomyocytes lacking ankyrin-B (Fig. 5). Neonatal cardiomyocytes were used because ankyrin-B-/- mice die at postnatal day 1 (1). Cardiomyocytes lacking 50% of ankyrin-B expression (ankyrin-B+/- myocytes) displayed no alteration in obscurin expression or localization compared with wild-type myocytes (Fig. 5, A and B). In fact, obscurin expression and localization were unaffected in neonatal cardiomyocytes completely lacking ankyrin-B expression (ankyrin-B-/- cardiomyocytes, Fig. 5C). Therefore, unlike other cardiac ankyrin-B protein partners, obscurin localization in primary cardiomyocytes is independent of ankyrin-B expression. These unexpected results suggest the exciting possibility that obscurin may represent the first protein that targets ankyrin-B protein complexes to specific membrane domains in cardiomyocytes.
FIGURE 5.
Ankyrin-B is not required for obscurin targeting in native cardiomyocytes. Subcellular localization of ankyrin-B and obscurin in ankyrin-B wild-type (A), ankyrin-B+/- (B), and ankyrin-B-/- cardiomyocytes (C). Scale bar represents 10 μm. Lower panels represent magnified views of the areas boxed in white. Note that loss of ankyrin-B does not affect the expression/localization of obscurin.
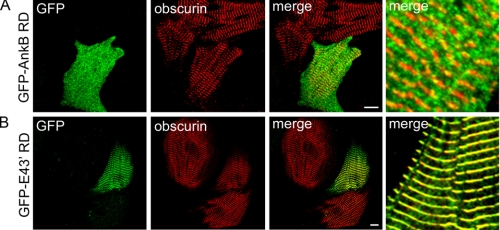
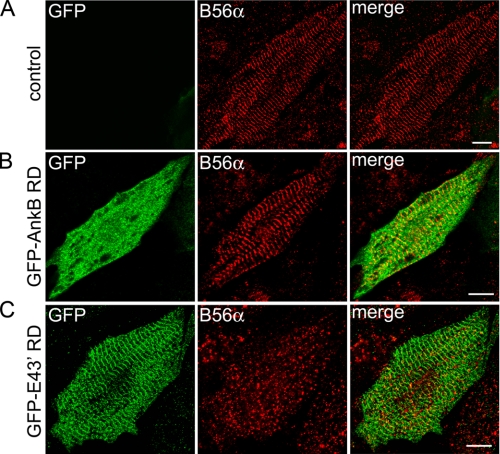
ANK2 Exon 43′ Targets Ankyrin-B RD to the Cardiac M-line—We evaluated the targeting of ankyrin-B RD by obscurin to the M-line using GFP-ankyrin-B RD (residues 1451-1840) containing or lacking E43′. Specifically, ankyrin-B RD±E43′ cDNAs fused in-frame with GFP were subcloned into the lentiviral vector pCDHI-MCS1 (System Biosciences). Pseudoviral particles were generated and concentrated to equal titer levels. Neonatal rat cardiomyocytes were infected with each viral construct, and immunofluorescent localization of GFP was evaluated by confocal microscopy. Myocytes infected with GFP-ankyrin-B RD lacking E43′ (GFP-AnkB RD) displayed a diffuse cytoplasmic distribution in contrast to M-line-localized obscurin (Fig. 6A, red). In striking contrast, GFP-ankyrin-B RD E43′ (GFP E43′ RD) displayed tightly localized staining to the primary cardiomyocyte M-line that co-localized with endogenous obscurin immunostaining (Fig. 6B). These findings clearly demonstrate that the association of ankyrin-B with obscurin via the two obscurin-binding domains is sufficient and required for targeting of ankyrin-B RD to the cardiac M-line. Furthermore, the GFP-ankyrin-B RD E43′ construct may act in a dominant-negative manner to inhibit M-line targeting of endogenous ankyrin-B, thereby establishing a potential tool to evaluate the role of obscurin binding for ankyrin-B targeting and function to/at the cardiac M-line.
FIGURE 6.
OBD1 regulates the targeting of ankyrin-B RD to the M-line. A, lentiviral expression of GFP-AnkB RD lacking exon 43′ that contains OBD1 is diffusely cytoplasmic in neonatal rat cardiomyocytes. Note the expression of this construct by immunofluorescent detection of GFP does not co-localize with obscurin (red) at M-lines (merge). B, lentiviral expression of GFP-AnkB RD with exon 43′ (GFP-E43′ RD) co-localizes with obscurin at M-lines (merge). Scale bar represents 10 μm.
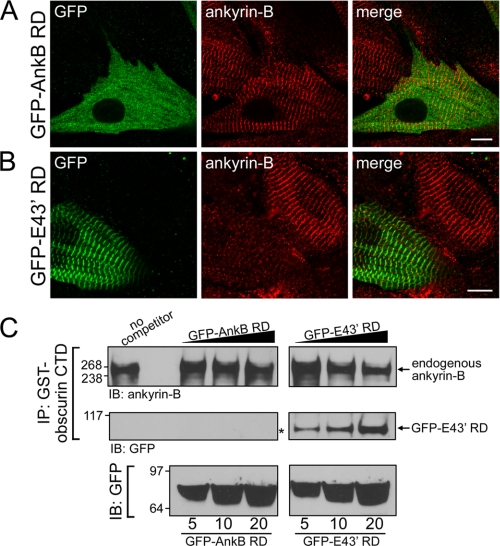
Obscurin Binding Is Required for Ankyrin-B E43′ M-line Targeting in Myocytes—We tested the requirement of obscurin binding for endogenous ankyrin-B M-line targeting using viral expression of GFP-ankyrin-B RD and GFP-ankyrin-B RD E43′ in neonatal rat cardiomyocytes. Specifically, we evaluated the ability of each viral construct to compete endogenous ankyrin-B expression from the cardiomyocyte M-line. Confocal imaging revealed no difference in expression or localization of endogenous ankyrin-B in nontransduced myocytes compared with GFP-ankyrin-B RD transduced myocytes (Fig. 7A). Expression of endogenous ankyrin-B was evaluated using an affinity-purified monoclonal ankyrin-B Ig that does not recognize RD sequence. In contrast, expression of GFP-ankyrin-B RD E43′ significantly reduced, and in most cases abolished, endogenous ankyrin-B expression at the cardiomyocyte M-line (Fig. 7B, note difference in endogenous ankyrin-B staining in infected versus uninfected myocyte). These data strongly implicate obscurin for endogenous ankyrin-B M-line targeting. Moreover, these results suggest that GFP-ankyrin-B RD E43′ acts in a dominant-negative manner to compete with endogenous ankyrin-B/obscurin interactions at the M-line of primary cardiomyocytes.
FIGURE 7.
Ankyrin-B RD with exon 43′ inhibits association of endogenous ankyrin-B with obscurin. A, immunolocalization of viral GFP-AnkB RD (lacking OBD1) with endogenous ankyrin-B. Expression of endogenous ankyrin-B was detected using the ankyrin-B mAb that was generated against SBD; therefore, this antibody will not recognize virally expressed GFP-AnkB RD. GFP-AnkB RD is cytoplasmic and does not co-localize with endogenous ankyrin-B at the M-line. B, immunolocalization of viral ankyrin-B containing OBD1 and -2 (GFP-E43′ RD) with endogenous ankyrin-B. Viral expression of GFP-E43′ RD decreases localization of endogenous ankyrin-B at the M-line. Scale bars represent 10 μm. C, immunoblot (IB) analysis of endogenous ankyrin-B associated with GST-obscurin CTD in the presence of competitors GFP-AnkB RD or GFP-E43′ RD. Heart lysate was incubated with a limited amount of GST-obscurin CTD and varying amounts of competitors (5, 10, and 20 μg). Immunoprecipitated (IP) complexes were probed for endogenous ankyrin-B (upper panels) and GFP (lower panels). Note that the level of endogenous ankyrin-B associated with obscurin CTD decreases in the presence of increasing amounts of GFP-E43′ RD (upper right panel) but not with GFP-AnkB RD (upper left panel). Correspondingly, there is an increase in GFP-E43′ RD associated with obscurin (lower right panel) as the amount of this competitor is increased (bottom right panel). In contrast, GFP-AnkB RD does not associate with obscurin (lower left panel, denoted by asterisk). Therefore, the association of endogenous ankyrin-B with obscurin is unaffected by GFP-AnkB RD (upper left panel).
Our findings at the single myocyte level (Fig. 7A and B) predict that expression of GFP-ankyrin-B RD E43′, but not GFP-ankyrin-B RD, would compete with endogenous ankyrin-B for obscurin interactions in biochemical assays. To test this hypothesis, we generated purified and immobilized GST-obscurin CTD on glutathione beads. The beads were incubated with detergent-soluble lysate from human left ventricle, and endogenous cardiac ankyrin-B association was assessed by immunoblot. As predicted from co-immunoprecipitation assays, GST-obscurin CTD strongly associates with endogenous ankyrin-B from human heart (Fig. 7C, top left). In parallel, we performed the binding experiments in the presence of increasing concentrations of GFP-ankyrin-B RD or GFP-ankyrin-B RD E43′ (generated by lentiviral expression in HEK293 cells). As shown in Fig. 7C, co-incubation of GST-obscurin CTD and cardiac lysates with GFP-ankyrin-B RD had no effect on obscurin/ankyrin-B binding activity (at all concentrations tested). In contrast, the identical experiment utilizing GFP-ankyrin-B RD E43′ showed a concentration-dependent decrease in association between GST-obscurin CTD and endogenous ankyrin-B (Fig. 7C, right). These data demonstrate that ankyrin-B RD E43′, but not ankyrin-B RD, associates with obscurin and competes with endogenous ankyrin-B for obscurin binding. Together, our immunofluorescence and biochemical findings demonstrate that obscurin binding is required for targeting ankyrin-B to the M-line of primary cardiomyocytes. Moreover, these studies identify a powerful new tool to dissect the function of obscurin/ankyrin-B interactions at the cardiac M-line.
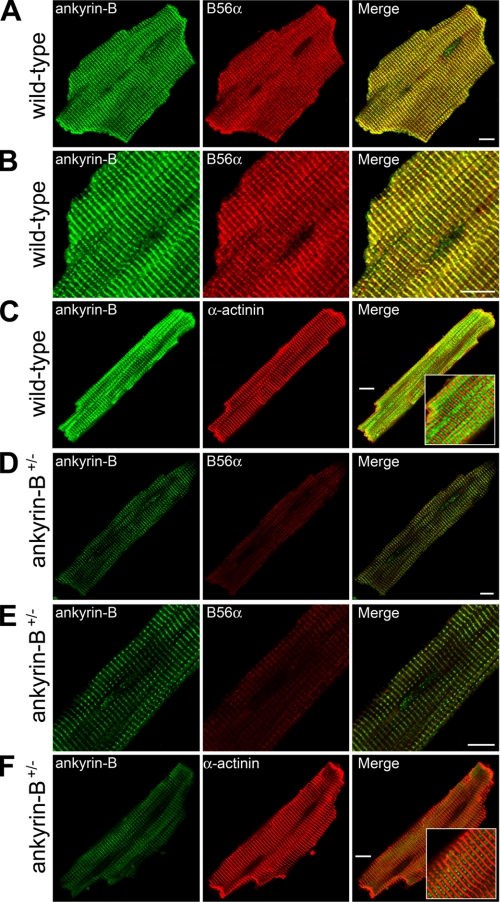
Obscurin-Ankyrin-B Interactions Are Required for Targeting B56α to the Cardiac M-line—We previously identified the regulatory subunit of protein phosphatase 2A (B56α) as a direct binding partner of ankyrin-B spectrin-binding domain in cardiomyocytes (see Fig. 1A) (10). Moreover, ankyrin-B co-immunoprecipitates the entire PP2A complex from cardiac lysates (10). Ankyrin-B and B56α were also found to co-localize over the M-lines of both neonatal and adult cardiomyocytes (10). To address whether obscurin is required for the subcellular targeting of ankyrin-B-associated B56α to the M-lines of cardiomyocytes, we first investigated the requirement of ankyrin-B expression for B56α M-line expression in adult cardiomyocytes. As expected, ankyrin-B and B56α are co-localized at the M-line of adult mouse cardiomyocytes (Fig. 8, A and B) as assessed by co-staining experiments with α-actinin (Fig. 8C, resident Z-line protein). In addition, adult cardiomyocytes derived from mice heterozygous for a null mutation in ankyrin-B (ankyrin-B+/-) display an approximate 50% reduction in ankyrin-B levels at the cardiac M-line (Fig. 8, D-E). These myocytes also display significant reduction in B56α M-line expression (Fig. 8, D-E). Analysis of α-actinin (Fig. 8F) and other critical myocyte structural or membrane proteins (2) revealed no difference in staining, thereby confirming that the prepared myocytes were viable. These data demonstrate a requirement of ankyrin-B expression for B56α M-line localization in adult cardiomyocytes.
FIGURE 8.
Ankyrin-B is required for B56α M-line targeting in primary adult cardiomyocytes. Subcellular localization of ankyrin-B and PP2A regulatory subunit B56α in ankyrin-B wild-type (A and B) and ankyrin-B+/- (D and E) adult cardiomyocytes. Note that loss of ankyrin-B results in clear loss of M-line localization of B56α. C and F, α-actinin staining was used to confirm cell viability following Langendorff isolation of myocytes (note that loss of ankyrin-B does not affect α-actinin expression or localization). Scale bars represent 10 μm.
Finally, we tested the requirement of obscurin/ankyrin-B interactions for B56α targeting in primary cardiomyocytes. Cardiomyocytes infected with either GFP-ankyrin-B RD or GFP-ankyrin-B RD E43′ dominant-negative virus were evaluated for B56α M-line expression/localization. Importantly, neither viral ankyrin-B RD construct contains the B56α-binding site previously identified in the spectrin-binding domain (10). As observed previously, GFP-ankyrin-B RD was expressed throughout the cytoplasm (Fig.9B), and endogenous ankyrin-B levels were unchanged (see Fig. 7A). Moreover, myocytes expressing GFP-ankyrin-B RD displayed no difference in B56α expression or distribution when compared with untreated myocytes (Fig. 9, A-B). In contrast, we observed a striking reduction in B56α M-line targeting in myocytes infected with GFP-ankyrin-B RD E43′ (Fig. 9C). In fact, GFP-ankyrin-B RD E43′ expression at the M-line nearly eliminated B56α M-line expression. These data support that obscurin binding is critical for the targeting of ankyrin-B to the cardiac M-line (Fig. 8B). Moreover, our data support a role for obscurin/ankyrin-B interactions in the M-line targeting of B56α, a key cardiac signaling molecule.
FIGURE 9.
Ankyrin-B/obscurin interaction is required for B56α M-line targeting in primary cardiomyocytes. Control cardiomyocytes (A) or cardiomyocytes transduced with equal multiplicities of infection of GFP-ankyrin-B RD (B) or GFP-ankyrin-B RD + E43′ (GFP-E43′ RD) (C) were immunostained with GFP and B56α-specific Igs and analyzed by confocal microscopy. Note that M-line expression of ankyrin-B associated protein B56α is reduced by GFP-E43′ RD (C) but not by GFP-AnkB RD (B). Scale bars represent 10 μm.
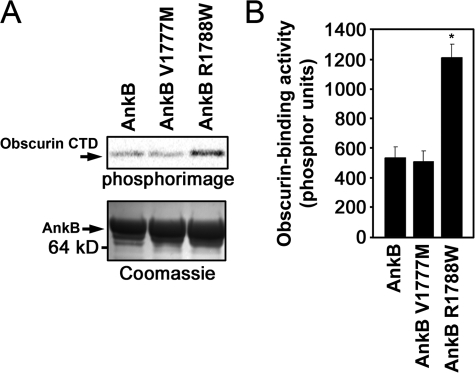
Ankyrin-B Human Arrhythmia Variant Modulates Ankyrin-B/Obscurin Interaction—Human ANK2 loss-of-function variants are associated with cardiac dysfunction and potentially fatal arrhythmia (2-4). To date, the molecular mechanisms underlying these variants are unresolved. Interestingly, two previously identified human ANK2 loss-of-function variants, V1777M and R1788W (4), are located in close proximity to the second obscurin-binding domain in ankyrin-B (see Figs. 1A and 4A). To assess whether these human mutations affect obscurin binding, we engineered the two disease variants into the ankyrin-B RD for binding assays with obscurin. Introduction of the two variants into ankyrin-B RD E43′ revealed no significant change in obscurin binding (data not shown). Consistent with earlier findings, ankyrin-B RD displayed minimal binding activity for obscurin (Fig. 10A). Similarly, ankyrin-B RD V1777M displayed minimal obscurin binding activity. In contrast, ankyrin-B RD R1788W displayed enhanced obscurin binding activity (∼2-fold) (Fig. 10, A and B). Evaluation of a recently identified ANK2 polymorphism in this region (S1791P) (31) revealed no difference in obscurin binding activity (data not shown).
FIGURE 10.
Ankyrin-B human arrhythmia disease mutation R1788W increases ankyrin/obscurin binding activity. A, ankyrin-B human mutation R1788W associated with arrhythmia displays increased binding activity for obscurin when introduced into ankyrin-B RD. Ankyrin-B mutation V1777M associates with obscurin similar to wild-type ankyrin-B RD. Upper panel, phosphorimage of obscurin CTD labeled with [35S]methionine bound to GST fusion proteins of ankyrin-B RD containing the missense variants V1777M or R1788W. Lower panel, Coomassie Blue stain of the corresponding protein gel that demonstrates equal loading of GST fusion proteins. B, quantification of obscurin binding activity (phosphor units). Error bars represent standard deviation (n = 3, p < 0.05).
DISCUSSION
Previous reports demonstrate a critical role of the ankyrin-B RD for ankyrin-B-dependent function in cardiomyocytes. For example, ankyrin-B lacking the RD is insufficient to rescue abnormal ion channel targeting and Ca2+ transients in ankyrin-B null or heterozygous cardiomyocytes (12). Moreover, eight of nine reported human ankyrin-B loss-of-function mutations associated with human arrhythmia reside within ankyrin-B RD (2-4). Despite the abundance of data implicating an essential role for RD in normal ankyrin-B function, its specific function has yet to be resolved.
Here we demonstrate that alternative splicing of the ankyrin-B RD modulates ankyrin-B M-line localization and function in primary cardiomyocytes. Novel ankyrin-B exon 43′ mediates interaction of ankyrin-B with the large Rho-GEF obscurin. Obscurin interacts with ankyrin-B in vivo and in vitro, and is co-localized with ankyrin-B at the M-line of neonatal and adult cardiomyocytes. Interestingly, our data demonstrate that ankyrin-B interaction with obscurin requires two unique motifs in the ankyrin-B RD. Our data demonstrate that obscurin acts upstream of ankyrin-B-dependent protein targeting, as ankyrin-B null cardiomyocytes display normal obscurin organization. In fact, obscurin binding activity is critical for recruitment of the ankyrin-B RD to the cardiac M-line. Specifically, exogenous expression of GFP-ankyrin-B RD dominant-negative virus containing both obscurin-binding motifs is sufficient to compete with endogenous ankyrin-B for obscurin binding at the cardiac M-line. Finally, our data demonstrate that the ankyrin-B/obscurin interaction is critical for recruitment of protein phosphatase 2A (PP2A) to the cardiac M-line of cardiomyocytes. Together, these new data provide the first evidence for the molecular basis of ankyrin-B and protein phosphatase 2A targeting and function at the cardiac M-line. Moreover, these data demonstrate that the ANK2 gene is subject to alternative splicing that gives rise to unique ankyrin-B polypeptides with diverse roles in cardiac function.
Our new findings offer additional insight into the complex mechanisms underlying ankyrin targeting in metazoans. Specifically, our data suggest that multiple targeting cues direct the subcellular localization of unique ankyrin-B polypeptides in cardiomyocytes. In cardiomyocytes, ankyrin-B is localized to both the Z-line (overlying the transverse tubule and sarcoplasmic reticulum) and the M-line (Fig. 3D). Interestingly, in adult cardiomyocytes the M-line population of ankyrin-B is more abundant than the Z-line population (2). Our new data demonstrate that obscurin recruits ankyrin-B with exon 43′, and subsequently B56α to the M-line of neonatal and adult cardiomyocytes. Correspondingly, human ANK2 transcripts containing exon 43′ are expressed 8-fold greater than transcripts lacking this exon (Fig. 2E). In mouse, Ank2 transcripts containing exon 43′ are 12-fold more abundant than those lacking this exon (Fig. 2H). Obscurin is an 800-kDa protein localized to the M-line of sarcomeres where it regulates myofibrillogenesis, including A-band formation, myosin incorporation, and M-line alignment (13-18). The OBSCN gene encodes the 800-kDa isoform in addition to other obscurin-related gene products, including the 900-kDa isoform and the smaller obscurin myosin light chain kinases (32). The structural requirements in obscurin for ankyrin binding activity are only present in the 800-kDa isoform (32). Interestingly, similar to ankyrin-B, this 800-kDa isoform is preferentially localized to the M-line of striated muscle tissue (33, 34). Based on these findings and our new data, we hypothesize that the primary population of ankyrin-B is targeted/organized over the M-line through interactions with 800-kDa obscurin.
The molecular cues governing the targeting of ankyrin-B lacking exon 43′ are currently unknown. Previous experiments in neonatal cardiomyocytes demonstrate that full-length ankyrin-B lacking E43′ is expressed at the M-line (6). A growing body of literature supports that targeting and stabilization of ankyrin polypeptides at distinct membrane domains are most likely mediated by multiple molecular cues, including cell adhesion molecules and cytoskeletal elements such as neurofascin and β-spectrin, respectively (9, 35-42). Although the ankyrin-B regulatory domain lacking E43′ was not targeted to the M-line, we cannot exclude the possibility that additional targeting/stabilization mechanisms interact with the full-length ankyrin-B polypeptide (minus E43′) to bring about its M-line localization. Future studies will be necessary to identify the molecular mechanisms governing the targeting of this ankyrin-B polypeptide (lacking E43′) in mature cardiomyocytes with a fully developed membrane structure (neonatal cardiomyocytes lack Z-line/transverse-tubule network).
In analogous situations, alternative splicing has been shown to regulate the subcellular localization of other ankyrin gene products. For example, multiple isoforms of ankyrin-R have been characterized in erythrocytes (43-45), brain (46, 47), kidney (48, 49), and muscle (50-53). In kidney cells, a 195-kDa isoform of ankyrin-R is distinctly localized to the Golgi complex (48). In both cardiac and skeletal muscles, small ankyrin-R isoforms (∼20 kDa) associate with the sarcoplasmic reticulum via unique N-terminal domains that form transmembrane-spanning domains (20, 23, 50, 53). Similar to ankyrin-R, there is an alternative isoform of ankyrin-G (119 kDa) in kidney and muscle that localizes to the Golgi complex (54). In addition, small alternative isoforms of ankyrin-G are selectively expressed at costameres in skeletal muscle (55, 56). In brain, two large alternative isoforms of ankyrin-G (270 and 480 kDa) express a unique serine-rich domain that facilitates the subcellular localization of these isoforms to axon initial segments (57).
Bennett and co-workers (11) previously demonstrated an intra-molecular interaction between the ankyrin-B RD and the membrane-binding domain by yeast two-hybrid and purified protein binding assays. This interaction was necessary for ankyrin-B-dependent targeting of InsP3 receptor in cardiomyocytes (11). Ankyrin-B RD intra-molecular interactions may play a key role in the regulation of ankyrin-B inter-molecular interactions in heart. Studies addressing the association of the structurally similar canonical 210-kDa ankyrin-R with the anion exchanger and β-spectrin suggest that intra-molecular interactions between the functional domains negatively regulate inter-molecular interactions. Specifically, an alternative ankyrin-R isoform lacking 161 residues in the C-terminal domain exhibits increased affinity for the anion exchanger and β-spectrin (43, 58). An obvious future direction will be to determine whether ankyrin-B intra-molecular interactions regulate ankyrin-B/obscurin inter-molecular interactions.
Myocytes lacking obscurin expression reveal a critical role for the large Rho-GEF in myofibrillogenesis (14, 16). Although this experimental system intuitively appears as an ideal model to evaluate the importance of obscurin for ankyrin-B localization and function, multiple groups (including Bloch and co-workers (14, 16)) have demonstrated that obscurin-deficient myocytes are devoid of critical myocyte components, including both M-line and A-band protein/structural components (by immunofluorescence/electron microscopy). Therefore, based on these caveats it would be difficult (if not impossible) to conclusively differentiate direct effects of obscurin loss on ankyrin-B targeting and function versus indirect effects because of defects in myofibrillogenesis using the obscurin-deficient myocyte. On the other hand, our new results demonstrate that the GFP-ankyrin-B RD E43′ virus will serve as an unexpected and powerful tool to evaluate the role of ankyrin-B targeting and ankyrin-B/obscurin function at the cardiomyocyte M-line. Specifically, future experiments using this virus will be important for identifying M-line-specific ankyrin-B interacting proteins as well as characterizing the role of obscurin- and ankyrin-B-dependent M-line phospho-regulation by the B56α-PP2A phosphatase complex.
Protein phosphatase 2A is a serine/threonine phosphatase comprised of a structural A subunit, a catalytic C subunit, and a regulatory B subunit (59). Although the A and C subunits constitute the core enzyme, the B subunit is responsible for substrate specificity, subcellular localization, and enzymatic activity of the entire PP2A holoenzyme (59). Although PP2A function at cardiac T-tubule and sarcolemmal domains has been extensively explored (60-64), it is primarily expressed at the cardiac M-line (Fig. 8, A and B) (10) where its function has not been characterized. Of particular relevance to our findings, transgenic mice that overexpress a mutant PP2A scaffolding subunit (PP2A/A) that cannot bind B subunits display mis-localized cardiac PP2A and dilated cardiomyopathy (65). At the M-line, obscurin/ankyrin-B sequestered PP2A may regulate the effects of the titin kinase domain that is also located at the M-line and associated with the assembly, maintenance, and re-organization of the sarcomere (66-71). As PP2A activity is increased in heart failure (72), and overexpression of PP2A catalytic subunit (PP2A/c) results in dilated cardiomyopathy accompanied by reduced contractility (73), it will be important in future studies to explore the role of obscurin/ankyrin-B interactions for local M-line PP2A-associated phospho-regulation in normal and diseased hearts.
Finally, our data demonstrate that a human ankyrin-B loss-of-function disease mutation (R1788W) significantly increases the binding activity of ankyrin-B lacking exon 43′ (Fig. 10). Additional future experiments will be necessary to determine the functional significance of these findings.
Acknowledgments
We acknowledge R. J. Bloch (University of Maryland) for the generous gift of obscurin antibodies, Jordan Miller (University of Iowa) for coordinating the procurement of human hearts, Madhu Singh for helpful advice on qt-PCR, and Michael Welsh (University of Iowa) for use of the PhosphorImager.
This work was supported, in whole or in part, by National Institutes of Health Grants HL084583 and HL083422 (to P. J. M.). This work was also supported by Pew Scholars Trust (to P. J. M.). The costs of publication of this article were defrayed in part by the payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 U.S.C. Section 1734 solely to indicate this fact.
Footnotes
The abbreviations used are: InsP3, inositol 1,4,5-trisphosphate; GFP, green fluorescent protein; GST, glutathione S-transferase; mAb, monoclonal antibody; pAb, polyclonal monoclonal antibody; qt-PCR, quantitative real time-PCR; RD, regulatory domain; CTD, C-terminal domain.
References
- 1.Scotland, P., Zhou, D., Benveniste, H., and Bennett, V. (1998) J. Cell Biol. 143 1305-1315 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Mohler, P. J., Schott, J. J., Gramolini, A. O., Dilly, K. W., Guatimosim, S., duBell, W. H., Song, L. S., Haurogne, K., Kyndt, F., Ali, M. E., Rogers, T. B., Lederer, W. J., Escande, D., Le Marec, H., and Bennett, V. (2003) Nature 421 634-639 [DOI] [PubMed] [Google Scholar]
- 3.Mohler, P. J., Splawski, I., Napolitano, C., Bottelli, G., Sharpe, L., Timothy, K., Priori, S. G., Keating, M. T., and Bennett, V. (2004) Proc. Natl. Acad. Sci. U. S. A. 101 9137-9142 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Mohler, P. J., Le Scouarnec, S., Denjoy, I., Lowe, J. S., Guicheney, P., Caron, L., Driskell, I. M., Schott, J. J., Norris, K., Leenhardt, A., Kim, R. B., Escande, D., and Roden, D. M. (2007) Circulation 115 432-441 [DOI] [PubMed] [Google Scholar]
- 5.Davis, J. Q., and Bennett, V. (1994) J. Biol. Chem. 269 27163-27166 [PubMed] [Google Scholar]
- 6.Mohler, P. J., Davis, J. Q., Davis, L. H., Hoffman, J. A., Michaely, P., and Bennett, V. (2004) J. Biol. Chem. 279 12980-12987 [DOI] [PubMed] [Google Scholar]
- 7.Mohler, P. J., Davis, J. Q., and Bennett, V. (2005) PLoS Biol. 3 e423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Cunha, S. R., Bhasin, N., and Mohler, P. J. (2007) J. Biol. Chem. 282 4875-4883 [DOI] [PubMed] [Google Scholar]
- 9.Dubreuil, R. R. (2006) J. Membr. Biol. 211 151-161 [DOI] [PubMed] [Google Scholar]
- 10.Bhasin, N., Cunha, S. R., Mudannayake, M., Gigena, M. S., Rogers, T. B., and Mohler, P. J. (2007) Am. J. Physiol. 293 H109-H119 [DOI] [PubMed] [Google Scholar]
- 11.Abdi, K. M., Mohler, P. J., Davis, J. Q., and Bennett, V. (2006) J. Biol. Chem. 281 5741-5749 [DOI] [PubMed] [Google Scholar]
- 12.Mohler, P. J., Gramolini, A. O., and Bennett, V. (2002) J. Biol. Chem. 277 10599-10607 [DOI] [PubMed] [Google Scholar]
- 13.Benian, G. M., Tinley, T. L., Tang, X., and Borodovsky, M. (1996) J. Cell Biol. 132 835-848 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Borisov, A. B., Sutter, S. B., Kontrogianni-Konstantopoulos, A., Bloch, R. J., Westfall, M. V., and Russell, M. W. (2006) Histochem. Cell Biol. 125 227-238 [DOI] [PubMed] [Google Scholar]
- 15.Kontrogianni-Konstantopoulos, A., Catino, D. H., Strong, J. C., Randall, W. R., and Bloch, R. J. (2004) Am. J. Physiol. 287 C209-C217 [DOI] [PubMed] [Google Scholar]
- 16.Kontrogianni-Konstantopoulos, A., Catino, D. H., Strong, J. C., Sutter, S., Borisov, A. B., Pumplin, D. W., Russell, M. W., and Bloch, R. J. (2006) FASEB J. 20 2102-2111 [DOI] [PubMed] [Google Scholar]
- 17.Borisov, A. B., Kontrogianni-Konstantopoulos, A., Bloch, R. J., Westfall, M. V., and Russell, M. W. (2004) J. Histochem. Cytochem. 52 1117-1127 [DOI] [PubMed] [Google Scholar]
- 18.Raeker, M. O., Su, F., Geisler, S. B., Borisov, A. B., Kontrogianni-Konstantopoulos, A., Lyons, S. E., and Russell, M. W. (2006) Dev. Dyn. 235 2018-2029 [DOI] [PubMed] [Google Scholar]
- 19.Armani, A., Galli, S., Giacomello, E., Bagnato, P., Barone, V., Rossi, D., and Sorrentino, V. (2006) Exp. Cell Res. 312 3546-3558 [DOI] [PubMed] [Google Scholar]
- 20.Bagnato, P., Barone, V., Giacomello, E., Rossi, D., and Sorrentino, V. (2003) J. Cell Biol. 160 245-253 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Borzok, M. A., Catino, D. H., Nicholson, J. D., Kontrogianni-Konstantopoulos, A., and Bloch, R. J. (2007) J. Biol. Chem. 282 32384-32396 [DOI] [PubMed] [Google Scholar]
- 22.Kontrogianni-Konstantopoulos, A., and Bloch, R. J. (2003) J. Biol. Chem. 278 3985-3991 [DOI] [PubMed] [Google Scholar]
- 23.Kontrogianni-Konstantopoulos, A., Jones, E. M., Van Rossum, D. B., and Bloch, R. J. (2003) Mol. Biol. Cell 14 1138-1148 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lowe, J. S., Palygin, O., Bhasin, N., Hund, T. J., Boyden, P. A., Shibata, E., Anderson, M. E., and Mohler, P. J. (2008) J. Cell Biol. 180 173-186 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Mohler, P. J., Rivolta, I., Napolitano, C., Lemaillet, G., Lambert, S., Priori, S. G., and Bennett, V. (2004) Proc. Natl. Acad. Sci. U. S. A. 101 17533-17538 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Cartegni, L., Chew, S. L., and Krainer, A. R. (2002) Nat. Rev. Genet. 3 285-298 [DOI] [PubMed] [Google Scholar]
- 27.Vandenbroucke, I. I., Vandesompele, J., Paepe, A. D., and Messiaen, L. (2001) Nucleic Acids Res. 29 E68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Mohler, P. J., Hoffman, J. A., Davis, J. Q., Abdi, K. M., Kim, C. R., Jones, S. K., Davis, L. H., Roberts, K. F., and Bennett, V. (2004) J. Biol. Chem. 279 25798-25804 [DOI] [PubMed] [Google Scholar]
- 29.Hopitzan, A. A., Baines, A. J., and Kordeli, E. (2006) Mol. Biol. Evol. 23 46-55 [DOI] [PubMed] [Google Scholar]
- 30.Mohler, P. J., Yoon, W., and Bennett, V. (2004) J. Biol. Chem. 279 40185-40193 [DOI] [PubMed] [Google Scholar]
- 31.Sherman, J., Tester, D. J., and Ackerman, M. J. (2005) Heart Rhythm 2 1218-1223 [DOI] [PubMed] [Google Scholar]
- 32.Fukuzawa, A., Idowu, S., and Gautel, M. (2005) J. Muscle Res. Cell Motil. 26 427-434 [DOI] [PubMed] [Google Scholar]
- 33.Bowman, A. L., Kontrogianni-Konstantopoulos, A., Hirsch, S. S., Geisler, S. B., Gonzalez-Serratos, H., Russell, M. W., and Bloch, R. J. (2007) FEBS Lett. 581 1549-1554 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Young, P., Ehler, E., and Gautel, M. (2001) J. Cell Biol. 154 123-136 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Dzhashiashvili, Y., Zhang, Y., Galinska, J., Lam, I., Grumet, M., and Salzer, J. L. (2007) J. Cell Biol. 177 857-870 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Ogawa, Y., Schafer, D. P., Horresh, I., Bar, V., Hales, K., Yang, Y., Susuki, K., Peles, E., Stankewich, M. C., and Rasband, M. N. (2006) J. Neurosci. 26 5230-5239 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Koticha, D., Maurel, P., Zanazzi, G., Kane-Goldsmith, N., Basak, S., Babiarz, J., Salzer, J., and Grumet, M. (2006) Dev. Biol. 293 1-12 [DOI] [PubMed] [Google Scholar]
- 38.Lustig, M., Zanazzi, G., Sakurai, T., Blanco, C., Levinson, S. R., Lambert, S., Grumet, M., and Salzer, J. L. (2001) Curr. Biol. 11 1864-1869 [DOI] [PubMed] [Google Scholar]
- 39.Kizhatil, K., Yoon, W., Mohler, P. J., Davis, L. H., Hoffman, J. A., and Bennett, V. (2007) J. Biol. Chem. 282 2029-2037 [DOI] [PubMed] [Google Scholar]
- 40.Das, A., Base, C., Dhulipala, S., and Dubreuil, R. R. (2006) J. Cell Biol. 175 325-335 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Lacas-Gervais, S., Guo, J., Strenzke, N., Scarfone, E., Kolpe, M., Jahkel, M., De Camilli, P., Moser, T., Rasband, M. N., and Solimena, M. (2004) J. Cell Biol. 166 983-990 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Yang, Y., Lacas-Gervais, S., Morest, D. K., Solimena, M., and Rasband, M. N. (2004) J. Neurosci. 24 7230-7240 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Hall, T. G., and Bennett, V. (1987) J. Biol. Chem. 262 10537-10545 [PubMed] [Google Scholar]
- 44.Lambert, S., Yu, H., Prchal, J. T., Lawler, J., Ruff, P., Speicher, D., Cheung, M. C., Kan, Y. W., and Palek, J. (1990) Proc. Natl. Acad. Sci. U. S. A. 87 1730-1734 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Lux, S. E., John, K. M., and Bennett, V. (1990) Nature 344 36-42 [DOI] [PubMed] [Google Scholar]
- 46.Lambert, S., and Bennett, V. (1993) J. Neurosci. 13 3725-3735 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Peters, L. L., Birkenmeier, C. S., Bronson, R. T., White, R. A., Lux, S. E., Otto, E., Bennett, V., Higgins, A., and Barker, J. E. (1991) J. Cell Biol. 114 1233-1241 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Beck, K. A., Buchanan, J. A., and Nelson, W. J. (1997) J. Cell Sci. 110 1239-1249 [DOI] [PubMed] [Google Scholar]
- 49.Drenckhahn, D., and Bennett, V. (1987) Eur J. Cell Biol. 43 479-486 [PubMed] [Google Scholar]
- 50.Birkenmeier, C. S., Sharp, J. J., Gifford, E. J., Deveau, S. A., and Barker, J. E. (1998) Genomics 50 79-88 [DOI] [PubMed] [Google Scholar]
- 51.Birkenmeier, C. S., White, R. A., Peters, L. L., Hall, E. J., Lux, S. E., and Barker, J. E. (1993) J. Biol. Chem. 268 9533-9540 [PubMed] [Google Scholar]
- 52.Gallagher, P. G., and Forget, B. G. (1998) J. Biol. Chem. 273 1339-1348 [DOI] [PubMed] [Google Scholar]
- 53.Zhou, D., Birkenmeier, C. S., Williams, M. W., Sharp, J. J., Barker, J. E., and Bloch, R. J. (1997) J. Cell Biol. 136 621-631 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Devarajan, P., Stabach, P. R., Mann, A. S., Ardito, T., Kashgarian, M., and Morrow, J. S. (1996) J. Cell Biol. 133 819-830 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Gagelin, C., Constantin, B., Deprette, C., Ludosky, M. A., Recouvreur, M., Cartaud, J., Cognard, C., Raymond, G., and Kordeli, E. (2002) J. Biol. Chem. 277 12978-12987 [DOI] [PubMed] [Google Scholar]
- 56.Hopitzan, A. A., Baines, A. J., Ludosky, M. A., Recouvreur, M., and Kordeli, E. (2005) Exp. Cell Res. 309 86-98 [DOI] [PubMed] [Google Scholar]
- 57.Zhang, X., and Bennett, V. (1998) J. Cell Biol. 142 1571-1581 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Davis, L. H., Davis, J. Q., and Bennett, V. (1992) J. Biol. Chem. 267 18966-18972 [PubMed] [Google Scholar]
- 59.Mumby, M. C., and Walter, G. (1993) Physiol. Rev. 73 673-699 [DOI] [PubMed] [Google Scholar]
- 60.Davare, M. A., Avdonin, V., Hall, D. D., Peden, E. M., Burette, A., Weinberg, R. J., Horne, M. C., Hoshi, T., and Hell, J. W. (2001) Science 293 98-101 [DOI] [PubMed] [Google Scholar]
- 61.Davare, M. A., Horne, M. C., and Hell, J. W. (2000) J. Biol. Chem. 275 39710-39717 [DOI] [PubMed] [Google Scholar]
- 62.Hall, D. D., Feekes, J. A., Arachchige Don, A. S., Shi, M., Hamid, J., Chen, L., Strack, S., Zamponi, G. W., Horne, M. C., and Hell, J. W. (2006) Biochemistry 45 3448-3459 [DOI] [PubMed] [Google Scholar]
- 63.Marx, S. O., Kurokawa, J., Reiken, S., Motoike, H., D'Armiento, J., Marks, A. R., and Kass, R. S. (2002) Science 295 496-499 [DOI] [PubMed] [Google Scholar]
- 64.Marx, S. O., Reiken, S., Hisamatsu, Y., Jayaraman, T., Burkhoff, D., Rosemblit, N., and Marks, A. R. (2000) Cell 101 365-376 [DOI] [PubMed] [Google Scholar]
- 65.Brewis, N., Ohst, K., Fields, K., Rapacciuolo, A., Chou, D., Bloor, C., Dillmann, W., Rockman, H., and Walter, G. (2000) Am. J. Physiol. 279 H1307-H1318 [DOI] [PubMed] [Google Scholar]
- 66.Mayans, O., van der Ven, P. F., Wilm, M., Mues, A., Young, P., Furst, D. O., Wilmanns, M., and Gautel, M. (1998) Nature 395 863-869 [DOI] [PubMed] [Google Scholar]
- 67.Gotthardt, M., Hammer, R. E., Hubner, N., Monti, J., Witt, C. C., McNabb, M., Richardson, J. A., Granzier, H., Labeit, S., and Herz, J. (2003) J. Biol. Chem. 278 6059-6065 [DOI] [PubMed] [Google Scholar]
- 68.Lange, S., Xiang, F., Yakovenko, A., Vihola, A., Hackman, P., Rostkova, E., Kristensen, J., Brandmeier, B., Franzen, G., Hedberg, B., Gunnarsson, L. G., Hughes, S. M., Marchand, S., Sejersen, T., Richard, I., Edstrom, L., Ehler, E., Udd, B., and Gautel, M. (2005) Science 308 1599-1603 [DOI] [PubMed] [Google Scholar]
- 69.Weinert, S., Bergmann, N., Luo, X., Erdmann, B., and Gotthardt, M. (2006) J. Cell Biol. 173 559-570 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Musa, H., Meek, S., Gautel, M., Peddie, D., Smith, A. J., and Peckham, M. (2006) J. Cell Sci. 119 4322-4331 [DOI] [PubMed] [Google Scholar]
- 71.Peng, J., Raddatz, K., Molkentin, J. D., Wu, Y., Labeit, S., Granzier, H., and Gotthardt, M. (2007) Circulation 115 743-751 [DOI] [PubMed] [Google Scholar]
- 72.Neumann, J., Eschenhagen, T., Jones, L. R., Linck, B., Schmitz, W., Scholz, H., and Zimmermann, N. (1997) J. Mol. Cell Cardiol. 29 265-272 [DOI] [PubMed] [Google Scholar]
- 73.Gergs, U., Boknik, P., Buchwalow, I., Fabritz, L., Matus, M., Justus, I., Hanske, G., Schmitz, W., and Neumann, J. (2004) J. Biol. Chem. 279 40827-40834 [DOI] [PubMed] [Google Scholar]