Abstract
Mutations in TREX1 have been linked to a spectrum of human autoimmune diseases including Aicardi-Goutières syndrome (AGS), familial chilblain lupus (FCL), systemic lupus erythematosus, and retinal vasculopathy and cerebral leukodystrophy. A common feature in these conditions is the frequent detection of antibodies to double-stranded DNA (dsDNA). TREX1 participates in a cell death process implicating this major 3′ → 5′ exonuclease in genomic DNA degradation to minimize potential immune activation by persistent self DNA. The TREX1 D200N and D18N dominant heterozygous mutations were identified in AGS and FCL, respectively. TREX1 enzymes containing the D200N and D18N mutations were compared using nicked dsDNA and single-stranded DNA (ssDNA) degradation assays. The TREX1WT/D200N and TREX1WT/D18N heterodimers are completely deficient at degrading dsDNA and degrade ssDNA at an expected ∼2-fold lower rate than TREX1WT enzyme. Further, the D200N- and D18N-containing TREX1 homo- and heterodimers inhibit the dsDNA degradation activity of TREX1WT enzyme, providing a likely explanation for the dominant phenotype of these TREX1 mutant alleles in AGS and FCL. By comparison, the TREX1 R114H homozygous mutation causes AGS and is found as a heterozygous mutation in systemic lupus erythematosus. The TREX1R114H/R114H homodimer has dysfunctional dsDNA and ssDNA degradation activities and does not detectibly inhibit the TREX1WT enzyme, whereas the TREX1WT/R114H heterodimer has a functional dsDNA degradation activity, supporting the recessive genetics of TREX1 R114H in AGS. The dysfunctional dsDNA degradation activities of these disease-related TREX1 mutants could account for persistent dsDNA from dying cells leading to an aberrant immune response in these clinically related disorders.
The TREX1 gene encodes the major 3′ → 5′ exonuclease detected in mammalian cells. TREX1 is a single open reading frame located on chromosome 3p21.31 and encodes a 314-amino acid polypeptide (1–3). TREX1 is closely related to TREX2 (located on chromosome Xq28) except TREX1 contains additional coding sequence for a C-terminal region not found in TREX2 (4). The N-terminal 242 amino acids of TREX1 contain the catalytically robust 3′ → 5′ exonuclease that is active on ssDNA2 and dsDNA (5) with the highest activity detected using a partial duplex DNA containing unpaired 3′ nucleotides (1–3). The C-terminal 72 amino acids contain a transmembrane region that localizes TREX1 to the endoplasmic reticulum in the perinuclear space of cells (6). Activation of a cell death pathway and treatment of cells with DNA-damaging agents cause TREX1 to relocate to the nucleus where it acts on DNA 3′ termini (6–8).
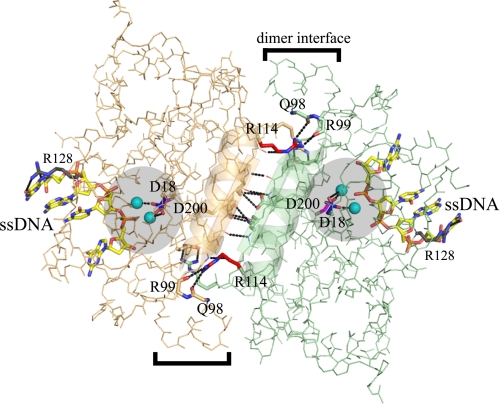
The structure of TREX1 catalytic domain with bound DNA reveals the protein-polynucleotide interactions that explain the preference for multiple unpaired 3′ termini in TREX1 exonuclease action and highlights the extensive interface contacts in the stable dimeric enzyme (9). A short ssDNA of approximately four nucleotides is accommodated in the TREX1 active site, and the Arg-128 interacting with the polynucleotide bases is proposed to destabilize dsDNA to provide ssDNA to the enzyme active site. A stable TREX1 dimer is generated through extensive interactions between the β3 strands and the α4 helices of each protomer at the interface (see Fig. 1). The hydrogen bonding network between protomers includes backbone contacts of the β3 strands and three pairs of side chain contacts from each protomer. The antiparallel α4 helices contribute hydrophobic packing interactions between the protomers. Symmetry in the TREX1 dimer positions the active sites of each protomer at opposite outer edges, providing open access for binding of two ssDNA polynucleotides. The dimeric structure of TREX1 and the closely related TREX2 (10) is unusual for 3′ → 5′ exonucleases and likely reflects the biological function of these enzymes.
FIGURE 1.
The TREX1 dimer with bound ssDNA. The TREX1 protomer backbones (orange and green) are shown. The Asp-18 (blue), Asp-200 (magenta), Arg-114 (red), and Arg-128 (gray) residue side chains (shown as sticks) are indicated in each of the TREX1 protomers. The active sites (highlighted circles) and dimer interface (brackets) are indicated. The Ca2+ divalent ions (cyan) coordinated by D18 and D200 (black dotted lines) and 4-mer ssDNA (as sticks) are shown bound in the active sites. The β3 strands and α4 helices at the dimer interface are shown as cartoons. The hydrogen bonding at the dimer interface (black dotted lines) is indicated. The Arg-114 of each protomer hydrogen bonds to the backbone carbonyl oxygens of Gln-98 and Arg-99 in the opposing protomer. The figure was prepared using the program PyMol (Delano Scientific).
Nucleases in mammals process DNA and RNA polynucleotides to prevent these macromolecules from inappropriately activating the immune system. The connection between TREX1 and immune activation was first indicated in the Trex1 null mice that develop inflammatory myocarditis consistent with autoimmune disease (11). Mutations in the human TREX1 gene have now been linked to the severe neurological brain disease Aicardi-Goutières syndrome (12), to a monogenic form of cutaneous lupus erythematosus named “familial chilblain lupus” (13–15), to systemic lupus erythematosus (16), and to retinal vasculopathy and cerebral leukodystrophy (17). The shared TREX1 genetics and similarities in these clinically distinct human disorders point to a common molecular etiology. Some AGS- and chilblain lupus-affected individuals show evidence of circulating antinuclear antibodies including those with antigenic specificity to ssDNA and dsDNA, highlighting the clinical overlap of these disorders (18–20). Defective TREX1 in humans might result in the failure to degrade ssDNA or dsDNA leading to immune activation and development of autoantibodies to these macromolecules. The source of this DNA could be genomic from the billions of cells that die daily in humans or from DNA replication and repair intermediates generated in the “normal” maintenance of genome integrity.
The TREX1 dimer structure, its stability, and the noncatalytic C-terminal region have implications for enzyme dysfunction dependent upon the allele-specific mutation. The TREX1 mutations identified in AGS include null alleles and missense mutations that map mostly to the catalytic core region of the enzyme, although mutations in the C-terminal region have been identified (18). AGS is mostly an autosomal recessive disorder, but the TREX1 D200N mutation causes autosomal dominant AGS (15). Similarly, the autosomal dominant FCL mutations c.375dupT (15) and D18N (13, 14) also map to the TREX1 catalytic core. TREX1 mutations located in the catalytic core reduce the ssDNA degradation activity to lower and dramatically varying levels (9, 14–16). The heterozygous TREX1 mutations that cause retinal vasculopathy and cerebral leukodystrophy are exclusively frameshifts localized to the C-terminal region that eliminate the transmembrane endoplasmic reticulum anchoring region and do not affect the catalytic activity of TREX1 (17).
The levels of ssDNA degradation activities in mutant TREX1 enzymes have provided limited correlation between diminished TREX1 function and human disease phenotype or severity. We have proposed that TREX1 degrades DNA by acting at nicked dsDNA generated by the NM23-H1 endonuclease during cell death (6), and others have proposed that TREX1 degrades ssDNA generated from processing of aberrant replication intermediates (8) or derived from endogenous retroelements (24). To establish a link between TREX1 3′ exonuclease dysfunction and human autoimmune disease phenotype, we designed a dsDNA degradation assay to measure the activity of TREX1 variants using a nicked dsDNA substrate. Using this assay, we show that TREX1 enzymes containing the dominant D200N and D18N mutations are defective in dsDNA degradation activity. Moreover, these TREX1 mutants are likely trapped in nonproductive enzyme-DNA complexes at the sites of nicked DNA, thus inhibiting the dsDNA degradation activity of the TREX1WT enzyme, supporting the dominant phenotypes of the TREX1 D200N and D18N mutations. Further, we show that the TREX1R114H/R114H homodimer degrades dsDNA ∼300-fold less efficiently than TREX1WT, and this dsDNA degradation dysfunction is not detected in the TREX1WT/R114H heterodimer. These TREX1 enzymatic data parallel the genetic findings of TREX1 homozygous R114H in AGS and heterozygous R114H in SLE. These results support a role for TREX1 in dsDNA degradation to prevent immune activation leading to autoimmune disease.
EXPERIMENTAL PROCEDURES
Materials—The synthetic 30-mer oligonucleotide 5′-ATACGACGGTGACAGTGTTGTCAGACAGGT-3′ with 5′ fluorescein was from Operon. Plasmids 1 (9.4 kb) and 2 (10.8 kb) are derivatives of the pMYB5 plasmid (New England Biolabs). Plasmid 1 contains one Nt.BbvCI restriction enzyme site, and plasmid 2 contains two Nt.BbvCI sites ∼800 nucleotides apart in opposite orientation. Plasmid 2 contains unique EcoRI and SacI restriction enzyme sites. The plasmids were purified from bacterial cultures and from restriction enzyme digests using Qiagen kits.
Enzyme Preparation—The human TREX1 enzymes contain the catalytic core amino acids 1–242. The TREX1 homodimers were first expressed in bacteria as N-terminal maltose-binding protein (MBP) fusions with a PreScission Protease recognition sequence between the MBP and TREX1. The pLM303 plasmid constructs were transformed into Escherichia coli BL21(DE3) Rosetta 2 cells (Novagen) for overexpression. The cells were grown to an A600 = 0.5 at 37 °C and quickly cooled on ice to 17 °C. After induction with 1 mm isopropyl-β-d-thiogalactopyranoside, the cells were allowed to grow for 15 h at 17 °C. The MBP-TREX1 fusion protein was bound to an amylose resin (New England Biolabs), washed, and the column resin was incubated at 4 °C overnight with PreScission Protease (GE Biosciences) to separate TREX1 from MBP. The TREX1 was collected from the column flow-through, dialyzed against 50 mm Tris-HCl (pH 7.5), 100 mm NaCl, 1 mm EDTA, and 10% glycerol, and purified to homogeneity using phosphocellulose chromatography (9).
To generate TREX1 heterodimers, the mutant TREX1 was cloned into pLM303X (14) and WT TREX1 was cloned into the pCDFDuet-1 (Novagen). Mutant TREX1 plasmids were produced using a PCR site-directed mutagenesis strategy (21) and confirmed by DNA sequencing. Expression from pLM303X generates MBP-TREX1MUT and expression from pCDFDuet-1 generates HisNusAHis-TREX1WT, with both fusion proteins containing a PreScission Protease sequence between the affinity tag and TREX1. Coexpression of pLM303X and pCDFDuet-1 plasmids in the same bacterial cell as described above generates a mixture of TREX1WT/WT homodimers, TREX1WT/MUT heterodimers, and TREX1MUT/MUT homodimers. Only the TREX1WT/MUT heterodimer contains both affinity tags. The TREX1WT/WT and TREX1WT/MUT were first recovered by chromatography using a nickel-nitrilotriacetic acid resin (Qiagen). The TREX1WT/MUT heterodimer was then bound to an amylose resin and washed, and the column resin was incubated at 4 °C overnight with PreScission Protease to separate TREX1WT/MUT heterodimer from the MBP and NusA. The TREX1WT/MUT was collected and purified to homogeneity using phosphocellulose chromatography as described above. Protein concentrations were determined by A280 using the molar extinction coefficient for human TREX1 protomer ε = 23,950 m-1 cm-1.
Exonuclease Assays—The exonuclease reactions contained 20 mm Tris-HCl (pH 7.5), 5 mm MgCl2, 2 mm dithiothreitol, 100 μg/ml bovine serum albumin, 50 nm fluorescein-labeled 30-mer oligonucleotide (ssDNA assays) or 10 μg/ml plasmid DNA (dsDNA assays), and TREX1 protein as indicated in the figure legends. For all assays, the TREX1 enzyme and variant mixtures were prepared on ice at 10 times the final concentrations and added to reactions to yield the indicated final concentrations. Incubations were for the indicated times at 25 °C. After incubation, the samples were removed, quenched by the addition of three volumes of cold ethanol, and dried in vacuo. For ssDNA assays, the reaction products were processed and quantified as described (9, 10). For dsDNA assays, the reaction products were resuspended in 20 μl of TAE-agarose gel running solution and electrophoresed on 0.8% agarose gels containing ethidium bromide. DNA was visualized using a FluorChem 8900 imaging system (Alpha Innotech). Activity was determined by calculating the time for TREX1WT (7.6 nm) to completely degrade the nicked polynucleotide of the dsDNA (30 μm as nucleotide) to dNMP as indicated by the loss of ethidium bromide staining material and the accumulation of the un-nicked ssDNA visible in the agarose gels.
RESULTS AND DISCUSSION
The TREX1 exonuclease degrades DNA polynucleotides to nucleoside monophosphates from available 3′ termini. We showed previously that TREX1 removes nucleotides from ssDNA and dsDNA oligonucleotide constructs (1, 3, 5) and from a nicked dsDNA plasmid generated by incubation with the NM23-H1 endonuclease (6). The TREX1 structure indicates that four nucleotides of ssDNA, not dsDNA, bind into the active sites of each protomer of the dimer (9, 22) consistent with the greater TREX1 activity detected on partial duplex DNAs containing 3′ terminal unpaired nucleotides (1, 3). These combined data illustrate TREX1 exonuclease action using a variety of DNA structures that contain accessible 3′ termini.
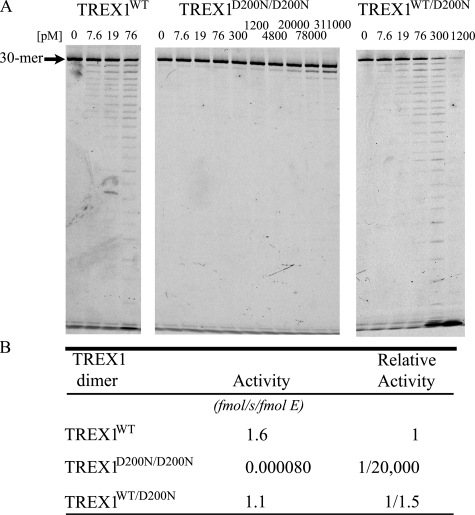
The TREX1 3′ Exonuclease Activities of Disease-related Mutant Enzymes Using ssDNA—Dominant mutant TREX1 D200N and D18N alleles have been identified in the heterozygous genotypes of individuals affected with the autoimmune disorders AGS (15) and FCL (13, 14). The Asp-200 and Asp-18 residues are two of the divalent metal ion Mg2+-coordinating aspartates in the TREX1 active site required for catalysis (Fig. 1). TREX1 is a homodimer, so TREX1 dimers in cells of the AGS heterozygous individual could be TREX1D200N/D200N and TREX1WT homodimers and TREX1WT/D200N heterodimers. Similarly, TREX1 dimers in cells of the FCL heterozygous individuals could be TREX1D18N/D18N and TREX1WT homodimers and TREX1WT/D18N heterodimers. These recombinant TREX1 variant dimers were prepared, and the ssDNA exonuclease activities were measured (Fig. 2 and Table 1) (14). The exonuclease activities of the TREX1D200N/D200N and TREX1D18N/D18N homodimers are reduced by more than 104-fold compared with TREX1WT, exhibiting almost no catalytic activity using our standard ssDNA 3′ → 5′ exonuclease activity assay. On the other hand, the activities of the TREX1WT/D200N and TREX1WT/D18N heterodimers are reduced by only 1.5- and 2.6-fold compared with TREX1WT (Fig. 2 and Table 1) (14). The ∼2-fold loss in activity of the TREX1WT/D200N and TREX1WT/D18N heterodimers suggests that the WT TREX1 protomer within the dimer containing a D200N or D18N protomer is apparently fully functional and unaffected when degrading small ssDNA polynucleotides. Thus, cells of heterozygous affected individuals carrying the TREX1 D200N and D18N dominant mutant alleles might be expected to contain ∼50% of the TREX1 activity as compared with cells from unaffected individuals with respect to ssDNA polynucleotide degradation activity, as we have demonstrated with patient cells carrying the D200N allele (15).
FIGURE 2.
The ssDNA exonuclease activities of TREX1WT and D200N variants. Standard exonuclease reactions (30 μl) were prepared with a fluorescein-labeled 30-mer oligonucleotide and dilutions of the recombinant wild type (TREX1WT), mutant (TREX1D200N/D200N) homodimer, and (TREX1WT/D200N) heterodimer were prepared at 10 times the final concentrations. Samples (3 μl) containing the TREX1 enzymes to yield the final indicated concentrations were added to reactions. The incubations were 20 min at 25 °C. The reaction products were subjected to electrophoresis on 23% urea-polyacrylamide gels (A) and quantified (B) as described under “Experimental Procedures.” The position of migration of the 30-mer is indicated. Relative activities of TREX1D200N/D200N and TREX1WT/D200N are compared with TREX1WT dimers. The relative activity was calculated as relative activity = 100 × [(fmol of dNMP released/s/fmol of mutant enzyme)/(fmol of dNMP released/s/fmol WT enzyme)].
TABLE 1.
Relative ssDNA exonuclease activities of TREX1WT and variants
The TREX1 R114H homozygous genotype is the most common missense mutation found in AGS (12, 18), and the R114H heterozygous genotype has been identified in a patient with SLE (16). The Arg-114 residue is positioned ∼15 Å away from the active site where the arginine side chain hydrogen bonds with the backbone carbonyl oxygens of Gln-98 and Arg-99 on a loop of the opposing protomer stabilizing the dimer interface (Fig. 1). Despite the considerable distance of Arg-114 from the TREX1 active site, the TREX1R114H/R114H homodimer exhibits a 34-fold reduced activity, and the TREX1WT/R114H heterodimer exhibits a 2.8-fold reduced activity compared with TREX1WT using the standard ssDNA exonuclease assay (Fig. 3 and Table 1). These data indicate that mutations at the dimer interface can decrease the catalytic competency of the TREX1 dimer when degrading ssDNA.
FIGURE 3.
The ssDNA exonuclease activities of TREX1WT and R114H variants. The standard exonuclease reactions (30 μl) were prepared with a fluorescein-labeled 30-mer oligonucleotide, and dilutions of the recombinant wild type (TREX1WT), mutant (TREX1R114H/R114H) homodimer, and (TREX1WT/R114H) heterodimer were prepared at 10 times the final concentrations. Samples (3 μl) containing the TREX1 enzymes to yield the final indicated concentrations were added to reactions. The incubations were 20 min at 25 °C. The reaction products were subjected to electrophoresis on 23% urea-polyacrylamide gels (A) and quantified (B) as described under “Experimental Procedures.” The position of migration of the 30-mer is indicated. Relative activities of TREX1R114H/R114H and TREX1WT/R114H are compared with TREX1WT dimers. The relative activity was calculated as relative activity = 100 × [(fmol of dNMP released/s/fmol of mutant enzyme)/(fmol of dNMP released/s/fmol WT enzyme)].
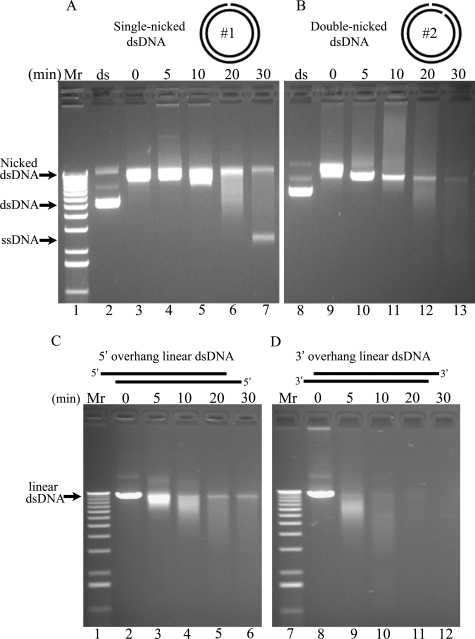
The TREX1 3′ Exonuclease Has dsDNA Degradation Activity—TREX1 initiates polynucleotide degradation from a nick in dsDNA. Our previous studies measuring TREX1 variant exonuclease activities using a small ssDNA polynucleotide have provided insights into residues affecting catalytic activity but have not revealed a clear correlation between diminished TREX1 activity and disease phenotype. We have proposed that TREX1 participates in a cell death pathway by degrading dsDNA at nicks generated by the NM23-H1 endonuclease (6). To test this possibility, a dsDNA degradation assay was developed to measure the 3′ exonuclease activities of TREX1 and variants using a nicked dsDNA plasmid (Fig. 4). Two different plasmids (plasmids 1 and 2) were incubated with the Nt.BbvCI restriction endonuclease nicking enzyme to introduce a single nick in one of the DNA strands of plasmid 1 or one nick in each of the two DNA strands positioned ∼800 nucleotides apart in plasmid 2. Incubation of the plasmids with the nicking enzyme results in conversion of the plasmids from Form I supercoiled dsDNA to Form II nicked dsDNA as indicated by the changed migration of the plasmid DNAs in the agarose gel (Fig. 4A, compare lanes 2 and 3, and B, compare lanes 8 and 9). Incubation of plasmid 1 with TREX1WT results in the nearly complete degradation of the nicked polynucleotide strand and the accumulation of the un-nicked ssDNA strand (Fig. 4A). Incubation of plasmid 2 with TREX1WT results in the complete degradation of both polynucleotide strands of the dsDNA (Fig. 4B). These data show that TREX1 recognizes a single nick in dsDNA to initiate complete degradation of the DNA polynucleotide.
FIGURE 4.
The TREX1 3′ exonuclease has dsDNA degradation activity. The supercoiled dsDNA plasmids 1 (A, lane 2) and 2 (B, lane 8) were nicked by incubating with Nt.BbvCI restriction enzyme (A, lane 3; and B, lane 9) and purified as described under “Experimental Procedures.” The plasmid 2 was linearized by incubating with EcoRI (C, lane 2) or SacI (D, lane 8) and purified. Exonuclease time course reactions (100 μl) were prepared containing nicked dsDNA plasmid 1 (A) or 2 (B) or linearized dsDNA plasmid 2 (C and D) and 7.6 nm TREX1WT. The samples (20 μl) were removed after incubation for the indicated times (A, lanes 4–7; and B, lanes 10–13). The reaction products were subjected to electrophoresis on agarose gels, and the amounts of dNMP excised by TREX1WT were estimated as described under “Experimental Procedures.” The positions of migration of Form I supercoiled dsDNA (dsDNA), Form II nicked dsDNA (Nicked dsDNA), Form III linear dsDNA (linear dsDNA), and circular ssDNA (ssDNA) are indicated. Lane 1 contains the 1-kb ladder (Invitrogen).
TREX1 degrades both polynucleotide strands of a linear duplex DNA. To further assess TREX1 exonuclease activity using dsDNA, TREX1WT enzyme was incubated with linear dsDNA plasmids and shown to degrade both polynucleotide strands of the dsDNA (Fig. 4). Plasmid 2 was digested with EcoRI or SacI to generate Form III linear dsDNA containing 5′ or 3′ overhangs. Incubation of TREX1WT with the linear dsDNA plasmids results in nearly complete degradation of the 5′ overhang DNA (Fig. 4C) and complete degradation of the 3′ overhang DNA (Fig. 4D). These data demonstrate the substantial 3′ exonuclease action of TREX1 at a nick and from the ends of dsDNA, which is similar to that measured using ssDNA oligomers as substrate (1, 3, 5, 9, 14–16).
TREX1WT/D200N and TREX1WT/D18N Heterodimer Exonuclease Activities Using dsDNA—The D200N and D18N protomers exhibit a dominant negative effect on the dsDNA degradation activity of the WT TREX1 protomer within the TREX1WT/D200N and TREX1WT/D18N heterodimers. The activities of the TREX1WT/D200N and TREX1WT/D18N enzymes were compared with TREX1WT by incubating the nicked dsDNA plasmid 1 with increased amounts of each enzyme (Fig. 5). The addition of 7.6 nm TREX1WT to reactions results in nearly complete degradation of the nicked polynucleotide strand of the plasmid DNA (Fig. 5, lane 3), and the addition of increased amounts of TREX1WT has little effect as apparent by the continued presence of the DNA band corresponding to the un-nicked ssDNA polynucleotide (Fig. 5, lanes 4–7). Additions of up to 10-fold higher concentrations of (76 nm) TREX1WT/D200N (Fig. 5, lanes 9–13) and TREX1WT/D18N (Fig. 5, lanes 15–19) heterodimers resulted in no detectible dsDNA degradation by these TREX1 mutant enzymes. In further attempts to determine the extent of reduced dsDNA degradation activity of the TREX1WT/D200N and TREX1WT/D18N heterodimers, additional reactions were performed using nicked dsDNA with enzyme concentrations as high as 230 nm for incubation times up to 5 h at 25 and 37°C with no evidence of dsDNA degradation activity by the TREX1WT/D200N and TREX1WT/D18N enzymes (data not shown). Also, the TREX1WT/D200N and TREX1WT/D18N heterodimer enzymes exhibit no detectible dsDNA degradation activity using the linear DNA plasmids 1 and 2 described in Fig. 4(C and D) (data not shown). These data indicate that the TREX1WT/D200N and TREX1WT/D18N heterodimers exhibit at least a 200-fold decreased level of dsDNA degradation activity relative to TREX1WT in contrast to the modest expected ∼2-fold level of reduced ssDNA degradation activity by these mutant heterodimer enzymes (Table 1). Moreover, these data show the dramatic differential impact on catalytic function that mutations in one TREX1 protomer can have on the opposing protomer within the TREX1 dimer dependent upon the nature of the specific mutation and on the structure of the DNA substrate.
FIGURE 5.
The TREX1WT/D200N and TREX1WT/D18N heterodimers have defective dsDNA degradation activities. The exonuclease reactions (20 μl) were prepared containing nicked dsDNA plasmid 1 (10 μg/ml = 1.6 nm nicks) and no enzyme (lanes 2, 8, and 14) or the indicated increased concentrations (in nm) of TREX1WT (lanes 3–7), TREX1WT/D200N (lanes 9–13), and TREX1WT/D18N (lanes 15–19). The reactions were 30 min, and the products were subjected to electrophoresis on agarose gels. Lane 1 (ds) contains the supercoiled dsDNA plasmid 1. The positions of migration of Form I supercoiled dsDNA (dsDNA), Form II nicked dsDNA (Nicked dsDNA), and circular ssDNA (ssDNA) are indicated.
The D200N- and D18N-containing TREX1 Mutant Enzymes Inhibit the TREX1WT dsDNA Degradation Activity—TREX1 enzymes containing D200N and D18N protomers inhibit the dsDNA degradation activity of TREX1WT. The AGS TREX1 D200N (15) and FCL TREX1 D18N (14) heterozygote patients likely have varying mixtures of mutant and WT TREX1 homodimers and mutant TREX1 heterodimers dependent upon differential allele-specific expression and subsequent dimer formation in various cell types. In light of the dramatic negative effect that the D200N- and D18N-containing protomers have on the dsDNA degradation activity within the TREX1 heterodimers, we examined the effects of the D200N and D18N mutant homo- and heterodimers on the TREX1WT dsDNA degradation activity. The TREX1WT enzyme was mixed with increased amounts of the TREX1D200N/D200N, TREX1WT/D200N, TREX1D18N/D18N, or TREX1WT/D18N enzymes and subsequently incubated with the nicked dsDNA plasmid 1 (Fig. 6). In these reactions the TREX1WT competes with the mutant TREX1 enzyme to degrade the nicked dsDNA plasmid. The amount of TREX1WT (76 nm) added in these reactions is 10-fold higher than the amount required to completely degrade the nicked polynucleotide of the dsDNA substrate (Fig. 5). Thus, in the absence of competing TREX1 mutant enzyme, the TREX1WT completely degrades the nicked polynucleotide of the dsDNA plasmid, as is apparent by the accumulation of the un-nicked polynucleotide ssDNA (Fig. 6, A and B, lane 3). The presence of increased amounts of the TREX1D200N/D200N (Fig. 6A, lanes 4–8), TREX1WT/D200N (Fig. 6A, lanes 9–13), TREX1D18N/D18N (Fig. 6B, lanes 4–8), and TREX1WT/D18N (Fig. 6B, lanes 9–13) results in decreased dsDNA degradation activity by the TREX1WT enzyme as evidenced by the increased amount of remaining nicked dsDNA. The TREX1D200N/D200N and TREX1D18N/D18N homodimer enzymes are more potent inhibitors of TREX1WT dsDNA degradation activity than are the corresponding TREX1WT/D200N and TREX1WT/D18N heterodimers (Fig. 6, A and B, compare lanes 4–8 with lanes 9–13). However, some level of TREX1WT inhibition is apparent in the presence of as little as 1/10 molar equivalent of any of the D200N- and D18N-containing TREX1 mutant enzymes (Fig. 6, A and B, compare lanes 4 and 9 with lane 3). The addition of increased amounts of the D200N- and D18N-containing TREX1 mutant enzymes results in decreased levels of dsDNA degradation with complete inhibition of the TREX1WT dsDNA degradation activity apparent in the presence of 1/4 molar equivalent of TREX1D200N/D200N (Fig. 6A, lane 5), 1/2 molar equivalent of TREX1WT/D200N (Fig. 6A, lane 11), 1/4 molar equivalent of TREX1D18N/D18N (Fig. 6B, lane 5), and 1/1 molar equivalent of TREX1WT/D18N (Fig. 6B, lane 13). Interestingly, the TREX1WT/D200N heterodimer exhibits a somewhat greater level of inhibition of the TREX1WT dsDNA degradation activity compared with the TREX1WT/D18N heterodimer (Fig. 6, compare A, lanes 9–13, with B, lanes 9–13). The potent inhibition of TREX1WT dsDNA degradation activity exhibited by D200N- and D18N-containing TREX1 protomers within TREX1 dimers (Fig. 5) and in enzyme mixtures containing TREX1WT with D200N- and D18N-containing mutant enzymes could explain the dominant phenotypes exhibited by the TREX1 D200N and D18N mutant alleles described in AGS and FCL (14, 15). Also, the more potent TREX1WT inhibition exhibited by the TREX1WT/D200N heterodimer relative to the TREX1WT/D18N heterodimer might contribute to the different clinical diagnosis of AGS for the TREX1 D200N and FCL for the TREX1 D18N mutant alleles.
FIGURE 6.
The D200N- and D18N-containing TREX1 mutant enzymes inhibit the TREX1WT dsDNA degradation activity. Exonuclease reactions (20 μl) were prepared containing nicked dsDNA plasmid 1 (10 μg/ml = 1.6 nm nicks) and no enzyme (A and B, lane 2), the indicated concentration of TREX1WT only (A and B, lane 3), or a mixture of TREX1WT with the indicated increased concentrations of TREX1D200N/D200N (A, lanes 4–8), TREX1WT/D200N (A, lanes 9–13), TREX1D18N/D18N (B, lanes 4–8), and TREX1WT/D18N (lanes 9–13). The reactions were 30 min, and the products were subjected to electrophoresis on agarose gels. Lane 1 contains the supercoiled dsDNA plasmid 1. The positions of migration of Form I supercoiled dsDNA (dsDNA), Form II nicked dsDNA (Nicked dsDNA), and circular ssDNA (ssDNA) are indicated.
TREX1R114H/R114H and TREX1WT/R114 Exonuclease Activities Using dsDNA—The TREX1R114H/R114H homodimer degrades dsDNA much less efficiently than ssDNA, and this dsDNA degradation dysfunction is not detected in the TREX1WT/R114H heterodimer. The TREX1 R114H mutation is the most common recessive allele identified in AGS (18), and our data show that the TREX1R114H/R114H homodimer enzyme has approximately a 34-fold lower ssDNA degradation activity relative to that detected in the TREX1WT enzyme (Fig. 3) (9). The dsDNA degradation activity of the TREX1R114H/R114H homodimer was measured using the nicked dsDNA substrate (Fig. 7). A 34-fold higher concentration of the TREX1R114H/R114H homodimer (260 nm) relative to the amount of TREX1WT enzyme (7.6 nm) was incubated in time course reactions with the nicked dsDNA plasmid. After 0.5 h incubation the TREX1WT enzyme completely degrades the nicked polynucleotide strand of the dsDNA plasmid (Fig. 7, lane 3). In contrast, there is no detectible degradation of the nicked dsDNA after 0.5 h of incubation with the TREX1R114H/R114H homodimer despite the presence of the 34-fold higher amount of enzyme (Fig. 7, lane 7). Incubation for extended times with the TREX1R114H/R114H homodimer enzyme results in detectible degradation of the nicked dsDNA (Fig. 7, lanes 8–12). However, even after 3 h of incubation, there is markedly less accumulation of the un-nicked ssDNA polynucleotide relative to that detected after 0.5 h of incubation with 34-fold less TREX1WT enzyme (Fig. 7, compare lane 3 with lane 12). Thus, the TREX1R114H/R114H homodimer does exhibit some detectible dsDNA degradation activity. However, this activity is estimated to be at least 300-fold lower than that detected in the TREX1WT enzyme. This dramatically reduced dsDNA degradation activity of the TREX1R114H/R114H homodimer results from mutations positioned at the TREX1 dimer interface supporting the requirement of the dimer structure in TREX1 dsDNA degradation activity. Contrasting the TREX1R114H/R114H homodimer activity, the TREX1WT/R114H heterodimer has dsDNA degradation activity similar to that of the TREX1WT enzyme (data not shown). These data indicate that the R114H mutation is required in both TREX1 protomers within the dimer to significantly disrupt the dsDNA degradation activity of TREX1.
FIGURE 7.
The TREX1R114H/R114H dsDNA degradation activity is more dysfunctional than the ssDNA exonuclease activity. Exonuclease time course reactions (200 μl) were prepared containing nicked dsDNA plasmid 1 (10 μg/ml = 1.6 nm nicks) and 7.6 nm TREX1WT (A) and 260 nm TREX1R114H/R114H (B). The samples (20 μl) were removed prior to enzyme addition (lanes 2 and 5) and after incubation for the indicated times (A, lanes 3 and 4; and B, lanes 7–12). The reaction products were subjected to electrophoresis on agarose gels. Lanes 1 and 5 (ds) contain the supercoiled dsDNA plasmid 1. The positions of migration of Form I supercoiled dsDNA (dsDNA), Form II nicked dsDNA (Nicked dsDNA), and circular ssDNA (ssDNA) are indicated.
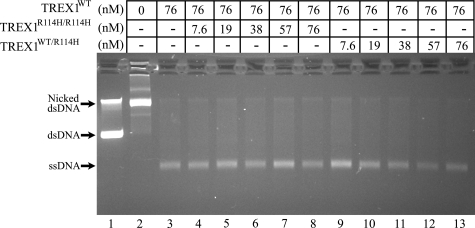
The TREX1R114H/R114H and TREX1WT/R114H enzymes do not inhibit the TREX1WT dsDNA degradation activity. A heterozygous TREX1 R114H mutation in a SLE-affected individual indicates that TREX1 dimers in cells of this individual are mixtures of TREX1WT, TREX1R114H/R114H, and TREX1WT/R114H (16), as would also be the case for parents of homozygous TREX1 R114H AGS-affected children (12, 18). The TREX1WT dsDNA degradation activity was measured in the presence of increased amounts of TREX1R114H/R114H and TREX1WT/R114H enzymes to determine whether these R114H-containing TREX1 enzymes might inhibit the TREX1WT activity (Fig. 8). Neither R114H-containing enzyme inhibits the TREX1WT activity, as is apparent from the complete degradation of the nicked polynucleotide strand and the accumulation of the un-nicked ssDNA in all reactions. Additional reactions were performed containing up to a 10-fold molar excess of TREX1R114H/R114H relative to TREX1WT with no evidence of inhibition of the TREX1WT dsDNA degradation activity (data not shown). These data are consistent with the recessive genetics of the TREX1 R114H allele in AGS where the TREX1R114H/R114H homodimer enzyme is defective in both ssDNA and dsDNA degradation activities. We propose that TREX1R114H/R114H homodimer enzyme present at undetermined levels in cells of heterozygous TREX1 R114H individuals contributes to failed ssDNA and dsDNA degradation, leading to immune activation and possibly SLE in some individuals.
FIGURE 8.
The TREX1R114H/R114H and TREX1WT/R114H enzymes do not inhibit the TREX1MWT dsDNA degradation activity. Exonuclease reactions (20 μl) were prepared containing nicked dsDNA plasmid 1 (10 μg/ml = 1.6 nm nicks) and no enzyme (lane 2), the indicated concentration of TREX1WT only (lane 3), or a mixture of TREX1WT with the indicated increased concentrations of TREX1R114H/R114H (lanes 4–8), and TREX1WT/R114H (lanes 9–13). The reactions were 30 min, and the products were subjected to electrophoresis on agarose gels. Lane 1 contains the supercoiled dsDNA plasmid 1. The positions of migration of Form I supercoiled dsDNA (dsDNA), Form II nicked dsDNA (Nicked dsDNA), and circular ssDNA (ssDNA) are indicated.
In conclusion, the activities of these TREX1 disease-related mutants and the inhibitory potential of these mutants on TREX1WT activity using the dsDNA degradation assay introduced in this work support a direct role for TREX1 in the degradation of dsDNA to prevent autoimmune disease. TREX1 resides in the perinuclear space of cells where it is anchored to the endoplasmic reticulum by the C-terminal hydrophobic region with the catalytic core protruding into cytosol. TREX1 is recruited into the nucleus upon induction of cell death or by treatment of cells with DNA-damaging agents (6, 8, 16). Translocation of TREX1 into the nucleus by the SET complex positions TREX1 to act in cell death pathways with the NM23-H1 endonuclease and perhaps other nucleases to degrade the polynucleotide chains of duplex DNA (7). The defective dsDNA degradation activities of the TREX1WT/D200N and TREX1WT/D18N heterodimers might be explained by the combination of an intact DNA binding process, analogous to that proposed for TREX2 (23), and a dysfunctional catalytic site caused by the D200N and D18N mutations. The TREX1 enzyme exhibits a distributive pattern of nucleotide excision using ssDNA substrates (1, 3), but the precise binding mechanism to dsDNA has not been determined. The heterodimer would be expected to bind DNA, identify the nick in the dsDNA, and successfully move the available 3′ terminus into the enzyme active site. The inability to perform chemistry of phosphodiester bond cleavage resulting from the D200N or D18N mutation might trap the TREX1 mutant enzyme onto the dsDNA in a nonproductive enzyme-DNA complex at the site of the nick. This result would also account for the inhibition of the TREX1WT dsDNA degradation activity detected in the presence of the D200N- or D18N-containing TREX1 mutant enzymes. Thus, the dominant genetics exhibited by the TREX1 D200N and D18N alleles is explained by the combination of TREX1 catalytic deficiency and DNA binding proficiency generating TREX1 mutant enzymes that fail to process nicks in dsDNA and block access to these sites by TREX1WT enzyme, resulting in failure to process the DNA.
This work was supported, in whole or in part, by National Institutes of Health Grant GM069962 (to F. W. P.). This work was also supported by Alliance for Lupus Research Grant 67692 (to F. W. P.) and American Cancer Society Grant RSG-04-187-01-GMC (to T. H.). The costs of publication of this article were defrayed in part by the payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 U.S.C. Section 1734 solely to indicate this fact.
Footnotes
The abbreviations used are: ssDNA, single-stranded DNA; dsDNA, double-stranded DNA; AGS, Aicardi-Goutières syndrome; FCL, familial chilblain lupus; SLE, systemic lupus erythematosus; MBP, maltose-binding protein; WT, wild type.
References
- 1.Mazur, D. J., and Perrino, F. W. (1999) J. Biol. Chem. 274 19655-19660 [DOI] [PubMed] [Google Scholar]
- 2.Hoss, M., Robins, P., Naven, T. J., Pappin, D. J., Sgouros, J., and Lindahl, T. (1999) EMBO J. 18 3868-3875 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Mazur, D. J., and Perrino, F. W. (2001) J. Biol. Chem. 276 17022-17029 [DOI] [PubMed] [Google Scholar]
- 4.Mazur, D. J., and Perrino, F. W. (2001) J. Biol. Chem. 276 14718-14727 [DOI] [PubMed] [Google Scholar]
- 5.Perrino, F. W., Miller, H., and Ealey, K. A. (1994) J. Biol. Chem. 269 16357-16363 [PubMed] [Google Scholar]
- 6.Chowdhury, D., Beresford, P. J., Zhu, P. C., Zhang, D., Sung, J. S., Demple, B., Perrino, F. W., and Lieberman, J. (2006) Mol. Cell 23 133-142 [DOI] [PubMed] [Google Scholar]
- 7.Chowdhury, D., and Lieberman, J. (2008) Annu. Rev. Immunol. 26 389-420 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Yang, Y. G., Lindahl, T., and Barnes, D. E. (2007) Cell 131 873-886 [DOI] [PubMed] [Google Scholar]
- 9.deSilva, U., Choudhury, S., Bailey, S. L., Harvey, S., Perrino, F. W., and Hollis, T. (2007) J. Biol. Chem. 282 10537-10543 [DOI] [PubMed] [Google Scholar]
- 10.Perrino, F. W., Harvey, S., McMillin, S., and Hollis, T. (2005) J. Biol. Chem. 280 15212-15218 [DOI] [PubMed] [Google Scholar]
- 11.Morita, M., Stamp, G., Robins, P., Dulic, A., Rosewell, I., Hrivnak, G., Daly, G., Lindahl, T., and Barnes, D. E. (2004) Mol. Cell. Biol. 24 6719-6727 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Crow, Y. J., Hayward, B. E., Parmar, R., Robins, P., Leitch, A., Ali, M., Black, D. N., van Bokhoven, H., Brunner, H. G., Hamel, B. C., Corry, P. C., Cowan, F. M., Frints, S. G., Klepper, J., Livingston, J. H., Lynch, S. A., Massey, R. F., Meritet, J. F., Michaud, J. L., Ponsot, G., Voit, T., Lebon, P., Bonthron, D. T., Jackson, A. P., Barnes, D. E., and Lindahl, T. (2006) Nat. Genet. 38 917-920 [DOI] [PubMed] [Google Scholar]
- 13.Lee-Kirsch, M. A., Gong, M. L., Schulz, H., Ruschendorf, F., Stein, A., Pfeiffer, C., Ballarini, A., Gahr, M., Hubner, N., and Linne, M. (2006) Am. J. Human Genet. 79 731-737 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lee-Kirsch, M. A., Chowdhury, D., Harvey, S., Gong, M., Senenko, L., Engel, K., Pfeiffer, C., Hollis, T., Gahr, M., Perrino, F. W., Lieberman, J., and Hubner, N. (2007) J. Mol. Med. 85 531-537 [DOI] [PubMed] [Google Scholar]
- 15.Rice, G., Newman, W. G., Dean, J., Patrick, T., Parmar, R., Flintoff, K., Robins, P., Harvey, S., Hollis, T., O'Hara, A., Herrick, A. L., Bowden, A. P., Perrino, F. W., Lindahl, T., Barnes, D. E., and Crow, Y. J. (2007) Am. J. Human Genet. 80 811-815 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lee-Kirsch, M. A., Gong, M., Chowdhury, D., Senenko, L., Engel, K., Lee, Y. A., deSilva, U., Bailey, S. L., Witte, T., Vyse, T. J., Kere, J., Pfeiffer, C., Harvey, S., Wong, A., Koskenmies, S., Hummel, O., Rohde, K., Schmidt, R. E., Dominiczak, A. F., Gahr, M., Hollis, T., Perrino, F. W., Lieberman, J., and Hubner, N. (2007) Nat. Genet. 39 1065-1067 [DOI] [PubMed] [Google Scholar]
- 17.Richards, A., van den Maagdenberg, A., Jen, J. C., Kavanagh, D., Bertram, P., Spitzer, D., Liszewski, M. K., Barilla-LaBarca, M. L., Terwindt, G. M., Kasai, Y., McLellan, M., Grand, M. G., Vanmolkot, K. R. J., de Vries, B., Wan, J., Kane, M. J., Mamsa, H., Schafer, R., Stam, A. H., Haan, J., Paulus, T., Storimans, C. W., van Schooneveld, M. J., Oosterhuis, J. A., Gschwendter, A., Dichgans, M., Kotschet, K. E., Hodgkinson, S., Hardy, T. A., Delatycki, M. B., Hajj-Ali, R. A., Kothari, P. H., Nelson, S. F., Frants, R. R., Baloh, R. W., Ferrari, M. D., and Atkinson, J. P. (2007) Nat. Genet. 39 1068-1070 [DOI] [PubMed] [Google Scholar]
- 18.Rice, G., Patrick, T., Parmar, R., Taylor, C. F., Aeby, A., Aicardi, J., Artuch, R., Montalto, S. A., Bacino, C. A., Barroso, B., Baxter, P., Benko, W. S., Bergmann, C., Bertini, E., Biancheri, R., Blair, E. M., Blau, N., Bonthron, D. T., Briggs, T., Brueton, L. A., Brunner, H. G., Burke, C. J., Carr, I. M., Carvalho, D. R., Chandler, K. E., Christen, H. J., Corry, P. C., Cowan, F. M., Cox, H., D'Arrigo, S., Dean, J., De Laet, C., De Praeter, C., Dery, C., Ferrie, C. D., Flintoff, K., Frints, S. G. M., Garcia-Cazorla, A., Gener, B., Goizet, C., Goutieres, F., Green, A. J., Guet, A., Hamel, B. C. J., Hayward, B. E., Heiberg, A., Hennekam, R. C., Husson, M., Jackson, A. P., Jayatunga, R., Jiang, Y. H., Kant, S. G., Kao, A., King, M. D., Kingston, H. M., Klepper, J., van der Knaap, M. S., Kornberg, A. J., Kotzot, D., Kratzer, W., Lacombe, D., Lagae, L., Landrieu, P. G., Lanzi, G., Leitch, A., Lim, M. J., Livingston, J. H., Lourenco, C. M., Lyall, E. G. H., Lynch, S. A., Lyons, M. J., Marom, D., McClure, J. P., McWilliam, R., Melancon, S. B., Mewasingh, L. D., Moutard, M. L., Nischal, K. K., Ostergaard, J. R., Prendiville, J., Rasmussen, M., Rogers, R. C., Roland, D., Rosser, E. M., Rostasy, K., Roubertie, A., Sanchis, A., Schiffmann, R., Scholl-Burgi, S., Seal, S., Shalev, S. A., Corcoles, C. S., Sinha, G. P., Soler, D., Spiegel, R., Stephenson, J. B. P., Tacke, U., Tan, T. Y., Till, M., Tolmie, J. L., Tomlin, P., Vagnarelli, F., Valente, E. M., Van Coster, R. N. A., Van der Aa, N., Vanderver, A., Vles, J. S. H., Voit, T., Wassmer, E., Weschke, B., Whiteford, M. L., Willemsen, M. A. A., Zankl, A., Zuberi, S. M., Orcesi, S., Fazzi, E., Lebon, P., and Crow, Y. J. (2007) Am. J. Human Genet. 81 713-72517846997 [Google Scholar]
- 19.Kolivras, A., Aeby, A., Crow, Y. J., Rice, G. I., Sass, U., and Andre, J. (2008) J. Cutaneous Pathol. 35 774-778 [DOI] [PubMed] [Google Scholar]
- 20.Kavanagh, D., Spitzer, D., Kothari, P. H., Shaikh, A., Liszewski, M. K., Richards, A., and Atkinson, J. P. (2008) Cell Cycle 7 1718-1725 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Higuchi, R. (1990) PCR Protocols: A Guide to Methods and Applications (Innis, M. A., Gelfand, D. H., Shinsky, J. J., and White, T. J., eds) pp. 177-183, Academic Press, San Diego, CA
- 22.Brucet, M., Querol-Audi, J., Serra, M., Ramirez-Espain, X., Bertlik, K., Ruiz, L., Lloberas, J., Macias, M. J., Fita, I., and Celada, A. (2007) J. Biol. Chem. 282 14547-14557 [DOI] [PubMed] [Google Scholar]
- 23.Perrino, F. W., de Silva, U., Harvey, S., Pryor, E., and Hollis, T. (2008) J. Biol. Chem. 283 21441-21452 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Stetson, D. B., Ko, J. S., Heidmann, T., and Medzhitov, R. (2008) Cell 134 587-598 [DOI] [PMC free article] [PubMed] [Google Scholar]