Ca2+-bound and Zn2+-bound forms of ALG-2 were crystallized and X-ray diffraction data were collected to 3.1 and 3.3 Å resolution, respectively.
Keywords: apoptosis, calcium-binding protein, penta-EF-hand
Abstract
ALG-2 (apoptosis-linked gene 2) is an apoptosis-linked calcium-binding protein with five EF-hand motifs in the C-terminal region. N-terminally truncated ALG-2 (des3-23ALG-2) was crystallized by the vapour-diffusion method in buffer consisting of either 50 mM MES pH 6.5, 12.5%(v/v) 2-propanol and 150 mM calcium acetate or 100 mM MES pH 6.0, 15%(v/v) ethanol and 200 mM zinc acetate. Crystals of the Ca2+-bound form belonged to space group P212121, with unit-cell parameters a = 54.8, b = 154.4, c = 237.7 Å, α = β = γ = 90°, and diffracted to 3.1 Å resolution. Crystals of the Zn2+-bound form belonged to space group P212121, with unit-cell parameters a = 52.8, b = 147.5, c = 230.7 Å, α = β = γ = 90°, and diffracted to 3.3 Å resolution. The structures of the Ca2+-bound form and the Zn2+-bound form were solved by the molecular-replacement method. Although both crystals contained eight ALG-2 molecules per asymmetric unit, the metal-ion locations and octameric arrangements were found to be significantly different.
1. Introduction
ALG-2 (apoptosis-linked gene 2) is a 22 kDa calcium-binding protein that contains five EF-hand motifs (EF1–EF5) in its C-terminal region and belongs to the penta-EF-hand (PEF) family, which includes conventional calpains, sorcin, grancalcin and peflin in mammals (for reviews, see Maki et al., 1997 ▶, 2002 ▶). With the exception of the calpain large subunit, PEF family proteins have various lengths of Gly/Pro-rich hydrophobic regions in their N-terminal regions (human ALG-2: 1-MAAYSYRPGPGAGPGPAAGAALP-23). ALG-2 and its interacting protein Alix/AIP1 have been suggested to function in apoptosis, development and cancer (Vito et al., 1996 ▶; Sadoul, 2006 ▶; Maki & Shibata, 2007 ▶). ALG-2 exists as a weak dimer in solution in the absence of Ca2+ (Lo et al., 1999 ▶) and forms a homodimer or a heterodimer with another PEF protein, peflin, through their EF5 regions (Kitaura et al., 2001 ▶, 2002 ▶). However, in the presence of Ca2+ ALG-2 undergoes a conformational change that exposes its hydrophobic surfaces and precipitates at high protein concentrations (Maki et al., 1998 ▶). The Ca2+-bound form of elastase-treated mouse ALG-2 (des1-20ALG-2) has been solved (PDB code 1hqv; Jia et al., 2001 ▶) and we have recently reported the structures of the Ca2+-bound form of N-terminally truncated human ALG-2 (des3-20ALG-2) and the Zn2+-bound form of full-length ALG-2 (Suzuki et al., 2008 ▶). Here, we report the crystallization and X-ray diffraction analysis of Ca2+-bound and Zn2+-bound forms of human ALG-2 with a deletion of amino-acid residues 3–23 (des3-23ALG-2; Fig. 1 ▶). Both crystals contain eight ALG-2 molecules in the asymmetric unit, but the metal-ion locations and octameric arrangements were found to differ significantly between crystals grown in the presence of Ca2+ or Zn2+.
Figure 1.

N-terminal sequences of mouse and human ALG-2. There are two amino-acid substitutions between mouse and human ALG-2 in the full-length sequence of 191 residues (Gly12 and Gln46 in mouse versus Ala12 and Thr46 in human). We have previously crystallized the full-length ALG-2 protein and the N-terminal deletion mutant des3-20ALG-2. In this study, we crystallized des3-23ALG-2.
2. Materials and methods
2.1. Expression and purification
Construction of an expression plasmid for untagged but N-terminally truncated human ALG-2 that lacks the Gly/Pro-rich hydrophobic region, designated ALG-2ΔN23, has been described previously (Satoh et al., 2002 ▶) and is renamed des3-23ALG-2 here (Fig. 1 ▶). The des3-23ALG-2 protein was expressed and purified using an affinity column in which a phospholipid scramblase 3 (PLSCR3) oligopeptide, a Ca2+-dependent ALG-2-interacting peptide, was immobilized and the bound protein in the presence of Ca2+ was eluted with a solution of the calcium chelator EGTA essentially as described previously (Shibata et al., 2008 ▶). After affinity purification, 2 ml of the ALG-2 protein solution was dialyzed first against 2 l of 2 mM Tris–HCl pH 7.5 containing 1 mM each of EDTA and EGTA and then twice against 2 l of 2 mM Tris–HCl pH 7.5 containing 2 µM each of EDTA and EGTA. Proteins were then concentrated to about 10 mg ml−1 using a vacuum centrifuge evaporator (Sakuma, Japan). Finally, the concentrated proteins were dialyzed against 10 mM Tris–HCl pH 7.5 containing 10 µM each of EDTA and EGTA.
2.2. Crystallization
Initial crystallization trials were performed with crystallization reagent kits from Hampton Research (Crystal Screens 1 and 2, PEG/Ion Screen, Natrix and MembFac), from Emerald BioStructures (Wizard I and II, Cryo I and II) and from Molecular Dimensions (Stura Footprint Screens) by the sitting-drop vapour-diffusion method at 293 K using an automatic crystallization robot system (Hiraki et al., 2006 ▶). Drops containing 0.5 µl protein sample (10 mg ml−1) and 0.5 µl reservoir solution were equilibrated against 180 µl reservoir solution. Crystals of des3-23ALG-2 were obtained using Wizard I condition No. 40 [100 mM MES pH 6.0, 10%(v/v) 2-propanol, 200 mM calcium acetate] and Wizard II condition No. 20 [100 mM MES pH 6.0, 15%(v/v) ethanol, 200 mM zinc acetate] and crystallization conditions were optimized manually by the hanging-drop vapour-diffusion method. In the final crystallization conditions, the Ca2+-bound form of des3-23ALG-2 was crystallized using 50 mM MES pH 6.5 containing 12.5%(v/v) 2-propanol and 150 mM calcium acetate (Fig. 2 ▶ a). The Zn2+-bound form of des3-23ALG-2 was crystallized under the same conditions using Wizard II condition No. 20 [100 mM MES pH 6.0, 15%(v/v) ethanol, 200 mM zinc acetate] (Fig. 2 ▶ b).
Figure 2.
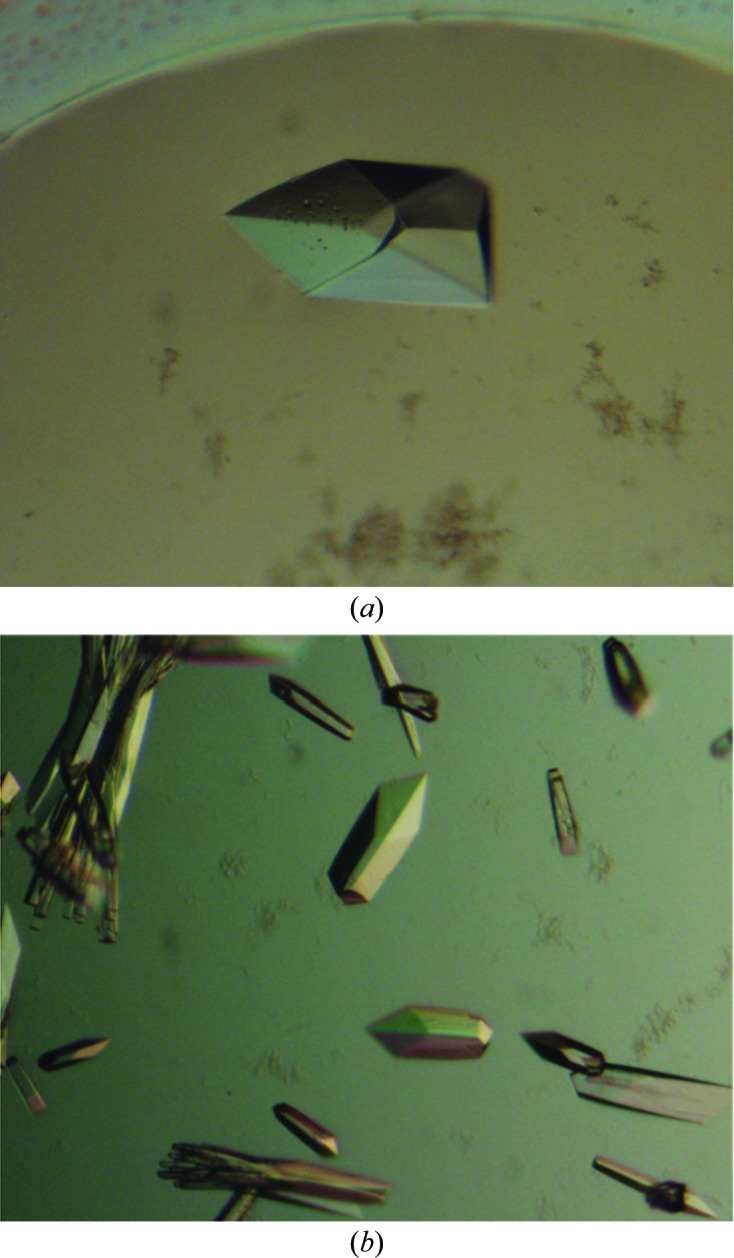
Photomicrographs of des3-23ALG-2 crystals. (a) Ca2+-bound form, (b) Zn2+-bound form.
2.3. Data collection and refinement
Data collection was performed on BL-5A at The Photon Factory (Tsukuba, Japan) and on BL41XU at SPring-8 (Harima, Japan). The crystals were soaked for a few seconds in reservoir solution containing 20% glycerol. Crystals were then picked up with nylon-fibre loops and flash-cooled in a nitrogen-gas stream. The diffraction data were indexed, integrated and scaled with the HKL-2000 program package (Otwinowski & Minor, 1997 ▶). The phase problem was solved by the molecular-replacement method with the program MOLREP (Vagin & Teplyakov, 1997 ▶) using the structure of elastase-treated N-terminally truncated mouse ALG-2 (PDB code 1hqv; Jia et al., 2001 ▶) as a search model. The X-ray data statistics are shown in Table 1 ▶. Both models were refined using the programs CNS (Brünger et al., 1998 ▶) and REFMAC5 (Murshudov et al., 1997 ▶). The models were manually adjusted using Coot (Emsley & Cowtan, 2004 ▶).
Table 1. Data-collection, processing and refinement statistics.
Values in parentheses are for the highest resolution shell.
| Ca2+-bound | Zn2+-bound | |
|---|---|---|
| PDB code | 2zrs | 2zrt |
| Data collection | ||
| Space group | P212121 | P212121 |
| Unit-cell parameters | ||
| a () | 54.8 | 52.8 |
| b () | 154.4 | 147.5 |
| c () | 237.7 | 230.7 |
| = = () | 90 | 90 |
| X-ray source | PF BL-5A | Spring-8 BL41XU |
| Wavelength () | 1.0000 | 1.2819 |
| Temperature (K) | 100 | 100 |
| Resolution () | 503.1 (3.23.1) | 503.3 (3.43.3) |
| Observed reflections | 263300 | 243748 |
| Unique reflections | 37803 | 27970 |
| Completeness (%) | 100.0 (98.4) | 99.5 (96.6) |
| R merge † (%) | 9.4 (30.9) | 10.2 (34.3) |
| I/(I) | 6.7 (4.6) | 8.0 (2.9) |
| Refinement | ||
| R work/R free ‡(%) | 22.8/28.0 | 16.9/22.1 |
| Average B factor (2) | 42.06 | 99.31 |
| R.m.s.d. bond lengths () | 0.008 | 0.006 |
| R.m.s.d. bond angles () | 1.138 | 0.890 |
| No. of atoms | 10956 | 11010 |
R
merge = 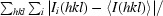
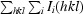 , where I
i(hkl) is the ith measurement of reflection indices hkl and I(hkl) is the mean intensity.
, where I
i(hkl) is the ith measurement of reflection indices hkl and I(hkl) is the mean intensity.
R
work = 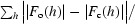
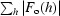 ; R
free was calculated using 10% of data excluded from refinement.
; R
free was calculated using 10% of data excluded from refinement.
3. Results and discussion
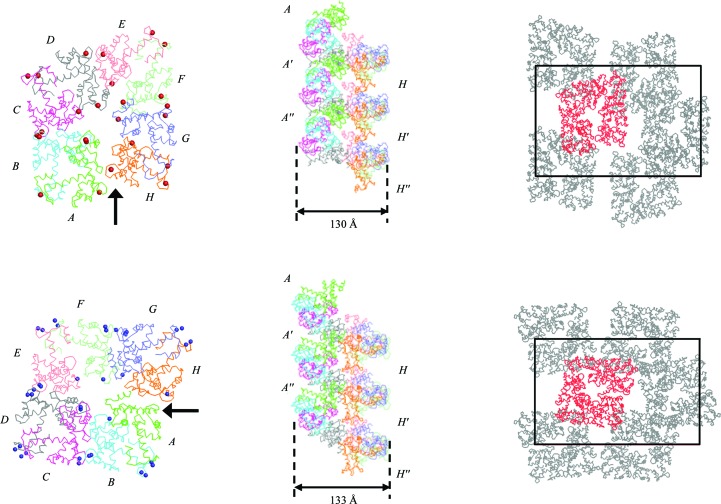
The crystals of des3-23ALG-2 grown in the buffer containing calcium acetate belonged to space group P212121, with unit-cell parameters a = 54.8, b = 154.4, c = 237.7 Å, α = β = γ = 90°, and diffracted to 3.1 Å resolution. Specific volume calculations based on the unit-cell parameters and the molecular weight (the Matthews coefficient; Matthews, 1968 ▶) suggested that there could be eight ALG-2 molecules per asymmetric unit, with a V M value of 2.8 Å3 Da−1 and a solvent content of 61.1%. The crystals grown in the buffer containing zinc acetate also belonged to space group P212121, with unit-cell parameters a = 52.8, b = 147.5, c = 230.7 Å, α = β = γ = 90°, and diffracted to 3.3 Å resolution. The Matthews coefficient suggests the presence of eight ALG-2 molecules in the asymmetric unit (V M = 2.8 Å3 Da−1; 56.5% solvent content). Although only seven ALG-2 molecules were obtained by molecular replacement with the Ca2+-bound structure of elastase-treated mouse ALG-2 (PDB code 1hqv; Jia et al., 2001 ▶) as a search model, the eighth ALG-2 molecule was assigned during the course of refinement. ALG-2 formed a homodimer and four homodimers (molecules AB, CD, EF and GH) are arranged in a pseudo-screw along the a axis (Fig. 3 ▶). A comparison of the resolved structures (PDB codes: Ca2+-bound form, 2zrs; Zn2+-bound form, 2zrt) revealed differences in the octameric arrangements (Fig. 3 ▶) and metal-ion locations (Table 2 ▶). According to the crystal packing (Fig. 3 ▶) and the solvent contents from V M calculations, the Zn2+-bound form seems to be better packed than the Ca2+-bound form. However, the overall B factor of the Ca2+-bound form (42.1 Å2) was lower than that of the Zn2+-bound form (99.3 Å2). This may explain why the crystals of the Ca2+-bound form diffracted to higher resolution than those of the Zn2+-bound form.
Figure 3.
Overall structure in the asymmetric unit (left) and crystal packing (right) of the Ca2+-bound form (top) and Zn2+-bound form (bottom) of des3-23ALG-2 viewed from the a axis. Three stacked octamers (middle) are viewed along the a axis from the direction of the arrows indicated in the left panels. In the left and middle panels, the des3-23ALG-2 molecules are coloured as follows: molecule A, green; molecule B, cyan; molecule C, magenta; molecule D, grey; molecule E, pink; molecule F, lime; molecule G, blue; molecule H, orange. Calcium and zinc ions are shown as red and blue spheres, respectively, in the left panels. In the right panels the des3-23ALG-2 molecules are displayed in grey, except for the molecules in asymmetric unit, which are coloured red. Unit cells are depicted by black boxes. Diameters of psuedo-screws are indicated by arrow-headed bars.
Table 2. Numbers of calcium ions and zinc ions at EF1, EF3 and EF5.
Calcium ions and zinc ions are found intermolecularly coordinated to the Asn106 residues of two adjacent ALG-2 molecules (between molecules B and C and between molecules F and G in the Ca2+-bound form and between molecules B and C and between molecules D and E in the Zn2+-bound form).
| Calcium ions | Zinc ions | |||||
|---|---|---|---|---|---|---|
| EF1 | EF3 | EF5 | EF1 | EF3 | EF5 | |
| A | 1 | 1 | 0 | 0 | 0 | 2 |
| B | 1 | 1 | 1 | 1 | 2 | 2 |
| C | 1 | 1 | 1 | 1 | 2 | 2 |
| D | 1 | 1 | 1 | 1 | 2 | 2 |
| E | 1 | 1 | 1 | 1 | 2 | 2 |
| F | 1 | 1 | 1 | 1 | 2 | 2 |
| G | 1 | 1 | 1 | 1 | 2 | 2 |
| H | 1 | 1 | 1 | 0 | 1 | 2 |
Using different recombinant proteins, we were able to solve the structures of the Ca2+-bound form of des3-20ALG-2 and the Zn2+-bound form of full-length ALG-2 at better resolutions (PDB codes: Ca2+-bound form, 2zn9; Zn2+-bound form, 2zn8; Suzuki et al., 2008 ▶). These crystals were obtained in buffer consisting of either 100 mM sodium cacodylate pH 6.5, 18%(w/v) PEG 8000 and 200 mM calcium acetate (Ca2+-bound form) or 100 mM sodium cacodylate pH 6.5, 25%(w/v) MPD (2-methyl-2,4-pentanediol) and 75 mM zinc acetate (Zn2+-bound form). They belonged to space groups P41 and C2221 and diffracted to 2.4 and 2.7 Å resolution, respectively. Jia et al. (2001 ▶) previously crystallized mouse ALG-2 (des1-20ALG-2) using 100 mM sodium cacodylate pH 6.5, 12–15% PEG 8000 and 100 mM calcium acetate (space group P4122) and determined the structure to 2.3 Å resolution. The organic precipitant may be involved in determining the Bravais lattice (ethanol and 2-propanol, primitive orthorhombic; PEG, primitive tetragonal; MPD, C-centred orthorhombic), as all the ALG-2 crystals described above were grown under similar conditions except for differences in the organic precipitant. Further truncation of the N-terminal sequence by three residues (21-AAP-23), which are located near the loop between EF3 and EF4 in the structures of the Ca2+-bound forms of both human des3-20ALG-2 (PDB code 2zn9) and mouse des1-20ALG-2 (PDB code 1hqv), may have decreased the diffraction quality of the crystals obtained. We investigated the locations of the calcium and zinc ions (Table 2 ▶). These metal ions were found at EF1, EF3 and EF5 in most ALG-2 molecules, but molecule A lacks zinc ions at EF1 and EF3, whereas molecule H lacks a zinc ion at EF1. Surprisingly, both ions were also found intermolecularly in locations near the Asn106 residues of two adjacent ALG-2 molecules (between molecules B and C and between molecules F and G in the Ca2+-bound form and between molecules B and C and between molecules D and E in the Zn2+-bound form). Furthermore, we observed anomalous signals from Zn atoms and one extra zinc ion was found in the vicinity of the coordinated zinc ions at EF3 and EF5, with the exception of EF3 in molecule H. The smaller ionic radius of zinc (0.74 Å) compared with calcium (0.99 Å) may explain the binding of two zinc ions to EF5. The previously solved structure of the Zn2+-bound form of full-length ALG-2 is very similar to that of the Ca2+-bound form of des3-20ALG-2, with the exception of EF5 in which the relative position of α8 (exiting the EF5 helix) of the Zn2+-bound form resembles that of the Ca2+-free form (Suzuki et al., 2008 ▶). Unfortunately, the resolutions of the structures of des3-23ALG-2 were not high enough to determine the metal-coordinating residues and quantitative comparisons with the previous structures could not be performed. Calcium ions bind to EF1 and EF3 of ALG-2 with a dissociation constant of 1.2 µM (Tarabykina et al., 2000 ▶). The binding ability of zinc ions to ALG-2 seems to be much weaker than that of calcium ions (Shibata et al., 2008 ▶), suggesting that zinc ions do not bind to ALG-2 under physiological conditions. In conclusion, the crystal packing of ALG-2 is influenced by both the organic precipitants used in crystallization and the degree of truncation of the N-terminal sequence of ALG-2, resulting in differences in the metal-ion-binding pattern in the octameric structures and the quality of X-ray diffraction.
Supplementary Material
PDB reference: ALG-2, Ca2+-bound form, 2zrs, r2zrssf
PDB reference: Zn2+-bound form, 2zrt, r2zrtsf
Acknowledgments
We thank Dr Hitomi (Nagoya University) and Dr Kato (KEK) for valuable suggestions. This work was partly supported by a Grant-in-Aid for Scientific Research (B) (to MM) and a Grant-in-Aid for JSPS Fellows (to HS) from JSPS.
References
- Brünger, A. T., Adams, P. D., Clore, G. M., DeLano, W. L., Gros, P., Grosse-Kunstleve, R. W., Jiang, J.-S., Kuszewski, J., Nilges, M., Pannu, N. S., Read, R. J., Rice, L. M., Simonson, T. & Warren, G. L. (1998). Acta Cryst. D54, 905–921. [DOI] [PubMed]
- Emsley, P. & Cowtan, K. (2004). Acta Cryst. D60, 2126–2132. [DOI] [PubMed]
- Hiraki, M. et al. (2006). Acta Cryst. D62, 1058–1065. [DOI] [PubMed]
- Jia, J., Tarabykina, S., Hansen, C., Berchtold, M. & Cygler, M. (2001). Structure, 9, 267–275. [DOI] [PubMed]
- Kitaura, Y., Matsumoto, S., Satoh, H., Hitomi, K. & Maki, M. (2001). J. Biol. Chem. 276, 14053–14058. [DOI] [PubMed]
- Kitaura, Y., Satoh, H., Takahashi, H., Shibata, H. & Maki, M. (2002). Arch. Biochem. Biophys. 399, 12–18. [DOI] [PubMed]
- Lo, K. W., Zhang, Q., Li, M. & Zhang, M. (1999). Biochemistry, 38, 7498–7508. [DOI] [PubMed]
- Maki, M., Kitaura, Y., Satoh, H., Ohkouchi, S. & Shibata, H. (2002). Biochim. Biophys. Acta, 1600, 51–60. [DOI] [PubMed]
- Maki, M., Narayana, S. V. & Hitomi, K. (1997). Biochem. J. 328, 718–720. [PMC free article] [PubMed]
- Maki, M. & Shibata, H. (2007). Calcium Binding Proteins, 2, 4–10.
- Maki, M., Yamaguchi, K., Kitaura, Y., Satoh, H. & Hitomi, K. (1998). J. Biochem. 124, 1170–1177. [DOI] [PubMed]
- Matthews, B. W. (1968). J. Mol. Biol. 33, 491–497. [DOI] [PubMed]
- Murshudov, G. N., Vagin, A. A. & Dodson, E. J. (1997). Acta Cryst. D53, 240–255. [DOI] [PubMed]
- Otwinowski, Z. & Minor, W. (1997). Methods Enzymol. 276, 307–326. [DOI] [PubMed]
- Sadoul, R. (2006). Biol. Cell, 98, 69–77. [DOI] [PubMed]
- Satoh, H., Nakano, Y., Shibata, H. & Maki, M. (2002). Biochim. Biophys. Acta, 1600, 61–67. [DOI] [PubMed]
- Shibata, H., Suzuki, H., Kakiuchi, T., Inuzuka, T., Yoshida, H., Mizuno, T. & Maki, M. (2008). J. Biol. Chem. 283, 9623–9632. [DOI] [PubMed]
- Suzuki, H., Kawasaki, M., Inuzuka, T., Okumura, M., Kakiuchi, T., Shibata, H., Wakatsuki, S. & Maki, M. (2008). Structure. 10.1016/j.str.2008.07.012. [DOI] [PubMed]
- Tarabykina, S., Moller, A. L., Durussel, I., Cox, J. & Berchtold, M. W. (2000). J. Biol. Chem. 275, 10514–10518. [DOI] [PubMed]
- Vagin, A. & Teplyakov, A. (1997). J. Appl. Cryst. 30, 1022–1025.
- Vito, P., Lacanà, E. & D’Adamio, L. (1996). Science, 271, 521–525. [DOI] [PubMed]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
PDB reference: ALG-2, Ca2+-bound form, 2zrs, r2zrssf
PDB reference: Zn2+-bound form, 2zrt, r2zrtsf