Abstract
Background
OTK18 is a C2H2 type zinc finger protein expressed by human macrophages following HIV infection. OTK18 possesses antiretroviral activity and its processing products accumulate in the cytoplasm of perivascular brain macrophages in advanced HIV encephalitis cases.
Objectives
Since the regulation of OTK18 expression in living patients following HIV-1 infection is unknown, our objective is to investigate the first cohort study on OTK18 protein levels in living patients.
Study Design
We assessed OTK18 levels in plasma and cerebrospinal fluid (CSF) in 44 living patients with or without HIV-1 infection with diverse demographic and clinical background. A novel high-sensitivity OTK18 ELISA system was developed to measure OTK18 levels in CSF and plasma using custom made biotinylated monoclonal antibodies against OTK18. The correlation of OTK18 levels with epidemiological parameters was statistically analyzed.
Results
Multiple linear regression modeling suggested that plasma OTK18 levels for HIV-1-positive subjects were only about one-sixth of that for HIV-1-negative subjects. Higher CD8 T-cell counts were associated with higher levels of OTK18. Using proportional odds logistic regression, we showed that HIV-1-positive patients have significantly lower OTK18 in CSF samples, but we did not observe significant correlation between CD8 T-cell counts and CSF OTK18 levels.
Conclusion
OTK18 levels in both plasma and CSF are significantly lower in HIV-1-positive subjects as compared to HIV-1-negatives. Plasma OTK18 levels are positively correlated to CD8 T-cell counts, independent of HIV-1 status.
Keywords: HIV-1, OTK18, CSF, CD4, CD8, plasma
1. Introduction
Human immunodeficiency virus-1 (HIV-1) infects and is disseminated throughout the body by CD4+ T-cells and mononuclear phagocytes (MP), which include infiltrating perivascular and resident tissue macrophages and microglia (Hussain and Lehner, 1995; Gendelman et al., 1997). Infection of brain MP plays a central role in the pathogenesis of HIV-1 associated dementia (HAD) and HIV-1 encephalitis (HIVE), the pathological correlate of HAD. Generally, HIV-1 replication is under tight control in the brain for years following the initial infection (Navia et al., 1986). What initiates the high level of viral replication seen late in the course of the disease is unknown. Neurological disease is likely to be the result of complex interactions between viral and host cellular factors.
OTK18, a protein produced during HIV-1 infection of MP, suppresses HIV-1 replication through the viral LTR (Carlson et al., 2004a) and may play an important role in the interaction of viral and host cell factors leading to HAD and HIVE. OTK18 contains 13 C2H2-type zinc fingers and is characterized as a repressive transcription factor (Saito et al., 1996). OTK18's repressive activity lies within amino acids 26−89 located within a conserved Krüppel associated box (KBAB), a repression domain encoded by a number of transcription factors. OTK18 gene expression is inducible by HIV-1 infection in macrophages but is tightly regulated and is only detectable in the cytoplasm of brain macrophages in severe HIVE brains (Carlson et al., 2004b). This observation suggests that HIV-1 induced OTK18 expression in HIVE brains might be used as a surrogate marker for HAD. In this paper we developed an ELISA for OTK18 detection to examine OTK18 levels in both plasma and CSF collected from uninfected and HIV+ patients. Our results indicate that OTK18 levels decrease in HIV+ patients in both the plasma and CSF. Furthermore decreased OTK18 levels correlate with decreased CD8+ cell count in the plasma independent of HIV infection.
2. Methods
2.1. Study Population
Twenty-three HIV-1-seronegative and twenty-one HIV-1-seropositive subjects paired plasma and CSF samples were obtained at University of California San Francisco. Paired CSF and plasma samples were derived from participants in 2 longitudinal cohort studies approved by the University of California-San Francisco Committee on Human Research. Informed consent was obtained from all subjects (and from the durable power of attorney of an individual with stage 2 ADC). The CDC stages of the HIV+ patients were all 0 except one case, which was 2. Subjects with active CNS diseases other than ADC were excluded (Spudich et al., 2005a).
2.2. Plasma and CSF Processing and Analysis
The CSF and plasma were collected at the same time and were obtained from a depository for general study purposes and were processed in standardized fashion as previously described (Price and Deeks, 2004). The specimens were kept frozen at −80°C and stored for less than 5 years. To avoid the repeated freezing of the samples, aliquots were made at the initial defrost. CSF and plasma were analyzed for HIV RNA concurrently. CSF cell counts, CD4+ and CD8+ T lymphocyte counts, as well as differential protein and albumin blood levels were measured by flow cytometry using routine clinical methods in the San Francisco General Clinical Laboratory. HIV-1 RNA levels were measured in cell-free CSF and plasma using Roche Amplicor HIV-1 Monitor assay (versions 1.0 and 1.5; Roche Diagnostic systems) with standard and ultrasensitive extraction methods, as appropriate, according to the manufacturer's guidelines. HIV-1 RNA concentrations were transformed to log10 values for all calculations (Spudich et al., 2005a; Spudich et al., 2005b).
2.3. OTK18 Monoclonal Antibody (mAb) Biotinylation
OTK18 mAbs were generated by S.C. injection of His-tagged OTK18 (1−178) truncated recombinant protein purified from E. coli as described (Carlson et al., 2004a). OTK18 monoclonal antibody 211−3.2−1 (5 mg) was biotinylated using EZ-Link Sulfo-NHS-Biotinylation kit (Pierce Biotechnology). The efficiency of the biotinylation was 3.72 moles biotin per mole of OTK18 mAb.
2.4. Luminescent OTK18 ELISA
Microtiter plates were coated with anti-OTK18 monoclonal antibody (211−3.1−1, 100ng/well), blocked with blocking buffer (TBST with 3% Blocker BSA in TBS, Pierce Biotechnology), and incubated with 100 μl of standards prepared in PBS or 100 μl of plasma or CSF to wells and add 20 μl of 6X blocking buffer (final is TBST with 1% Blocker BSA) overnight 4°C. After washing, 100 μl of biotinylated antibody (211−3.2−1 at 1.6 μg/ml) was added and incubated for 2 hours at room temperature. After washing, 100 μl of streptavidin-HRP (R & D Systems, diluted 1/200 in blocking buffer) and incubated for 20 minutes at room temperature in darkness. After washing, 100 μl of SuperSignal ELISA Pico Chemiluminescent Substrate (Pierce Biotechnology) to each well, and luminescence signal was quantified for 5 seconds using Microplate Luminometer LB 96V (Perkin Elmer).
2.5. Z’ factor calculation
Z’ factor was calculated as per Zhang and others (Z’ = 1 – [(3σc+ + 3σc–)/(μc+ – μc–)], where σc+ and σc– are the standard deviations of positive and negative controls, and (μc+ and μc– are the mean values of positive and negative controls) (Zhang et al., 1999).
2.6. Statistical Analyses
Pearson's correlation coefficient and scatter plot were first used to get an overview of the relationships between response and predictors. Multiple linear and logistic regressions (Kleinbaum et al., 1997; Hosmer, 2000)were applied to analyze plasma and CSF data, respectively, to study the effects of predictors on the level of OTK18 (Hosmer, 2000; Kleinbaum et al., 1997). The procedures of stepwise model selection were used for both models to choose the “best” model to explain the relationship between response and predictors. Additional statistical techniques were applied to the two samples to deal with observations below the detectable limit (BDL), ranging between 0 and 10. For plasma data, there was one BDL observation, and we replaced its value with the average which was 5. We then transformed the response into a logarithm scale in order to normalize the distribution. For CSF data, there were 14 BDL observations, and we categorized them into one level of response and modeled the data using logistic regression.
3. Results
3.1. Development of the OTK18 ELISA
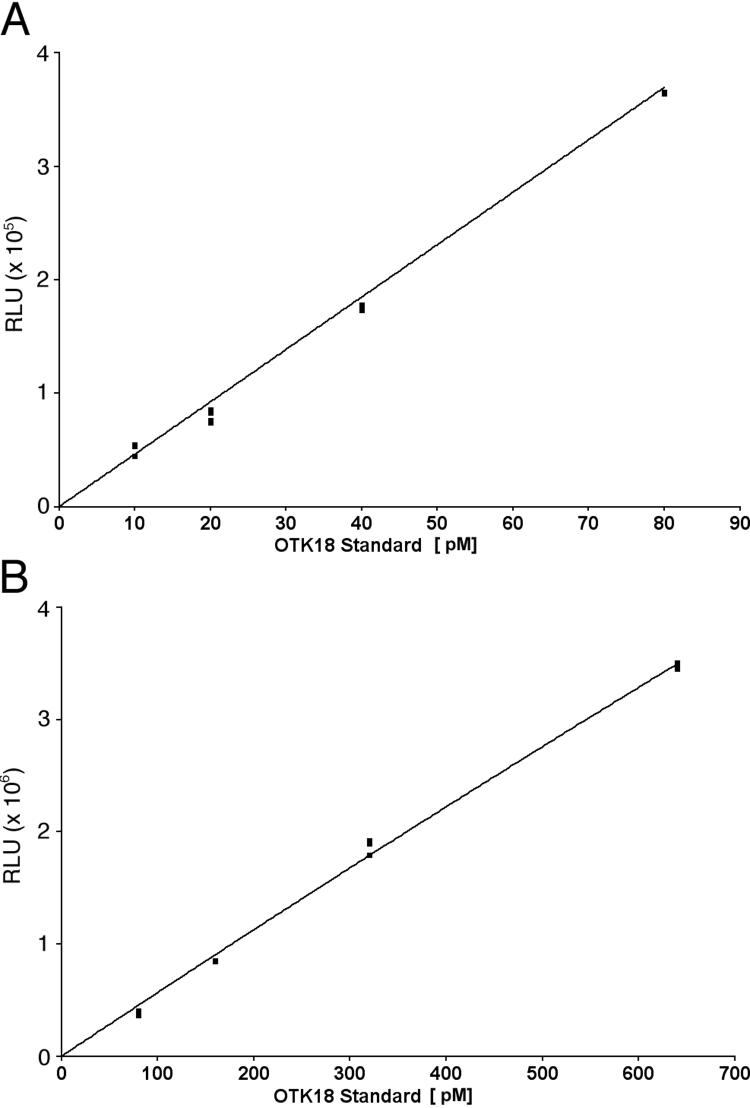
The luminescent ELISA assay for quantification of OTK18 in plasma and CSF was optimized with purified OTK18 standard protein. The assay was performed against a standard curve of OTK18(1−178) with increasing concentrations of both capturing antibody and detection antibody. The resulting standard curve is biphasic and for the purposes of OTK18 quantification is split into a low and a high standard curve (Figure 1A and B, respectively). The standard curve can typically quantitate OTK18 levels from a 10 to 640 pM range with a signal to noise ratio of 1.68 to 73.13 and a Z’ factor of .07 to .97. These results indicate that this ELISA assay provides sensitivity and specificity sufficient for the clinical study.
Figure 1. OTK18 ELISA Standard Curves.
Breakdown of the biphasic standard curve into a low (A) and a high (B) standard curves which have R2 value of 0.998 and 0.999, respectively. RLU: relative intensity unit.
3.2. Plasma OTK18 levels and demographic data analysis
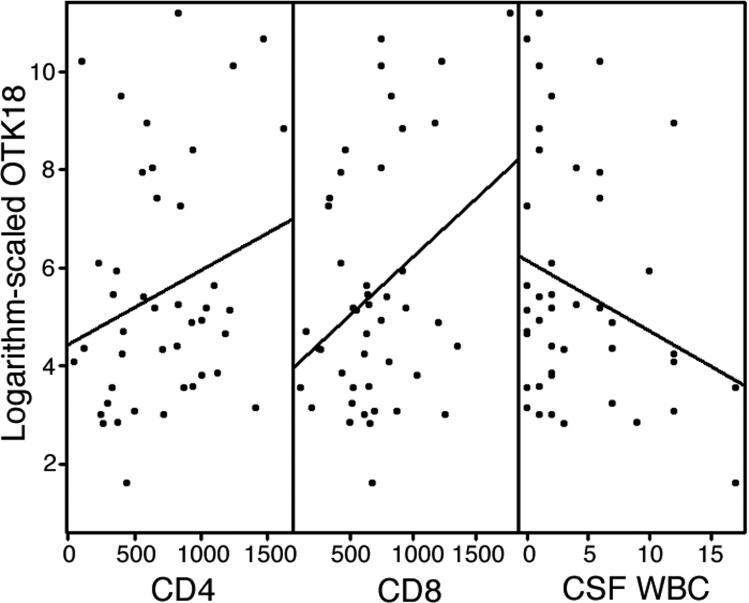
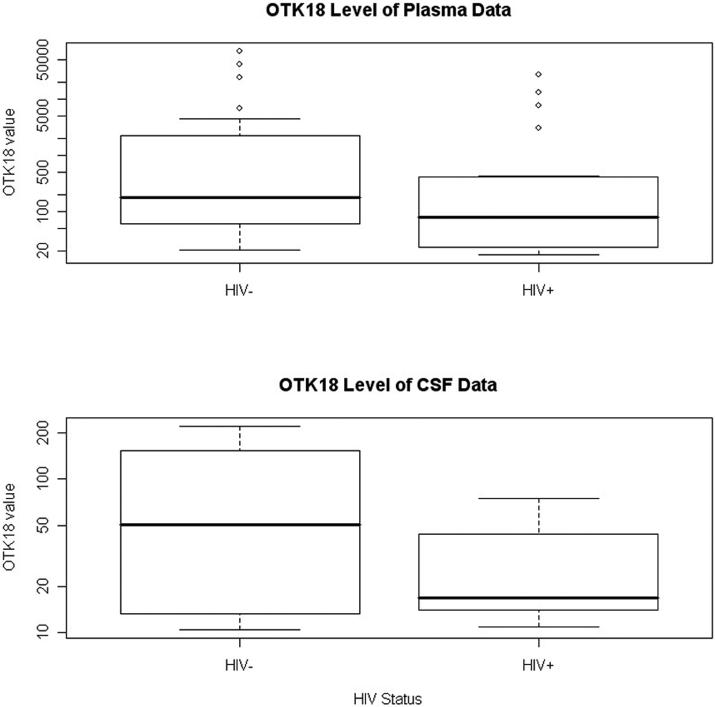
The mean age (SD) for the 44 subjects is 42.9 (7.6) years. There are 37 males and 7 females. Table 4 shows the summary statistics of age and gender for HIV+ and HIV- subjects separately. Pearson's correlation coefficient and scatter plot were used to create an overview of the relationship between response and predictors. These methods show that log2(OTK18) is correlated with HIV status, CD4 value, CD8 value and WBC value and are presented as scatter plots (See Table 1 and Figure 2). Both CD4 and CD8 show positive relationships with OTK18 while HIV status and WBC have negative relationships with OTK18. In particular, for HIV+ vs. HIV- the mean values of OTK18 are 141.1 vs. 407.5 pM and the medians are 77.5 vs. 177.7 pM, respectively (Figure 3).
Table 4.
Demographic information
| HIV+ | HIV− | |
|---|---|---|
| Sample size | 21 | 23 |
| Age: mean±SD | 41.2±5.3 | 44.5±9.0 |
| Gender: # male (%) | 19 (90.5%) | 18 (78.3%) |
Table 1.
Correlation matrix between response and predictor variables for plasma dataset.
| Age | Gender | HIV | ART | CD4 | CD8 | WBC | P-CSF | OTK18 | |
|---|---|---|---|---|---|---|---|---|---|
| Age | 1 | −.03 | −.26 | −.02 | 0.18 | −.07 | −.15 | 0.12 | −.07 |
| Gender | 1 | −.17 | −.02 | 0.26 | −.08 | −.17 | −.34 | −.02 | |
| HIV | 1 | NA | −.67 | 0.26 | 0.61 | 0.03 | −.22 | ||
| ART | 1 | −.49 | −.35 | −.03 | 0.45 | 0.08 | |||
| CD4 | 1 | −.03 | −.56 | −.16 | 0.24 | ||||
| CD8 | 1 | 0.11 | 0.07 | 0.33 | |||||
| WBC | 1 | 0.15 | −.28 | ||||||
| P-CSF | 1 | −.08 | |||||||
| OTK18 | 1 |
ART: currently under antiretroviral treatment, P-CSF; Protein CSF.
Figure 2. Scatter plots of logarithm-scaled OTK18 with the values of CD4, CD8 and WBC.
A linear regression line is also drawn for each plot.
Figure 3. Boxplot of OTK18 with the status of HIV for plasma and CSF datasets.
Please note that the y-axes are displayed in a logarithm scale for both plots.
Next, we applied multiple linear regression with stepwise model selection for analysis of plasma data. After adjusting for age (categorized as two groups: ≥45 and <45) and gender, we found HIV status and CD8 value to be the only predictors to significantly explain OTK18 measurements (Table 2). In particular, OTK18 levels of HIV+ subjects are estimated to be about 16.4% (95% C.I. is from 3.6% to 76.5%) of OTK18 levels of HIV− subjects. OTK18 levels increase by about 30% when CD8 counts increase by 100 on average. The p-values of HIV status and CD8 counts in the model are 0.02 and 0.007, respectively.
Table 2.
Results of Linear Regression Analysis on plasma data after stepwise model selection and controlling for the effects of age (categorized as two groups: ≥45 and <45) and gender.
| Predictor | Parameter (95% C.I.) | P value |
|---|---|---|
| HIV (+ vs. −) | 0.164 ( (0.036, 0.735) | 0.020 |
| CD8 | 1.003 (1.0009, 1.005) | 0.007 |
3.3. CSF OTK18 levels and demographic data analysis
For the CSF samples, the histogram of OTK18 shows that OTK18 distribution is a bimodal function on the logarithm scale and that the cutoff is around 3.4 (30 pM). In addition, over one-third of OTK18 observations have missing values due to BDL. A common statistical technique is to model these BDL observations together as a single level of the response. Due to these reasons, we categorized the values of OTK18 into three ordered levels: ≥30, <30 and ≥10, and <10, with the respective number of observations for each level at 13, 17 and 14. Fitting a proportional odds model through logistic regression, we found that the status of HIV is the only predictor that can significantly predict the CSF OTK18 level, after adjusting for age and gender. The chi-square value for testing the proportional odds assumption indicates this model adequately fits the data. Our results show that the estimated cumulative odds ratio of having higher OTK18 levels for HIV+ and HIV- subjects is 0.152 and the 95% C.I. is (0.042, 0.557). Thus, the odds for HIV+ subjects to have OTK18 level ≥30 are 15.2% of that for HIV− subjects on average. In particular, for HIV+ vs. HIV- the mean values of OTK18 are 27.2 vs. 84.7 pM, and the medians are 16.9 vs. 50.5 pM, respectively (Figure 3). Table 3 shows the frequency of subjects with different HIV status and the categorized OTK18 levels. Specifically, we can see that HIV+ subjects are much less likely to have an OTK18 level of 30 or higher (14%) than are subjects who are HIV− (43%) (p = 0.03 by Chi-square test). Please note the predictor of CD8 value is not significant for this model (p-value = 0.40 in the model).
Table 3.
Frequency of subjects with different HIV status and CSF OTK18 level.
| HIV− (%) | HIV+ (%) | |
|---|---|---|
| OTK18 < 30 | 13 (57%) | 18 (86%) |
| OTK18 ≥ 30 | 10 (43%) | 3 (14%) |
4. Discussion
Our results indicate that plasma OTK18 levels correlate best with CD8 counts and HIV status, while CSF levels correlate best with HIV status. This study indicates that CD8+ T-cells may play an additional role in the expression of OTK18 regulation. Antigen-specific CD8+ cytotoxic T-cells serve a prominent role in the control of HIV infection in both brain and peripheral tissues (Sewell et al., 2000). Since we have observed OTK18 upregulation in brain perivascular macrophages in advanced HIV encephalitis (Carlson et al., 2004b), it is possible that CD8+ T-cells infiltrate into the brain perivascular region and locally stimulate macrophages to express OTK18. It will be of interest to examine the neuropathological correlation of OTK18 expression in brain macrophages and CD8+ T-cells in HIV encephalitic cases.
Correlation of OTK18 levels with HIV status was predicted since OTK18 was originally cloned as HIV-1-inducible C2H2 type zinc finger protein from monocyte-derived macrophages (Carlson et al., 2004a). However, we did not predict that OTK18 levels would be negatively correlated with HIV status. These results suggest additional regulation for OTK18 expression in other peripheral tissues in vivo. We have previously shown that OTK18 mRNA is expressed in multiple tissues, including kidney, liver, heart, skeletal muscle, placenta, but not in peripheral blood leukocytes (Carlson et al., 2004b). Thus it is possible that OTK18 is normally secreted from other tissues and is down-regulated upon chronic HIV infection. In addition, multi-institutional international Human Plasma Proteome Project identified 9052 proteins in plasma samples, of which 196 are zinc finger proteins, accounting 2.2% of total proteins in plasma (Omenn et al., 2005; Ping et al., 2005). This report strongly supports our data that zinc finger proteins can be detected in human plasma. Further investigation is necessary to identify peripheral OTK18 secretion and its regulation by HIV-1 infection.
Acknowledgment
We would like to thank Drs. J. Anderson and H. Gendelman for suggestions and critical reading of the manuscript, and Russell Swan and Meg Maquart for manuscript editing. This work is supported in part by NIH Grants R01 AI5089401, R01 MH072539, and NCRR P20RR15635 (T.I.).
References
- Carlson K, Leisman G, Limoges J, Pohlman G, Horiba M, Buescher J, Gendelman H, Ikezu T. Molecular Characterization of a Putative Anti-Retroviral Transcriptional Factor, OTK18. J Immunol. 2004a;172:381–391. doi: 10.4049/jimmunol.172.1.381. [DOI] [PubMed] [Google Scholar]
- Carlson KA, Limoges J, Pohlman GD, Poluektova LY, Langford D, Masliah E, Ikezu T, Gendelman HE. OTK18 expression in brain mononuclear phagocytes parallels the severity of HIV-1 encephalitis. J Neuroimmunol. 2004b;150:186–198. doi: 10.1016/j.jneuroim.2004.01.021. [DOI] [PubMed] [Google Scholar]
- Gendelman HE, Persidsky Y, Ghorpade A, Limoges J, Stins M, Fiala M, Morrisett R. The neuropathogenesis of the AIDS dementia complex. Aids. 1997;11:S35–45. [PubMed] [Google Scholar]
- Hosmer DW, Lemeshow S. Applied Logistic Regression. Wiley Series in Probability and Statistics. 2000 [Google Scholar]
- Hussain LA, Lehner T. Comparative investigation of Langerhans’ cells and potential receptors for HIV in oral, genitourinary and rectal epithelia. Immunology. 1995;85:475–484. [PMC free article] [PubMed] [Google Scholar]
- Kleinbaum D, Kupper L, Muller K, Nizam A. Applied Regression Analysis and Other Multivariable Methods. 3rd Edition Duxbury Press; Pacific Grove, CA: 1997. [Google Scholar]
- Navia BA, Jordan BD, Price RW. The AIDS dementia complex: I. Clinical features. Ann Neurol. 1986;19:517–524. doi: 10.1002/ana.410190602. [DOI] [PubMed] [Google Scholar]
- Omenn GS, States DJ, Adamski M, Blackwell TW, Menon R, Hermjakob H, Apweiler R, Haab BB, Simpson RJ, Eddes JS, Kapp EA, Moritz RL, Chan DW, Rai AJ, Admon A, Aebersold R, Eng J, Hancock WS, Hefta SA, Meyer H, Paik YK, Yoo JS, Ping P, Pounds J, Adkins J, Qian X, Wang R, Wasinger V, Wu CY, Zhao X, Zeng R, Archakov A, Tsugita A, Beer I, Pandey A, Pisano M, Andrews P, Tammen H, Speicher DW, Hanash SM. Overview of the HUPO Plasma Proteome Project: results from the pilot phase with 35 collaborating laboratories and multiple analytical groups, generating a core dataset of 3020 proteins and a publicly-available database. Proteomics. 2005;5:3226–3245. doi: 10.1002/pmic.200500358. [DOI] [PubMed] [Google Scholar]
- Ping P, Vondriska TM, Creighton CJ, Gandhi TK, Yang Z, Menon R, Kwon MS, Cho SY, Drwal G, Kellmann M, Peri S, Suresh S, Gronborg M, Molina H, Chaerkady R, Rekha B, Shet AS, Gerszten RE, Wu H, Raftery M, Wasinger V, Schulz-Knappe P, Hanash SM, Paik YK, Hancock WS, States DJ, Omenn GS, Pandey A. A functional annotation of subproteomes in human plasma. Proteomics. 2005;5:3506–3519. doi: 10.1002/pmic.200500140. [DOI] [PubMed] [Google Scholar]
- Price RW, Deeks SG. Antiretroviral drug treatment interruption in human immunodeficiency virus-infected adults: Clinical and pathogenetic implications for the central nervous system. J Neurovirol. 2004;10(Suppl 1):44–51. doi: 10.1080/753312752. [DOI] [PubMed] [Google Scholar]
- Saito H, Fujiwara T, Takahashi EI, Shin S, Okui K, Nakamura Y. Isolation and mapping of a novel human gene encoding a protein containing zinc-finger structures. Genomics. 1996;31:376–379. doi: 10.1006/geno.1996.0062. [DOI] [PubMed] [Google Scholar]
- Sewell AK, Price DA, Oxenius A, Kelleher AD, Phillips RE. Cytotoxic T lymphocyte responses to human immunodeficiency virus: control and escape. Stem Cells. 2000;18:230–244. doi: 10.1634/stemcells.18-4-230. [DOI] [PubMed] [Google Scholar]
- Spudich SS, Huang W, Nilsson AC, Petropoulos CJ, Liegler TJ, Whitcomb JM, Price RW. HIV-1 chemokine coreceptor utilization in paired cerebrospinal fluid and plasma samples: a survey of subjects with viremia. J Infect Dis. 2005a;191:890–898. doi: 10.1086/428095. [DOI] [PubMed] [Google Scholar]
- Spudich SS, Nilsson AC, Lollo ND, Liegler TJ, Petropoulos CJ, Deeks SG, Paxinos EE, Price RW. Cerebrospinal fluid HIV infection and pleocytosis: Relation to systemic infection and antiretroviral treatment. BMC Infect Dis. 2005b;5:98. doi: 10.1186/1471-2334-5-98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang JH, Chung TD, Oldenburg KR. A Simple Statistical Parameter for Use in Evaluation and Validation of High Throughput Screening Assays. J Biomol Screen. 1999;4:67–73. doi: 10.1177/108705719900400206. [DOI] [PubMed] [Google Scholar]