Abstract
Defects in frataxin result in Friedreich ataxia, a genetic disease characterized by early onset of neurodegeneration, cardiomyopathy, and diabetes. Frataxin is a conserved mitochondrial protein that controls iron needed for iron-sulfur cluster assembly and heme synthesis and also detoxifies excess iron. Studies in vitro have shown that either monomeric or oligomeric frataxin delivers iron to other proteins, whereas ferritin-like frataxin particles convert redox-active iron to an inert mineral. We have investigated how these different forms of frataxin are regulated in vivo. In Saccharomyces cerevisiae, only monomeric yeast frataxin (Yfh1) was detected in unstressed cells when mitochondrial iron uptake was maintained at a steady, low nanomolar level. Increments in mitochondrial iron uptake induced stepwise assembly of Yfh1 species ranging from trimer to >24-mer, independent of interactions between Yfh1 and its major iron-binding partners, Isu1/Nfs1 or aconitase. The rate-limiting step in Yfh1 assembly was a structural transition that preceded conversion of monomer to trimer. This step was induced, independently or synergistically, by mitochondrial iron increments, overexpression of wild type Yfh1 monomer, mutations that stabilize Yfh1 trimer, or heat stress. Faster assembly kinetics correlated with reduced oxidative damage and higher levels of aconitase activity, respiratory capacity, and cell survival. However, deregulation of Yfh1 assembly resulted in Yfh1 aggregation, aconitase sequestration, and mitochondrial DNA depletion. The data suggest that Yfh1 assembly responds to dynamic changes in mitochondrial iron uptake or stress exposure in a highly controlled fashion and that this may enable frataxin to simultaneously promote respiratory function and stress tolerance.
A reduction in the levels of frataxin is responsible for Friedreich ataxia, an autosomal recessive disease affecting ∼1/40,000 individuals with early onset and progressive course (1). Biochemical and biophysical studies have shown that frataxin binds iron and can promote iron delivery to different iron-binding proteins as well as detoxify iron. In either monomeric or oligomeric form, frataxin can serve as a Fe2+ donor for ferrochelatase (2, 3), the iron-sulfur cluster (ISC)2 scaffold protein, IscU/Isu1 (4, 5), and the [4Fe-4S]2+ enzyme, aconitase (6). Moreover, frataxin assembles into particles that convert redox-active iron to a stable mineral, which remains soluble within the protein and is inactive in the generation of free radicals (2, 5, 7, 8). The crystal structure of yeast frataxin trimer has suggested that this form of the protein may be able to perform both iron delivery and iron detoxification (9). Electron microscopy reconstructions of a 24-subunit oligomer of yeast frataxin have further revealed that trimer is the building block of larger frataxin oligomers, which have a hollow cavity where an iron core can be progressively accumulated (10). There are striking similarities in the structural organization of frataxin oligomers and ferritin, consistent with functional similarities between these two iron-storage proteins (2, 7). Accordingly, expression of mitochondrial ferritin can rescue the phenotype of frataxin-depleted yeast and human cells (11, 12).
The biochemical properties of frataxin can explain the biological functions in which this protein has been implicated in vivo, both in humans and model systems. The ability to bind Fe2+ and deliver it to such proteins as the ISC scaffold IscU/Isu1, aconitase, and ferrochelatase can account for the roles played by frataxin in ISC enzyme biogenesis (13–16), aconitase activation (6, 17), and heme synthesis (18, 19), as well as cellular iron balance (20, 21) and energy production (22, 23). In addition, the ability to convert potentially toxic iron to a redox-inactive mineral explains why frataxin is involved in oxidative stress tolerance, mitochondrial and nuclear DNA integrity, tumor suppression, as well as cellular and organism longevity (20, 24–34).
Other known multifunctional proteins accomplish different biological roles through various mechanisms that may include changes in oligomeric state (35). Similarly, frataxin appears to use self-assembly to deliver iron to other proteins or detoxify excess iron. However, little is still known about this process. Assembly of the yeast frataxin homologue (Yfh1) in vitro requires the presence of Fe2+ and proceeds in an iron concentration-dependent manner, according to the progression α → α3 → α6 → α12 → α24 → α48 (36, 37). Although one group reported that Yfh1 oligomerization is dispensable in vivo (38), we later showed that mutations affecting Yfh1 assembly are asymptomatic in unstressed cells but become harmful during stress exposure and cause synthetic lethality in cells lacking copper-zinc superoxide dismutase (34). Iron-dependent assembly has also been characterized for the Escherichia coli orthologue, CyaY, and shown to be required for interaction of CyaY with the ISC assembly proteins IscU and IscS in vitro (8, 39). Unlike the yeast or E. coli orthologues, human frataxin assembles at physiological iron concentrations during expression in E. coli or in human heart but not in vitro (40, 41), indicating that different mechanisms govern frataxin assembly in different species. In the monocellular eukaryote, Saccharomyces cerevisiae, frataxin most likely carries out iron delivery and detoxification concurrently, and indeed these functions are interdependent and difficult to separate (34, 37, 42). We therefore used S. cerevisiae as a model to begin to elucidate regulation of frataxin assembly in vivo.
EXPERIMENTAL PROCEDURES
S. cerevisiae Expression Constructs and Strains—The mutant yfh1-Y73A and yfh1-T118A/V120A alleles were created by PCR, cloned into low copy plasmid YCplac22-YFH1 (34) under the control of the natural YFH1 promoter, and sequenced completely. The YFH1, yfh1-Y73A, and yfh1-T118A/V120A alleles were also cloned in the EcoRI-SalI sites of a TRP1-based pRS404 integration vector under the control of the strong glyceraldehyde-3-phosphate dehydrogenase (GPD) promoter (43) yielding plasmids pRSGPD-YFH1, pRSGPD-YFH1[Y73A], and pRSGPD-YFH1[T118A/V120A]. Strain yfh1Δ[YFH1][ρ+] (MATα ura3-52 lys2–801amber ade2-101ochre trp1-Δ63 his3-Δ200 leu2-Δ1 yfh1Δ::HIS3 + YCp50-YFH1-URA3 [ρ+]) is a fully complemented derivative of strain yfh1Δ (34). Strains wild type (WT), Y73A, and T118A/V120A were obtained by transformation of strain yfh1Δ[YFH1][ρ+] with vectors YCplac22-YFH1, YCplac22-YFH1[Y73A], or YCplac22-YFH1[T118A/V120A], whereas strains WT*, Y73A*, and T118A/V120A* were obtained by transformation of strain yfh1Δ[YFH1][ρ+] with vectors pRSGPD-YFH1, pRSGPD-YFH1[Y73A], or pRSGPD-YFH1[T118A/V120A], followed by counter selection with 5′-fluoroorotic acid to eliminate the YCp50-YFH1-URA3 plasmid (34).
Mitochondria Isolation and Fractionation Procedures—The cells were preconditioned at 30 °C in 50 ml of YPGal medium (1% yeast extract, 2% peptone, 2% galactose, 0.5% glucose) or 160 ml of SD medium (6.7% bacto-yeast nitrogen base, 2% glucose, supplemented with amino acids and other growth requirements) for ∼12 h. These cultures were used to inoculate larger cultures (500 ml at an A600 = 0.4 for YPGal cultures, and 1,500 ml at an A600 = 0.1 for SD cultures), and the incubation was continued at 30 °C. After 5 h of growth, iron (100 or 500 μm from a 100 mm FeCl3 stock solution) was added, and growth continued at 30 or 37 °C as required. Aerobic conditions were maintained by using a culture volume ≤⅕ of the volume of the culturing flask and by shaking the culture at 225 rpm (34). The cells were harvested by centrifugation at 3,000 × g for 20 min at 4 °C, washed with 500 ml of ice-cold water, and resuspended in 16 ml of MST buffer (210 mm mannitol, 70 mm sucrose, 5 mm Tris-HCl, pH 7.5, with 1 tablet/50 ml of complete protease inhibitor mixture (Roche Applied Science) and 2 mm phenylmethylsulfonyl fluoride) and aliquoted into two 50-ml tubes. About 4 ml of 425–600 μm acid-washed glass beads (Sigma) were added to each tube, and the cells were lysed by vortexing three times for 2 min with 2-min intervals on ice. All of the subsequent steps were carried out at 4 °C. Glass beads and unbroken cells were removed by centrifugation at 1,000 × g for 1 min, and mitochondria were isolated from the supernatant by centrifugation at 14,000 × g for 20 min. The pellets were resuspended in 800 μl of HN100 buffer (10 mm HEPES-KOH, pH 7.4, 100 mm NaCl, with protease inhibitors) and immediately frozen in dry ice/ethanol. For size exclusion chromatography, each mitochondrial preparation was thawed on ice and immediately sonicated on ice, six times for 10 s with a Branson Sonifier 250 (with output control set at 5 and duty cycle at 50%), with a 1-min interval after each 10-s period. This step was immediately followed by ultracentrifugation at 100,000 × g for 30 min at 4 °C, and the supernatant (typically 250 μl at a protein concentration of ∼20 mg/ml for YPGal cultures or ∼5 mg/ml for SD cultures) immediately loaded on a 10 mm × 30 cm column packed with Superdex 75 support (GE Healthcare). Fractions (0.5 ml) from the column were collected, and aliquots (20 and 35 μl from YPGal and SD cultures, respectively) were analyzed by SDS/PAGE and Western blotting. In some experiments, the mitochondria were disrupted by sonication as described above and diluted in HN100 buffer to a protein concentration of 4.5 mg/ml. The samples of these disrupted mitochondria were treated with 100 mm sodium carbonate, 1% dodecyl maltoside, or 1% Triton X-100 in HN100 buffer for 30 min at 4 °C. Each sample was subsequently centrifuged at 20,800 × g for 20 min at 4 °C, the supernatant was carefully transferred to a new tube, and the sediment was resuspended in an equal volume of Laemmli buffer. The sediment was further heated at 100 °C for 5 min and centrifuged for 1 min at 20,800 × g. Equal volumes of the total, supernatant, and sediment samples were then analyzed by SDS-PAGE and Western blotting. For blue native PAGE, the samples of disrupted mitochondria were incubated in HN100 buffer in the presence of 1% dodecyl maltoside for designated times. After centrifugation at 10,000 × g for 20 min at 4 °C, the supernatant was analyzed by blue native PAGE on 3–12% Bis-Tris gels (Invitrogen) per the manufacturer's protocol and subsequently detected by Western blotting with a polyclonal anti-Yfh1 antibody. In other experiments, the samples of disrupted mitochondria were incubated in the presence of 0.01–0.1% SDS and centrifuged at 5,000 × g for 20 min at 4 °C. The supernatants were analyzed by native PAGE (3–12% Bis-Tris) followed by Western blotting or Prussian blue staining. Polyclonal antisera against recombinant purified forms of Yfh1, Nfs1, and Isu1 were generated in rabbits; anti-Aco1 polyclonal antiserum was a generous gift of A. L. Bulteau (Université Paris, Paris, France).
Aconitase Activity, Protein Carbonylation, and Mitochondrial Iron Content—Mitochondria were suspended in MST buffer at a protein concentration of 25 mg/ml. Aliquots of these samples were frozen immediately, whereas additional aliquots were first subject to a 15-min aerobic incubation or a 5-min treatment with 100 μm H2O2 (from a 8.8 m solution) (Sigma) at room temperature and immediately frozen. These samples were subsequently used for (i) detection of protein carbonyls (100 μg and total protein) using an OxiBlot kit (Chemicon) per manufacturer's protocol; (ii) in-gel aconitase activity assays (150 μg of total protein) (44); and (iii) measurement of mitochondrial iron content (10–600 μg of total protein, depending on growth conditions) (45). Protein concentration was measured by the BCA kit (Pierce).
Measurements of Petite Formation and Cell Viability—All of the experiments were started with freshly streaked frozen stocks grown on YPD plates (2% peptone, 1% yeast extract, 2% dextrose) for 5 days (34). The incubations were carried out aerobically as described above. The cultures were initially synchronized to late logarithmic phase (A600 =∼1) in SD medium at 30 °C and diluted in fresh SD to A600 = 0.1. The cultures were allowed to reach A600 = 0.3 (∼5 h), at which point 100 μm FeCl3 or 500 μm H2O2 was added, and incubation continued at 37 °C. After plating on YPD and YPG (like YPD but with 2% glycerol instead of dextrose), the cells were scored for viability or respiratory capacity (33).
RESULTS
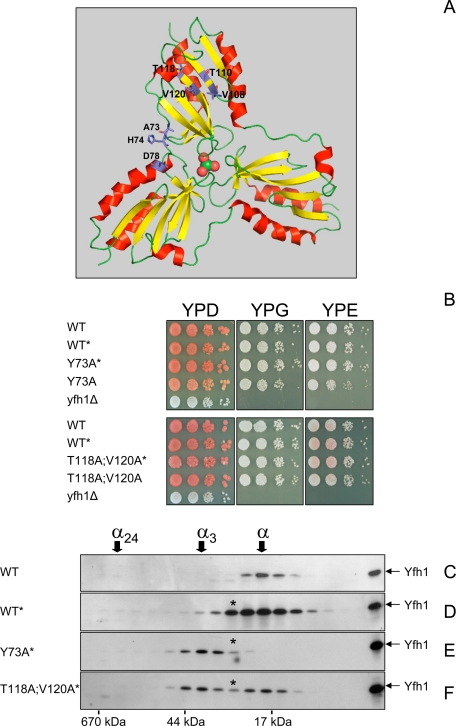
Mutations That Enhance Yfh1 Oligomerization Hinder Yfh1 Precursor Maturation—A previous study identified amino acid substitutions (Y73A, H74A, D78A, V108A/T110A, and T118A/V120A) that enabled the Yfh1 monomer to form stable trimer at physiological iron concentrations during expression in E. coli (9) (Fig. 1A). In an attempt to genetically manipulate the oligomeric state of frataxin in yeast, we expressed two Yfh1 proteins with stabilizing mutations, Yfh1-Y73A and Yfh1-T118A/V120A, in a yeast frataxin knock out strain (yfh1Δ). Frataxin is synthesized in the cytoplasm as a precursor polypeptide that is imported by mitochondria and processed to mature form by mitochondrial processing peptidase (46). The precursor forms of the Yfh1-Y73A and Yfh1-T118A/V120A proteins were initially expressed by use of a low copy plasmid and the natural YFH1 promoter to yield strains Y73A and T118A/V120A. The ∼20-kDa mature form of Yfh1 was nearly undetectable by Western blotting in these strains, whereas it was readily detected in an isogenic WT strain (not shown). Unlike the yfh1Δ strain, however, Y73A and T118A/V120A grew nearly as well as WT on solid rich medium supplemented with fermentable or nonfermentable carbon sources at 30 °C (Fig. 1B), indicating that despite being present at very low levels, the Yfh1-Y73A and Yfh1-T118A/V120A proteins were functional. Analyses with purified yeast mitochondrial processing peptidase or isolated yeast mitochondria showed that the precursor form of the Yfh1-Y73A protein was proteolytically cleaved with slower kinetics as compared with the wild type Yfh1 precursor (supplemental Fig. S1). However, the mature forms derived from the wild type and mutant precursors were equally stable over time in isolated mitochondria (supplemental Fig. S1). This suggested that improving processing kinetics might be sufficient to increase the steady-state levels of mature Yfh1-Y73A. We therefore expressed mutant and wild type Yfh1 precursors under the control of the strong GPD promoter (43). A yfh1Δ strain bearing one chromosomally integrated copy of the yfh1-Y73A allele (strain Y73A*) contained levels of mature Yfh1 comparable with those present in WT, whereas strains bearing one integrated copy of wild type YFH1 or the yfh1-T118A/V120A allele (strains WT* and T118A/V120A*) contained ∼ 5 and ∼2 times more mature Yfh1 protein than WT (Fig. 1, C–F). The WT*, Y73A*, and T118A/V120A* strains were indistinguishable from WT on solid rich medium supplemented with fermentable or nonfermentable carbon sources (Fig. 1B).
FIGURE 1.
Mutations that stabilize Yfh1 trimer are functional in yeast. A, structure of iron-loaded Yfh1 trimer in ribbon representation with one iron atom (green sphere) and three water molecules (red spheres) in the central channel (9). Residues involved in trimer stabilization are shown as sticks. The figures were generated with PyMOL. B, freshly streaked frozen stocks were grown for ∼12 h in 10 ml of SD medium at 30 °C. 10-fold serial dilutions were plated on solid rich (YP) medium with dextrose (D), glycerol (G), or ethanol (E) as the carbon source and grown for 5 days at 30 °C. Respiring strains are pigmented on YPD plates because of the endogenous ade2–101ochre mutation. C–F, yeast cultures were started from freshly streaked glycerol stocks, preconditioned in YPGal at 30 °C for ∼12 h, diluted into fresh YPGal, and grown for ∼20 h at 30 °C, after which mitochondria were isolated. The soluble mitochondrial fractions (∼5 mg total protein) were analyzed by size exclusion chromatography on a Superdex 75 column. This column gives excellent resolution of purified Yfh1 monomer (α, ∼14 kDa) and trimer (α3, ∼42 kDa), which are eluted with apparent molecular masses of ∼20 and ∼40 kDa, respectively, whereas 24-mer (α24, ∼336 kDa) is eluted at the size exclusion limit of the column. Fractions comprising the entire fractionation range of the column were analyzed by Western blotting with anti-Yfh1 antiserum. Recombinant Yfh1 (residues 52–174) indicates the position of the mature form of the protein on SDS-PAGE. The asterisk denotes the α* intermediate.
Oligomerization of Yfh1 Can Be Genetically Manipulated in Vivo—To analyze the assembly state of Yfh1 in conditions of low mitochondrial iron uptake, the strains described above were grown in liquid rich medium supplemented with galactose (YPGal), a fermentable carbon source that does not repress mitochondrial biogenesis and allows yeast to use respiration (47). The YPGal medium prepared in our laboratory normally contains ∼12 μm iron, a concentration sufficient to partially repress the plasma membrane high affinity iron transport system and limit cellular iron influx (48). The cultures were preconditioned in YPGal for 12 h, diluted into fresh YPGal, and harvested after ∼20 h, corresponding to the early exponential phase of growth. The mitochondria were isolated, and the total mitochondrial iron content was determined as a measure of mitochondrial iron uptake. Mitochondria consistently contained ∼3 nmol of iron/mg of protein (Table 1). To assess the assembly state of Yfh1, the soluble mitochondrial fraction (100,000 × g supernatant) was analyzed by size exclusion chromatography followed by Western blotting. Monomer was the only species detected in WT (Fig. 1C). Similarly, monomer was the predominant species detected in WT*, although lower levels of trimer and larger oligomers were also present in this strain (Fig. 1D). In contrast, trimer was the predominant form present in Y73A*, whereas monomer could not be detected (Fig. 1E). Equivalent levels of monomer and trimer were observed in T118A/V120A* (Fig. 1F).
TABLE 1.
Mitochondrial iron uptake during different growth conditions
The iron concentration was determined as described under “Experimental Procedures.” PMS, post mitochondrial supernatant; Medium, post-culture medium after cells were harvested by centrifugation. —, not determined.
|
Strain
|
YPGal 30 °C
|
YPGal 30 °C + 100 μm Fe
|
YPGal 30 °C + 500 μm Fe
|
S.D.
|
S.D. 37 °C + 100 μm iron
|
|||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Mitochondria | Mitochondria | PMS | Medium | Mitochondria | PMS | Medium | 30 °C Mitochondria | 37 °C Mitochondria | Mitochondria | PMS | Medium | |
| nmol/mg | nmol/mg | nmol/mg | μm | nmol/mg | nmol/mg | μm | nmol/mg | nmol/mg | μnmol/mg | nmol/mg | μm | |
| WT | 3.6 ± 0.4 (7)a | 5.2 ± 0.2 (6)a | 3.5 ± 0.0 (2) | 93 (1) | 15.7 ± 2.0 (6)a | 13.7 ± 3.0 (4) | 422 (2) | 21.6 ± 1.3 (3)a | 24.7 ± 3.3 (5)a | 2.5 ± 0.4 (8)a | 79.3 ± 5.1 (8) | 48 (2) |
| WT* | 3.0 ± 0.7 (7) | 5.3 ± 1.0 (6) | 3.5 ± 0.0 (2) | 88 (1) | 15.5 ± 2.3 (6) | 13.8 ± 4.2 (4) | 425 (2) | 21.9 ± 1.2 (3) | 15.9 ± 1.8 (3) | 2.5 ± 0.0 (2) | 88.2 ± 1.6 (2) | 44 (1) |
| Y73A* | 2.8 ± 0.4 (7) | 4.4 ± 0.3 (6) | 3.6 ± 0.0 (2) | 90 (1) | 14.9 ± 0.4 (6) | 12.3 ± 1.9 (4) | 424 (2) | 19.5 ± 0.8 (3) | 21.1 ± 1.0 (5) | 3.0 ± 0.3 (8) | 60.5 ± 19 (8) | 46 (2) |
| T118A/V120A* | — | — | — | — | — | — | — | — | — | 2.6 ± 0.1 (2) | 96.5 ± 5.3 (2) | 45 (1) |
The number of measurements obtained from at least two independent mitochondrial isolations, except for T118A,V120A*, for which only one isolation was analyzed.
The Y73A Mutation Does Not Alter the Biochemical Properties of Yfh1—To assess possible effects of the Y73A mutation on the functions of Yfh1, we used discontinuous assays that measure iron uptake, iron oxidation, or iron delivery independent of the kinetics of Yfh1 assembly (2, 49) (supplemental Table S1). At the end of a 60-min iron-binding reaction at a saturating iron to Yfh1 ratio of 75:1, there was a modest reduction in iron uptake (∼80% of wild type). In addition, oxygen consumption measurements revealed a final Fe2+/O2 ratio intermediate between wild type Yfh1 and buffer, indicating that the Y73A mutation decreased but did not abolish the ferroxidase activity of Yfh1, which can be explained by the proximity of residue 73 to the ferroxidation site of Yfh1 (9). On the other hand, Fe2+ delivery assays did not reveal any significant differences compared with wild type. We concluded that the main effect of the Y73A mutation on Yfh1 was to stabilize a conformation that promoted assembly of monomer to trimer. This, coupled with the virtual absence of monomeric Yfh1 in the Y73A* strain, provided a system to assess the functional role of the Yfh1 trimer in vivo.
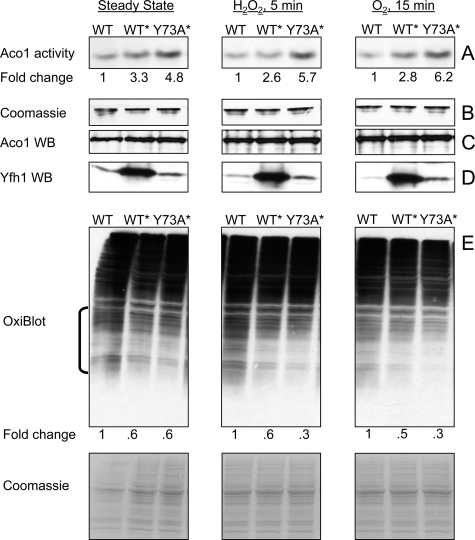
Stabilization of Yfh1 Trimer Enhances Mitochondrial Aconitase Activity and Limits Protein Carbonylation—Mitochondria were isolated from the WT, WT*, and Y73A* strains under the conditions described above and used to determine aconitase activity, as a measure of the mitochondrial anti-oxidant capacity and iron chaperone function of frataxin (6, 50), and protein carbonylation, as a measure of oxidative damage. In freshly isolated mitochondria, aconitase (Aco1) activity was consistently ∼3- and ∼5-fold higher in WT* and Y73A*, respectively, as compared with WT (Fig. 2A) and remained higher upon exposure of isolated mitochondria to hydrogen peroxide or atmospheric oxygen (Fig. 2A). Because the three strains contained equal levels of Aco1 protein as determined by Western blotting (Fig. 2C), the higher levels of Aco1 activity exhibited by WT* and Y73A* reflected the presence of a larger fraction of the active, [4Fe-4S]2+ form of the protein. In the same mitochondria preparations used to measure Aco1 activity, protein carbonyls were slightly but consistently lower in WT* and Y73A* relative to WT, both at steady state and upon exposure to hydrogen peroxide or atmospheric oxygen (Fig. 2E). These results did not correlate with the mitochondrial iron content, which was ∼3 nmol/mg protein in all three strains (Table 1), nor with the absolute levels of Yfh1 (Fig. 2D), which were similar between WT and Y73A* and at least 5-fold higher in WT* mitochondria (∼0.04, 0.06, and 0.25 nmol of Yfh1/mg of mitochondrial protein, as determined by densitometry relative to a known amount of purified Yfh1). On the other hand, the increasing levels of Aco1 activity (Y73A* > WT* > WT) (Fig. 2A) and the decreasing levels of protein carbonylation (Y73A* < WT* < WT) (Fig. 2E) appeared to correlate with the levels of trimer present in each of the three strains at steady state (Y73A* > WT* > WT) (Fig. 1, C–E). It appeared that under steady state conditions insufficient to drive oligomerization of wild type Yfh1 (as seen in WT), the constitutive presence of trimer enhanced detoxification of labile iron, resulting in higher proportions of active Aco1 and lower levels of protein carbonylation in Y73A* and WT* compared with WT. Monomeric frataxin could have contributed to this effect in WT*.
FIGURE 2.
Constitutive presence of trimer enhances aconitase activity and limits protein oxidation. Yeast cultures were grown and mitochondria isolated as in Fig. 1 (C–F). A, in-gel aconitase activity was measured in untreated mitochondria (Steady State) or upon exposure of mitochondria to 100 μm H2O2 or atmospheric oxygen. A fixed amount of total protein (150 μg) was analyzed in each case. Fold change shows the average increase in aconitase activity in WT* and Y73A* relative to WT as determined by ImageQuant 5.0 densitometric analysis of three independent experiments, one shown. The relative densitometric intensity of the aconitase band for WT, WT*, and Y73A* was (means ± standard deviation): Steady State, 11.0 ± 7.0, 36.0 ± 15.7 (p = 0.032), 53.0 ± 13.6 (p = 0.004); H2O2 5 min, 10.8 ± 8.5, 28.0 ± 17.7 (p = 0.101), 61.2 ± 17.4 (p = 0.005); O2 15 min, 10.0 ± 5.0, 28.0 ± 6.3 (p = 0.009), 62.0 ± 11.0 (p = 0.0009). In all cases, the p value was determined versus WT by the t test for equal variances. B, after aconitase activity was measured, the same gel was stained with Coomassie Blue to verify equal protein loading. C and D, aliquots of the mitochondrial preparations analyzed above (20 and 100 μg of total protein for Aco1 and Yfh1 detection, respectively) were analyzed by SDS-PAGE and Western blotting (WB) with specific antisera. E, aliquots of the same mitochondrial preparations (100 μg of total protein) were analyzed with OxiBlot kit to detect carbonylated proteins. ImageQuant 5.0 densitometric analysis was performed on the region denoted by the bracket, after which the membrane was stained with Coomassie Blue to verify equal protein loading (only a portion of the stained membrane is shown). Fold change represents the average decrease in protein carbonylation in WT* and Y73A* relative to WT as determined from three independent experiments, one shown. The relative densitometric intensity of the membrane region analyzed for WT, WT*, and Y73A* was (mean ± standard deviation): Steady State, 45.2 ± 2.1, 27.8 ± 1.6 (p = 0.0002), 27.1 ± 3.4 (p = 0.0007); H2O2 5 min: 51.2 ± 8.0, 31.7 ± 7.7 (p = 0.020), 17.0 ± 2.5 (p = 0.001); O2 15 min, 54.7 ± 6.2, 27.3 ± 5.6 (p = 0.002), 18.1 ± 6.0 (p = 0.0009). The p value was determined as above.
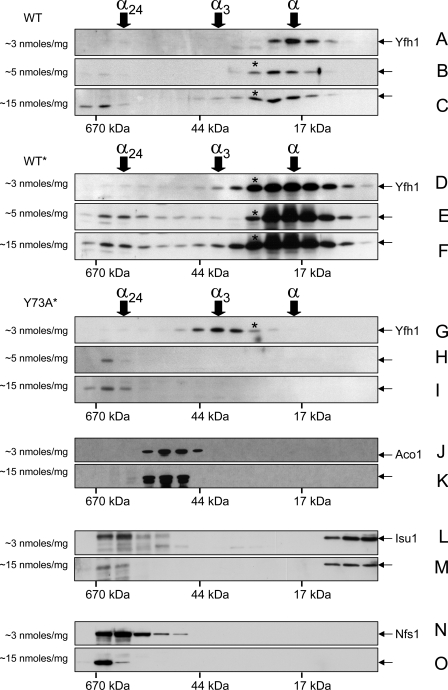
Yfh1 Oligomerizes in Response to Increments in Mitochondrial Iron Uptake—To assess Yfh1 assembly during limited increases in mitochondrial iron uptake, WT, WT*, and Y73A* cultures were preconditioned in YPGal for ∼12 h, diluted into fresh YPGal supplemented with 100 or 500 μm iron, and harvested after ∼20 h. The strains grew at the same rate, and their cytoplasmic and mitochondrial iron levels increased in parallel upon iron supplementation, although most added iron remained in the culture medium (Table 1). This result was consistent with repression of the high affinity iron transport system during culture preconditioning, which limited cellular iron uptake during the subsequent iron supplementation. Accordingly, increments in mitochondrial iron content were relatively small, from ∼3 to ∼5 and 15 nmol/mg of protein (Table 1). Nevertheless, they were associated with significant changes in the Yfh1 distribution in all three strains. In WT, when mitochondrial iron levels increased from 3 to 5 nmol/mg protein, monomer was shifted to a slightly larger species intermediate in size between monomer and trimer (hereafter denoted α*) (Fig. 3, A versus B); larger levels of α* and lower but discrete levels of trimer as well as species ≥24-mer were detected when mitochondrial iron levels increased further to ∼15 nmol/mg protein (Fig. 3C). In WT*, the α* and trimer already present at the initial mitochondrial iron concentration of ∼3 nmol/mg protein (Fig. 3D) were converted to larger oligomers when iron levels increased to ∼5 nmol/mg protein (Fig. 3E); additional formation of α*, trimer, and larger species occurred when mitochondrial iron levels increased to ∼15 nmol/mg protein (Fig. 3F). In Y73A*, trimer and α* were fully converted to species ≥24-mer when mitochondrial iron concentration increased from ∼3 to either ∼5 or 15 nmol/mg of protein (Fig. 3, H and I). Under these conditions, the three strains contained equal levels of Aco1 protein, but Aco1 activity was consistently higher in Y73A* before or after exposure of isolated mitochondria to hydrogen peroxide or atmospheric oxygen (not shown). Thus, the behavior of Yfh1 was consistent with iron-dependent assembly (36, 37), with a faster assembly rate in Y73A* that correlated with a higher proportion of active Aco1.
FIGURE 3.
Iron-dependent assembly of Yfh1 analyzed by size exclusion chromatography. Yeast cultures were started from freshly streaked glycerol stocks, preconditioned in YPGal at 30 °C for ∼12 h, and further grown in YPGal without iron supplementation (A, D, G, J, L, and N) or in the presence of 100 μm (B, E, and H) or 500 μm (C, F, I, K, M, and O) FeCl3 for ∼20 h at 30 °C. The mitochondria were isolated and soluble mitochondrial fractions (5 mg of total protein) analyzed by size exclusion chromatography as in Fig. 1 (C and D). The membranes analyzed in A and C were probed sequentially with antibodies against Yfh1, Nfs1, and Isu1. The membranes analyzed in G and I were probed sequentially with antibodies against Yfh1 and Aco1. A, D, and G are the same as C, D, and E in Fig. 1. The average iron concentrations measured in the mitochondrial preparations analyzed by size exclusion chromatography are shown on the left side of each panel (see also Table 1).
When mitochondrial iron content increased from ∼3 to 15 nmol/mg protein, the molecular mass distributions of Aco1 and monomeric Isu1 did not change (Fig. 3, J versus K and L versus M). Conversely, Nfs1 and the low levels of Isu1 initially present in ∼80–670-kDa fractions shifted to the highest of these fractions (Fig. 3, L versus M and N versus O), possibly reflecting interactions of the Nfs1-Isu1 complex with iron-loaded Yfh1 oligomers, as observed for bacterial frataxin in vitro (8). Thus, the broad molecular mass shift exhibited by Yfh1, from ∼17 to >670 kDa, appeared to be largely independent of interactions with its main protein partners.
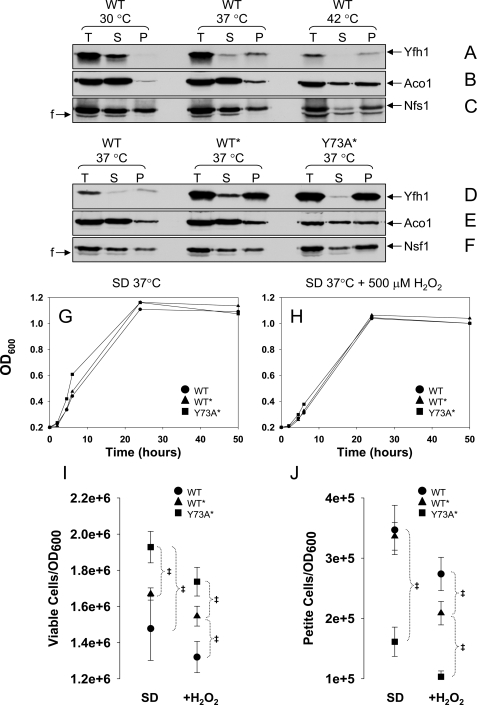
Yfh1 Forms Fast Sedimenting Complexes in Response to Heat Stress—Next we asked whether the conformational adjustment required for assembly of wild type Yfh1 could be induced independent of an increase in mitochondrial iron uptake. Heat shock enhances yeast oxygen consumption, causing an increase in the generation of reactive oxygen species in mitochondria (51, 52). To test possible effects of heat shock on Yfh1 assembly, the WT strain was grown in synthetic defined medium with glucose as the carbon source (SD) at 30, 37, or 42 °C for 24 h, after which cells were harvested and mitochondria isolated. Only very low levels of Yfh1 were detected in the 100,000 × g soluble mitochondrial fraction from cells grown at 37 °C (not shown). Even when mitochondria were lysed by sonication and separated into supernatant and pellet by table top centrifugation (20,000 × g for 20 min), wild type Yfh1 was mostly soluble in mitochondria from cells grown at 30 °C but largely insoluble in mitochondria from cells grown at 37 or 42 °C (Fig. 4A). In the same mitochondrial preparations, there was also a temperature-dependent increase in the relative amounts of Aco1 and Nfs1 detected in the insoluble fraction, although not as pronounced as for Yfh1 (Fig. 4, B and C). On the other hand, similar mitochondrial iron levels were present in WT mitochondria at 30 and 37 °C (Table 1). It appeared that wild type Yfh1 formed quickly sedimenting complexes in a temperature-dependent manner.
FIGURE 4.
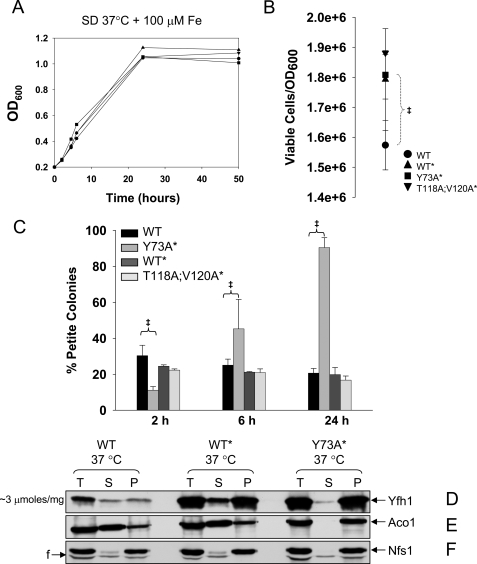
Yfh1 solubility decreases in response to heat stress. A–F, yeast cultures were started from freshly streaked glycerol stocks, preconditioned in SD at 30 °C for ∼12 h, diluted into fresh SD, and grown for ∼24 h at different temperatures, after which the mitochondria were isolated. Total mitochondrial lysates (T; 4.5 mg/ml total protein) were prepared by sonication and separated into supernatant (S) and pellet (P). Equal amounts (by volume) of the T, S, and P fractions were analyzed by SDS/PAGE and Western blotting. In D only, to enable simultaneous detection of Yfh1 in the WT, WT*, and Y73A* strains despite their different levels of Yfh1 expression, the amounts of T, S, and P fractions analyzed for WT* and Y73A* were 5- and 2.5-fold lower, respectively, than those analyzed for WT. The membrane analyzed in B–D was probed sequentially with antibodies against Aco1 and Nfs1, and the membrane analyzed in F and G was probed with antibodies against Aco1 and Nfs1. F, fragment of Nfs1 (65). G and H, yeast cultures were started from freshly streaked glycerol stocks, synchronized to late logarithmic phase in SD at 30 °C for ∼12 h, diluted into fresh SD in the absence or presence of 500μm H2O2, and shifted to 37 °C. The data points are the averages from two independent growth curves. I and J, at different times, aliquots of the cultures were diluted to A600 = 0.0002, equal volumes (150μl) were plated on YPD or YPG plates, and the colonies were counted after 5 days at 30 °C. I and J show the number of viable and nonrespiring cells, respectively, expressed per A600 = 1(1 A600 is ∼1–2 × 107 cells/ml). The data show the means ± standard deviation of four (WT and Y73A*) or two (WT* and T118A/V120A*) independent experiments, with two plates per experiment scored for each strain analyzed. ‡, p ≤ 0.02 as determined by the t test for equal variances.
The Rate of Frataxin Assembly Influences Heat Stress Tolerance—During growth in SD at 37 °C, WT* and Y73A* accumulated ∼20 and 10-fold higher levels of mature Yfh1, respectively, relative to WT (Fig. 4D), probably as a result of improved kinetics of Yfh1 precursor import and processing caused by induction of heat shock proteins (53). Despite the different protein levels, the ratio of insoluble versus soluble (P versus S) Yfh1 was similar between WT and WT* and slightly higher in Y73A* (Fig. 4D). The ratio of insoluble versus soluble Aco1 or Nfs1 was also slightly higher in Y73A* (Fig. 4, E and F). Moreover, the ratios of insoluble versus soluble Yfh1, Aco1, and Nfs1 present in Y73A* at 37 °C were similar to those present in WT at 42 °C (Fig. 4, B and C versus E and F). It appeared that Yfh1, Aco1, and Nfs1 became prone to aggregation in a temperature-dependent manner, as seen in WT, and that this behavior was enhanced in the Y73A* strain.
We analyzed whether Y73A* continued to exhibit a higher degree of stress tolerance relative to WT and WT* during growth in SD at 37 °C, as observed under less stressful conditions in YPGal at 30 °C. WT, WT*, and Y73A* grew at similar rates, reaching the stationary phase at ∼24 h (Fig. 4G). During growth in SD, entrance into the stationary phase requires that yeast cells be able to shift to respiration (diauxic shift) and maintain a high metabolic rate (54, 55). Under these conditions, Y73A* cultures contained a higher proportion of viable cells (i.e. stationary cells able to resume growth after transfer to rich medium) (Fig. 4I), and a smaller proportion of petite cells (i.e. cells that have lost respiratory capacity because of a loss of mitochondrial DNA integrity) (Fig. 4J). The same was true in the presence of 500 μm H2O2 (Fig. 4, H–J), which potentiates oxidative stress during heat shock (51). These results were obtained with two independent isolates for each strain, indicating that in Y73A* faster kinetics of Yfh1 assembly consistently enhanced yeast stress tolerance.
Rate-limiting Step in Frataxin Assembly Is Critical for Mitochondrial DNA Maintenance—We stressed our strains further by use of growth conditions that resulted in a rapid and large increase in mitochondrial iron influx. WT, WT*, and Y73A* cultures were synchronized at 37 °C for 12 h in SD medium, which only contained ∼0.2 μm iron and was not expected to repress the plasma membrane high affinity iron transport system (48). The cultures were then diluted into fresh SD medium supplemented with 100 μm FeCl3 and harvested after another 24 h, corresponding to entry in the stationary phase. In all strains, mitochondrial iron levels raised from ∼20 nmol to ∼3 μmol/mg protein, a >100-fold increase (Table 1). The strains exhibited similar growth rates (Fig. 5A), and overall similar cell viabilities at 24 h (Fig. 5B). After 2 h of growth in SD supplemented with iron, Y73A* accumulated less petite colonies as compared with WT and WT* (11% versus 30 and 24% respectively; Fig. 5C), in agreement with the results in the absence of iron supplementation. However, Y73A* cultures subsequently showed a striking phenotypic change characterized by the accumulation of a large proportion of petites, reaching 50% of the total viable cells after 6 h and nearly 100% after 24 h (Fig. 5C). Thus, the Y73A mutation was for the first time associated with a detrimental effect, primarily on mitochondrial DNA integrity. Although the ratio of insoluble versus soluble (P versus S) Yfh1 was similar in all strains (Fig. 5D), the fraction of insoluble Aco1 and Nfs1 was enhanced in Y73A* relative to WT and WT* (Fig. 5, E and F). This was especially evident in the case of Aco1, which was mostly soluble in WT and WT* and completely insoluble in Y73A* (Fig. 5E). Aco1 binding to mitochondrial DNA is required for mitochondrial DNA integrity (56), and therefore, sequestration or aggregation of Aco1 could have been sufficient to cause the dramatic accumulation of petite colonies in the Y73A* strain.
FIGURE 5.
Deregulated assembly of Yfh1 causes Aco1 sequestration and loss of mitochondrial DNA integrity. A, yeast cultures were performed as in Fig. 4G except that 100 μm FeCl3 was added during growth at 37 °C. The data points are the averages of two independent growth curves. B and C, equal numbers of cells from the cultures described above, after 24 h (B) or 2, 6, and 24 h (C) of growth, were plated and scored for viability (B) or respiratory capacity (C) as in Fig. 4 (I and J). The data show the means ± standard deviation of four (WT and Y73A*) or two (WT* and T118A/V120A*) independent experiments, with two plates per experiment scored for each strain analyzed. ‡, p ≤ 0.02. D–F, cultures were performed as above, and mitochondria were isolated. T, S, and P fractions were prepared and analyzed as in Fig. 4. In D only, the amounts of T, S, and P fractions analyzed for WT* and Y73A* were 5- and 2.5-fold lower, respectively, than those analyzed for WT. The membrane analyzed in B and C was probed sequentially with antibodies against Aco1 and Nfs1.
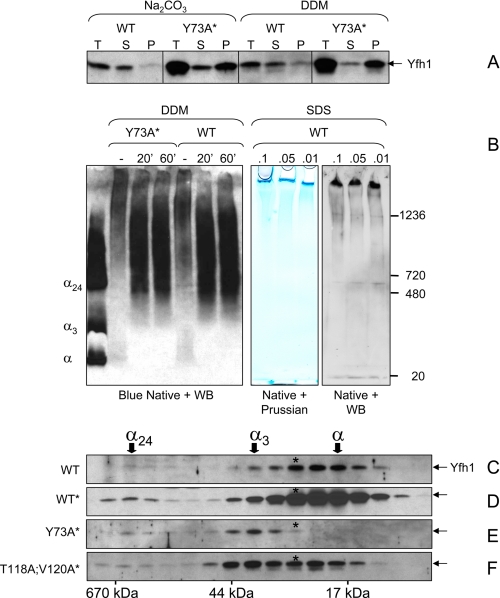
Polymerization of Wild Type Yfh1 Occurs via Stepwise Assembly—We attempted to characterize the insoluble Yfh1 species described above. WT and Y73A* mitochondria were broken by sonication and incubated with sodium carbonate or the non-ionic detergents dodecyl maltoside or Triton X-100, and mitochondrial lysates were clarified by tabletop centrifugation. Most Yfh1 present in WT mitochondria was solubilized upon treatment with either sodium carbonate or mild detergents, whereas the vast majority of Yfh1 present in Y73A* mitochondria remained in the sediment upon either treatment (Fig. 6A and not shown for Triton X-100). Blue native PAGE followed by Western blotting showed that the detergent-solubilized fraction of Yfh1 from either WT or Y73A* mitochondria was assembled into species ≥24-mer (Fig. 6B, left panel). Thus, in the case of WT, insoluble Yfh1 appeared to consist at least in part of homopolymers that were peripherally associated with mitochondrial membranes. Similar homopolymers were present in Y73A*, although this strain also contained a larger fraction of Yfh1 that was irreversibly aggregated. Native PAGE was further performed on WT or WT* mitochondria upon treatment with low concentrations of SDS followed by tabletop centrifugation. Analysis of duplicate gels by Western blotting or Prussian blue staining revealed high molecular mass Yfh1 complexes at the top of the gel that appeared to co-migrate with iron (Fig. 6B, right panel; not shown for WT*), probably corresponding to the polymers of iron-loaded frataxin particles previously observed in vitro and in vivo (57, 58). Attempts to affinity purify these complexes and analyze their composition failed because of their limited solubility.
FIGURE 6.
Polymerization of Yfh1 in response to heat stress. A, mitochondria were isolated under the same conditions used in Fig. 5 (A–C), and the total mitochondrial lysates (T) were extracted with 100 mm Na2CO3 or 1% dodecyl maltoside for 30 min on ice prior to separation into supernatant (S) and pellet (P). B, total mitochondrial lysates (4.5 mg of total protein/ml) were left untreated or extracted with 1% dodecyl maltoside for 20 or 60 min on ice. After centrifugation at 10,000 × g for 20 min at 4 °C, aliquots of the supernatants were analyzed by blue native PAGE and Western blotting (WB) with anti-Yfh1 antibody. Purified monomer, trimer, and 24-mer (α, α3, and α24) were used as molecular mass standards. In another experiment, total mitochondrial lysates were incubated in the presence of low concentrations of SDS followed by centrifugation at 5,000 × g for 20 min. Two identical sets of samples from the resulting supernatants were analyzed by native PAGE (3–12% Bis-Tris) on the same gel. One half of the gel was stained with Prussian blue for iron detection, and the second half was analyzed by Western blotting for Yfh1 detection. C and F, yeast cells were preconditioned in SD and further grown in 1,500 ml of SD in the presence of 100μm FeCl3 for ∼24 h at 37 °C. The mitochondria were isolated, and a soluble mitochondrial fraction (1.5 mg of total protein) was analyzed by size exclusion chromatography and Western blotting as described in the legend of Fig. 3 (C–F). Because of the very low levels of soluble Yfh1 present in mitochondria under the conditions used in these experiments, long exposures were required for Yfh1 detection resulting in high backgrounds.
To gain insight into the mechanism of Yfh1 polymerization, we analyzed by size exclusion chromatography the distribution of the residual Yfh1 present in the soluble mitochondrial fraction (i.e. after the insoluble polymers had been removed by ultracentrifugation at 100,000 × g). In WT and WT*, soluble Yfh1 consisted primarily of monomer and lower levels of α* and trimer (Fig. 6, C and D). This distribution was nearly identical to that observed when mitochondrial iron content was in the low nanomolar range (Fig. 3, C and F), indicating that stress-induced polymerization of Yfh1 occurred via the same α → α* → α3 transition observed in unstressed cells. Trimer was the predominant species present in the soluble fraction derived from Y73A* mitochondria (Fig. 6E), suggesting that the Y73A mutation had accelerated Yfh1 assembly until free iron had become limiting and/or the accumulation of Yfh1 aggregates had inhibited further trimer assembly.
DISCUSSION
Our results indicate that Yfh1 assembly in vivo is activated upon increments in mitochondrial iron uptake. Such increments may transiently increase the fraction of labile iron in the mitochondrial matrix, which may induce Yfh1 self-assembly as observed in vitro (36, 37). Given that the iron binding capacity of Yfh1 increases from 2 atoms/subunit in the monomer (59) to >50 atoms/subunit in Yfh1 oligomers (7), iron-dependent assembly probably serves to rapidly expand the mitochondrial iron buffering capacity independent of de novo Yfh1 protein synthesis. This idea is consistent with current knowledge that mitochondrial iron influx is proportional to iron availability in the culture medium (17) or the cytoplasm (60) and that mitochondrial iron efflux is mainly controlled by mitochondrial ISC synthesis (61, 62). Thus, rapid increases in cellular iron availability and mitochondrial iron uptake, such as those induced in this study, may temporarily exceed the existing rate of mitochondrial iron incorporation into ISC. Under these conditions, Yfh1 assembly may help to detoxify excess iron until iron uptake returns to basal levels or a new balance between iron uptake and iron efflux is established.
In all conditions leading to assembly, a species intermediate in size between monomer and trimer, denoted α*, consistently preceded or accompanied the appearance of trimer, suggesting that formation of α* is an obligate step for trimer formation. Moreover, during iron increments in either WT or WT* mitochondria, monomer was consistently present in excess relative to trimer, further suggesting that the α → α* → α3 transition is the rate-limiting step in the assembly reaction of wild type Yfh1. The α* intermediate could represent Yfh1 dimer or an assembly-competent form of the monomer. The possibility that Yfh1 monomer undergoes a conformational change to form trimer is supported by limited but significant differences in the structure of Yfh1 monomer before and after assembly into trimer. Both forms of the protein share the typical α/β sandwich fold of frataxin with two α helices (α2, α3) packed against a five-strand (β1-β5) β sheet (9, 63). However, in the monomer helix α2 has seven turns, whereas in the trimer the first two turns of helix α2 unwind, enabling interaction between the N terminus of one subunit and the β sheet of another (Fig. 1A). Helix α2 contains an extended acidic patch involved in iron binding as well as ferroxidase activity and iron core nucleation (34, 38, 42, 59). Iron binding to one or more sites on helix α2 may disrupt interactions needed to maintain the α-helical conformation, which may result in partial unwinding of helix α2 leading to oligomerization. In agreement with this idea, point mutations within or adjacent to the region of helix α2 that undergoes unwinding (D78A, H74A, or Y73A; Fig. 1A) enable Yfh1 to assemble in the presence of iron concentrations lower than those needed to activate assembly of the wild type monomer (this study).3 Similarly, the T118A/V120A mutations may stabilize interactions of the β-sheet of one subunit with the tip of the N-terminal region of another subunit, which may in turn lead to the unwinding of helix α2.
The importance of the rate-limiting α → α* → α3 transition was underscored by the dramatic loss of mitochondrial DNA integrity exhibited by Y73A* during heat shock coupled with high mitochondrial iron uptake, which appeared to result from Yfh1 hyperpolymerization leading to aconitase sequestration (56). In addition, mutations that expedite trimer assembly interfered with Yfh1 precursor processing (supplemental Fig. S1), probably by inducing a protein fold incompatible with efficient cleavage by mitochondrial processing peptidase, whose activity depends on substrate conformation (64). Therefore, the rate-limiting step in Yfh1 assembly responds both to structural requirements for efficient maturation of the protein precursor as well as the need to prevent Yfh1 aggregation during mitochondrial iron overload. The data suggest that careful pharmacological modulation of this step may be useful to enhance frataxin function in Friedreich ataxia or other conditions with altered mitochondrial iron balance or oxidative stress tolerance.
Supplementary Material
Acknowledgments
We thank A. L. Bulteau for anti-Aco1 antibody and H. Li for help in generating anti-Nfs1 and anti-Isu1 antibodies.
This work was supported, in whole or in part, by National Institutes of Health Grant AG15709. This work was also supported by the Friedreich Ataxia Research Alliance/Muscular Dystrophy Association. The costs of publication of this article were defrayed in part by the payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 U.S.C. Section 1734 solely to indicate this fact.
The on-line version of this article (available at http://www.jbc.org) contains supplemental Fig. S1 and Table S1.
Footnotes
The abbreviations used are: ISC, iron-sulfur cluster; GPD, glyceraldehyde-3-phosphate dehydrogenase; WT, wild type.
O. Gakh, D. Y. Smith IV, and G. Isaya, unpublished data.
References
- 1.Campuzano, V., Montermini, L., Moltò, M. D., Pianese, L., Cossée, M., Cavalcanti, F., Monros, E., Rodius, F., Duclos, F., Monticelli, A., Zara, F., Cañizares, J., Koutnikova, H., Bidichandani, S. I., Gellera, C., Brice, A., Trouillas, P., De Michele, G., Filla, A., De Frutos, R., Palau, F., Patel, P. I., Di Donato, S., Mandel, J. L., Cocozza, S., Koenig, M., and Pandolfo, M. (1996) Science 271 1423–1427 [DOI] [PubMed] [Google Scholar]
- 2.Park, S., Gakh, O., O'Neill, H. A., Mangravita, A., Nichol, H., Ferreira, G. C., and Isaya, G. (2003) J. Biol. Chem. 278 31340–31351 [DOI] [PubMed] [Google Scholar]
- 3.Yoon, T., and Cowan, J. A. (2004) J. Biol. Chem. 279 25943–25946 [DOI] [PubMed] [Google Scholar]
- 4.Yoon, T., and Cowan, J. A. (2003) J. Am. Chem. Soc. 125 6078–6084 [DOI] [PubMed] [Google Scholar]
- 5.Kondapalli, K. C., Kok, N. M., Dancis, A., and Stemmler, T. L. (2008) Biochemistry 47 6917–6927 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bulteau, A. L., O'Neill, H. A., Kennedy, M. C., Ikeda-Saito, M., Isaya, G., and Szweda, L. I. (2004) Science 305 242–245 [DOI] [PubMed] [Google Scholar]
- 7.Nichol, H., Gakh, O., O'Neill, H. A., Pickering, I. J., Isaya, G., and George, G. N. (2003) Biochemistry 42 5971–5976 [DOI] [PubMed] [Google Scholar]
- 8.Layer, G., Ollagnier-de Choudens, S., Sanakis, Y., and Fontecave, M. (2006) J. Biol. Chem. 281 16256–16263 [DOI] [PubMed] [Google Scholar]
- 9.Karlberg, T., Schagerlof, U., Gakh, O., Park, S., Ryde, U., Lindahl, M., Leath, K., Garman, E., Isaya, G., and Al-Karadaghi, S. (2006) Structure 14 1535–1546 [DOI] [PubMed] [Google Scholar]
- 10.Schagerlöf, U., Elmlund, H., Gakh, O., Nordlund, G., Hebert, H., Lindahl, M., Isaya, G., and Al-Karadaghi, S. (2008) Biochemistry 47 4948–4954 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Campanella, A., Isaya, G., O'Neill, H., Santambrogio, P., Cozzi, A., Arosio, P., and Levi, S. (2004) Hum. Mol. Genet. 13 2279–2288 [DOI] [PubMed] [Google Scholar]
- 12.Zanella, I., Derosas, M., Corrado, M., Cocco, E., Cavadini, P., Biasiotto, G., Poli, M., Verardi, R., and Arosio, P. (2008) Biochim. Biophys. Acta 1782 90–98 [DOI] [PubMed] [Google Scholar]
- 13.Muhlenhoff, U., Richhardt, N., Ristow, M., Kispal, G., and Lill, R. (2002) Hum. Mol. Genet. 11 2025–2036 [DOI] [PubMed] [Google Scholar]
- 14.Duby, G., Foury, F., Ramazzotti, A., Herrmann, J., and Lutz, T. (2002) Hum. Mol. Genet. 11 2635–2643 [DOI] [PubMed] [Google Scholar]
- 15.Seznec, H., Simon, D., Monassier, L., Criqui-Filipe, P., Gansmuller, A., Rustin, P., Koenig, M., and Puccio, H. (2004) Hum. Mol. Genet. 13 1017–1024 [DOI] [PubMed] [Google Scholar]
- 16.Martelli, A., Wattenhofer-Donze, M., Schmucker, S., Bouvet, S., Reutenauer, L., and Puccio, H. (2007) Hum. Mol. Genet. 16 2651–2658 [DOI] [PubMed] [Google Scholar]
- 17.Foury, F. (1999) FEBS Lett. 456 281–284 [DOI] [PubMed] [Google Scholar]
- 18.Zhang, Y., Lyver, E. R., Knight, S. A., Lesuisse, E., and Dancis, A. (2005) J. Biol. Chem. 280 19794–19807 [DOI] [PubMed] [Google Scholar]
- 19.Schoenfeld, R. A., Napoli, E., Wong, A., Zhan, S., Morin, D., Buckpitt, A. R., Taroni, F., Lonnerdal, B., Ristow, M., Puccio, H., and Cortopassi, G. A. (2005) Hum. Mol. Genet. 14 3787–3799 [DOI] [PubMed] [Google Scholar]
- 20.Babcock, M., de Silva, D., Oaks, R., Davis-Kaplan, S., Jiralerspong, S., Montermini, L., Pandolfo, M., and Kaplan, J. (1997) Science 276 1709–1712 [DOI] [PubMed] [Google Scholar]
- 21.Foury, F., and Talibi, D. (2001) J. Biol. Chem. 276 7762–7768 [DOI] [PubMed] [Google Scholar]
- 22.Wilson, R. B., and Roof, D. M. (1997) Nat. Genet. 16 352–357 [DOI] [PubMed] [Google Scholar]
- 23.Ristow, M., Pfister, M. F., Yee, A. J., Schubert, M., Michael, L., Zhang, C. Y., Ueki, K., Michael, M. D., 2nd, Lowell, B. B., and Kahn, C. R. (2000) Proc. Natl. Acad. Sci. U. S. A. 97 12239–12243 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Emond, M., Lepage, G., Vanasse, M., and Pandolfo, M. (2000) Neurology 55 1752–1753 [DOI] [PubMed] [Google Scholar]
- 25.Schultz, J. B., Dehmer, T., Schols, L., Mende, H., Hardt, C., Vorgerd, M., Burk, K., Matson, W., Dichgans, J., Beal, M. F., and Bogdanov, M. B. (2000) Neurology 55 1719–1721 [DOI] [PubMed] [Google Scholar]
- 26.Wong, A., Yang, J., Cavadini, P., Gellera, C., Lonnerdal, B., Taroni, F., and Cortopassi, G. (1999) Hum. Mol. Genet. 8 425–430 [DOI] [PubMed] [Google Scholar]
- 27.Hart, P. E., Lodi, R., Rajagopalan, B., Bradley, J. L., Crilley, J. G., Turner, C., Blamire, A. M., Manners, D., Styles, P., Schapira, A. H., and Cooper, J. M. (2005) Arch. Neurol. 62 621–626 [DOI] [PubMed] [Google Scholar]
- 28.Shoichet, S. A., Baumer, A. T., Stamenkovic, D., Sauer, H., Pfeiffer, A. F., Kahn, C. R., Muller-Wieland, D., Richter, C., and Ristow, M. (2002) Hum. Mol. Genet. 11 815–821 [DOI] [PubMed] [Google Scholar]
- 29.Al-Mahdawi, S., Pinto, R. M., Varshney, D., Lawrence, L., Lowrie, M. B., Hughes, S., Webster, Z., Blake, J., Cooper, J. M., King, R., and Pook, M. A. (2006) Genomics 88 580–590 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ristow, M., Mulder, H., Pomplun, D., Schulz, T. J., Muller-Schmehl, K., Krause, A., Fex, M., Puccio, H., Muller, J., Isken, F., Spranger, J., Muller-Wieland, D., Magnuson, M. A., Mohlig, M., Koenig, M., and Pfeiffer, A. F. (2003) J. Clin. Investig. 112 527–534 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Anderson, P. R., Kirby, K., Orr, W. C., Hilliker, A. J., and Phillips, J. P. (2008) Proc. Natl. Acad. Sci. U. S. A. 105 611–616 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Vazquez-Manrique, R. P., Gonzalez-Cabo, P., Ros, S., Aziz, H., Baylis, H. A., and Palau, F. (2006) FASEB J. 20 172–174 [DOI] [PubMed] [Google Scholar]
- 33.Karthikeyan, G., Santos, J. H., Graziewicz, M. A., Copeland, W. C., Isaya, G., van Houten, B., and Resnick, M. A. (2003) Hum. Mol. Genet. 12 3331–3342 [DOI] [PubMed] [Google Scholar]
- 34.Gakh, O., Park, S., Liu, G., Macomber, L., Imlay, J. A., Ferreira, G. C., and Isaya, G. (2006) Hum. Mol. Genet. 15 467–479 [DOI] [PubMed] [Google Scholar]
- 35.Jeffery, C. J. (1999) Trends Biochem. Sci. 24 8–11 [DOI] [PubMed] [Google Scholar]
- 36.Adamec, J., Rusnak, F., Owen, W. G., Naylor, S., Benson, L. M., Gacy, A. M., and Isaya, G. (2000) Am. J. Hum. Genet. 67 549–562 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Wang, T., and Craig, E. A. (2008) J. Biol. Chem. 283 12674–12679 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Aloria, K., Schilke, B., Andrew, A., and Craig, E. A. (2004) EMBO Rep. 5 1096–1101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Bou-Abdallah, F., Adinolfi, S., Pastore, A., Laue, T. M., and Chasteen, N. D. (2004) J. Mol. Biol. 341 605–615 [DOI] [PubMed] [Google Scholar]
- 40.O'Neill, H. A., Gakh, O., and Isaya, G. (2005) J. Mol. Biol. 345 433–439 [DOI] [PubMed] [Google Scholar]
- 41.Adinolfi, S., Trifuoggi, M., Politou, A. S., Martin, S., and Pastore, A. (2002) Hum. Mol. Genet. 11 1865–1877 [DOI] [PubMed] [Google Scholar]
- 42.Foury, F., Pastore, A., and Trincal, M. (2007) EMBO Rep. 8 194–199 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Mumberg, D., Muller, R., and Funk, M. (1995) Gene (Amst.) 156 119–122 [DOI] [PubMed] [Google Scholar]
- 44.Tong, W. H., and Rouault, T. A. (2006) Cell Metab. 3 199–210 [DOI] [PubMed] [Google Scholar]
- 45.Molik, S., Lill, R., and Muhlenhoff, U. (2007) Methods Cell Biol. 80 261–280 [DOI] [PubMed] [Google Scholar]
- 46.Branda, S. S., Cavadini, P., Adamec, J., Kalousek, F., Taroni, F., and Isaya, G. (1999) J. Biol. Chem. 274 22763–22769 [DOI] [PubMed] [Google Scholar]
- 47.Velagapudi, V. R., Wittmann, C., Lengauer, T., Talwar, P., and Heinzle, E. (2006) Process Biochem. 41 2170–2179 [Google Scholar]
- 48.Felice, M. R., De Domenico, I., Li, L., Ward, D. M., Bartok, B., Musci, G., and Kaplan, J. (2005) J. Biol. Chem. 280 22181–22190 [DOI] [PubMed] [Google Scholar]
- 49.Park, S., Gakh, O., Mooney, S. M., and Isaya, G. (2002) J. Biol. Chem. 277 38589–38595 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Gardner, P. R. (2002) Methods Enzymol. 349 9–23 [DOI] [PubMed] [Google Scholar]
- 51.Christos Moraitis, B. P. G. C. (2004) Yeast 21 313–323 [DOI] [PubMed] [Google Scholar]
- 52.Sugiyama, K., Izawa, S., and Inoue, Y. (2000) J. Biol. Chem. 275 15535–15540 [DOI] [PubMed] [Google Scholar]
- 53.Neupert, W., and Herrmann, J. M. (2007) Annu. Rev. Biochem. 76 723–749 [DOI] [PubMed] [Google Scholar]
- 54.Herman, P. K. (2002) Curr. Opin. Microbiol. 5 602–607 [DOI] [PubMed] [Google Scholar]
- 55.Kaeberlein, M., McVey, M., and Guarente, L. (2001) Sci. Aging Knowledge Environ. 2001, pe1. [DOI] [PubMed]
- 56.Chen, X. J., Wang, X., and Butow, R. A. (2007) Proc. Natl. Acad. Sci. U. S. A. 104 13738–13743 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Cavadini, P., O'Neill, H. A., Benada, O., and Isaya, G. (2002) Hum. Mol. Genet. 33 217–227 [DOI] [PubMed] [Google Scholar]
- 58.Popescu, B. F., Pickering, I. J., George, G. N., and Nichol, H. (2007) J. Inorg. Biochem. 101 957–966 [DOI] [PubMed] [Google Scholar]
- 59.Cook, J. D., Bencze, K. Z., Jankovic, A. D., Crater, A. K., Busch, C. N., Bradley, P. B., Stemmler, A. J., Spaller, M. R., and Stemmler, T. L. (2006) Biochemistry 45 7767–7777 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Chen, O. S., and Kaplan, J. (2000) J. Biol. Chem. 275 7626–7632 [DOI] [PubMed] [Google Scholar]
- 61.Chen, O. S., Hemenway, S., and Kaplan, J. (2002) Proc. Natl. Acad. Sci. U. S. A. 99 12321–12326 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Chen, O. S., Crisp, R. J., Valachovic, M., Bard, M., Winge, D. R., and Kaplan, J. (2004) J. Biol. Chem. 279 29513–29518 [DOI] [PubMed] [Google Scholar]
- 63.He, Y., Alam, S. L., Proteasa, S. V., Zhang, Y., Lesuisse, E., Dancis, A., and Stemmler, T. L. (2004) Biochemistry 43 16254–16262 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Taylor, A. B., Smith, B. S., Kitada, S., Kojima, K., Miyaura, H., Otwinowski, Z., Ito, A., and Deisenhofer, J. (2001) Structure 9 615–625 [DOI] [PubMed] [Google Scholar]
- 65.Adam, A. C., Bornhovd, C., Prokisch, H., Neupert, W., and Hell, K. (2006) EMBO J. 25 174–183 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.