Abstract
The number of distinct signaling pathways that can transactivate the epidermal growth factor receptor (EGFR) in a single cell type is unclear. Using a single strain of human mammary epithelial cells, we found that a wide variety of agonists, such as lysophosphatidic acid (LPA), uridine triphosphate, growth hormone, vascular endothelial growth factor, insulin-like growth factor-1 (IGF-1), and tumor necrosis factor-α, require EGFR activity to induce ERK phosphorylation. In contrast, hepatocyte growth factor can stimulate ERK phosphorylation independent of the EGFR. EGFR transactivation also correlated with an increase in cell proliferation and could be inhibited with metalloprotease inhibitors. However, there were significant differences with respect to transactivation kinetics and sensitivity to different inhibitors. In particular, IGF-1 displayed relatively slow transactivation kinetics and was resistant to inhibition by the selective ADAM-17 inhibitor WAY-022 compared with LPA-induced transactivation. Studies using anti-ligand antibodies showed that IGF-1 transactivation required amphiregulin production, whereas LPA was dependent on multiple ligands. Direct measurement of ligand shedding confirmed that LPA treatment stimulated shedding of multiple EGFR ligands, but paradoxically, IGF-1 had little effect on the shedding rate of any ligand, including amphiregulin. Instead, IGF-1 appeared to work by enhancing EGFR activation of Ras in response to constitutively produced amphiregulin. This enhancement of EGFR signaling was independent of both receptor phosphorylation and PI-3-kinase activity, suggestive of a novel mechanism. Our studies demonstrate that within a single cell type, the EGFR autocrine system can couple multiple signaling pathways to ERK activation and that this modulation of EGFR autocrine signaling can be accomplished at multiple regulatory steps.
The extracellular milieu is a rich mixture of small molecules, each conveying a particular type of information. Endocrine hormones integrate organ functions across the organism; cytokines and growth factors reflect the inflammatory or proliferative status of a tissue; matrix molecules convey information about location; and other small molecules, such as amino acids, sugars, lipids, and electrolytes, can reflect the activities of neighboring cells. Any given cell must integrate all of these various sources of information to make the appropriate decision to proliferate, die, or remain quiescent. Failure of a cell to appropriately interpret and respond to its informational context can contribute to many aspects of cancer, from hyperproliferation to inappropriate motility to resistance to apoptosis. Thus, it is important to not only study the activity of individual signaling pathways but also to investigate the mechanisms by which they are integrated to give rise to final cellular outcomes.
An important example of information integration is the ability of multiple stimuli to activate the ERK pathway indirectly by stimulating epidermal growth factor receptor (EGFR)2 signaling, a process known as transactivation (1). For example, activation of G-protein-coupled receptors (GPCRs), such as the endothelin, lysophosphatidic acid (LPA), and calcium receptors (1–4, 6–8), can result in autophosphorylation of the EGFR and subsequent activation of the MAPK cascade. Similar results have been reported following activation of the tumor necrosis factor receptor system (9) and insulin-like growth factor receptor (IGFR) systems (10). EGFR transactivation has been proposed to be mediated by the activation of metalloproteases that release EGFR ligands proteolytically (4). Although the general features of transactivation have been described for numerous cell and receptor types, the molecular mechanisms responsible for activation of ligand shedding are still unclear. Nevertheless, the process is critically important in the action of many hormones. For example, vascular smooth muscle cell hypertrophy induced by angiotensin II is mediated through EGFR transactivation (11), as is the motility and proliferation of multiple types of cancer cells (12).
Despite the importance of EGFR transactivation in pathological processes, very little is known about its role in normal cell physiology. The complexity of the EGFR system is a significant barrier to this understanding, because it obscures the mechanisms by which a transactivating factor might act. For example, normal human mammary epithelial cells (HMEC) express four different EGFR autocrine ligands: transforming growth factor-α (TGF-α), heparin-binding EGF-like growth factor (HB-EGF), amphiregulin (AR), and epiregulin (EPR) (13). These cells also express HER2 and HER3 that can form heterodimers, depending on the activating ligand (14–16). Understanding transactivation in this context requires an understanding of the cell type, the levels of the different receptors, and how the different ligands are regulated. Because few experimental systems are amenable to this level of analysis, most of our knowledge of transactivation is restricted to specific combinations of cell type and transactivating ligand without integration across receptor classes and signaling mechanisms. One consequence of this complexity is uncertainty as to whether multiple mechanisms of transactivation can function in concert in the same cell.
Over the last decade, we have been building a systems-level model of the EGFR system in HMEC (17). Using this model cell system, we and our collaborators have built mathematical models of EGFR dynamics and shown their utility in predicting the effect of perturbations, such as HER2 overexpression (18, 19). Proteome analysis of these cells has been performed to define the repertoire of receptors and signaling molecules that they express (20) as well as the pattern of proteins that are phosphorylated in response to EGF (21). We have also investigated the regulation of ligand processing and activation in these cells (22, 23). However, we still know little about how ligand processing is regulated and how different ligands participate in EGFR transactivation. Therefore, we designed a systematic study to define the range of signals that can induce EGFR transactivation and the role it might play in regulating these cells.
In this investigation, we have used a wide variety of hormones and stimuli known to activate the ERK pathway and have determined whether they operate by transactivating EGFR. Surprisingly, we found that this was the case for the majority of inputs. These signals include GPCRs, receptor tyrosine kinases (RTKs), and cytokine receptors. However, the mechanisms by which different receptors transactivate EGFR are distinct with respect to metalloprotease utilization, ligand targeting, and kinetics. We additionally uncovered a novel mechanism of apparent transactivation by IGF-1 acting through its own receptor, in which there is no increase in ligand shedding but rather a sensitization of the EGFR to constitutively released amphiregulin. The end result is still modulation of autocrine signaling through the EGFR. These results suggest that EGFR autocrine signaling integrates signals from a surprising diversity of different receptors within a single cell type and that there are multiple points in the pathway at which signal integration can occur.
EXPERIMENTAL PROCEDURES
Antibodies and Materials—Rabbit polyclonal anti-EGFR (1005) was obtained from Santa Cruz Biotechnology. Anti-AKT (Pan), anti-phospho-AKT (Ser473), anti-ERK, and anti-phospho-p44/42 MAPK (Thr202/Tyr204) (E10) antibodies were obtained from Cell Signaling Technology. Goat anti-rabbit and goat anti-mouse horseradish peroxidase-conjugated antibodies were obtained from Jackson Immunoresearch Laboratories. Vascular endothelial cell-derived growth factor (VEGF) was obtained from R & D Systems. LPA and LY294002 were purchased from BIOMOL Research Laboratories Inc. Hepatocyte growth factor/scatter factor (HGF), AG 1517, and GM6001 (Galardin/Ilomastat) were purchased from Calbiochem. Insulin and insulin-like growth factor 1 (IGF-1) were from Sigma. Human EGF was obtained from Peprotech, Inc. All capture antibodies, biotinylated detection antibodies, and antigens for the sandwich ELISAs were purchased from R&D Systems (Minneapolis, MN) and suspended in water to 500 μg of protein/ml, aliquoted, and lyophilized. Anti-ligand antibodies against TGF-α, epiregulin, and amphiregulin were obtained from R&D Systems. CRM197 was purchased from EMD Biosciences (San Diego, CA). Monoclonal antibody 13A9 was a generous gift from Genentech, Inc. (San Francisco, CA).
Batimastat (BB-94; (4-(N-hydroxyamino)-(2R)-isobutyl-(3S)-(thienylthiomethyl)succinyl)-l-phenylalanine-N-methylamide) was custom-synthesized by Kimia Corp. (Santa Clara, CA). WAY-022 was a generous gift from Jay Gibbons (Wyeth Ayerst, Pearl River, NY) to Bob Coffey. Galardin (GM6001) and Batimastat have similarly broad activities against multiple matrix metalloproteases (MMPs) as well as ADAM family proteases but differ in potency, with Galardin being more potent by a factor of 10 (24, 25).
Cell Culture and Treatment—HMEC 184A1-1 were maintained at 37 °C in 95% air, 5% CO2 in DFCI-1 medium as previously described (26). New cultures were started from stocks approximately every 12 weeks. Overnight incubation in low serum (0.1%) medium, either Ham's F-12 medium (0.3 mm Ca2+) or DFCI-1 (0.8 mm Ca2+), was used to serum-deplete cells. Serum-deprived HMEC 184A1-1 monolayers were incubated with protease or kinase inhibitors for 0.5 h or with blocking antibodies for 2–18 h prior to the indicated treatments.
Proliferation Assays—HMEC 184A1-1 cells in 12-well plates were seeded at 1.0 × 105 cells/well in DFCI-1 medium containing 1% fetal bovine serum, allowed to attach for 4 h, and serum-deprived for 18–24 h in F-12 minimal medium. Cell proliferation was determined by direct cell counts using a Coulter counter at 24-h intervals. In addition, MTT assays were conducted once a day for 3 days. MTT (Sigma) was added to a final concentration of 0.5 μg/ml. Following 4 h of incubation at 37 °C in the dark, the MTT crystals were solubilized in 100 μl of acid-isopropyl alcohol (0.04 n HCl in isopropyl alcohol), and quadruplicates of 100 μl were collected and clarified by high speed centrifugation. The supernatants were subjected to optical density analysis. Approximately 100 μl of solution were added to a 96-well microplate and read at 570 nm with background subtraction at 630 nm. In parallel experiments, cell number was determined directly by a Coulter counter, yielding very similar results.
Immunoblot Analysis of p42/44 ERK and AKT Phosphorylation—HMEC 184A1-1 cells were seeded into 100-mm dishes in DFCI-1 medium, cultured to ∼80% confluence, and serum-deprived for 18 h. Cells were pretreated with inhibitors for the time periods indicated in each figure legend and then stimulated with agonists for 15 min. After stimulation, cultures were washed twice with ice-cold phosphate-buffered saline and lysed in MT-G buffer (20 mm HEPES, pH 8.0, 1% Triton X-100, 10% glycerol, 150 mm NaCl, 2 mm Na3OV4, 1 mm phenylmethylsulfonyl fluoride, 1% aprotinin, protease inhibitor mixture), and cell lysates were then cleared by centrifugation. Approximately 20 μg of total protein of each sample was loaded onto a 10% BisTris gel, transferred to polyvinylidene difluoride membrane, and probed for phospho-ERK. p42/44 ERK phosphorylation was detected by immunoblotting with a 1:2,000 dilution of mouse monoclonal phosphospecific p42/44 ERK antibody. AKT phosphorylation was determined using an anti-phospho-AKT antibody purchased from Cell Signaling Technology that was used at a 1:1000 dilution. Horseradish peroxidase-conjugated goat anti-mouse secondary antibody was used at a 1:5,000 dilution. Quantification of bands was performed using a Roche Applied Science LumiImager and associated image analysis software. After quantification, membranes were stripped and reprobed using rabbit anti-total p42/44 ERK antibody or rabbit anti-total AKT antibody to confirm equal protein loading.
Phosphoprotein and Ligand ELISAs—Phosphorylated ERK1 was measured in cell lysates using phospho-ERK1 (Thr202/Tyr204) and phospho-AKT (Pan) (Ser473) sandwich ELISA kits from R&D Systems in a 96-well format with slight modifications. Briefly, protein was isolated from HMEC using MTG lysis buffer. Approximately 40 μg of total protein was diluted to a 1:1 ratio with a lysis buffer supplemented with 8 m urea. The lysate samples were vortexed and incubated at room temperature for 1 h and then centrifuged at 2,000 × g for 5 min, and the supernatant was collected in a fresh tube. The samples were diluted to a final urea concentration of 1 m. The same samples were used to evaluate the levels of phosphorylated EGFR using the STAR phospho-EGFR ELISA kit from Millipore. For determination of AR levels, medium was collected from each dish and then analyzed directly for AR using the AR ELISA kit from R&D Systems.
The levels of activated Ras were determined by using a pull-down assay kit based on the Raf-RBD-glutathione S-transferase fusion protein (Cytoskeleton, Inc.). In brief, cells were rinsed and lysed in detergent solution containing a protease inhibitor mixture (provided with kit). A cell scraper was used to transfer the cells to a microcentrifuge tube, and the lysate was clarified by centrifugation. Approximately 500 μg of total protein was incubated with 15 μg of Raf1-RBD beads for 1 h at 4 °C on a rotator. The beads were rinsed with a wash buffer twice, suspended in SDS sample buffer, heated to 95 °C for 5 min, and then run on a 12% SDS gel and transferred to polyvinylidene difluoride membrane and probed with an anti-Ras antibody. Horseradish peroxidase-conjugated goat anti-mouse secondary antibody was used at a 1:5,000 dilution. Quantification of bands was performed using a Roche Applied Science LumiImager and associated image analysis software.
Quantitative ELISA Microarrays—ELISA microarray chips were prepared and processed essentially as described previously (1, 2). To facilitate sample throughput and reproducibility, 16 identical chips were printed on each slide, using a hydrophobic barrier to separate the samples, as described previously (3). Each of the capture antibodies was printed four times on each chip, along with Cy3-labeled protein that served as an orientation marker. All antigen standards were combined prior to serially diluting 3-fold in 0.1% casein to create 11 standards that spanned a 2,187-fold concentration range. Prior to analysis, conditioned medium was diluted 10% and 21-fold and analyzed in duplicate. To avoid any potential slide-related bias in signal intensity, duplicates were placed in different positions on different slides. Sample positioning was also blocked based on treatment group so that typically no more than two samples of the same treatment group were located on a single slide.
The biotin signal was amplified using the biotinyltyramide amplification procedure followed by incubation with streptavidin conjugated to Cy3 (1). The fluorescent images for the slides were obtained using a ScanArray Express HT laser scanner, and the spot intensity was quantified using ScanArray Express (PerkinElmer Life Sciences). Standard curves were generated, and sample antigen concentrations were calculated using the Protein Microarray Analysis Tool (ProMAT), a custom freeware program developed specifically for this use (available on the World Wide Web) (1, 4). Statistical differences between all treatment groups were initially determined by analysis of variance and then delineated using Fisher's protected least significant difference test using StatView 5.0.1 software (SAS Institute). A significance level of 0.05 was used in all cases.
RESULTS
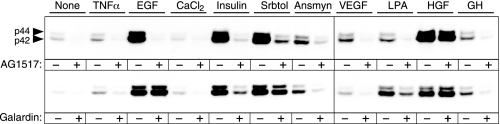
Activation of the MAPK Pathway—Proximal signaling through the EGFR involves activation of EGFR tyrosine kinase and the resultant activation of the Ras-Raf-MEK-MAPK pathway via scaffolding intermediates, such as Grb-Sos (for a review, see Ref. 27). Transactivation of the EGFR is defined operationally as phosphorylation of p42/44 ERK in response to an unrelated agonist that requires EGFR kinase activity and the proteolytic release of EGFR ligands. To characterize the range of transactivating signals operative in HMEC, we selected a panel of agonists that have previously been shown to induce EGFR transactivation in other cell types. These agonists include insulin/IGF-1, HGF, TNFα, VEGF, LPA, GH, sorbitol, and anisomycin; activated RTKs (including IGFR, c-Met, Flt, and GH receptor); GPCRs (Edg/LPAR); and stress response pathways (TNFα, sorbitol, and anisomycin). As shown in Fig. 1, treating HMEC with this panel of agonists induces phosphorylation of ERK. The ligands EGF, insulin, sorbitol, anisomycin, and HGF each produced a strong increase in ERK1/2 phosphorylation within 15 min of treatment, as detected by anti-phospho-ERK antibodies. Identical results were obtained with 10 nm IGF-1 and 100 nm insulin (data not shown), implicating the IGFR rather than the insulin receptor. The GPCR ligand LPA and VEGF, GH, and TNFα also stimulated ERK phosphorylation but to a lesser and more variable extent. Pretreatment of HMEC with the highly specific and potent EGFR kinase inhibitor AG 1517 (28) completely blocked the increase in ERK phosphorylation in response to EGF, insulin/IGF, anisomycin, VEGF, LPA, GH, and TNFα, whereas sorbitol-induced ERK phosphorylation was only partially inhibited (Fig. 1A). The same results were also obtained using the EGFR kinase inhibitor AG 1478 (data not shown). HGF was not significantly affected by either of the EGFR kinase inhibitors, demonstrating that the loss of ERK activation was not caused by a nonspecific effect of the inhibitors.
FIGURE 1.
Dependence of EGFR activity and proteolytic activity for activation of ERK by multiple stimuli. HMEC 184A1–1 cells were treated with the indicated agonists for 15 min: TNFα (10 ng/ml), EGF (1 ng/ml), CaCl2 (2.0 mm), insulin (10 nm), sorbitol (Srbtol) (0.5 m), anisomycin (50 ng/ml), VEGF (100 ng/ml), LPA (20 μm), HGF (20 ng/ml), and GH (500 ng/ml) in the absence or presence of inhibitors, as indicated below. Cell lysates (normalized to 20 μgof protein) were separated by SDS-PAGE, transferred to polyvinylidene difluoride membranes, and phosphorylated ERK visualized using anti-phospho-ERK1/2 antibodies, as described under “Experimental Procedures.” Top, effect of the EGFR kinase inhibitor AG1517. Cells were pretreated with the specific EGFR inhibitor AG1517 at a concentration of 500 nm for 15 min prior to agonist addition. The concentration of AG1517 was based on preliminary experiments showing it to be the minimum concentration that completely blocked EGF-induced EGFR phosphorylation (data not shown). Results are typical of triplicate samples from three independent experiments. Bottom, effect of the metalloprotease inhibitor Galardin (also known as Ilomastat or GM6001). The inhibitor was added at a final concentration of 10 μm 15 min prior to the agonist addition. Results are typical of triplicate samples from three independent experiments.
The most frequently implicated mechanism for transactivation of the EGFR is the cleavage and release of EGF family ligands from the cell surface by activation of cell surface metalloproteases (2, 29–31). This general mechanism was tested by using the metalloprotease inhibitor Galardin (32) to inhibit proteolytic cleavage of membrane-bound EGF-like ligands. Using a concentration of Galardin that has previously been shown to block shedding-mediated transactivation (31), we observed a significant decrease in ERK phosphorylation in response to GPCR agonists (LPA) and RTK agonists (insulin/IGF-1) but no significant effect on EGF- or HGF-induced ERK activation. Galardin also substantially inhibited ERK phosphorylation in response to VEGF, GH, and anisomycin. In contrast, ERK phosphorylation in response to the osmotic stressor sorbitol was only partially inhibited by Galardin, implicating both EGFR-dependent and EGFR-independent mechanisms in the osmotic stress response.
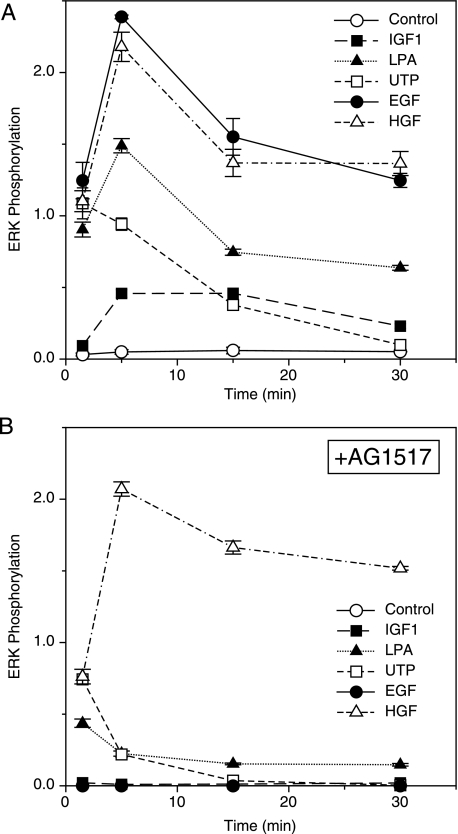
Kinetics of Induction—In addition to indirect activation of ERK through the EGFR axis, several of the receptors studied have been implicated in the direct activation of the Ras-Raf-MEK-MAPK pathways (25, 30). Because the kinetics of ERK activation could potentially differentiate between direct and indirect agonists, we used a quantitative ELISA to measure the total increase in ERK phosphorylation over time, with time points ranging from 1.5 to 30 min after agonist addition. As shown in Fig. 2A, a marked increase in ERK phosphorylation was observed as early as 1.5 min after the addition of IGF-1, LPA, UTP (which activates purinergic GPCRs), EGF, or HGF. The UTP-mediated ERK activation appeared to peak by 1.5 min, with no further increase and a gradual return to basal levels by 30 min. ERK phosphorylation in response to IGF-1, LPA, EGF, and HGF appeared to peak at 5 min, followed by a gradual decline, but remained substantially above control levels at 30 min. Interestingly, AG 1517 completely inhibited ERK activation at all time points after either EGF or IGF-1 treatment (Fig. 2B) but was only partially effective at the 1.5-min time point for UTP and LPA. This suggests that ERK phosphorylation in response to these two GPCR agonists may have an initial, transient direct component, followed by a subsequent component dependent on EGFR transactivation. HGF-induced ERK phosphorylation was insensitive to AG 1517 treatment at all time points, consistent with an EGFR-independent mode of action.
FIGURE 2.
Kinetics of ERK phosphorylation in response to multiple agonists. A, cells were serum-deprived for 18 h and then treated with the indicated agonists: control (DMSO alone; ○), IGF-1 (1 nm; ▪), LPA (20 μm; ▴) UTP (100 μm; □), EGF (1 ng/ml; •), and HGF (20 ng/ml; ▵). Cell lysates were harvested on ice at the indicated times after agonist addition. Changes in the level of ERK phosphorylation were quantified by ELISA as described under “Experimental Procedures.” Results are presented as ng/ml pERK/40 μgof total protein ± S.D. (n = 3). B, same as A, with the addition of the tyrosine kinase inhibitor, AG 1517, at 500 nm for 1 h prior to the addition of agonists.
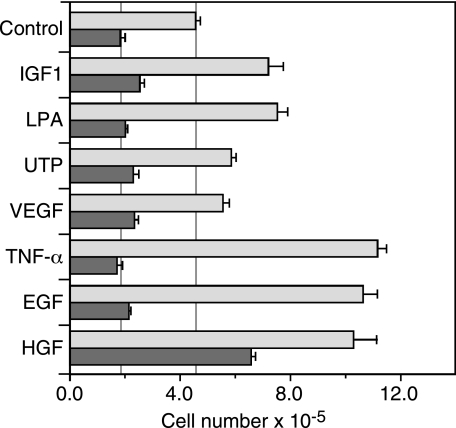
Coupling of Proliferation to EGFR Transactivation—Since EGFR activation is required for the proliferation of the 184A1 and 184A1–1 HMEC lines (33, 34), we investigated whether those extracellular stimuli that transactivate the EGFR in HMEC could induce a proliferative response in the absence of exogenous EGF and, if so, whether this response was dependent on EGFR kinase activity. We rendered the HMEC cell line 184A1-1 quiescent by overnight culture in serum-free medium and then treated cells with either 100 ng/ml EGF, 20 μm LPA, 1 nm IGF-1, 100 μm UTP, 10 ng/ml TNFα, 100 ng/ml VEGF, or 20 ng/ml HGF. Proliferation was determined by direct cell count at 24-h intervals for 3 days after agonist addition.
As shown in Fig. 3, EGF, HGF, and TNFα stimulated HMEC proliferation by 2.3–2.5-fold. The GPCR-coupled agonists LPA and UTP stimulated a statistically significant 1.4–1.7-fold increase in proliferation over the 3-day period (p ≤ 0.01), as did also stimulation with IGF-1. VEGF had little effect on proliferation (1.2-fold increase, p = 0.06). The proliferative responses to EGF, UTP, LPA, IGF-1, and TNFα were dependent on EGFR kinase activity, as shown by the significant inhibition observed with the EGFR kinase inhibitor AG1517. AG1517 did reduce proliferation of control cells by about 50%, but this effect can be attributed to inhibition of the basal levels of EGFR activation seen in the presence of constitutive ligand shedding (see below). Proliferation in response to HGF was resistant to AG1517, with a >3-fold increase in cell proliferation in response to HGF plus AG1517, demonstrating that HGF exerts its effects independent of the EGFR and that the effects of AG1517 are relatively specific.
FIGURE 3.
HMEC proliferation in response to multiple agonists is dependent on EGFR kinase activity. HMEC were plated at 30% confluence in serum-free DFCI medium supplemented with the indicated agonists or 0.1% DMSO (control), in the absence (light gray bars) or presence (dark gray bars) of the EGFR inhibitor AG1517. Cell number was estimated by MTT assay on day 1 and 3 after stimulation, as described under “Experimental Procedures.” Results are presented as the ratio of MTT absorbance on day 3/day 1 (mean ± S.D., n = 3). Vertical lines indicate the values of control cells as a basis for comparisons.
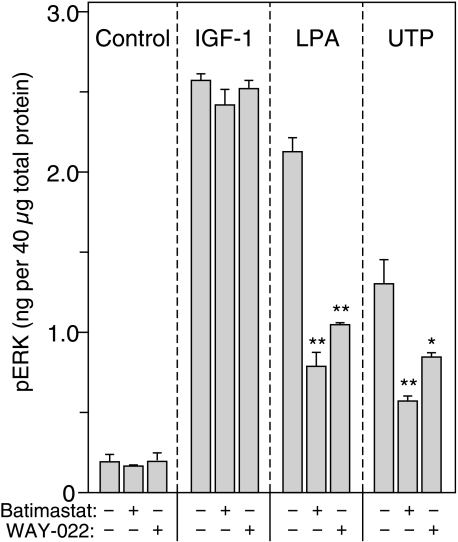
Differential Protease Specificity—The protease inhibitor Galardin (GM6001) used in Fig. 1 is a relatively broad-spectrum metalloprotease inhibitor with activity against both MMPs and members of the ADAM family (25, 32). The ADAM proteases have been implicated as the primary effectors of regulated proteolysis in numerous systems (2, 29, 35, 36), and proteomic analysis of HMEC indicates that these cells express both ADAM 17 and ADAM 10 (13). To determine which of these ADAMs was most likely responsible for ligand release in HMEC cells, we compared the effects of various protease inhibitors on ERK activation. Although Galardin was effective against IGF-1-induced ERK phosphorylation (Fig. 1), Batimastat, a chemically unrelated inhibitor of MMPs and ADAM family metalloproteases (24), was ineffective against IGF-1 when used at a concentration that we have previously shown to completely block ligand shedding (37) (Fig. 4A). However, Batimastat substantially blocked ERK phosphorylation in response to LPA or UTP. Since neither Batimastat nor Galardin can distinguish between MMPs and ADAM family proteases, we tested the effects of WAY-022, a selective inhibitor of ADAM 17 (38). WAY-022 was highly effective in blocking ERK activation in response to either LPA or UTP-induced transactivation of the EGFR, implicating ADAM 17 as the metalloprotease responsible for regulated proteolysis in response to these GPCR agonists. In contrast, WAY-022 was as ineffective at blocking IGF-1-induced ERK phosphorylation as Batimastat (Fig. 4A). Kinetic experiments showed that metalloprotease inhibitors were effective at even the earliest time points of stimulation (1.5 min; data not shown). These results suggest that GPCRs work through ligand shedding via an ADAM 17-mediated pathway, whereas IGFR-mediated transactivation uses a different mechanism.
FIGURE 4.
Effect of metalloprotease inhibitors on transactivation. HMEC in serum-free medium were treated with either the metalloprotease inhibitor Batimastat (10 μm) or the selective inhibitor of TACE/ADAM-17 WAY-022 (1 μm) for 1 h and then stimulated with IGF-1 (1 nm), LPA (20 μm), or UTP (100 μm) for 5 min. An ELISA was then used to quantify ERK phosphorylation, as described under “Experimental Procedures.” Values are mean ± S.D., n = 3. The asterisks denote statistically significant differences between control and inhibitor treatment within each agonist group, as determined by one-tailed Student's t test. *, p ≤ 0.05; **, p ≤ 0.01. The concentrations of inhibitors used have previously been shown to significantly inhibit the shedding of EGFR ligands by cells (37, 38).
Identification of Potential Ligands Involved in EGFR Transactivation—Proteomic analyses of HMEC indicate that they produce the EGFR ligands HB-EGF, AR, TGFα, and EPR (13), of which any one singly or in combination could potentially mediate EGFR transactivation in these cells. We used CRM-197, an antagonist of HB-EGF (39), to determine whether HB-EGF was an important mediator of LPA- or IGF-1-mediated transactivation. CRM-197 at the concentration used (10 μg/ml) binds specifically to membrane-anchored HB-EGF and blocks all of its biological activity (39). However, this inhibitor had no effect on ERK phosphorylation in response to any of the agonists tested (Fig. 5A), suggesting that HB-EGF is not a relevant shed ligand with the tested stimuli.
FIGURE 5.
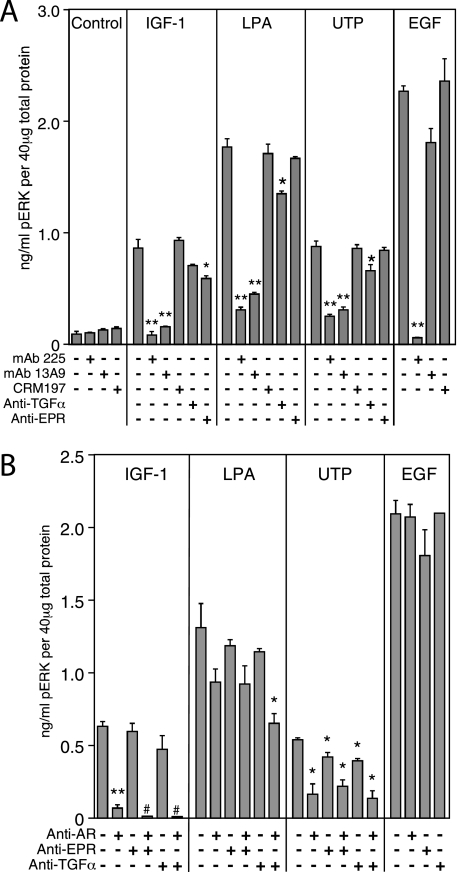
Effect of antagonistic antibodies against either the EGFR or its ligands on stimulation of ERK phosphorylation. A, following overnight incubation in serum-free medium, HMEC were pretreated with the following inhibitors at a concentration of 10 μg/ml for 1 h: anti-EGFR mAb 225 (which blocks all ligand binding); anti-EGFR mAb 13A9 (which blocks TGFα and AR but not EGF or HB-EGF); CRM-197 (a specific inhibitor of HB-EGF binding), and specific neutralizing antibodies for either TGFα or epiregulin. The cells were then stimulated with IGF-1 (1 nm), LPA (20 μm), UTP (100 μm), or EGF (1 ng/ml) for 5 min. ELISAs were used to quantify ERK phosphorylation, as described under “Experimental Procedures.” Values are mean ± S.D. (n = 3). The asterisks denote statistically significant differences between control and inhibitor treatment within each agonist group, as determined by one-tailed Student's t test. *, p ≤ 0.05; **, p ≤ 0.01. B, combinations of neutralizing antibodies to AR, TGFα, and EPR were used, each at a concentration of 10 μg/ml. All other conditions are the same as in A. The phospho-ERK levels were below the ELISA limits of detection for all control samples (not shown) and for IGF-1-stimulated cells treated with anti-AR and a second antibody.
To determine which of the other EGFR ligands might be contributing to EGFR transactivation, we measured ERK phosphorylation in the presence of a series of anti-ligand antibodies. As shown in Fig. 5A, the anti-EGFR antibody mAb 13A9, which blocks TGFα and AR-mediated activation of the EGFR but not EGF or HB-EGF binding (43, 44), was generally as effective as mAb 225 in blocking EGFR transactivation by either LPA, UTP, or IGF-1. This finding provides further evidence that HB-EGF is not the transactivating ligand in HMEC, in concordance with the ineffectiveness of CRM-197. Anti-EPR antibody had no effect on either LPA- or UTP-mediated transactivation but did produce a slight decrease in ERK phosphorylation in response to IGF-1. The anti-TGFα antibody had a modest, statistically significant inhibitory effect against LPA and UTP (but not IGF-1). However, the extent of inhibition in the presence of anti-TGFα antibodies was insufficient to account entirely for the inhibitory effects of mAb 13A9.
We tested combinations of the individual antibodies against AR, EPR, and TGFα, as shown in Fig. 5B. The anti-AR antibody produced a very significant decrease in ERK phosphorylation in response to IGF-1, whereas neither anti-EPR nor anti-TGFα had a statistically significant effect. In contrast, no single antibody, not even the anti-AR antibody, produced a major decrease in LPA-induced ERK phosphorylation; rather, each individual antibody produced a partial reduction in ERK phosphorylation. Combining anti-AR and anti-TGFα produced a significant inhibition of ERK phosphorylation in response to LPA, suggesting that both ligands contribute to LPA-mediated transactivation. UTP-mediated ERK phosphorylation showed a statistically significant decrease in response to either anti-AR, anti-TGFα, or anti-EPR antibodies, although the anti-AR antibody was clearly the most effective whether used singly or in combination (Fig. 5B). The maximum inhibition of the UTP response by combined anti-TGFα and anti-AR antibodies was to 25% of controls, compared with inhibition to 11% of the control value by anti-AR in IGF-1-stimulated cells.
These results implicate AR as the major ligand responsible for transactivating the EGFR in response to IGF-1 treatment, whereas the LPA and UTP receptors appear to act through multiple ligands. Interestingly, both GPCRs maintain a substantial level of ERK phosphorylation activity even in the presence of combined anti-ligand antibodies, suggesting that there may be additional ligand-independent effects of GPCR activation.
Ligand Production in Response to Stimulation—To verify that the EGFR ligands implicated in the specific antibody experiments were actually produced by HMEC cells and responsive to agonist stimulation, we used a quantitative ELISA microarray to measure basal and induced levels of AR, HB-EGF, and TGFα in conditioned medium from HMEC cells stimulated with either IGF-1, LPA, UTP, or HGF (Table 1). The available antibodies against EPR are not suitable for use in an ELISA.3 In concordance with the metalloprotease inhibitor studies, LPA treatment induced a 5- and 1.6-fold increase in HB-EGF and TGFα production and a minor (1.3-fold) increase in AR release. HGF also significantly increased HB-EGF and TGFα production, although it does not use the EGFR pathway for ERK activation. UTP, despite being a GPCR agonist like LPA, stimulated a minor increase in AR production but not HB-EGF or TGFα release. Surprisingly, IGF-1 had no significant effect on the release of any ligand. Of particular note are the absolute levels of AR, HB-EGF, and TGFα under both basal and stimulated conditions. AR is obviously the dominant EGFR ligand in HMEC with levels 10–100-fold higher than TGFα or HB-EGF. The constitutive levels of AR production are higher than the stimulated levels of TGFα and HB-EGF combined.
TABLE 1.
Quantitative ELISA Microarray to Measure Basal and Induced Levels of AR, HB-EGF, and TGFα
Cells were maintained overnight in basal medium and then incubated for 1 h in fresh basal medium plus 10 μg/ml 225 mAb prior to the addition of the indicated stimulus. After a 3-h incubation, the medium was removed and assayed for the levels of the indicated growth factor using the protein microarray described under “Experimental Procedures.” Results are expressed ± S.D.
| Stimulus | AR | HB-EGF | TGFα |
|---|---|---|---|
| pg/ml | pg/ml | pg/ml | |
| None | 1180 ± 470 | 11 ± 8 | 110 ± 5 |
| IGF-1 (1 nm) | 1190 ± 510 | 10 ± 2 | 117 ± 16 |
| LPA (20 μm) | 1410 ± 610 | 57 ± 11a | 180 ± 18a |
| UTP (100 μm) | 1290 ± 600 | 11 ± 3 | 100 ± 16 |
| HGF (0.3 nm) | 1260 ± 470 | 67 ± 8a | 145 ± 1a |
Significantly different by both analysis of variance and Fisher's protected least significant difference analysis.
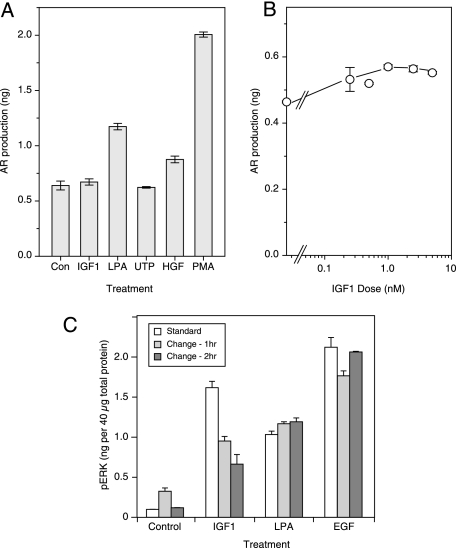
To verify the lack of increased AR production in response to IGF-1, we used an ELISA in a 96-well format. Cells were provided fresh medium together with different agonists for 2 h, followed by the analysis of the culture medium for AR levels. In the case of the untreated cells, the amount of AR in the medium had already reached a substantial level by 2 h (Fig. 6A). The addition of PMA, which is known to elicit a strong shedding response in HMEC (40), stimulated more than a 3-fold increase in AR levels in the medium, whereas LPA and HGF induced an ∼2- and 1.5-fold increase, respectively (Fig. 6A). UTP and IGF-1 had no apparent effect on AR production, consistent with the results obtained from the micro-ELISA assay. When we used increasing doses of IGF-1, we did see a small effect on AR production after 2 h (Fig. 6B). However, the maximum increase was only about 25% at 5 nm IGF-1, which is far less than the severalfold increase observed in the case of LPA or PMA.
FIGURE 6.
Stimulation of amphiregulin release from HMEC by different agonists. A, cells were seeded at ∼300,000 cells/35-mm dish and then serum-deprived the following day. Following an 18-h incubation in low serum medium, the cells were treated with IGF-1, LPA, UTP, HGF, or PMA for 2 h. Medium was collected from each dish and then analyzed for amphiregulin release by ELISA. B, HMEC were incubated overnight in serum-free medium and then changed to fresh medium containing IGF-1 at concentrations ranging from 0 to 10 nm for 2 h. The conditioned medium was collected and analyzed for amphiregulin release using an ELISA, as described under “Experimental Procedures.” C, HMEC were incubated overnight in serum-free medium before adding IGF-1 (10 nm), LPA (20 μm), or EGF (10 ng/ml) for 5 min (white bars). Alternately, the medium was changed before agonist addition for 1 h (gray bar) or 2 h (dark gray bar). The results are means from three samples ± S.D.
Because the increase in AR production following IGF-1 stimulation is small relative to the magnitude of ERK activation, it seemed likely that a mechanism other than induced ligand shedding must be involved. However, any alternate mechanism must explain why ERK activation by IGF-1 is dependent on both the presence of AR and EGFR activity. One possibility is that IGF-1 binding to its receptor could amplify EGFR signaling arising from constitutively released AR.
Our normal experimental protocol is to change cells to fresh medium the evening before an experiment and then add stimulants in a small aliquot. This protocol would allow AR to accumulate in the medium before the cells were stimulated. If IGF-1 is sensitizing the EGFR to constitutively produced AR, then we should see a reduction in its effect if we changed cells to fresh medium shortly before adding IGF-1. To test this idea, we grew cells at low density and either stimulated them with IGF-1, LPA, or EGF using our normal protocol or changed them to fresh medium for 1 or 2 h to allow any prebound ligand to dissociate. As shown in Fig. 6C, changing the medium had little or no effect on cells treated with either LPA or EGF. In the case of IGF-1-treated cells, however, changing the medium significantly reduced the level of ERK phosphorylation, consistent with the hypothesis that IGF-1 works by increasing cell sensitivity to constitutively produced AR.
Enhancement of EGFR Signaling by IGF-1—To critically test the hypothesis that IGF-1 treatment increases the sensitivity of cells to EGFR ligands, we measured EGFR phosphorylation, Ras activity, and ERK phosphorylation simultaneously as a function of EGF dose in the presence or absence of IGF-1. We measured EGFR and ERK phosphorylation directly using specific ELISAs, whereas Ras activity was measured by affinity isolation using an RBD-glutathione S-transferase fusion protein (41). It is known that ligand binding activates ERK by first inducing EGFR phosphorylation and then Ras activation, which controls the Raf-MEK-ERK cascade (42). By examining the effect of IGF-1 treatment at three different levels of the EGFR signaling pathway, we should see the level at which IGF-1 exerts its effects.
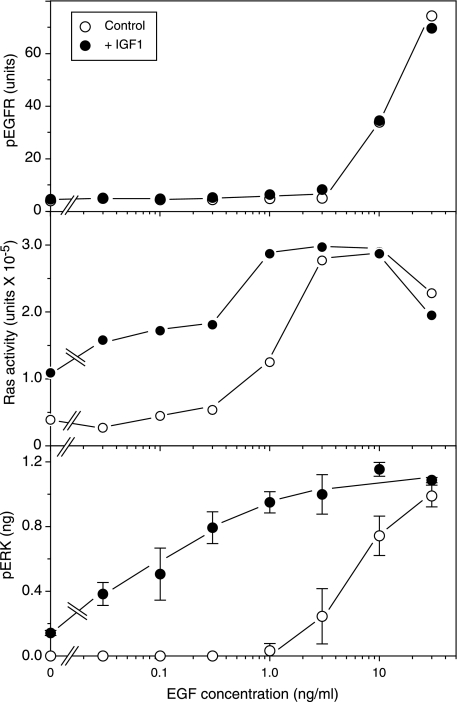
As shown in Fig. 7 (bottom), the addition of EGF alone had little effect on ERK phosphorylation below a concentration of 3 ng/ml. In the presence of 10 nm IGF-1, however, an increase was observed at 0.03 ng/ml, an ∼30-fold increase in sensitivity. Similar results were obtained using AR as an agonist rather than EGF (data not shown). An enhancement was also seen when Ras activation was evaluated as well (Fig. 7, middle). In the absence of IGF-1, there was little activity below 1 ng/ml EGF. In the presence of IGF-1, however, there was an increase of Ras activity at 0.03 ng/ml EGF. The increase in both Ras and ERK activity seen by the addition of IGF-1 alone required constitutive autocrine signaling, since it could be completely blocked with EGFR-blocking antibodies (data not shown; also see below).
FIGURE 7.
Sensitization of the response of HMEC to EGFR activation by treatment with IGF-1. HMEC were incubated overnight in serum-free medium and treated with (closed circle) or without (open circle) IGF-1 (10 nm) and EGF at the indicated final concentration, and cells were harvested 5 min later. ELISA assays were used to quantify EGFR phosphorylation (top) or ERK phosphorylation (bottom) as described under “Experimental Procedures.” Alternately, cells were lysed, and activated Ras was isolated using RBD-glutathione S-transferase bead affinity isolation. The amount of isolated Ras was then quantified by Western blot analysis, followed by densitometry. The amount of Ras from each sample is expressed in arbitrary densitometry units.
To determine whether the IGF-1-induced sensitization of signaling was at the level of EGFR activity, we measured the levels of EGFR tyrosine phosphorylation as a function of EGF concentration in the presence and absence of IGF-1. The level of receptor phosphorylation remained minimal below an EGF concentration of 3 ng/ml, above which it increased sharply (Fig. 7, bottom). This corresponds approximately to the dose required for induction of ERK phosphorylation in the absence of IGF-1. However, there was no significant dose-dependent change in EGFR phosphotyrosine levels when IGF-1 was added. The small difference that was observed at 3 ng/ml EGF was not reproducible in replicate experiments (data not shown). These data suggest that IGF-1 exerts its mechanism of action downstream of the EGFR but upstream of Ras.
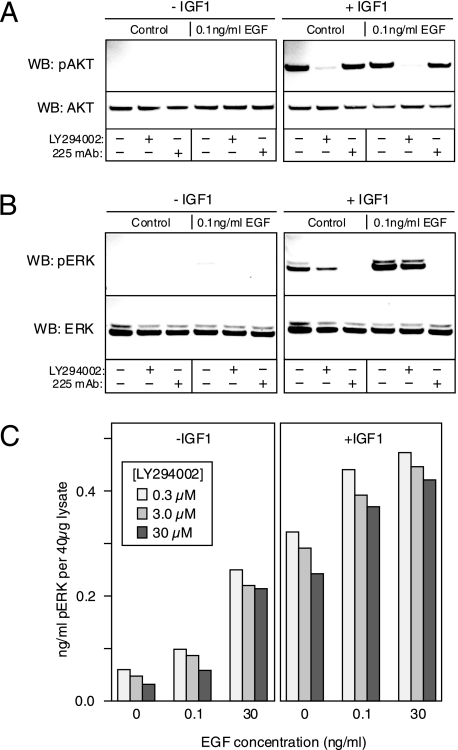
Activation of PI3K Is Not Involved in the Sensitization Activity of IGF-1—One potential mechanism by which the IGF-1 could enhance the activity of the EGFR is by stimulating the PI3K pathway. It has been shown recently that activation of PI3K can greatly stimulate EGFR signaling at low ligand concentrations by recruiting the adaptor protein Gab1 to the plasma membrane (43). To determine whether IGF-1 could be affecting this mechanism, we first determined whether IGF-1 treatment could activate PI3K using phosphorylation of AKT as a surrogate end point. As shown in Fig. 8A, IGF-1 is capable of stimulating AKT phosphorylation equivalently in the presence or absence of 0.1 ng/ml EGF, and furthermore, the presence of mAb 225 has no effect on the level of AKT phosphorylation in response to IGF-1. As expected, treatment with LY294002 substantially inhibited AKT phosphorylation in response to IGF-1. These results clearly indicate that IGF-1 treatment can activate the PI3K-AKT pathway independent of the EGFR.
FIGURE 8.
Sensitization of the EGFR response by IGF-1 is independent of PI3K activity. A, HMEC were pretreated with LY294002 (30 μm) or mAb 225 (10 μg/ml) or vehicle (DMSO) for 1 h and then stimulated with or without 0.1 ng/ml of EGF for 10 min in either the absence (left) or presence (right) of 10 nm IGF-1. The cells were then solubilized, and the extract was evaluated by Western blot (WB) analysis using antibodies against AKT or phospho-AKT as indicated. B, same as A, except the antibodies used were specific for ERK and phospho-ERK. C, HMEC were treated with the indicated concentrations of LY294002 for 1 h prior to stimulation with the indicated concentration of EGF for 5 min in the presence or absence of IGF-1. Cells were then extracted, and the levels of phospho-ERK were quantified by ELISA. Shown are averages of duplicate samples.
We next examined whether activation of PI3K was required for the increase in ERK phosphorylation observed in response to IGF-1. As seen in Fig. 8B, the PI3K inhibitor LY294002 had only a minor effect on ERK phosphorylation in response to IGF-1 alone. The addition of IGF1 greatly enhanced ERK phosphorylation stimulated by 0.1 ng/ml EGF, but although this response was completely blocked by mAb 225, LY294002 had little effect. This was true over a wide range of LY294002 concentrations, as shown by the results of a quantitative phospho-ERK ELISA (Fig. 8C). Essentially identical results were obtained using wortmannin as a PI3K inhibitor (data not shown). These results suggest that IGF-1 enhances EGFR signaling independent of PI3K activity.
DISCUSSION
In this study, we have demonstrated that activation of the ERK pathway in HMEC by a large variety of distinct ligand-receptor pairs was mediated through transactivation of the EGFR. Factors that could transactivate the EGFR acted through both GPCR and RTKs as well as through general cell stressors, such as osmotic shock (sorbitol) and inhibition of protein synthesis (anisomycin). Transactivation of the EGFR has been well documented for several GPCRs, most notably endothelin, LPA, and β-adrenergic receptors (2, 7, 8, 44, 45) as well as for IGF receptors (10, 46), but this is the first report demonstrating that such a breadth of transactivation mechanisms can operate simultaneously in a single cell type. The general paradigm for GPCR-induced transactivation involves the activation of membrane-associated metalloproteases of either the ADAM or MMP families. The metalloproteases then cleave membrane-tethered ligands of the EGF family, such as TGFα, HB-EGF, or AR. The newly soluble ligands then bind to receptors of the EGFR family, inducing ligand-dependent dimerization and activation (1, 4, 12). Downstream signaling from the activated EGFR receptors is presumed to be the same from this point as it is after adding exogenous ligands.
To determine how such a wide range of different agonists could stimulate the same pathway, we focused on two that could stimulate GPCRs (LPA and UTP) and one that activated an RTK (IGF-1). We found that LPA, UTP, and IGF-1 induction of ERK phosphorylation or cell proliferation was inhibited by EGFR kinase inhibitors or ligand-blocking antibodies, implicating ligand-dependent EGFR activation in the action of these agents. Activation of c-Met by HGF provided a useful contrast, since neither HGF-induced proliferation nor ERK phosphorylation was affected by inhibiting the EGFR, indicating that c-Met activates the MAPK cascade by an independent mechanism.
Although there was a general correspondence between the ability of various agonists to induce ERK phosphorylation and the ability of the same agonists to stimulate a proliferative response (e.g. EGF and HGF were potent as both mitogens and activators of the MAPK pathway), there were some notable exceptions. For example, sorbitol induced rapid phosphorylation of ERK but was antiproliferative, and TNFα induced a strong mitogenic response without producing a robust activation of MAPK. Clearly, ERK activation alone is insufficient to drive a full mitogenic response, and coordinated activation of other pathways, such as the PI3K pathway, may substantially modify the proliferative response.
The mechanisms of EGFR transactivation in HMEC appear to diverge at the level of regulated proteolysis, in that three different metalloprotease inhibitors with differing specificities produced different results. For example, the induction of ERK phosphorylation in response to LPA, IGF-1, TNFα, VEGF, and GH were all significantly inhibited by Galardin, whereas Batimastat, which appears to be less potent, proved ineffective against IGF-1 but highly effective against LPA- and UTP-induced ERK phosphorylation. This distinction held when WAY-022, a selective inhibitor of ADAM 17 with minimal activity against ADAM 10, was used. WAY-022 was highly effective at inhibiting the LPA and UTP responses but ineffective against IGF-1.
These results indicate that the two GPCR agonists rely on activation of ADAM 17 to induce proteolytic release of EGFR ligand, whereas IGF-1 relies predominantly on an alternate mechanism of transactivation that is dependent on other metalloproteases. Ligand antibody studies supported this notion, with AR antibodies being potent inhibitors of IGF-1-induced transactivation, whereas combinations of TGFα and EPR antibodies were the most effective inhibitors of GPCR-induced transactivation.
It should be noted that antibodies against high affinity ligands, such as TGFα, are poorly effective in blocking autocrine signaling. As we have previously shown, micromolar concentrations of anti-TGFα antibodies would be required to effectively block autocrine signaling mediated by this ligand, because it never enters the bulk medium prior to being captured by the EGFR (47). Thus, the significant inhibition we observe in LPA-mediated transactivation in response to the addition of relatively low concentrations of anti-TGFα antibodies (10 μg/ml) is quite striking. In contrast, low affinity ligands, such as AR, can be blocked much more effectively by antibodies, because they enter the bulk medium, where they can be captured (48). Inhibitors that bind to membrane-anchored proteins, such as mAb 225, against the EGFR or CRM197 against HB-EGF are also very effective, because they can persistently accumulate on the cell surface during the entire incubation period. The ability of anti-AR antibodies alone to completely inhibit IGF-1 transactivation of the EGFR strongly suggests that AR is the only ligand involved in this process.
The simplest explanation for the differences between IGF-1 and the other agonists would have been that it specifically induced the shedding of AR. However, direct measurements of ligand release did not support this idea. Instead, it appears that IGF-1 has little effect on the rate of AR shedding, suggesting that it operates through an alternate mechanism.
In contrast to the weak effect of IGF-1 on ligand shedding, LPA had a pronounced effect on the shedding rate of all three ligands that we could measure. Our antibodies against EPR were unsuitable for use in an ELISA, so we cannot evaluate the level at which HMEC produce this ligand. Based on the observed inhibitory effect of anti-EPR antibodies on GPCR-mediated transactivation, we suspect that it is an important ligand in the biology of these cells. UTP did not induce a significant release of any ligand, but it is a very labile agonist and probably could not persist during the time required for our ligand-shedding assays (2–4 h). Although HGF did not transactivate the EGFR, it was a surprisingly effective inducer of ligand shedding, probably because of its ability to directly stimulate ERK activation. It has been shown previously that EGFR ligand shedding is coupled to ERK activation (49). Conversely, although IGF-1 is a good transactivation agent, it has little if any ability to stimulate ligand shedding. These results directly demonstrate that ligand shedding is not necessarily coupled to EGFR transactivation but is at variance with a previous report that IGF-1 is a strong stimulator of HB-EGF shedding in HEK-293 and MEF cells (46). Our finding on the lack of IGF-1-induced shedding might be explained by differences in cell types (epithelial cells in our studies versus mesenchymal HEK-293 and MEF cells in Ref. 46). Alternately, it could be because IGF-1 can induce a robust ERK response in HEK-293 or MEF cells independently of the EGFR and thus could act in a similar fashion as HGF.
The inability of IGF-1 treatment to stimulate a significant increase in AR release was initially puzzling. However, we noticed that the constitutive level of AR shedding in HMEC was ∼400 pg/h/106 cells. This is 10- and 100-fold higher than the basal level of TGFα and HB-EGF production, respectively. Despite the high rate of constitutive AR release, there was little indication that the EGFR system was constitutively active; thus, we reasoned that some process must be inhibiting AR activity. If IGF-1 treatment released this inhibitory mechanism, then the constitutively released AR would be able to activate the ERK pathway. Importantly, this “sensitization” mechanism would appear similar to a ligand-shedding mechanism with respect to its dependence on both metalloprotease and EGFR activity.
Sensitization of cells to constitutively produced AR could occur by an increase in ligand accessibility to the receptor or by increasing the amount of signal produced by each occupied receptor. Our data were consistent with the latter hypothesis. In the presence of IGF-1, small amounts of EGF caused a very large increase in ERK phosphorylation. The sensitization occurred downstream of the EGFR itself but upstream of Ras activation, because IGF-1 did not change the degree of EGFR self-phosphorylation in response to EGF but did enhance the ability of the EGFR to activate Ras. In the absence of added EGF, IGF-1 appears to require the presence of accumulated AR, because changing to fresh extracellular medium or adding anti-AR antibodies significantly reduced the ability of IGF-1 to stimulate ERK phosphorylation. This demonstrates that IGF-1 induces “transactivation” of the EGFR by enhancing constitutive autocrine signaling by a postreceptor mechanism.
An attractive candidate mechanism for IGF-1 action was activation of the PI3K pathway. It is known that stimulation of the IGFR can induce strong activation of the PI3K pathway (10) and that PI3K activity enhances signaling through the EGFR at low ligand concentrations (43). However, blocking PI3K activity by either LY294002 or wortmannin had no effect on the ability of IGF-1 to enhance signaling through the EGFR, although it did block its ability to stimulate AKT phosphorylation. The actual mechanism by which IGF-1 acts to increase EGFR activity is unknown, but it could involve an increase in substrate availability for receptor phosphorylation or binding or suppression of an inhibitory mechanism. Studies are currently in progress to explore these possibilities.
In contrast to IGF-1, LPA appeared to activate the EGFR by stimulating the shedding of multiple ligands. Although the induction of HB-EGF shedding by LPA was very substantial, blocking HB-EGF availability had little effect on cellular responses. This is probably because of the very small amounts of HB-EGF produced by these cells relative to other ligands, such as TGFα. The actual mechanism by which the LPA receptor and other membrane receptors activate ADAMs and MMPs is currently unclear, although there is evidence in the literature for both PKC-dependent (50, 51) and c-Src-dependent (5, 52–54) pathways. We found that inhibitors of PKC, such as staurosporine, had no effect on LPA-induced ligand shedding or ERK activation, but the Src family kinase inhibitor PP2 inhibited both IGF-1- and LPA-induced ERK phosphorylation.4 These data implicate Src family kinases as mediators of ligand shedding in HMEC.
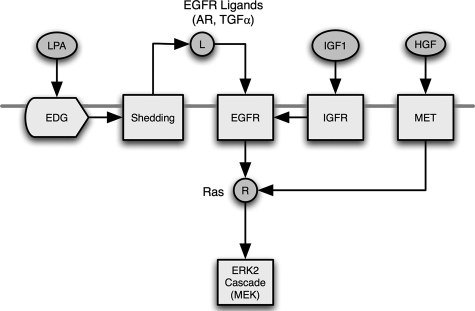
The results presented here suggest that a broad variety of different receptor types and stressors activate the ERK pathway in mammary epithelial cells through transactivation of the EGFR. This transactivation requires shedding of autocrine ligands that activate the EGFR, but different receptor types appear to stimulate transactivation at different points in the EGFR signaling pathway (see Fig. 9). A unique finding in our study was that IGF-1 appears to transactivate the EGFR by increasing its sensitivity to constitutively produced AR rather than by stimulating ligand shedding. This leads to a different perspective on EGFR transactivation itself. Instead of considering it a “stimulus-response” process in which stimulation of one signaling pathway simply activates the EGFR, transactivation is better regarded as a perturbation of a steady-state autocrine circuit. In this model, autocrine signaling occurs at a low, continuous level under normal conditions because of the balance between constitutive ligand production (positive) and receptor desensitization (negative). The addition of a transactivating agonist alters the steady state balance between the positive and negative aspects of the autocrine circuit, resulting in a change in ERK activity. Seen in this way, transactivation can be considered as a perturbation in “signaling homeostasis” of the EGFR autocrine system. It is known that the EGFR is a central regulator of epithelial cell proliferation and differentiation. Its ability to serve as a broad-spectrum integrator of multiple cellular stimuli could explain its importance in epithelial cell biology.
FIGURE 9.
Summary of the multiple pathways for EGFR transactivation and ERK activation in HMEC. Modulators of G-protein-coupled receptors, such as LPA, stimulate the shedding of EGFR ligands, such as AR or TGFα, which in turn activate the EGFR. Alternately, IGF1 working through its receptor stimulates the activity of the EGFR through an unknown mechanism upstream of Ras. HGF operating through its own receptor can activate Ras and ERK independently of the EGFR.
This work was supported, in whole or in part, by National Institutes of Health, NCI, Grant CA 46413 (to R. J. C.). This work was also supported by the Biomolecular Systems Initiative Laboratory Directed Research and Development Program at the Pacific Northwest National Laboratory, a multiprogram national laboratory operated by Battelle for the United States Department of Energy under Contract DE-AC05-76RL01830 and by Gastrointestinal Special Program of Research Excellence Grant P50 95103 (to R. J. C.). The costs of publication of this article were defrayed in part by the payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 U.S.C. Section 1734 solely to indicate this fact.
Footnotes
The abbreviations used are: EGFR, epidermal growth factor receptor; GPCR, G-protein-coupled receptor; RTK, receptor tyrosine kinase; ADAM, a disintegrin and metalloprotease; IGF-1, insulin-like growth factor 1; IGFR, insulin-like growth factor receptor; LPA, lysophosphatidic acid; HMEC, human mammary epithelial cell(s); TGFα, transforming growth factor α; EGF, epidermal growth factor; HB-EGF, heparin-binding EGF-like growth factor; AR, amphiregulin; EPR, epiregulin; VEGF, vascular endothelial cell-derived growth factor; HGF, hepatocyte growth factor/scatter factor; GH, growth hormone; mAb, monoclonal antibody; MAPK, mitogen-activated protein kinase; ERK, extracellular signal-regulated kinase; MMP, matrix metalloprotease; MTT, 3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide; BisTris, 2-[bis(2-hydroxyethyl)amino]-2-(hydroxymethyl)propane-1,3-diol; ELISA, enzyme-linked immunosorbent assay; PI3K, phosphatidylinositol 3-kinase.
R. Zangar, unpublished observations.
K. Rodland, unpublished observations.
References
- 1.Gschwind, A., Zwick, E., Prenzel, N., Leserer, M., and Ullrich, A. (2001) Oncogene 20 1594–1600 [DOI] [PubMed] [Google Scholar]
- 2.Schafer, B., Gschwind, A., and Ullrich, A. (2004) Oncogene 23 991–999 [DOI] [PubMed] [Google Scholar]
- 3.Kue, P. F., Taub, J. S., Harrington, L. B., Polakiewicz, R. D., Ullrich, A., and Daaka, Y. (2002) Int. J. Cancer 102 572–579 [DOI] [PubMed] [Google Scholar]
- 4.Prenzel, N., Zwick, E., Daub, H., Leserer, M., Abraham, R., Wallasch, C., and Ullrich, A. (1999) Nature 402 884–888 [DOI] [PubMed] [Google Scholar]
- 5.Xi, S., Zhang, Q., Dyer, K. F., Lerner, E. C., Smithgall, T. E., Gooding, W. E., Kamens, J., and Grandis, J. R. (2003) J. Biol. Chem. 278 31574–31583 [DOI] [PubMed] [Google Scholar]
- 6.Vacca, F., Bagnato, A., Catt, K. J., and Tecce, R. (2000) Cancer Res. 60 5310–5317 [PubMed] [Google Scholar]
- 7.Daub, H., Wallasch, C., Lankenau, A., Herrlich, A., and Ullrich, A. (1997) EMBO J. 16 7032–7044 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Tomlins, S. A., Bolllinger, N., Creim, J., and Rodland, K. D. (2005) Exp. Cell Res. 308 439–445 [DOI] [PubMed] [Google Scholar]
- 9.Chen, W. N., Woodbury, R. L., Kathmann, L. E., Opresko, L. K., Zangar, R. C., Wiley, H. S., and Thrall, B. D. (2004) J. Biol. Chem. 279 18488–18496 [DOI] [PubMed] [Google Scholar]
- 10.Roudabush, F. L., Pierce, K. L., Maudsley, S., Khan, K. D., and Luttrell, L. M. (2000) J. Biol. Chem. 275 22583–22589 [DOI] [PubMed] [Google Scholar]
- 11.Ohtsu, H., Dempsey, P. J., Frank, G. D., Brailoiu, E., Higuchi, S., Suzuki, H., Nakashima, H., Eguchi, K., and Eguchi, S. (2006) Arterioscler. Thromb. Vasc. Biol. 26 e133–e137 [DOI] [PubMed] [Google Scholar]
- 12.Fischer, O. M., Hart, S., Gschwind, A., and Ullrich, A. (2003) Biochem. Soc. Trans. 31 1203–1208 [DOI] [PubMed] [Google Scholar]
- 13.Jacobs, J. M., Rong, L.-R., Mottaz, H. M., Anderson, D. J., Moore, R. J., Chen, W.-N. U., Auberry, K. J., Strittmatter, E. F., Monroe, M. E., Thrall, B. D., Camp, D. G. I., and Smith, R. D. (2004) J Proteome Research 3 68–75 [DOI] [PubMed] [Google Scholar]
- 14.Klapper, L. N., Glathe, S., Vaisman, N., Hynes, N. E., Andrews, G. C., Sela, M., and Yarden, Y. (1999) Proc. Natl. Acad. Sci. U. S. A. 96 4995–5000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Stortelers, C., van der Woning, S. P., Jacobs-Oomen, S., Wingens, M., and van Zoelen, E. J. (2003) J. Biol. Chem. 278 12055–12063 [DOI] [PubMed] [Google Scholar]
- 16.Wolf-Yadlin, A., Kumar, N., Zhang, Y., Hautaniemi, S., Zaman, M., Kim, H. D., Grantcharova, V., Lauffenburger, D. A., and White, F. M. (2006) Mol. Syst. Biol. 2 54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Wiley, H. S., Shvartsman, S. Y., and Lauffenburger, D. A. (2003) Trends Cell Biol. 13 43–50 [DOI] [PubMed] [Google Scholar]
- 18.Hendriks, B. S., Orr, G., Wells, A., Wiley, H. S., and Lauffenburger, D. A. (2005) J. Biol. Chem. 280 6157–6169 [DOI] [PubMed] [Google Scholar]
- 19.Shankaran, H., Wiley, H. S., and Resat, H. (2006) Biophys. J. 90 3993–4009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Liu, T., Qian, W. J., Chen, W. N., Jacobs, J. M., Moore, R. J., Anderson, D. J., Gritsenko, M. A., Monroe, M. E., Thrall, B. D., Camp, D. G., II, and Smith, R. D. (2005) Proteomics 5 1263–1273 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Zhang, Y., Wolf-Yadlin, A., Ross, P. L., Pappin, D. J., Rush, J., Lauffenburger, D. A., and White, F. M. (2005) Mol. Cell Proteomics 4 1240–1250 [DOI] [PubMed] [Google Scholar]
- 22.Dong, J., Opresko, L. K., Chrisler, W., Orr, G., Quesenberry, R. D., Lauffenburger, D. A., and Wiley, H. S. (2005) Mol. Biol. Cell 16 2984–2998 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Monine, M. I., Berezhkovskii, A. M., Joslin, E. J., Wiley, H. S., Lauffenburger, D. A., and Shvartsman, S. Y. (2005) Biophys. J. 88 2384–2390 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Brown, P. D. (1998) Breast Cancer Res. Treat. 52 125–136 [DOI] [PubMed] [Google Scholar]
- 25.Solorzano, C. C., Ksontini, R., Pruitt, J. H., Auffenberg, T., Tannahill, C., Galardy, R. E., Schultz, G. P., MacKay, S. L., Copeland, E. M., III, and Moldawer, L. L. (1997) Shock 7 427–431 [DOI] [PubMed] [Google Scholar]
- 26.Band, V., and Sager, R. (1989) Proc. Natl. Acad. Sci. U. S. A. 86 1249–1253 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Dreesen, O., and Brivanlou, A. H. (2007) Stem Cell Rev. 3 7–17 [DOI] [PubMed] [Google Scholar]
- 28.Al-Obeidi, F. A., and Lam, K. S. (2000) Oncogene 19 5690–5701 [DOI] [PubMed] [Google Scholar]
- 29.Lee, D. C., Sunnarborg, S. W., Hinkle, C. L., Myers, T. J., Stevenson, M. Y., Russell, W. E., Castner, B. J., Gerhart, M. J., Paxton, R. J., Black, R. A., Chang, A., and Jackson, L. F. (2003) Ann. N. Y. Acad. Sci. 995 22–38 [DOI] [PubMed] [Google Scholar]
- 30.Pierce, K. L., Tohgo, A., Ahn, S., Field, M. E., Luttrell, L. M., and Lefkowitz, R. J. (2001) J. Biol. Chem. 276 23155–23160 [DOI] [PubMed] [Google Scholar]
- 31.Gschwind, A., Hart, S., Fischer, O. M., and Ullrich, A. (2003) EMBO J. 22 2411–2421 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Santiskulvong, C., and Rozengurt, E. (2003) Exp. Cell Res. 290 437–446 [DOI] [PubMed] [Google Scholar]
- 33.Stampfer, M. R., and Bartley, J. C. (1988) Cancer Treat. Res. 40 1–24 [DOI] [PubMed] [Google Scholar]
- 34.Stampfer, M. R., and Yaswen, P. (1993) Cancer Surv. 18 7–34 [PubMed] [Google Scholar]
- 35.Bercherer, J. D., and Blobel, C. P. (2003) Curr. Top. Dev. Biol. 54 2031–2044 [DOI] [PubMed] [Google Scholar]
- 36.Yan, Y., Shirakabe, K., and Werb, Z. (2002) J. Cell Biol. 158 221–226 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Dong, J., Opresko, L. K., Dempsey, P. J., Lauffenburger, D. A., Coffey, R. J., and Wiley, H. S. (1999) Proc. Natl. Acad. Sci. U. S. A. 96 6235–6240 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Merchant, N. B., Rogers, C. M., Trivedi, B., Morrow, J., and Coffey, R. J. (2005) Surgery 138 415–421 [DOI] [PubMed] [Google Scholar]
- 39.Mitamura, T., Higashiyama, S., Taniguchi, N., Klagsbrun, M., and Mekada, E. (1995) J. Biol. Chem. 270 1015–1019 [DOI] [PubMed] [Google Scholar]
- 40.Dong, J., and Wiley, H. S. (2000) J. Biol. Chem. 275 557–564 [DOI] [PubMed] [Google Scholar]
- 41.de Rooij, J., and Bos, J. L. (1997) Oncogene 14 623–625 [DOI] [PubMed] [Google Scholar]
- 42.McKay, M. M., and Morrison, D. K. (2007) Oncogene 26 3113–3121 [DOI] [PubMed] [Google Scholar]
- 43.Sampaio, C., Dance, M., Montagner, A., Edouard, T., Malet, N., Perret, B., Yart, A., Salles, J. P., and Raynal, P. (2008) Mol. Cell. Biol. 28 587–600 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Hart, S., Fischer, O. M., Prenzel, N., Zwick-Wallasch, E., Schneider, M., Hennighausen, L., and Ullrich, A. (2005) Biol. Chem. 386 845–855 [DOI] [PubMed] [Google Scholar]
- 45.Zwick, E., Hackel, P. O., Prenzel, N., and Ullrich, A. (1999) Trends Pharmacol. Sci. 20 408–412 [DOI] [PubMed] [Google Scholar]
- 46.El-Shewy, H. M., Kelly, F. L., Barki-Harrington, L., and Luttrell, L. M. (2004) Mol. Endocrinol. 18 2727–2739 [DOI] [PubMed] [Google Scholar]
- 47.Lauffenburger, D. A., Oehrtman, G. T., Walker, L., and Wiley, H. S. (1998) Proc. Natl. Acad. Sci. U. S. A. 95 15368–15373 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.DeWitt, A., Iida, T., Lam, H. Y., Hill, V., Wiley, H. S., and Lauffenburger, D. A. (2002) Dev. Biol. 250 305–316 [PubMed] [Google Scholar]
- 49.Fan, H., and Derynck, R. (1999) EMBO J. 18 6962–6972 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Diaz-Rodriguez, E., Montero, J. C., Esparis-Ogando, A., Yuste, L., and Pandiella, A. (2002) Mol. Biol. Cell 13 2031–2044 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Montero, J. C., Yuste, L., Diaz-Rodriguez, E., Esparis-Ogando, A., and Pandiella, A. (2002) Biochem. J. 362 211–221 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Shah, B. H., Farshori, M. P., Jambusaria, A., and Catt, K. J. (2003) J. Biol. Chem. 278 19118–19126 [DOI] [PubMed] [Google Scholar]
- 53.Wu, W., Graves, L. M., Gill, G. N., Parsons, S. J., and Samet, J. M. (2002) J. Biol. Chem. 277 24252–24257 [DOI] [PubMed] [Google Scholar]
- 54.Zhang, Q., Thomas, S. M., Xi, S., Smithgall, T. E., Siegfried, J. M., Kamens, J., Gooding, W. E., and Grandis, J. R. (2004) Cancer Res. 64 6166–6173 [DOI] [PubMed] [Google Scholar]