Abstract
Raf kinases are essential for regulating cell proliferation, survival, and tumorigenesis. However, the mechanisms by which Raf is activated are still incompletely understood. Phosphorylation plays a critical role in Raf activation in response to mitogens. The present study characterizes phosphorylation of Ser338, a crucial event for Raf-1 activation. Here we report that mutation of Lys375 to Met diminishes phosphorylation of Ser338 on both wild type Raf-1 in cells treated with epidermal growth factor (EGF) or 12-O-tetradecanoylphorbol-13-acetate (TPA) and a constitutively active mutant in which Tyr340/Tyr341 are replaced by 2 aspartic acids, a conserved substitution present in natural B-Raf. The loss of Ser338 phosphorylation in these Raf mutants is not engendered by a mutation-induced conformational change, inasmuch as mutation of another site (Ser471 to Ala) in the activation segment also abolishes Ser338 phosphorylation, whereas both the kinase-dead mutants of Raf-1 are phosphorylated well by active Pak1. Furthermore, our data demonstrate that EGF-stimulated phosphorylation of Ser338 is inhibited by Sorafenib, a Raf kinase inhibitor, but not by the MEK inhibitor U0126. Interestingly, a kinase-dead mutation and Sorafenib also markedly reduce phosphorylation of Ser445 on B-Raf, a site equivalent to Raf-1 Ser338. Finally, our data reveal that Ser338 is phosphorylated on inactive Raf-1 by an active mutant of Raf-1 when they are dimerized in cells and that artificial dimerization of Raf-1 causes Ser338 phosphorylation, accompanied by activation of ERK1/2. Altogether, our data suggest that Ser338 on Raf-1 is autophosphorylated in response to mitogens.
The Raf family of serine/threonine protein kinases consists of three isoforms, A-Raf, B-Raf, and Raf-1 (C-Raf), all of which serve as immediate downstream effectors of Ras (1). In response to extracellular stimuli, activated Ras-GTP relays signals to Raf, which in turn evokes a serine/threonine phosphorylation cascade through sequential phosphorylation of MEK1/2,3 ERK1/2, and further downstream effectors to elicit a variety of cellular responses. Raf-1 is ubiquitously expressed in mammalian cells, whereas A-Raf and B-Raf exhibit more tissue-specific expression profiles (2). Likewise, knocking out individual genes for these Raf isoforms creates different phenotypes (3). Deletion of the Raf-1 gene in an inbred background causes death of mice in midgestation, and its knock-out in an outbred strain results in general growth retardation, developmental abnormalities, and death shortly after birth (4–6). Similarly, deletion of the B-Raf gene is embryonic lethal due to defects in neuroepithelial differentiation and maturation and in the maintenance of endothelial cells (7). Elimination of A-Raf causes intestinal and neurological defects, but the pups are born alive (8).
Activating ras mutations are found in 30% of all human cancers (9). In some cancers such as breast cancer, even if Ras is not mutated, amplification of growth factor receptors (e.g. epidermal growth factor receptor), or alterations in other positive or negative regulators of Ras also lead to higher than normal levels of Ras activity (10). As a result, the Raf/MEK/ERK pathway is constitutively activated, leading to increased cell proliferation and invasion. The B-Raf mutants have been frequently identified, whereas oncogenic mutations for Raf-1 and A-Raf have not yet been reported in human cancers (11, 12).
Activation of Raf-1 has served as a framework for the other two isoforms, but still remains incompletely understood. Raf-1 activity is tightly controlled by both intramolecular and extramolecular interactions. In the resting state, Ser259 on Raf-1 is phosphorylated and bound by 14-3-3, which possibly promotes the intramolecular interaction between the amino-terminal regulatory domain and the carboxyl-terminal kinase domain, preventing the spontaneous association of Raf-1 with Ras-GTP and thus keeping it inactive (13, 14). When cells are provoked by mitogenic ligands, Ras-GTP binds to a site aminoterminal proximate to the 14-3-3 binding site, thereby destabilizing the interaction between 14-3-3 and the phosphoylated Ser259 on Raf-1 and allowing its dephosphorylation by protein phosphatase 2A (15–18). Unlike phosphophorylation of Ser259, 14-3-3 binding to phosphophorylation of Ser621 on Raf-1 is absolutely required for Raf kinase activity (19–22). Removal of 14-3-3 from Raf-1 greatly inhibits Raf kinase activity, and this inhibition can be relieved by adding bacterially expressed recombinant 14-3-3 (22). These studies strongly suggest that 14-3-3 acts primarily to maintain the active conformation of Raf-1.
In response to mitogens, phosphorylation of both Ser338 and Tyr341 occurs in the junction region between the aminoterminal negative domain and the carboxyl-terminal kinase domain (23–29). The Src family has been implicated in the phosphorylation of Tyr340 (in insect Sf9 cells) and Tyr341 (in mammalian cells) (23, 24, 27–29). Interestingly, phosphorylation of Tyr341 is barely detectable unless v-Src is coexpressed with Raf-1. Nevertheless, like phosphorylation of Ser338, substitution of Tyr341 with alanine or phenylalanine abolishes Raf-1 activation, whereas its mutation to aspartic acid increases Raf kinase activity. Phosphorylation of Ser338 and Tyr341 dissociates the intramolecular interaction (30–33) and the extramolecular interaction of Raf-1 with RKIP (34).
The p21-activated protein kinases (Pak), mammalian homologues of yeast Ste20-like serine/threonine protein kinases, are activated by direct binding of Rac and Cdc42 GTPases (35). Members of the Pak family appear to mediate cross-talk between Ras/Raf/MEK and Rac/Cdc42 (36–40). However, it is controversial as to if Pak is a bona fide Ser338 kinase mediating growth factors to activate Raf-1 (28). We have demonstrated that activation of Cdc42/Rac/Pak by the microtubule depolymerizing drug nocodazole results in Ser338 phosphorylation and activation of Raf-1, independent of Ras (41). In contrast, the Cdc42/Rac/Pak module does not mediate EGF-induced activation of Raf-1 (41). Therefore, a true kinase phosphorylating Ser338 during the course of growth factor-induced Raf-1 activation still remains elusive.
A number of studies have shown that phosphorylation of Ser471, Thr491, Ser494, Ser497, and Ser499 (42–44) occurs during Raf-1 activation. These phosphorylation sites are located between subdomains VII and VIII (activation segment) in the Raf-1 catalytic domain. The biological importance of their phosphorylation is supported by the isolation at a high frequency from human cancers of a B-Raf oncogene containing activating mutations of V599D or V599E (12, 45–55). The valine exists in all three Raf isoforms, next to the phosphorylation site in the activation segment (e.g. Thr598/Val599 in B-Raf and Thr491/Val492 in Raf-1) (44, 56). Chong et al. (44) have shown that phosphorylation of Thr491 and Ser494 is involved in the activation of Raf-1. In addition, Ser497 and Ser499 are phosphorylated by protein kinase C (42, 43). Mutation of these double sites partially blocks Raf activation by phorbol 12,13-dibutyrate (TPA), but it does not affect growth factor-induced activation (57, 58).
To search for an additional kinase that phosphorylates Ser338 in response to growth factors, we first asked if this site is autophosphorylated. Thus, we mutated Lys375 to methionine (K375M) at the ATP binding site of the catalytic domain of Raf-1 and examined the Ser338 phosphorylation state. To our surprise, the effect of EGF/TPA on Ser338 phosphorylation was severely impaired by this kinase-dead mutation. The inhibition might not be attributed to conformational changes, as the ability of this mutant to be phosphorylated by Pak1 was as great as that of the wild type Raf-1. A second mutant converting Ser471 to Ala in the activation segment behaved the same as the K375M mutant. Interestingly, when the equivalent lysine in B-Raf (Lys482) or Raf-1 Y340D/Y341D (Lys375), a constitutively active mutant of Raf-1, was changed to methionine, Ser445 and Ser338 phosphorylation was also diminished. Furthermore, phosphorylation of these two sites was inhibited by Sarofenib, a Raf inhibitor. Therefore, our results suggest that Ser338 on Raf-1 is autophosphorylated in response to EGF or TPA. In addition, our results demonstrate that Raf-1 dimerization plays a role in phosphorylation of Ser338.
EXPERIMENTAL PROCEDURES
Materials—TPA was purchased from Sigma. EGF and U0126 were from Calbiochem. The anti-Myc monoclonal antibody 9E10.2 was purchased from American Tissue Culture Collection. The anti-GST monoclonal antibody, anti-Raf E10 monoclonal antibody, and anti-Raf C12 polyclonal antibody were from Santa Cruz. Phospho-MEK, phospho-ERK1/2, and their non-phosphorylated forms were from Cell Signaling Technology. Glutathione (GSH)-Sepharose was purchased from GE Healthcare. The rat monoclonal antibody against phospho-Ser338 Raf-1 was from Millipore. The anti-EE epitope antibody was a gift from Dr. A. Makkinje. Sorafenib was purchased from LC Laboratories. The dimerization drug AP1510 was provided by ARIAD Pharmaceuticals, Inc.
Plasmid DNA and Site-directed Mutagenesis—Plasmid DNA for Myc-Raf, GST-Raf, FKBP12-Myc-Raf, Pak1, MEK1, and Ras were used as previously described (41, 62). The mutation of K482M on B-Raf and mutation of S621A on Raf-1 was made using the QuikChange kit manufactured by Stratagene. Other Raf mutants tagged by the FLAG epitope were constructed in pCMV5.
Transfections, Immunoprecipitation, and Western Blot—Human embryonic kidney 293 cells (HEK293T) were cultured in Dulbecco's modified Eagle's medium supplemented with 10% fetal bovine serum. Transfection of plasmid DNA was carried out using the calcium phosphate precipitation method (41). The Pak1 siRNA sequence was selected using the Dhamacon program (sense, UGUCUGAUGAGGAGAUCUUdTdT). Double-stranded RNA oligonucleotides were synthesized by Dhamacon and transfected with Oligofectamine according to the protocol provided by the manufacturer (Invitrogen). Forty-eight h post-transfection, cells were serum-starved in Dulbecco's modified Eagle's medium containing 0.1% fetal bovine serum for 16–20 h, treated with TPA for 15 min or EGF for 10 min, and then lysed in a lysis buffer (25 mm Tris-HCl, pH 7.8, 100 mm NaCl, 1 mm EDTA, 1 mm EGTA, 1 mm Na3VO4, and 25 mm β-glycerolphosphate, 1 mm dithiothreitol, 1% Nonidet P-40 and protease inhibitors) (41). Cell debris was removed by centrifugation at 14,000 × g at 4 °C for 15 min and protein concentrations in cell lysates were measured using Bio-Rad Protein Assay kit.
For Western blotting, cell extracts were separated by 8% SDS-PAGE and electrophoretically transferred to Immobilon (Millipore). The membranes were blocked, incubated with specific antibodies, and then with a horseradish peroxidase-conjugated second antibody. Immunoreactive bands were visualized by the enhanced chemiluminescence (ECL) detection system.
For immunoprecipitation, proteins of interests were first normalized by Western blotting. Cell lysates containing equal amounts of specific proteins were then incubated with protein A/G-agarose (Santa Cruz Biotechnology) pre-absorbed with specific antibodies at 4 °C overnight. The precipitates were washed, as described (41). For purification of recombinant GST-Raf, the lysates were incubated with GSH beads and washed as immunoprecipitation.
Raf-1 Kinase Assay—Raf-1 kinase activity was measured by a coupled enzyme assay in which bacterially expressed recombinant GST-MEK1 and a kinase-dead mutant of ERK2 were sequentially added to the Raf preparation in the presence of [γ-32P]ATP (100 μm, 4000 cpm/pmol), as described previously (41). The reaction was stopped by the addition of a SDS-PAGE sample buffer, and the labeled mixture was resolved by 8% SDS-PAGE, transferred to Immobilon, and visualized by autoradiography. The radiolabeled ERK2 bands were excised and quantified by liquid scintillation counting.
RESULTS
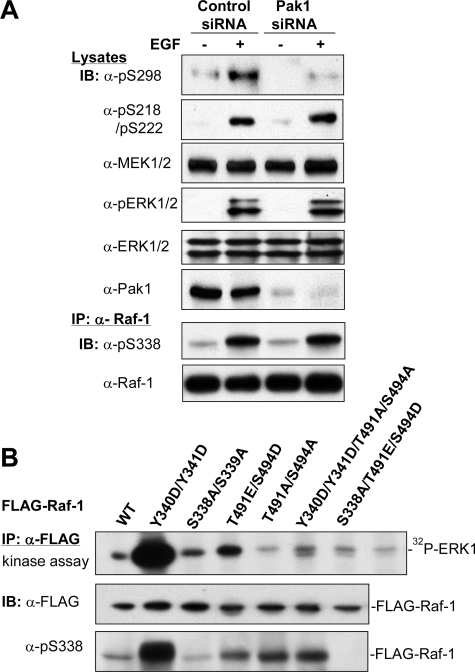
EGF-induced Activation of Raf-1 Is Independent of Pak1—To ascertain if Pak kinases are the bona fide enzymes that phosphorylate Ser338 in response to mitogens, we transfected siRNA for Pak1 into HEK293T cells and then examined the effects of EGF on phosphorylation of Raf-1, MEK1/2, and ERK1/2. The results revealed that phosphorylation of Raf at Ser338 and MEK at Ser218/Ser222, sites phosphorylated by active Raf, as well as ERK phosphorylation, were not affected. In contrast, phosphorylation of MEK at Ser298, a specific site phosphorylated by Pak1 (36, 60), was diminished by Pak1 siRNA (Fig. 1A). Next, we made a series of mutations to mimic or disable phosphorylation in two sets of sites, Ser338 and Tyr341, the negative region between the amino-terminal regulatory and carboxyl kinase domains, and Ser471/Ser494, in the activation segment, and tested the impacts of phosphorylation of these sites on Raf kinase activity. We transfected these mutants into HEK293T cells and carried out an in vitro kinase assay. As shown in Fig. 1B, mutation of Y340D/Y341D gave rise to the greatest activation of Raf-1, concurrently with a dramatic increase in Ser338 phosphorylation. Second, the T491E/S494D mutant also significantly increased kinase activity. In contrast, mutation of S338A brought down the kinase activity of the Raf-1 T491E/S494D mutant to the basal level. Likewise, the T491A/S494A mutation also greatly diminished the activity of Raf Y340D/Y341D, regardless of residual phosphorylation of Ser338. These results suggest that phosphorylation of both groups are necessary for Raf activation.
FIGURE 1.
Effects of Ser338 phosphorylation on Raf-1 activation. A, knockdown of Pak1 by siRNA suppresses EGF-stimulated phosphorylation of MEK1 at Ser298, but not that of MEK at Ser218/Ser222 or Raf-1 at Ser338. The siRNA for human Pak1 or scrambled siRNA was transfected into HEK293T cells. Forty eight h later, the cells were starved in 0.1% serum overnight prior to treatment with EGF (10 ng/ml) for 10 min. Cell lysates were prepared and blotted (IB) with antibodies against phospho-Ser298 MEK1, phospho-Ser218/Ser222 MEK1/2, total MEK1/2, phospho-ERK1/2, total ERK1/2, and Pak1, respectively. The lower part shows immunoblots for phospho-Ser338 and Raf-1 (E10) after immunoprecipitation (IP) of endogenous Raf-1. B, the effect of Ser338 phosphorylation on Raf kinase activity. Raf variants tagged with the FLAG epitope, as indicated, were transfected into HEK293T cells. After 2 days, recombinant Raf was immunoprecipitated with the FLAG antibody and assayed for the in vitro kinase activity by sequential incubation with bacterially expressed and purified GST-MEK1 and kinase-dead ERK1. The autoradiogram shows 32P-incorporated ERK1. Aliquots of Raf-1 IP were probed with anti-phospho-Ser338 and anti-FLAG antibodies, respectively.
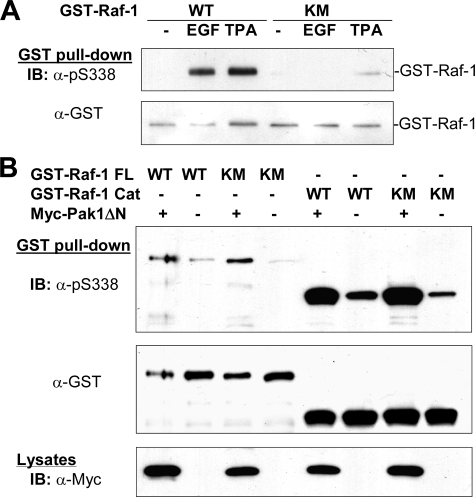
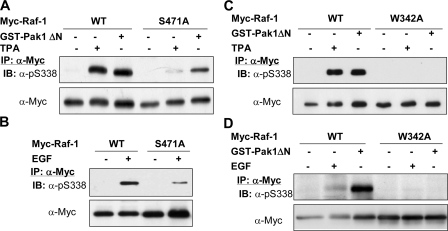
Different Effects of Kinase-dead Mutations of Raf-1 on EGF-induced and Pak1-mediated Phosphoyrlation of Ser338—To determine whether Ser338 phosphorylation requires an intact kinase domain structure, we transfected a kinase-dead mutant, K375M, and wild type Raf expressed as a GST fusion protein into HEK293T cells and examined Ser338 phosphorylation in response to EGF or TPA. As shown in Fig. 2A, Ser338 phosphorylation was markedly impeded by the Lys375 to Met mutation. The failure of Ser338 to be phosphorylated might be accounted for by several reasons; for example, the mutation may disturb the tertiary structure of Raf-1, so as to interfere with its phosphorylation by an upstream kinase; second, the site is possibly phosphorylated by a downstream kinase; and third, it is autophosphorylated. To test these possibilities, we first transfected GST-Raf-1, full-length or carboxyl kinase domain, into HEK293T cells with or without an active Pak mutant, and examined Ser338 phosphorylation. As shown in Fig. 2B, both the wild type and K375M mutant of Raf-1 were equally phosphorylated at Ser338 by Pak1. In parallel, we used another mutant, S471A, a site further downstream from Ser338 as compared with K375M mutant. The mutation of Ser471 to Ala has been shown to abolish Raf kinase activity (61). Our result revealed that phosphorylation of Ser338 on this mutant was also impeded in response to TPA and EGF (Fig. 3, A and B). Conversely, when an active mutant of Pak1 was cotransfected with the S471A mutant (Fig. 3A), Ser338 was phosphorylated as potently as it was on the wild type Raf-1. Finally, our data showed that mutation of Trp342 to Ala completely abrogated Ser338 phosphorylation induced by the active Pak1, TPA, and EGF (Fig. 3, C and D), suggesting that this residue is required for Ser338 phosphorylation under both scenarios.
FIGURE 2.
Requirement of Raf kinase activity for EGF-induced Ser338 phosphorylation. A, the mutation of Lys375 to Met (KM) on Raf-1 abolishes EGF-induced phosphorylation of Ser338. HEK293T cells transiently expressing wild type (WT) and GST-K375M mutant of Raf-1 were treated with or without EGF (10 ng/ml, 10 min) or TPA (1 μm, 15 min). Recombinant Raf-1 was purified with GSH beads and blotted with anti-phospho-Ser338 and anti-GST antibodies. B, phosphorylation of Ser338 on kinase-dead mutants of Raf-1 by Pak1. Full-length Raf-1, wild type (GST-Raf-1 FL WT), and K375M mutant (GST-Raf-1 FL KM), and catalytic kinase domain, wild type (GST-Raf-1 Cat WT), and K375M (GST-Raf-1 Cat KM), were expressed in HEK293T cells with the active kinase domain of Pak1 (Myc-Pak1 ΔN) or an empty plasmid, as indicated. Immunoblotting was carried out using antibodies against phospho-Ser338 and GST after GSH pull-down. Expression of Pak1 was monitored by anti-Myc blot.
FIGURE 3.
Different phosphorylation of Ser338 on Raf-1 S471A and W342A mutants. A and B, full-length Myc-Raf-1 variants, wild type (WT), and S471A mutant were transfected with or without the catalytic domain of Pak1 (GST-Pak1 ΔN) into HEK293T cells. Two days later, the cells without cotransfection of Pak1 were stimulated with TPA (1 μm, 15 min) (A) or EGF (10 ng/ml, 10 min) (B). Recombinant Raf-1 was immunoprecipitated (IP) with anti-Myc antibody and blotted (IB) with anti-phospho-Ser338 and anti-Myc antibodies. C and D, full-length Myc-Raf-1 variants, wild type and W342A, were expressed with or without the active kinase mutant of Pak1 (GST-Pak1 ΔN). The cells were treated the same as described in A and B. Recombinant Raf-1 was immunoprecipitated and blotted with antibodies, as indicated.
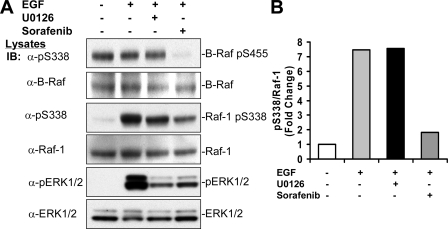
In addition, we tested if Ser338 phosphorylation was attributed to downstream kinases. Thus, HEK293T cells were starved and treated with or without the MEK inhibitor U0126 or the Raf inhibitor Sarofenib prior to incubation with EGF. Using anti-phospho-Ser338 antibody, we could detect phosphorylation of both Raf-1 and B-Raf (Fig. 4A). Although phosphorylation of Ser338 was stimulated by EGF, phosphorylation of Ser445, an equivalent site, was constitutive, consistent with previous findings (27, 32). Interestingly, we found that whereas U0126 did not affect EGF-stimulated Ser338 phosphorylation on Raf-1 or constitutive phosphorylation of Ser445 on B-Raf, Sarofenib greatly impaired phosphorylation of both (Fig. 4). These results suggest that Ser338 or Ser445 are not phosphorylated by MEK1/2 or ERK1/2, downstream of Raf, but might be autophosphorylated.
FIGURE 4.
Ser338 phosphorylation was suppressed by the Raf inhibitor Sarofenib, but not by the MEK inhibitor U0126. A, HEK293T cells were pretreated with or without Sarofenib (25μm, 12 h), or U0126 (10μm) for 30 min, and then treated with EGF (10 ng/ml) for 10 min. Cell lysates were blotted with antibodies against phospho-Ser338, total Raf-1 (C12), total B-Raf, phospho-ERK1/2, and total ERK1/2, respectively. B, the degree of suppression of Raf Ser338 phosphorylation was assessed by scan densitometry. Anti-phospho-Ser338 signals in A were scanned and divided by the scan units of total Raf-1 signals and the ratio was expressed as -fold of untreated cells. IB, immunoblot.
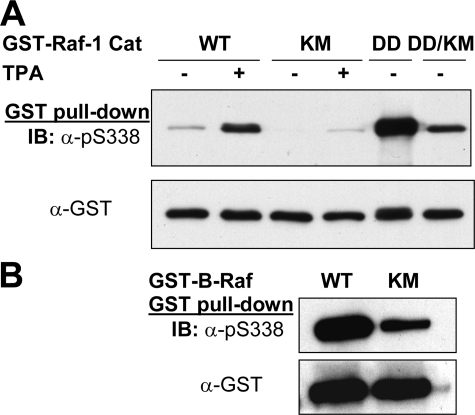
To further test if Ser338 on Raf-1 and Ser445 on B-Raf are autophosphorylated, we mutated Lys375 to Met on Raf-1 Y340D/Y341D and Lys482 to Met on B-Raf and transfected them into HEK293T cells. As shown in Fig. 5A, when the catalytic kinase domain containing the Y340D/Y341D substitution was expressed, Ser338 was highly phosphorylated in the absence of any treatment, which was severalfold greater than that of wild type Raf-1 C activated by TPA. It was then significantly reduced by the Lys375 to Met mutation. Of note, the phosphorylation signal was still equal to that of wild type Raf-1C induced by TPA. As compared with wild type B-Raf, Ser445 phosphorylation of the K482M mutant was markedly reduced (Fig. 5B). The residual signal in both B-Raf K482M and Raf-1C Y340D/Y341D/K375M is probably attributed to cross-reaction of the phosphoantibody to the 2 acidic residues or mutation renders them a better substrate for other upstream kinase such as Pak.
FIGURE 5.
Phosphorylation of Ser338 on Raf-1 Y340D/Y341D or Ser445 on B-Raf is diminished by the kinase-dead KM mutation. A, Raf-1. Catalytic domain variants of Raf-1, wild type (WT), K375M mutant (KM), Y340D/Y341D mutant (DD), and Y340D/Y341D-K375M mutant (DD/KM), were transfected into HEK293T cells, purified by GSH affinity purification after treating the cells with or without TPA, and blotted with antibodies against phospho-Ser338 and GST. B, B-Raf. Lysine 482, an essential residue for ATP binding, was mutated to methionine. The mutant was expressed as a GST fusion protein in HEK293T cells, as opposed to wild type B-Raf, purified and blotted (IB) as described in A.
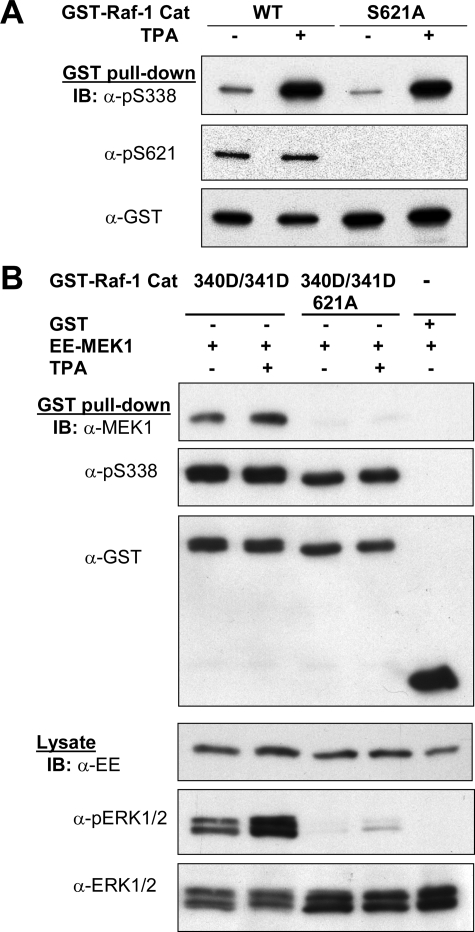
The Effect of Ser621 Phosphorylation on Ser338 Phosphorylation—The binding of 14-3-3 to phosphophorylated Ser621 on Raf-1 is essential for Raf kinase activity toward MEK1. To test if Ser621 exerts a similar effect on Ser338 phosphorylation, we converted Ser621 to Ala in the context of the Raf kinase domain, wild type or Y340D/Y341D mutant, and transfected them into HEK293T cells. As shown in Fig. 6A, TPA stimulated Ser338 phosphorylation equally well on both wild type and S621A mutant Raf-1. When Raf-1C Y340D/Y341D was cotransfected with MEK1, it was almost fully active in terms of its ability to stimulate phosphorylation of ERK1/2 and bind to MEK1, whereas Raf-1C Y340D/Y341D/S621A was inactive and unable to bind MEK1 (Fig. 6B). Interestingly, phosphorylation of Ser338 was nearly the same in both Raf-1C at variants. Thus, the result distinguishes the structural requirement for MEK kinase activity and Ser338 kinase activity and also indicates that Ser338 phosphorylation is not sufficient for Raf activation.
FIGURE 6.
Phosphorylation of Ser338 is essential but not sufficient for Raf-1 activation. A, HEK293T cells were transfected with the catalytic domain variants of Raf-1, wild type (WT), and S621A mutant and then stimulated with or without TPA. Recombinant Raf-1 purified by GSH pull-down was blotted with anti-phospho-Ser338, phospho-Ser621, and GST, respectively. B, Raf-1 catalytic domain variants, wild type, Y340D/Y341D, or Y340D/Y341D/S621A, were transfected together with EE-tagged MEK1, purified, as described in A, and sequentially blotted (IB) with anti-EE, phospho-Ser338, and GST antibodies. Cell lysates were blotted with antibodies, as indicated.
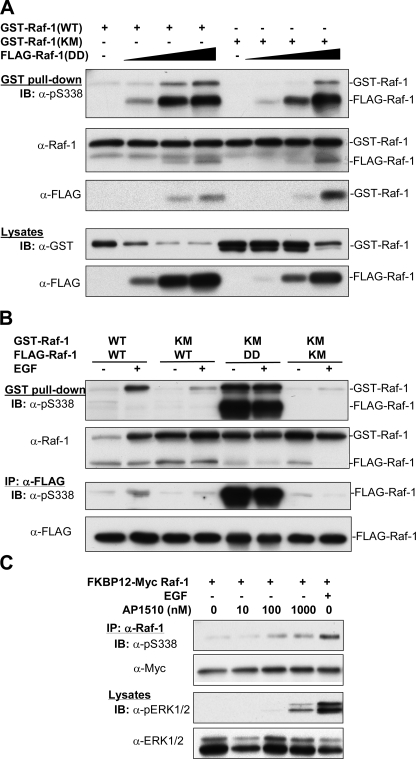
The Effect of Raf-1 Dimerization on Ser338 Phosphorylation—We and others have previously shown that Raf-1 exist as homodimer or heterodimer with B-Raf that promotes their activation (62, 63). We then evaluated if the dimerization played a role in the regulation of Ser338 phosphorylation. In this regard, we cotransfected GST-Raf-1, wild type, and the K375M mutant with FLAG-Raf-1 Y340D/Y341D at different doses and purified Raf complexes by GSH-Sepharose beads. Fig. 7A shows that phosphorylation of Ser338 on both Raf variants increased as more Raf Y340D/Y341D bound, despite that the K375M mutant showed less phosphorylation than the wild type. Next, we coexpressed GST-Raf-1, wild type (WT), or K375M mutant with FLAG-Raf-1, WT, K375M, or the Y340D/Y341D mutant and sequentially purified GST-Raf-1 and FLAG-Raf-1 after treatment of the cells with or without EGF. Our results showed different degrees of Ser338 phosphorylation of GST-Raf-1, depending on the combination of FLAG-Raf-1 (Fig. 7B). The greatest phosphorylation was observed on GST-Raf-1 when it was coexpressed with wild type FLAG-Raf-1 in cells treated with EGF or on the K375M GST-Raf-1 coexpressed with the FLAG-Raf-1 Y340D/Y341D mutant. Then the phosphorylation decreased on GST-Raf-1 K375M when cotransfected with wild type and the K375M FLAG-Raf-1. Again, we observed the dimerization of these two versions of Raf-1. When the copurified FLAG-Raf was assayed on Ser338 phosphorylation, most were barely detectible except that high amounts of phosphorylation were found on the FLAG-Raf Y340D/Y341D mutant. Obviously, this difference is due to the fact that a small portion of FLAG-Raf-1 was pulled down by GST-Raf-1 and that the specific activity of Ser338 phosphorylation on the DD mutant was much greater than that of the wild type Raf-1. When Ser338 phosphorylation was examined on FLAG immunoprecipitates with anti-FLAG antibody, we observed that Ser338 phosphorylation on wild type Raf was suppressed by coexpression of the kinase-dead mutant (lower panel of Fig. 7B).
FIGURE 7.
Effects of Raf-1 dimerization on Ser338 phosphorylation. A, plasmids encoding GST-Raf-1, wild type (WT), or kinase-dead mutant (K375M)(5 μg) was transfected alone or together with the plasmids encoding FLAG-Raf Y340D/Y341D (DD) at different doses (0.5, 1, and 2.5 μg) into HEK293T cells. Two days later, cell extracts were blotted with anti-GST and FLAG antibodies, respectively. The amounts of cell extracts were adjusted according to GST signals (lower panel) for purification of GST-Raf by GSH beads. Dimerized Raf proteins were blotted with anti-phospho-Ser338, Raf-1 (C12), and FLAG antibodies, respectively. B, GST-Raf-1 variants, wild type (WT), or K375M (KM), were coexpressed with FLAG-tagged Raf-1 variants, WT, KM, or DD, into HEK293T cells. The recombinant GST-Raf and FLAG-Raf were purified by GSH beads and anti-FLAG immunoprecipitation (IP), respectively, after starvation of the cells followed by incubation with EGF, as described in the legend to Fig. 2. The dimerization of Raf were examined by Western blot with antibodies, as indicated. C, FKBP12-Myc-Raf-1 was transiently expressed in HEK293T cells for 2 days. The cells were starved overnight and treated with different doses of AP1510, as indicated, for 1 h, or EGF (10 ng/ml) for 10 min. Raf was immunoprecipitated and blotted with anti-phospho-Ser338 and Myc antibodies. Crude extracts were blotted with anti-phospho-ERK1/2 and total ERK1/2 antibodies.
Finally, we transfected FKBP12-Myc-Raf-1 into HEK293T cells and incubated the cells with the dimerization drug AP1510 that induces the dimerization of FKBP12. Fig. 7C shows that Ser338 phosphorylation was progressively enhanced, as doses of the drug were increased. In keeping with this, ERK phosphorylation was also increased, although the effect of the drug was not as potent as that of EGF. Collectively, these results suggest that dimerization stimulates Ser338 phosphorylation on Raf-1.
DISCUSSION
In the present study, we have shown that mutations of Lys375, essential for ATP binding site, to Met, and Ser471 to Ala in the activation segment both abolish Ser338 phosphorylation of Raf-1 in response to EGF or TPA, whereas these mutants are still phosphorylated well by Pak1. Likewise, mutation of Lys482 to Met on B-Raf and Lys375 to Met on the Raf-1 Y340D/Y341D mutant diminishes phosphorylation of Ser445 and Ser338, respectively. The phosphorylation is also suppressed by treating the cells with the Raf inhibitor Sarofenib. Thus, these results suggest that Ser338 is autophosphorylated during the course of Raf-1 activation induced by mitogens such as EGF and TPA, whereas Ser445 is autophosphorylated in B-Raf to maintain its higher basal activity. Our results show that the phosphorylation of Ser621, the carboxyl 14-3-3 binding site, is necessary for MEK kinase activity of Raf-1, but dispensable for Ser338 phosphorylation. Finally, our data indicate that dimerization of Raf-1 plays a role in Ser338 phosphorylation. Therefore, the present study provides novel insights into the mechanism of Ser338 phosphorylation of Raf-1.
Phosphorylation has been known to play an important role in Raf activation after Ras-GTP binding. In the past, tremendous efforts have been made to determine the phosphorylation sites and to identify the responsible kinases. The crucial phosphorylation sites for Raf activation in response to growth factors include Ser338/Tyr341 and Thr491/Ser494 and Ser471 (64). The kinases responsible for their phosphorylation still remain elusive.
Pak family has been shown to regulate the ERK pathway via phosphorylating MEK1 at Ser298 (36, 60, 65, 66) and Raf-1 at Ser338 (36–40). Although some of previous studies have demonstrated that Pak phosphorylation of MEK1 Ser298 promotes the ability of Raf-1 to phosphorylate and activate MEK1/2 (36, 60), others do not observe a significant effect of Ser298 phosphorylation on MEK1 activation by growth factors when Pak1 expression is silenced or Ser298 is mutated to Ala (65, 67). Consistent with these latter findings, the present study shows that knockdown of Pak1 by siRNA does not affect the ability of EGF to stimulate MEK phosphorylation at Ser218/Ser222 and ERK activation, whereas it diminishes phosphorylation of MEK1 at Ser298, the site phosphorylated by Pak, under the same condition. One possible explanation is that although suppression of Pak1 expression may reduce MEK1 activation, it does not affect MEK2 (59). The latter may be sufficient to offset the effect of reduced MEK1 activation.
Our studies and those of others (36–40) have shown that Pak1/2 can phosphorylate Ser338 under some circumstances, but they do not participate in growth factor-invoked Ser338 phosphorylation and activation of Raf-1. To test if this site is autophosphorylated in response to growth factor, we created two kinase-dead mutants (K375M and S471A) of Raf-1 and assessed phosphorylation of Ser338. Our results showed that the mutation abolished the phosphorylation induced by EGF and TPA. This could be accounted for by several reasons. Using the specific MEK inhibitor U0126, we ruled out that it is phosphorylated by MEK or ERK downstream of Raf-1. This is also not due to disturbance of the tertiary structure of Raf-1 caused by the mutation, as the mutants are phosphorylated well by Pak1. One could argue that overexpressed Pak might be promiscuous and it is not sensitive enough to discern the structural difference between wild type and mutant Raf-1. This argument is not supported by our observation that the W342A mutation blocks Ser338 phosphorylation by Pak1. Interestingly, Ser445 on B-Raf, a site equivalent to Raf-1 Ser338, has been shown to be constitutively phosphorylated (27, 32). In the present study, we mutated Lys482 on B-Raf to Met, equivalent to the Raf-1 K375M mutation and found that the mutation markedly impaired Ser445 phosphorylation. The same result was also observed with the mutation of Lys375 to Met on Raf-1 Y340D/Y341D. Most strikingly, phosphorylation of Ser338 and Ser445 on endogenous Raf-1 and B-Raf, respectively, was greatly suppressed by the Raf inhibitor Sarofenib. Therefore, after excluding kinases downstream of Raf-1, our data suggest that Ser338 is autophosphorylated.
If Ser338 is autophosphorylated when Raf-1 is activated by mitogens such as EGF and TPA, the next question is how this is initiated. To address this, we tested a series of mutations and found that Ser338 phosphorylation appears to be independent of phosphorylation of Thr491/Ser494 and Ser621. Zhu et al. (61) have identified a new phosphorylation site, Ser471, responsive to EGF. When this site is converted to Ala or Asp, Raf-1 loses its activity, which is compensated by mutation of T491E/S494D. This finding suggests that phosphorylation of Ser471 configures an active conformation, instead of creating an electrostatic effect of negative charge. It is possible to test if phosphorylation of Ser471 initiates Ser338 phosphorylation given that the Ser471 kinase has been identified. Another possible mechanism is that Ser338 phosphorylation is primed by phosphorylation of Tyr341. Mason et al. (27) have described that when Asp447/Asp448 on B-Raf are replaced by two tyrosines, the level of Ser445 phosphorylation drops dramatically and it is restored by EGF, an event that is accompanied by changes in B-Raf kinase activity. Similarly to natural B-Raf, Ser338 is highly phosphorylated in the Raf Y340D/Y341D mutant. Taken together, these results suggest that phosphorylation of Ser338 may ensue after that of Tyr341 during the course of Raf-1 activation by EGF.
Several lines of evidence suggest that dimerization plays a role in the regulation of Raf kinase activity. First, we and others have previously shown that Raf-1 is invariably dimerized before and after EGF treatment and that artificial dimerization can induce Raf activation (62, 68). Second, Ras has been reported to form a dimmer, which is required for the activation of Raf-1 (69), and third, it has recently been documented that Ras-stimulated heterodimerization between B-Raf and Raf-1 results in an increase in the latter activity (63). Intriguingly, Garnett et al. (70) have shown that several B-Raf mutants isolated from human cancer cells do not display increased kinase activity per se, but they stimulate ERK activation and Raf-1 activity in vivo. Their results suggest that B-Raf via dimerizing with Raf-1 can stimulate phosphorylation and activation of the latter. In the same study, however, they do not find a significant increase in Raf-1 Ser338 phosphorylation when cotransfected with the B-Raf mutants. Our present study show that Ser338 phosphorylation of the kinase-dead K375M mutant of Raf-1 is greatly suppressed, but not completely abolished in the cells treated with mitogens. The residual phosphorylation might be attributed to dimerization of the K375M mutant with endogenous Raf-1. In addition, Ser338 phosphorylation is increased when the mutant is cotransfected with active Raf-1 Y340D/Y431D. Thus, the results argue that homodimerization at least in part plays a role in mitogen-stimulated Ser338 phosphorylation. They also suggest different mechanisms by which heterodimerization and homodimerization regulates Raf-1 activation and phosphorylation.
Of note, when two versions of Raf were coexpressed and purified, we only observed a small amount of Raf associated with each other, instead of a stoichiometric level. The low level of association may suggest that the dimerization is mediated by a scaffold protein that is a limiting factor or that the dissociation rate (Kd) is high. In keeping with this, we could not show a direct Raf-Raf interaction by the yeast two-hybrid system or using purified Raf proteins (data not shown). By comparing the degree of Ser338 phosphorylation and kinase activity of the Y340D/Y341D mutant with those of wild type Raf-1, we estimated that the majority of wild type Raf was not activated when cells were treated with EGF. This is in line with the general view that only 5–10% of Raf is activated by mitogens (64). It is not clear if only dimerized Raf can be activated by EGF or TPA. This question will be answered when the dimerization site is identified. Also, at this moment, we cannot exclude the possibility of cis-autophosphorylation.
Thus far, all existing data have documented that autophosphorylation of Raf exerts a negative effect on kinase activity (59). In light of our results obtained in vivo, we propose that autophosphorylation could have both negative and positive effects on Raf kinase activity, probably depending on other factors involved such as timing. For example, the conclusion on the negative effect of autophosphorylation is based on the result of in vitro incubation of Raf with ATP after isolation of activated Raf-1. We believe that autophosphorylation of Ser338 is an early event during Raf-1 activation. One perplex is that we could not observe changes in Ser338 phosphorylation when Raf-1 is incubated in vitro with ATP. An explanation is that it requires an additional activating factor that might have been missed during the purification or phosphorylation of other sites such as Tyr341 or Ser471. This will be our next task in elucidating the mechanism of Raf activation.
Acknowledgments
We thank Dr. Kun-Liang Guan for providing B-Raf and MEK constructs, Dr. A. Makkinje for anti-EE antibody, and Dr. Guri Tzivion for constructive suggestions. We also thank ARIAD Pharmaceuticals, Inc. for the gift of the dimerization drug AP1510.
This work was supported, in whole or in part, by National Institutes of Health Grants GM057959 and CA118918 (to Z. L.). The costs of publication of this article were defrayed in part by the payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 U.S.C. Section 1734 solely to indicate this fact.
Footnotes
The abbreviations used are: MEK, mitogen-activated protein kinase and extracellular-regulated kinase kinase; ERK, extracellular-regulated kinase; EGF, epidermal growth factor; GST, glutathione S-transferase; PAK, p21-activated protein kinase; TPA, 4β-12-O-tetradecanoylphorbol-13-acetate; HEK, human embryonic kidney; siRNA, small interfering RNA.
References
- 1.Kolch, W. (2000) Biochem. J. 351 289–305 [PMC free article] [PubMed] [Google Scholar]
- 2.Storm, S. M., Cleveland, J. L., and Rapp, U. R. (1990) Oncogene 5 345–351 [PubMed] [Google Scholar]
- 3.Hagemann, C., and Rapp, U. R. (1999) Exp. Cell Res. 253 34–46 [DOI] [PubMed] [Google Scholar]
- 4.Wojnowski, L., Stancato, L. F., Zimmer, A. M., Hahn, H., Beck, T. W., Larner, A. C., Rapp, U. R., and Zimmer, A. (1998) Mech. Dev. 76 141–149 [DOI] [PubMed] [Google Scholar]
- 5.Mikula, M., Schreiber, M., Husak, Z., Kucerova, L., Ruth, J., Wieser, R., Zatloukal, K., Beug, H., Wagner, E. F., and Baccarini, M. (2001) EMBO J. 20 1952–1962 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Huser, M., Luckett, J., Chiloeches, A., Mercer, K., Iwobi, M., Giblett, S., Sun, X. M., Brown, J., Marais, R., and Pritchard, C. (2001) EMBO J. 20 1940–1951 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Wojnowski, L., Zimmer, A. M., Beck, T. W., Hahn, H., Bernal, R., Rapp, U. R., and Zimmer, A. (1997) Nat. Genet. 16 293–297 [DOI] [PubMed] [Google Scholar]
- 8.Pritchard, C. A., Bolin, L., Slattery, R., Murray, R., and McMahon, M. (1996) Curr. Biol. 6 614–617 [DOI] [PubMed] [Google Scholar]
- 9.Barbacid, M. (1987) Annu. Rev. Biochem. 56 779–827 [DOI] [PubMed] [Google Scholar]
- 10.Downward, J. (2003) Nat. Rev. Cancer 3 11–22 [DOI] [PubMed] [Google Scholar]
- 11.Dhillon, A. S., Hagan, S., Rath, O., and Kolch, W. (2007) Oncogene 26 3279–3290 [DOI] [PubMed] [Google Scholar]
- 12.Michaloglou, C., Vredeveld, L. C., Mooi, W. J., and Peeper, D. S. (2008) Oncogene 27 877–895 [DOI] [PubMed] [Google Scholar]
- 13.Morrison, D. K., Heidecker, G., Rapp, U. R., and Copeland, T. D. (1993) J. Biol. Chem. 268 17309–17316 [PubMed] [Google Scholar]
- 14.Michaud, N. R., Fabian, J. R., Mathes, K. D., and Morrison, D. K. (1995) Mol. Cell. Biol. 15 3390–3397 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Rommel, C., Radziwill, G., Lovric, J., Noeldeke, J., Heinicke, T., Jones, D., Aitken, A., and Moelling, K. (1996) Oncogene 12 609–619 [PubMed] [Google Scholar]
- 16.Abraham, D., Podar, K., Pacher, M., Kubicek, M., Welzel, N., Hemmings, B. A., Dilworth, S. M., Mischak, H., Kolch, W., and Baccarini, M. (2000) J. Biol. Chem. 275 22300–22304 [DOI] [PubMed] [Google Scholar]
- 17.Kubicek, M., Pacher, M., Abraham, D., Podar, K., Eulitz, M., and Baccarini, M. (2002) J. Biol. Chem. 277 7913–7919 [DOI] [PubMed] [Google Scholar]
- 18.Dhillon, A. S., Meikle, S., Yazici, Z., Eulitz, M., and Kolch, W. (2002) EMBO J. 21 64–71 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Muslin, A. J., Tanner, J. W., Allen, P. M., and Shaw, A. S. (1996) Cell 84 889–897 [DOI] [PubMed] [Google Scholar]
- 20.Roy, S., McPherson, R. A., Apolloni, A., Yan, J., Lane, A., Clyde-Smith, J., and Hancock, J. F. (1998) Mol. Cell. Biol. 18 3947–3955 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Thorson, J. A., Yu, L. W., Hsu, A. L., Shih, N. Y., Graves, P. R., Tanner, J. W., Allen, P. M., Piwnica-Worms, H., and Shaw, A. S. (1998) Mol. Cell. Biol. 18 5229–5238 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Tzivion, G., Luo, Z., and Avruch, J. (1998) Nature 394 88–92 [DOI] [PubMed] [Google Scholar]
- 23.Fabian, J. R., Daar, I. O., and Morrison, D. K. (1993) Mol. Cell. Biol. 13 7170–7179 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Marais, R., Light, Y., Paterson, H. F., and Marshall, C. J. (1995) EMBO J. 14 3136–3145 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Diaz, B., Barnard, D., Filson, A., MacDonald, S., King, A., and Marshall, M. (1997) Mol. Cell. Biol. 17 4509–4516 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.King, A. J., Sun, H., Diaz, B., Barnard, D., Miao, W., Bagrodia, S., and Marshall, M. S. (1998) Nature 396 180–183 [DOI] [PubMed] [Google Scholar]
- 27.Mason, C. S., Springer, C. J., Cooper, R. G., Superti-Furga, G., Marshall, C. J., and Marais, R. (1999) EMBO J. 18 2137–2148 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Chiloeches, A., Mason, C. S., and Marais, R. (2001) Mol. Cell. Biol. 21 2423–2434 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.King, A. J., Wireman, R. S., Hamilton, M., and Marshall, M. S. (2001) FEBS Lett. 497 6–14 [DOI] [PubMed] [Google Scholar]
- 30.Qin, H., Carr, H. S., Wu, X., Muallem, D., Tran, N. H., and Frost, J. A. (2005) J. Biol. Chem. 280 7603–7613 [DOI] [PubMed] [Google Scholar]
- 31.Tran, N. H., and Frost, J. A. (2003) J. Biol. Chem. 278 11221–11226 [DOI] [PubMed] [Google Scholar]
- 32.Tran, N. H., Wu, X., and Frost, J. A. (2005) J. Biol. Chem. 280 16244–16253 [DOI] [PubMed] [Google Scholar]
- 33.Chong, H., and Guan, K. L. (2003) J. Biol. Chem. 278 36269–36276 [DOI] [PubMed] [Google Scholar]
- 34.Park, S., Rath, O., Beach, S., Xiang, X., Kelly, S. M., Luo, Z., Kolch, W., and Yeung, K. C. (2006) FEBS Lett. 580 6405–6412 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Bagrodia, S., and Cerione, R. A. (1999) Trends Cell Biol. 9 350–355 [DOI] [PubMed] [Google Scholar]
- 36.Frost, J. A., Steen, H., Shapiro, P., Lewis, T., Ahn, N., Shaw, P. E., and Cobb, M. H. (1997) EMBO J. 16 6426–6438 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Eblen, S. T., Slack, J. K., Weber, M. J., and Catling, A. D. (2002) Mol. Cell. Biol. 22 6023–6033 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Sun, H., King, A. J., Diaz, H. B., and Marshall, M. S. (2000) Curr. Biol. 10 281–284 [DOI] [PubMed] [Google Scholar]
- 39.Chaudhary, A., King, W. G., Mattaliano, M. D., Frost, J. A., Diaz, B., Morrison, D. K., Cobb, M. H., Marshall, M. S., and Brugge, J. S. (2000) Curr. Biol. 10 551–554 [DOI] [PubMed] [Google Scholar]
- 40.Li, W., Chong, H., and Guan, K. L. (2001) J. Biol. Chem. 276 34728–34737 [DOI] [PubMed] [Google Scholar]
- 41.Zang, M., Waelde, C. A., Xiang, X., Rana, A., Wen, R., and Luo, Z. (2001) J. Biol. Chem. 26 25157–25165 [DOI] [PubMed] [Google Scholar]
- 42.Kolch, W., Heidecker, G., Kochs, G., Hummel, R., Vahidi, H., Mischak, H., Finkenzeller, G., Marme, D., and Rapp, U. R. (1993) Nature 364 249–252 [DOI] [PubMed] [Google Scholar]
- 43.Carroll, M. P., and May, W. S. (1994) J. Biol. Chem. 269 1249–1256 [PubMed] [Google Scholar]
- 44.Chong, H., Lee, J., and Guan, K. L. (2001) EMBO J. 20 3716–3727 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Davies, H., Bignell, G. R., Cox, C., Stephens, P., Edkins, S., Clegg, S., Teague, J., Woffendin, H., Garnett, M. J., Bottomley, W., Davis, N., Dicks, E., Ewing, R., Floyd, Y., Gray, K., Hall, S., Hawes, R., Hughes, J., Kosmidou, V., Menzies, A., Mould, C., Parker, A., Stevens, C., Watt, S., Hooper, S., Wilson, R., Jayatilake, H., Gusterson, B. A., Cooper, C., Shipley, J., Hargrave, D., Pritchard-Jones, K., Maitland, N., Chenevix-Trench, G., Riggins, G. J., Bigner, D. D., Palmieri, G., Cossu, A., Flanagan, A., Nicholson, A., Ho, J. W., Leung, S. Y., Yuen, S. T., Weber, B. L., Seigler, H. F., Darrow, T. L., Paterson, H., Marais, R., Marshall, C. J., Wooster, R., Stratton, M. R., and Futreal, P. A. (2002) Nature 417 949–954 [DOI] [PubMed] [Google Scholar]
- 46.Rajagopalan, H., Bardelli, A., Lengauer, C., Kinzler, K. W., Vogelstein, B., and Velculescu, V. E. (2002) Nature 418 934. [DOI] [PubMed] [Google Scholar]
- 47.Brose, M. S., Volpe, P., Feldman, M., Kumar, M., Rishi, I., Gerrero, R., Einhorn, E., Herlyn, M., Minna, J., Nicholson, A., Roth, J. A., Albelda, S. M., Davies, H., Cox, C., Brignell, G., Stephens, P., Futreal, P. A., Wooster, R., Stratton, M. R., and Weber, B. L. (2002) Cancer Res. 62 6997–7000 [PubMed] [Google Scholar]
- 48.Yuen, S. T., Davies, H., Chan, T. L., Ho, J. W., Bignell, G. R., Cox, C., Stephens, P., Edkins, S., Tsui, W. W., Chan, A. S., Futreal, P. A., Stratton, M. R., Wooster, R., and Leung, S. Y. (2002) Cancer Res. 62 6451–6455 [PubMed] [Google Scholar]
- 49.Pollock, P. M., Harper, U. L., Hansen, K. S., Yudt, L. M., Stark, M., Robbins, C. M., Moses, T. Y., Hostetter, G., Wagner, U., Kakareka, J., Salem, G., Pohida, T., Heenan, P., Duray, P., Kallioniemi, O., Hayward, N. K., Trent, J. M., and Meltzer, P. S. (2003) Nat. Genet. 33 19–20 [DOI] [PubMed] [Google Scholar]
- 50.Pollock, P. M., and Meltzer, P. S. (2002) Cancer Cell 2 5–7 [DOI] [PubMed] [Google Scholar]
- 51.Lang, J., Boxer, M., and MacKie, R. (2003) Hum. Mutat. 21 327–330 [DOI] [PubMed] [Google Scholar]
- 52.Kimura, E. T., Nikiforova, M. N., Zhu, Z., Knauf, J. A., Nikiforov, Y. E., and Fagin, J. A. (2003) Cancer Res. 63 1454–1457 [PubMed] [Google Scholar]
- 53.Cohen, Y., Libster, D., Da'as, N., Amir, G., and Polliack, A. (2003) Hematol. J. 4 151–153 [DOI] [PubMed] [Google Scholar]
- 54.Singer, G., Oldt, R., 3rd, Cohen, Y., Wang, B. G., Sidransky, D., Kurman, R. J., and Shih Ie, M. (2003) J. Natl. Cancer Inst. 95 484–486 [DOI] [PubMed] [Google Scholar]
- 55.Tannapfel, A., Sommerer, F., Benicke, M., Katalinic, A., Uhlmann, D., Witzigmann, H., Hauss, J., and Wittekind, C. (2003) Gut 52 706–712 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Zhang, B. H., and Guan, K. L. (2000) EMBO J. 19 5429–5439 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Schonwasser, D. C., Marais, R. M., Marshall, C. J., and Parker, P. J. (1998) Mol. Cell. Biol. 18 790–798 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Barnard, D., Diaz, B., Clawson, D., and Marshall, M. (1998) Oncogene 17 1539–1547 [DOI] [PubMed] [Google Scholar]
- 59.Dhillon, A. S., von Kriegsheim, A., Grindlay, J., and Kolch, W. (2007) Cell Cycle 6 3–7 [DOI] [PubMed] [Google Scholar]
- 60.Coles, L. C., and Shaw, P. E. (2002) Oncogene 21 2236–2244 [DOI] [PubMed] [Google Scholar]
- 61.Zhu, J., Balan, V., Bronisz, A., Balan, K., Sun, H., Leicht, D. T., Luo, Z., Qin, J., Avruch, J., and Tzivion, G. (2005) Mol. Biol. Cell 16 4733–4744 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Luo, Z., Tzivion, G., Belshaw, P. J., Vavvas, D., Marshall, M., and Avruch, J. (1996) Nature 383 181–185 [DOI] [PubMed] [Google Scholar]
- 63.Weber, C. K., Slupsky, J. R., Kalmes, H. A., and Rapp, U. R. (2001) Cancer Res. 61 3595–3598 [PubMed] [Google Scholar]
- 64.Leicht, D. T., Balan, V., Kaplun, A., Singh-Gupta, V., Kaplun, L., Dobson, M., and Tzivion, G. (2007) Biochim. Biophys. Acta 1773 1196–1212 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Beeser, A., Jaffer, Z. M., Hofmann, C., and Chernoff, J. (2005) J. Biol. Chem. 280 36609–36615 [DOI] [PubMed] [Google Scholar]
- 66.Slack-Davis, J. K., Eblen, S. T., Zecevic, M., Boerner, S. A., Tarcsafalvi, A., Diaz, H. B., Marshall, M. S., Weber, M. J., Parsons, J. T., and Catling, A. D. (2003) J. Cell Biol. 162 281–291 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Park, E. R., Eblen, S. T., and Catling, A. D. (2007) Cell. Signal. 19 1488–1496 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Farrar, M. A., Alberol, I., and Perlmutter, R. M. (1996) Nature 383 178–181 [DOI] [PubMed] [Google Scholar]
- 69.Inouye, K., Mizutani, S., Koide, H., and Kaziro, Y. (2000) J. Biol. Chem. 275 3737–3740 [DOI] [PubMed] [Google Scholar]
- 70.Garnett, M. J., Rana, S., Paterson, H., Barford, D., and Marais, R. (2005) Mol. Cell 20 963–969 [DOI] [PubMed] [Google Scholar]