This paper describes how four general optimization techniques, growth-rate modulation, fine screening, seeding and additive screening, have been adapted for automation in a medium-throughput crystallization service facility.
Keywords: crystallization, automation, Phoenito experiments
Abstract
The use of crystallization robots for initial screening in macromolecular crystallization is well established. This paper describes how four general optimization techniques, growth-rate modulation, fine screening, seeding and additive screening, have been adapted for automation in a medium-throughput crystallization service facility. The use of automation for more challenging optimization experiments is discussed, as is a novel way of using both the Mosquito and the Phoenix nano-dispensing robots during the setup of a single crystallization plate. This dual-dispenser technique plays to the strengths of both machines.
1. Introduction
One of the major benefits of the structural genomics era has been the development of automation tools for macromolecular crystallization that have migrated from dedicated structural genomics projects into the home laboratory (Hughes & Ng, 2007 ▶). Three classes of robots are in common use for automated crystallization: liquid-handling machines that create bespoke crystallization screens, sub-microlitre dispensing robots, which are used to create experimental plates, and imaging systems that record the progress of crystallization experiments (Berry et al., 2006 ▶). There are a number of machines available commercially within each class; for example, the Oryx (Douglas Instruments), Phoenix (Art Robbins Industries), Mosquito (TTP) and Honeybee 963 (Genomic Solutions) are popular choices for machines that are used to set up experimental crystallization plates (Berry et al., 2006 ▶).
The Bio21 Collaborative Crystallization Centre (C3) is a shared crystallization facility in Melbourne that has five initial academic and government partners [The Commonwealth Scientific and Industrial Research Organization (CSIRO), The Walter and Eliza Hall Institute of Medical Research (WEHI), St Vincent’s Institute (SVI), The Burnet Institute (BI) and The Victorian College of Pharmacy (VCP)]. The core of this facility is housed at the CSIRO laboratory in Parkville and has sufficient equipment and IT infrastructure to set up and image crystallization experiments for researchers from the five partner institutes as well as other scientists from around Australia. The automation in the core facility consists of a TECAN EVO 100 (Tecan), a Phoenix, a Mosquito, two Minstrel Imagers HT and two incubators (Rigaku), along with the CrystalTrak database application (Rigaku) and some custom software. The vast majority of the plates set up in the centre are sitting-drop vapour-diffusion experiments using a two-subwell 96-well sitting-drop plastic plate (Innovaplate SD-2, Innovadyne), with the total volume of the crystallization droplets ranging from 200 to 800 nl.
The rationale for setting up the centre was to provide a small-volume (sub-microlitre) crystallization screening service for the local structural biology community. Since then, the centre has extended its services to include robotic optimization. This addition required the creation of protocols that could simplify optimization down to a limited number of robust techniques which would be suitable for the available automation. Given that the role of C3 is as a service provider to over 70 individual researchers, these optimization protocols have to be customizable, as few (if any) of the users of C3 agree as to what is the best path from an initial hit to a well diffracting crystal. This negates the implementation of a single optimization path as has been shown to be very successful elsewhere (Leulliot et al., 2005 ▶). Optimization of initial crystal hits can require significant ingenuity and creativity (see, for example, Hassell et al., 2007 ▶); however, most optimization techniques are a variant of one of four basic protocols: modulation of the rate of crystallization, directed or fine screening, seeding and additive screening (Bergfors, 2007 ▶). These four cornerstones of crystal-growth optimization are quite straightforward; it is the infinite variation in their application which makes optimization such a challenge. Modulation of the rate of crystallization (generally slowing down crystal growth) can be achieved by manipulating the concentration of the crystallant or protein, adjusting the ratio of the protein solution to the crystallant solution, changing the experimental setup or a combination of all of these techniques (Luft et al., 2007 ▶; Walter et al., 2005 ▶). In fine screening, chemicals that were presumed to positively affect crystallization in the initial screening are recombined to create directed screens that sample a small part of crystallization chemical space more closely than did the initial experiments (Shaw-Stewart & Baldock, 1999 ▶). Seeding is the optimization process by which crystal nucleation is decoupled from crystal growth within the crystallization experiment (Stura & Wilson, 1992 ▶; Bergfors, 2003 ▶). In additive optimization, a collection of single or pooled chemicals are tested for their effect on a single crystallization condition (Cudney et al., 1994 ▶).
Irrespective of whether the experiment is a screening experiment or optimization, the core functionality of C3 is to set up low-volume crystallization experiments. To ensure that the centre can always perform this service for its users, CSIRO invested in a second protein-dispensing robot. The initial robot was a Phoenix; a different robot, a Mosquito, was chosen as the second protein-dispensing robot, which provides complementary functionality as well as the required core ability to set up low-volume crystallization drops. Having the two robots has allowed the development of crystallization protocols that utilize the strengths of both machines for a single crystallization plate. For seeding and additive optimization experiments, a protocol is used (dubbed the ‘Phoenito’ protocol) that starts on the Phoenix robot then moves to the Mosquito robot.
As we are often asked about the relative pros and cons of different low-volume dispensers, it is clear that there is little literature to source this type of information. The relative strengths and weaknesses of the two dispensers in the centre are quite different and a discussion of these is presented.
2. Discussion
2.1. The C3 centre
To date (from the second quarter of 2006 to July 2008), close to 4000 96-well crystallization plates have been set up and imaged using less than two full-time equivalent employees. Most of the crystallization plates set up have been screening plates, with JCSG+ and PACT (Newman et al., 2005 ▶) being the two most commonly requested screens. Which initial screens are used seems to be quite subjective: for instance, C3 offers two standard collections of 8 × 96-well initial screens, one made up of commercially available screens (the commercial_set, which consists of Crystal Screen HT, Index, Grid Screen MPD and Grid Screen Ammonium Sulfate from Hampton Research, Wizard I and II and Expanded Precipitant Synergy from Emerald BioSystems and The PACT Suite and The Anion Suite from Qiagen) and one which was designed in-house and produced for C3 by Emerald BioSystems (the c3 screen). About 75% of the conditions in the two collections are identical. Researchers from one partner institute (SVI) use the commercial_set almost exclusively, whereas researchers from another institute (WEHI) use the c3 screen. The high use of JCSG+ and PACT may have more to do with suggestions from the staff of C3 than any intrinsic properties of those two screens. Overall, about 130 of each of the two eight-plate collections have been set up, compared with 300 instances of JCSG+. PACT has been set up (independently of the commercial collection) about 110 times, as has the PEG/Ion HT Screen from Hampton Research. The users of the facility may score the images of the plates they have requested through the web-based version of CrystalTrak (CTweb; Rigaku). As the centre has no robust automatic scoring function and manual scoring cannot be enforced, there is no straightforward method of determining the relative success rates of the screens. The centre has no control over the quality of the samples submitted, nor is there any control of the scoring performed by the users of the centre. Therefore, our success metrics are based more on whether samples are set up as the user requested and in a timely manner. An additional metric is the number of users that have come back to use the centre again. This assumes that repeat users are those that are satisfied with the service.
2.2. Optimization techniques at C3
2.2.1. Modulation of the rate of crystal nucleation and growth
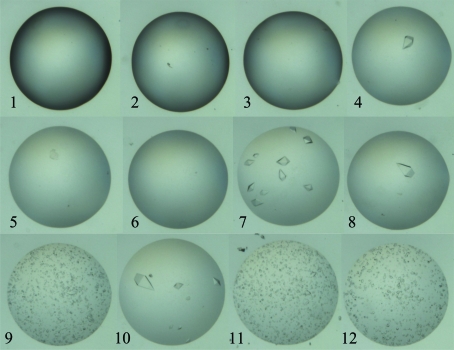
The rate at which crystals nucleate and grow can be most trivially altered by changing the drop size of the crystallization experiment: larger drops equilibrate more slowly with the reservoir in a vapour-diffusion experiment (Carter et al., 2005 ▶; Newman et al., 2007 ▶). Changing the ratio of the components within the droplet will also alter the rate of equilibration (Dunlop & Hazes, 2003 ▶). Both of these techniques are used at C3. Limitations of the CrystalTrak database software and the Phoenix robot require that for any one set of 96 droplets the size and ratio of the protein and crystallant solutions are kept constant, unlike the elegant DVR/T optimizations described by Luft et al. (2007 ▶). Another method of altering the kinetics of equilibration in use at C3 is to set up identical crystallization droplets which consist of the crystallant pre-mixed with the protein solution over reservoirs that contain a gradient of salt concentration (Newman, 2005 ▶; see Fig. 1 ▶).
Figure 1.
Thaumatin (T7638, Sigma–Aldrich) at 8 mg ml−1 in 0.5 M sodium potassium tartrate, 0.05 M bis-tris propane pH 6.5 was set up in 200 nl droplets over reservoirs containing linearly increasing concentrations of sodium chloride [from 0.7 M (droplet 1) to 2.5 M (droplet 12)]; images were collected 5 d after the droplets were set up. There appears to be a nonlinear response to this linear gradient; the growth of large single crystals in droplet 8 (1.845 M) and droplet 10 (2.172 M) interspersed with the growth of numerous microcrystals in droplet 9 (2.009 M) was observed repeatedly (data not shown). The absence of crystals in droplets 5 and 6 may arise from a lack of spontaneous nucleation in those droplets over the timeframe of the experiment.
2.2.2. Fine screening
The initial screens in use at C3 are commercial screens, bought in 10 ml aliquots, which have been transferred to 96 deep-well blocks (Axygen P-2ML-SQ). These source blocks are placed on the Phoenix robot and used to fill the experimental plates as part of the standard Phoenix protocol for setting up a crystallization experiment. One obvious way to accommodate fine screening is to create optimization or fine screens in the same 96 deep-well blocks as the initial screens. The same dispensing protocols may then be used for the optimization screens as were used for the initial screening. The optimization and reservoir-design functionalities of CrystalTrak are used to design optimization screens, which can be simple gradients or more complicated ‘Crystools’ such as random optimizations (Segelke & Rupp, 1998 ▶; Segelke, 2001 ▶). The recipe-generation tool from CrystalTrak is used to translate the design (which is in units of concentration) into a recipe, in which the concentration information is converted into the equivalent information but now in terms of specific stock concentrations and the associated required volumes. The CrystalChef application (Spocksoft software) reads the recipe generated by CrystalTrak, generates a Tecan workfile, prompts the user to place the correct stocks on the Tecan deck and starts a Tecan run. A 96 deep-well block containing 1 ml of each condition generally takes 1 h or less to produce. The block is then sealed and mixed before being used on the Phoenix in a standard plate-setup protocol.
2.2.3. Seeding
Seeding in C3 is performed using the Mosquito robot. A typical Mosquito seeding protocol uses 150 nl protein, 15 nl seed stock and 135 nl reservoir, yielding a droplet in which the seed stock is 5% of the total drop volume. The protocol in use at C3 requires that the protein component of the droplet be dispensed, then the seeds aspirated, the reservoir solution aspirated and the resultant volume dispensed on top of the existing protein drop. Both the protein (if dispensed using the Mosquito) and the seed stock are pre-aliquoted into separate columns of a V-bottomed microtitre plate (651101, Greiner Bio-one) and sealed with a strip of tape (Scotch ‘Magic Tape’ 810) to minimize evaporation. The type of tape used to cover the wells is important, as the Mosquito tips have problems piercing some thicker or more plastic tapes. The tape is also a convenient way of distinguishing used rows of a V-plate from the clean rows, as the same V-plate can be used for 12 different samples.
2.2.4. Additive screening
The Phoenix is used to create additive-optimization experiments. The additive screen is stored in a 96 deep-well block and 10 ml of the ‘base condition’ (that is, the promising crystallization condition, which will be found in every well of the additive optimization) is made up by hand. The Tecan robot is used to aliquot 50 µl of this base condition into each reservoir of a 96-well crystallization plate, which is then transferred to the Phoenix. A protocol is used which aspirates 6 µl of the additive screen and mixes this into the reservoir solution, thus creating 96 unique conditions. The reservoir solution is now nominally 90% base condition and 10% additive. These mixed reservoirs are then used to create the crystallization droplet in a standard screening-like experiment, yielding a drop which has a 10:9:1 (protein:reservoir:additive) ratio.
This same protocol used in seeding (above) can be used to introduce small aliquots of an expensive additive screen directly into the crystallization droplet, bypassing the need to pre-mix the additive into the reservoir solution.
2.3. Optimization at C3
For optimization, over 100 96-well fine screens have been produced and microseeding has been used in 124 crystallization plates; 132 additive screens have been run [39 using the Silver Bullets screen, 46 using Additive Screen HT and 13 using Detergent Screen HT (all from Hampton Research) and 34 using the OptiSalt Suite from Qiagen]. Less than five optimizations have been set up over a salt gradient to modulate the kinetics of the experiment. As this latter technique has not been widely promoted, it could be that most users do not know that this optimization strategy exists, rather than it being unpopular.
There are a number of different approaches to seeding, from macroseeding, which requires a large amount of manual dexterity, to various types of microseeding, in which a small aliquot of a seed stock needs to be added to the crystallization droplet: this is often introduced via a horsehair wand or an acupuncture needle (Bergfors, 2003 ▶). Recently, microseeding has also been shown to be very powerful in initial screening (D’Arcy et al., 2007 ▶). Most of the current crystallization robots can reliably dispense 100 nl, or perhaps 50 nl (results from a trial performed at the BioXhit/SPINE Course on High-Throughput Macromolecular Crystallization, Amsterdam, 2005, see http://www.bioxhit.org; Berry et al., 2006 ▶), but struggle with smaller volumes. For a microseeding experiment, the seed-stock volume should be a minor fraction of the total drop volume; in a 200 nl crystallization drop even 50 nl is 25% of the volume. However, aspiration of small volumes rarely has the same problem. The Mosquito robot is well suited to seeding experiments, as the multi-aspirate function allows one to perform serial aspirations and then dispense the cumulative volume, efficiently side-stepping the problem of low-volume dispensing. Certainly, other robots can also be used for seeding: the matrix-seeding experiments performed by D’Arcy et al. (2007 ▶) used the Oryx 8 robot from Douglas Instruments.
Additive screening using the Phoenix robot (described above) works well for some of the less expensive additive screens (for example, The Optisalt Suite from Qiagen or Additive Screen 96 from Hampton Research), but would be prohibitively expensive for a 96-well detergent optimization or the Silver Bullet screen from Hampton Research (McPherson & Cudney, 2006 ▶). In these cases, we use the Phoenito technique (described below) to reduce the costs of the experiment.
2.4. Robots
The two dispensing robots both have strong points and weaknesses. The Phoenix is relatively cheap to run, as the only consumables are the nano-dispensing tip and the Teflon tips of the 96 syringes, neither of which have to be replaced often. In our experience, a good nano-tip will provide three months of use (a few hundred plates) and the Teflon tips need replacing every six months or thereabouts. Currently, there is an issue with quality control of the nano-tips, which means that about one in three nano-tip assemblies do not work or do not work well enough for the production of crystallization plates. However, given a good nano-tip, the Phoenix produces exquisite droplets and requires only about 5 µl of additional protein over and above that which is dispensed into the plate. One of the downsides of the Phoenix is the requirement to wash both dispensers: this means that the footprint of the machine is significantly more than just the machine itself, as the system includes water containers and pumps. The washing slows down each protocol overall, although it does not greatly influence the time that the nascent protein droplets are exposed. The requirement for washing also can lead to anxiety over how much to wash and a concern if all contaminants will be removed by the aqueous wash cycle. The C3 centre uses standard protocols that include a wash at the beginning as well as at the end of each run, so that the tips are in the same state (i.e. just washed) each time the machine is used. A more serious issue is the inability of the Phoenix nano-tip to dispense some protein solutions successfully. This appears to be related in part to the viscosity of the protein solution; protein dispensing becomes unreliable if the samples contain more than 10% glycerol. Other factors which may to be related to poor nano-tip dispensing are samples that contain highly glycosylated proteins or very hydrophobic components (peptide ligands, for example). The protein samples do need to be either spun or filtered before use, as the fine capillary of the nano-tip can become blocked.
The Mosquito’s positives include a very tight footprint, high speed and the use of positive-displacement disposable tips, which remove any doubts about contamination as well as enabling the aspiration and dispensing of very viscous liquids. At C3, even protein samples that have gelled have been successfully set up with the Mosquito. Furthermore, the use of disposable tips allows this machine to be essentially maintenance-free, which would be even more of a positive if C3 were a hands-on user facility rather than a service. The column-wise dispensing of the Mosquito allows the set up of ‘mirrored’ drops which can be inverted over a pre-filled plate to give a hanging-drop experiment. The less endearing features of the Mosquito include the cost of the tips, which currently cost about 14 cents each, adding over $27 (AUD) to the cost of setting up a two-subwell 96-well crystallization plate, which is a significant fraction of the total cost of the plate, excluding the cost of the sample. Another issue is having to aliquot out the protein sample into eight wells for set up: this means that setting up a crystallization experiment on the Mosquito will invariably require more protein sample than the same experiment set up on the Phoenix. Table 1 ▶ shows a comparison of the amount of sample required for the two machines. Finally, the Mosquito requires that the reservoirs of the crystallization plate be pre-filled with reservoir solution.
Table 1. A comparison of the volume of sample that C3 requires its users to provide for the Phoenix and the Mosquito robots.
For the Phoenix, these numbers were generated from the equation [drop size × (No. of plates × 96) × 1.2] + (No. of plates × 5) + 10. The (No. of plates × 5) is required as each plate is a new aspiration step, which requires an overfill and pre-dispense to prime the tip. The constant 10 is to ensure that there is enough liquid in the tubes to enable aspiration for the last plate. The factor of 1.2 is included as the variation in dispensing different protein samples can lead to drops being 20% bigger than the requested volume. For the Mosquito, the quantity required was generated from the following: the maximum of [(1.1 × drop size) or 5] × 8 + 2 (for the first plate in a set) and 1.6 × drop size × 96 (for each additional plate in that set). The maximum of [(1.1 × drop size) or 5] is required as there has to be a sufficient pool of protein in the well of the V-plate to ensure reliable tip filling. The constant 2 is required to counteract the losses in manually pipetting protein from the original sample tube to the V-plate.
| No. of plates of 96 drops of 100 nl each | Phoenix (µl) (rounded up) | Mosquito (µl) (rounded up) |
|---|---|---|
| 1 | 27 | 42 |
| 2 | 44 | 58 |
| 3 | 60 | 74 |
| 4 | 77 | 90 |
| 5 | 93 | 106 |
| 6 | 110 | 122 |
| 7 | 127 | 138 |
| 8 | 143 | 154 |
| 9 | 159 | 170 |
| 10 | 176 | 186 |
2.5. The Phoenito experiment
In a Phoenito experiment, the crystallization plate, the deep-well block containing the screening solutions and the protein sample are loaded onto the deck of the Phoenix. The seed stock or additive screen are loaded into a V-bottomed 96-well plate and placed on the Mosquito. The Phoenix part of the protocol transfers crystallant into the reservoirs of the crystallization plate and dispenses the protein into the sample wells. The crystallization plate is then transferred to the Mosquito, where a multi-aspirate protocol is run, completing the droplets with seeds/additives and crystallant. The plate is then removed and sealed. For 96 drops, the elapsed time from the first dispense of a protein droplet to sealing the plate is approximately 3 min.
The advantages of the Phoenito experiment for seeding are that only the seed stock (which is generally available in relatively large quantities) needs to be aliquoted out into eight wells and the seed stock may be anything: crushed crystals or powdered horsehair or other ‘lumpy’ heterologous nucleants (D’Arcy et al., 2003 ▶; Thakur et al., 2007 ▶). The advantage of the Phoenito experiment for an additive screen is that one can access 5:4:1 ratios of protein:reservoir:additive and still have small droplets. Hampton Research recommends this 5:4:1 ratio for their detergent additive screens (http://www.hamptonresearch.com, product insert for Detergent Screen HT) and using a Phoenito protocol this ratio has been set up in a total drop volume of 200 nl (100 nl protein:80 nl reservoir:20 nl detergent). Fig. 2 ▶ shows images from a Phoenito seeding experiment. In this experiment, each of the 96 reservoirs of an SD-2 plate were filled with the same solution; the top subwells all contain identical unseeded droplets and the bottom subwells all contain identical seeded droplets. After 19 h, there were no crystals in the 96 unseeded droplets, whereas 68 of the 96 seeded droplets contained crystals. Although some crystals did eventually grow in the unseeded droplets, the set of seeded droplets always yielded more crystals than the unseeded drops (data not shown).
Figure 2.
No seeding (a) versus seeding (b) for the protein trypsin. Trypsin (T1246, Sigma–Aldrich) at 50 mg ml−1 in 10 mM CaCl2, 1 mM benzamidine was set up in droplets consisting of (a) 300 nl protein solution and 200 nl reservoir solution or (b) 300 nl protein solution, 195 nl reservoir solution and 5 nl seed stock. The reservoir consists of 25%(w/v) PEG 3350, 0.2 M ammonium sulfate, 0.1 M bis-tris pH 5.5 in each case. The seed stock consisted of crystals of the trypsin–CaCl2–benzamidine complex crushed in the reservoir solution using a seed-bead protocol (Luft & DeTitta, 1999 ▶). The coloured border around each droplet is a user-selected scoring: any shade on the palette of yellow to red indicates that the user scored that drop as crystalline or containing crystals. The scoring scheme describes the colours as follows: lemon, crystalline; dark yellow, crystals*; orange, crystals**; red, crystals***. The images were collected 19 h after the plate was set up.
Both instruments can be used independently to run additive experiments, but with limitations: for example, Silver Bullet additive screens have been run on the Phoenix alone, but given that the recommended ratio of protein:reservoir:additive is 2:1:1 (McPherson & Cudney, 2006 ▶), the minimum drop size for the droplets is 400 nl. Using the Phoenix to perform the Silver Bullet screens also means that a minimum of 1.2 µl of each additive is used per optimization, compared with less than 200 nl for the same screen run with a Phoenito protocol. Using the Mosquito to perform the entire additive experiment uses more of the expensive tips, as well as using significantly more protein than the protocol that uses both machines. Seeding experiments can be performed on the Mosquito alone, but with a concomitant need for more protein than in a Phoenito experiment.
3. Conclusion
We have successfully adapted four common optimization techniques to the automation available in the Bio21 Collaborative Crystallization Centre, which are general techniques rather than specific implementations of an optimization strategy. A novel protocol has been developed that uses both a Phoenix and a Mosquito robot, which effectively plays to the strengths of these two machines whilst avoiding their weaknesses.
Acknowledgments
Thanks to the users of the Bio21 C3 and to the State Government of Victoria for providing the initial funds to start up the centre. Thanks to Patricia Pilling for designing the initial CSIRO screen, upon which the c3 screen is based. We would like to thank the initial reviewers and particularly the co-editor of the paper for their comments, which led to a significantly improved manuscript.
References
- Bergfors, T. (2003). J. Struct. Biol.142, 66–76. [DOI] [PubMed]
- Bergfors, T. (2007). Methods Mol. Biol.363, 131–151. [DOI] [PubMed]
- Berry, I. M., Dym, O., Esnouf, R. M., Harlos, K., Meged, R., Perrakis, A., Sussman, J. L., Walter, T. S., Wilson, J. & Messerschmidt, A. (2006). Acta Cryst. D62, 1137–1149. [DOI] [PubMed]
- Carter, D. C., Rhodes, P., McRee, D. E., Tari, L. W., Dougan, D. R., Snell, G., Abola, E. & Stevens, R. C. (2005). J. Appl. Cryst.38, 87–90.
- Cudney, R., Patel, S., Weisgraber, K., Newhouse, Y. & McPherson, A. (1994). Acta Cryst. D50, 414–423. [DOI] [PubMed]
- D’Arcy, A., Mac Sweeney, A. & Haber, A. (2003). Acta Cryst. D59, 1343–1346. [DOI] [PubMed]
- D’Arcy, A., Villard, F. & Marsh, M. (2007). Acta Cryst. D63, 550–554. [DOI] [PubMed]
- Dunlop, K. V. & Hazes, B. (2003). Acta Cryst. D59, 1797–1800. [DOI] [PubMed]
- Hassell, A. M. et al. (2007). Acta Cryst. D63, 72–79.
- Hughes, R. C. & Ng, J. D. (2007). Cryst. Growth Des.7, 2226–2238.
- Leulliot, N., Trésaugues, L., Bremang, M., Sorel, I., Ulryck, N., Graille, M., Aboulfath, I., Poupon, A., Liger, D., Quevillon-Cheruel, S., Janin, J. & van Tilbeurgh, H. (2005). Acta Cryst. D61, 664–670. [DOI] [PubMed]
- Luft, J. R. & DeTitta, G. T. (1999). Acta Cryst. D55, 988–993. [DOI] [PubMed]
- Luft, J. R., Wolfley, J. R., Said, M. I., Nagel, R. M., Lauricella, A. M., Smith, J. L., Thayer, M. H., Veatch, C. K., Snell, E. H., Malkowski, M. G. & DeTitta, G. T. (2007). Protein Sci.16, 715–722. [DOI] [PMC free article] [PubMed]
- McPherson, A. & Cudney, B. (2006). J. Struct. Biol.156, 387–406. [DOI] [PubMed]
- Newman, J. (2005). Acta Cryst. D61, 490–493. [DOI] [PubMed]
- Newman, J., Egan, D., Walter, T. S., Meged, R., Berry, I., Ben Jelloul, M., Sussman, J. L., Stuart, D. I. & Perrakis, A. (2005). Acta Cryst. D61, 1426–1431. [DOI] [PubMed]
- Newman, J., Xu, J. & Willis, M. C. (2007). Acta Cryst. D63, 826–832. [DOI] [PubMed]
- Segelke, B. W. (2001). J. Cryst. Growth, 232, 553–562.
- Segelke, B. W. & Rupp, B. (1998). Am. Crystallogr. Assoc. Meet. Ser.25, 78.
- Shaw-Stewart, P. D. & Baldock, P. F. M. (1999). J. Cryst. Growth, 196, 665–673.
- Stura, E. A. & Wilson, I. A. (1992). Crystallization of Nucleic Acids and Proteins: A Practical Approach, edited by A. Ducruix & R. Giegé, pp. 99–126. Oxford University Press.
- Thakur, A. S., Robin, G., Guncar, G., Saunders, N. F., Newman, J., Martin, J. L. & Kobe, B. (2007). PLoS ONE, 2, e1091. [DOI] [PMC free article] [PubMed]
- Walter, T. S. et al. (2005). Acta Cryst. D61, 651–657. [DOI] [PMC free article] [PubMed]