Abstract
Recent studies using SOCS family knock-out mice have suggested that SOCS proteins have multiple biological functions in addition to their role as negative regulators of JAK-STAT signaling. To explore these other functions of this family of proteins, we used yeast two-hybrid screening to find proteins interacting with human SOCS-3. We identified the transcriptional factor DP-1 as a SOCS-3-interacting protein involved in regulation of the cell cycle. Immunoprecipitation-Western blot assay showed that this interaction between these endogenous proteins occurred in cells both in vitro and in vivo. SOCS-3 interacted with the C-terminal region of DP-1, and amino acids 156–172 of SOCS-3 were required for this interaction. Confocal microscopy revealed that SOCS-3 and DP-1 were primarily colocalized in the cytoplasm. SOCS-3 inhibited E2F/DP-1 transcriptional activity under the cyclin-E promoter and actually inhibited cell cycle progression and cell growth under E2F/DP-1 control. In contrast, DP-1 almost completely eliminated the inhibitory action of SOCS-3 on LIF-stimulated STAT-3 transcriptional activity in JAK-STAT signaling. Interestingly, the alternative regulatory action of SOCS-3 and DP-1 was dramatically eliminated by each siRNA. Taken together, these findings demonstrate that SOCS-3 acts as a negative regulator of the cell cycle progression under E2F/DP-1 control by interfering with heterodimer formation between DP-1 and E2F and also that DP-1 plays an important role in controlling JAK-STAT signaling.
Members of the family of suppressor of cytokine signaling (SOCS),2 designated SOCS-1 to SOCS-7 and CIS, are induced by stimulation via several kinds of cytokines and growth factors (1–3). These proteins regulate JAK-STAT signaling in a classical negative feedback loop of the signaling cascade (4, 5). SOCS family proteins also act as an important regulator of cell differentiation, as evidenced by the following findings: SOCS-1 suppresses muscle differentiation (6); SOCS-2 regulates neuronal differentiation (7); SOCS-3 induces myoblast differentiation (8); and SOCS-3 and SOCS-5 are involved in T helper cell differentiation (9, 10). In addition, these proteins are thought to strongly contribute to the development and progression of several kinds of tumors such as hepatocellular carcinoma (11–13), chronic myeloid leukemia (14), ovarian and breast carcinoma (15), and so on (16–21). These observations suggested to us that SOCS family proteins might exhibit multipotential functions as regulators of cell differentiation and tumor cell growth besides being a negative regulator of JAK-STAT signaling. To explore this possibility, we considered that identification of SOCS-interacting proteins would be an extraordinarily good strategy. Thus, using the yeast two-hybrid screening system, we sought to identify presently proteins interacting with human SOCS-3. As a result, we found DP-1, a transcriptional factor for cell cycle regulation, to be such a SOCS-3-interacting protein.
DP-1 was first identified as a partner protein of E2F-1, the latter being a transcriptional factor for regulating the cell cycle (22, 23). Interestingly, a recent study (24) demonstrated that knocking out DP-1 in mice results in embryonic lethality, due to a failure in the development of extra-embryonic tissues. This finding suggests that DP-1 plays an important role in cell development and morphogenesis. DP-1/E2F family proteins contain two well-characterized DNA binding and heterodimerization domains. Three DP subunits and 8 E2F subunits presently have been characterized. For example, recently, a new member of the DP family, DP-3, was reported (25, 26). As for DP-1 isoforms, we also previously identified 2 new isoforms (DP-1α and -β) of DP-1 (27). DP-3 and DP-1α exhibited an inhibitory effect on E2F/DP-1 transcriptional activity (25–27). Although DP-2 has a clear nuclear localization signal (NLS) and is localized in nuclei (28), DP-1 and -3 have no clear NLS; and some of these proteins reside in the cytoplasm. The functions of these cytoplasmic DPs are not yet clear. On the other hand, the 8 members of the E2F family can be classified into 4 subfamilies by their properties and structures. Perhaps many combinations of DP and E2F are involved in the regulation of gene expression in vivo. Although many studies (29–31) have demonstrated that the retinoblastoma protein (pRb) negatively regulates the transcriptional activity of E2F and tumor progression, the molecular mechanisms responsible for negative regulation of the DP family in transcription and tumor progression have not been well characterized.
Therefore, we investigated the possibility of an inhibitory action of SOCS-3 on DP-1 function to understand the mechanism of SOCS family involvement in the regulation of tumor cell growth and cell differentiation. Interestingly, we found that the SOCS-3 inhibited E2F/DP-1 transcriptional activity by interfering with heterodimer formation between DP-1 and E2F and was actually able to retard the cell cycle progression and cell growth. These findings suggest a novel function of human SOCS-3 as a potent regulator of cell cycle progression.
EXPERIMENTAL PROCEDURES
Plasmids and Constructs—Human SOCS-3 cDNA was amplified from a human testis cDNA library (BD Biosciences) by PCR using specific primers (forward, 5′-ATGGTCACCCACAGCAAGTTTCCC-3′; reverse, 5′-TTAAAGCGGGGCATCGTACTGGTC-3′) and Pyrobest DNA polymerase (Takara). The PCR conditions were as follow: preincubation at 98 °C for 20 s and then 28 cycles of denaturation at 98 °C for 15 s, annealing at 70 °C for 20 s, and extension at 72 °C for 2 min 30 s. The PCR products were electrophoresed on 1.5% agarose gels, and fragments were extracted using a QIAEX II kit (Qiagen). These extracted fragments were amplified by PCR under the same conditions using specific primers containing EcoRI and BamHI sites. The PCR products were digested with EcoRI and BamHI and cloned into the same sites of pGBT9 (BD Biosciences). For generation of Flag-SOCS-3 (WT), SOCS-3 cDNA was digested by EcoRI and SalI from SOCS-3pGBT9 and cloned into the EcoRI and XhoI sites of Flag-pcDNA3 (Invitrogen). cDNA fragments of SOCS-3 deletions were amplified from full-length SOCS-3 cDNA by PCR using specific primers and Pyrobest DNA polymerase (Takara). GFP-DP-1 and 6xMyc-DP-1 expression plasmids were previously described (27). hDP-2 cDNA also was amplified from a human testis cDNA library (BD Biosciences) by PCR using specific primers (forward, 5′-ATGATTATAAGCACACCACAGAGACTAACCAGTTCAGG-3′; reverse, 5′-GAAACGTAGGCTTTCTCTTGTCTTTATTCTGGGGAG-3′). hDP-3 cDNA was kindly provided by Dr. W. F. Chen. hDP-2 and hDP-3 cDNAs were amplified by PCR using specific primers containing EcoRI and XhoI sites. The PCR products were digested and cloned into the same sites of pcDNA3–6xMyc. Cyclin-E-Luc reporter plasmid was kindly provided by Dr. M. Hatakeyama (32). The complete coding sequences of all constructs were verified by sequencing.
Yeast Two-hybrid Screen—Yeast cultures, the preparation of yeast selection media, and yeast transformations were carried out according to the manufacturer's protocol (BD Biosciences). In the first step of the two-hybrid library screening procedure, 100 ng of pGBT9-hSOCS-3 plasmid (bait) was used to transform AH109 yeast strain containing 4 reporter genes, i.e. lacZ, MEL1, HIS3, and ADE2. The transformants were mated with a pretransformed human skeletal muscle cDNA library in yeast strain Y190 (BD Biosciences). In the second step, the transformants were selected for growth on media lacking tryptophan, leucine, histidine, and adenine and containing glucose as the carbon source. Interactors were selected by plating on the same medium containing the substrate of β-galactosidase. Positive clones were grown in S.D. (-Leu) medium. Plasmids were extracted from Leu+ colonies and used to transform Escherichia coli (DH5α). E. coli clones containing pACT2-cDNA were selected by PCR. The plasmids were amplified, purified, and sequenced. The sequence data were matched to the NCBI data base.
Cell Culture and Transfection of HEK 293 Cells—Human embryonic kidney (HEK) 293 cells were cultured at 37 °C with 5% CO2 in Dulbecco's modified Eagle's medium (DMEM; Sigma) containing 100 units/ml penicillin, 100 μg/ml streptomycin, and 10% fetal calf serum (Thermo Trace) as previously described (27). For the luciferase and cell growth assays, plasmids and siRNAs were introduced into HEK 293 cells using Lipofectamine 2000 (Invitrogen). In other experiments, Polyfect Transfection Reagent (Qiagen) was also used for plasmid transfection. For establishing HEK 293 cells stably expressing Flag-SOCS-3, the cells were cultured at 90% confluence in 6-well dishes and then transfected with 4 μg of plasmid expressing Flag-SOCS-3. One day later, the transfected cells were subcultured and grown in the presence of 1 mg/ml of G418 (Invitrogen). After 2 weeks, single G418-resistant colonies were obtained by serial dilution in 96-well dishes. These colonies were then maintained in medium containing 1 mg/ml of G418 and analyzed individually for expression of Flag-SOCS3.
Immunoprecipitation and Immunoblotting Assays—HEK 293 cells (1 × 107) were transfected with Flag-SOCS-3 and 6xMyc-DP-1 expression plasmids using Polyfect (Qiagen) and then cultured for 24 h. The transfected cells were lysed in TNE buffer (10 mm Tris-HCl, pH 7.9, 150 mm NaCl, 1 mm EDTA) containing 0.1% Nonidet P-40 and a protease inhibitor mixture (Roche Applied Science). C57BL/6JJcl mouse (12–14 weeks old)-derived lung and testis were homogenized in TNE buffer using a pellet pestle (Kontes). After centrifugation at 13,500 rpm for 10 min at 4 °C, the supernatants (5 mg of total proteins) were used for each immunoprecipitation (IP) assay. Proteins in the cell lysate or tissue extracts were precipitated for 1 h at 4 °C with anti-Flag M2-agarose (Sigma) or anti-SOCS-3 polyclonal antibody (IBL) bound to protein A-Sepharose (Sigma). The immunocomplexes were washed five times with TNE buffer. After having been washed, the antigen of the immunocomplexes obtained with anti-Flag M2-agarose was eluted using Flag peptide (Sigma), and the supernatant was suspended in 2× SDS-PAGE sample buffer and boiled for 2 min at 98 °C. The immunocomplexes formed with anti-SOCS-3 polyclonal antibody bound to protein A-Sepharose were directly suspended in 2× SDS-PAGE sample buffer and boiled. The samples were separated by SDS-PAGE and analyzed by immunoblotting using mouse anti-Flag M2 monoclonal antibody (Sigma, 1:1,000 dilution), mouse anti-Myc monoclonal antibody 9E10 (Santa Cruz Biotechnology, 1:1,000 dilution), anti-SOCS-3 antibody or anti-DP-1 monoclonal antibody (Genetex, 1:1,000 dilution) as first antibodies. After four washes with TBST, incubation with secondary antibody was performed using peroxidase-conjugated goat anti-rabbit IgG (for anti-SOCS-3, abcam, 1:2,000 dilution) or peroxidase-conjugated rabbit anti-mouse IgG (for other monoclonal antibodies, DAKO Japan, 1:1,000 dilution). Finally, after four more washes with TBST, the immunoblots were apposed to Hyperfilm (GE Healthcare), which was then exposed for 15 s to 2 min.
Immunostaining and Confocal Fluorescence Microscopy—HEK 293 cells were divided into several 35-mm glass bottom culture dishes (MatTec Corp.) and transfected with the GFP-empty, GFP-DP-1, and/or Flag-SOCS-3 plasmids. Transfected cells were grown for 24 h at 37 °C in DMEM containing 10% fetal bovine serum. The cells were then washed three times with PBS (-) and fixed in 4% paraformaldehyde/PBS for 30 min at room temperature. They were then again washed three times with PBS (-). For the detection of Flag-SOCS-3, after the wash with PBS (-), the cells were incubated with anti-Flag M2 antibody (Sigma) in blocking solution at 1:1000 dilution for 1 h. They were subsequently washed five times with 0.1% Triton X-100/PBS (PBST) and thereafter incubated with Cy-5-labeled anti-mouse IgG (CHEMICON International) in blocking solution at 1:1000 dilution for 1 h. Finally, the cells were washed five times with PBST, and the coverslip was mounted with Vectashield (Vector Lab, Inc.). Samples were examined using an LSM 510 META confocal imaging system (Carl Zeiss). HEK 293 cells stably expressed Flag-SOCS-3 were also cultured in 35-mm glass bottom dishes and fixed by the same methods. The fixed cells were then incubated with anti-SOCS-3 polyclonal antibody (IBL) and anti-DP-1 monoclonal antibody SPM178 (Genetex) in blocking solution at 1:1000 dilution for 1 h. They were subsequently washed five times with PBST and thereafter incubated with Cy-5-labeled anti-mouse IgG (CHEMICON International, 1:1000 dilution) and Alexa Fluor 488-labeled anti-rabbit IgG (Molecular Probes, 1:2000 dilution) in blocking solution for 1 h. The samples were then examined using an FV-1000 confocal imaging system (Olympus).
Luciferase Assay—HEK 293 cells were cultured in 24-well microplates and transfected with 100 ng of Cyclin-E-Luc or APRE-Luc reporter plasmids, 10 ng of pRL-tk-Luc internal control plasmid, and the desired expression plasmids (100 ng of each expression plasmid). The total amount of plasmid DNA was kept constant by balancing with empty expression plasmids. Transfected cells were cultured for 24 h. For the APRE-Luc assay, cells were incubated for 6 h with or without 10 ng/ml recombinant hLIF (Sigma). Luciferase activity was detected with a Veritas microplate luminometer (Promega) and Dual-Luciferase Reporter Assay System (Promega). Luciferase assays were previously described (27).
Chromatin Immunoprecipitation (ChIP)—HEK 293 cells were cultured in 12-well microplates and transfected with expression plasmids and siRNA. Cells were fixed by directly adding formaldehyde to the medium (final concentration 1%). 15 min later, the fixed cells were washed with ice-cold PBS containing protease inhibitors. The cells were solubilized in lysis buffer, and then chromatin was sheared to an average DNA fragment size of 200–500 bp by sonication. ChIP assays were performed using One Day ChIP assay kit (Diagenode) according to the manufacturer's protocol. Antibodies used for immunoprecipitation included anti-DP-1 monoclonal antibody SPM178 (Genetex) and anti-E2F monoclonal antibody (Upstate). With the use of Prime STAR (Takara), the eluted DNA was amplified by PCR with the following primers for the cyclin-E promoter (encompassing the 3 major E2F binding sites; forward, 5′-CGCCCGCCGTGTTTACATTCCAC-3′; reverse, 5′-CGAGGCGCAGGGACGGGGAATC-3′).
Flow Cytometry Analysis—Analysis of the cell cycle was carried out using flow cytometry. HEK 293 cells were cultured in 10-cm dishes and transfected with the desired plasmids. 24 h later, the cells were harvested and then stained with CycleTEST PLUS (BD Biosciences). Subsequently, flow cytometric analysis was carried out in a FACSCalibur flow cytometer (BD Biosciences) using CellQuest software. Cell cycle analysis was performed with FlowJo (Tree Star).
Measurement of Cell Growth—HEK 293 cells were transfected with the indicated expression plasmids, and then cultured for 2 days in 10% fetal bovine serum containing DMEM. Cell growth was examined by performing a CellTiter-Glo Luminescent Cell Viability Assay (Promega).
Short-interfering RNA (siRNA) Transfection—The following siRNA oligonucleotides were used to interrupt the expression of endogenous and exogenous SOCS-3 or DP-1: SOCS3 Stealth Select RNAi (catalog number, HSS113313, Invitrogen), DP-1 Stealth Select RNAi (catalog number, HSS144254, Invitrogen), and Stealth RNAi Negative Control Med GC Duplex 3 (catalog number, 12935-113, Invitrogen). HEK 293 cells were transfected with these siRNAs according to the manufacturer's protocol (Invitrogen) with the aid of Lipofectamine 2000 (Invitrogen).
RESULTS
Identification of DP-1 as a SOCS-3-interacting Protein—Using yeast two-hybrid screening and a human testis cDNA library, we sought proteins that could interact with SOCS-3. We initially found that 28 of 76 clones were the C-terminal portion of the transcription factor DP-1. By this screening, four types of DP-1 clones (103–410, 192–410, 238–410, 240–410 amino acids) were identified. These results suggest that the C-terminal domain of DP-1 (240–410 amino acids) was the region interacting with SOCS-3.
SOCS-3 Interacts with DP-1 in Cells in Vitro and in Vivo—The interaction between SOCS-3 and DP-1 in HEK 293 cells was investigated by conducting an immunoprecipitation-Western blot assay. The cell lysates were examined for the expression of Flag-SOCS-3 and 6× Myc-DP-1 proteins (Fig. 1B, c and d). The lysates were first immunoprecipitated with Flag antibody beads, which were then washed and examined by Western blotting assay with a Flag or Myc antibody. As shown in Fig. 1, SOCS-3 bound DP-1 (Fig. 1Ba, lane 1). In addition, DP-1 also interacted with SOCS-1 and SOCS-2 (data not shown). Further, using deletion mutants of SOCS-3 (Fig. 1A), we determined the critical region in SOCS-3 required for interaction with DP-1. SOCS-3 having an N- or C-terminal truncation, CΔ188 and NΔ45, were able to bind DP-1 (Fig. 1Ba, lanes 3 and 4), but CΔ141 could not (Fig. 1Ba, lane 2). Therefore, we created a further deletion between 141 and 188 and examined the ability of this mutant to bind DP-1. Mutant Δ156–172 was not able to bind to DP-1 (Fig. 1Ba, lane 5). In addition, although we prepared C-terminally truncated forms of DP-1 to address the domain interacting with SOCS-3, these proteins were expressed at low levels, making such an assessment impossible (data not shown).
FIGURE 1.
Interaction of SOCS-3 with DP-1 in cells in vitro and in vivo. A, schematic structure of SOCS-3 proteins analyzed in this study. B, interaction of Flag-SOCS-3 with 6xMyc-DP-1 was analyzed using the immunoprecipitation-Western blotting assay. HEK 293 cells were transiently transfected with plasmids expressing either Flag-SOCS-3 or 6×Myc-DP-1. The immunoprecipitates (a and b) or cell lysates (c and d) were blotted with anti-Flag (b and c) or anti-Myc (a and d) antibodies. C, interaction of SOCS-3 with endogenous DP-1 in cells in vitro and in vivo was analyzed using the immunoprecipitation-Western blotting assay. Lysates of HEK 293 cells (lane 1), HEK 293 cells stably expressing Flag-SOCS-3 (lane 2) and extracts prepared in TNE buffer by homogenization of lung and testis excised from a normal C57BL/6JJcl male mouse (12–14 weeks old, lanes 3 and 4) were used for immunoprecipitation-Western blotting. D, interaction of Flag-SOCS-3 with 6xMyc-DP-1 (WT, lane 1), DP-1 (α, lane 2), DP-1 (β, lane 3), DP-2 (WT, lane 4), and DP-3 (WT, lane 5), was analyzed using the immunoprecipitation-Western blotting assay.
Next, to confirm this interaction in cells in vitro as well as in vivo, we used HEK 293 cells stably expressing Flag-SOCS-3 and mouse lung or testis. As shown in Fig. 1C, we observed the interactions between SOCS-3 and DP-1 in a stable-expressed HEK 293 transfectant of Flag-SOCS-3 as well as in mouse lung and testis extracts (Fig. 1Ca, lanes 2–4).
Furthermore, it was of interest to us to examine whether SOCS-3 also would be able to interact with other DP molecules (DP-1 α, β, DP-2, and DP-3). Therefore, we investigated this point using the immunoprecipitation assay. As shown in Fig. 1D, we observed that DP-3 also was able to interact with SOCS3 (Fig. 1Da, lane 5) but that DP-1 α, β, and DP-2 was not (Fig. 1Da, lanes 2–4). These results strongly suggest that SOCS-3 is able to interact with DP-1 in cells in vitro and in vivo and that the 156–172 region of SOCS-3 and the C terminus of DP-1 are required for this interaction.
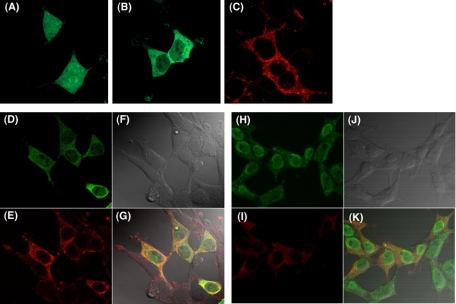
Co-localization of SOCS-3 and DP-1—Although we found that SOCS-3 interacted with DP-1 in HEK 293 cells, it is important to observe visually this cellular interaction. Therefore, we used confocal microscopy to examine the localization of DP-1 and SOCS-3 and their interaction. HEK 293 cells were transiently transfected with green fluorescent protein (GFP)-fused DP-1 or Flag-tagged SOCS-3 expression plasmid or both. The localization of each protein was observed at 18 h after the transfection. When these expression plasmids were used separately, although GFP was expressed throughout the cells (Fig. 2A), GFP-DP-1 was predominantly located in the cytoplasm (Fig. 2B). Several studies (28, 33, 34) also have demonstrated that DP-1 overexpressed in HEK 293 cells in the cytoplasm. On the other hand, Flag-SOCS-3 alone was also strongly expressed in the cytoplasm (Fig. 2C). In addition, we observed that both proteins were co-localized in the cytoplasm after co-transfection (Fig. 2G, yellow to orange fluorescence); thus the expression pattern of these proteins remained unaltered (Fig. 2, D–G). Next, to confirm this co-localization we used HEK 293 cells stably expressing Flag-SOCS-3. As shown in Fig. 2, H–K, although endogenously expressed DP-1 was mainly expressed in the nucleus (Fig. 2H), the co-localization was observed in the cytoplasm (Fig. 2K). These results suggest that the interacting event between SOCS-3 and DP-1 occurred in the cytoplasm of HEK 293 cells.
FIGURE 2.
Immunofluoresence analysis for the localization of SOCS-3 and DP-1. HEK 293 cells were transiently transfected with pEGFP-C2 empty plasmid (A) pEGFP-C2-DP-1 (B), or pcDNA3-Flag-SOCS-3 (C). HEK 293 cells were also co-transfected with pEGFP-C2-DP-1 and pcDNA3-Flag-SOCS-3 (D–G). DP-1 was detected by GFP fluorescence (D), whereas SOCS-3 was detected with anti-Flag antibody and Cy5-conjugated secondary antibody (E). DIC image (F), and merged GFP/Cy5 fluorescence (G) are also shown. HEK 293 cells stably expressing Flag-SOCS-3 were also examined (H–K). DP-1 was detected with anti-DP-1 monoclonal antibody and Cy5-labeled secondary antibody (H), whereas SOCS-3 was detected with anti-SOCS-3 polyclonal antibody and Alexa Fluor 488-labeled anti-rabbit IgG (I). The DIC image (J) and merged Alexa Fluor 488/Cy5 fluorescence (K) are also shown. Yellow-to-orange staining indicates co-localized SOCS-3 and DP-1 (G and K).
SOCS-3 Inhibits the Transcriptional Activity of DP-1/E2F—Because it was of interest to us to explore whether SOCS-3 actually would be able to regulate the transcriptional activity of DP-1, we next investigated the effect of SOCS-3 on this transcriptional activity in HEK 293 cells. As shown in a previous study (35), because expression of the endogenous DP-1 is widely observed in several cell lines and the level of DP-1 is considered to be sufficient to exhibit E2F/DP-1 transcriptional activity, using the Cyclin-E-Luc assay we explored the effect of SOCS-3 on endogenously expressed DP-1 in HEK 293 cells. First, we examined whether the E2F/DP-1 transcriptional activity in the cells depended on endogenously expressed DP-1. As shown in Fig. 3A, lane 2, Cyclin-E-Luc activity was dramatically inhibited by DP-1 siRNA. These observations suggest that endogenous DP-1 plays an important role in E2F/DP-1 transcriptional activity in the cells. Therefore, we next addressed whether SOCS-3 could actually inhibit E2F/DP-1-dependent transcriptional activity in an endogenous DP-1-driven Cyclin-E-Luc activity. As expected, SCOS-3 (WT) expression inhibited about 50% of the transcriptional activity (Fig. 3A, lane 5). However, such inhibitory effect was not observed with SOCS-3 (Δ156–172), in which the DP-1-binding region is absent (Fig. 3A, lane 8). Importantly, because this inhibitory action of SOCS-3 was eliminated completely by SOCS-3 siRNA (Fig. 3A, lane 6), these results strongly suggest that the inhibitory effect of SOCS-3 on endogenous DP-1-dependent transcriptional activity is specific to SOCS-3.
FIGURE 3.
SOCS-3 inhibits the transcriptional activity of DP-1/E2F. A, HEK 293 cells were transiently transfected with the indicated expression plasmids and siRNA together with reporter (Cyclin-E-Luc) and internal control (pRL-tk-Luc) plasmids. After transfection, the cells were incubated for 24–30 h, and then cell extracts were prepared. Firefly luciferase activity (Cyclin-E-Luc) from triplicate samples was determined and normalized against Renilla luciferase (pRL-tk-Luc) activity. Effects of siRNA were analyzed by Western blotting with anti-DP-1 antibody (B) and anti-SOCS-3 antibody (C). D, ChIP assays for binding of DP-1/E2F-1 to the cyclin-E promoter. HEK 293 cells were transiently transfected with the indicated expression plasmids and siRNA and then were fixed and subjected to the ChIP assay. PCR amplification of DNA precipitated with anti-DP-1 (lanes 3–7) and anti-E2F-1 (lanes 8–12) using primers for the cyclin-E promoter. Input lane (lane 1) shows products after PCR amplification of chromatin DNA before the addition of an antibody. PCR products were separated by 2% agarose gel electrophoresis and visualized by ethidium bromide staining.
In addition, to support the contribution of endogenous DP-1 and exogenous SOCS-3, we investigated the expression of both proteins by Western blotting. As shown in Fig. 3B, the endogenous DP-1 expression in HEK 293 cells was almost completely inhibited by DP-1 siRNA. Also, the expression of exogenous SOCS-3 at the protein level in SOCS-3-expressing cells was inhibited by SOCS-3 siRNA (Fig. 3C). Next, using the ChIP assay, we explored the interference of SOCS-3 with the binding of DP-1/E2F-1 to the cyclin-E promoter. As shown in Fig. 3D, SOCS3 clearly inhibited this DNA binding activity (Fig. 3D, lanes 4 and 9). Together, these results suggest to us the possibility that SOCS-3 may retard cell cycle progression by inhibiting DP-1-elicited transcriptional activity of the cyclin-E gene.
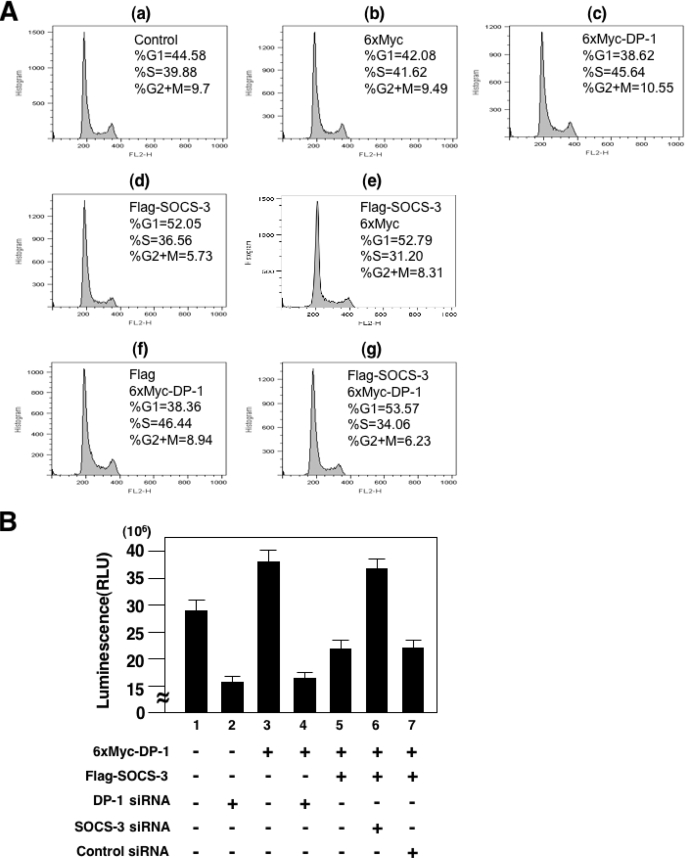
SOCS-3 Inhibits the Cell Cycle Progression of DP-1-transfected Cells—To verify the above-mentioned possibility, we used flow cytometry to determine whether or not SOCS-3 could retard the E2F/DP-1-dependent cell cycle progression. We transfected HEK 293 cells with the appropriate expression plasmids and then analyzed the distribution of cells in the various phases of the cell cycle, as described previously (27). As shown in Fig. 4A, DP-1 enhanced the proportion of HEK 293 cells that had passed from G1 to S phase (Fig. 4A, c and f). In contrast, SOCS-3 clearly retarded cell cycle progression (Fig. 4A, d, e, and g). Furthermore, we examined cell growth by conducting a luminescent cell viability assay, which is a homogeneous method for determining the number of viable cells. As we expected, the stimulated cell growth of DP-1-transfected cells was clearly eliminated by DP-1 siRNA (Fig. 4B, lane 4). When the cells were co-transfected with SOCS-3 and DP-1 expression plasmids, although the DP-1-mediated cell growth was markedly inhibited by SOCS-3 (Fig. 4B, lane 5), importantly, this inhibitory action of SOCS-3 was blocked by SOCS-3 siRNA (Fig. 4B, lane 6). These results suggest that SOCS-3 acts as a negative regulator of the cell cycle and cell growth under E2F/DP-1 control.
FIGURE 4.
SOCS-3 inhibits cell cycle progression under E2F/DP-1 control. A, HEK 293 cells were transfected with the indicated expression plasmids, cultured for 24 h, harvested, and stained with propidium iodide using CycleTEST PLUS. Flow cytometric analysis was carried out in a FACSCalibur flow cytometer. Cell cycle analysis was performed using FlowJo. B, HEK 293 cells were transiently transfected or not with the indicated plasmids and siRNA, and then cultured for 2 days in DMEM containing 10% fetal bovine serum. The number of viable cells was measured using the CellTiter-Glo assay system based on quantification of the ATP present. Each value is the average of triplicate transfections.
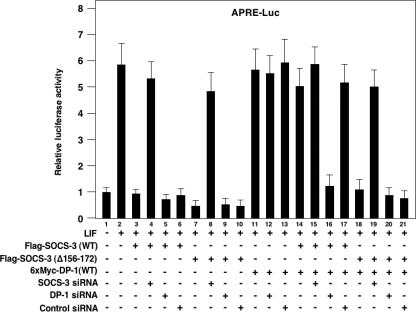
DP-1 Eliminates the Negative Regulation of SOCS-3 in STAT Signaling—Finally, to confirm definitively this interaction between SOCS-3 and DP-1, in reverse, we explored whether DP-1 could eliminate the negative regulation of SOCS-3 in JAK-STAT signaling. To explore this, we examined whether DP-1 could block the SOCS-3 inhibition of LIF-stimulated STAT3 transcriptional activity by conducting a luciferase assay with a reporter plasmid bearing the APRE promoter and luciferase gene. SOCS-3 (WT) dramatically inhibited the LIF-stimulated STAT3 transcriptional activity in HEK 293 cells (Fig. 5, lane 3). This inhibition was almost completely blocked by SOCS-3 siRNA (Fig. 5, lane 4). Importantly, DP-1 clearly eliminated the inhibitory action of SOCS-3 toward LIF-stimulated STAT3 transcriptional activity in SOCS-3 (WT)-transfected cells (Fig. 5, lane 14). In addition, as we expected, this elimination by DP-1 was blocked by DP-1 siRNA (Fig. 5, lane 16). However, such elimination was not observed for SOCS-3 (Δ156–172)-mediated inhibition of the STAT3 transcriptional activity, because SOCS-3 (Δ156–172) is not able to bind to DP-1 (Fig. 5, lane 18). These results strongly indicate that DP-1 was able to block the SOCS-3 action as a negative regulator of JAK-STAT signaling.
FIGURE 5.
DP-1 eliminates the inhibitory action of SOCS-3 toward LIF-stimulated STAT3 transcriptional activity. HEK 293 cells were transiently transfected with various combinations of Flag-SOCS-3 (WT), Flag-SOCS-3 (Δ156–172), 6×Myc-DP-1 (WT), and siRNA in the presence of the APRE-Luc and pRL-tk-Luc reporter plasmids. The cells were incubated in the presence or absence of 10 ng/ml hLIF for 6 h, and then the cell extracts were prepared. Firefly luciferase activity (APRE-Luc) from triplicate samples was determined and normalized against Renilla luciferase (pRL-tk-Luc) activity.
DISCUSSION
Herein, using the two-hybrid system, we identified DP-1 as a SOCS-3-interacting protein. Because E2F/DP-1 plays an important role in the G1-to-S phase transition in the cell cycle (22–24), we suspected that SOCS-3 may regulate cell cycle progression under E2F/DP-1 control via interaction with DP-1. Although it is well known that retinoblastoma tumor suppressor protein (Rb) regulates negatively E2F function by interacting directly with this transcriptional protein (29–31, 36–38), to our knowledge, a DP-1-interacting protein that is able to regulate cell cycle progression under E2F/DP-1 control has not previously been demonstrated. Our present study is the first one to show that SOCS-3 acts as a negative regulator of the cell cycle under E2F/DP-1 control by interacting with DP-1. Interestingly, these findings also suggest the possibility that human SOCS-3 may regulate tumor cell growth and cell differentiation via this novel mechanism.
We found using an immunoprecipitation assay that SOCS-3 interacted with DP-1 both in cells in vitro as well as in vivo (Fig. 1, B and C). In addition, using a series of deletion mutants of SOCS-3, we determined that the 156–172 amino acid region of SOCS-3 was required for interaction with DP-1 (Fig. 1B). Although the function of this region located between SH2 and SOCS-box domains is not yet known, this region is considered to be a part of the domain interacting with DP-1. It is well known that DP-1 transcriptional activity is regulated by interaction with p53 (39) or ARF (40, 41) and also that E2F is controlled by interaction with pRb (29–31, 36–38). However, because the cell cycle is a series of complicated cellular events regulated by many kinds of signaling molecules, it was of much interest to us to identify novel factors interacting with E2F/DP-1 and also to demonstrate the regulatory mechanism involving E2F/DP-1 in detail. Therefore, we investigated here the regulatory mechanism of SOCS-3 with respect to E2F/DP-1 control of the cell cycle. Several recent studies have shown that the decreased expression of SOCS-1, -2, and -3 by hypermethylation of its gene promoter is closely related to aggravation of some cancers (11–21). Although these observations suggested to us some regulatory action of SOCS-1 for development and progression of tumor cells, the molecular mechanism underlying the inhibitory action of SOCS family proteins have not yet been demonstrated in detail. We found that SOCS-3 inhibited the transcriptional activity of E2F/DP-1 via its interaction with DP-1 and consequently retarded the cell cycle and cell growth under E2F/DP-1 control (Figs. 3 and 4), though SOCS-3 did not completely inhibit the transcriptional activity of E2F/DP-1 (Fig. 3A). Importantly, these inhibitions of SOCS-3 were almost completely blocked by SOCS-3 siRNA (Figs. 3 and 4B). As shown in Fig. 2, the localization of both DP-1 and SOCS-3 co-overexpressed in HEK 293 cells almost was the same as that of each overexpressed alone; and, importantly, both proteins were detected together in the cytoplasm. Because we observed the co-localization of DP-1 and SOCS-3 in the cytoplasm (Fig. 2, G and K), we suspect that SOCS-3 probably may inhibit the transcriptional activity by interacting with DP-1 located in the cytoplasm and promote the proteolysis of DP-1 via the SOCS-box. This possibility is supported by our data from the ChIP assay showing that SOCS-3 clearly inhibited DNA binding activity of DP-1/E2F-1 at the cyclin-E promoter (Fig. 3D).
Interestingly, because SOCS-1 and SOCS-2 also were able to interact with DP-1 and to inhibit its transcriptional activity,3 both SOCS proteins also may act as novel negative regulator of E2F/DP-1 via direct interaction with DP-1. Recently, Qiao et al. (26) identified a novel DP subclass, DP-3, that is able to inhibit E2F-1 transcriptional activity. Therefore, our interest was to address whether SOCS-3 also is able to interact with DP-3. Consequently, we proved that SOCS-3 directly is able to interact with DP-3 (Fig. 1D). Therefore, although it is very important to explore the regulatory action of SOCS-3 for DP-3, these results suggest to us a novel mechanism of SOCS protein-mediated regulation of tumor cell growth and development.
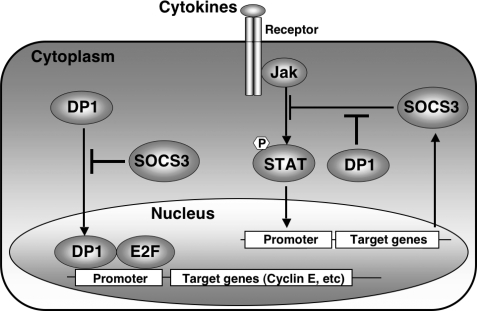
On the other hand, to confirm our finding that human SOCS-3 was an interacting protein of DP-1, approaching the question from another angle, we investigated whether DP-1 could eliminate the inhibitory action of SOCS-3 toward LIF-stimulated STAT3 transcriptional activity in JAK-STAT signaling. Consequently we observed that DP-1 almost completely eliminated such inhibitory action of SOCS-3 in this signaling pathway (Fig. 5). This elimination by DP-1 was dramatically blocked by DP-1 siRNA. As described above, the cytoplasmic DP-1 may interact with endogenous SOCS-3 induced by cytokines or growth hormones and act as its modulator. These observations lead us to speculate that SOCS-3 and DP-1 may alternatively regulate each other in local sites of tissues having inflammatory reaction such as tumor and rheumatoid arthritis (Fig. 6).
FIGURE 6.
Model illustrating the regulatory mechanisms between SOCS-3 and DP-1 for JAK-STAT signaling and cell cycle progression. Several kinds of cytokines induce SOCS-3 expression via the JAK-STAT signaling system, and then the endogenous SOCS-3 inhibits the transcriptional activity of E2F/DP-1 by binding to cytoplasmic DP-1. Consequently SOCS-3 acts as a negative regulator of cell cycle progression under E2F/DP-1 control. Acting differently, DP-1 eliminates the inhibitory action of endogenous SOCS-3 toward JAK-STAT signaling.
Also, interestingly, it has been shown that SOCS-3 is deeply involved in both initiation and development of allergic diseases such as atopic dermatitis and asthma (42). Therefore, DP-1 may act as a potent modulator of allergic diseases(s) regulated by SOCS-3. Importantly, it has been shown that ubiquitinated DP-1 is degraded by the proteasome system (43). Also, SOCS-3 is known to recruit ECS (Elongin B/C-Cul2/Cul5-SOCS-box protein) E3 ligase, resulting in accelerated proteasomal degradation of target proteins and SOCS-3 (44–47). Therefore, to understand how each of these factors actually exhibits its regulatory action in the cytoplasm, in further experiments, it will be very important for us to address in detail the degradation events of both factors in the cytoplasm.
In conclusion, we demonstrated that human SOCS-3 interacted with DP-1 and regulated cell cycle progression under E2F/DP-1 control (Fig. 6). Thus, these findings provide us with new insights into the role of SOCS-3 in cell growth and cell differentiation besides its function as a negative regulator of JAK-STAT signaling.
Acknowledgments
We thank Dr. A. Yoshimura for the kind gift of the APRE-Luc reporter plasmid and helpful comments. We also thank Dr. M. Hatakeyama for the gift of the Cyclin-E-Luc reporter plasmid and Dr. W. F. Chen for the gift of the hDP-3 expression plasmid.
This work was supported by a grant-in-aid for scientific research from the Ministry of Education, Science, Sports, and Culture of Japan and by the OSAKA Cancer Research Foundation. Additional support came from a Nihon University Individual Research Grant. The costs of publication of this article were defrayed in part by the payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 U.S.C. Section 1734 solely to indicate this fact.

Author's Choice—Final version full access.
Footnotes
The abbreviations used are: SOCS, suppressor of cytokine signaling; CIS, cytokine-inducible SH2 domain-containing protein; JAK, Janus kinase; STAT, signal transducers and activators of transcription; GH, growth hormone; dn, dominant negative; ECS, Elongin B/C-Cul2/Cul5-SOCS-box protein; siRNA, short interfering RNA; LIF, leukemia inhibitory factor; DIC, differential interference contrast; WT, wild type; PBS, phosphate-buffered saline; ChIP, chromatin immunoprecipitation assay; GFP, green fluorescent protein; DMEM, Dulbecco's modified Eagle's medium.
S. Hanazawa, unpublished data.
References
- 1.Endo, T. A., Masuhara, M., Yokouchi, M., Suzuki, R., Sakamoto, H., Mitsui, K., Matsumoto, A., Tanimura, S., Ohtsubo, M., Misawa, H., Miyazaki, T., Leonor, N., Taniguchi, T., Fujita, T., Kanakura, Y., Komiya, S., and Yoshimura, A. (1997) Nature 387 921-924 [DOI] [PubMed] [Google Scholar]
- 2.Naka, T., Narazaki, M., Hirata, M., Matsumoto, T., Minamoto, S., Aono, A., Nishimoto, N., Kajita, T., Taga, T., Yoshizaki, K., Akira, S., and Kishimoto, T. (1997) Nature 387 924-929 [DOI] [PubMed] [Google Scholar]
- 3.Starr, R., Willson, T. A., Viney, E. M., Murray, L. J., Rayner, J. R., Jenkins, B. J., Gonda, T. J., Alexander, W. S., Metcalf, D., Nicola, N. A., and Hilton, D. J. (1997) Nature 387 917-921 [DOI] [PubMed] [Google Scholar]
- 4.Aaronson, D. S., and Horvath, C. M. (2002) Science 296 1653-1655 [DOI] [PubMed] [Google Scholar]
- 5.O'Shea, J. J., Gadina, M., and Schreiber, R. D. (2002) Cell 109, (suppl.), S121-S131 [DOI] [PubMed] [Google Scholar]
- 6.Inaba, M., Saito, H., Fujimoto, M., Sumitani, S., Ohkawara, T., Tanaka, T., Kouhara, H., Kasayama, S., Kawase, I., Kishimoto, T., and Naka, T. (2005) Biochem. Biophys. Res. Commun. 328 953-961 [DOI] [PubMed] [Google Scholar]
- 7.Turnley, A. M., Faux, C. H., Rietze, R. L., Coonan, J. R., and Bartlett, P. F. (2002) Nat. Neurosci. 5 1155-1162 [DOI] [PubMed] [Google Scholar]
- 8.Spangenburg, E. E. (2005) J. Biol. Chem. 280 10749-10758 [DOI] [PubMed] [Google Scholar]
- 9.Inoue, H., and Kubo, M. (2004) Expert Rev. Mol. Med. 6 1-11 [DOI] [PubMed] [Google Scholar]
- 10.Ozaki, A., Seki, Y., Fukushima, A., and Kubo, M. (2005) J. Immunol. 175 5489-5497 [DOI] [PubMed] [Google Scholar]
- 11.Miyoshi, H., Fujie, H., Moriya, K., Shintani, Y., Tsutsumi, T., Makuuchi, M., Kimura, S., and Koike, K. (2004) J. Gastroenterol. 39 563-569 [DOI] [PubMed] [Google Scholar]
- 12.Niwa, Y., Kanda, H., Shikauchi, Y., Saiura, A., Matsubara, K., Kitagawa, T., Yamamoto, J., Kubo, T., and Yoshikawa, H. (2005) Oncogene 24 6406-6417 [DOI] [PubMed] [Google Scholar]
- 13.Okochi, O., Hibi, K., Sakai, M., Inoue, S., Takeda, S., Kaneko, T., and Nakao, A. (2003) Clin. Cancer Res. 9 5295-5298 [PubMed] [Google Scholar]
- 14.Hatirnaz, O., Ure, U., Ar, C., Akyerli, C., Soysal, T., Ferhanoglu, B., Ozcelik, T., and Ozbek, U. (2007) Am. J. Hematol. 82 729-730 [DOI] [PubMed] [Google Scholar]
- 15.Sutherland, K. D., Lindeman, G. J., Choong, D. Y., Wittlin, S., Brentzell, L., Phillips, W., Campbell, I. G., and Visvader, J. E. (2004) Oncogene 23 7726-7733 [DOI] [PubMed] [Google Scholar]
- 16.Chan, M. W., Chu, E. S., To, K. F., and Leung, W. K. (2004) Biotechnol. Lett. 26 1289-1293 [DOI] [PubMed] [Google Scholar]
- 17.Galm, O., Yoshikawa, H., Esteller, M., Osieka, R., and Herman, J. G. (2003) Blood 101 2784-2788 [DOI] [PubMed] [Google Scholar]
- 18.He, B., You, L., Uematsu, K., Zang, K., Xu, Z., Lee, A. Y., Costello, J. F., McCormick, F., and Jablons, D. M. (2003) Proc. Natl. Acad. Sci. U. S. A. 100 14133-14138 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Oshimo, Y., Kuraoka, K., Nakayama, H., Kitadai, Y., Yoshida, K., Chayama, K., and Yasui, W. (2004) Int. J. Cancer 112 1003-1009 [DOI] [PubMed] [Google Scholar]
- 20.Tokita, T., Maesawa, C., Kimura, T., Kotani, K., Takahashi, K., Akasaka, T., and Masuda, T. (2007) Int. J. Oncol. 30 689-694 [PubMed] [Google Scholar]
- 21.Weber, A., Hengge, U. R., Bardenheuer, W., Tischoff, I., Sommerer, F., Markwarth, A., Dietz, A., Wittekind, C., and Tannapfel, A. (2005) Oncogene 24 6699-6708 [DOI] [PubMed] [Google Scholar]
- 22.Girling, R., Partridge, J. F., Bandara, L. R., Burden, N., Totty, N. F., Hsuan, J. J., and La Thangue, N. B. (1993) Nature 365 468. [DOI] [PubMed] [Google Scholar]
- 23.Helin, K., Wu, C. L., Fattaey, A. R., Lees, J. A., Dynlacht, B. D., Ngwu, C., and Harlow, E. (1993) Genes Dev. 7 1850-1861 [DOI] [PubMed] [Google Scholar]
- 24.Kohn, M. J., Bronson, R. T., Harlow, E., Dyson, N. J., and Yamasaki, L. (2003) Development 130 1295-1305 [DOI] [PubMed] [Google Scholar]
- 25.Milton, A., Luoto, K., Ingram, L., Munro, S., Logan, N., Graham, A. L., Brummelkamp, T. R., Hijmans, E. M., Bernards, R., and La Thangue, N. B. (2006) Oncogene 25 3212-3218 [DOI] [PubMed] [Google Scholar]
- 26.Qiao, H., Di Stefano, L., Tian, C., Li, Y. Y., Yin, Y. H., Qian, X. P., Pang, X. W., Li, Y., McNutt, M. A., Helin, K., Zhang, Y., and Chen, W. F. (2007) J. Biol. Chem. 282 454-466 [DOI] [PubMed] [Google Scholar]
- 27.Ishida, H., Masuhiro, Y., Fukushima, A., Argueta, J. G., Yamaguchi, N., Shiota, S., and Hanazawa, S. (2005) J. Biol. Chem. 280 24642-24648 [DOI] [PubMed] [Google Scholar]
- 28.Magae, J., Wu, C. L., Illenye, S., Harlow, E., and Heintz, N. H. (1996) J. Cell Sci. 109 1717-1726 [DOI] [PubMed] [Google Scholar]
- 29.Bracken, A. P., Ciro, M., Cocito, A., and Helin, K. (2004) Trends Biochem. Sci. 29 409-417 [DOI] [PubMed] [Google Scholar]
- 30.DeGregori, J., Kowalik, T., and Nevins, J. R. (1995) Mol. Cell. Biol. 15 4215-4224 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Stevens, C., and La Thangue, N. B. (2003) Arch. Biochem. Biophys. 412 157-169 [DOI] [PubMed] [Google Scholar]
- 32.Yamada, M., Kondo, T., Ashizawa, S., Takebayashi, T., Higashi, H., and Hatakeyama, M. (2002) Cytokine 17 91-97 [DOI] [PubMed] [Google Scholar]
- 33.Allen, K. E., de la Luna, S., Kerkhoven, R. M., Bernards, R., and La Thangue, N. B. (1997) J. Cell Sci. 110 2819-2831 [DOI] [PubMed] [Google Scholar]
- 34.Verona, R. Moberg, K. Estes, S. Starz, M. Vernon, J. P., and Lees, J. A. (1997) Mol. Cell. Biol. 17 7268-7282 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Wu, C. L., Zukerberg, L. R., Ngwu, C., Harlow, E., and Lees, J. A. (1995) Mol. Cell. Biol. 15 2536-2546 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Hamel, P. A., Gill, R. M., Phillips, R. A., and Gallie, B. L. (1992) Mol. Cell. Biol. 12 3431-3438 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Hiebert, S. W., Chellappan, S. P., Horowitz, J. M., and Nevins, J. R. (1992) Genes Dev. 6 177-185 [DOI] [PubMed] [Google Scholar]
- 38.Zamanian, M., and La Thangue, N. B. (1992) EMBO J. 11 2603-2610 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Sorensen, T. S., Girling, R., Lee, C. W., Gannon, J., Bandara, L. R., and La Thangue, N. B. (1996) Mol. Cell. Biol. 16 5888-5895 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Datta, A., Nag, A., and Raychaudhuri, P. (2002) Mol. Cell. Biol. 22 8398-8408 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Datta, A., Sen, J., Hagen, J., Korgaonkar, C. K., Caffrey, M., Quelle, D. E., Hughes, D. E., Ackerson, T. J., Costa, R. H., and Raychaudhuri, P. (2005) Mol. Cell. Biol. 25 8024-8036 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Seki, Y., Inoue, H., Nagata, N., Hayashi, K., Fukuyama, S., Matsumoto, K., Komine, O., Hamano, S., Himeno, K., Inagaki-Ohara, K., Cacalano, N., O'Garra, A., Oshida, T., Saito, H., Johnston, J. A., Yoshimura, A., and Kubo, M. (2003) Nat. Med. 9 1047-1054 [DOI] [PubMed] [Google Scholar]
- 43.Magae, J., Illenye, S., Chang, Y. C., Mitsui, Y., and Heintz, N. H. (1999) Oncogene 18 593-605 [DOI] [PubMed] [Google Scholar]
- 44.Kamura, T., Sato, S., Haque, D., Liu, L., Kaelin, W. G., Jr., Conaway, R. C., and Conaway, J. W. (1998) Genes Dev. 12 3872-3881 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Kile, B. T., Schulman, B. A., Alexander, W. S., Nicola, N. A., Martin, H. M., and Hilton, D. J. (2002) Trends Biochem. Sci. 27 235-241 [DOI] [PubMed] [Google Scholar]
- 46.Orr, S. J., Morgan, N. M., Elliott, J., Burrows, J. F., Scott, C. J., McVicar, D. W., and Johnston, J. A. (2007) Blood 109 1061-1068 [DOI] [PubMed] [Google Scholar]
- 47.Zhang, J. G., Farley, A., Nicholson, S. E., Willson, T. A., Zugaro, L. M., Simpson, R. J., Moritz, R. L., Cary, D., Richardson, R., Hausmann, G., Kile, B. J., Kent, S. B., Alexander, W. S., Metcalf, D., Hilton, D. J., Nicola, N. A., and Baca, M. (1999) Proc. Natl. Acad. Sci. U. S. A. 96 2071-2076 [DOI] [PMC free article] [PubMed] [Google Scholar]