Abstract
The 26 S proteasome is an energy-dependent protease that degrades proteins modified with polyubiquitin chains. It is assembled from two multi-protein subcomplexes: a protease (20 S proteasome) and an ATPase regulatory complex (PA700 or 19 S regulatory particle) that contains six different AAA family subunits (Rpt1 to -6). Here we show that binding of PA700 to the 20 S proteasome is mediated by the COOH termini of two (Rpt2 and Rpt5) of the six Rpt subunits that constitute the interaction surface between the subcomplexes. COOH-terminal peptides of either Rpt2 or Rpt5 bind to the 20 S proteasome and activate hydrolysis of short peptide substrates. Simultaneous binding of both COOH-terminal peptides had additive effects on peptide substrate hydrolysis, suggesting that they bind to distinct sites on the proteasome. In contrast, only the Rpt5 peptide activated hydrolysis of protein substrates. Nevertheless, the COOH-terminal peptide of Rpt2 greatly enhanced this effect, suggesting that proteasome activation is a multistate process. Rpt2 and Rpt5 COOH-terminal peptides cross-linked to different but specific subunits of the 20 S proteasome. These results reveal critical roles of COOH termini of Rpt subunits of PA700 in the assembly and activation of eukaryotic 26 S proteasome. Moreover, they support a model in which Rpt subunits bind to dedicated sites on the proteasome and play specific, nonequivalent roles in the asymmetric assembly and activation of the 26 S proteasome.
The 26 S proteasome is an energy-dependent protease that degrades proteins modified with polyubiquitin chains (1–3). It comprises two multiprotein subcomplexes: a cylindrical protease (20 S proteasome) consisting of four axially stacked heteroheptameric rings and a 20-subunit ATPase regulatory complex (PA700 or 19 S regulatory particle (4, 5). The 20 S proteasome contains six catalytically active subunits whose active sites reside in a luminal chamber circumscribed by the two inner rings (6). Access of substrates to these sequestered sites is achieved via narrow pores in the center of the two outer rings. In isolated 20 S proteasomes, substrate traffic through these pores is blocked by occlusions formed of NH2-terminal tails of outer ring subunits (7–9). However, binding of PA700 to the 20 S proteasome appears to conformationally remove the occlusions, thereby creating a path for substrate transit to the catalytic sites, a process manifested as increased proteasome activity (8, 10, 11). In the absence of a crystal structure of the 26 S proteasome, evidence in support of this “gating” model of 26 S proteasome activation and details about the mechanism of holoenzyme assembly are provided by biochemical and genetic studies of the 26 S proteasome and its component subcomplexes and by structural studies with related proteasome-regulator complexes (1, 12). For example, insights into possible mechanisms of 26 S proteasome assembly and activation have been obtained from studies of PA28, an alternate 20 S proteasome regulator (13). Like PA700, this heptameric ring-shaped complex binds to the outer rings of the 20 S proteasome and activates proteolysis (14–16). PA28 binding requires the extreme COOH termini of its constituent subunits and is necessary but insufficient for proteasome activation, which requires a second PA28 domain termed the “activation loop” (11, 16–18). An x-ray crystal structure of a PA26 (the PA28 ortholog in trypanosomes)-proteasome complex identified the cognate binding sites of the PA28 COOH termini in pockets between adjacent subunits of the heteroheptameric outer ring of the 20 S proteasome. This structure revealed the conformational rearrangements of the NH2-terminal tails of 20 S proteasome subunits required for proteasome activation (19, 20). Despite the significant insights provided by this model, there are appreciable differences in the mechanisms by which PA28 and PA700 interact with and activate the proteasome. For example, PA700 binding to the proteasome requires ATP, whereas PA28 binding does not (14, 15, 21, 22). Moreover, 20 S proteasome-PA28 binding involves interactions between heptameric rings, whereas the interface of 20 S proteasome and PA700 involves an asymmetrical interaction between the heteroheptameric ring of proteasome α subunits and a heterohexameric ring of AAA (ATPases associated with various cellular activities) family member subunits of PA700 (denoted Rpt1 to -6) (23–27). In addition to the 26 S proteasome, interactions between cylinder-shaped proteases and AAA protein ring complexes occur elsewhere in nature and include the eubacterial ClpP-B/X and HslUV systems and the archeabacterial 20 S proteasome-PAN system (12). Recently, Goldberg and co-workers (28) have exploited the architecturally simpler nature of the latter system, which features interacting rings of homomeric subunit composition, to define important structural elements involved in proteasome activation. Their results reveal that, like PA28, PAN binds to the proteasome via COOH termini of its constituent subunits, but unlike PA28, these termini are sufficient for proteasome activation. Cryoelectron microscopic imaging of COOH-terminal PAN peptides demonstrated that these peptides bind to pockets between adjacent 20 S subunits and directly promote gate opening (29).
We previously established an in vitro system for ATP-dependent assembly and activation of the mammalian 26 S proteasome from purified 20 S and PA700 subcomplexes (11, 25). The purpose of the present work was to define the relative roles of individual AAA subunits in the ATP-dependent assembly and activation of mammalian 26 S proteasome and to determine features of these subunits that govern interactions with cognate binding sites on the 20 S proteasome. Our results demonstrate that specific Rpt subunits interact with dedicated binding sites on the heteroheptameric outer rings of 20 S proteasome and support a general model in which different AAA subunits play distinct roles in 26 S proteasome function.
EXPERIMENTAL PROCEDURES
Proteins—26 S proteasome, 20 S proteasome, and PA700 were purified from bovine red blood cells, as described previously (22, 30, 11, 31). Recombinant SUMO3 and SUMO-Rpt peptide fusion proteins were expressed in E. coli and purified to homogeneity using standard recombinant methodology. Amino acid sequences corresponding to variants of the COOH termini of Rpt proteins were inserted into SUMO protein by amplifying the whole pET28a-SUMO cassette with primers containing the appropriate sequences. Proteins of the resulting His-Smt3-fusion constructs were expressed at 15 °C overnight in BL21(DE3) cells (Invitrogen) and purified by affinity chromatography using Ni2+-nitrilotriacetic acid beads (Qiagen).
The cDNA of human Rpt proteins were purchased from Open Biosystems. The Rpt5 and Rpt2 cDNA were PCR-amplified and inserted into pET-28a vector (Novagen) utilizing NdeI and BamHI restriction enzymes. The COOH-terminal deletion mutant, Rpt5 (-C3), was generated by introducing termination codon Rpt5 cDNA at Tyr437 using the QuikChange site-directed mutagenesis kit (Stratagene). Recombinant Rpt proteins were expressed as NH2-terminal His-tagged fusion proteins in Bl21(DE3) cells (Invitrogen) at 25 °C for 3 h. The recombinant proteins were purified by His trap fast protein affinity column chromatography.
Proteasome Activity and Activation Assays—Proteasome activity was measured by measuring rates of hydrolysis of Suc-Leu-Leu-Val-Tyr-7-amino-4-methylcourmarin (AMC), Suc-Leu-Leu-Glu-AMC, and benzyloxycarbonyl-Val-Leu-Arg-AMC, as described previously (32). Assays, in a final volume of 50 μl, contained 45 mm Tris-HCl (pH 8.0 at 37 °C), 5 mm dithiothreitol, 50 μm substrate, and concentrations of 20 S proteasome and/or other components specified in legends for individual experiments. The rate of AMC production was measured continuously by florescence (360 nm excitation/460 nm emission) for 21 min at 37 °C in a Biotek Microplate fluorescence plate reader. For 20 S proteasome activation by PA700, samples were preincubated at 37 °C with 0.2 mm ATP and 5 mm MgCl2 for 30 min prior to the addition of substrate. Proteasome activity against methyl-14C-labeled proteins, including casein and carboxymethylated titin (domain I27), was determined by the production of acid-soluble peptides, as described previously (33).
Carboxypeptidase Treatment of PA700—CbpA (carboxypeptidase A) was purchased from Sigma. CbpA was incubated with PA700 at 25 °C as described in the figure legends for specific experiments. Control experiments document that CbpA had no direct effect on the activity of 20 S proteasome. For experiments measuring whole mass values of PA700 subunits, purified PA700 (7 mg) was incubated with or without 1.6 milliunits of CbpA at 25 °C for 10 min. CbpA was removed from the sample by glycerol density gradient centrifugation prior to HPLC.
Glycerol Density Gradient Centrifugation—Glycerol density gradient centrifugation was conducted as described in detail previously (32).
Native PAGE—Native PAGE was conducted as described previously using 4% polyacrylamide (22, 34).
Peptide Synthesis—Peptides corresponding to the COOH termini of human Rpt subunits of PA700 were synthesized using Fmoc (N-(9-fluorenyl)methoxycarbonyl) chemistry and purified by HPLC by the Protein Core Facility at the University of Texas Southwestern Medical Center. The sequences were as follows: Rpt1, SATPRYMTYN; Rpt2, GTPEGLYL; Rpt3, KDEQEHEFYK; Rpt4, LESKLDYKPV; Rpt5, KKKANLQYYA; Rpt6, KNMSIKKLWK; Rpt5(-C3), KKKANLQ; Rpt2(-C3), QEGTPEG.
Pull-down Assays—50 μg of His-tagged SUMO variants or recombinant Rpt2 and Rpt5 proteins were incubated with 10 μg of 20 S proteasome at 37 °C for 15 min in 50 mm Tris-HCl, pH 8.0, and 1 mm β-mercaptoethanol. The bound protein was immobilized on 25 μl of nickel beads for 1 h at 4 °C. Beads were washed with 50 mm Tris-HCl and 1 mm β-mercaptoethanol. Proteins bound to nickel beads were eluted with buffer containing 250 mm imidazole. The proteins were separated by gel electrophoresis and blotted with anti-20 S antibody.
Mass Spectrometry—Subunits of CbpA-treated and -untreated PA700 were separated by HPLC through a Jupiter C4 column (Phenomenex) with a 5–95% acetonitrile gradient containing 0.05% trifluoroacetic acid. Peaks containing Rpt subunits were identified by Western blotting and silver staining. Time-of-flight spectra were obtained to determine the whole mass of the indicated subunits. HPLC fractions were dried and resuspended in 50% acetonitrile with 1% formic acid. Proxeon nanotips were used to infuse the samples into a QStar XL mass spectrometer (Applied Biosystems, Framingham, MA). Spectra were acquired with mass range m/z = 500–2000. The molecular weights of proteins were calculated using the Baysesian Protein Reconstruct tool of the Analyst QS1.1 software.
Chemical Cross-linking—Chemical cross-linking was performed with peptides containing 3,4-dihydroxyphenylalanine (Dopa) as described previously (35, 36). Peptides included biotin-G-(Dopa)-GSKKKANLQYYA (designated Dopa-Rpt5), biotin-G-(Dopa)-GSKKKANLQ (designated Dopa-Rpt5-C3), and biotin-G-(Dopa)-GTPEGLYL (designated Dopa-Rpt2). Peptides were incubated with purified 20 S proteasome in Buffer H (20 mm Tris-HCl, pH 7.6, 20 mm NaCl, 1 mm EDTA, 2 mm β-mercaptoethanol, 10% glycerol) for 10 min at room temperature. Cross-linking was activated by the addition of 10 mm NaIO4 for 30 s. Samples were quenched by the addition of SDS-PAGE buffer (for one-dimensional analysis) or 50 mm β-mercaptoethanol for two-dimensional PAGE. For two-dimensional PAGE and identification of the cross-linked species, the non-cross-linked peptide was removed by multiple passes through a Microcon YM-100 centrifugal filter initially in Buffer H and then in Buffer H with 400 mm NaCl and 0.05% Tween 20. The cross-linked sample was then enriched by binding to monomeric avidin beads in Buffer H with 400 mm NaCl and 0.05% Tween 20. Beads were washed with the same buffer and finally with Buffer H. Bound samples were eluted by incubation in isoelectric focusing sample buffer (7 m urea, 2 m thiourea, 4% CHAPS, 65 mm dithiothreitol, Pharmolytes, pH 3–10, and bromphenol blue).
Two-dimensional PAGE Analysis of Cross-linked Products—Isoelectric focusing was carried out using Bio-Rad Ready Strip IPG, pH 3–10, on an IPGphor isoelectric focusing system. The second dimension was resolved by 15% SDS-PAGE. Western blotting for 20 S proteasome subunits was conducted by standard methods with antibodies against subunits α7 (C8), mouse monoclonal antibody (clone MCP72) (BioMol), and α4, mouse monoclonal antibody (clone MCP79) (BioMol). Cross-linked products were detected using neutravidin-HRP (Pierce). Silver staining was conducted with Silver Quest (Invitrogen), and the region corresponding to the cross-linked product was excised for mass spectrometric analysis (see supplemental materials for details).
RESULTS
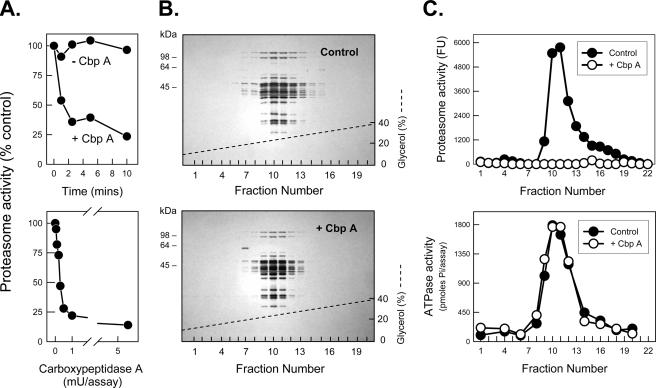
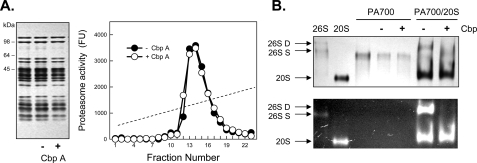
Carboxypeptidase Treatment of PA700 Inhibits Its Binding to and Activation of the Proteasome—We previously established an in vitro assay for ATP-dependent assembly and activation of the 26 S proteasome from purified isolated 20 S and PA700 subcomplexes (11, 25). To define the role of COOH-terminal residues of AAA subunits on PA700 binding to and activation of the 20 S proteasome we treated purified PA700 with several carboxypeptidases. CbpA inhibited PA700-dependent activation of 20 S proteasome (Fig. 1A) but had no detectable effect on structural or other functional characteristics of PA700, such as the general subunit pattern after SDS-PAGE, native size as judged by glycerol density gradient centrifugation or native PAGE (Figs. 1B and 2B), ATPase activity (Fig. 1C), or deubiquitylating activity (data not shown). CbpA inactivated PA700 equally well in the presence or absence of ATP (data not shown). In contrast, CbpA did not alter activity or structural features of purified intact 26 S proteasome (Fig. 2A). These results suggest that the COOH terminus of one or more PA700 subunits is required for 26 S proteasome assembly and activation and that these residues are inaccessible to and protected from CbpA when in contact with 20 S proteasome. To demonstrate directly that changes in proteasome activity monitored 26 S proteasome assembly, we analyzed the binding of CbpA-treated and -untreated PA700 to the 20 S proteasome. Pretreatment of PA700 with CbpA inhibited formation and corresponding activation of 26 S proteasome (Fig. 2B). Thus, the loss of PA700-dependent proteasome activation resulted from lack of PA700 binding to 20 S proteasome.
FIGURE 1.
Carboxypeptidase A inhibits PA700 stimulation of 20 S proteasome activity. A (top), PA700 (5 pmol) was incubated at 25 °C in the presence and absence of 13 milliunits of pancreatic CbpA. At the indicated times, PA700 was assayed for ATP-dependent stimulation of 20 S proteasome-catalyzed hydrolysis of Suc-Leu-Leu-Val-Tyr-AMC. Proteasome activity stimulated by untreated PA700 was assigned a value of 100% (control), and other activities are expressed as a percentage of that value. Untreated PA700 stimulated proteasome activity by 44-fold in this experiment. A (bottom), PA700 (5 pmol) was incubated with the indicated concentrations of CbpA for 5 min at 25 °C and then assayed for proteasome stimulatory activity as above. Untreated PA700 stimulated proteasome activity by 49-fold in this experiment. B and C, PA700 was incubated for 10 min with and without 10 milliunits of CbpA, as indicated. Samples were then subjected to 10–40% glycerol density gradient centrifugation, as described previously (32). Fractions were either subjected to SDS-PAGE and stained with Coomassie Blue (B) or assayed for proteasome stimulatory activity, expressed as fluorescent units (Fu), and ATPase activity (C). The dashed line denotes the glycerol concentration gradient from 10 to 40%.
FIGURE 2.
Carboxypeptidase A inhibits assembly and activation of the 26 S proteasome. A (left), SDS-PAGE of purified 26 S proteasome without incubation or after a 10-min incubation with (+) and without (-) CbpA. A (right), 26 S proteasome was incubated in the presence and absence of CbpA and subjected to glycerol density gradient centrifugation. Fractions were assayed for proteasome activity by hydrolysis of Suc-Leu-Leu-Val-Tyr-AMC and expressed as fluorescent units (Fu). B, purified PA700 was preincubated with (+, lanes 5 and 7) and without (-, lanes 4 and 6) CbpA and then incubated with purified 20 S proteasome in the presence of ATP. After 30 min, samples were subjected to native PAGE, as described previously (32). Gels were stained with Coomassie Blue (top) or overlaid with Suc-Leu-Leu-Val-Tyr-AMC, incubated for 10 min, and exposed to UV light (bottom). Purified 26 S proteasome (lane 1), 20 S proteasome (lane 2), and PA700 were electrophoresed as standards. 26S-D, doubly capped 26 S proteasome; 26S-S, singly capped 26 S proteasome.
Carboxyl Termini of Rpt2 and Rpt5 Are Selectively Modified by CbpA—To identify the structural basis for the CbpA-catalyzed loss of proteasome-activating activity of PA700, we performed mass analysis of PA700 subunits after incubation of PA700 in the presence and absence of CbpA. PA700 subunits were separated by HPLC; the Rpt proteins were identified in the eluted peaks by Western blotting and subjected to mass analysis, as described under “Experimental Procedures.” Each Rpt subunit of untreated PA700 yielded a mass expected for the respective mature protein except for Rpt5, whose observed mass was ∼2000 daltons lower (Table 1). CbpA treatment of PA700 sufficient for greater than 95% reduction of proteasome activating activity caused selective alterations of masses of specific Rpt subunits. Thus, CpbA treatment did not alter the masses of Rpt1, Rpt3, Rpt4, and Rpt6 but reduced the masses of Rpt2 and Rpt5 by 405 and 400 daltons, respectively. These altered masses are equivalent to the loss of the three COOH-terminal residues (LYL for Rpt2 and YYA for Rpt5) of each subunit. In a separate analysis, the mass of Rpt2 was examined after 2 and 10 min of CbpA treatment, times at which PA700 had lost ∼20 and 90%, respectively, of its proteasome activating activity (Fig. S1). The mass of Rpt2 was reduced by 386 and 439 daltons after 2 and 10 min, respectively, values equivalent to the successive loss of the two COOH-terminal residues (YL) and three COOH-terminal residues (LYL). These results demonstrate that the COOH termini of Rpt subunits are differentially processed under conditions during which PA700 loses its ability to activate the proteasome and indicate a special role of two of the six AAA subunits in proteasome binding and activation.
TABLE 1.
Carboxypeptidase A treatment of PA700 selectively alters the mass of two Rpt subunits PA700 was incubated in the presence or absence of carboxypeptidase A for 10 min. PA700 subunits were separated by HPLC. Rpt subunits were identified and subjected to mass determination as described under “Experimental Procedures.” Predicated mass values include known NH2-terminal modifications for each subunit (M, methionine; Acetyl, acetylation; Myr, myristoylation).
| Subunit | Predicted mass | Observed mass without CbpA | Observed mass with CbpA | COOH terminus |
|---|---|---|---|---|
| Da | Da | Da | ||
| Rpt1 | 48,502 (–M) | 48,504 ± 15 | 48,503 ± 15 | -TYN |
| Rpt2 | 49,290 (–M, +Myr) | 49,279 ± 15 | 48,874 ± 15 | -LYL |
| Rpt3 | 47,408 (+Acetyl) | 47,409 ± 10 | 47,409 ± 10 | -FYK |
| Rpt4 | 43,943 (–M, +Acetyl) | 44086 ± 10 | 44,085 ± 15 | -KPV |
| Rpt5 | 49,358 (+Acetyl) | 47,294 ± 15 | 46,894 ± 10 | -YYA |
| Rpt6 | 45,537 (–M, +Acetyl) | 45,538 ± 10 | 45,539 ± 15 | -LWK |
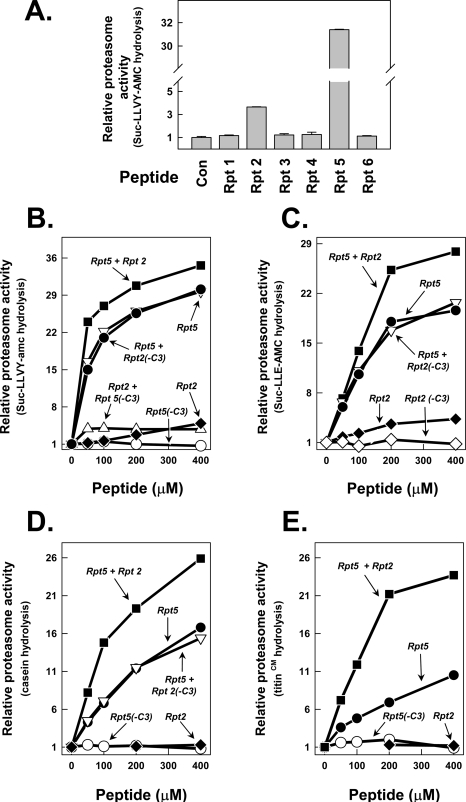
COOH-terminal Peptides of Rpt2 and Rpt5 Activate the Proteasome—To determine the relative roles of COOH termini of individual Rpt subunits in 26 S proteasome assembly and activation, we tested the ability of each to activate 20 S proteasome hydrolysis of peptide substrates. To this end, we synthesized 8–10-residue peptides corresponding to the COOH terminus of each Rpt protein. Only COOH-terminal peptides corresponding to Rpt2 and Rpt5 stimulated 20 S proteasome activity (Fig. 3A), but the magnitude of proteasome activation by these peptides differed significantly; the Rpt5 peptide stimulated proteasome activity up to 30-fold, whereas the maximal stimulation by the Rpt2 peptide seldom exceeded 7-fold. Nevertheless, the stimulatory effect of each peptide was highly specific because respective peptides lacking the last three amino acids, denoted Rpt2(-C3) and Rpt5(-C3), and peptides with similar compositions but different sequences had no stimulatory effect (Fig. 3B and Fig. S2) (data not shown). Proteasome activation was achieved similarly with different peptide substrates hydrolyzed by each of the three distinct proteasome catalytic sites (Fig. 3C and Fig. S2). We next expressed and purified recombinant proteins representing either each of the same COOH-terminal peptides of the Rpt proteins fused to the COOH terminus of SUMO or selected full-length Rpt proteins. The SUMO-Rpt peptide fusion proteins selectively stimulated 20 S proteasome activity with features similar to those promoted by the isolated peptides. Thus, only SUMO fused to COOH-terminal peptides of Rpt2 and Rpt5 peptides activated proteasome function (Fig. S3). Purified full-length recombinant Rpt2 and Rpt5 proteins also stimulated proteasome activity against peptide substrates, although evaluation of the selectivity of these effects must await purification and assay of the remaining intact Rpt proteins (Fig. S3) (data not shown). Effects on proteasome activation by isolated peptides, SUMO-peptide fusion proteins, and Rpt proteins did not depend on and were not influenced by ATP (data not shown). In sum, these results demonstrate that the COOH termini of the Rpt2 and Rpt5 subunits of PA700 are sufficient to activate proteasome function directly and do so selectively among the six Rpt subunits.
FIGURE 3.
COOH-terminal peptides of Rpt2 and Rpt5 selectively stimulate 20 S proteasome activity. Peptides corresponding to the COOH-terminal residues of AAA subunits of PA700 (Rpt1 to -6) were synthesized and purified as described under “Experimental Procedures.” A, indicated peptides (400 μm) were incubated with 20 nm purified bovine 20 S proteasome; proteasome activity against Suc-Leu-Leu-Val-Tyr-AMC was measured as described under “Experimental Procedures.” Proteasome activity in the absence of peptide was set at a relative value of 1.0, and other activities are expressed as a -fold increase of that value. B and C, individual peptides at the indicated concentrations were incubated with 20 nm 20 S proteasome for assay of proteasome activation as in A. Where a second peptide is indicated, the concentration of the second peptide is 400 μm. B, Suc-Leu-Leu-Val-Tyr-AMC substrate; C, Suc-Leu-Leu-Glu-AMC substrate. D and E, Rpt peptides, as described for B and C, were assayed for stimulation of 20 S proteasome against casein (D) and carboxymethylated (CM) titin (E). All values are means of triplicate assays. Similar results were obtained in at least three independent experiments.
To determine whether Rpt2 and Rpt5 peptides acted independently, redundantly, or synergistically, we examined their combined effects on proteasome activity. The addition of Rpt2 and Rpt5 peptides produced approximately additive effects on the stimulation of peptide substrate hydrolysis (Fig. 3, B and C). These additive effects were abolished when either of the peptides lacked its COOH-terminal three residues. These results suggest that the Rpt2 and Rpt5 peptides bound to different sites on the 20 S proteasome.
COOH-terminal Peptides of Rpt2 and Rpt5 Stimulate Proteasome Hydrolysis of Protein Substrates Differentially and Synergistically—Proteasome hydrolysis of peptide substrates should be a sensitive monitor of the status of gate openness, since these substrates have insignificant size or conformational restraints to hinder their passage to catalytic sites. In fact, yeast mutant 20 S proteasomes that lack the NH2-terminal tail of the α3 subunit required for proper gate closure feature increased peptidase activity (8, 10). The 20 S proteasome also can degrade certain unstructured nonubiquitylated proteins when activated under unique assay conditions (1, 10, 31, 37) or by intact PA700 (22). Surprisingly, however, at least one such protein substrate is not degraded by the α3 “gateless” mutant (38). To determine the relative roles of Rpt2 and Rpt5 in proteasome activation of protein substrate hydrolysis, we repeated the analysis described above, using protein substrates with disordered structures such as casein and carboxymethylated titin I27. The COOH-terminal peptide of Rpt5 stimulated proteasome activity against these substrates by up to 15-fold; this effect was abolished with a peptide lacking the three COOH-terminal residues (Fig. 3, D and E). In contrast, the Rpt2 peptide had no detectable stimulatory effect against protein substrates but, surprisingly, increased the stimulatory effect of the Rpt5 by 2–4-fold in various experiments. This synergistic effect was abolished when either peptide lacked its three COOH-terminal residues. These results provide additional support for the independent sites of action of the Rpt peptides but indicate that they function coordinately to promote the degradation of protein substrates.
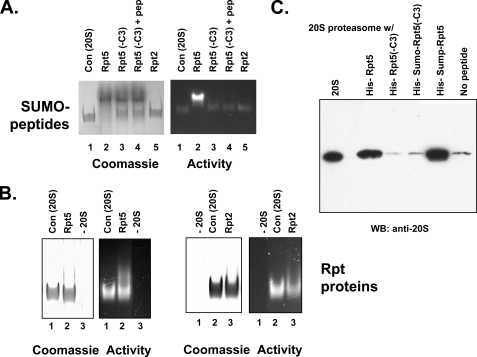
Rpt2 and Rpt5 Peptides Bind to 20 S Proteasome—To further define the relationship between proteasome activation and proteasome binding of Rpt peptides, we conducted several types of analysis. First, we determined the effects of these molecules on migration of 20 S proteasome during nondenaturing PAGE. Both SUMO-Rpt5 peptide and recombinant Rpt5 retarded migration of 20 S proteasome as detected by Coomassie Blue staining and by proteasome activity after substrate overlay (Fig. 4, A and B). In contrast, SUMO-Rpt5(-C3) had no effect on the migration of 20 S proteasome. To further characterize these effects, we incubated 20 S proteasome with SUMO-Rpt5(-C3) and the activating Rpt5 peptide. No alteration in migration of 20 S proteasome was observed, showing that formation of a proteasome-SUMO complex and not proteasome activation per se caused altered migration (Fig. 4A). SUMO-Rpt2 protein did not affect 20 S proteasome migration detectably, whereas recombinant Rpt2 protein modestly but reproducibly retarded migration (Fig. 4B). These latter results may reflect lower binding affinity of Rpt2 to the proteasome, as suggested by the lower magnitude of proteasome activation by Rpt2 molecules compared with the corresponding molecules of Rpt5. In sum, these data demonstrate a close correlation between binding and activation of proteasome by various Rpt COOH-terminal peptides.
FIGURE 4.
Binding of COOH-terminal Rpt peptides to the 20 S proteasome. A and B, purified 20 S proteasome and recombinant proteins were incubated at 37 °C for 15 min and then subjected to native PAGE. A, SUMO-Rpt COOH-terminal peptide fusion proteins, produced as described under “Experimental Procedures,” were incubated at 300 μm with 650 nm 20 S proteasome. Lane 1, 20 S proteasome alone; lane 2, 20 S proteasome and SUMO-Rpt5; lane 3, 20 S proteasome and SUMO-Rpt5(-C3); lane 4, 20 S proteasome and SUMO-Rpt5(-C3) and Rpt5 peptide (100 μm); lane 5, 20 S proteasome and SUMO-Rpt2. The gel was overlaid with Suc-Leu-Leu-Val-Tyr-AMC for identification of proteasome activity (right) and stained with Coomassie Blue (left). B, recombinant Rpt proteins, produced as described under “Experimental Procedures,” were incubated at 5 μm with 450 nm 20 S proteasome and analyzed as in A. Left pair of panels, Rpt5(lanes 1–3). Lane 1, 20 S proteasome alone; lane 2, 20 S proteasome and Rpt5; lane 3, Rpt5 alone. Right pair of panels, Rpt2 (lanes 1–3). Lane 1, Rpt2 alone; lane 2, 20 S proteasome alone; lane 3, 20 S proteasome and Rpt2. Gels were either assayed for proteasome activity or stained with Coomassie Brilliant Blue as above. C, purified 20 S proteasome (285 nm) was incubated with indicated His-tagged recombinant proteins (11 μm for Rpt5 and Rpt5-C3 and 90 μm for SUMO-Rpt5 and SUMO-Rpt5-C3) and subjected to pull-down on nickel beads as described under “Experimental Procedures.” Bound and eluted material was Western blotted for 20 S proteasome. Lane 1 (20 S) shows 5% of the input 20 S proteasome; the remaining lanes show 20 S proteasome pulled down by the indicated samples.
Second, we performed pull-down experiments using recombinant Rpt5 and the SUMO-Rpt5 peptide fusion proteins with purified 20 S proteasome. 20 S proteasome was pulled down with intact Rpt5 and SUMO-Rpt5 peptide fusion proteins but not with their -3C variants (Fig. 4C). This result further demonstrates the differential and selective binding of these intact and truncated COOH-terminal peptides to the 20 S proteasome and indicates that lack of proteasome activation by the corresponding -3C variants is a consequence of their failure to bind the proteasome.
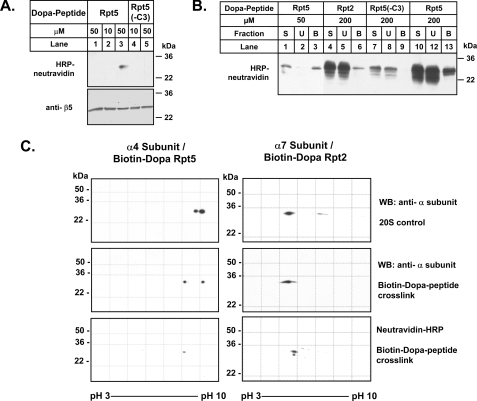
Rpt5 and Rpt2 Peptide Selectively Cross-link to Distinct α Subunits of the 20 S Proteasome—The Rpt subunits of PA700 are arranged in a heterohexameric ring of fixed order (Rpt1-Rpt2-Rpt6-Rpt4-Rpt5-Rpt3-) (39). Likewise, the outer ring of the 20 S proteasome to which Rpt subunits bind is composed of a heteroheptameric ring of α subunits in fixed order (α1-α2-α3-α4-α5-α6-α7). To determine whether the COOH termini of the Rpt subunits interact with specific α subunits, we synthesized COOH-terminal Rpt5 and Rpt2 peptides containing a Dopa residue for covalent cross-linking and a biotin residue for detection and identification of cross-linked products (35, 36). As a negative control, we also synthesized the biotin-Dopa-Rpt5 peptide lacking the three COOH-terminal residues (-C3). Both the biotin-Dopa-Rpt5 and biotin-Dopa-Rpt2 peptide activated the 20 S proteasome similarly to their non-biotin-Dopa counterparts, whereas the biotin-Dopa-Rpt5(-C3) variant did not (data not shown). Cross-linking of the biotin-Dopa-Rpt5 peptide with 20 S proteasome revealed a single band of the size characteristic of 20 S proteasome subunits, as detected by Western blotting of the biotin tag (Fig. 5A, lane 3). This band was absent when cross-linking was conducted with the Rpt5(-C3) peptide (Fig. 5A, lane 5) or when cross-linking of the full-length Dopa peptide was not induced (Fig. 5A, lane 1). Thus, under these conditions, only the activating peptide cross-linked and did so to one or a selected few proteasome subunits.
FIGURE 5.
Dopa-Rpt activating peptides interact with specific α subunits of the 20 S proteasome. A, purified 20 S proteasome (200 nm) was incubated at the indicated concentrations with biotinylated Dopa-containing Rpt5 and Rpt5(-C3) peptides. Cross-linking was performed as described under “Experimental Procedures.” Lane 1, a control reaction in which cross-linking was not induced with oxidant. Western blot analysis (WB) after SDS-PAGE shows the cross-linked product detected by Neutravidin-HRP (top) and a loading control for 20 S proteasome β5 subunit (bottom). B, samples of cross-linking reactions, performed as in A, were applied to monomeric avidin beads. S, starting cross-linked material; U, unbound fraction; B, bound fraction. In each case, 20% of total fraction was applied to the gel. Western blot analysis after SDS-PAGE shows the cross-linked product detected by neutravidin-HRP. C, two-dimensional PAGE analysis of enriched Dopa peptide-20 S proteasome cross-linked products. Western blotting of 20 S proteasome control (top) and Dopa peptide-20 S proteasome cross-link for the indicated Rpt peptides and proteasome subunits using subunit-specific antibodies (middle). Western blotting for the indicated cross-linked proteasome subunit using neutravidin-HRP (bottom).
To identify the proteasome subunit(s) cross-linked to the Rpt5 peptide, we enriched the cross-linked product by affinity chromatography on monomeric avidin-agarose beads. The enriched fraction was separated by two-dimensional PAGE. Western blotting of enriched cross-linked and control samples with subunit-specific anti-proteasome antibodies (Fig. 5, B and C) (data not shown) gave a preliminary identification of the cross-linked product as α4 (PSMA7). This identification was confirmed by the precise overlay of the anti-α4 and biotin blots (Fig. 5C). Silver staining revealed the appearance of several unique closely migrating spots in gels of proteasomes subjected to cross-linking with intact Rpt5 peptide (Fig. S4). These spots were coincident with spots detected upon Western blotting with HRP-neutravidin and absent from analogous gels of uncross-linked 20 S proteasome. To confirm this finding, we excised these spots (and analogous positions in the control gel) and subjected them to mass spectrometry. Only peptides corresponding to the α4 subunit were identified unambiguously in the gel of cross-linked protein; no α4 subunit peptides were identified in the sample from control gels (Fig. S4). These results indicate that the Rpt5 peptide bound to the 20 S proteasome at or in close proximity to the α4 subunit.
We repeated this analysis for the Rpt2 peptide. Because the apparent binding affinity of the Rpt2 peptide for 20 S proteasome appeared lower than that of the Rpt5 peptide, increased peptide concentration (200 μm) was required to detect cross-linked products (Fig. 5B). Although this variation produced a number of nonspecific cross-linked bands with both activating and nonactivating (i.e. -3C) peptides, subsequent enrichment of the products by monomeric avidin affinity chromatography resulted in a single major cross-linked band with the activating peptide (Fig. 5B). Analysis similar to that described above indicated that the major HRP-neutravidin-positive spot after two-dimensional PAGE represents the α7 subunit (PSMA3) (Fig. 5C) (data not shown). Thus, the two functionally distinct Rpt peptides appear to bind to the proteasome at distinct and dedicated sites. These results are consistent with a model of 26 S assembly in which PA700 binds to the 20 S proteasome with a specific predetermined orientation.
DISCUSSION
Despite the lack of a detailed atomic structure of the 26 S proteasome, genetic, biochemical, and imaging data indicate that the AAA subunits of PA700 (Rpt1 to -6) form a heterohexameric ring that abuts directly to the outer rings of the cylindrical 20 S proteasome (26). Previous results have demonstrated that PA700 binding is mechanistically linked to proteasome activation, probably via PA700-induced opening of the gate that allows entry of substrates to the degradation chamber of the 20 S proteasome (8, 10, 11, 40), but the structural details of this interaction have been lacking. While our work was in progress, Goldberg and co-workers (28) demonstrated the critical roles of the COOH termini of Rpt2 and Rpt5 subunits in PA700 binding and activation of the proteasome. The results reported here confirm and extend those results in several important ways and provide new insights to the mechanisms of 26 S proteasome assembly and activation. First, carboxypeptidase-inhibited binding of PA700 to the proteasome occurs after modification of only two of the six Rpt subunits (Rpt2 and Rpt5) that constitute the proteasome interaction surface. Our current data cannot exclude the possibility that binding of PA700 could depend on only one of these subunits. Second, a COOH-terminal peptide of either of the subunits involved in binding was sufficient to stimulate the peptidase activity of the proteasome, whereas COOH-terminal peptides of the other Rpt subunits were incompetent for activation. These results further highlight the nonequivalent roles of the AAA subunits of PA700 function and are consistent with a tight mechanistic link between binding and activation. The remarkable effects of these short peptides also demonstrate that other domains of ATPase subunits are not essential for binding and activation per se. Importantly, our results provide new details about the relative roles of Rpt2 and Rpt5 subunits in proteasome activation and indicate that they have either additive or synergistic effects depending on the nature of the substrate (see below). Finally, our results show that Rpt2 and Rpt5 have dedicated binding sites on the heterohexameric outer ring of the 20 S proteasome and identify the interacting subunits as α7 and α4, respectively. These assignments assume that the peptides are cross-linked to the same subunits that bind the COOH termini. Cross-linking, however, could occur to subunits distinct from but in close proximity to those with which the peptides bind, especially if the binding sites reside in pockets between adjacent α subunits (see below). In either case, our results provide evidence for a general model in which binding of 20 S proteasome and PA700 requires dedicated and invariant interactions between specific subunits of each ring. Moreover, the two inter-ring subunit interactions identified here (Rpt2-α7 and Rpt5-α4) satisfy the general topological requirements imposed by the relative positions of the intra-ring subunit pairs, which are each ∼180° apart. Therefore, these results provide reference points for the registration of the inter-ring interactions of the remaining subunits (see Fig. 6). The possible functional and structural roles of these additional interactions are unknown, and we assume that the COOH termini of the remaining Rpt subunits also bind to specific sites on the 20 S proteasome to affect processes required for 26 S proteasome function.
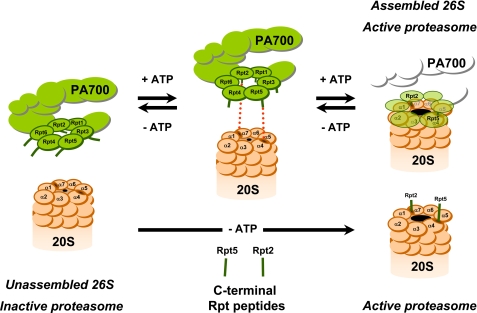
FIGURE 6.
Model for PA700 binding to and activation of the proteasome via COOH termini of Rpt2 and Rpt5 subunits. ATP binding to Rpt subunits of PA700 promotes conformational changes that optimize registration of COOH termini of Rpt2 and Rpt5 for interaction with cognate binding sites at proteasome subunits α7 and α4, respectively. Binding of these subunits induces conformational changes in α subunits of proteasome, resulting in gate opening and an increase in substrate access to catalytic sites. Peptides corresponding to the COOH termini of Rpt2 and/or Rpt5 can bind directly to their respective sites on α subunits of the proteasome to promote gate opening and proteasome activation by the same mechanism. Because no conformational change of PA700 is required for peptide binding, proteasome activation is ATP-independent. The nomenclature for 20 S proteasome subunits corresponds to that of Groll et al. (9).
The recent studies by Cheng and co-workers (29) have revealed important mechanistic details of how the COOH-terminal peptide of PAN, the homohexameric AAA regulator of the archaeal 20 S proteasome, can bind to and activate the proteasome. Cryoelectron microscopy of the PAN COOH-terminal peptide-archaeal proteasome complex shows that the peptide closely interacts with residues Gly34, Leu81, and Lys66 of one α subunit and extends NH2-terminally to an adjacent α subunit (29). Peptide binding correlates with conformational changes in the positions of the α subunits that result in rotation of a reverse loop containing Pro17. This loop was identified previously as a critical element in the mechanism of PA28-induced proteasome activation by gate opening (19, 20). In the latter case, an interaction between Pro17 of the proteasome, and the activation loop of PA28 was required to stabilize the open gate conformation. Remarkably, the structurally similar rotation of the Pro17 loop and concomitant gate opening caused by binding of the PAN COOH-terminal peptide occurs without the involvement or need of other structural elements of the PAN protein.
These elegant images describe the structural consequences of the binding of seven identical PAN peptides to seven identical sites on the archeal 20 S proteasome. This condition, however, differs from that which must exist upon binding of either the intact PAN or PA700 complexes to their respective 20 S proteasomes. In the case of PAN, the COOH termini of only six identical subunits can interact with the seven possible identical binding sites of the homoheptameric archeal 20 S proteasome. In the case of PA700, six different COOH termini interact with seven different potential binding sites of the heteroheptameric eukaryotic 20 S proteasome. Our data highlight the unique functional roles of two PA700 subunits when they interact with dedicated binding sites on the 20 S proteasome. Thus, although the precise structural consequences of the asymmetrical interactions between the heterohexameric and heteroheptameric rings required for 26 S proteasome assembly and activation remain unclear, it is reasonable to assume that the Rpt peptides function by a generally similar mechanism as that indicated for PAN peptides. Indeed, the latter mechanism provides additional insight for interpretation of our selective cross-linking data by suggesting an orientation for the binding of the peptide between adjacent α subunits. Thus, the position of the Dopa residue in the Rpt peptides used for our experiments may be closer to and therefore more likely to cross-link with the α subunit adjacent to that which binds the COOH-terminal residues directly. If this speculation is correct, the COOH termini of Rpt2 and Rpt5 peptides would bind to α1 and α5, respectively. Both of these subunits contain the Gly34 residue identified as critical for PAN peptide binding. The α5 subunit also features the Lys66 and Leu81 residues, which are highly conserved among α subunits of most archaeal 20 S proteasomes and most α subunits in various eukaryotic proteasomes. In contrast, the α1 subunit of bovine and yeast proteasome contain His66 and Tyr66, and Met81 and Pro81, respectively. Although the significance of these latter features with respect to our subunit binding assignments remains to be determined, we note that recent cryoelectron microscopic images of the 26 S proteasome reveal three prominent densities at the interface of 20 S proteasome and PA700 (41). These links include one at the interface of the α1 and α7 subunits and another at the α5 subunit and are consistent with our interpretation of cross-linking data presented above. Moreover, the α1, α7, α4, α5, and α6 subunits underwent radial displacements substantially greater than other α subunits. These asymmetric conformational changes may be linked to the rotation of the reverse turn described above involved in gate opening and highlight the likely different structural changes among different α subunits of the proteasome upon binding of PA700.
Our results show that although Rpt2 and Rpt5 is each sufficient to activate the proteasome's hydrolysis of short peptide substrates, they do so to different degrees, and their combined effect is approximately additive. Such results are consistent with separate sites of action for the Rpt peptides, as demonstrated by cross-linking, and suggest that gating is not a simple two-state process (i.e. closed or open) but instead might feature graded responses that reflect variable pore size or shape in response to binding of different individual and combined Rpt peptides. In this model, Rpt5 would activate the proteasome to a greater degree than Rpt2 because it induces a larger or more optimally shaped open pore. Likewise, pore size or shape would be proportionally greater in response to both Rpt peptides than to either alone. In all cases, the rate of peptide substrate entry would be proportional to pore size. This model might also explain the surprising synergistic effect of the Rpt peptides on hydrolysis of protein substrates if such substrates were too large to pass through Rpt2-opened pores but not Rpt5-opened pores. Thus, the Rpt2 peptide would be incompetent to stimulate hydrolysis of proteins but would enhance the effect of the Rpt5 peptide on this process. This speculative explanation is consistent with the differential degrees of radial displacement among α subunits when bound to PA700, as described above.
Although our results indicate that Rpt2 and Rpt5 are each sufficient for proteasome activation, they do not exclude roles for the other Rpt subunits in this process. Thus, Rpt2 and Rpt5 may provide sufficient affinity for PA700 binding and promote proteasome activation, but once bound, other PA700 subunits may also participate in proteasome activation. We note that in occasional experiments, we observed proteasome activation by a COOH-terminal peptide of Rpt1 but found this effect difficult to reproduce consistently; similar results were discussed by Smith and colleagues (28). Perhaps subtle differences in experimental conditions can affect binding and/or activation by this peptide. In contrast, other Rpt subunits may play roles in other 26 S proteasome functions. Differential roles of distinct AAA subunits in proteasome function have been suggested previously based on different cellular phenotypes caused by subunit-specific mutations (42).
The Rpt peptides and SUMO fusion proteins activate the proteasome in an ATP-independent fashion that presumably reflects their conformationally unrestrained ability to bind to cognate sites on the proteasome. In contrast, proteasome binding and activation by intact PA700 depends strictly on ATP binding to one or more of its AAA subunits. ATP binding probably promotes conformational changes in PA700 that alter the spatial orientation of the Rpt COOH termini for optimal interaction with the proteasome (Fig. 6). Additional biochemical and structural studies will be required to further define the basis of ATP-dependent binding of PA700 to the proteasome and to establish the relative roles of individual AAA subunits in proteasome binding and activation.
Supplementary Material
Acknowledgments
We thank Kerry Wooding for preparation and analysis of the Rpt2 recombinant protein.
This work was supported, in whole or in part, by National Institutes of Health Grant R01 DK46181. This work was also supported by Welch Foundation Grant I-500 (to G. N. D.). The costs of publication of this article were defrayed in part by the payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 U.S.C. Section 1734 solely to indicate this fact.
The on-line version of this article (available at http://www.jbc.org) contains supplemental Figs. S1–S4.
Footnotes
The abbreviations used are: SUMO, small ubiquitin-like modifier; AMC, 7-amino-4-methylcourmarin; HPLC, high pressure liquid chromatography; CHAPS, 3-[(3-cholamidopropyl)dimethylammonio]-1-propanesulfonic acid; HRP, horseradish peroxidase; Dopa, 3,4-dihydroxyphenylalanine.
References
- 1.Coux, O., Tanaka, K., and Goldberg, A. L. (1996) Annu. Rev. Biochem. 65 801-847 [DOI] [PubMed] [Google Scholar]
- 2.Voges, D., Zwickl, P., and Baumeister, W. (1999) Annu. Rev. Biochem. 68 1015-1068 [DOI] [PubMed] [Google Scholar]
- 3.Pickart, C. M., and Cohen, R. E. (2004) Nat. Rev. Mol. Cell Biol. 5 177-187 [DOI] [PubMed] [Google Scholar]
- 4.Gillette, T. G., and DeMartino, G. N. (2007) Cell 129 659-662 [DOI] [PubMed] [Google Scholar]
- 5.DeMartino, G. N., and Slaughter, C. A. (1999) J. Biol. Chem. 274 22123-22126 [DOI] [PubMed] [Google Scholar]
- 6.Baumeister, W., Walz, J., Zühl, F., and Seemüller, E. (1998) Cell 92 367-380 [DOI] [PubMed] [Google Scholar]
- 7.Bochtler, M., Ditzel, L., Groll, M., Hartmann, C., and Huber, R. (1999) Annu. Rev. Biophys. Biomol. Struct. 28 295-317 [DOI] [PubMed] [Google Scholar]
- 8.Groll, M., Bajorek, M., Kohler, A., Moroder, L., Rubin, D. M., Huber, R., Glickman, M. N., and Finley, D. (2001) Nat. Struct. Biol. 11 1062-1067 [DOI] [PubMed] [Google Scholar]
- 9.Groll, M., Ditzel, L., Lowe, J., Stock, D., Bochtler, M., Bartunik, H. D., and Huber, R. (1997) Nature 386 463-471 [DOI] [PubMed] [Google Scholar]
- 10.Köhler, A., Cascio, P., Leggett, D. S., Woo, K. M., Goldberg, A. L., and Finley, D. (2001) Mol. Cell 7 1143-1152 [DOI] [PubMed] [Google Scholar]
- 11.Ma, C.-P., Vu, J. H., Proske, R. J., Slaughter, C. A., and DeMartino, G. N. (1994) J. Biol. Chem. 269 3539-3547 [PubMed] [Google Scholar]
- 12.Groll, M., Bochtler, M., Brandstetter, H., Clausen, T., and Huber, R. (2005) Chembiochem. 6 222-256 [DOI] [PubMed] [Google Scholar]
- 13.Rechsteiner, M., Realini, C., and Ustrell, V. (2000) Biochem. J. 345 1-15 [PMC free article] [PubMed] [Google Scholar]
- 14.Ma, C.-P., Slaughter, C. A., and DeMartino, G. N. (1992) J. Biol. Chem. 267 10515-10523 [PubMed] [Google Scholar]
- 15.Dubiel, W., Pratt, G., Ferrell, K., and Rechsteiner, M. (1992) J. Biol. Chem. 267 22369-22377 [PubMed] [Google Scholar]
- 16.Gray, C. W., Slaughter, C. A., and DeMartino, G. N. (1994) J. Mol. Biol. 236 7-15 [DOI] [PubMed] [Google Scholar]
- 17.Song, X., von Kampen, J., Slaughter, C. A., and DeMartino, G. N. (1997) J. Biol. Chem. 272 27994-28000 [DOI] [PubMed] [Google Scholar]
- 18.Zhang, Z., Clawson, A., Realini, C., Jensen, C. C., Knowlton, J. R., Hill, C. P., and Rechsteiner, M. (1998) Proc. Natl. Acad. Sci. U. S. A. 95 2807-2811 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Forster, A., Masters, E. I., Whitby, F. G., Robinson, H., and Hill, C. P. (2005) Mol. Cell 18 589-599 [DOI] [PubMed] [Google Scholar]
- 20.Whitby, F. G., Masters, E. I., Kramer, L., Knowlton, J. R., Yao, Y., Wang, C. C., and Hill, C. P. (2001) Nature 408 115-120 [DOI] [PubMed] [Google Scholar]
- 21.Smith, D. M., Kafri, G., Cheng, Y., Ng, D., Wala, T., and Goldberg, A. L. (2005) Mol. Cell 20 687-698 [DOI] [PubMed] [Google Scholar]
- 22.Liu, C. W., Li, X., Thompson, D., Wooding, K., Chang, T., Tang, Z., Yu, H., Thomas, P. J., and DeMartino, G. N. (2006) Mol. Cell 24 39-50 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Beyer, A. (1997) Protein Sci. 6 2043-2058 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ogura, T., and Wilkinson, A. J. (2001) Genes Cells 6 575-597 [DOI] [PubMed] [Google Scholar]
- 25.DeMartino, G. N., Moomaw, C. R., Zagnitko, O. P., Proske, R. J., Ma, C.-P., Afendis, S. J., Swaffield, J. C., and Slaughter, C. A. (1994) J. Biol. Chem. 269 20878-20884 [PubMed] [Google Scholar]
- 26.Glickman, M. H., Rubin, D. M., Coux, O., Wefes, I., Pfeifer, G., Cjeka, Z., Baumeister, W., Fried, V., and Finley, D. (1998) Cell 94 615-623 [DOI] [PubMed] [Google Scholar]
- 27.Glickman, M. H., Rubin, D. M., Fried, V. A., Fischer, J. E., and Finley, D. (1998) Mol. Cell Biol. 18 3149-3162 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Smith, D. M., Chang, S. C., Park, S., Finley, D., Cheng, Y., and Goldberg, A. L. (2007) Mol. Cell 27 731-744 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Rabl, J., Smith, D. M., Yu, Y., Chang, S. C., Goldberg, A. L., and Cheng, Y. (2008) Mol. Cell 30 360-368 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.DeMartino, G. N. (2005) Methods Enzymol. 398 295-306 [DOI] [PubMed] [Google Scholar]
- 31.McGuire, M. J., McCullough, M. L., Croall, D. E., and DeMartino, G. N. (1989) Biochim. Biophys. Acta 995 181-186 [DOI] [PubMed] [Google Scholar]
- 32.Koulich, E., Li, X., and DeMartino, G. N. (2008) Mol. Biol. Cell 19 1072-1082 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.McGuire, M. J., and DeMartino, G. N. (1986) Biochim. Biophys. Acta 873 279-289 [DOI] [PubMed] [Google Scholar]
- 34.Elsasser, S., Schmidt, M., and Finley, D. (2005) Methods Enzymol. 398 353-363 [DOI] [PubMed] [Google Scholar]
- 35.Burdine, L., Gillette, T. G., Lin, H. J., and Kodadek, T. (2004) J. Am. Chem. Soc. 126 11442-11443 [DOI] [PubMed] [Google Scholar]
- 36.Liu, B., Burdine, L., and Kodadek, T. (2006) J. Am. Chem. Soc. 128 15228-15235 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Mykles, D. L., and Haire, M. F. (1991) Arch. Biochem. Biophys. 288 543-551 [DOI] [PubMed] [Google Scholar]
- 38.Bajorek, M., Finley, D., and Glickman, M. H. (2003) Curr. Biol. 13 1140-1144 [DOI] [PubMed] [Google Scholar]
- 39.Ferrell, K., Wilkinson, C. R. M., Dubiel, W., and Gordon, C. (2000) Trends Biochem. Sci. 25 83-88 [DOI] [PubMed] [Google Scholar]
- 40.Hoffman, L., Pratt, G., and Rechsteiner, M. (1992) J. Biol. Chem. 267 22362-22368 [PubMed] [Google Scholar]
- 41.da Fonseca, P. C., and Morris, E. P. (2008) J. Biol. Chem. 283 23305-23314 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Rubin, D. M., Glickman, M. H., Larsen, C. N., Druvakumar, S., and Finley, D. (1998) EMBO J. 17 4909-4919 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.