Abstract
Fatty-acid synthase (FAS) is up-regulated in a broad range of cancers, including those of the breast, prostate, and ovaries. In tumor cells, the inhibition of FAS elicits cell cycle arrest and apoptosis, so it is considered a potential drug target for oncology. Results from this study show that inhibition of FAS, by either knockdown with small interfering RNA or inhibition with the small molecule drug orlistat, leads to activation of the receptor-mediated apoptotic cascade (caspase-8-mediated) and ultimately to cell death. However, knockdown of two enzymes upstream of FAS, acetyl-CoA carboxylase-α and ATP-citrate lyase, fails to activate caspase-8 or to elicit apoptosis in tumor cells, even though palmitate synthesis was suppressed. Using differential gene analysis, we traced the unique apoptotic effect of FAS inhibition to up-regulation of DDIT4 (DNA damage-inducible transcript 4), a stress-response gene that negatively regulates the mTOR pathway. These findings indicate that suppression of palmitate synthesis is not sufficient for eliciting tumor cell death and suggest that the unique effect of inhibition of FAS results from negative regulation of the mTOR pathway via DDIT4.
Eukaryotic fatty-acid synthase (FAS)2 synthesizes palmitate, the precursor of long chain fatty acids (1). FAS is up-regulated in a wide range of tumors (2–7), with levels increasing as tumor grade and severity increase (3, 4). The up-regulation of FAS is associated with poor prognosis, so the enzyme has become recognized in recent years as a target for anti-tumor therapy (2, 5, 6). In this regard, the targeted inhibition of FAS by the obesity drug orlistat or analogs of cerulenin blocks tumor proliferation and induces apoptosis in cultured cells (8–11) and also suppresses growth of xenografts in mice (8, 12, 13). Inhibition of FAS has no effect on the survival of normal differentiated cells in vitro and displays no signs of toxicity in vivo (8, 14, 15). This tumor selectivity stems from the increased reliance of tumor cells on de novo fatty acid synthesis to satisfy their metabolic needs, whereas normal cells obtain most lipids from the dietary supply (16).
Up-regulation of FAS in tumors represents an overall activation of genes involved in lipogenesis (17). Lipogenic enzymes that function upstream of FAS such as acetyl-CoA carboxylase-α (ACC-α) and ATP-citrate lyase (ACL) are elevated in cancer and, like FAS, have been implicated as targets for tumor treatment, suggesting that palmitate suppression can halt tumorigenesis (supplemental Fig. 1) (17–23), yet there are also other hypotheses on how inhibition of FAS elicits tumor cell death. Recent evidence has linked the inhibition of FAS to endoplasmic reticulum stress (24), the generation of reactive oxygen species (25), and ceramide accumulation (26). Nevertheless, an understanding of how inhibition of FAS leads to apoptosis remains elusive.
Here, we show that inhibition of FAS activates caspase-8 and induces tumor apoptosis but that knockdown of ACC-α or ACL is without effect, even though their knockdown suppresses palmitate production. These findings indicate that suppression of palmitate biosynthesis alone is not sufficient to elicit tumor cell death and reveal that inhibition of FAS has effects on tumor cells that extend beyond lipid biosynthesis. We traced these FAS-specific effects to its unique ability to up-regulate the stress-response gene DDIT4 (DNA damage-inducible transcript 4; also known as REDD1/RTP801/Dig2), which negatively regulates the mTOR (mammalian target of rapamycin) pathway.
EXPERIMENTAL PROCEDURES
Cell Lines—The MDA-MB-435 cell line was obtained from Janet Price (University of Texas Southwestern Medical Center). MCF-7, LNCaP, PC-3, and Hs58.Fs cells were purchased from American Type Culture Collection (Manassas, VA). MDA-MB-435 and MCF-7 tumor cells were maintained in minimum Eagle's medium with Earle's salts (Mediatech, Inc., Herndon, VA) supplemented with 10% fetal bovine serum (Irvine Scientific, Santa Ana, CA), 2 mm l-glutamine (Invitrogen), minimum Eagle's medium vitamins (Invitrogen), nonessential amino acids (Mediatech, Inc.), and antibiotics (Omega Scientific, Inc., Tarzana, CA). LNCaP and PC-3 tumor cells were maintained in RPMI 1640 medium (Mediatech, Inc.) supplemented with 10% fetal bovine serum and antibiotics. Human fibroblast Hs58.Fs cells were maintained in Dulbecco's modified Eagle's medium (Irvine Scientific) supplemented with 10% fetal bovine serum and antibiotics. All cells were grown at 37 °C under a humidified 5% CO2 atmosphere.
Gene Silencing Using Small Interfering RNA—All small interfering RNAs (siRNAs) were purchased from Dharmacon (Lafayette, CO). SMARTpool siRNA contains a pool of four siRNA sequences directed against the target gene. The siRNA sequences targeting FAS (SMARTpool M-003954-02) were designed and pooled as described previously (9). Fibroblast and tumor cells were plated at 3.91 × 103 and 7.81 × 103 cells/cm2, respectively, and grown for 24–48 h prior to transfection with 1–100 nm FAS SMARTpool, FAS duplex 9 (D-003954-09), caspase-8 SMARTpool (M-003466-00), caspase-9 SMARTpool (M-003309-00), ACC-α SMARTpool (M-004551-02), ACL SMARTpool (M-004915-00), DDIT4 duplex 9 (J-010855-09), DDIT4 duplex 10 (J-010855-10), DDIT4 duplex 11 (J-010855-11), non-silencing control pool (D-001206-13), non-silencing control duplex 1 (D-001810-01), and non-silencing control duplex 2 (D-001210-02) siRNAs. Cells were transfected in Opti-MEM I (Invitrogen) using Lipofectamine 2000 reagent (Invitrogen) according to the manufacturer's instructions. After 5 h, cells were placed in normal culture medium and grown for an additional 42–91 h.
Inhibition of FAS with Orlistat—Orlistat was extracted from Xenical™ capsules (Roche Applied Science) and stored as described previously (9). Fibroblast and tumor cells were plated at 3.91 × 103 and 7.81 × 103 cells/cm2, respectively, and grown for 24–48 h. The medium was subsequently removed, and cells were washed with phosphate-buffered saline (PBS), refed with fresh medium containing orlistat (0–50 μm), and grown for an additional 24–72 h.
Measuring Fatty Acid Biosynthesis—In vivo fatty acid synthesis was measured according to the method described by Lee et al. (27). MDA-MB-435 tumor cells were transfected with siRNA targeting FAS or ACC-α or non-silencing control siRNA for 48 h, washed with medium once, and labeled for 24 h in glutamine-free minimum Eagle's medium containing 0.5 g/liter [U-13C]glucose (Cambridge Isotope Laboratories, Andover, MA) and 2.0 g/liter unlabeled glucose (Sigma). Labeled cells were harvested using a cell scraper, rinsed with PBS, and centrifuged at 2000 rpm for 5 min. The cell pellet was saponified with 1 ml of 30% KOH/ethanol (1:1, v/v) at 70 °C overnight. Neutral lipids were removed by petroleum ether extraction. The aqueous layer was acidified, and fatty acids were recovered by another petroleum ether extraction. The petroleum ether layer was backwashed with water and evaporated to dryness. Fatty acids were methylated with 0.5 n HCl in methanol (Supelco, Bellefonte, PA) for gas chromatography/mass spectrometry analysis.
Fatty acid methyl esters were analyzed on the Trace GC/Trace MS Plus system (Thermo Electron Corp., Waltham, MA) using an Rtx-5MS column (fused silica, 15 m × 0.25 mm × 0.25 μm; (Restek, Bellefonte). Gas chromatography conditions were as follows. The helium flow rate was 2 ml/min, and the oven temperature was programmed from 180 °C (1 min) to 210 °C at 3 °C/min. The interface temperature was maintained at 250 °C and the source temperature at 200 °C. Mass spectra were obtained using electron ionization at –70 eV. Palmitate, stearate, and oleate were monitored at m/z 270, 298, and 296, respectively.
Mass isotopomer distribution was determined after correcting the contribution of labeling arising from natural abundances of carbon (13C), oxygen (17O, 18O), and hydrogen (2H) (28). The 13C enrichment of acetyl units and the de novo synthesis of fatty acids were determined from the distribution of mass isotopomers of palmitate (27). De novo synthesis of palmitate produces palmitate with two, four, or six 13C atoms (m2, m4, and m6). Thus, the enrichment of acetyl units was calculated from the m4/m2 or m6/m4 ratio using the formula m4/m2 = (n – 1)/2*(p/q) and m6/m4 = (n – 2)/3*(p/q), where n is the number of acetyl units in palmitate (n = 8), p is the labeled fraction, and q is the unlabeled fraction (p + q = 1). De novo synthesis was then calculated by dividing the observed by the predicted mass isotopomer fraction, i.e. m2/(8*p*q7) or m4/(28*p2*q6). The effects of knockdown on glucose uptake were excluded based on the ratio of [13C]acetyl-CoA to the naturally labeled [12C]acetyl-CoA. No changes were observed in the fraction of the acetyl-CoA pool labeled by 13Cinany experiments.
Measuring DNA Fragmentation—DNA fragmentation was measured using the Cell Death Detection ELISAplus kit (Roche Applied Science) according to the manufacturer's instructions.
Western Blot Analysis—Floating and adherent cell populations were harvested using a cell scraper and rinsed with PBS. Pellets were lysed in 2× SDS sample buffer, passed 10 times through an 18-gauge needle, boiled for 5 min, and stored at –80 °C. Insoluble material was removed by centrifugation at 14,000 rpm for 15 min at 4 °C. Protein was separated by SDS-PAGE; transferred onto nitrocellulose; stained with 0.05% Ponceau S (Sigma) to ensure equivalent protein loading per lane; and probed overnight at 4 °C with anti-caspase-8, anti-caspase-9, anti-ACC, anti-ACL, anti-eukaryotic initiation factor 4E (eIF4E), anti-phospho-eIF4E (Ser209) (Cell Signaling Technology, Beverly, MA), anti-FAS (BD Transduction Laboratories), anti-FLICE-like inhibitory protein (FLIP; Dave-2; Axxora LLC, San Diego, CA), and anti-actin (Millipore) antibodies. Immunoreactivity was detected using peroxidase-conjugated anti-mouse or anti-rabbit IgG and visualized by enhanced chemiluminescence. Relative band density was determined using AlphaImager Version 4.1.0 software (Alpha Innotech Corp., San Leandro, CA).
Assessing the Activity of Caspase-3/7—The activity of caspase-3 and caspase-7 was measured using the Apo-ONE® homogeneous caspase-3/7 assay kit (Promega Corp., Madison, WI). Cells were incubated with the caspase-3/7 substrate benzyloxycarbonyl-DEVD-R110, and fluorescence intensity (Ex485 nm/Em538 nm) was measured using a fluorescent plate reader (Molecular Devices, Sunnyvale, CA). The amount of benzyloxycarbonyl-DEVD-R110 cleavage and fluorescence directly corresponded to caspase-3/7 activity.
Gene Analysis—MDA-MB-435 tumor cells were treated with orlistat, EtOH, FAS, ACC-α, ACL, or non-silencing control siRNA on four separate occasions to generate biologically independent measurements for each treatment. Whole genome expression was examined 48 h following treatment. Total RNA was isolated from adherent cells using the Qiagen RNeasy kit and hybridized to the HumanRef-8 v2 Expression BeadChip (>22,000 gene transcripts; Illumina, San Diego, CA) according to the manufacturer's instructions. BeadChips were developed as described previously (29).
Expression data were filtered to identify genes expressed on the BeadChip at >0.99 confidence (10,859 genes) and normalized per chip using nonlinear normalize.quantiles to remove array-to-array variability. Knockdown of the target genes were verified, leading to removal of one ACL treatment group from further analysis due to lack of gene suppression. Genes differentially expressed between treatments were identified using the GeneSpring GX 7.3.1 software package (Agilent Technologies, Palo Alto, CA). Briefly, chip normalized normalize.quantiles data were imported into GeneSpring GX 7.3.1 and per gene normalized to the appropriate biological control for each treatment replicate. Genes that significantly changed in response to orlistat or siRNA targeting FAS, ACC-α, or ACL compared with controls were identified using one-way analysis of variance with a Benjamini and Hochberg false discovery rate of 0.05. Genes with a p value ≤0.05 and with 1.5-fold changes occurring in three or more biological replicates for a given treatment were then overlapped using Venn diagrams to identify a set of genes modified solely by ACC-α and ACL siRNAs and those regulated by FAS siRNA and orlistat.
Measuring Sensitivity to TRAIL—His6-FLAG-tagged recombinant soluble TRAIL (tumor necrosis factor-related apoptosis-inducing ligand) was obtained from John Reed (Burnham Institute for Medical Research) (30). MCF-7 tumor cells were exposed to either orlistat (0–25 μm) or 5-(tetradecyloxy)-2-furoic acid (25 μm; Sigma) for 24 h or transfected with 25 nm FAS, ACC-α, ACL, or non-silencing control siRNA for 48 h. TRAIL sensitization was subsequently determined by incubating the cells with TRAIL (200–500 ng/ml) for 4 h and measuring the level of DNA fragmentation as described above.
RESULTS
Knockdown of FAS Is Unique in Its Ability to Induce Tumor Cell Apoptosis—RNA interference was used to silence the expression of FAS, ACC-α, and ACL and to assess their effects on apoptosis in a panel of tumor cells lines (MDA-MB-435 and MCF-7 mammary carcinoma cells and LNCaP and PC-3 prostate cancer cells). Off-target effects are a major concern when using siRNA-mediated gene silencing, especially when followed by gene expression analysis (31). We minimized off-target activity by using pools of four siRNA sequences (SMART-pools) (31). Measurements of protein expression showed that the targeted SMARTpool siRNAs knocked down between 65 and 90% of cellular FAS, ACC-α, and ACL; the non-silencing control siRNA had no impact on the expression of these enzymes when compared against Lipofectamine 2000 transfection alone (Fig. 1A, right panels, and supplemental Fig. 2). Transfection with 25 nm FAS siRNA induced apoptosis in all four tumor cell lines (Fig. 1A). In contrast, knockdown of ACL failed to induce tumor cell death in MDA-MB-435, MCF-7, and LNCaP cells and had only minor apoptotic activity in PC-3 cells; knockdown of ACC-α also failed to induce DNA fragmentation. These results are consistent with the conclusion that knockdown of enzymes upstream of FAS is not sufficient to elicit an apoptotic response in tumor cells.
FIGURE 1.
Knockdown of FAS, but not ACC-α or ACL, induces tumor cell apoptosis. A, cell death was monitored by measuring the level of DNA fragments present within immunocaptured nucleosomes (left panel). Tumor cells were exposed to 25 nm siRNA targeting FAS, ACC-α, or ACL or non-silencing control siRNA for 72 h (MDA-MB-435 and PC-3 cells) or 96 h (MCF-7 and LNCaP cells). Cells were lysed, and DNA fragmentation was measured by enzyme-linked immunosorbent assay. Values are normalized to Lipofectamine 2000 (mock)-transfected controls and are the means ± S.E. of at least three replicates per treatment. Knockdown of FAS, ACC-α, and ACL was verified by Western blotting (MDA-MB-435 cells (right panels) and MCF-7, PC-3, and LNCaP cells (supplemental Fig. 1)). B, the de novo synthesis of palmitate and the enrichment of acetyl-CoA were measured by gas chromatography/mass spectrometry in MDA-MB-435 cells transfected with 25 nm siRNA targeting either FAS or ACC-α or a non-silencing control and labeled with [U-13C]glucose. Values are the means ± S.E. of two independent experiments.
Anti-tumor effects of FAS inhibition are proposed to be mediated by suppression of palmitate production (32), so it is surprising that knockdown of ACC-α and ACL had little effect on tumor cell death. To confirm that knockdown of the target enzymes resulted in suppression of palmitate biosynthesis, we compared the effect of knockdown of ACC-α or FAS on palmitate biosynthesis in MDA-MB-435 cells (Fig. 1B). Cells were transfected with 25 nm siRNA targeting FAS or ACC-α, and 48 h later, cells were labeled with [U-13C]glucose for 24 h. The enrichment of [13C]acetyl-CoA and [13C]palmitate was assessed by gas chromatography/mass spectrometry. As expected, knockdown of FAS or ACC-α blocked palmitate biosynthesis to comparable levels (49 and 55%, respectively). A similar level of palmitate inhibition following knockdown of ACL was reported previously (22, 23). The suppression of palmitate was not due to differences in the uptake or utilization of glucose because the ratio of [13C]acetyl-CoA to naturally occurring [12C]acetyl-CoA was unchanged in all experiments (Fig. 1B). Because knockdown of FAS or ACC-α leads to an equivalent reduction in de novo palmitate biosynthesis, we concluded that suppression of fatty acid biosynthesis by itself is not sufficient to induce tumor cell apoptosis. It has also been suggested that inhibition of FAS induces apoptosis by increasing the concentration of malonyl-CoA, a substrate of FAS. If this were the case, one might expect that simultaneous inhibition of ACC-α, which produces malonyl-CoA, would rescue tumor cells from apoptosis. However, inhibition of neither ACC-α nor ACL could rescue cells from the apoptotic effects of knockdown of FAS (supplemental Fig. 3).
Inhibition of FAS Induces Tumor Cell Apoptosis by Activating Caspase-8—To determine which apoptotic pathways are activated by inhibition of FAS, we examined the effects of FAS knockdown on the activation of caspase-8 (receptor-mediated apoptosis) and caspase-9 (stress-induced apoptosis) (33). MDA-MB-435 cells were incubated either with a titration range of orlistat (0–50 μm) for 48 h or with 100 nm FAS siRNA for 72 h. Cell extracts were separated by SDS-PAGE and probed with antibodies specific for either full-length or cleaved (activated) caspase-8 and caspase-9. Inhibition of FAS by both FAS siRNA and orlistat induced activation of caspase-8 but failed to activate caspase-9 (Fig. 2A).
FIGURE 2.
Knockdown of FAS induces caspase-8-dependent cell death. A, the effect of mock, non-silencing control, or FAS siRNA (100 nm; left panel) and orlistat (0–50 μm; right panel) on the cleavage of caspase-8 and caspase-9 was analyzed in MDA-MB-435 cells by Western blotting. Camptothecin (1.4 μm) served as a positive control for induction of the proteolytic processing of caspase-9. Equivalent protein loading per lane was ensured by staining blots with Ponceau. B and C, MDA-MB-435 cells were cotransfected with 50 nm FAS siRNA together with 50 nm non-silencing control, caspase-8, or caspase-9 siRNA. DNA fragmentation (B) and caspase-3/7 activity (C, left panel) were measured 72 h after transfection. Transfection with 100 nm non-silencing control siRNA was used to indicate the basal level of apoptosis occurring in response to transfection. Knockdown of caspase-8 and caspase-9 was verified by Western blotting (C, right panels). Values are the means ± S.E. of at least three replicates per treatment. D, the effect of 100 nm siRNA targeting FAS, ACC-α, or ACL on cleavage of caspase-8 was measured in MDA-MB-435 cells 96 h after transfection by Western blotting using antibody against caspase-8. E, Hs58.5s fibroblasts were exposed to mock, non-silencing control, or FAS siRNA (100 nm) for 96 h and assessed for cleavage of caspase-8. Camptothecin served as a positive control. RFLU, relative fluorescence units.
Rescue experiments were conducted to verify that the apoptotic effects of FAS inhibition are elicited solely by caspase-8. Cells were cotransfected with siRNA targeting caspase-8 and caspase-9 along with FAS. Cell death was gauged by measuring DNA fragmentation (Fig. 2B). Knockdown of caspase-8 rescued cells from FAS knockdown by 64%. Correspondingly, we observed a decrease in the activity of the executioner caspase-3/7 in response to the same treatment (Fig. 2C) (33). Knockdown of caspase-9 with siRNA failed to block apoptosis or the activation of caspase-3/7 (Fig. 2, B and C). Collectively, these findings demonstrate that ablation of FAS induces apoptosis through caspase-8. In contrast to the effects of knockdown of FAS, knockdown of ACC-α or ACL was without effect on the activation of caspase-8 (Fig. 2D). Notably, the apoptotic effect is unique to tumor cells because it does not occur in normal fibroblasts (Fig. 2E). The unique effect of FAS ablation on caspase-8 was also observed in the MCF-7, LNCaP, and PC-3 tumor cell lines (supplemental Fig. 2).
Inhibition of FAS Up-regulates the Expression of DDIT4—To better understand the link between inhibition of FAS and activation of caspase-8, we compared changes in gene expression induced by knockdown of FAS with those induced by knockdown of ACC-α or ACL. Genome-wide expression patterns were monitored in response to the knockdowns in the MDA-MB-435 cells.
Three filtering criteria were applied to identify gene expression changes unique to inhibition of FAS: (i) only genes modified by at least at least 1.5-fold were considered; (ii) target genes had to be affected both by the siRNA targeting FAS and by treatment with orlistat; and (iii) target genes could not be modified by knockdown of ACC-α or ACL (meaning a change of <1.5-fold) because they are not linked to apoptosis. Three genes that met these criteria were identified (supplemental Table 1): GPX8 (also known as LOC493869), a sec24 homolog, and DDIT4.
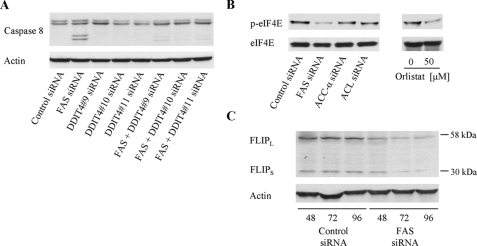
We chose to test the hypothesis that DDIT4 is a link between FAS and caspase-8 because this gene was up-regulated the most by ablation of FAS and because there is evidence in the literature showing that DDIT4 negatively regulates the mTOR pathway (34, 35). To test whether up-regulation of DDIT4 is necessary for the activation of caspase-8 in response to FAS inhibition, we treated MDA-MB-435 cells with siRNA targeting FAS and DDIT4. This experiment showed that knockdown of DDIT4 prevented activation of caspase-8 (Fig. 3A). Thus, DDIT4 is necessary for the activation of caspase-8 in response to FAS inhibition.
FIGURE 3.
Knockdown of FAS induces activation of caspase-8, expression of DDIT4, suppression of eIF4E phosphorylation, and reduction in the anti-apoptotic protein FLIP. A, MDA-MB-435 cells were cotransfected with 25 nm FAS duplex 9 siRNA together with 25 nm non-silencing control duplex 1 or DDIT4 duplex 9–11 siRNAs. Caspase-8 cleavage was measured 96 h after transfection by Western blotting. Transfection with 50 nm non-silencing control siRNA was used to indicate the basal level of caspase-8 cleavage occurring in response to siRNA transfection. B, the effect of non-silencing control siRNA or siRNA targeting FAS, ACC-α, or ACL (25 nm; left panel) and orlistat (0–50 μm; right panel) on phosphorylation and the expression levels of the translation protein eIF4E (p-eIF4E and eIF4E) were measured by Western blotting in MDA-MB-435 cells 96 h after transfection. C, MDA-MB-435 cells were transfected with non-silencing control or FAS siRNA (25 nm) and analyzed 48–96 h post-transfection for changes in the expression of the anti-apoptotic protein FLIP. Actin was used as a loading control.
Inhibition of FAS Reduces the Expression of eIF4E and the Antiapoptotic Protein FLIP—DDIT4 is reportedly a negative regulator of the mTOR pathway, a pathway that controls protein biosynthesis through activation of the rate-limiting translation initiation factor eIF4E (36, 37). We found that knockdown of FAS and orlistat reduced the phosphorylation of eIF4E (Fig. 3B), showing that inhibition of FAS negatively regulates protein translation. Knockdown of ACC-α or ACL had no effect on eIF4E phosphorylation, confirming that down-regulation of protein translation is specific to the inhibition of FAS. Suppression of protein translation by mTOR inhibition has been linked to down-regulation of the synthesis of FLIP, an anti-apoptotic protein that blocks activation of caspase-8 (38). We also found that inhibition of FAS reduced the expression of both isoforms of FLIP, FLIPL and FLIPS (Fig. 3C).
Inhibition of FAS Sensitizes Tumor Cells to TRAIL-induced Apoptosis—Interestingly, down-regulation of FLIP sensitizes tumor cells to TRAIL (39), a member of the tumor necrosis factor family that activates caspase-8. We tested whether inhibition of FAS could be exploited as a means to sensitize tumor cells to TRAIL. MCF-7 tumor cells that are resistant to TRAIL were used as a model (30). Pretreatment of MCF-7 cells for 24 h with increasing concentrations of orlistat sensitized the cells to TRAIL (Fig. 4A). Very little apoptosis occurred in cells exposed to TRAIL along with vehicle (data not shown). Similar observations were observed using siRNA to inhibit FAS (Fig. 4B). In contrast, neither small molecule inhibition nor siRNA knockdown of ACC-α or ACL sensitized the tumor cells to TRAIL (Fig. 4B).
FIGURE 4.
Inhibition of FAS sensitizes MCF-7 tumor cells to TRAIL. A, MCF-7 cells were treated with a concentration range of orlistat (0–25 μm) for 24 h prior to the addition of 200 ng/ml His6-FLAG-tagged recombinant TRAIL or a PBS control. After 4 h, cell death was monitored by measuring the level of DNA fragmentation. Values are the means ± S.E. of four replicates per treatment normalized to TRAIL or PBS treatment alone. B, TRAIL sensitization was monitored in MCF-7 cells transfected with 25 nm siRNA targeting FAS, ACC-α, or ACL or non-silencing control siRNA for 48 h or treated with 25 μm orlistat or an ACC-α inhibitor (5-(tetradecyloxy)-2-furoic acid (TOFO)) for 24 h. DNA fragmentation was measured 4 h after the addition of TRAIL (200–500 ng/ml); values were normalized to background levels. TRAIL sensitization was calculated by dividing treatment + TRAIL by treatment + PBS. Values are the means ± S.E. of eight replicates per treatment from two independent experiments.
DISCUSSION
The salient conclusions of this study are that inhibition of FAS, but not other enzymes involved in fatty acid biosynthesis, (i) induces apoptosis in tumor cells by activating caspase-8; (ii) up-regulates the stress-response gene DDIT4, which inhibits the mTOR pathway; and (iii) sensitizes tumor cells to the apoptotic protein TRAIL.
FAS is required for both tumor cell growth and survival (9, 11), yet the mechanistic links between FAS and tumor cell death have not been unraveled. Here, we have shown that knockdown of FAS or its inhibition with orlistat induces tumor cell apoptosis through activation of caspase-8, with no detectable role for caspase-9. Hence, the central mechanism of tumor cell apoptosis in response to inhibition of FAS is activation of caspase-8. Knockdown of FAS leads to activation of caspase-8 in both breast and prostate carcinoma cells and in both less aggressive (MCF-7 and LNCaP) and more aggressive (MDA-MB-435 and PC-3) tumor cell lines (40–43).
Perhaps most significantly though, knockdown of ACL or ACC-α failed to induce caspase-8-mediated apoptosis, even though their inhibition reduced palmitate production to the same level as knockdown of FAS. These findings are consistent with the conclusion that a blockade of fatty acid biosynthesis alone is not sufficient to induce tumor cell apoptosis. Indeed, prior work showed that inhibition of ACL can inhibit tumor growth but that this effect results from inhibition of cell cycle progression, not through tumor cell apoptosis (22). Similarly, a blockade of ACC-α by 5-(tetradecyloxy)-2-furoic acid fails to induce tumor cell apoptosis even though it inhibits palmitate biosynthesis (13, 44). Altogether then, there is a growing body of evidence that separates the apoptotic effects of FAS inhibitors from the de novo synthesis of palmitate. Therefore, the simplest interpretation of our findings is that a reduction in the de novo synthesis of palmitate alone is not sufficient for activation of caspase-8 or for tumor cell apoptosis.
Our observations support a model whereby inhibition of FAS up-regulates DDIT4 and, as a result, interferes with mTOR signaling. DDIT4 is involved in a series of binding interactions that ultimately lead to mTORC1. DDIT4 binds 14-3-3, preventing 14-3-3 from sequestering TSC2 in a bound complex (46). Unbound TSC2 is therefore free to bind TSC1, and their complex acts as a negative regulator of Rheb, a positive upstream regulator of mTORC1 (47, 48). Consequently, inhibition of FAS negatively regulates the mTORC1 signaling pathway (Fig. 5).
FIGURE 5.
Connections between FAS, caspase-8, and TRAIL. The inhibition of FAS induces the transcription of the stress-response gene DDIT4 (also known as REDD1/RTP801/Dig2). DDIT4 binds to 14-3-3, causing dissociation of TSC2 from the 14-3-3 complex. TSC2 is then able to complex with TSC1 and reduce the activity of Rheb, which activates mTORC1 (mTOR, Raptor, and mLST8). Consequently, protein translation (indicated by phosphorylation of eIF4E) and the level of FLIP are reduced.
The connection between FAS and mTOR probably also explains why inhibition of the enzyme elicits apoptosis through caspase-8. The inhibition of mTOR suppresses protein translation, thereby reducing biosynthesis of FLIP, which blocks the activation of caspase-8 (38). Hence, the reduction in mTOR signaling caused by knockdown of FAS or by inhibition of FAS with orlistat releases a natural inhibition of caspase-8.
The down-regulation of FLIP was found previously to sensitize tumor cells to TRAIL (39), which probably explains how inhibition of FAS sensitizes tumor cells to TRAIL. TRAIL triggers apoptosis through engagement of death receptors that trigger apoptosis by recruiting caspase-8 (49–51). Because the effects of TRAIL are rather selective for tumor cells, this protein is considered an attractive target for cancer treatment. Currently, recombinant TRAIL and TRAIL receptor agonists are being tested in clinical trials and are showing encouraging anti-tumor activity (52). However, the potential of TRAIL as a chemotherapeutic agent is limited due to the emergence of TRAIL resistance (53). Our findings suggest that such resistance could potentially be avoided or overcome by sensitizing tumor cells to this agent by combining it with an inhibitor of FAS. A similar strategy could also be applied to drug development programs aimed at inhibiting the mTOR pathway (54).
Supplementary Material
This work was supported, in whole or in part, by National Institutes of Health Grants CA108959, RR020843, and HL080718. This work was also supported by Department of Defense Grant DAMD17-02-0693 and California Breast Cancer Research Program Grants CA6982713 and CA69306 (to J. W. S.) and Grant 8FB-0107 (to L. M. K.). The costs of publication of this article were defrayed in part by the payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 U.S.C. Section 1734 solely to indicate this fact.
The on-line version of this article (available at http://www.jbc.org) contains supplemental Figs. 1–3 and Table 1.
Footnotes
The abbreviations used are: FAS, fatty-acid synthase; ACC-α, acetyl-CoA carboxylase-α; ACL, ATP-citrate lyase; siRNA, small interfering RNA; PBS, phosphate-buffered saline; eIF4E, eukaryotic initiation factor 4E; FLIP, FLICE-like inhibitory protein.
References
- 1.Wakil, S. J. (1989) Biochemistry 28 4523–4530 [DOI] [PubMed] [Google Scholar]
- 2.Alo, P. L., Visca, P., Marci, A., Mangoni, A., Botti, C., and Di Tondo, U. (1996) Cancer 77 474–482 [DOI] [PubMed] [Google Scholar]
- 3.Swinnen, J. V., Roskams, T., Joniau, S., Van Poppel, H., Oyen, R., Baert, L., Heyns, W., and Verhoeven, G. (2002) Int. J. Cancer 98 19–22 [DOI] [PubMed] [Google Scholar]
- 4.Piyathilake, C. J., Frost, A. R., Manne, U., Bell, W. C., Weiss, H., Heimburger, D. C., and Grizzle, W. E. (2000) Hum. Pathol. 31 1068–1073 [DOI] [PubMed] [Google Scholar]
- 5.Innocenzi, D., Alo, P. L., Balzani, A., Sebastiani, V., Silipo, V., La Torre, G., Ricciardi, G., Bosman, C., and Calvieri, S. (2003) J. Cutan. Pathol. 30 23–28 [DOI] [PubMed] [Google Scholar]
- 6.Gansler, T. S., Hardman, W., III, Hunt, D. A., Schaffel, S., and Hennigar, R. A. (1997) Hum. Pathol. 28 686–692 [DOI] [PubMed] [Google Scholar]
- 7.Rashid, A., Pizer, E. S., Moga, M., Milgraum, L. Z., Zahurak, M., Pasternack, G. R., Kuhajda, F. P., and Hamilton, S. R. (1997) Am. J. Pathol. 150 201–208 [PMC free article] [PubMed] [Google Scholar]
- 8.Kridel, S. J., Axelrod, F., Rozenkrantz, N., and Smith, J. W. (2004) Cancer Res. 64 2070–2075 [DOI] [PubMed] [Google Scholar]
- 9.Knowles, L. M., Axelrod, F., Browne, C. D., and Smith, J. W. (2004) J. Biol. Chem. 279 30540–30545 [DOI] [PubMed] [Google Scholar]
- 10.Kuhajda, F. P., Jenner, K., Wood, F. D., Hennigar, R. A., Jacobs, L. B., Dick, J. D., and Pasternack, G. R. (1994) Proc. Natl. Acad. Sci. U. S. A. 91 6379–6383 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Pizer, E. S., Chrest, F. J., DiGiuseppe, J. A., and Han, W. F. (1998) Cancer Res. 58 4611–4615 [PubMed] [Google Scholar]
- 12.Pizer, E. S., Wood, F. D., Heine, H. S., Romantsev, F. E., Pasternack, G. R., and Kuhajda, F. P. (1996) Cancer Res. 56 1189–1193 [PubMed] [Google Scholar]
- 13.Pizer, E. S., Thupari, J., Han, W. F., Pinn, M. L., Chrest, F. J., Frehywot, G. L., Townsend, C. A., and Kuhajda, F. P. (2000) Cancer Res. 60 213–218 [PubMed] [Google Scholar]
- 14.De Schrijver, E., Brusselmans, K., Heyns, W., Verhoeven, G., and Swinnen, J. V. (2003) Cancer Res. 63 3799–3804 [PubMed] [Google Scholar]
- 15.Orita, H., Coulter, J., Lemmon, C., Tully, E., Vadlamudi, A., Medghalchi, S. M., Kuhajda, F. P., and Gabrielson, E. (2007) Clin. Cancer Res. 13 7139–7145 [DOI] [PubMed] [Google Scholar]
- 16.Kuhajda, F. P. (2000) Nutrition 16 202–208 [DOI] [PubMed] [Google Scholar]
- 17.Swinnen, J. V., Vanderhoydonc, F., Elgamal, A. A., Eelen, M., Vercaeren, I., Joniau, S., Van Poppel, H., Baert, L., Goossens, K., Heyns, W., and Verhoeven, G. (2000) Int. J. Cancer 88 176–179 [DOI] [PubMed] [Google Scholar]
- 18.Szutowicz, A., Kwiatkowski, J., and Angielski, S. (1979) Br. J. Cancer 39 681–687 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Turyn, J., Schlichtholz, B., Dettlaff-Pokora, A., Presler, M., Goyke, E., Matuszewski, M., Kmiec, Z., Krajka, K., and Swierczynski, J. (2003) Horm. Metab. Res. 35 565–569 [DOI] [PubMed] [Google Scholar]
- 20.Milgraum, L. Z., Witters, L. A., Pasternack, G. R., and Kuhajda, F. P. (1997) Clin. Cancer Res. 3 2115–2120 [PubMed] [Google Scholar]
- 21.Thupari, J. N., Pinn, M. L., and Kuhajda, F. P. (2001) Biochem. Biophys. Res. Commun. 285 217–223 [DOI] [PubMed] [Google Scholar]
- 22.Hatzivassiliou, G., Zhao, F., Bauer, D. E., Andreadis, C., Shaw, A. N., Dhanak, D., Hingorani, S. R., Tuveson, D. A., and Thompson, C. B. (2005) Cancer Cell 8 311–321 [DOI] [PubMed] [Google Scholar]
- 23.Bauer, D. E., Hatzivassiliou, G., Zhao, F., Andreadis, C., and Thompson, C. B. (2005) Oncogene 24 6314–6322 [DOI] [PubMed] [Google Scholar]
- 24.Little, J. L., Wheeler, F. B., Fels, D. R., Koumenis, C., and Kridel, S. J. (2007) Cancer Res. 67 1262–1269 [DOI] [PubMed] [Google Scholar]
- 25.Chajes, V., Cambot, M., Moreau, K., Lenoir, G. M., and Joulin, V. (2006) Cancer Res. 66 5287–5294 [DOI] [PubMed] [Google Scholar]
- 26.Bandyopadhyay, S., Zhan, R., Wang, Y., Pai, S. K., Hirota, S., Hosobe, S., Takano, Y., Saito, K., Furuta, E., Iiizumi, M., Mohinta, S., Watabe, M., Chalfant, C., and Watabe, K. (2006) Cancer Res. 66 5934–5940 [DOI] [PubMed] [Google Scholar]
- 27.Lee, W. N., Bassilian, S., Guo, Z., Schoeller, D., Edmond, J., Bergner, E. A., and Byerley, L. O. (1994) Am. J. Physiol. 266 E372–E383 [DOI] [PubMed] [Google Scholar]
- 28.Wittmann, C., and Heinzle, E. (1999) Biotechnol. Bioeng. 62 739–750 [DOI] [PubMed] [Google Scholar]
- 29.Knowles, L. M., and Smith, J. W. (2007) BMC Genomics 8 168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Hyer, M. L., Croxton, R., Krajewska, M., Krajewski, S., Kress, C. L., Lu, M., Suh, N., Sporn, M. B., Cryns, V. L., Zapata, J. M., and Reed, J. C. (2005) Cancer Res. 65 4799–4808 [DOI] [PubMed] [Google Scholar]
- 31.Jackson, A. L., Bartz, S. R., Schelter, J., Kobayashi, S. V., Burchard, J., Mao, M., Li, B., Cavet, G., and Linsley, P. S. (2003) Nat. Biotechnol. 21 635–637 [DOI] [PubMed] [Google Scholar]
- 32.Pizer, E. S., Wood, F. D., Pasternack, G. R., and Kuhajda, F. P. (1996) Cancer Res. 56 745–751 [PubMed] [Google Scholar]
- 33.Fuentes-Prior, P., and Salvesen, G. S. (2004) Biochem. J. 384 201–232 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Sofer, A., Lei, K., Johannessen, C. M., and Ellisen, L. W. (2005) Mol. Cell. Biol. 25 5834–5845 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Corradetti, M. N., Inoki, K., and Guan, K. L. (2005) J. Biol. Chem. 280 9769–9772 [DOI] [PubMed] [Google Scholar]
- 36.Gingras, A. C., Kennedy, S. G., O'Leary, M. A., Sonenberg, N., and Hay, N. (1998) Genes Dev. 12 502–513 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Kleijn, M., Scheper, G. C., Voorma, H. O., and Thomas, A. A. (1998) Eur. J. Biochem. 253 531–544 [DOI] [PubMed] [Google Scholar]
- 38.Panner, A., James, C. D., Berger, M. S., and Pieper, R. O. (2005) Mol. Cell. Biol. 25 8809–8823 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Siegmund, D., Hadwiger, P., Pfizenmaier, K., Vornlocher, H. P., and Wajant, H. (2002) Mol. Med. 8 725–732 [PMC free article] [PubMed] [Google Scholar]
- 40.Price, J. E., Polyzos, A., Zhang, R. D., and Daniels, L. M. (1990) Cancer Res. 50 717–721 [PubMed] [Google Scholar]
- 41.Shafie, S. M., and Liotta, L. A. (1980) Cancer Lett. 11 81–87 [DOI] [PubMed] [Google Scholar]
- 42.Kurebayashi, J., McLeskey, S. W., Johnson, M. D., Lippman, M. E., Dickson, R. B., and Kern, F. G. (1993) Cancer Res. 53 2178–2187 [PubMed] [Google Scholar]
- 43.Koshida, K., Konaka, H., Imao, T., Egawa, M., Mizokami, A., and Namiki, M. (2004) Int. J. Urol. 11 1114–1121 [DOI] [PubMed] [Google Scholar]
- 44.Zhou, W., Simpson, P. J., McFadden, J. M., Townsend, C. A., Medghalchi, S. M., Vadlamudi, A., Pinn, M. L., Ronnett, G. V., and Kuhajda, F. P. (2003) Cancer Res. 63 7330–7337 [PubMed] [Google Scholar]
- 45.Deleted in proof
- 46.DeYoung, M. P., Horak, P., Sofer, A., Sgroi, D., and Ellisen, L. W. (2008) Genes Dev. 22 239–251 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Inoki, K., Li, Y., Xu, T., and Guan, K. L. (2003) Genes Dev. 17 1829–1834 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Tee, A. R., Manning, B. D., Roux, P. P., Cantley, L. C., and Blenis, J. (2003) Curr. Biol. 13 1259–1268 [DOI] [PubMed] [Google Scholar]
- 49.Sprick, M. R., Weigand, M. A., Rieser, E., Rauch, C. T., Juo, P., Blenis, J., Krammer, P. H., and Walczak, H. (2000) Immunity 12 599–609 [DOI] [PubMed] [Google Scholar]
- 50.Kischkel, F. C., Lawrence, D. A., Chuntharapai, A., Schow, P., Kim, K. J., and Ashkenazi, A. (2000) Immunity 12 611–620 [DOI] [PubMed] [Google Scholar]
- 51.Bodmer, J. L., Holler, N., Reynard, S., Vinciguerra, P., Schneider, P., Juo, P., Blenis, J., and Tschopp, J. (2000) Nat. Cell Biol. 2 241–243 [DOI] [PubMed] [Google Scholar]
- 52.Carlo-Stella, C., Lavazza, C., Locatelli, A., Vigano, L., Gianni, A. M., and Gianni, L. (2007) Clin. Cancer Res. 13 2313–2317 [DOI] [PubMed] [Google Scholar]
- 53.LeBlanc, H. N., and Ashkenazi, A. (2003) Cell Death Differ. 10 66–75 [DOI] [PubMed] [Google Scholar]
- 54.Faivre, S., Kroemer, G., and Raymond, E. (2006) Nat. Rev. Drug Discov. 5 671–688 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.