Abstract
Escherichia coli thioredoxin binds to a unique flexible loop of 71 amino acid residues, designated the thioredoxin binding domain (TBD), located in the thumb subdomain of bacteriophage T7 gene 5 DNA polymerase. The initial designation of thioredoxin as a processivity factor was premature. Rather it remodels the TBD for interaction with DNA and the other replication proteins. The binding of thioredoxin exposes a number of basic residues on the TBD that lie over the duplex region of the primer-template and increases the processivity of nucleotide polymerization. Two small solvent-exposed loops (loops A and B) located within TBD electrostatically interact with the acidic C-terminal tail of T7 gene 4 helicase-primase, an interaction that is enhanced by the binding of thioredoxin. Several basic residues on the surface of thioredoxin in the polymerase-thioredoxin complex lie in close proximity to the TBD. One of these residues, lysine 36, is located proximal to loop A. The substitution of glutamate for lysine has a dramatic effect on the binding of gene 4 helicase to a DNA polymerase-thioredoxin complex lacking charges on loop B; binding is decreased 15-fold relative to that observed with wild-type thioredoxin. This defective interaction impairs the ability of T7 DNA polymerase-thioredoxin together with T7 helicase to mediate strand displacement synthesis. This is the first demonstration that thioredoxin interacts with replication proteins other than T7 DNA polymerase.
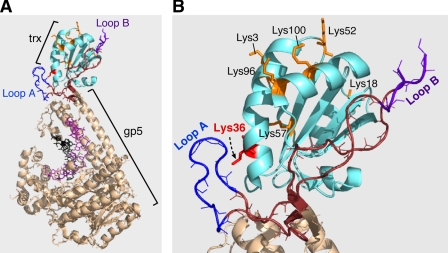
Bacteriophage T7 encodes its own replicative DNA polymerase, the product of gene 5 of the phage (1). The gene 5 protein (gp5) is a nonprocessive DNA polymerase, dissociating from the primer-template after the polymerization of 1-20 nucleotides (2). Upon infection of Escherichia coli gp5 forms a one-to-one complex with the host thioredoxin (trx)2 (3). The binding of trx to gp5 increases the processivity to ∼800 nucleotides per binding event (3, 4). The crystal structure of gp5/trx in complex with a primer-template and a nucleoside triphosphate reveals trx bound to a unique 71-amino acid loop lying between helices H1 and H2 in the thumb subdomain of gp5 (5). This thioredoxin binding domain (TBD) is not present in other members of this family such as E. coli DNA polymerase I and TaqDNA polymerase (5, 6). The largely hydrophobic interaction of trx with gp5 provides for considerable stability (Kd = 5 nm) (2). A number of basic residues in the TBD lie over the duplex portion of the primer-template within the DNA binding crevice and presumably account for the increase in processivity via electrostatic interactions (7, 8). This requirement of trx as a processivity factor for T7 DNA polymerase provides one explanation for its essential function in T7 growth; T7 does not grow in E. coli cells lacking trx (9, 10).
The crystal structure of the complex also reveals two small, solvent-exposed basic loops (loops A and B) in the TBD (Fig. 1A) (5, 11). Recent studies have shown that both T7 gene 4 helicase-primase (gp4) and T7 gene 2.5 ssDNA-binding protein (gp2.5) bind to the gp5/trx complex by interaction with these two loops. Both gp4 and the gp2.5 have flexible and highly acidic C-terminal tails that bind to these loops (11, 12). The binding of gp5/trx to the C-terminal tail of the helicase provides a mechanism for the assembly of additional DNA polymerases at the replication fork (12). The presence of additional polymerases within the replisome provides for increased processivity and may also facilitate primer initiation.
FIGURE 1.
Structure of gp5/trx. A, crystal structure of T7 gp5 (wheat) complexed with trx (cyan) bound to primer-template and an incoming nucleotide at 2.3 Å resolution. The DNA is depicted as sticks with the primer and template in dark pink and black, respectively. trx binds to the unique 76-amino acid segment at the tip of the thumb (TBD) (brown, blue, and purple). B, enlargement of the TBD that indicates the position of the two basic loops A (blue) and B (purple) in position with the trx (cyan). The basic residues on the surface of trx are indicated in orange. In this study, lysine at position 36 (shown in red) is changed to glutamate. It is located proximally, 3 Å, to loop A of TBD of gp5.
The precise role of each of the two loops in the TBD in binding to the gene 4 helicase and gp2.5 ssDNA-binding protein is unclear. When the basic charges in loop A were eliminated by substituting alanine for His-276, Lys-278, and Arg-281, the helicase and gp2.51 still bound to gp5/trx (2-fold less) but with decreased affinity (11). A similar result (3-fold reduction in attaining steady state) was found when the charges were reduced in loop B alone by substituting alanine for Lys-302, Lys-304, Arg-307, and 8 Arg-310. However, when the charges were eliminated in both loops, there was essentially no binding of gp4 or gp2.52 to gp5/trx (11). Thus it would appear that the C-terminal tails of these proteins interact with both loops. However, the possibility exists that during DNA synthesis each loop plays a distinctive role. It is important to note that these results were all obtained in the absence of DNA. When gp5/trx is bound to a primer-template the helicase binds to gp5/trx via a different mode that does not involve the C-terminal tail or loops A and B (11, 12). This nonelectrostatic mode gives rise to a very stable association of the proteins. The binding of gp2.5, however, still occurs through the electrostatic interaction (12).
The interaction of the primer-template, thioredoxin, gp4, and gp2.5 with portions of the TBD is quite remarkable. Clearly this unique insert in the thumb subdomain plays a pivotal role in the assembly of the replisome. Our designation of E. coli thioredoxin as a processivity factor was premature. Thioredoxin does increase the processivity, but it is more appropriate to consider its role in the remodeling of the TBD upon binding. Most likely the increased processivity arises from the proper positioning of basic residues within the TBD so that they contact the primer-template. In this regard it is interesting, as noted above, that thioredoxin increases the binding of both gp4 and gp2.5 to the TBD (12). In these studies gp4 bound to gp5 with relatively low affinity (Kd = 370 nm), whereas it binds to gp5/trx with a 4-fold higher affinity of Kd = 90 nm. Similar studies with gp2.5 show that gp5 alone binds to gp2.5 with a affinity of Kd = 1600 nm, which is about 10-fold stronger in the presence of trx (Kd = 130 nm). The observation that gp4 or gp2.5 does not bind strongly to trx alone (Kd = 130 μm, 500 μm respectively) supports a role of trx in the creation and/or the proper positioning of loops A and B. However, examination of the crystal structure of gp5/trx reveals the presence of several solvent-exposed residues, predominantly lysine residues, on the surface of trx (Fig. 1B) (13, 14). The location of some of these residues in proximity to loops A and B in the TBD raises the possibility that the C-terminal tail of gp4 or gp2.5 could bind to both thioredoxin and loops A and B in a synergistic manner in leading strand synthesis and primer extension. To examine this possibility, we have eliminated several of the positively charged residues on thioredoxin, confirmed that they bind to gp5, and determined the affinity of the resulting gp5/trx to gp4. One residue, lysine 36, in proximity to loop A, has a particularly interesting effect on the binding of gp4 to the gp5/trx complex.
EXPERIMENTAL PROCEDURES
Mutagenesis of trx, Protein Expression, and Purification—Site-directed point mutants of trx were constructed using PCR with plasmid ptrx-3 harboring the gene for trx (trxA). The mutagenesis, using a “Megaprimer” method, requires two separate PCRs using PfuTurbo DNA polymerase (2, 15). The construct has Lys-36 of trx replaced with glutamate. The identity of the construct was confirmed by sequencing. trx variant was overproduced in E. coli strain A307(DE3) that does not express trxA and then purified using procedures described previously (2, 7).
DNA Polymerase Assay—DNA polymerase activity was measured by procedures modified from those described previously (16-18). When DNA polymerase activity was measured using M13 ssDNA as a template, the reaction (10 μl) contained 50 mm Tris-HCl (pH 7.5), 10 mm MgCl2, 5 mm DTT, 50 mm NaCl, 10 nm M13 mGP1-2 ssDNA annealed to a 24-nt oligonucleotide, 500 μm each of dATP, dCTP, dGTP, and [3H]dTTP (2 cpm/pmol), 50 μg/ml bovine serum albumin, and the indicated amounts of trx and gp5. trx was preincubated with gp5, and the reaction was initiated by adding primer-template DNA and 0.5 mm of each of the four deoxyribonucleoside triphosphates. Reaction mixtures were incubated at 37 °C for 3 min, and the reactions were stopped by addition of 5 μl of 0.25 m EDTA (pH 7.5). The incorporation of [3H]TMP was measured on DE81 filter disks as described (2).
Strand Displacement Assay—Strand displacement DNA synthesis was measured by using circular M13 containing a preformed replication fork (19). The replication fork was constructed by annealing M13 mGP1-2 ssDNA to an oligonucleotide (5′-36TAATTCGTAATCATCATGGTCATAGCTGTTTCCT-3′) with 36 bases forming a 5′-tail and 30 bases complementary to the M13 ssDNA template. The oligoribonucleotide was then extended by T7 DNA polymerase to obtain fully duplex DNA. Strand displacement DNA synthesis was carried out in a reaction mixture (10 μl) containing 10 nm dsDNA, 40 mm Tris-HCl (pH 7.5), 10 mm MgCl2, 5 mm DTT, 50 mm potassium glutamate, and 500 μm each dCTP, dGTP, dTTP, and 0.05 μCi [α-33P]dATP, 10 nm gp4 (hexamer), 4 μm trx, and 2.5-20 nm of gp5. gp5, trx, and gp4 were incubated on ice for 15 min, and reactions were initiated by transferring to 37 °C. After 10 min, the reaction was stopped by the addition of EDTA to a final concentration of 80 mm. DNA synthesis was monitored by the amount of [α-33P]dAMP incorporated into DNA (2). To visualize the products of the DNA synthesis, the DNA products were denatured and analyzed by electrophoresis in an alkaline 0.6% agarose gel.
Processivity Assay—Processivity assays were carried out using M13 ssDNA with a 24-nt primer using procedures modified from those described previously (16, 17, 20). DNA synthesis reactions contained 40 mm Tris-HCl (pH 7.5), 10 mm MgCl2, 5 mm DTT, 50 mm NaCl, 500 μm each of dATP, dGTP, and dTTP, 0.05 μCi of [α-32P]dCMP, 20 nm primed M13 ssDNA, 2 nm gp5, and 200 nm trx. Reactions were incubated at 37 °C. Aliquots (10 μl) were removed from the reaction at the indicated times and stopped by the addition of EDTA to a final concentration of 100 mm. Reaction products were subjected to electrophoresis on a 0.6% alkaline-agarose gel. Gels were dried and exposed to a PhosphorImager Fuji BAS 1000 bioimaging analyzer (Fuji Photo Co., Tokyo, Japan).
Surface Plasmon Resonance (SPR)—SPR analysis was performed using the BIAcore-3000 instrument (Uppsala, Sweden). Protein coupling via primary amine groups to the carboxymethyl-5 chip (CM-5) was performed according to the manufacturer's instructions at a flow rate of 5 μl/min. gp4 was coupled to the matrix at 125 μg/ml in 10 mm sodium acetate (pH 5.0) and 10 mm MgCl2 (12). A control flow cell was activated and blocked in the absence of protein to subtract the response units (RU) from the nonspecific interactions and bulk refractive index. Binding studies were performed at room temperature at a flow rate of 40 μl/min in 20 mm Hepes (pH 7.5), 10 mm MgCl2, 5 mm DTT, 250 mm potassium glutamate, and 0.005% (v/v) Tween 20. The chip surface was regenerated using two injections of 150 μl of the above buffer containing 1 m NaCl at 100 μl/min. gp5/trx was reconstituted by incubating gp5 with a 40-fold excess of trx at 20 °C for 10 min.
To investigate the binding of gp5/trx to gp4 in the presence of primer-template, biotinylated DNA was coupled to a streptavidin-coated chip as described previously (11, 12). A template strand was used with a biotin group attached to 5′-end and an annealed primer (11). The template DNA was coupled at a concentration of 0.25 μm in HBS-EP buffer (10 mm Hepes (pH 7.4), 150 mm NaCl, and 0.005% (v/v) Tween 20) at a flow rate of 10 μl/min. Binding studies of gp5/trx were performed at a concentration of 0.25 μm in 20 mm Hepes (pH 7.4), 5 mm MgCl2, 2.5 mm DTT, 200 mm potassium glutamate, and 1% (w/v) glycerol at a flow rate of 10 μl/min. gp4 was injected over the chip in the above buffer containing 0.1 mm ddGTP and 2 mm dTTP. A flow cell blocked with biotin was used as a control to measure nonspecific interaction and bulk refractive index of the sample buffer containing gp4. The chip surface was stripped of bound proteins by sequential injections of 150 μl of 1 m NaCl at a flow rate of 100 μl/min.
RESULTS
trx is the only host protein that is essential for T7 DNA replication and hence phage growth (9, 10). The increase in processivity of T7 gp5 DNA polymerase conferred by trx may be sufficient in and of itself to explain its essential nature. However, the role of trx in the electrostatic binding of gp4 and gp2.5, and to loops A and B in the TBD of gp5 may be equally important.
The observation that gp5/trx binds gp4 and gp2.5 with a higher affinity than does gp5 alone could be explained by the ability of trx itself to bind these proteins or it could reflect a conformational change in loops A and B making them more accessible to gp4 and gp2.5 (12). trx alone binds these proteins extremely weakly and cannot account for the increased binding observed (12). However, the presence of several positively charged lysine residues on the surface of trx prompted us to examine their role in the enhancement of binding induced by trx (Fig. 1B). Initially we substituted glutamate for lysines at positions 3, 18, 36, 57, 96, and 100, purified the altered proteins, and examined their ability to bind to gp5 (Fig. 1B). In addition we also constructed mutant trx proteins containing multiple substitutions (K3E/K18E, K18E/K96E/K100E, and K36E/K57E). All of these altered proteins bound to gp5 normally as measured by their ability to stimulate DNA synthesis catalyzed by gp5 on primed M13 ssDNA, a measure of processivity (data not shown). Using surface plasmon resonance, we examined the ability of gp5 bound to each of these altered trxs to bind gp4 (data not shown). Of all the altered trxs, only constructs having glutamate substituted for lysine at position 36 had a significant effect on binding to gp4 (see below). This lysine residue lies within proximity of 3 Å from the basic residues in loop A of TBD (Fig. 1B). Consequently, trx-K36E and wild-type trx (trx-wt) were purified to apparent homogeneity using standard methods, and their biochemical properties were compared (2).
Polymerase Activity of gp5/trx Complexes—A prerequisite to examining the effect of substituting glutamate for lysine 36 on interactions with gp4 is to ensure that the interaction of the altered trx with gp5 does not differ from that of wild-type trx. The ability of trx to increase the processivity of nucleotide polymerization by gp5 provides an excellent screen for the normal interaction of the two proteins. Earlier experiments have shown that this stimulation of DNA synthesis results from an increase in processivity, presumably because of the elimination of many rate-limiting cycles of dissociation and re-association of the polymerase (2).
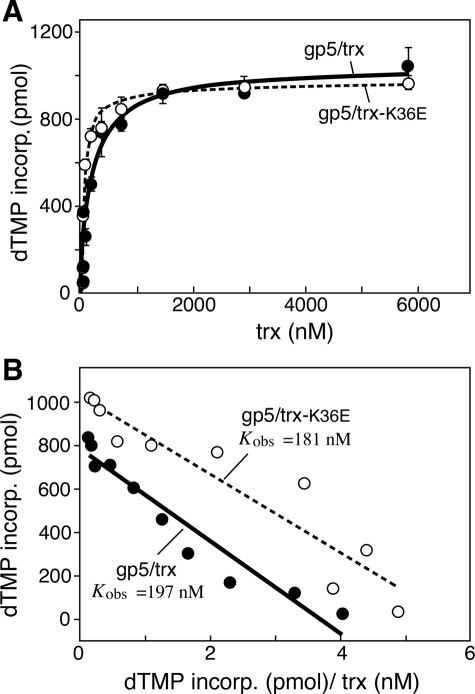
The polymerase activity of gp5/trx-K36E was compared with gp5/trx-wt on primed M13 ssDNA (Fig. 2A) (25). The specific activities of trx-wt and trx-K36E in complex with gp5 protein from the linear range of the data are presented in Table 1. trx-K36E in complex with gp5-wt exhibits very similar activity compared with trx-wt as determined by specific activity and binding affinity. Binding affinity (Kobs), on titrating trx to gp5 as shown by Scatchard plots, shows that although gp5/trx binds to M13 ssDNA with Kobs = 197 (±15) nm, for gp5/trx-K36E the Kobs = 181 (±22) nm (Fig. 2B and Table 2).
FIGURE 2.
Binding affinity of gp5 proteins to thioredoxins in the presence of M13 ssDNA. gp5-trx complexes were formed using A, gp5, with increasing amounts of trx (solid circles) or trx(K36E) (open circles). The polymerase activity of the complexes was measured as described under “Experimental Procedures.” DNA polymerase reactions (10 μl) contained 50 mm Tris-HCl (pH 7.5), 10 mm MgCl2, 5 mm DTT, 50 mm NaCl, 500 μm each of dATP, dCTP, dGTP, and [3H]dTTP (2 cpm/pmol), 50 μg/ml bovine serum albumin, 10 nm primed M13 ssDNA, and 10 nm gp5 or gp5-loop A or gp5-loop B or gp5-loop AB. Increasing amounts of trx or trx-K36E were added to each reaction as indicated, and the reactions were carried out at 37 °C. The amount of DNA synthesis for each reaction was determined by the amount of [3H]TMP incorporated over 3 min. B, data in A were used to generate Scatchard plots for binding affinity of the gp5-trx and gp5-trx-K36E complexes. The observed equilibrium constant (Kobs) for each complex was determined as the negative slope of the corresponding plot.
TABLE 1.
Specific activity of gp5 variants with combinations of trx and trx-K36E
DNA polymerase activities were determined as described under “Experimental Procedures.” Reaction mixtures were incubated with 20 nm primed M13 ssDNA with [gp5] in linear range of 4 nm and trx in 100-fold excess over [gp5]. DNA polymerase activities were measured by the incorporation of [3H]TMP over 3 min. Specific activities were determined as nanomoles of TMP incorporated per ng of protein per min. Comparison of specific activities of reconstituted gp5 variants with trx and trx-K36E is shown.
|
dTMP incorporated
|
|||
|---|---|---|---|
| trx | trx-K36E | ||
| nmol/min/ng protein | |||
| gp5 | 20.11 ± 1.0 | 19.38 ± 3.0 | |
| gp5-loop A | 15.73 ± 3.0 | 14.10 ± 1.0 | |
| gp5-loop B | 3.79 ± 0.8 | 4.10 ± 0.7 | |
| gp5-loop AB | 4.16 ± 0.9 | 3.40 ± 0.2 | |
TABLE 2.
Binding affinity of gp5 variants with combinations of trx and trx-K36E
DNA polymerase activities were determined under “Experimental Procedures.” Reaction mixtures were incubated with 10 nm primed M13 ssDNA with [gp5] in linear range of 4 nm and titrating [trx] in the range from 1 to 6000 nm. DNA polymerase activities were measured by the incorporation of[3H]TMP at 37 °C over 3 min. Binding affinity was determined by using titration curves as shown in Fig. 2A for each gp5/trx complex to generate Scatchard plots. The observed equilibrium constant (Kobs) for each complex was determined as the negative slope of the corresponding plot. Comparison of bindings affinities (Kobs) of reconstituted gp5 variants with trx and trx-K36E is shown.
| trx | trx-K36E | |
|---|---|---|
| nm | nm | |
| gp5 | 197.9 ± 15 | 181.5 ± 22 |
| gp5-loop A | 247.5 ± 52 | 234.4 ± 22 |
| gp5-loop B | 134.4 ± 17 | 157.0 ± 18 |
| gp5-loop AB | 154.1 ± 16 | 149.1 ± 12 |
Because interactions of trx with the other proteins of the replisome are through the TBD of gp5, we have examined gp5-trx complexes in which loops A and B within the TBD have been altered. In gp5-loop A and gp5-loop B, the lysines in loop A or B, respectively, have been substituted with alanine so as to eliminate the charge in each of these two loops (12). In gp5-loop AB, the charges in both loops A and B have been eliminated. To quantitatively assess the interaction between a given gp5 and either trx-wt or trx-K36E, the Kobs between each gp5 and each trx was determined as described previously (26). gp5-wt, gp5-loop A, gp5-loop B, and gp5-loop AB appear to interact with trx and trx-K36E equally well (Table 2). However, the specific activity for gp5-loop B and gp5-loop AB is reduced with trx-wt compared with that with gp5-wt or gp5-loop A (Table 1). As shown previously, the reduction in specific activity is caused by the pausing of gp5/trx as it encounters secondary structures along the M13 DNA during DNA replication, which can be resolved by E. coli SSB protein (11, 22). Thus, trx-K36E does not impose any additional restrictions on the interface with the TBD of gp5.
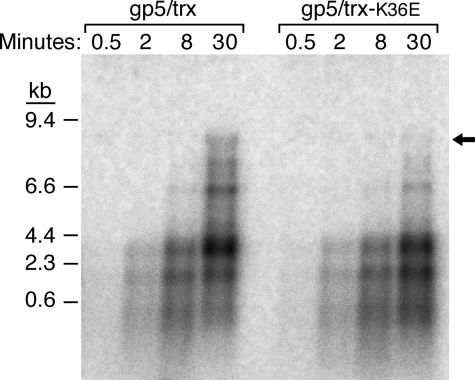
Processivity—gp5 has a low processivity, incorporating only a few nucleotides for each binding event (2). trx binds tightly to the polymerase and dramatically stimulates its processivity (2, 3). We compared the processivity of gp5/trx-K36E to gp5/trx-wt using an M13 ssDNA template annealed to a 5′-32P-labeled oligonucleotide (2). The products of DNA synthesis at various times were analyzed on an alkaline-agarose gel (Fig. 3). Both wild-type trx and trx-K36E increase the processivity of gp5 as evidenced by the high molecular weight DNA products formed over the course of 30 min of incubation. Both gp5/trx complexes catalyze the synthesis of full-length M13 DNA (Fig. 3, indicated by arrow), although the level supported by trx-K36E is somewhat reduced compared with trx-wt. The multiple bands observed have been shown previously to arise from pausing at secondary structures in the DNA template and can be resolved upon addition of E. coli SSB protein or gp2.5 ssDNA-binding protein (11).
FIGURE 3.
Processivity of gp5/trx and gp5/trx-K36E on M13 ssDNA. Processivity assays were carried out on M13 ssDNA primed with a 5′-32P-labeled 24-nt primer in the absence of E. coli SSB protein as described under “Experimental Procedures.” DNA synthesis reactions (50 μl) contained 40 mm Tris-HCl (pH 7.5), 10 mm MgCl2, 5 mm DTT, 50 mm NaCl, 500 μm each of dATP, dGTP, and dTTP, and 0.05 μCi of [α-33P]dCMP, 20 nm primed M13 ssDNA, 2 nm gp5, and 200 nm trx. Reactions were incubated at 37 °C. Aliquots (10 μl) were removed from the reaction at the indicated times and stopped by addition of EDTA to a final concentration of 125 mm EDTA. Reaction products were subjected to electrophoresis on a 0.6% alkaline-agarose gel. The arrow indicates full-length DNA product.
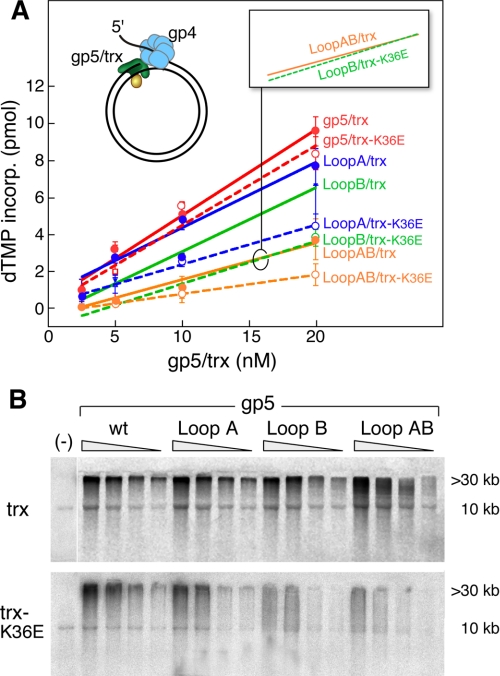
Strand Displacement DNA Synthesis—During leading strand DNA synthesis, the helicase domain of gp4 assembles on the lagging strand as a hexamer and unwinds the duplex DNA to expose the ssDNA template for the leading strand DNA polymerase. To coordinate these activities, a physical interaction between the helicase domain of gp4 and the polymerase occur (21). To study this interaction between gp5/trx and gp4, we measured strand displacement DNA synthesis using a primer-template consisting of a circular M13 dsDNA with a 36-nt ssDNA tail on one strand. This DNA construct, depicted in Fig. 4, resembles a replication fork with the 5′-ssDNA providing a site for the assembly of the hexameric gp4. Strand displacement synthesis was measured by the amount of DNA synthesized (Fig. 4A), and the products of that synthesis were analyzed on alkaline-agarose gels (Fig. 4B). In agreement with previous studies, in the absence of gp4 there is no strand displacement synthesis (Fig. 4B, 1st lane) (11). However, in the presence of gp4, this stimulation results in the formation of DNA products >30 kilobases as revealed by alkaline-agarose gel electrophoresis (Fig. 4B).
FIGURE 4.
Leading strand DNA synthesis. The ability of gp5/trx complexes and gp4 to catalyze strand displacement DNA synthesis was examined using a circular duplex DNA with a preformed replication fork. A, efficiency of strand displacement DNA synthesis by the complex of trx and trx-K36E with gp5, gp5-loop A, gp5-loop B, and gp5-loop AB was determined. The M13 dsDNA with a replication fork was prepared as described under “Experimental Procedures.” The reaction (10 μl) contained 10 nm DNA template, 40 mm Tris-HCl (pH 7.5), 10 mm MgCl2, 5 mm DTT, 50 mm potassium glutamate, and 500 μm each dCTP, dGTP, and dTTP, 0.05 μCi of [α-33P]dATP, 10 nm gp4 (hexamer), 4 μm trx, and 2.5-20 nm of gp5. After 10 min of incubation at 37 °C, the reaction was stopped by EDTA, and the amount of [α-32P]dAMP incorporated into the DNA was measured as described under “Experimental Procedures.” The solid lines represent trx-wt, and broken lines represent trx-K36E. Red lines depict gp5-wt; blue lines depict gp5-loop A; green lines depict gp5-loop B, and yellow lines depict gp5-loop AB. Error bars were derived from two independent experiments. B, products of the reaction described in A were analyzed by electrophoresis on 0.6% alkaline-agarose gel. The products corresponding to the full-length M13 dsDNA (10 kb) and of strand displacement DNA synthesis (>30 kb) are indicated. Synthesis in the absence of gp4 (lane 1) is shown using 20 nm gp5 and 4 μm trx (lane 1). wt, wild type.
trx interacts with gp5-loop A and supports leading strand synthesis almost 80% as well as with gp5-wt (Fig. 4A, red and blue solid lines), whereas gp5-loop B/trx exhibits 1.5-fold less synthesis (Fig. 4A, green solid line). However, the products are similar to those obtained with gp5/trx. In the case of gp5-loop AB/trx, there is 3-fold less synthesis than with wild-type gp5/trx (Fig. 4A, yellow solid line versus red solid line) (12). The amount of the product is less, and there is a wide distribution of product size in the lower molecular weight range particularly at the lower concentrations of protein. The decrease in polymerase activity by gp5-loop B and gp5-loop AB on the primed M13 ssDNA can be overcome by E. coli SSB protein upon removal of secondary structure (11). However, in leading strand synthesis gp4 removes any potential secondary structures ahead of the polymerase. gp5-loop B mediates leading strand synthesis nearly as well as gp5-wt. This observation is consistent with the interpretation that its decreased activity on primed M13 ssDNA (Table 1) is only a result of encountering secondary structure rather than perturbing the intrinsic polymerization activity of gp5/trx.
Based on recent studies of protein affinities measured by surface plasmon resonance, gp5/trx and gp5-loop AB/trx bound equally well to gp4 and formed stable complexes in the presence of DNA (11). This result is also supported by the binding affinity between trx and gp5-wt, gp5-loop A, gp5-loop B, and gp5-loop AB in the presence of M13 ssDNA (Table 2). Furthermore, single molecule studies have also shown that the interaction of gp4 with gp5-loop A, gp5-loop B, and gp5-loop AB are similar to that of gp5-wt in leading strand synthesis, where the processivity is only reduced but nucleotide polymerization was not affected (data not shown). All these results taken together suggest that the alteration of loops A and B does not affect the ability of these enzymes to catalyze strand displacement synthesis (11).
gp5/trx-K36E catalyzes strand displacement synthesis with gp4 helicase almost as well as does the trx-wt (Fig. 4, A, broken lines, and B, bottom panel). The amount of synthesis is nearly the same, and an equal amount of high molecular weight product is formed. However, when in complex with any of the polymerases containing alterations in loops A or B or both, strand displacement synthesis is severely affected. gp5-loop A/trx-K36E synthesizes 2-fold less DNA, and the amount of high molecular product is reduced (Fig. 4, A, blue broken line, and B, bottom panel). In the case of gp5-loop B and gp5-loop AB, the differences are even more striking (Fig. 4, A, green and yellow broken lines, and B, bottom panel). Not only is the amount of DNA synthesized considerably less, but the amount of high molecular DNA products is significantly less with essentially none observed at low concentrations of the polymerase. It is noteworthy that the rates of strand displacement synthesis of gp5-loop B/trx-K36E and gp5-loop AB/trx are similar (Fig. 4A, green broken line and yellow solid line, inset). Thus the charge contribution from lysine 36 of trx is not only sufficient to mimic the role of the three charged residues of loop A of gp5 but the consequence of removal of this charge on trx is even more drastic than all three residues.
Binding of gp4 to gp5-trx Complexes—To investigate the physical interaction of gp4 with gp5/trx, we determined the affinity of gp4 to gp5/trx using surface plasmon resonance (SPR) (Fig. 5). gp4 was immobilized through amine groups to the carboxymethyl (CM5) sensor chip. Different trxs were incubated with each gp5 variant and then injected over the immobilized gp4. At the start of the injection, there is a rapid association as seen by the sharp rise in the RU, immediately followed by a rapid decrease of RU as the free concentration of gp5/trx declines at the end of the injection. This rapid decrease is followed by a slower decrease of RU. The later phase is indicative of the formation of a stable complex. In confirmation of previous studies, the binding of gp5/trx to gp4 depends on the C-terminal tail of gp4, as evidenced by the lack of any binding to gp4 lacking the terminal 17 amino acids (data not shown) (12, 21).
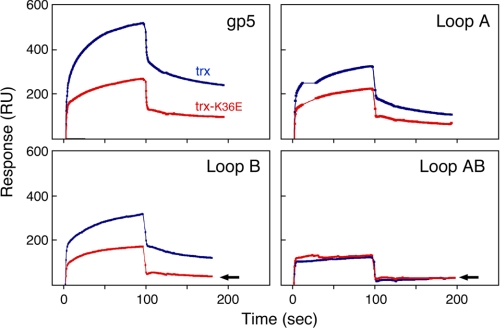
FIGURE 5.
Binding of gp5/trx complexes to g4. gp4 is immobilized on the surface of the CM5 chip via amine coupling, and gp5/trx variants are flowed over the surface. 1000 response units of gp4 were coupled to the chip. Binding studies were conducted as described under “Experimental Procedures.” The concentration of gp5 variants in the flow buffer is 0.2 μm and that of trx and trx-K36E is 8 μm (ratio 1:40). A control cell (no coupled gp4) is used as background to subtract for the RU resulting from any nonspecific interaction and bulk refractive index. Red indicates complexes with trx-K36E, and blue indicates complexes with trx-wt.
trx-K36E in complex with gp5 binds to gp4 to form a stable complex in the later phase of binding (Fig. 5, top left panel). However, the amount of stable complex is approximately half of that formed by gp5/trx and gp4. This reduction in binding was not sufficient to affect leading strand synthesis (Fig. 4). trx-K36E has reduced binding affinity to the variants of gp5-loop A and gp5-loop B but with a slight additive effect (Fig. 5). The reduced binding observed follows the same pattern observed when leading strand synthesis was measured (Fig. 4). With gp5-loop AB, there is no stable complex formed with trx-K36E, analogous to that seen with gp5-loop AB/trx (Fig. 5, bottom right panel). This inhibition of formation of a stable complex with gp4 is reflected in the drastic reduction in processivity during leading strand synthesis. Strikingly, the weak binding of gp5-loop B/trx-K36E to gp4 is comparable with the weak binding of trx or trx-K36E to gp5-loop AB, an analogy that was also made with regard to strand displacement synthesis (Fig. 5, indicated by arrows). Thus the charge contribution made by loop A of the gp5 TBD in the interface with gp4 can apparently be replaced by lysine 36 of trx.
Binding of gp4 to gp5-trx DNA Complexes—When T7 gp5/trx is bound to a primer-template in a polymerizing mode, it interacts with gp4 via a different binding mode (11, 12). This interaction is not electrostatic in that neither the C-terminal tail of gp4 or loops A and B in the TBD are required. The interaction is far more stable than the electrostatic mode. Although it seemed unlikely that substitution of trx-K36E for trx in the gp5-trx complex would affect this binding mode, we examined the interaction of gp4 with gp5/trx-K36E bound to a primer-template. We formed a stable complex of gp5/trx, a primer-template, and a ddNTP, as described previously (5, 11, 12). gp5/trx forms a stable complex with the primer-template in which the primer strand is terminated by 2′,3′-dideoxynucleotide (ddGMP in this experiment), provided the next dNTP specified by the template (dTTP in this experiment) is present. The primer-template used here has one orientation, where the biotin is attached to the 5′-end of the primer-template that is coupled to the streptavidin coated on the surface of the chip (Fig. 6).
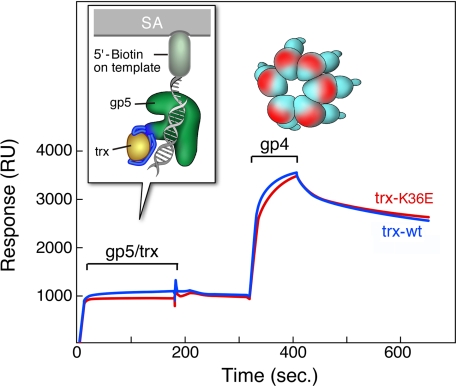
FIGURE 6.
Binding of gp4 to gp5-trx complexes bound to DNA. A primer-template with biotin at the 5′-end of the template strand is immobilized on an SA-sensor chip. gp5/trx-wt or gp5/trx-K36E is injected, followed by gp4. Binding studies were carried out as described under “Experimental Procedures.” One hundred response units of the biotinylated primer-template is coupled to the surface. gp5/trx-wt or gp5/trx-K36E is injected at concentrations of 0.2 μm gp5 and 8 μm trx and trx-K36E (ratio 1:40) in flow buffer containing 1 mm dTTP and 10 μm ddGTP, at saturating conditions of gp5/trx and primer-template. The 100 RU resulting from the coupling of primer-template is subtracted from base line. gp4 is injected at a concentration of 0.7 μm (monomer) in flow buffer containing 0.1 mm ddGTP and 2 mm dTTP. The start and end of the injections are indicated. Red indicate complexes, with trx-K36E and blue indicate complexes with trx-wt.
Both gp5/trx and gp5/trx-K36E formed a highly stable complex with the primer-template attached to the BIAcore chip (Fig. 6). The ratio of RU of gp5/trx or gp5/trx-K36E to the attached primer-template corresponds to a 1:1 binding under saturating condition (12). gp4 binds tightly to either the gp5/trx-DNA or gp5/trx-K36E-DNA complex with no apparent difference between the two. We illustrated previously that gp4 alone does not interact directly with the immobilized DNA (12).
DISCUSSION
Processivity is defined as the number of nucleotides synthesized by the DNA polymerase per single binding event. In bacteriophage T7, binding of trx to gp5 increases the processivity of gp5 on M13 ssDNA several hundredfold (2). The polymerization stalls when it encounters secondary structures on ssDNA, and this pausing can be prevented by binding of single-stranded DNA proteins (2, 12, 22). A replisome consisting of gp5/trx, gp4, and gp2.5 mediate coordinated DNA synthesis in vitro, where leading and lagging strand synthesis proceed at identical rates (24).
The precise mechanism by which trx confers processivity on T7 DNA polymerase is not known. In contrast to E. coli DNA polymerase III, trx is not known to encircle the DNA as a clamp (24). The crystal structure of gp5/trx in complex with the primer-template is believed to be that of the enzyme in the nonprocessive mode (11). It is postulated that in the processive mode, the TBD bound to trx will swing down onto the duplex portion of the primer-template to prevent dissociation from the DNA prior to completion of the polymerization cycle. The binding of trx to the TBD exposes a number of positively charged residues that are in position to contact the duplex portion of the primer-template and suggests that an electrostatic interaction between the TBD and DNA may also be involved. Genetic and biochemical studies support these models (8, 25, 26).
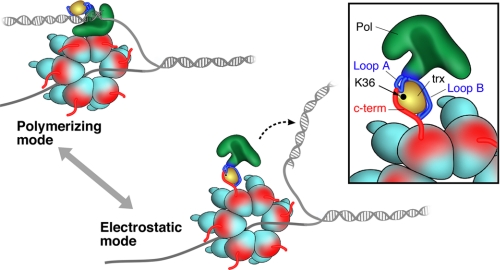
Recent studies indicate gp5/trx binds to gp4 in two modes: a tight polymerizing mode, and a weaker electrostatic mode (12). These two modes are depicted in the schematic presented in Fig. 7. In the polymerizing mode, the interaction does not require the C-terminal tail of gp4 (11, 12). In the second mode, in the absence of primer-template, the avidity of the interaction shifts to a lower affinity, where the gp5/trx and gp4 interaction involves the TBD of gp5 and C-terminal tail of gp4. This latter interaction is maximal when trx is bound to gp5 (12). We have postulated that the binding of trx to the TBD leads to a structural change that creates the two basic loops that interact with gp4 (12). The stable association of gp5/trx-gp4 during leading strand DNA synthesis provides for a relatively high processivity of ∼5 kb, far above the processivity of 800 bp provided by gp5/trx alone (2). After condensation of a nucleotide, a realignment of the primer-template must occur to accommodate the next incoming nucleotide. It is during this transient phase that the polymerase releases from the primer and is dependent on contacts of the TBD with the duplex portion of the primer-template and on contacts with the helicase. We postulate that when the contacts fail and gp5/trx does dissociate from the primer-template it can remain bound to the replisome via its electrostatic interaction with the C-terminal tail of subunits of the hexameric helicase, eventually returning to the primer-template without entering solution. We illustrated that this incident occurred at an average of 5 kb (2). The ability of the polymerase to recycle to the primer after dissociation gives rise to the high processivity greater than 17 kb observed by single molecule techniques, when leading strand synthesis was measured by a single copy of primase and helicase in the absence of any polymerase from solution (4). However, if excess gp5/trx is present in solution then a different gp5/trx may assume DNA synthesis, thus essentially exchanging with the original polymerase (12, 27). This model explains the inability of a DNA trap to stop DNA synthesis, whereas exogenous gp5/trx can exchange with the replicating polymerase without affecting processivity (27).
FIGURE 7.
Two modes of binding of gp5/trx with gp4. In the polymerizing mode, gp5-trx and gp4 form a tight complex that is independent of the C-terminal tail of gp4. The complex synthesizes an average of 5 kb of DNA before gp5/trx dissociates from the DNA. In the electrostatic mode, gp5/trx forms an unstable complex with gp4 via the C-terminal tail. This latter mode provides a mechanism for the exchange of the DNA polymerase at the replication fork without affecting the processivity. The inset displays the acidic C-terminal tail of gp4 electrostatically interacting with the basic residues in loop A and B of the TBD and lysine 36 of trx.
The processivity of the gp5/trx does not affect the active site of trx that is otherwise a cofactor in reducing disulfide bonds in many proteins (28). The physical studies of the gp5/trx in the presence of primer-template show that the binding affinity of gp5/trx-K36E remains the same compared with gp5/trx-wt (Fig. 6). This comparison remains unchanged even in the presence of gp4, which is evident by the formation of an identical additive complex. The normal interaction of trx-K36E with gp5 provides assurance that the interactions we describe with trx-K36E are not because of an alteration of the interaction of the altered trx with the TBD of gp5 (Fig. 2 and Table 2).
Both loops A and B of the TBD of gp5 are required for optimal interaction with the C terminus of gp4. Lysine 36 is located on the surface of trx and at a distance of 3 Å from loop A of the TBD (Fig. 1B and Fig. 7, inset). The likelihood of lysine 36 contributing to the interaction with gp4 is suggested by the strand displacement studies. Strand displacement synthesis mediated by gp5/trx is dependent on the interaction of gp5/trx with T7 helicase. When trx-K36E was substituted for trx-wt, the resulting gp5/trx-K36E could not interact with gp4 to generate high molecular weight species of DNA on circular M13 DNA molecules. Indeed, surface plasmon resonance studies revealed that gp5-loop A/trx-K36E could not form a stable complex with gp4, whereas gp5-loop A/trx-wt does. Furthermore, the dramatic effects observed when gp5-loop B and trx-K36E were used in combination suggest strongly that lysine 36 can partially compensate for the loss of charges in loop A (Fig. 5). In gp5-loop A three positively charged residues, His-276, Lys-278, and Arg-281 were changed to alanine. Thus the reversal of charge on lysine 36 has essentially the same impact on binding to gp4 as does neutralization of three residues in loop A. The overall reduction in leading strand synthesis because of the reversal of charge of lysine 36 to glutamate implies that this residue has even more effect than that of loop A (Fig. 4B, compare top panel versus bottom panel). An alternative explanation is that the lysine 36 substitution impairs the ability of loop A to interact with gp4; however, the data do not support this. In these studies a number of controls show that trx-K36E interacts with gp5 in a manner similar to that observed for the interaction with trx-wt and that the reconstituted gp5/trx-K36E binds to a primer template and catalyzes processive DNA synthesis essentially as does gp5/trx-wt.
In this study we have identified the first interaction of trx with any replication protein other than gp5, the T7 DNA polymerase. In addition to interacting with gp4, we suspect that lysine 36 will also electrostatically interact with the acidic C-terminal tail of gp2.5 ssDNA-binding protein. The C-terminal tails of both gp4 and gp2.5 are in contact with loops A and B of the TBD. Of the two loops in the TBD our earlier evidence (11) as well as the results presented here reveal a greater contribution of loop B to gp4 relative to loop A. In the earlier studies we were not aware of the interaction of gp4 with lysine 36, and hence removing the charges in loop A would only partially prevent the binding of gp4 to this region of gp5, which includes both loop A and lysine 36. In fact, if the charges on loop A are eliminated and the positive charge of lysine 36 is reversed, then the effect on binding to gp4 is greater than that obtained by removal of the charges in loop B. Thus, it appears that loop B is more essential in leading strand synthesis and loop A provides the conformational changes required for this event. One can speculate that differences in the affinity of these sites for trx, gp4, and gp2.5 provides for a subtle control of events at the replications, events we have not yet dissected.
Acknowledgments
We thank all the members, particularly Boriana Marintcheva and Masateru Takahashi, from the Richardson lab for help in scientific discussions and Steven Moskowitz (Advanced Medical Graphics, Boston) for help with figure preparation.
This work was supported, in whole or in part, by National Institutes of Health Grant GM 54397 (USPHS). The costs of publication of this article were defrayed in part by the payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 U.S.C. Section 1734 solely to indicate this fact.
Footnotes
The abbreviations used are: trx, thioredoxin; ssDNA, single-stranded DNA; dsDNA, double-stranded DNA; SPR, surface plasmon resonance; TBD, thioredoxin binding domain; nt, nucleotide; DTT, dithiothreitol; RU, response unit.
S. M. Hamdan and C. C. Richardson, unpublished results.
References
- 1.Richardson, C. C. (1983) Cell 270 315-317 [DOI] [PubMed] [Google Scholar]
- 2.Tabor, S., Huber, H. E., and Richardson, C. C. (1987) J. Biol. Chem. 262 16212-16223 [PubMed] [Google Scholar]
- 3.Huber, H. E., Tabor, S., and Richardson, C. C. (1987) J. Biol. Chem. 262 16224-16232 [PubMed] [Google Scholar]
- 4.Lee, J. B., White, R. K., Hamden, S. M., Ix, X. S., Richardson, C. C., and van Oji, A. M. (2006) Nature 439 621-624 [DOI] [PubMed] [Google Scholar]
- 5.Double, S., Tabor, S., Long, A., Richardson, C. C., and Ellenberger, T. (1998) Nature 391 251-258 [DOI] [PubMed] [Google Scholar]
- 6.Beese, L. S., Derbyshire, V., and Steitz, T. A. (1993) Science 260 352-355 [DOI] [PubMed] [Google Scholar]
- 7.Bedford, E., Tabor, S., and Richardson, C. C. (1997) Proc. Natl. Acad. Sci. U. S. A. 94 479-484 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Yang, X., and Richardson, C. C. (1997) J. Biol. Chem. 272 6599-6606 [DOI] [PubMed] [Google Scholar]
- 9.Mark, D. F., and Richardson, C. C. (1976) Proc. Natl. Acad. Sci. U. S. A. 73 780-784 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Modrich, P., and Richardson, C. C. (1975) J. Biol. Chem. 250 5515-5522 [PubMed] [Google Scholar]
- 11.Hamdan, S. M., Johnson, D. E., Tanner, N. A., Lee, J.-B., Qimron, E., Tabor, S., van Oijen, A. M., and Richardson, C. C. (2007) Mol. Cell 27 538-549 [DOI] [PubMed] [Google Scholar]
- 12.Hamdan, S., Marintcheva, B., Cook, T., Lee, S.-L., Tabor, S., and Richardson, C. C. (2005) Proc. Natl. Acad. Sci. U. S. A. 102 5096-6010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Katti, S. K., and LeMaster, D. M. (1990) J. Mol. Biol. 212 167-184 [DOI] [PubMed] [Google Scholar]
- 14.Dyson, H. J., Gippert, G. P., Case, D. A., Holmgren, A., and Wright, P. E. (1990) Biochemistry 29 4129-4136 [DOI] [PubMed] [Google Scholar]
- 15.Sarkar, G., and Sommer, S. S. (1990) BioTechniques 8 404-407 [PubMed] [Google Scholar]
- 16.Kelman, Z., Hurwitz, J., and O'Donnell, M. (1998) Structure (Lond.) 6 121-125 [DOI] [PubMed] [Google Scholar]
- 17.Kumar, J. K., Kremsdorf, R., Tabor, S., and Richardson, C. C. (2001) J. Biol. Chem. 276 46151-46159 [DOI] [PubMed] [Google Scholar]
- 18.Bryant, F. R., Johnson, K. A., and Benkovic, S. J. (1983) Biochemistry 22 3537-3546 [DOI] [PubMed] [Google Scholar]
- 19.Delagoutte, E., and von Hippel, P. H. (2001) Biochemistry 40 4459-4477 [DOI] [PubMed] [Google Scholar]
- 20.Bambara, R. A., Uyemura, D., and Choi, T. (1978) J. Biol. Chem. 253 413-423 [PubMed] [Google Scholar]
- 21.Notarnicola, S. M., Mulcahy, H. L., Lee, J., and Richardson, C. C. (1997) J. Biol. Chem. 272 18425-18433 [DOI] [PubMed] [Google Scholar]
- 22.Johnson, D. E., and Richardson, C. C. (2003) J. Biol. Chem. 278 23762-23772 [DOI] [PubMed] [Google Scholar]
- 23.Lee, J., Chastain, P. D., II, Kusakabe, T., Griffith, J. D., and Richardson, C. C. (1998) Mol. Cell 1 1001-1010 [DOI] [PubMed] [Google Scholar]
- 24.Cannistraro, V. J., and Taylor, J.-S. (2004) J. Biol. Chem. 279 18288-18295 [DOI] [PubMed] [Google Scholar]
- 25.Himawan, J. S., and Richardson, C. C. (1992) Proc. Natl. Acad. Sci. U. S. A. 89 9774-9778 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Himawan, J. S., and Richardson, C. C. (1996) J. Biol. Chem. 271 19999-20008 [DOI] [PubMed] [Google Scholar]
- 27.Johnson, D. E., Takahashi, M., Hamdan, S. M., Lee, S. J., and Richardson, C. C. (2007) Proc. Natl. Acad. Sci. U. S. A. 104 5312-5317 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Huber, H. E., Russel, M., Model, P., and Richardson, C. C. (1986) J. Biol. Chem. 261 15006-15012 [PubMed] [Google Scholar]