Summary
During cytochrome c maturation (Ccm), the DsbA-dependent thio-oxidative protein-folding pathway is thought to introduce a disulfide bond into the heme-binding motif of apocytochromes c. This disulfide bond is believed to be reduced through a thio-reductive pathway involving the Ccm components CcdA (DsbD), CcmG and CcmH. Here, we show in Rhodobacter capsulatus that in the absence of DsbA cytochrome c levels were decreased and CcdA or CcmG or the putative glutathione transporter CydDC were not needed for Ccm. This decrease was not due to overproduction of the periplasmic protease DegP as a secondary effect of DsbA absence. In contrast, CcmH was absolutely necessary regardless of DsbA, indicating that compensatory thio-redox interactions excluded it. Remarkably, the double (DsbA-CcmG) and triple (DsbA-CcmG-CcdA) mutants produced cytochromes c at lower levels than the DsbA-null mutants, unless they contained a CcmG derivative (CcmG*) lacking its thio-reductive activity. Purified CcmG* can bind apocytochrome c in vitro, revealing for the first time a thiol-independent, direct interaction between apocytochrome c and CcmG. Furthermore, elimination of the thio-redox components does not abolish cytochrome c production, restricting the number of Ccm components essential for heme-apocyt c ligation per se during Ccm.
Keywords: cytochrome c maturation, thio-oxidation, thio-reduction, Rhodobacter capsulatus, photosynthetic and respiratory electron transfer
Introduction
The cytochromes (cyts) c are membrane-attached or soluble ubiquitous electron carrier proteins essential for energy metabolisms of almost all organisms. They contain Fe-protoporphyrin IX (heme), which is stereo-specifically and covalently attached through two thioether bonds between the heme vinyl groups and the cysteine sulfhydryls of the apocyt c C1XXC2H motif. The covalent heme-apocyt c ligation is carried out by a post-translation and post-export process called cyt c maturation (Ccm), which involves in most gram-negative bacteria ten essential components (CcmABCDEFGHI and CcdA (or DsbD) (Barker and Ferguson, 1999; Thony-Meyer, 2000). During Ccm, it is thought that the cysteine thiols of apocyt c C1XXC2H motif are first oxidized by the DsbA-dependent extracytoplasmic thio-oxidative protein-folding pathway (Fig. 1) (Allen et al., 2003b; Kranz et al., 1998; Thony-Meyer, 2002). Subsequent reduction of this disulfide bond then becomes a prerequisite for heme ligation (Daltrop et al., 2002). The reduction is accomplished by a thio-reductive pathway consisting of CcdA, CcmG and CcmH (Monika et al., 1997). In Rhodobacter capsulatus, CcdA transfers electrons from cytoplasmic thioredoxins to CcmG (Katzen and Beckwith, 2000; Katzen et al., 2002). CcmG is believed to be re-oxidized by shuttling its electrons to CcmH, which then transfers them to apocyts c (Fig. 1) (Fabianek et al., 1999; Monika et al., 1997).
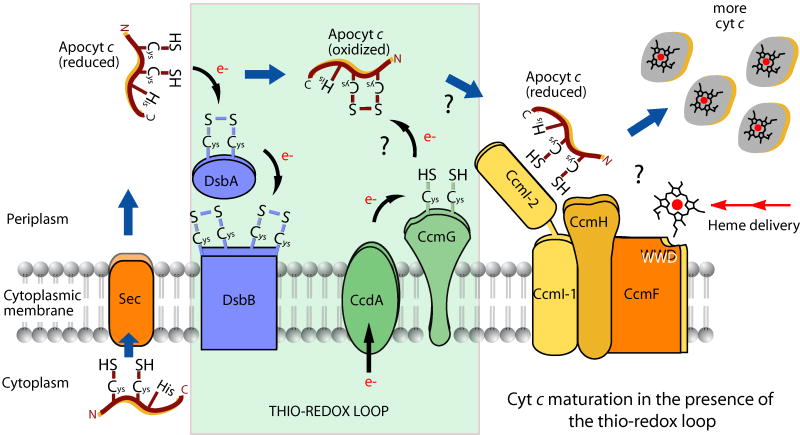
Fig. 1. Production of cyts c in the presence of the Ccm thio-redox loop.
In a wild type background, the cysteine thiols of the C1XXC2H motif of reduced apocyts c, secreted through the general secretion pathway (Sec), are oxidized via the DsbA and DsbB dependent thio-oxidation pathway. Oxidized apocyts c are then reduced by the CcdA and CcmG dependent thio-reduction pathway, and conveyed to the CcmFHI heme ligation components that use heme from the heme delivery pathway (double arrows) to yield mature cyts c. Thick arrows show the route followed by apocyts c, and Cys, His, N and C refer to cysteine, histidine, N-terminal and C-terminal of apocyts c, respectively, and WWD the tryptophan rich motif of CcmF component of Ccm.
Formation of a disulfide bond in the apocyt c C1XXC2H motif occurs in vitro, and is considered to be an intermediate of Ccm (Daltrop et al., 2002). However, whether this is always the case in vivo is unclear. Initially, DsbA and DsbB mutants in E. coli were identified as unable to produce cyts c (Metheringham et al., 1996). Recent works in E. coli (Allen et al., 2003a; Kojima et al., 2005) and other bacteria (Erlendsson and Hederstedt, 2002: Deshmukh et al., 2003) indicated that DsbA- and DsbB-null mutants are Ccm proficient. Furthermore, the lack of DsbA or DsbB functions suppresses the cyt c deficiency of CcdA-null mutants in B. subtilis or R. capsulatus (Deshmukh et al., 2003; Erlendsson and Hederstedt, 2002), demonstrating that the thio-reductive branch becomes dispensable in the absence of the thio-oxidative pathway. Therefore, why a thio-redox loop (i. e., thio-oxidation followed by thio-reduction) occurs at the apocyt c heme binding motifs during Ccm remains unclear (Fig. 1). Here, we examined cyt c production without such a thio-redox loop in R. capsulatus. We found that mutants lacking DsbA produced less cyts c, but this decrease was not due to the overproduction of periplasmic protease DegP that occurs in the absence of DsbA in R. capsulatus (Onder et al., 2008). Moreover, when the DsbA-dependent thio-oxidative pathway was compromised, only CcmH, but neither CcdA nor CcmG, was required for heme-apocyt c ligation. The compensatory thio-redox interactions were also independent of the glutathione transporter CydDC (Pittman et al., 2005), unlike in E. coli (Poole et al., 1994). Remarkably though, the DsbA-CcmG double and DsbA-CcmG-CcdA triple mutants produced consistently lower levels of cyt c than the DsbA-null mutants, unless they contained a CcmG derivative devoid of its thio-reductive activity. This finding revealed a second role of CcmG acting as an apocyt c holdase during Ccm in addition to its thio-reductive function. Our findings show that the thio-redox pathway is important, but not essential, for cyt c production, and reduce the complex Ccm apparatus to those that are absolutely required for heme-apocyt c ligation per se in R. capsulatus.
Results
R. capsulatus mutants lacking DsbA produce lower amounts of cyts c
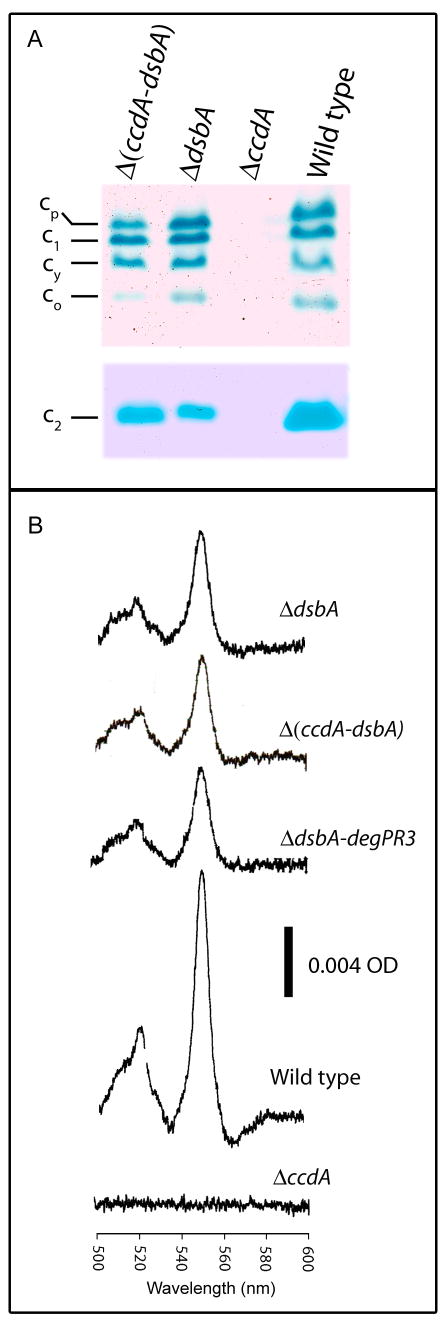
R. capsulatus mutants lacking DsbA are proficient for Ccm, unlike those lacking CcdA (Deshmukh et al., 2000). To further assess the role of DsbA during Ccm, we compared the total amounts of periplasmic soluble and membrane-associated cyts c produced in a wild type strain of R. capsulatus with an isogenic DsbA-null mutant (Fig. 1). Qualitative SDS-PAGE/TMBZ analyses showed quasi-wild type like cyts c profiles in DsbA-null mutants (Fig. 2A) (Deshmukh et al., 2003). Although this staining technique is very useful to monitor the overall cyts c profiles, the band intensities are time-dependent and may not be directly proportional to cyts c amounts. Thus, we used optical redox difference spectroscopy to obtain more quantitative estimations. Ascorbate-reduced minus ferricyanide-oxidized spectra recorded between 500 and 600 nm using membranes and membrane supernatants revealed that supernatant fractions of a DsbA-null mutant contained twice less (∼ 40 % per 0.5 mg /ml of total proteins) 550 nm absorbing materials relative to a wild type strain (Fig. 2B). In R. capsulatus, this absorbance reflects almost exclusively the characteristic α-band of cyt c2 (Daldal et al., 1986). Using membrane fractions representing the pool of membrane-associated cyts c that comprises of the cyts c1, cy, co and cp (Davidson and Daldal, 1987; Gray et al., 1994; Koch et al., 1998; Zannoni and Daldal, 1993) similar spectral data were also obtained (not shown). Furthermore, optical spectra acquired with a Ccm proficient mutant lacking both CcdA and DsbA revealed that the amounts of cyts c produced were similar to those seen in the absence of DsbA (Fig. 2B, top and bottom two spectra). We therefore concluded that, although the absence of DsbA did not abolish covalent heme-apocyt c ligation per se, it decreased cyt c production.
Fig. 2. Ccm thio-redox loop and related mutants.
(A) Cyt c profiles of the Ccm thio-redox loop mutants. Chromatophore membranes were prepared from R. capsulatus wild type (MT1131), ΔccdA (MD1), ΔdsbA (MD20) and Δ(ccdA-dsbA) (MD21) strains grown by respiration in minimal medium. Approximately 100 μg of total proteins were separated by SDS-PAGE, and cyts c profiles were visualized by TMBZ staining as described in Material and Methods. Upper and lower panels correspond to the membrane and soluble fractions, respectively. The membrane bound cyts cp, c1, cy, co and the periplasmic cyt c2 are indicated on the left. (B) Cyt c2 levels in the Ccm thio-redox loop mutants. Ascorbate-reduced minus ferricyanide-oxidized optical difference spectra of soluble fractions (protein concentration of 0.5 mg/ml) obtained from R. capsulatus wild type, ΔccdA, ΔdsbA, Δ(ccdA-dsbA) and ΔdsbA-degPR3 strains, as described in Materials and Methods and Table 2.
Neither the overproduction of DegP in the absence of DsbA nor the lack of CydDC affects cyt c production in R. capsulatus
Recently, it was reported that an E. coli mutant lacking the periplasmic protease DegP accumulates the heterologously expressed Bradyrhizobium japonicum apocyt c550 when the Ccm system is defective (Gao and O'Brian, 2007). In an earlier work, we found that R. capsulatus mutants lacking DsbA overproduce DegP, and that DsbA derivatives containing low amounts of DegP are viable (Onder et al., 2008). We therefore investigated whether the decreased amounts of cyts c in mutants lacking DsbA or DsbA CcdA were due to the overproduction of DegP. Comparison of a DsbA mutant (overproducing DegP) with its derivative harboring very low amounts of DegP (degPR3) showed that both strains produced similar amounts of cyts c (Fig. 2B, middle spectrum). Similar data were also obtained with DegP derivatives of other Ccm mutants (Table 1). Thus, the observed decrease of cyt c amounts in the absence of DsbA or the entire thio-redox pathway was unrelated to DegP overproduction in this mutant background.
Table 1. Ps growth properties and cyt c2 contents of R. capsulatus thio-redox mutants.
| Pathway | Strain | Plasmid | 1Med A | Med A* | MPYE | MPYE* | Cyt c2 |
|---|---|---|---|---|---|---|---|
| 2Ps | Ps | Ps | Ps | (%) | |||
| Wild Type | + | + | + | + | 100 | ||
| Thio- oxidation mutants | |||||||
| ΔdsbA | + | + | + | + | ∼44 | ||
| ΔdsbA-degPR3 | + | + | + | + | ∼40 | ||
| Thio-reduction mutants | |||||||
| ΔccdA or ΔccmG or ΔccmH | - | - | - | - | 0 | ||
| Thio-redox loop mutants | |||||||
| Δ(ccdA-dsbA) | + | + | + | + | ∼50 | ||
| Δ(ccmG-dsbA) | - | + | - | - | ∼10-15 | ||
| Δ(ccmH-dsbA) | - | - | - | - | 0 | ||
| ΔccmG-Rev2 | - | + | - | - | ∼10-15 | ||
| Δ(ccmG-dsbA) degPR3 | - | + | - | - | ∼10 | ||
| Δ(ccdA-ccmG-dsbA) | - | + | - | - | ∼10-15 | ||
| Δ(ccdA-ccmH-dsbA) | - | - | - | - | 0 | ||
| ΔccmG | pCcmGWT | + | + | + | + | 100 | |
| Δ(ccmG-dsbA) | pCcmGWT | + | + | - | + | ∼65 | |
| ΔccmG | pCcmGC75S or | ||||||
| pCcmGC78S or | - | - | - | - | 0 | ||
| pCcmGC75S/78S | |||||||
| Δ(ccmG-dsbA) | pCcmGC75S | s | + | - | + | ∼40-50 | |
| pCcmGC78S | - | + | - | + | ∼40-50 | ||
| pCcmGC75S/C78S | + | + | - | + | ∼48 | ||
| Δ(ccmG-dsbA)-degPR3 | pCcmGWT | + | + | + | + | ∼70 | |
| pCcmGC75S/C78S | + | + | + | + | ∼41 | ||
| ΔccmH | pCcmHWT | + | + | + | + | 100 | |
| pCcmHC42S/C45S | - | - | - | - | 0 | ||
| Δ(ccmH-dsbA) | pCcmHWT | + | + | + | + | ∼90 | |
| pCcmHC42S/C45S | - | - | - | - | 0 | ||
MedA and MPYE refer to minimal and enriched growth medium, respectively,
indicates that they were supplemented with 0.33 mM cysteine and 0.165 mM cystine.
Ps designate photosynthetic growth conditions, and +, s and – refer to normal, slow and no growth, respectively. All strains are Res+ in both Med A* and MPYE*, but those lacking DsbA often exhibit variable degrees of Res or Ps growth defects (slow to no growth) in the absence of the thio-reactive supplements.
Earlier work indicated that the E. coli putative ABC-type glutathione transporter CydDC (Pittman et al., 2005) is a component required for the bd-type hydroquinone oxidase assembly and for cyt c production (Georgiou et al., 1987; Poole et al., 1994). We found that R. capsulatus mutants lacking CydC or CydD, or their derivatives lacking DsbA or DsbA CcdA (Table 2) also did not contain any active hydroquinone oxidase (Zhang & Daldal, unpublished data). However, all CydDC derivatives still produced cyts c, unlike their E. coli counterparts (Fig. 3A). Thus, in R. capsulatus, the putative ABC-type gluthatione transporter CydDC was not involved in cyt c production, and the compensatory thio-redox interactions seen between DsbA and CcdA did not rely on the periplasmic reductive power that might be due the CydDC transporter.
Table 2. Bacterial strains and plasmids used in this study.
| Strain or plasmid | Description | Relevant phenotype | Source or reference |
|---|---|---|---|
| Strains | |||
| E. coli | |||
| HB101 | F- Δ(gpt-proA)62 araC14 leuB6(Am)
supE44 galK2(Oc) lacY1 Δ(mcrC-mrr) rpsL20(Strr) xylA5 mtl-1 thi-1 |
(Sambrook, 2001) | |
| XL1-Blue | F′∷Tn10 proA+B+ lacIq Δ(lacZ)M15/recA1 endA1
gyrA96 (Nalr) thi hsdR17 (rK- mK+) supE44 relA1 lac |
Stratagene | |
| R. capsulatus | |||
| MT1131a | crtD121 Rifr | Wild type, Res+, Ps+, cyts c+ | (Scolnik et al., 1980) |
| MD1 | Δ(ccdA∷spe) | Res+/Ps-, cyts c- | (Deshmukh et al., 2000) |
| MD14 | ccmH∷spe ccmF+ | Res+/Ps-, cyts c- | (Deshmukh et al., 2002) |
| MD20 | Δ(dsbA∷kan) | Res+/Ps+ on MedA, Res-/Ps+ on MPYE, cyts c+ | (Deshmukh et al., 2003) |
| MD20R3 | Δ(dsbA∷kan) degPR3 | Res+/Ps+, cyts c+, low DegP | (Onder et al., 2008) |
| MD21 | Δ(ccdA∷spe) Δ(dsbA∷kan) | Res+/Ps+ on MedA, Res-/Ps+ on MPYE, cyts c+ | (Deshmukh et al., 2003) |
| MD11 | Δ(ccmG∷kan) | Res+/Ps-, cyts c- | (Sanders et al., 2007) |
| SB1003-ΔhelX | Δ(ccmG∷kan) | Res+/Ps-, cyts c- | (Beckman and Kranz, 1993) |
| ST21 | Δ(ccmG∷kan) Δ(dsbA∷spe) | Res+/Ps- on MPYE and MPYE*, Ress/Ps- on MedA, Res+/Ps+ on MedA* | This work |
| ST38 | Δ(dsbA∷spe) degPR3 | Res+/Ps+, cyts c+, low DegP | This work |
| ST40 | Δ(ccmG∷kan) Δ(dsbA∷spe) degPR3 | Res+/Ps- on MPYE and MPYE*, Res+/Ps- on MedA, Res+/Ps+ on MedA*, low DegP | This work |
| ST34 | Δ(ccmG∷kan) Δ(dsbA∷spe) Δ(ccdA∷gen) | Res+/Ps- on MPYE and MPYE*, Ress/Ps- on MedA, Res+/Ps+ on MedA* | This work |
| ST27 | ccmH∷spe Δ(dsbA∷kan) | Res+/Ps-, cyts c- | This work |
| ST36 | ccmH∷spe Δ(dsbA∷kan) Δ(ccdA∷gen) | Res+/Ps-, cyts c- | This work |
| MD11Ri or SB1003-ΔhelXRi
(i=1-10) |
Δ(ccmG∷kan)-Revi(i.e., dsbAi) | Res+/Ps- on MPYE and MPYE*, Ress/Ps- on MedA
Res+/Ps+ on MedA* |
This work |
| STD1 | Δ(cydD∷gen) | Res+/Ps+, cyts c+ | This work |
| STD3 | Δ(cydD∷gen) Δ(dsbA∷kan) | Res+/Ps+, cyts c+ | This work |
| STD5 | Δ(cydD∷gen) Δ(dsbA∷kan) Δ(ccdA∷spe) | Res+/Ps+, cyts c+ | This work |
| Plasmids | |||
| pBluescript | pBluescript II KS(+) (pBSK) | Ampr | Stratagene |
| pRK2013 | Kanr, helper | (Ditta et al., 1985) | |
| pRK415 | Broad host-range vector with E. coli lacZ promoter | Tetr | (Keen et al., 1988) |
| pCHB500 | Broad host-range vector with R. capsulatus cycA promoter | Tetr | (Benning and Somerville, 1992) |
| pHP45Ω-Spc | Ω-spectinomycin (spe) cassette | Sper | (Prentki and Krisch, 1984) |
| pBS-DsbAwt | Phosphorylated 700 bp PCR product with dsbA cloned into the SmaI site of pBSK | Ampr | (Deshmukh et al., 2003) |
| pTC4-1K | 4.2 kb insert carrying dsbA interrupted by kanamycin casette in pRK415 | Tetr, Kanr | (Deshmukh et al., 2003) |
| pK6 | ccdA with engineered BglII site at position 749 in pBluescript | Ampr | (Deshmukh et al., 2000) |
| pCS1540 | 570 bp PCR product containing ccmG cloned into XbaI-KpnI site of pCHB500 | Tetr | (Sanders et al., 2005a) |
| pCS1545 | cysteineless ccmG-C75S/C78S allele of ccmG in pCS1540 | Tetr | (Sanders et al., 2005a) |
| pYZ1 | 2.82 kb fragment carrying ccmFH operon expressed from G488A up-promoter mutation in pBluescript | Ampr | (Deshmukh et al., 2002) |
| pHX1 | pRK415 with a DNA fragment containing the Δ(ccmG∷kan) [previously called Δ(helX∷kan)] | Tetr, Kanr | (Katzen et al., 2002) |
| pQE60 | Expression vector (Qiagen) | Ampr, PT5 | Qiagen |
| pQE60-helX | R. capsulatus ccmG without its signal peptide cloned at the NcoI and BamHI sites of pQE60 to yield a C-terminal His tagged derivative of CcmG | Ampr, PT5 | J. F. Collet, unpublished |
| pCS905 | pET-3a derivative (Novagen) with T7 promoter region replaced by a DNA fragment encoding lacI and the tac promoter region | Ampr, Ptac, LacI+ | (Sanders et al., 2001) |
| pCS1302 | pCS905 derivative, Strep-tag II sequence (IBA) fused to GFP, rendering GFP replaceable by cloning any gene of interest in frame into NdeI and BamHI sites | Ampr, Ptac, LacI+, Strep-GFP+ | Sanders et al., submitted |
| pCS1726 | R. capsulatus cycA derivative encoding the mature form of cyt c2 with a N-terminal Strep-tag cloned into pCS1302 using NdeI and BamHI sites | Ampr, Ptac, LacI+, Strep-cyt c2+ | Sanders et al., submitted |
| pCS1757 | R. capsulatus cycA derivative encoding the mature form of cyt c2 with a N-terminal Strep-tag and a C-terminal 6xHis-tag cloned into pCS1302 using NdeI and BamHI sites | Ampr, Ptac, LacI+, Strep-cyt c2-6xHis+ | This work |
| pCS1554 | Cysteine 75 and 78 of R. capsulatus ccmG in pQE60-helX mutated to serines | Ampr, PT5 | This work |
| pST6 | ccmH-Strep from pST8 cloned into the XbaI-KpnI sites of pCHB500 | Tetr | This work |
| pST7 | PCR amplified 0.48 kb ccmH fragment from pYZ1 cloned into the XbaI-KpnI sites of pBluescript | Ampr | This work |
| pST8 | PCR amplified and 3′ end Strep-tagged 0.53 kb ccmH fragment from pST7 cloned into the XbaI-KpnI sites of pBSK | Ampr | This work |
| pST14 | ccmH-C42S/C45S allele in pCHB500 | Tetr | This work |
| pKZ66 | A 4.8 kb PCR fragment containing cydDC cloned into BamHI-EcoRI sites of pBSK | Ampr | This work |
| pKZ68 | pKZ66 with Genr cartridge replacing the 1.2 kb PstI fragment in cydC | Ampr Genr | This work |
| pKZ69 | XbaI-KpnI fragment of pKZ68 containing Δ(cydC∷gen) in pRK415 | Tetr Genr | This work |
| pKZ7 | HindIII digested and religated pKZ66 without its BbsI site downstream from cydC | Ampr | This work |
| pKZ71 | pKZ7 with Genr cartridge replacing the 320 bp BbsI fragment in cydD | Ampr Genr | This work |
| pKZ72 | XbaI-KpnI fragment of pKZ71 containing Δ(cydD∷gen) in pRK415 | Tetr, Genr | This work |
| pCHB∷Gm | pCHB500 carrying a Genr cassette (gen)driven by the cycA promoter | Tetr, Genr | K. Zhang and F. Daldal |
| pBS-STA3 | The Sper cartridge of pHP45Ω-Spe cloned into the BglII-SfiI sites of pBS-DsbAwt | Ampr, Sper | This work |
| pSTA1 | 2.1 kb XbaI-KpnI fragment of pBS-STA3 cloned into the same sites of pRK415 | Tetr, Sper | This work |
| pST1 | Phosphorylated 1.2 kb PCR fragment of Genr cassette from pCHB∷Gm cloned into pBluescript | Genr, Ampr | This work |
| pST3 | BamHI fragment of pST1 cloned into the BglII sites of pK6 | Genr, Ampr | This work |
| pST4 | Δ(ccdA∷gen) from pST3 cloned into pRK415 | Genr, Tetr | This work |
| pCS1543 | ccmG-C75S allele of ccmG in pCS1540 | Tetr | This work |
| pCS1544 | ccmG-C78S allele of ccmG in pCS1540 | Tetr | This work |
R. capsulatus MT1131 is a derivative of SB1003 and referred to as wild type with respect to its growth properties and cyts c profile.
refers to minimal (MedA) and enriched (MPYE) growth medium supplemented with 0.33 mM cysteine and 0.165 mM cystine.
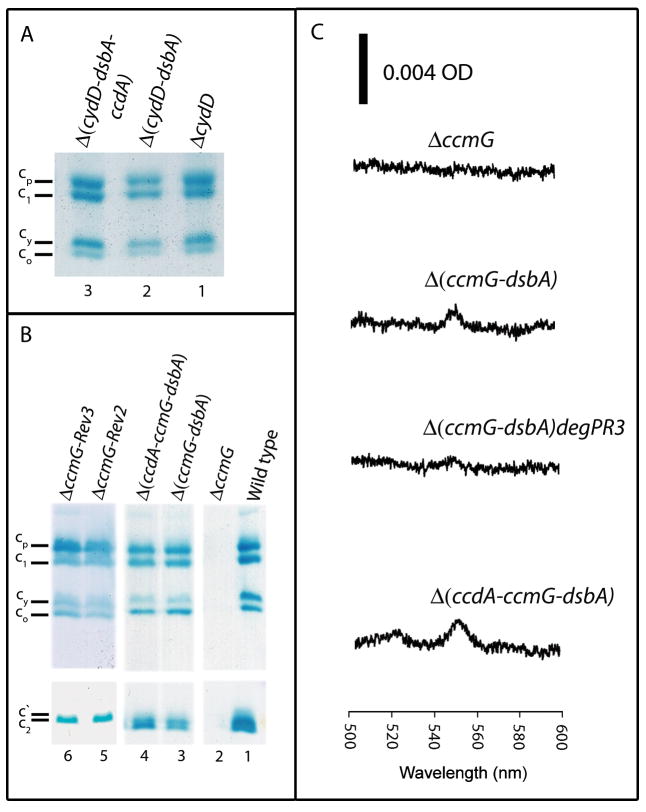
Fig. 3. Cyt c profiles of R. capsulatus thio-redox mutants.
The cyts c production in the putative glutathione transporter mutant ΔcydD (STD1) and its derivatives Δ(cydD-dsbA) (STD3) and Δ(cydD-dsbA-ccdA) (STD5) (A), in the wild type (MT1131) and Ccm thio-reduction mutant ΔccmG (MD11), its Ps+ revertants (ΔccmG-Rev2) (MD11R2) and ΔccmG-Rev3) (MD11R3) and its derivatives Δ(ccmG-dsbA) (ST21) and Δ(ccdA-ccmG-dsbA) (ST34) (B), grown by respiration in minimal medium were determined by SDS-PAGE/TMBZ as described in Fig. 2 and Materials and Methods. Different cyts c are indicated on the left. (C) Cyt c2 levels in the Ccm thio-reduction mutant CcmG and its derivatives: Ascorbate-reduced minus ferricyanide-oxidized optical difference spectra of soluble fractions (protein concentration of 0.5 mg/ml) obtained from R. capsulatus strains ΔccmG, Δ(ccmG-dsbA), Δ(ccmG-dsbA)degPR3 (ST40) and Δ(ccdA-ccmG-dsbA) (Table 2) were determined as in Fig. 2.
Compensatory thio-redox interactions during Ccm is not restricted to CcdA and DsbA
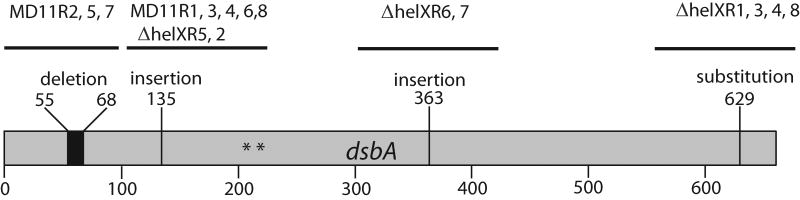
CcmG and CcmH are also thought to be parts of the Ccm thio-reductive branch (Deshmukh et al., 2003). Therefore, we probed whether the compensatory thio-redox interactions seen between DsbA and CcdA also extended to CcmG and CcmH. Like CcdA-null mutants, CcmG-null or CcmH-null mutants cannot grow under photosynthetic conditions (Ps-) due to the absence of cyts c (Beckman et al., 1992). However, unlike the CcdA-null mutants that revert readily to Ps+ by DsbA inactivation (Deshmukh et al., 2003), CcmG-null mutants required supplementation of the growth medium with thiol-reactive reagents (0.33 mM L-cysteine and 0.165 mM L-cystine) in the case of MD11, and without supplement in the case of an SB1003 derivative on enriched medium, to obtain Ps+ revertants. In contrast, CcmH-null mutants never yielded any Ps+ revertants under the conditions tested. Analyses of several Ps+ revertants of CcmG-null mutants indicated that they all produced cyts c (e. g., ccmG-Rev2 and -Rev3 in Fig. 3B), and that their Ps+ growth was suppressed upon introduction of a wild type allele of dsbA in trans. DNA sequence analyses revealed that the revertants carried chromosomal mutations inactivating dsbA. These mutations included a deletion encompassing the base pairs 55 to 68 in MD11R2, 5 and 7, a G insertion at position 135 in MD11R1, 3, 4, 6, 8, and ΔhelXR5, 2, an A insertion at position 363 in ΔhelXR6 and 7, and a T to G substitution at position 629 in ΔhelXR1, 3, 4 and 8 (Fig. 4 and Table 2).
Fig. 4. Ps+ revertants of R. capsulatus CcmG-null mutants are located in DsbA.
Ps+ revertants from a CcmG-null mutant (MD11Ri from MT1131 or ΔhelXRi from SB1003-ΔhelX) (Table 2) were isolated as described in the text, and DNA sequence of the chromosomal dsbA locus of each mutant was determined after PCR amplification. The mutant names, positions and type of the mutations found are indicated on top of the dsbA gene, and * indicates the active site cysteine residues of DsbA.
Indeed, construction of CcmG-null DsbA-null and CcmH-null DsbA-null double mutants confirmed that the loss of DsbA suppressed the Ccm deficiency of CcmG-null, but not that of CcmH-null, mutants (Table 2). The DsbA-null CcmG-null double mutants became Ps+ in minimal medium supplemented with thiol-reactive reagents and produced cyts c (Fig. 3B and C). In contrast, the DsbA-null CcmH-null double mutants remained Ps-, independent of thiol-reactive reagents addition to the growth medium, and produced no cyt c (Fig. 5A and B). Thus, the compensatory thio-redox interactions seen between DsbA and CcdA also occurred between DsbA and CcmG, but not between DsbA and CcmH. In agreement with these findings, the triple mutant lacking DsbA CcdA CcmG (but not DsbA CcdA CcmH) (Table 2) was Ps+ in thiol-reactive reagents supplemented minimal medium and exhibited cyts c profiles similar to those seen with the CcmG-null DsbA-null double mutant (Fig. 3B and C). We concluded that heme-apocyt c ligation occurred in the absence of the Ccm thio-redox loop encompassing DsbA, DsbB, CcdA and CcmG (Fig. 1).
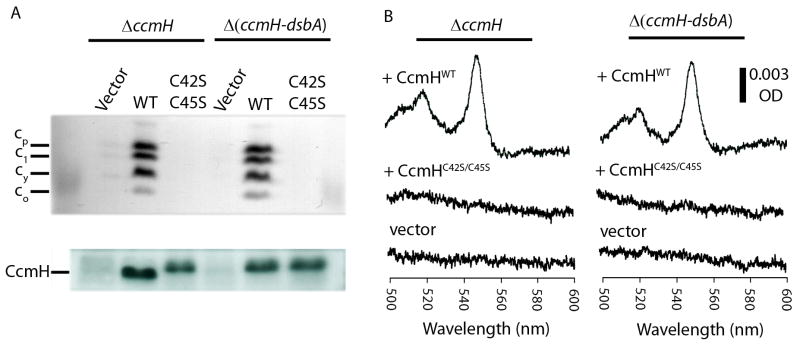
Fig. 5. Cyt c profiles and CcmH immunoblots of wild type and cysteineless CcmH mutants.
Cyts c profiles of R. capsulatus ΔccmH (MD14) and Δ(ccmH-dsbA) (ST27) strains harboring empty plasmid (vector) or wild type (WT/pST6) or cysteineless (C42S C45S/pST14) alleles of ccmH grown in minimal medium were determined by (A) SDS-PAGE/TMBZ analyses (top panel) as in Fig. 2. The same preparations were also subjected to SDS-PAGE using 50 μg total proteins per lane, and the amounts of CcmH were detected by immunoblot analyses (lower panel) using anti-CcmH polyclonal antibodies, as described in Materials and Methods. (B) Cyt c2 levels in the CcmH-null mutant and its DsbA-null derivatives: Ascorbate-reduced minus ferricyanide-oxidized optical difference spectra of soluble fractions (protein concentration of 0.5 mg/ml) obtained from R. capsulatus ΔccmH (left) and Δ(ccmH-dsbA) (right) mutants harboring empty plasmid (vector) or wild type (+ CcmHWT) or cysteineless (+ CcmHC42S/C45S) alleles of ccmH were determined as described in Fig. 2.
An additional role for CcmG as an apocyt c holdase during Ccm
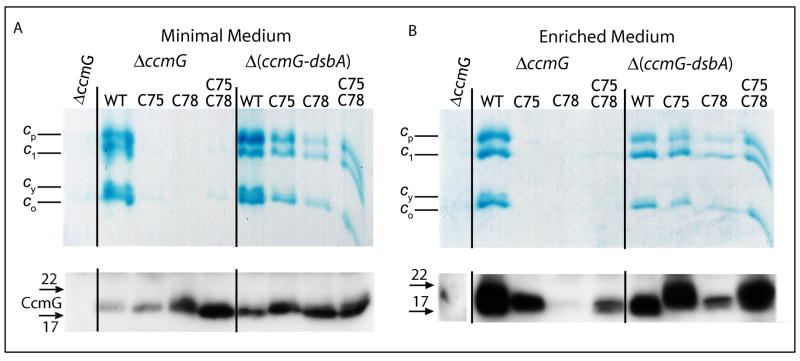
The observation that the CcmG-null DsbA-null double mutants required thiol-reactive supplements to grow under Ps conditions, unlike the CcdA-null DsbA-null mutants, suggested that absence of CcmG hampered Ccm efficiency more severely than CcdA. Examination of the optical redox difference spectra indicated that the CcmG-null DsbA-null double or CcdA-null CcmG-null DsbA-null triple mutants indeed contained much less (∼ 10-15 % of wild type) (Fig. 3C) 550 nm absorbing materials than the CcdA-null DsbA-null double mutants (∼ 40-50 % of wild type) (Fig. 2B). Since the only difference between these thio-redox loop mutants was the absence of CcmG, we probed whether the decrease of cyt c production was linked to the absence of the CcmG protein per se. CcmG has two active site cysteines (C75 and C78 in R. capsulatus) (Monika et al., 1997), which face the periplasm and accept electrons from CcdA (Katzen and Beckwith, 2000; Katzen et al., 2002). Substitution of these active site cysteines with serine yielded the CcmGC75S, CcmGC78S and CcmGC75S/C78S (the latter dubbed here as CcmG*) mutants. As expected, these mutants were unable to complement a CcmG-null strain for Ps growth in any growth medium, regardless of thiol-reactive supplement They also did not produce any cyt c that can be detected by SDS-PAGE/TMBZ (Fig. 6A and B, top panels) or optical redox difference spectra analyses (Fig. 7A) (Table 1). Immunoblot data indicated that all strains (except CcmGC78S in enriched medium) produced mutant forms of CcmG at wild type or higher amounts (Fig. 6A and B, lower panels). We also noted that the amounts of CcmG were variable in various mutants grown under different conditions, and this point deserves future studies.
Fig. 6. Cyt c profiles and CcmG-immunoblots of cysteineless CcmG mutants.
Cyts c profiles of R. capsulatus ΔccmG and Δ(ccmG-dsbA) strains harboring wild type (pCcmGWT (WT)) or different cysteineless (pCcmGC75S (C75S), pCcmGC78S (C78S) and pCcmGC75S/C78S (C75S C78S)) alleles of ccmG grown in either minimal (A) or enriched (B) medium were determined by SDS-PAGE/TMBZ analyses (top panels), as described in Fig. 2. The same preparations were subjected to SDS-PAGE using 50 μg total proteins per lane, and the amounts of CcmG were detected by immunoblot analyses (lower panels) using anti-CcmG polyclonal antibodies, as described in Materials and Methods.
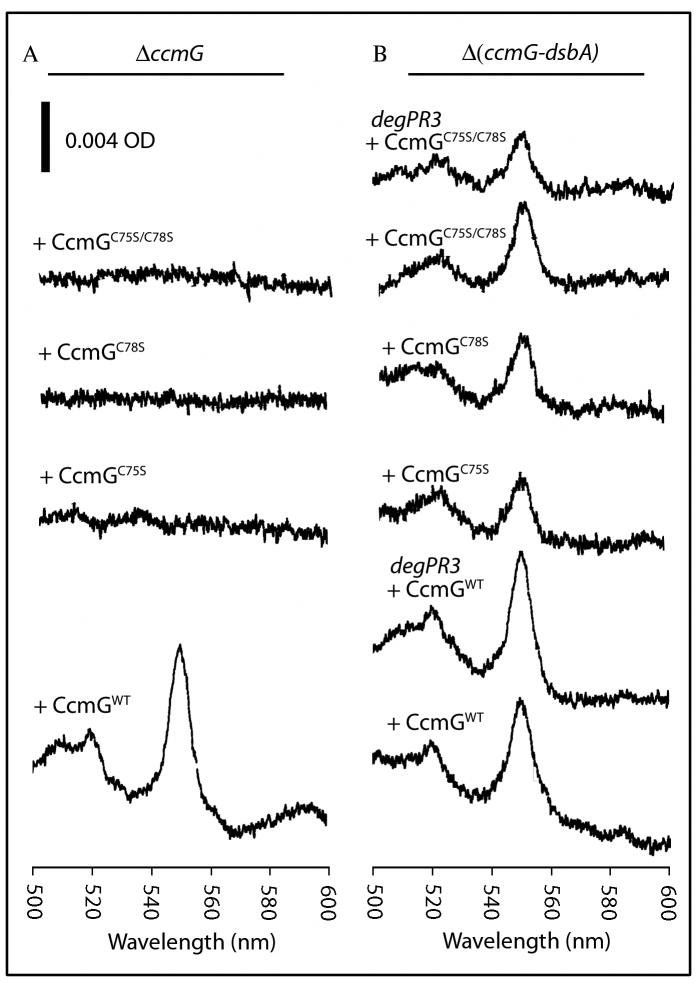
Fig. 7. Cyt c2 levels in R. capsulatus cysteineless CcmG mutants and its DsbA-null derivatives.
Ascorbate-reduced minus ferricyanide-oxidized optical difference spectra of soluble fractions (0.5 mg of total proteins/ml) prepared from R. capsulatus ΔccmG (A) and Δ(ccmG-dsbA) or Δ(ccmG-dsbA)degPR3 (B) strains harboring (+ pCcmGWT (WT)) or different cysteineless (+ pCcmGC75S (C75S), + pCcmGC78S (C78S) and + pCcmGC75S/C78S (C75S C78S)) alleles of ccmG, grown on enriched medium, were determined as described in Fig. 2. In B, the spectra labeled degPR3 were from a CcmG-null DsbA-null mutant carrying the Δ(ccmG-dsbA)degPR3 mutations lowering the levels of DegP in this background (Onder et al., 2008).
Interestingly, the cysteine-less derivative of CcmG complemented even in the absence of thiol-reactive supplements the DsbA-null CcmG-null mutant for Ps growth in minimal medium. Moreover, it also produced more cyts c as shown both by SDS-PAGE/TMBZ (Fig. 6A and B) and optical redox difference spectra analyses (Fig. 7B) (Table 1). Furthermore, the DsbA-null CcmG-null mutant carrying the CcmG* derivative was, like the DsbA-null or the DsbA-null CcdA-null mutants, Ps+ in minimal medium regardless of any thiol-reactive supplement, and contained the highest amounts of cyts c among the CcmG mutants (Fig. 6). Optical spectra again showed that the amounts of the 550 nm absorbing materials found in the DsbA-null CcmG-null mutants harboring CcmG* were comparable (∼ 40-50 % of wild type) to those found in the DsbA-null or DsbA-null CcdA-null mutants (Fig. 7B). Thus, the presence of CcmG* devoid of its thio-reductive activity increased drastically cyt c production in the absence of DsbA, indicating that CcmG had an additional, thio-reduction-independent function referred to as apocyt c holdase.
Similar studies were also conducted with CcmH and its cysteine-less derivatives. R. capsulatus CcmH has two functionally essential cysteine residues at positions 42 and 45 facing the periplasm (Monika et al., 1997). CcmH mutants lacking them (CcmHC42S, CcmHC45S and CcmHC42S/C45S, the latter dubbed CcmH*) did not complement for Ps growth the CcmH-null or DsbA-null CcmH-null mutants, and produced no cyt c (Fig. 5A and B) (Table 1) (unlike in E. coli, see (Robertson et al., 2008)) although they had similar amounts of wild type or mutant CcmH proteins, as indicated by immunoblot analyses (Fig. 5A, lower panel). Overall comparison of the data obtained with CcmH and CcmG indicated that the two thio-reductive components behaved very differently in respect to the DsbA- and CcdA-dependent thio-redox loop, suggesting that their reductive roles during Ccm might be different.
R. capsulatus apocyt c2 physically interacts with CcmG* in vitro
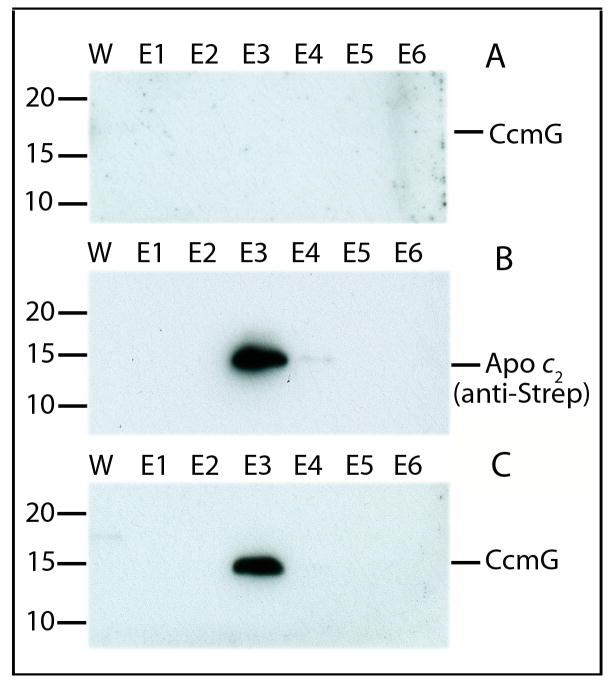
The additional, thioredoxin-independent apocyt c holdase function of CcmG during Ccm was investigated in vitro by probing the direct physical interactions between purified apocyt c and partially purified CcmG*. A C-teminally His epitope-tagged mature form of R. capsulatus CcmG* was expressed in E. coli, and partially purified by affinity chromatography. Similarly, a double (N-terminal His and C-terminal Strep) epitope-tagged derivative of R. capsulatus apocyt c2 devoid of its signal sequence was expressed in E. coli cytoplasm, and tandem affinity purified using His-Bind and then Strep-Tactin affinity columns, as described in Experimental Procedures. As the apocyt c2 was highly unstable, use of tandem affinity tags facilitated the isolation of intact apocyt c2. Purified apocyt c2 and CcmG* containing extracts (50 and 40 μg total proteins, respectively) were incubated together for two hours at 4 °C, and apocyt c2 was re-isolated using Strep-Tactin affinity chromatography. Column fractions were probed for the presence of apocyt c2 using anti-Strep and anti-His (not shown) antibodies, as well as for CcmG using anti-CcmG antibodies (Fig. 8). A similar mock experiment using partially purified CcmG* but no apocyt c2 was also conducted to show that CcmG* itself was not retained by the Strep-Tactin affinity column (Fig. 8A). However, when an incubation mixture containing both CcmG* and apocyt c2 were loaded onto the same column, and eluted with desthiobiotin (DTB), apparently CcmG* was retained by the column as both apocyt c2 and CcmG* were co-eluted in the same fractions (mainly E3) (Fig. 8B and C). We therefore concluded that a CcmG* lacking its active site cysteines can recognize and bind apocyt c2 in vitro as an apocyt c holdase.
Fig. 8. Co-purification of CcmG* with apocyt c2.
Purified apocyt c2 (50 μg) and CcmG* enriched extracts (40 μg) were incubated in the presence of 20 mM Tris-HCl pH = 7.9, 50 mM NaCl, 50 μg DTB and 10 mM imidazole for two hrs at 4 °C and loaded onto a StrepTactin® Sepharose affinity chromatography column, washed, and eluted with 2.5 mM DTB as described in Experimental Procedures. Aliquots (300 μl of 1 ml total) from various purification steps (W: column wash, and E1 to E6: DTB elution fractions 1-6) were precipitated with ice cold Acetone/Methanol and analyzed by SDS-PAGE and immunoblots. Antibodies against CcmG and Strep-tag are as indicated on the right, and molecular weight markers (in kDa) are as shown on the left of each panel. Panel A illustrates that CcmG*, which does not contain a Strep-tag, is not retained by the StrepTactin® Sepharose column in the absence of apocyt c2. Panels B and C shows that in the presence of apocyt c2, CcmG* is retained by the column and co-eluted with apocyt c2, documenting that the two proteins interact with each other under the conditions used.
Discussion
This work was initiated in order to assess whether the compensatory thio-redox interactions previously seen between DsbA and CcdA mutants (Deshmukh et al., 2003) also extended to other thio-reductive Ccm components. By doing so we hoped to dissect apart the Ccm components involved in preparing apocyt c as an adequate substrate from those catalyzing the subsequent heme-apocyt c ligation per se. Searches for spontaneous Ps+ revertants of CcmG-null and CcmH-null strains, and construction of appropriate double and triple knock-out mutants, demonstrated for the first time that the DsbA-dependent compensatory thio-redox interactions involved solely CcdA and CcmG, and not CcmH (or CcmI, data not shown). Earlier, it was observed that Ps+ revertants of CcmG-null mutants, unlike CcdA-null mutants, could only be obtained if the growth medium was supplemented with thiol-reactive chemicals (Monika et al., 1997). Similarly, we noted that CcmG DsbA and CcdA CcmG DsbA knock-out mutants were Ps+ only upon supplementation with thiol-reactive reagents. The basis for this redox constraint is unclear. Thus, in the absence of CcmG (in contrast to that of CcdA), R. capsulatus cells might be metabolically more limited to reduce their growth environment to initiate anoxygenic photosynthesis.
Although the cysteine-less derivatives (Fabianek et al., 1999; Monika et al., 1997) of the unusual thioredoxin-like CcmH (Di Matteo et al., 2007) were indeed Ccm deficient, the thio-reductive function of CcmH could not be compensated by a thio-oxidative defect. These findings limited the thio-redox loop to DsbA, DsbB, CcdA and CcmG (Fig. 1) establishing that these components were not essential for cyt c production in vivo. Moreover, they simplified the number of Ccm components required for heme-apocyt c ligation per se (Fig. 9). Importantly, the data also suggested that both CcmG and CcmH might have additional function(s) distinct from their known thio-reductive roles (Monika et al., 1997). In respect to CcmH, a possibility is that its oxidized form might interact with CcmG-reduced apocyt c during its transfer to heme ligation complex containing CcmF. Indeed, CcmH is known to co-immunoprecipitate with CcmF (Ren et al., 2002), and its overproduction together with CcmF bypasses the Ccm deficiency of CcmI-null mutants (Deshmukh et al., 2002; Sanders et al., 2005a). Furthermore, our recent data indicate that CcmH is a component of a complex also containing CcmF and CcmI in R. capsulatus membranes (Sanders et al., submitted).
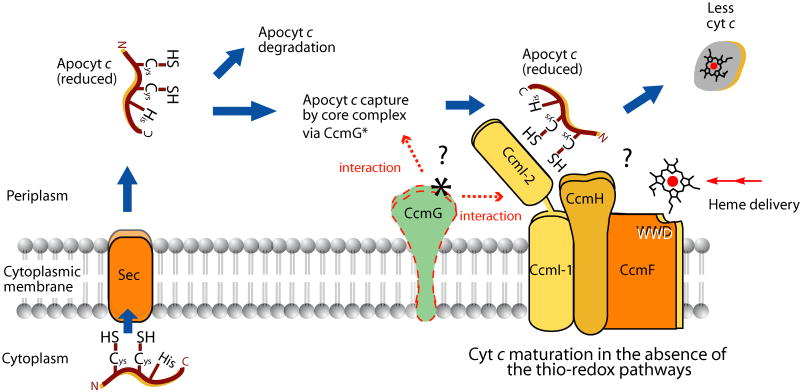
Fig. 9. Production of cyts c in the absence of the Ccm thio-redox loop.
In the absence of the DsbA-DsbB, CcdA and CcmG thio-redox loop shown in Fig. 1, reduced apocyts c, although more prone to degradation, are matured at lower levels by the CcmFHI ligation components. The presence of a thio-reduction inactive CcmG derivative (CcmG* shown in dashed line) increases the amounts of mature cyts c, by acting as an apocyt c holdase improving delivery of the apocyts c to the CcmFHI heme ligation components. Thick arrows show the route followed by the apocyts c, dashed arrows indicate the interactions of apocyts c and heme ligation components via CcmG*, and Cys, His, N, C and WWD are as described in Fig. 1.
Unlike other thioredoxins, CcmG is highly specific for Ccm (Edeling et al., 2002; Fabianek et al., 1998; Monika et al., 1997) with an active site unusually acidic, proposed to provide its substrate specificity (Edeling et al., 2004). Our finding that the cysteine-less CcmG derivative CcmG* produced high amounts of cyts c and exhibited Ps growth regardless of the thio-reactive supplements, indicated that CcmG protein per se also had a thioredoxin-independent role(s) referred to as apocyt c holdase during Ccm (Fig. 9). Earlier, the active site cysteines of E. coli CcmG was found to be important, but not essential, for Ccm in the presence of reducing agents (Fabianek et al., 1998). In the light of the compensatory thio-redox interactions seen between DsbA and CcmG, this finding could also be interpreted as counteracting the DsbA oxidative power, and not necessarily as an additional activity of CcmG. The significant increase of cyts c production in the presence of CcmG* in the absence of DsbA, as seen here, established unambiguously a thioredoxin-independent role for the CcmG protein per se (Fig. 9). This additional function of CcmG was further supported by the fact that purified apocyt c and CcmG* devoid of its cysteines directly interact with each other in vitro. Growing evidence suggests that CcmG acts as a holdase, conveying apocyt c to the heme ligation core complex. The crystal structure of B. subtilis CcmG homologue ResA revealed a cavity, close to its active site cysteines, speculated to form a binding surface for apocyt c (Colbert et al., 2006; Crow et al., 2004). Consistent with this proposal, we observed that overproduction of CcmG or apocyt c (Sanders et al., 2007) in addition to CcmF and CcmH also increased cyt c production in R. capsulatus mutants lacking CcmI (Deshmukh et al., 2002; Sanders et al., 2005a).
It should be emphasized that DsbA is not essential for Ccm, but its absence decreases cyt c production irrespective of the thio-reductive pathway (i. e., CcdA and CcmG). Although this observation could rationalize why the Ccm thio-redox loop might be conserved evolutionarily, why it occurs remains unknown. That the thio-redox loop reactions might protect some apocyts c intermediates (Sanders et al., 2005a), or improve the fidelity of stereo-specific heme ligation (Sanders et al., 2005b) has been suggested. Unfolded apocyts c might be more vulnerable to degradation unless protected by chaperones such as CcmG* (depicted as dashed outline in Fig. 9). Formation of a disulfide bond at the apocyt c C1XXC2H motif, although requires subsequent reduction by the thio-reductive pathway, might be beneficial by promoting partial folding and decreasing degradation of apocyt c. Consequently, in the absence of the thio-redox loop and CcmG*, cyt c production would be much lower than native amounts. Absence of the Ccm thio-redox loop might also compromise stereo-specific heme ligation (Sanders et al., 2005b), yielding non-native cyts c that are prone to degradation. Indeed the DsbA, CcdA DsbA, CcmG* DsbA or CcdA CcmG* DsbA mutants produced lower amounts of cyts c than a wild type strain of R. capsulatus. In summary, this study established that the Ccm thio-redox loop components DsbA, CcdA and CcmG are not essential for cyt c production, hence decreasing the number of Ccm components absolutely required for heme-apocyt c ligation. Furthermore, it revealed a second, thio-reduction independent role of CcmG, which is critical for cyt c production, and raised the possibility that the oxidized form of CcmH might be required for transferring CcmG-reduced apocyt c to a heme ligation complex containing CcmF and CcmI. Future studies would hopefully better define the interactions between apocyt c and the essential Ccm components during cyt c production in bacteria.
Experimental Procedures
Bacterial strains, plasmids, and growth conditions
The bacterial strains and plasmids used in this work are described in Table 2. R. capsulatus strains were grown at 35 °C on enriched (Daldal et al., 1986) or minimal (Sistrom, 1960) media, supplemented with appropriate antibiotics (2.5, 10, 10, and 1 μg/ml tetracycline, kanamycin, spectinomycin and gentamycin, respectively), either chemoheterotrophically (Res growth) or photoheterotrophically (Ps growth) in anaerobic jars with H2 + CO2 generating gas packs from BBL Microbiology Systems (Cockeyville, MD). All growth media were supplemented with 0.33 mM L-cysteine and 0.165 mM L-cystine when appropriate. E. coli strains were grown on Luria Bertani (LB) broth supplemented with appropriate antibiotics (12.5, 50, 10, 100 and 12 μg/ml tetracycline, kanamycin, spectinomycin, ampicilin and gentamycin, respectively) as needed.
Molecular Genetics Techniques
Standard molecular genetic techniques were performed as described (Sambrook, 2001). Plasmid pBS-DsbAwt (Deshmukh et al., 2003) carrying the R. capsulatus wild type dsbA was digested with BglII and SfiI to replace its 0.5 kb internal fragment with a 2.0 kb DNA polymerase blunted BamHI spectinomycin resistance cassette from pHP45Ωspec to yield pBS-STA3 harboring Δ(dsbA∷spe) (Table 2). The XbaI-KpnI fragment of pBS-STA3 was then cloned into the respective sites of pRK415 to yield pSTA1. A 1.2 kb gentamycin resistance cassette under the control of the cyt c2 gene promoter (PcycA-gen) was first amplified from pCHB∷Gm (Table 2) with M13-forward and M13-reverse primers, phosphorylated using T4 polynucleotide kinase and ligated into the EcoRV-digested pBluescript to yield pST1 (Table 2). To obtain a Δ(ccdA∷PcycA-gen) allele pST1 was restricted with BamHI and the 1.2 kb fragment harboring PcycA-gen ligated into BglII restricted pK6, eliminating a 0.3 kb internal fragment of ccdA. The 2.7 kb insert from the resulting plasmid pST3 was then transferred into the BamHI site of pRK415 to yield pST4. Plasmids pCS1540 and pCS1545 containing wild type ccmG and the ccmG* variant with both cysteines (C75 and C78) to serine substitutions, respectively, were constructed as described (Sanders et al., 2005a) (Table 2). To obtain pCS1543 and pCS1544, the C75 or C78 of ccmG in pCS1540 were substituted with serine using the QuikChange® XL Site-Directed Mutagenesis Kit according to the manufacturer's instructions (Stratagene, La Jolla, CA) and the mutagenic primers HelX-C75Ss (5′-CTT CTG GGC TTC CTG GAG CGC GCC CTG TCG-3′) and HelX-C75Sas (5′-ACC CGA CAG GGC GCG CTC CAG GAA GCC CAG-3′) for pCS1543, and HelX-C78Ss (5′-TCC TGG TGC GCG CCC AGT CGG GTC GAA CA-3′) and HelX-C78Sas (5′-GGA TGT TCG ACC CGA CTG GGC GCG CAC CAG-3′) for pCS1544 (Table 2). PCR-amplification of ccmH using pYZ1 as a template and CcmH/XbaI-Fwd (5′- CTC TCT AGA GTT GCG GCG GAG TGA GCG ATG CTG AAA CG-3′) and CcmH/KpnI-Rev (5′-GAG GGT ACC CTA GTC CTT CAG CAG GTC TTT CAG CCG CGC CTC-3′) as primers generated a 0.48 kb DNA fragment, which was digested with XbaI and KpnI and ligated into the respective sites of pBluescript II KS(+) (pBSK) to yield pST7 (Table 2). Plasmid pST8 was constructed by engineering a Strep-tag sequence at the 3′-end of ccmH via PCR amplification using pST7 as a template and the primers CcmH-StrepC/XbaI-Fwd (5′-CTC TCT AGA GTT GCG GCG GAG TGA GCG ATG CTG AAA CG-3′) and CcmH-StrepC/KpnI-Rev (5′- GAG GGT ACC CTA TTT TTC GAA CTG CGG GTG GCT CCA AGC GCT GTC CTT CAG CAG GTC TTT CAG CCG CGC CTC-3′). The 0.51 kb DNA fragment thus generated was cut with XbaI and KpnI and ligated into the same sites of pBSK. Plasmid pST8 was digested with XbaI and KpnI and the fragment carrying ccmH-Strep was ligated into the respective sites of pCHB500 to yield pST6. The cysteine-less derivative of CcmH was obtained by substituting C42 and C45 of ccmH (with an in-frame Strep-tag sequence at its 3′-end) with serine as described above by using pST8 as a template and CcmH-C42S/C45Ss (5′-TCG CAG GTG CTG CGC AGC CCG GTC AGT CAG GGC GAG A-3′) and CcmH-C42S/C45Sas (5′-GAT ATT CTC GCC CTG ACT GAC CGG GCT GCG CAG CAC CTG C-3′) as mutagenic primers to yield pST11 (Table 2). Plasmid pST11 was digested with XbaI and KpnI, and the fragment carrying mutant ccmH was ligated into the respective sites of pCHB500 to yield pST14. A 4.8 kb PCR fragment encompassing cydDC genes was amplified with primers CydDCF (5′- GCA CTT GCT CGT CGA ACA GGA TCC AG-3′) and CydDCR (5′- CCG GAA TTC GAA ACA GAC GCG CGA CA-3′), digested with BamHI and EcoRI (introduced sites in the primers are underlined) and cloned into respective sites of pBluescript to yield pKZ66. In order to create deletion-insertion allele of cydC, a 1.2 kb PstI fragment of pKZ66 was replaced with 1.2 kb Gentamycin resistance cartridge from pCHB∷Gm yielding pKZ68. XbaI-KpnI fragment from pKZ68 carrying cydD+ΔC∷Gm allele was inserted into the corresponding sites of pRK415 to yield pKZ69. Plasmid pKZ66 was digested with HindIII and ligated back, eliminating BbsI site immediately downstream of cydC to yield pKZ7. To create deletion-insertion allele of cydD, a 320 bp BbsI fragment from pKZ7 was excised, the remaining vector was made blunt and ligated to Gm cartridge from pCHB∷Gm to yield pKZ71. XbaI-KpnI fragment of pKZ71 was then cloned into XbaI-KpnI site of pRK415 to create pKZ72 (Table 2). Plasmid pQE60-helX was obtained by cloning into pQE60 (Qiagen, Valencia, CA) R. capsulatus ccmG lacking its N-terminal signal peptide/transmembrane helix by using the NcoI and BamHI sites. Plasmid pCS1554 expressing the C-terminally His tagged mature form of CcmG* was obtained by mutating to serine the cysteines 75 and 78 of ccmG in pQE60-helX using the primers HelX-C75S/C78Ss (5′-CTT CTG GGC TTC CTG GAG CGC GCC CAG TCG GGT CGA ACA-3′) and HelX-C75S/C78Sas (5′-GGA TGT TCG ACC CGA CTG GGC GCG CTC CAG GAA GCC CAG-3′) according to the instruction of the QuikChange® XL Site-Directed Mutagenesis Kit from Stratagene (La Jolla, CA). The mature form of R. capsulatus cyt c2 gene (cycAmat) was PCR-amplified using the primers CS24 (5′-GTT GAC AAT TAA TCA TCG GC-3′) and RccycACtHis6BamHIr (5′-GCC GGA TCC TAG TGA TGG TGA TGG TGA TGT TTC ACG ACC GAG GCC AGA TAG GC-3′) and pCS1726 (Sanders et al., submitted) as a template. The 0.5 kb fragment thus generated was digested with NdeI and BamHI, and cloned into the same sites of pCS1302 (Sanders et al., submitted) to yield pCS1757 that carries cycAmat with an in frame Strep-tag sequence at its 5′-end and a 6xHis-tag at its 3′-end. The expression of the double-tagged cycAmat derivative is controlled by a Ptac-lac promoter-operator system in E. coli.
Automated DNA sequencing with the Big-Dye™ terminator cycle sequencing kit (AmpliTaq FS; Applied Biosystems) was performed according to the manufacturer by using the M13-forward and M13-reverse primers to verify all plasmids constructed in this study. DNA sequence analyses, homology searches and genome sequence comparisons were done using MacVector (IBI; Kodak) and BLAST software packages (Altschul et al., 1990).
Conjugal transfers of plasmids from E. coli to R. capsulatus and chromosomal allele replacement via interposon mutagenesis using the gene transfer agent (Yen et al., 1979) were carried out as described earlier (Daldal et al., 1986). Briefly, the mutant strain ST40 (ccmG∷kan dsbA∷spe degPR3) mutant was obtained using pHX-1 (ccmG∷kan) as a donor and ST38 (dsbA∷spe, degPR3), which is a SpecR derivative of MD20R3 (Onder et al., 2008) as a recipient strain. The CcmH-null DsbA-null double mutant ST27, and CcdA-null CcmH-null DsbA-null triple mutant ST36, were constructed using pTC4-1K (dsbA∷kan) and pST4 (ccdA∷gen) as donors and MD14 (ccmH∷spe) and ST27 (ccmH∷spe dsbA∷kan) as recipient strains, respectively.
Biochemical techniques
Intracytoplasmic membrane vesicles (chromatophore membranes) were prepared in 50 mM MOPS (pH 7.0) containing 100 mM KCl for spectral, or 1 mM KCl for SDS-PAGE, analyses as described earlier (Gray et al., 1994). All preparations contained 10 mM EDTA, 1 mM PMFS and 0.1 mM ε-amino-caproic acid as protease inhibitors. Protein concentrations were determined according to (Lowry et al., 1951) and SDS-PAGE was used as in (Schagger and von Jagow, 1987) using 16.5 % polyacrylamide gels. Cyts c were revealed based on the endogenous peroxidase activity of covalently attached heme by using 3,3′,5,5′-tetramethylbenzidine (TMBZ) (Thomas et al., 1976). For CcmG and CcmH immunodetection, protein samples were separated using 15% SDS-PAGE (Laemmli, 1970). Following electrophoresis, gels were electroblotted onto Immobilon-P PVDF membranes (Millipore, Billerica, MA). CcmG and CcmH were detected using anti-CcmG and anti-CcmH polyclonal antibodies (Monika et al., 1997), ECL anti-rabbit IgG/horseradish peroxidase-conjugated secondary antibodies from Amersham-Pharmacia (Piscataway, NJ) and SuperSignal West Pico Chemiluminescent substrate from Pierce (Rockford, IL).
Expression and purification of apocytochrome c2 and CcmG*
E. coli strains containing pCS1757 (for apocyt c2) or pCS1554 (for CcmG*) were grown in Luria Broth (LB) liquid medium (four liters each) containing 100 μg/ml ampicillin at 37 °C with shaking (200 rpm) until an OD600 of approximately 0.8. Cultures were then induced under the same growth conditions for two hours by adding isopropyl-thio-β-D-galactoside (IPTG) to a final concentration of 10 mM. Cells were harvested by centrifugation for 10 min at 10,000 g and resuspended in Tris Buffer (50 mM Tris-HCl pH 8.0, 100 mM NaCl and 1mM PMSF), and disrupted on ice by using a French Pressure cell as described in (Daldal et al., 1986). Cell lysates were centrifuged at 25,571 g for 1 hour, followed by at 117,734 g for 1 hour. Cleared supernatants were filtered through a 0.45 μm filter and applied to a His·bind purification chromatography column (His·Bind Kit from Novagen, Inc.) following the manufacturer's instructions. Final concentrations of 25 and 100 mM imidazole in the washing and elution buffers, respectively, were used, and column fractions monitored by SDS-PAGE and Western Blot analyses. His-bind chromatography fraction containing apocyt c2 were pooled, concentrated by ultrafiltration (3,000 MWCO, Amicon Ultra, Millipore) to eliminate imidazole, diluted ten fold in Strep Wash buffer before loading onto the StrepTactin® Sepharose affinity column, and purified according to manufacturer's instruction (IBA, Inc.). Direct interactions between purified apocyt c2 and CcmG* were probed by mixing 50 μg of His and Strep purified apocyt c2 with 40 μg of CcmG* enriched extracts and incubating for two hours at 4 °C, in the presence of 20 mM Tris-HCl pH = 7.9, 50 mM NaCl, 50 μg DTB and 10 mM imidazole (final concentrations). After incubation, the reaction mixture was applied to the Strep-Tactin affinity chromatography column and purification was performed according to the manufacturer's instructions (IBA, Inc). Column fractions were subjected to SDS-PAGE and Western Blot analyses, and apocyt c2 was detected with anti-His and anti-Strep antibodies whereas CcmG was identified with CcmG antisera.
Spectroscopic analysis
Optical absorbance redox difference spectra were recorded using a Hitachi U-3210 UV/vis spectrophotometer between 500-600 nm. Chromatophore membrane supernatants were centrifuged at 120000 × g for 2 hours to eliminate membrane debris. The supernatants thus obtained were adjusted with 50 mM MOPS/100 mM KCl to a protein concentration of 0.5 mg/ml and oxidized by adding a crystal of potassium ferricyanide, and subsequently reduced by either solid sodium ascorbate or sodium dithionite to measure the total amounts of reduced cyts c.
Chemicals
All chemicals were of reagent grade and obtained from commercial sources.
Acknowledgments
This work was supported by grants from DOE 91ER20052 and NIH GM 38237 to F. D. The authors thank to Drs. R. G. Kranz for providing anti-CcmG and anti-CcmH antibodies, J. F. Collet for plasmid pQE60-helX, and K. Zhang and M. Deshmukh for construction of various pKZ plasmids and MD strains, respectively.
References
- Allen JW, Barker PD, Ferguson SJ. A cytochrome b562 variant with a c-type cytochrome CXXCH heme-binding motif as a probe of the Escherichia coli cytochrome c maturation system. J Biol Chem. 2003a;278:52075–52083. doi: 10.1074/jbc.M307196200. [DOI] [PubMed] [Google Scholar]
- Allen JW, Daltrop O, Stevens JM, Ferguson SJ. C-type cytochromes: diverse structures and biogenesis systems pose evolutionary problems. Philos Trans R Soc Lond B Biol Sci. 2003b;358:255–266. doi: 10.1098/rstb.2002.1192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ. Basic local alignment search tool. J Mol Biol. 1990;215:403–410. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- Barker PD, Ferguson SJ. Still a puzzle: why is haem covalently attached in c-type cytochromes? Structure. 1999;7:281–290. doi: 10.1016/s0969-2126(00)88334-3. [DOI] [PubMed] [Google Scholar]
- Beckman DL, Trawick DR, Kranz RG. Bacterial cytochromes c biogenesis. Genes Dev. 1992;6:268–283. doi: 10.1101/gad.6.2.268. [DOI] [PubMed] [Google Scholar]
- Beckman DL, Kranz RG. Cytochromes c biogenesis in a photosynthetic bacterium requires a periplasmic thioredoxin-like protein. Proc Natl Acad Sci U S A. 1993;90:2179–2183. doi: 10.1073/pnas.90.6.2179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benning C, Somerville CR. Isolation and genetic complementation of a sulfolipid-deficient mutant of Rhodobacter sphaeroides. J Bacteriol. 1992;174:2352–2360. doi: 10.1128/jb.174.7.2352-2360.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Colbert CL, Wu Q, Erbel PJ, Gardner KH, Deisenhofer J. Mechanism of substrate specificity in Bacillus subtilis ResA, a thioredoxin-like protein involved in cytochrome c maturation. Proc Natl Acad Sci U S A. 2006;103:4410–4415. doi: 10.1073/pnas.0600552103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crow A, Acheson RM, Le Brun NE, Oubrie A. Structural basis of Redox-coupled protein substrate selection by the cytochrome c biosynthesis protein ResA. J Biol Chem. 2004;279:23654–23660. doi: 10.1074/jbc.M402823200. [DOI] [PubMed] [Google Scholar]
- Daldal F, Cheng S, Applebaum J, Davidson E, Prince RC. Cytochrome c(2) is not essential for photosynthetic growth of Rhodopseudomonas capsulata. Proc Natl Acad Sci U S A. 1986;83:2012–2016. doi: 10.1073/pnas.83.7.2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Daltrop O, Allen JW, Willis AC, Ferguson SJ. In vitro formation of a c-type cytochrome. Proc Natl Acad Sci U S A. 2002;99:7872–7876. doi: 10.1073/pnas.132259099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davidson E, Daldal F. Primary structure of the bc1 complex of Rhodopseudomonas capsulata. Nucleotide sequence of the pet operon encoding the Rieske cytochrome b, and cytochrome c1 apoproteins. J Mol Biol. 1987;195:13–24. doi: 10.1016/0022-2836(87)90323-8. [DOI] [PubMed] [Google Scholar]
- Deshmukh M, Brasseur G, Daldal F. Novel Rhodobacter capsulatus genes required for the biogenesis of various c-type cytochromes. Mol Microbiol. 2000;35:123–138. doi: 10.1046/j.1365-2958.2000.01683.x. [DOI] [PubMed] [Google Scholar]
- Deshmukh M, May M, Zhang Y, Gabbert KK, Karberg KA, Kranz RG, Daldal F. Overexpression of ccl1-2 can bypass the need for the putative apocytochrome chaperone CycH during the biogenesis of c-type cytochromes. Mol Microbiol. 2002;46:1069–1080. doi: 10.1046/j.1365-2958.2002.03212.x. [DOI] [PubMed] [Google Scholar]
- Deshmukh M, Turkarslan S, Astor D, Valkova-Valchanova M, Daldal F. The dithiol:disulfide oxidoreductases DsbA and DsbB of Rhodobacter capsulatus are not directly involved in cytochrome c biogenesis, but their inactivation restores the cytochrome c biogenesis defect of CcdA-null mutants. J Bacteriol. 2003;185:3361–3372. doi: 10.1128/JB.185.11.3361-3372.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Di Matteo A, Gianni S, Schinina ME, Giorgi A, Altieri F, Calosci N, Brunori M, Travaglini-Allocatelli C. A strategic protein in cytochrome c maturation: three-dimensional structure of CcmH and binding to apocytochrome c. J Biol Chem. 2007;282:27012–27019. doi: 10.1074/jbc.M702702200. [DOI] [PubMed] [Google Scholar]
- Ditta G, Schmidhauser T, Yakobson E, Lu P, Liang XW, Finlay DR, Guiney D, Helinski DR. Plasmids related to the broad host range vector, pRK290, useful for gene cloning and for monitoring gene expression. Plasmid. 1985;13:149–153. doi: 10.1016/0147-619x(85)90068-x. [DOI] [PubMed] [Google Scholar]
- Edeling MA, Guddat LW, Fabianek RA, Thony-Meyer L, Martin JL. Structure of CcmG/DsbE at 1.14 A resolution: high-fidelity reducing activity in an indiscriminately oxidizing environment. Structure. 2002;10:973–979. doi: 10.1016/s0969-2126(02)00794-3. [DOI] [PubMed] [Google Scholar]
- Edeling MA, Ahuja U, Heras B, Thony-Meyer L, Martin JL. The acidic nature of the CcmG redox-active center is important for cytochrome c maturation in Escherichia coli. J Bacteriol. 2004;186:4030–4033. doi: 10.1128/JB.186.12.4030-4033.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Erlendsson LS, Hederstedt L. Mutations in the thiol-disulfide oxidoreductases BdbC and BdbD can suppress cytochrome c deficiency of CcdA-defective Bacillus subtilis cells. J Bacteriol. 2002;184:1423–1429. doi: 10.1128/JB.184.5.1423-1429.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fabianek RA, Hennecke H, Thony-Meyer L. The active-site cysteines of the periplasmic thioredoxin-like protein CcmG of Escherichia coli are important but not essential for cytochrome c maturation in vivo. J Bacteriol. 1998;180:1947–1950. doi: 10.1128/jb.180.7.1947-1950.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fabianek RA, Hofer T, Thony-Meyer L. Characterization of the Escherichia coli CcmH protein reveals new insights into the redox pathway required for cytochrome c maturation. Arch Microbiol. 1999;171:92–100. doi: 10.1007/s002030050683. [DOI] [PubMed] [Google Scholar]
- Gao T, O'Brian MR. Control of DegP-dependent degradation of c-type cytochromes by heme and the cytochrome c maturation system in Escherichia coli. J Bacteriol. 2007;189:6253–6259. doi: 10.1128/JB.00656-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Georgiou CD, Fang H, Gennis RB. Identification of the cydC locus required for expression of the functional form of the cytochrome d terminal oxidase complex in Escherichia coli. J Bacteriol. 1987;169:2107–2112. doi: 10.1128/jb.169.5.2107-2112.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gray KA, Grooms M, Myllykallio H, Moomaw C, Slaughter C, Daldal F. Rhodobacter capsulatus contains a novel cb-type cytochrome c oxidase without a CuA center. Biochemistry. 1994;33:3120–3127. doi: 10.1021/bi00176a047. [DOI] [PubMed] [Google Scholar]
- Katzen F, Beckwith J. Transmembrane electron transfer by the membrane protein DsbD occurs via a disulfide bond cascade. Cell. 2000;103:769–779. doi: 10.1016/s0092-8674(00)00180-x. [DOI] [PubMed] [Google Scholar]
- Katzen F, Deshmukh M, Daldal F, Beckwith J. Evolutionary domain fusion expanded the substrate specificity of the transmembrane electron transporter DsbD. Embo J. 2002;21:3960–3969. doi: 10.1093/emboj/cdf405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keen NT, Tamaki S, Kobayashi D, Trollinger D. Improved broad-host-range plasmids for DNA cloning in gram-negative bacteria. Gene. 1988;70:191–197. doi: 10.1016/0378-1119(88)90117-5. [DOI] [PubMed] [Google Scholar]
- Koch HG, Hwang O, Daldal F. Isolation and characterization of Rhodobacter capsulatus mutants affected in cytochrome cbb3 oxidase activity. J Bacteriol. 1998;180:969–978. doi: 10.1128/jb.180.4.969-978.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kojima N, Yamanaka M, Ichiki S, Sambongi Y. Unexpected elevated production of Aquifex aeolicus cytochrome c555 in Escherichia coli cells lacking disulfide oxidoreductases. Biosci Biotechnol Biochem. 2005;69:1418–1421. doi: 10.1271/bbb.69.1418. [DOI] [PubMed] [Google Scholar]
- Kranz R, Lill R, Goldman B, Bonnard G, Merchant S. Molecular mechanisms of cytochrome c biogenesis: three distinct systems. Mol Microbiol. 1998;29:383–396. doi: 10.1046/j.1365-2958.1998.00869.x. [DOI] [PubMed] [Google Scholar]
- Laemmli UK. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature. 1970;227:680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- Lowry OH, Rosebrough NJ, Farr AL, Randall RJ. Protein measurement with the Folin phenol reagent. J Biol Chem. 1951;193:265–275. [PubMed] [Google Scholar]
- Metheringham R, Tyson KL, Crooke H, Missiakas D, Raina S, Cole JA. Effects of mutations in genes for proteins involved in disulphide bond formation in the periplasm on the activities of anaerobically induced electron transfer chains in Escherichia coli K12. Mol Gen Genet. 1996;253:95–102. doi: 10.1007/pl00013815. [DOI] [PubMed] [Google Scholar]
- Monika EM, Goldman BS, Beckman DL, Kranz RG. A thioreduction pathway tethered to the membrane for periplasmic cytochromes c biogenesis; in vitro and in vivo studies. J Mol Biol. 1997;271:679–692. doi: 10.1006/jmbi.1997.1227. [DOI] [PubMed] [Google Scholar]
- Onder O, Turkarslan S, Sun D, Daldal F. Overproduction or absence of the periplasmic protease DegP severely compromises bacterial growth in the absence of the dithiol:disulfide oxidoreductase DsbA. Mol Cell Proteomics. 2008 doi: 10.1074/mcp.M700433-MCP200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pittman MS, Robinson HC, Poole RK. A bacterial glutathione transporter (Escherichia coli CydDC) exports reductant to the periplasm. J Biol Chem. 2005;280:32254–32261. doi: 10.1074/jbc.M503075200. [DOI] [PubMed] [Google Scholar]
- Poole RK, Gibson F, Wu G. The cydD gene product, component of a heterodimeric ABC transporter, is required for assembly of periplasmic cytochrome c and of cytochrome bd in Escherichia coli. FEMS Microbiol Lett. 1994;117:217–223. doi: 10.1111/j.1574-6968.1994.tb06768.x. [DOI] [PubMed] [Google Scholar]
- Prentki P, Krisch HM. In vitro insertional mutagenesis with a selectable DNA fragment. Gene. 1984;29:303–313. doi: 10.1016/0378-1119(84)90059-3. [DOI] [PubMed] [Google Scholar]
- Ren Q, Ahuja U, Thony-Meyer L. A bacterial cytochrome c heme lyase. CcmF forms a complex with the heme chaperone CcmE and CcmH but not with apocytochrome c. J Biol Chem. 2002;277:7657–7663. doi: 10.1074/jbc.M110979200. [DOI] [PubMed] [Google Scholar]
- Robertson IB, Stevens JM, Ferguson SJ. Dispensable residues in the active site of the cytochrome c biogenesis protein CcmH. FEBS Lett. 2008 doi: 10.1016/j.febslet.2008.07.052. [DOI] [PubMed] [Google Scholar]
- Sambrook J, Russel DW. Molecular cloning: a laboratory manual. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press; 2001. [Google Scholar]
- Sanders C, Wethkamp N, Lill H. Transport of cytochrome c derivatives by the bacterial Tat protein translocation system. Mol Microbiol. 2001;41:241–246. doi: 10.1046/j.1365-2958.2001.02514.x. [DOI] [PubMed] [Google Scholar]
- Sanders C, Deshmukh M, Astor D, Kranz RG, Daldal F. Overproduction of CcmG and CcmFH(Rc) fully suppresses the c-type cytochrome biogenesis defect of Rhodobacter capsulatus CcmI-null mutants. J Bacteriol. 2005a;187:4245–4256. doi: 10.1128/JB.187.12.4245-4256.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanders C, Turkarslan S, Daldal F. Periplasmic oxidative folding and cytochrome c maturation: A mechanistic view of stereo-selective heme attachment. In: Van der Est A, Bruce D, editors. Photosynthesis: Fundamental Aspects to Global Perspectives. 2005b. [Google Scholar]
- Sanders C, Boulay C, Daldal F. Membrane-spanning and periplasmic segments of CcmI have distinct functions during cytochrome c Biogenesis in Rhodobacter capsulatus. J Bacteriol. 2007;189:789–800. doi: 10.1128/JB.01441-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schagger H, von Jagow G. Tricine-sodium dodecyl sulfate-polyacrylamide gel electrophoresis for the separation of proteins in the range from 1 to 100 kDa. Anal Biochem. 1987;166:368–379. doi: 10.1016/0003-2697(87)90587-2. [DOI] [PubMed] [Google Scholar]
- Scolnik PA, Walker MA, Marrs BL. Biosynthesis of carotenoids derived from neurosporene in Rhodopseudomonas capsulata. J Biol Chem. 1980;255:2427–2432. [PubMed] [Google Scholar]
- Sistrom WR. A requirement for sodium in the growth of Rhodopseudomonas spheroides. J Gen Microbiol. 1960;22:778–785. doi: 10.1099/00221287-22-3-778. [DOI] [PubMed] [Google Scholar]
- Thomas PE, Ryan D, Levin W. An improved staining procedure for the detection of the peroxidase activity of cytochrome P-450 on sodium dodecyl sulfate polyacrylamide gels. Anal Biochem. 1976;75:168–176. doi: 10.1016/0003-2697(76)90067-1. [DOI] [PubMed] [Google Scholar]
- Thony-Meyer L. Haem-polypeptide interactions during cytochrome c maturation. Biochim Biophys Acta. 2000;1459:316–324. doi: 10.1016/s0005-2728(00)00167-5. [DOI] [PubMed] [Google Scholar]
- Thony-Meyer L. Cytochrome c maturation: a complex pathway for a simple task? Biochem Soc Trans. 2002;30:633–638. doi: 10.1042/bst0300633. [DOI] [PubMed] [Google Scholar]
- Yen HC, Hu NT, Marrs BL. Characterization of the gene transfer agent made by an overproducer mutant of Rhodopseudomonas capsulata. J Mol Biol. 1979;131:157–168. doi: 10.1016/0022-2836(79)90071-8. [DOI] [PubMed] [Google Scholar]
- Zannoni D, Daldal F. The role of c-type cytochromes in catalyzing oxidative and photosynthetic electron transport in the dual functional plasmamembrane of facultative phototrophs. Arch Microbiol. 1993;160:413–423. doi: 10.1007/BF00245301. [DOI] [PubMed] [Google Scholar]